Abstract
Switchgrass (Panicum virgatum L.) is a perennial C4 grass native to North America that is being developed as a feedstock for cellulosic ethanol production. Industrial nitrogen fertilizers enhance switchgrass biomass production but add to production and environmental costs. A potential sustainable alternative source of nitrogen is biological nitrogen fixation. As a step in this direction, we studied the diversity of nitrogen-fixing bacteria (NFB) associated with native switchgrass plants from the tallgrass prairie of northern Oklahoma (United States), using a culture-independent approach. DNA sequences from the nitrogenase structural gene, nifH, revealed over 20 putative diazotrophs from the alpha-, beta-, delta-, and gammaproteobacteria and the firmicutes associated with roots and shoots of switchgrass. Alphaproteobacteria, especially rhizobia, predominated. Sequences derived from nifH RNA indicated expression of this gene in several bacteria of the alpha-, beta-, delta-, and gammaproteobacterial groups associated with roots. Prominent among these were Rhizobium and Methylobacterium species of the alphaproteobacteria, Burkholderia and Azoarcus species of the betaproteobacteria, and Desulfuromonas and Geobacter species of the deltaproteobacteria.
INTRODUCTION
Switchgrass (Panicum virgatum L.) is a warm-season C4 grass that is native to the tallgrass prairies of North America, and it has been targeted for development as a bioenergy crop by the U.S. Department of Energy (1). Its features, such as perenniality, adaptation to diverse edaphic conditions, wide geographic distribution, growth on acidic soils, and mutualistic associations with soil microorganisms, make it an attractive choice for cultivation on marginal lands that are poorly suited to food crops (2–4).
Although switchgrass is thrifty in its use of N to produce biomass compared to other crops (2, 5), addition of N fertilizer enhances growth and generally is needed to maintain switchgrass productivity over multiple years (6, 7). However, application of N fertilizer increases production costs, reduces the energy balance of biomass production, and can be harmful to the environment through release of N-containing gases to the atmosphere and soluble N compounds to groundwater, which can lead to eutrophication (8). Biological nitrogen fixation has the potential to reduce the use and negative consequences of industrial N fertilizer and to put the nascent biofuels industry on a more sustainable path. Biological nitrogen fixation has been demonstrated in association with switchgrass (9, 10). However, to the best of our knowledge, the bacteria responsible for such activity are unknown, as is the diversity of potentially nitrogen-fixing bacteria associated with this species.
Nitrogen-fixing bacteria (NFB) associate with many grasses, including maize (11–13), rice (14), sugarcane (15), and Miscanthus species (16). These diazotrophs are endophytic, living between plant cells, and/or epiphytic, living on the surface of plant organs, and most do not elicit plant defense responses (17). However, the extent to which diazotrophic bacteria contribute to the N economy of grasses remains unclear.
Most soil microbes remain uncultured and largely uncharacterized, in part because appropriate culture conditions have not been found. Therefore, we used a culture-independent approach, based on PCR amplification and sequencing of nifH DNA (18, 19), to assess the diversity of potential diazotrophs associated with switchgrass. nifH has been used by many researchers to study total and functional diversity of bacteria in various environments (15, 20–22), and phylogenies based on nifH and 16S rRNA generally are congruent (23, 24). Additionally, PCR amplification of cDNA derived from nifH RNA was used to determine whether any bacteria express this gene in association with switchgrass. The results of this work indicate diverse and abundant NFB associated with switchgrass and active expression of the nifH nitrogen fixation gene in some of these bacteria.
MATERIALS AND METHODS
Plant material.
Switchgrass (Panicum virgatum L.) plants were harvested from separate locations in the tallgrass prairie (Oklahoma, USA) in April and July 2010. GPS coordinates of the sampling sites were between 36°38′38″N to 36°48′48″N latitude and 96°10′26″W to 96°26′23″W longitude (see Table S1 in the supplemental material). Plants were uprooted, and loosely attached soil was removed from roots by vigorous shaking. Roots and shoots were rinsed with tap water at the collection site to remove soil particles and were transferred to the laboratory on dry ice. Samples were stored at −80°C within 24 h of collection.
Nucleic acid extraction.
Shoot and root pieces were pulverized in liquid nitrogen using a cryo-mill (6870 freezer mill; SPEX SamplePrep, USA). DNA was extracted from 0.5 g pulverized tissue using a modified cetyltrimethylammonium bromide (CTAB) method (25). Total RNA was extracted from 100 mg of pulverized tissue using an RNeasy plant minikit (Qiagen, USA) by following the manufacturer's instructions. Genomic DNA was removed by DNaseI treatment (Turbo DNase; Ambion, USA), followed by column purification of RNA using an RNeasy MinElute cleanup kit (Qiagen, USA). RNA was quantified using a Nanodrop