Abstract
Soil and water are suggested to represent pivotal niches for the transmission of Listeria monocytogenes to plant material, animals, and the food chain. In the present study, 467 soil and 68 water samples were collected in 12 distinct geological and ecological sites in Austria from 2007 to 2009. Listeria was present in 30% and 26% of the investigated soil and water samples, respectively. Generally, the most dominant species in soil and water samples were Listeria seeligeri, L. innocua, and L. ivanovii. The human- and animal-pathogenic L. monocytogenes was isolated exclusively from 6% soil samples in regions A (mountainous region) and B (meadow). Distinct ecological preferences were observed for L. seeligeri and L. ivanovii, which were more often isolated from wildlife reserve region C (Lake Neusiedl) and from sites in proximity to wild and domestic ruminants (region A). The higher L. monocytogenes detection and antibiotic resistance rates in regions A and B could be explained by the proximity to agricultural land and urban environment. L. monocytogenes multilocus sequence typing corroborated this evidence since sequence type 37 (ST37), ST91, ST101, and ST517 were repeatedly isolated from regions A and B over several months. A higher L. monocytogenes detection and strain variability was observed during flooding of the river Schwarza (region A) and Danube (region B) in September 2007, indicating dispersion via watercourses.
INTRODUCTION
The genus Listeria comprises the species L. monocytogenes, L. ivanovii, L. seeligeri, L. innocua, L. welshimeri, and L. grayi, highly adapted to soil, water, and vegetation (1, 2). Recently, nine novel species and a subspecies, most of them isolated from natural environments, were introduced: L. rocourtiae, L. marthii, L. weihenstephanensis, L. fleischmannii sp. nov., L. fleischmannii subsp. coloradonensis subsp. nov., L. floridensis sp. nov, L. aquatica sp. nov., L. cornellensis sp. nov., L. grandensis sp. nov., and L. riparia sp. nov. (3, 4, 5, 6, 7, 8). Some Listeria species (L. monocytogenes, L. ivanovii, and L. seeligeri) harbor a gene cluster, Listeria pathogenicity island 1 (LIPI-1), that plays a cardinal role in Listeria virulence (9, 10). L. monocytogenes is pathogenic to both humans and animals, and L. ivanovii is pathogenic to animals, particularly ruminants. L. ivanovii possesses the separate Listeria pathogenicity island 2 (LIPI-2), which encodes phosphocholinesterases for efficient utilization of phospholipids in ruminant erythrocytes. This may explain the susceptibility of ruminants to L. ivanovii infection (11).
L. monocytogenes is transmitted to the consumer mainly via contaminated ready-to-eat foods (12, 13). The presence and potential persistence of Listeria spp. in food processing facilities are often caused by environmental recontamination at the farm or plant level (14, 15, 16). In order to unravel the transfer of L. monocytogenes between niches, molecular subtyping is essential both in outbreak clarification and in the management of contamination events in food business operations (17, 18). However, source tracking of L. monocytogenes often remains challenging due to its claimed ubiquity and adaptation to harsh environmental conditions (19, 20, 21). Since the Welshimer and Donker-Voet (22) and Weis and Seeliger (2) publications, most authors have hypothesized that the primary habitat of Listeria spp. is soil and decaying vegetation (23). In more recent decades, only a few studies have dealt with the occurrence of Listeria in uncultivated natural environments (24, 25).
Important factors that have been speculated to influence the occurrence of L. monocytogenes in soil are soil microbiota, fauna, soil composition, temperature, pH, moisture, and strain motility (2, 26, 27, 28). However, if further insight is to be gained into the unique ecological behavior of L. monocytogenes, it is necessary to map globally occurring genotypes and strains from environmental habitats (29, 30).
To add more insight to this issue, the objective of this study was to analyze the occurrence of L. monocytogenes in soil samples from areas with different soil compositions and, as a novel approach, to compare samples at different altitudes. The latter comparison was based on the hypothesis that L. monocytogenes as a cold-adapted organism might have a greater chance of surviving in soil that is exposed to frost conditions for almost half the year. A second objective was to characterize L. monocytogenes isolates by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) to estimate globally widely distributed strains. Soil can act as a reservoir for antibiotics produced by other soil microbiota (such as Streptomyces and Nocardia). Resistance to these compounds could potentially contribute to the survival of L. monocytogenes in this niche (31). Therefore, as a third field of inquiry, we investigated the antibiotic resistance of L. monocytogenes strains isolated during the survey.
MATERIALS AND METHODS
Sampling and description of sampling areas.
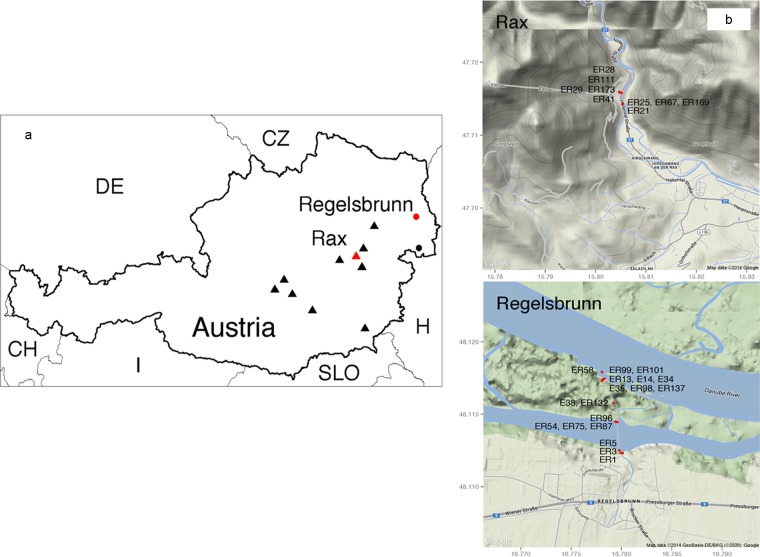
In this study, 467 soil and 68 water samples were collected from 12 areas in Austria between 2007 and 2009. Samples were collected from various distinct soil types (humus, sand, and clay). The sampling areas comprised 10 sites located in the eastern Alps (of different soil compositions, here identified as regions A and D through L) at different altitudes (0 to 500 m, 500 to 1,000 m, and ≥1,500 m), two flat-land sampling areas in the east of the country located in the Donauauen National Park adjacent to the River Danube (a humus-rich and wet region [region B]), and an area close to Lake Neusiedl (a sandy and dry region [region C]) straddling the Austrian-Hungarian border (Fig. 1).
FIG 1.
Sampling locations for Listeria detection in Austrian soil and water samples visualized in Google Maps. (a) Sampling areas A (Rax) and B (Regelsbrunn) are marked with a red triangle and red circle, respectively. Sampling areas C (Lake Neusiedl) or D to L (Eastern Alps) are labeled with black circles or black triangles, respectively. (b) L. monocytogenes-positive sampling locations in regions A and B are marked with red circles. L. monocytogenes isolation codes are abbreviated with E or ER. Further L. monocytogenes isolate characteristics are listed in Table 4.
Within the selected areas A to C, samples were collected on six separate occasions from discrete sampling points, including different altitudes in region A, during 2007 and 2008. The areas D to L were included to gain more insight into the presence of Listeria spp. at higher altitudes in 2009. Soil samples weighing between 50 to 100 g were taken from the surface down to a depth of 5 cm. They were transferred into stomacher bags (Seward Inc., West Sussex, United Kingdom) using sterile shovels. Approximately 500-ml volumes of water were taken aseptically in polypropylene bottles (Nalgene; Thermo Fisher Scientific, Waltham, MA, USA) from rivers and ponds located in areas A to G and I to K. All samples were transported to the laboratory in standardized cooling boxes at 4°C and investigated immediately on arrival. The dominant soil character, the pH value, and the moisture content were recorded.
Isolation of Listeria spp.
Pathogenic and apathogenic Listeria spp. were isolated after selective enrichment in buffered Listeria enrichment broth (BLEB; Merck KgA, Darmstadt, Germany) and two selective agar media: Oxoid chromogenic Listeria agar (OCLA; Oxoid Ltd., Hampshire, United Kingdom) and Palcam agar (Biokar Diagnostics). Specifically, 25-g portions of each soil sample were added to 225 ml BLEB and homogenized for 180 s in a Stomacher 400 (Seward Inc.). The 500-ml water samples were filtrated through three sterile analytical filters (Nalgene; Thermo Fisher Scientific) with pore sizes of 1 mm to 0.45 μm. The filters were enriched in 100 ml BLEB. The BLEB enrichments were incubated for 48 h at 30°C. Subsequently, 100-μl aliquots of BLEB enrichment were streaked onto OCLA and Palcam agars and incubated for 24 to 48 h at 37°C.
Identification and confirmation of Listeria spp.
Up to three Listeria colonies were streaked onto Rapid′L. mono agar (bioMérieux, Marcy l'Etoile, France) for purification and species differentiation. For further confirmation based on the PCR technique, Listeria colonies were dispersed in 100 μl of 0.1 M Tris-HCl buffer (Sigma-Aldrich, St. Louis, MO, USA). Additionally, the whole agar surface was swabbed and dispersed in 1 ml of 0.1 M Tris-HCl buffer (Sigma-Aldrich). DNA isolation was performed applying Chelex 100 resin (Bio-Rad, Hercules, CA, USA) according to the method of Walsh et al. (32). Isolates were confirmed by PCR detection of the hly gene, encoding the virulence factor listeriolysin O, and the highly conserved 23S rRNA genes of Listeria spp. (33). Subsequently, Listeria species were differentiated by multiplex PCR targeting the invasion-associated protein (iap) gene (34). Biochemical identification of Listeria spp. was performed by applying the API Listeria system (bioMérieux).
AMR testing.
The antimicrobial resistance (AMR) of L. monocytogenes isolated from soil samples was tested by applying the commercially available Micronaut-S Listeria MIC microtiter plate assay (Merlin; Sifin diagnostics GmbH, Berlin, Germany). A panel of 15 antimicrobials at the concentrations indicated in parentheses were included in the assay: amoxicillin-clavulanic acid (AMC; 0.125/2 to 16/2 μg/ml), ampicillin (AMP; 0.125 to 16 μg/ml), cefotaxime (CTX; 8 to 64 μg/ml), ceftriaxone (CRO; 1 to 64 μg/ml), ciprofloxacin (CIP; 0.24 to 4 μg/ml), clarithromycin (CLR; 0.25 to 8 μg/ml), erythromycin (ERY; 0.25 to 8 μg/ml), gentamicin (GEN; 0.5 to 16 μg/ml), imipenem (IMP; 0.06525 to 4 μg/ml), linezolid (LIZ; 0.5 to 16 μg/ml), penicillin G (0.0625 to 8 μg/ml), rifampin (RAM; 0.125 to 8 μg/ml), tetracycline (TET; 1 to 16 μg/ml), trimethoprim-sulfamethoxazole (T/S; 0.25/4.75 to 2/38 μg/ml), and vancomycin (VAN; 1 to 32 μg/ml). L. monocytogenes isolates were grown on Mueller-Hinton agar (Oxoid) for 24 h at 37°C incubation. The overnight cultures were suspended in sterile saline solution (0.85% NaCl) to achieve a turbidity of a McFarland standard of 0.5 and then diluted 1:100 before use. The breakpoints for MICs were determined according to the actual Eucast (http://www.eucast.org/clinical_breakpoints/; accessed 3 March 2014) and Clinical and Laboratory Standards Institute (CLSI) standards for 2010 (35).
Subtyping and epidemiological analysis.
Confirmed Listeria isolates were incubated overnight in brain heart infusion (BHI; Merck KgA) at 37°C. Subsequently, isolates were cryoconserved in 15% glycerol (Merck KgA) at −80°C in the Listeria collection of the Institute of Milk Hygiene, Milk Technology and Food Science (Vetmeduni, Vienna, Austria).
L. monocytogenes PCR serogroups were characterized by applying a multiplex PCR targeting the genes lmo0737, lmo1118, ORF2819, ORF2110, and Listeria-specific prs, published by Doumith et al. (36) and amended by Leclercq et al. (37), for PCR IVb-VI.
The PFGE analysis of L. monocytogenes isolated in this study followed the PulseNet protocol (http://www.cdc.gov/pulsenet/pathogens/) with the minor modifications that samples were digested, applying 50 U AscI and ApaI for 4 h at 37°C and 25°C incubation temperatures, respectively. Restricted samples were separated in a 1% (wt/vol) SeaKem gold agarose gel in 0.5× Tris-borate EDTA (TBE) buffer at 6 V/cm on a Chef DR III system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). A linear ramping factor with pulse times from 4.0 to 40.0 s at 14°C and an included angle of 120° was applied for 22.5 h. The gels were stained with ethidium bromide (Sigma-Aldrich), digitally photographed with Gel Doc 2000 (Bio-Rad Laboratories, Inc.), and normalized as TIFF images (BioNumerics 6.6 software; Applied Maths NV, Sint-Martens-Latem, Belgium) applying the PFGE global standard Salmonella enterica serovar Braenderup H9812.
PCR-restriction fragment length polymorphism (RFLP) for detection of point mutations in the 733-bp fragment of the inlA gene followed the protocol published by Rousseaux et al. (38). Thereby, 1 μl of amplified DNA was digested with 10 U AluI (1 h at 37°C) and separated on a 2% (wt/vol) agarose gel containing 3.5 μl SYBR Safe DNA gel stain (Invitrogen, Eugene, OR, USA).
The presence or absence of L. monocytogenes stress survival islet 1 (SSI-1) was screened by PCR, targeting the intergenic region between lmo0443 and lmo0449, according to Ryan et al. (39). The polymorphism in the actA gene, resulting in a 268-bp or 385-bp product, was determined by a PCR protocol published by Jaradat et al. (40).
MLST based on the seven housekeeping loci abcZ (ABC transporter), bglA (beta glucosidase), cat (catalase), dapE (succinyl diaminopimelate desuccinylase), dat (d-amino acid aminotransferase), ldh (l-lactate dehydrogenase), and lhkA (histidine kinase) was performed according to the method of Ragon et al. (41). An allele number was assigned for each housekeeping gene, and sequence types (STs) were determined and compared using the Institut Pasteur Listeria monocytogenes MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Lmono.html).
An allelic profile-based comparison, applying a minimum spanning tree (MST) and the Institut Pasteur online tool, was performed to define the relationships among strains at the microevolutionary level. Clonal complexes (CCs) were defined as groups of STs differing by only one housekeeping gene from another member of the group (41).
Statistical analysis and map design.
t tests and chi square (χ2) tests were performed in IBM SPSS (version 19.0; SPSS Inc., Chicago, IL, USA) to determine the statistical significance (P < 0.05) of the difference of the distributions in prevalence between various sample categories and parameters (Listeria isolation, pH value, moisture content, region, and season).
Sampling locations were georeferenced and inserted into maps applying the ggmap package (42), an open-source tool for spatial visualization with Google Maps within the freely available statistical computing environment R (43).
RESULTS
Occurrence of Listeria spp. in Austrian environmental samples.
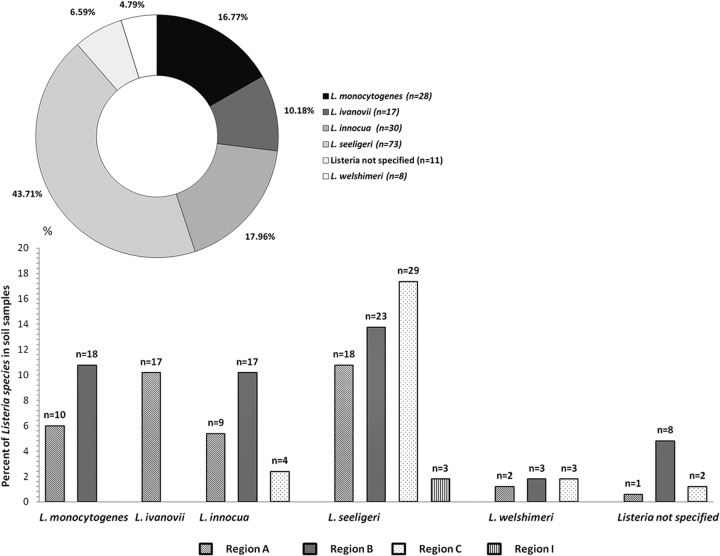
During the investigation of 467 soil samples, 30% (n = 140) were determined to be positive for Listeria spp., of which 28 samples (6%) were L. monocytogenes positive. Twenty-five soil samples contained mixed Listeria species, most frequently L. monocytogenes and L. innocua (Table 1). The distribution of confirmed Listeria in soil samples according to species and region is depicted in Fig. 2. Listeria was isolated from 26.5% of water samples. The predominant species in soil and water samples were L. seeligeri (regions A, B, C, and I), L. innocua (regions A, B, and C), and L. ivanovii (regions A and C). While the human- and animal-pathogenic L. monocytogenes was exclusively isolated from soil samples in regions A and B near the Schwarza and Danube rivers (Fig. 1), the animal pathogen L. ivanovii was isolated mainly in region A (mountain) and in water samples in regions B and C (see Table 5). The characteristics of Listeria-positive and -negative soil samples according to region, altitude, and dominant soil characteristics are shown in Tables 2 and 3, respectively. The influence of moisture, pH, and soil type on the isolation of Listeria spp. from soil samples revealed a significant difference (P ≤ 0.001). Listeria spp. were more frequently isolated from soil samples with low moisture content (22.96%; range, 2% to 80%), neutral pH (average mean, 7.44; range, 3.43 to 9.90), and soil types consisting of a mixture of sand and humus. A possible seasonal effect was observed, whereby the lowest Listeria isolation rates (3.33%; P ≤ 0.001) occurred in July.
TABLE 1.
Differentiation of Listeria species isolated from soil samples, according to region and altitude
| Region | Altitude (m) | No. of single-species samples |
Mixed-species samples |
||||||
|---|---|---|---|---|---|---|---|---|---|
| L. monocytogenes | L. ivanovii | L. innocua | L. seeligeri | Unspecified Listeria | L. welshimeri | Total no. | Speciesa identified (no. of samples) | ||
| A | 0–500 | 9 | 13 | 7 | 6 | 0 | 0 | 5 | M + IN (1), M + IV (1), SE + IV (3) |
| B | 0–500 | 18 | 0 | 17 | 23 | 8 | 3 | 13 | M + IN (8), M + SE (2), M + NS (1), M + IN + SE + WE (1), IN + WE (1) |
| C | 0–500 | 0 | 0 | 4 | 29 | 2 | 3 | 4 | SE + WE (3), IN + SE (1) |
| A | 500–1,000 | 1 | 3 | 2 | 12 | 1 | 2 | 3 | SE + WE (2), SE + IV (1) |
| A | ≥1,500 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| D to L | ≥1,500 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
Abbreviations: M, L. monocytogenes; IN, L. innocua; IV, L. ivanovii; SE, L. seeligeri; NS, unspecified Listeria; WE, L. welshimeri.
FIG 2.
Distribution and percentage of Listeria spp., according to species and region.
TABLE 5.
Characteristics of L. ivanovii isolates detected in soil and water samples between 2007 and 2008
| Isolate code(s) | Sample type | Region and date (mo, yr) of isolation | Altitude (m) | Sampling site(s) | API codea | AscIb | ApaIc |
|---|---|---|---|---|---|---|---|
| ER 24 | Soil | A (9, 2007) | 0–500 | A4 | 3350 | SIV1 | IV1 |
| ER 36, ER 41, ER 42, ER43 | Soil | A (10, 2007) | 0–500 | A4, A14, A15 | 3350 | SIV1 | IV1 |
| ER 66, ER 110, ER 69, ER 70 | Soil | A (11, 2007) | 0–500 | A5, A11, A28 | 3350 | SIV1 | IV1 |
| ER 180, ER 185, ER 186 | Soil | A (8, 2008) | 500–1,000 | A26, A30 | 3350 | SIV1 | IV1 |
| ER 30 | Soil | A (9, 2007) | 0–500 | A14 | 3370 | SIV2 | IV2 |
| ER 35, ER 37, ER 38 | Soil | A (10, 2007) | 0–500; ≥1,500 | A1, A5 | 3370 | SIV2 | IV2 |
| ER 40 | Soil | A (10, 2007) | 0–500 | A8 | 3370 | SIV2 | IV2 |
| ER 26 | Soil | A (9, 2007) | 0–500 | A7 | 3330 | SIV3 | IV3 |
| ER 122 | Water | C (10, 2007) | 0–500 | C | 3330 | WIV4 | IV4 |
| ER 123 | Water | B (11, 2007) | 0–500 | B | 3330 | WIV3 | IV3 |
| ER 171, 172 | Soil | A (8, 2008) | 0–500 | A11 | 3350 | SIV5 | IV5 |
API code, biochemical profile for species identification: 3330 and 3370, ribose positive; 3350, ribose negative; 3330, glucose-1-phosphate negative.
AscI, PFGE type after restriction digest with the enzyme AscI.
ApaI, PFGE type after restriction digest with the enzyme ApaI.
TABLE 2.
Characteristics of Listeria-negative soil samples according to region, altitude, and dominant soil characteristics
| Region | Altitude (m) | No. of samples | No. (%) of samples negative for Listeria | Avg % moisture content (range) | Avg pH (range) | Dominant soil characteristic(s) |
|---|---|---|---|---|---|---|
| A | 0–500 | 60 | 30 (50.00) | 20.30 (5.00–43.00) | 7.56 (6.88–8.29) | Humus; sand/clay |
| B | 0–500 | 101 | 47 (46.53) | 26.26 (11.00–48.00) | 7.47 (6.47–8.65) | Humus/clay; sand |
| C | 0–500 | 92 | 58 (63.04) | 12.00 (1.00–49.00) | 7.74 (6.27–9.90) | Humus/sand; sand/clay |
| A | 500–1,000 | 62 | 44 (70.97) | 27.97 (5.00–46.00) | 7.42 (5.79–7.97) | Humus |
| A | ≥1,500 | 62 | 61 (98.39) | 48.35 (16.00–75. 00) | 6.73 (4.22–7.85) | Humus |
| D to L | ≥1,500 | 90 | 87 (96.67) | 45.06 (11.20–80.00) | 5.26 (3.43–7.71) | Humus; sand/humus |
TABLE 3.
Characteristics of Listeria-positive soil samples according to region, altitude, and dominant soil characteristics
| Region(s) | Altitude (m) | No. of samples | No. (%) of samples positive for Listeria | Avg % moisture content (range) | Avg pH (range) | Dominant soil characteristic(s) |
|---|---|---|---|---|---|---|
| A | 0–500 | 60 | 30 (50.00) | 18.67 (3.00–37.00) | 7.53 (6.63–8.15) | Sand/humus; sand |
| B | 0–500 | 101 | 54 (53.47) | 28.45 (16.00–51.00) | 7.5 (6.71–8.36) | Sand/humus; humus/clay |
| C | 0–500 | 92 | 34 (36.96) | 16.40 (2.00–31.00) | 7.53 (5.55–8.82) | Sand/humus; humus |
| A | 500–1,000 | 62 | 18 (29.03) | 22.53 (6.00–46.00) | 7.24 (5.05–7.97) | Humus |
| A | ≥1,500 | 62 | 1 (1.61) | 56.00 | 6.30 | Humus |
| D to L | ≥1,500 | 90 | 3 (3.33) | 33.00 (13.00–45.00) | 5.84 (4.92–7.45) | Humus; sand |
Listeria spp. were most frequently isolated from water samples with an average mean pH of 7.94 (range, 7.2 to 8.87). Listeria occurrence in soil samples was highest, with 25.27% (n = 118), at altitudes between 0 to 500 m, followed by 3.85% (n = 19) at altitudes of 500 to 1,000 m. Listeria spp. (L. seeligeri and L. ivanovii) were isolated at low counts at altitudes of >1,500 m (0.86%; n = 4).
Higher isolation rates of L. monocytogenes and L. innocua in regions A and B matched the rising water level and flooding of the Danube and Schwarza rivers in September 2007 (http://ehyd.gv.at/; for the Danube water level, the measuring point was at Wildungsmauer [HZBRNR 207373]; for the Schwarza water level, the measuring point was at Gloggnitz [Adlerbrücke; HZBNR 208710]). L. seeligeri was most frequently isolated from soil and water samples in region C (Lake Neusiedl), a waterfowl nature reserve.
Epidemiological investigation.
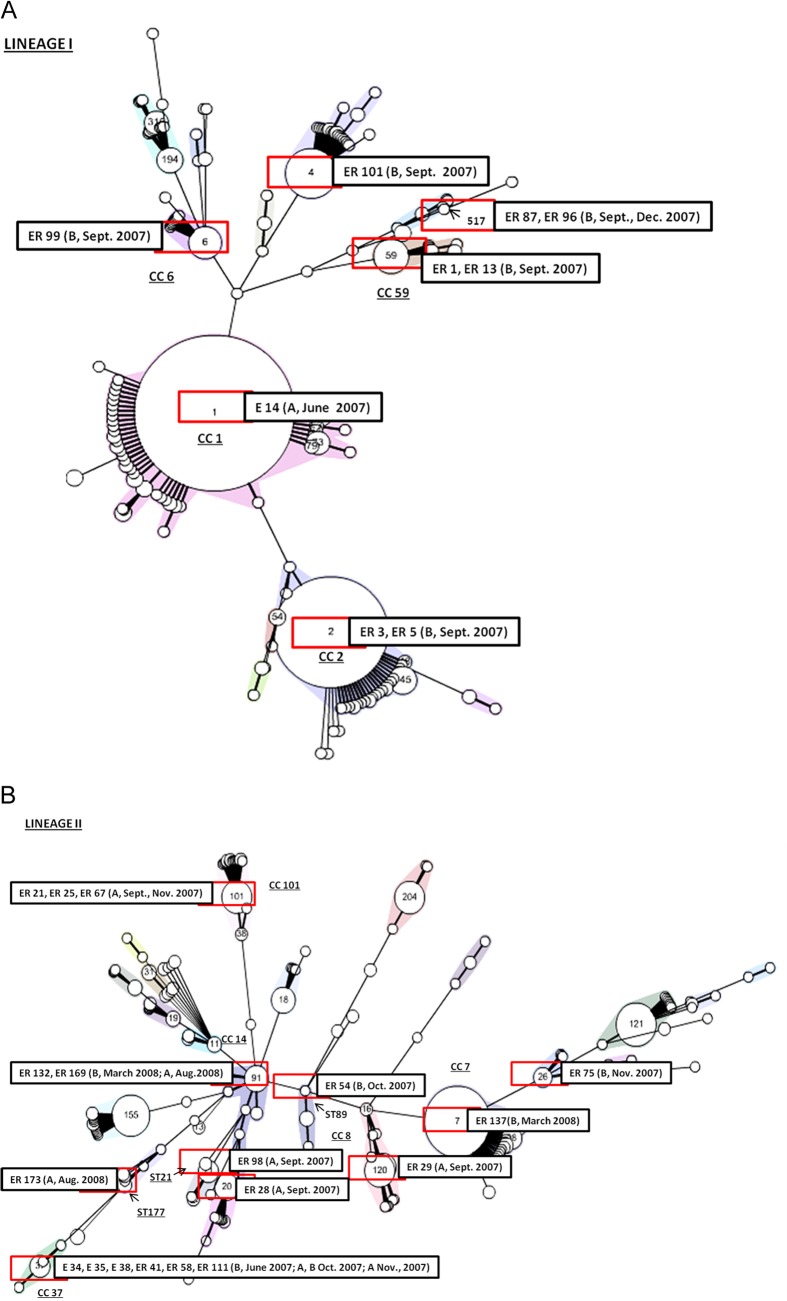
Twenty-seven L. monocytogenes isolates were used for further subtyping (Table 4). The multiplex serogroup PCR assay demonstrated that most of the L. monocytogenes isolates (66.67%) clustered in serogroups 1/2a and 3a (genetic lineage II), followed by 33.33% assigned to genetic lineage I (serogroups 4b, 4e, 4d, 1/2b, and 3b). All L. monocytogenes isolates were typeable with both macrorestriction enzyme analyses applying AscI and ApaI. The discriminatory power of PFGE typing was higher than that of MLST typing, resulting in 19 AscI and ApaI profiles (SOM1 to SOM19) and 16 multilocus sequence types (STs), respectively (Table 4). Interestingly, MLST analysis revealed that L. monocytogenes ST37 was predominant in Austrian soil samples and was repeatedly isolated from regions A and B in June and October 2007. Furthermore, ST91 was isolated from regions A and B in March and August 2008, and ST517 was prevalent in region B soil samples in September and December 2007. The highest diversity of L. monocytogenes genotypes was observed during the flooding of the River Danube in September 2007 (ST2, ST4, ST6, ST21, ST59, and ST517). Minimum spanning tree analysis of L. monocytogenes lineages I and II isolated in this study, in comparison with the Institut Pasteur strain collection based on identical allelic abcz types, is depicted in Fig. 3A and B.
TABLE 4.
Characteristics of L. monocytogenes isolates detected in soil samples between 2007 and 2008
| Isolate code(s) | Region(s) and date(s) (mo, yr) of isolation | STa | PCR serogroups | AscIb | ApaIc | inlA typed | SSI-1e | actAf (268 bp) |
|---|---|---|---|---|---|---|---|---|
| ER 3, ER 5 | B (9, 2007) | 2 | 4b, 4d, 4e | SOM1, SOM2 | SOM1, SOM2 | 2 | − | − |
| ER 101 | B (9, 2007) | 4 | 4b, 4d, 4e | SOM3 | SOM3 | 2 | − | − |
| E 14 | A (6, 2007) | 1 | 4b, 4d, 4e | SOM4 | SOM4 | 2 | − | − |
| ER 99 | B (9, 2007) | 6 | 4b, 4d, 4e | SOM5 | SOM5 | 2 | − | + |
| ER 87, ER 96 | B (9, 2007); B (12, 2007) | 517 | 4b, 4d, 4e | SOM8 | SOM8 | 2 | + | + |
| ER 1, ER 13 | B (9, 2007) | 59 | 1/2b, 3b | SOM6, SOM7 | SOM6, SOM7 | 2 | − | + |
| E 34, E 35, E 38, ER 41, ER 58, ER 111 | B (6, 2007); A, B (10, 2007); A (11, 2007) | 37 | 1/2a, 3a | SOM9 | SOM9 | 1 | − | − |
| ER 29 | A (9, 2007) | 120 | 1/2a, 3a | SOM10 | SOM10 | 3 | + | − |
| ER 137 | B (3, 2008) | 7 | 1/2a, 3a | SOM11 | SOM11 | 5 | + | − |
| ER 75 | B (11, 2007) | 26 | 1/2a, 3a | SOM12 | SOM12 | 4 | + | − |
| ER 173 | A (8, 2008) | 177 | 1/2a, 3a | SOM13 | SOM13 | 5 | − | − |
| ER 54 | B (10, 2007) | 89 | 1/2a, 3a | SOM14 | SOM14 | 1 | − | + |
| ER 98 | B (9, 2007) | 21 | 1/2a, 3a | SOM15 | SOM15 | 3 | − | + |
| ER 132, ER 169 | B (3, 2008); A (8, 2008) | 91 | 1/2a, 3a | SOM16, SOM17 | SOM16, SOM17 | 3 | − | − |
| ER 21, ER 25, ER 67 | A (9, 2007); A (11, 2007) | 101 | 1/2a, 3a | SOM18 | SOM18 | 6 | − | + |
| ER 28 | A (9, 2007) | 20 | 1/2a, 3a | SOM19 | SOM19 | 1 | − | − |
ST, MLST sequence type.
AscI, PFGE type after restriction digest with the enzyme AscI.
ApaI, PFGE type after restriction digestion with the enzyme ApaI.
inlA, PCR restriction fragment length polymorphism (RFLP) type for the detection of point mutations in the 733-bp fragment of the inlA gene; RFLP types 1 and 4 with truncation in inlA gene.
Presence or absence of SSI-1 (stress survival islet 1), in L. monocytogenes intergenic region lmo0443–lmo0449.
Polymorphism in the actA gene, producing a fragment of 268 bp instead of the expected 385 bp.
FIG 3.
Multilocus sequence typing of Listeria monocytogenes isolated from Austrian soil samples, showing genetic lineage I (A) or II (B). The sequence types were clustered according to the abcz housekeeping gene using a minimum spanning tree (MST) tool available from the Institut Pasteur MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/). Numbers within circles denote the corresponding ST. L. monocytogenes strains were grouped into clonal complexes (CC; underlined; randomly colored), defined as groups of profiles differing by no more than one gene from at least one other profile of the group. Strains with no CC designation correspond to genotypes that are not closely related to any other genotype (singletons) (41, 57). L. monocytogenes soil isolates from Austria, including region and date of isolation, are included in each MST.
Interestingly, comparison of the isolates recovered in this study with MLST isolates stored on the Institut Pasteur MLST database showed that the majority of genetic lineage I and II isolates were comparable to isolates recovered from compost samples commercially available in Austria in 2009 (ST1, ST4, ST6, ST7, ST20, ST26, ST37, ST59, ST91, and ST517). ST59 soil isolates were comparable to L. monocytogenes isolated from deer in Austria. Isolates representing ST1, ST2, and ST91 were also isolated from food in Austria. ST89, ST101, ST120, and ST177 were not present in the Institut Pasteur MLST database among Austrian isolates.
Truncation in the 733-bp fragment of the inlA fragment of L. monocytogenes soil isolates was found among ST26 (RFLP type 4), ST20, ST37, and ST89 (all RFLP type 1). RFLP type 2 was found solely among genetic lineage I isolates (Table 4). The stress survival islet (SSI-1) inserted into intergenic region lmo0443 to lmo0449 in L. monocytogenes was present in ST517 (lineage I; PCR serogroups 4b, 4d, and 4e) and in ST7, ST26, and ST120 (lineage II). Additionally, a polymorphism in the actA gene producing a fragment of 268 bp instead of the expected 385 bp was observed for L. monocytogenes ST6, ST21, ST59, ST89, ST101, and ST517. A larger amount of lineage II strains harbored at least one of the previously described targets suspected to induce environmental and host adaptation. ST517 and ST89 (environment-associated isolates according to the Institut Pasteur MLST database) and ST26 (most often isolated from wild animals) harbored two of the three investigated genetic modifications (Table 4).
AMR testing of 26 L. monocytogenes isolates showed broad susceptibility to the majority of tested antibiotics. Nevertheless, the highest antibiotic resistance rates observed among the panel of L. monocytogenes strains (88.46% and 65.38%) were against cefotaxime (CRO; MIC, >2 μg/ml) and erythromycin (ERY; MIC, >1 μg/ml), respectively. Further, 35%, 12%, and 6% of L. monocytogenes test strains were resistant to ceftriaxone (CTX; MIC, >2 μg/ml), ciprofloxacin (CIP; MIC, >1 μg/ml), and linezolid (LIZ; MIC, >4 μg/ml). L. monocytogenes isolates from region A (assigned to ST37 and ST91) were found to be multiresistant to four antibiotics (CRO, ERY, CTX, and CIP). L. monocytogenes ST1, ST2, and ST6 isolates were resistant to third-generation cephalosporins CRO and CTX, and the macrolide ERY. L. monocytogenes ST7 was resistant to CRO, ERY, and the fluoroquinolone CIP. ST20 was resistant to CRO, ERY, and LIZ.
Surprisingly, aside from L. monocytogenes, we found 22 L. ivanovii isolates in our study (Table 5). The analysis resulted in five PFGE profiles each when digested with both restriction enzymes (AscI and ApaI). Similar to what occurred with L. monocytogenes, the PFGE L. ivanovii profiles SIV1 (54.55% of isolates) and SIV2 (22.73% of isolates) were exclusively isolated from region A during September 2007 and August 2008. The PFGE profile SIV3 was isolated from soil in region A (September 2007) and from water in region B (November 2007) (Table 5).
DISCUSSION
Analysis of a total of 467 soil and 68 water samples from 12 sampling areas in Austria, in national parks or mountain summits, collected between the years 2007 and 2009 permitted further insight into the distribution of saprophytic Listeria species. Sampling contrasting geological and ecological areas such as mountainous regions in the eastern Alps, a steppe landscape, and a mixed forest and meadow area has shown that the pathogenic Listeria prevalence was higher in lower altitudes.
The overall Listeria prevalence in this study was high in uncultivated Austrian soil (30.0%) compared with those in reports from other authors, who describe prevalences ranging between 17.7% and 28% when testing samples originating from sample sites in different continents and in different climates (2, 25, 26, 44). While nonpathogenic Listeria species were recoverable from sampling areas A to C and I, L. monocytogenes was present in only 6% of soil samples from regions A and B, with a peak of positive findings for the month of September 2007.
Other researchers detected higher L. monocytogenes isolation rates (7 to 17%) in the natural environment (25, 26, 45). Many factors were assumed to contribute to a biased Listeria detection. These include fewer freezing and thawing cycles before sampling, proximity to water, higher water storage capacities of soil, a high percentage of clay in the soil texture, the absence of endogenous soil microbiota, and the presence of protozoa and nematodes as vectors (23, 26, 43). A recent study indicated, controversially, shorter survival capacities of L. monocytogenes in soil models with high clay content (45). Listeria persistence in soil was also assumed to be facilitated by strain motility and low temperatures (<8°C) (31). Therefore, outcomes of the different studies might be difficult to compare. Statistical evaluation of our results demonstrated that soil texture (sand/humus), average mean moisture (22.96%), and pH (7.44) have had a distinct influence on Listeria isolation in soil. Nevertheless, the impact of these factors should not be overrated since Listeria isolation was possible from almost all soil types and along the range of pH values (4.92 to 8.82) and moisture contents (3 to 56%) described. Welshimer and Donker-Voet (22) described a seasonality effect with high Listeria isolation rates during spring (86%), explained by higher humidity and remnants of decaying vegetation. In the current study, Listeria detection was possible during all seasons. Interestingly, Listeria isolation seemed to decrease to nearly zero at higher altitudes, independently from the location tested (regions A and D to L). This falsified our initial hypothesis that cold adaptation might result in a higher isolation frequency in such remote areas. The higher Listeria detection rates at lowland sampling points, located at altitudes of 0 to 500 m, could be explained by the proximity to farm/agricultural land and urban environments. Other authors have supposed an impact of wastewater effluents, watercourses, fertilized fields, wildlife feeding grounds and grazing ruminants on pastures on the prevalence of L. monocytogenes (29, 46, 47). This suspicion is corroborated by the MLST types found in our study, since L. monocytogenes ST517, ST37, ST91, and ST101 could be repeatedly isolated from regions A and B over several months. L. monocytogenes ST37 isolates are known to be related to urban, farm, and natural environments. ST517 is an MLST type that was isolated solely from Austrian compost samples and has not been confirmed elsewhere. ST101 and ST91 are globally distributed strains, the majority of which were isolated from animals and cheese products.
An interesting observation was drawn from the sampling during and after the severe weather period in September 2007, in which flooding of the Danube (sampling area B) and Schwarza (sampling area A) rivers occurred. Listeria contamination rates of soil and groundwater were increased, and the highest L. monocytogenes strain variability was observed in this period (ST2, ST4, ST6, ST21, ST59, and ST517). Globally distributed L. monocytogenes lineage I strains ST1, ST2, ST4, and ST59 are highly correlated with human sources and food, whereas ST21 was most frequently isolated from rodents and birds. ST59 soil isolates were comparable to L. monocytogenes isolated from deer in Austria. Surprisingly, L. monocytogenes genetic lineages I and II isolated in this study were comparable to isolates from commercially available compost samples in Austria, which were investigated in 2009 and indicated a high prevalence of these genotypes between the years 2007 and 2009 in soil (ST1, ST4, ST6, ST7, ST20, ST26, ST37, ST59, ST91, and ST517). ST89, ST101, ST120, and ST177 were not present in the Institut Pasteur MLST database among the MLST types that were reported from Austrian isolates.
The pattern of recovery of the other pathogenic Listeria species in soil, L. ivanovii, mimicked to some extent the isolation pattern for L. monocytogenes. Using PFGE, the 22 L. ivanovii isolates could be differentiated into five subtypes, whereas isolates of PFGE types SIV1 and SIV2 were recurrently isolated between 2007 and 2008 (Table 5) in region A. This observation could also indicate the impact of the flooding season, since abundance of L. ivanovii was speculated to occur due to fecal shedding by wildlife or extensively farmed domestic ruminants (2, 44, 48, 49). Similarly, the high abundance of L. seeligeri (88.33%) in the Lake Neusiedl nature reserve (region C), which is an arid, sandy endorheic environment comprising small, stagnant lakes and decaying water plants, may be associated with the enormous amount of resident wildlife, mainly waterfowl.
For more insight into the nature of the L. monocytogenes isolates identified in this study, we analyzed some genetic traits that were associated either with virulence attenuation (inlA) or with adapation (actA, SSI-1). Special features of L. monocytogenes suspected to induce environmental and mammalian host adaptation were found in 66.67% of all MLST types. A truncation in the 733-bp fragment of the inlA gene was solely found among lineage II strains of ST20, ST26, ST37, and ST89. According to the MLST database, these MLST types are most often isolated from the environment and animals. These findings are consistent with those of Orsi et al., 2011 (50), who described a positive selection for 1/2a and 1/2c strains with premature stop codons (PMSCs) important for environmental survival. The L. monocytogenes stress survival islet 1 (SSI-1), which encodes proteins allowing L. monocytogenes cells to adapt to hostile conditions, such as acid stress, was present in ST517 (lineage I) and in ST7, ST26, and ST120 (lineage II) (39). ST7 and ST120 are globally distributed strains, the majority of which have been isolated from animals (rodents and wild and domestic ruminants), human clinical cases, food, and feed. ST26 is overrepresented among isolates from animals. The actA gene polymorphism (producing a 268-bp instead of a 385-bp fragment), which is correlated with enhanced virulence properties and higher biofilm formation in food processing plants, was observed in ST6, ST21, ST59, ST89, ST101, and ST517 isolates (51, 52). Isolates of ST26, ST89, and ST517 (environmental and animal-associated strains) were frequently isolated in our study and harbor two of the three investigated features, suggesting a selective advantage in natural habitats and animal hosts. Even in Austrian soil samples, L. monocytogenes sequence types that were comparable to the worldwide most prevalent clonal complexes (CCs) among clinical, animal, environmental, and food isolates were detected (19, 53).
Generally, L. monocytogenes isolated from food, food processing environments, and clinical sources have been considered to be susceptible to a broad range of antibiotics. Resistance to clindamycin, linezolid, ciprofloxacin, ampicillin, trimethoprim-sulfamethoxazole, erythromycin, vancomycin, and tetracycline is usually confined to a small number of strains (54, 55).
As soil can act as a reservoir for antibiotics produced by other soil microbiota (such as Streptomyces and Nocardia), resistance to these compounds could potentially contribute to the survival of Listeria spp. in this niche (31, 56). The current study showed that 88.46% of isolates were resistant to third-generation cephalosporins (cefotaxime) and 65.38% to the macrolide antibiotic erythromycin. ST1, ST2, ST6, ST7, ST20, ST37, and ST91, with a high prevalence worldwide (41, 57) and also isolated during our study, were found to be multiresistant to cephalosporins (cefotaxime and ceftriaxone), the macrolide erythromycin, the fluoroquinolone ciprofloxacin, and the oxazolidinone linezolid. A high AMR to the cephalosporin cefoxitin (98%) and moderate resistance to ciprofloxacin (7%) among L. monocytogenes isolates from food and the environment have also been reported from Canada (27). Interestingly, a certain level of resistance to antibiotics has also been shown for isolates recovered from wildlife (58). These authors demonstrated resistant strains from carcasses of bisons hunted in North Dakota, USA. Our observations of a high degree of antibiotic resistance to an array of synthetic antibiotics, unexpected in natural samples, suggest that even isolates of L. monocytogenes recovered from uncultivated soil might to some extent originate from niches strongly associated with anthropogenic and livestock influences.
Data from this study support the hypothesis that Listeria species are widely distributed in nature. Soil is a crucial niche for the persistence of globally distributed L. monocytogenes isolates, which can harbor AMR. High numbers of L. monocytogenes can be dispersed via watercourses and larger rivers with a proximity to the urban environment and wastewater effluents. A higher environmental contamination was found to be associated with periods of prolonged rainfall and flooding. Distinct ecological preferences were observed for L. seeligeri and L. ivanovii, which were more often isolated from waterfowl wildlife reserves and from sites in proximity to grazing wild and domestic ruminants. The number of Listeria species was nearly zero at higher altitudes, most likely due to the lower human and animal population densities.
ACKNOWLEDGMENTS
The work was supported by the European Union-funded Integrated Project Biotracer (contract 036272) under the 6th RTD Framework.
We acknowledge the Institut Pasteur for providing the MLST database of L. monocytogenes in the Genotyping of Pathogens and Public Health Platform, with special thanks to Sylvain Brisse and Jana Haase. Additionally, we thank Petra Apfalter for providing the sample information for L. monocytogenes MLST types isolated from compost. We thank Cameron McCulloch for assistance with the manuscript.
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.Hawker J, Norman B, Blair I, Reintjes R, Weinberg J, Ekdahl K. (ed). 2012. Communicable disease control and health protection handbook, 3rd ed, section 3.46 John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 2.Weis J, Seeliger HPR. 1975. Incidence of Listeria monocytogenes in nature. Appl. Microbiol. 30:29–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertsch D, Rau J, Eugster MR, Haug MC, Lawson PA, Lacroix C, Meile L. 2012. Listeria fleischmannii sp. nov., isolated from cheese. 63:526–532 Int. J. Syst. Evol. Microbiol. 10.1099/ijs.0.036947-0 [DOI] [PubMed] [Google Scholar]
- 4.den Bakker HC, Manuel CS, Fortes ED, Wiedmann M, Nightingale KK. 2013. Genome sequencing identifies Listeria fleischmannii subsp. coloradensis subsp. nov., a novel Listeria fleischmannii subspecies isolated from a ranch in Colorado. Int. J. Syst. Evol. Microbiol. 63:3257–3268. 10.1099/ijs.0.048587-0 [DOI] [PubMed] [Google Scholar]
- 5.den Bakker HC, Warchocki S, Wright EM, Allred AF, Ahlstrom C, Manuel CS, Stasiewicz MJ, Burrell A, Roof S, Strawn L, Fortes ED, Nightingale KK, Kephart D, Wiedmann M. 5 March 2014. Five new species of Listeria (L. floridensis sp. nov., L. aquatica sp. nov., L. cornellensis sp. nov., L. riparia sp. nov., and L. grandensis sp. nov.) from agricultural and natural environments in the United States. Int. J. Syst. Evol. 10.1099/ijs.0.052720-0 [DOI] [PubMed] [Google Scholar]
- 6.Graves LM, Helsel LO, Steigerwalt AG, Morey RE, Daneshvar MI, Roof SE, Orsi RH, Fortes ED, Milillo SR, Den Bakker HC. 2010. Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 60:1280–1288. 10.1099/ijs.0.014118-0 [DOI] [PubMed] [Google Scholar]
- 7.Lang Halter E, Neuhaus K, Scherer S. 2012. Listeria weihenstephanensis sp. nov., isolated from the water plant Lemna trisulca of a German fresh water pond. Int. J. Syst. Evol. Microbiol. 63:641–647. 10.1099/ijs.0.036830-0 [DOI] [PubMed] [Google Scholar]
- 8.Leclercq A, Clermont D, Bizet C, Grimont PA, Le Fleche-Mateos A, Roche SM, Buchrieser C, Cadet-Daniel V, Le Monnier A, Lecuit M, Allerberger F. 2010. Listeria rocourtiae sp. nov. Int. J. Syst. Evol. Microbiol. 60:2210–2214. 10.1099/ijs.0.017376-0 [DOI] [PubMed] [Google Scholar]
- 9.Gouin E, Mengaud J, Cossart P. 1994. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 62:3550–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portnoy DA, Chakraborty T, Goebel W, Cossart P. 1992. Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 60:1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Zorn B, Domínguez-Bernal G, Suárez M, Ripio M, Vega Y, Novella S, Vázquez-Boland J. 1999. The smcL gene of Listeria ivanovii encodes a sphingomyelinase C that mediates bacterial escape from the phagocytic vacuole. Mol. Microbiol. 33:510–523. 10.1046/j.1365-2958.1999.01486.x [DOI] [PubMed] [Google Scholar]
- 12.Allerberger F, Wagner M. 2010. Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infect. 16:16–23. 10.1111/j.1469-0691.2009.03109.x [DOI] [PubMed] [Google Scholar]
- 13.Todd ECD, Notermans S. 2011. Surveillance of listeriosis and its causative pathogen, Listeria monocytogenes. Food Control 22:1484–1490. 10.1016/j.foodcont.2010.07.021 [DOI] [Google Scholar]
- 14.Boerlin P, Piffaretti J. 1991. Typing of human, animal, food, and environmental isolates of Listeria monocytogenes by multilocus enzyme electrophoresis. Appl. Environ. Microbiol. 57:1624–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelbícová T, Karpíšková R. 2012. Outdoor environment as a source of Listeria monocytogenes in food chain. Czech J. Food Sci. 30:83–88 http://www.agriculturejournals.cz/publicFiles/56595.pdf [Google Scholar]
- 16.Holch A, Webb K, Lukjancenko O, Ussery D, Rosenthal BM, Gram L. 2013. Genome sequencing identifies two nearly unchanged strains of persistent Listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl. Environ. Microbiol. 79:2944–2951. 10.1128/AEM.03715-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malley TJ, Stasiewicz MJ, Grohn YT, Roof S, Warchocki S, Nightingale K, Wiedmann M. 2013. Implementation of statistical tools to support identification and management of persistent Listeria monocytogenes contamination in smoked fish processing plants. J. Food Prot. 76:796–811. 10.4315/0362-028X.JFP-12-236 [DOI] [PubMed] [Google Scholar]
- 18.Stessl B, Fricker M, Fox E, Karpiskova R, Demnerova K, Jordan K, Ehling-Schulz M, Wagner M. 2014. Collaborative survey on the colonization of different types of cheese-processing facilities with Listeria monocytogenes. Foodborne Pathog. Dis. 11:8–14. 10.1089/fpd.2013.1578 [DOI] [PubMed] [Google Scholar]
- 19.Haase JK, Didelot X, Lecuit M, Korkeala H, Achtman M. 2014. The ubiquitous nature of Listeria monocytogenes clones: a large-scale multilocus sequence typing study. Environ. Microbiol. 16:405–416. 10.1111/1462-2920.12342 [DOI] [PubMed] [Google Scholar]
- 20.Knabel SJ, Reimer A, Verghese B, Lok M, Ziegler J, Farber J, Pagotto F, Graham M, Nadon CA, Gilmour MW. 2012. Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 50:1748–1751. 10.1128/JCM.06185-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomonaco S, Verghese B, Gerner-Smidt P, Tarr C, Gladney L, Joseph L, Katz L, Turnsek M, Frace M, Chen Y. 2013. Novel epidemic clones of Listeria monocytogenes, United States, 2011. Emerg. Infect. Dis. 19:147–150. 10.3201/eid1901.121167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welshimer H, Donker-Voet J. 1971. Listeria monocytogenes in nature. Appl. Microbiol. 21:516–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivant A, Garmyn D, Piveteau P. 2013. Listeria monocytogenes, a down-to-earth pathogen. Front. Cell. Infect. Microbiol. 3:87. 10.3389/fcimb.2013.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowe MJ, Jackson ED, Mori JG, Bell CR. 1997. Listeria monocytogenes survival in soil and incidence in agricultural soils. J. Food Prot. 60:1201–1207 [DOI] [PubMed] [Google Scholar]
- 25.Sauders BD, Overdevest J, Fortes E, Windham K, Schukken Y, Lembo A, Wiedmann M. 2012. Diversity of Listeria species in urban and natural environments. Appl. Environ. Microbiol. 78:4420–4433. 10.1128/AEM.00282-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapin T, Masiello S, Wiedmann M, Bergholz P, Strawn L. 2013. The spatio-temporal distribution and geographical predictors of Listeria species in natural areas and the produce pre-harvest environment of New York State, paper 4563. Int. Assoc. Food Prot. 2013 Annu. Meet., 28 to 31 July 2013 https://iafp.confex.com/iafp/2013/webprogram/Paper4563.html [Google Scholar]
- 27.Kovacevic J, Sagert J, Wozniak A, Gilmour MW, Allen KJ. 2013. Antimicrobial resistance and co-selection phenomenon in Listeria spp. recovered from food and food production environments. Food Microbiol. 34:319–327. 10.1016/j.fm.2013.01.002 [DOI] [PubMed] [Google Scholar]
- 28.Locatelli A, Spor A, Jolivet C, Piveteau P, Hartmann A. 2013. Biotic and abiotic soil properties influence survival of Listeria monocytogenes in soil. PLoS One 8:e75969. 10.1371/journal.pone.0075969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyautey E, Lapen DR, Wilkes G, McCleary K, Pagotto F, Tyler K, Hartmann A, Piveteau P, Rieu A, Robertson WJ. 2007. Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the South Nation River watershed, Ontario, Canada. Appl. Environ. Microbiol. 73:5401–5410. 10.1128/AEM.00354-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiedmann M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524–531 [PubMed] [Google Scholar]
- 31.McLaughlin HP, Casey PG, Cotter J, Gahan CG, Hill C. 2011. Factors affecting survival of Listeria monocytogenes and Listeria innocua in soil samples. Arch. Microbiol. 193:775–785. 10.1007/s00203-011-0716-7 [DOI] [PubMed] [Google Scholar]
- 32.Walsh PS, Metzger DA, Higushi R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513 [PubMed] [Google Scholar]
- 33.Border P, Howard J, Plastow G, Siggens K. 1990. Detection of Listeria species and Listeria monocytogenes using polymerase chain reaction. Lett. Appl. Microbiol. 11:158–162. 10.1111/j.1472-765X.1990.tb00149.x [DOI] [PubMed] [Google Scholar]
- 34.Bubert A, Hein I, Rauch M, Lehner A, Yoon B, Goebel W, Wagner M. 1999. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl. Environ. Microbiol. 65:4688–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clinical and Laboratory Standards Institute. 2010. M45-A Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Approved guideline Clinical and Laboratory Standards Institute, Wayne, PA: [DOI] [PubMed] [Google Scholar]
- 36.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819–3822. 10.1128/JCM.42.8.3819-3822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leclercq A, Chenal-Francisque V, Dieye H, Cantinelli T, Drali R, Brisse S, Lecuit M. 2011. Characterization of the novel Listeria monocytogenes PCR serogrouping profile IVb-v1. Int. J. Food Microbiol. 147:74–77. 10.1016/j.ijfoodmicro.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 38.Rousseaux S, Olier M, Lemaitre JP, Piveteau P, Guzzo J. 2004. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70:2180–2185. 10.1128/AEM.70.4.2180-2185.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan S, Begley M, Hill C, Gahan CG. 2010. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 109:984–995. 10.1111/j.1365-2672.2010.04726.x [DOI] [PubMed] [Google Scholar]
- 40.Jaradat Z, Schutze G, Bhunia A. 2002. Genetic homogeneity among Listeria monocytogenes strains from infected patients and meat products from two geographic locations determined by phenotyping, ribotyping and PCR analysis of virulence genes. Int. J. Food Microbiol. 76:1–10. 10.1016/S0168-1605(02)00050-8 [DOI] [PubMed] [Google Scholar]
- 41.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. 10.1371/journal.ppat.1000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahle D, Wickham H. 2013. ggmap: a package for spatial visualization with Google Maps and OpenStreetMap, version 2.3. http://cran.r-project.org/web/packages/ggmap/index.html
- 43.Core Team R. 2013. R: a language and environment for statistical computing, version 3.0.2. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/ [Google Scholar]
- 44.Ivanek R, Groehn YT, Wells MT, Lembo AJ, Jr, Sauders BD, Wiedmann M. 2009. Modeling of spatially referenced environmental and meteorological factors influencing the probability of Listeria species isolation from natural environments. Appl. Environ. Microbiol. 75:5893–5909. 10.1128/AEM.02757-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Locatelli A, Depret G, Jolivet C, Henry S, Dequiedt S, Piveteau P, Hartmann A. 2013. Nation-wide study of the occurrence of Listeria monocytogenes in French soils using culture-based and molecular detection methods. J. Microbiol. Methods 93:242–250. 10.1016/j.mimet.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 46.Cooley MB, Quiñones B, Oryang D, Mandrell RE, Gorski L. 2014. Prevalence of Shiga toxin producing Escherichia coli, Salmonella enterica and Listeria monocytogenes at public access watershed sites in a California central coast agricultural region. Front. Cell. Infect. Microbiol. 4:30. 10.3389/fcimb.2014.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strawn LK, Grohn YT, Warchocki S, Worobo RW, Bihn EA, Wiedmann M. 2013. Risk factors associated with Salmonella and Listeria monocytogenes contamination of produce fields. Appl. Environ. Microbiol. 79:7618–7627. 10.1128/AEM.02831-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacGowan AP, Bowker K, McLauchlin J, Bennett PM, Reeves DS. 1994. The occurrence and seasonal changes in the isolation of Listeria spp. in shop bought food stuffs, human faeces, sewage and soil from urban sources. Int. J. Food Microbiol. 21:325–334. 10.1016/0168-1605(94)90062-0 [DOI] [PubMed] [Google Scholar]
- 49.Reischer G, Kollanur D, Vierheilig J, Wehrspaun C, Mach R, Sommer R, Stadler H, Farnleitner A. 2011. Hypothesis-driven approach for the identification of fecal pollution sources in water resources. Environ. Sci. Technol. 45:4038–4045. 10.1021/es103659s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301:79–96. 10.1016/j.ijmm.2010.05.002 [DOI] [PubMed] [Google Scholar]
- 51.den Bakker HC, Didelot X, Fortes ED, Nightingale KK, Wiedmann M. 2008. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evol. Biol. 8:277. 10.1186/1471-2148-8-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meloni D, Mazza R, Piras F, Lamon S, Consolati SG, Mureddu A, Mazzette R. 2012. The biofilm formation ability of Listeria monocytogenes isolated from meat, poultry, fish and processing plant environments is related to serotype and pathogenic profile of the strains. Vet. Sci. Dev. 2:e12 [Google Scholar]
- 53.Cantinelli T, Chenal-Francisque V, Diancourt L, Frezal L, Leclercq A, Wirth T, Lecuit M, Brisse S. 2013. “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 51:3770–3779. 10.1128/JCM.01874-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conter M, Paludi D, Zanardi E, Ghidini S, Vergara A, Ianieri A. 2009. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 128:497–500. 10.1016/j.ijfoodmicro.2008.10.018 [DOI] [PubMed] [Google Scholar]
- 55.Granier SA, Moubareck C, Colaneri C, Lemire A, Roussel S, Dao TT, Courvalin P, Brisabois A. 2011. Antimicrobial resistance of Listeria monocytogenes isolates from food and the environment in France over a 10-year period. Appl. Environ. Microbiol. 77:2788–2790. 10.1128/AEM.01381-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Troxler R, Von Graevenitz A, Funke G, Wiedemann B, Stock I. 2000. Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 6:525–535. 10.1046/j.1469-0691.2000.00168.x [DOI] [PubMed] [Google Scholar]
- 57.Chenal-Francisque V, Lopez J, Cantinelli T, Caro V, Tran C, Leclercq A, Lecuit M, Brisse S. 2011. Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 17:1110. 10.3201/eid/1706.101778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Q, Sherwood J, Logue C. 2007. Antimicrobial resistance of Listeria spp. recovered from processed bison. Lett. Appl. Microbiol. 44:86–91. 10.1111/j.1472-765X.2006.02027.x [DOI] [PubMed] [Google Scholar]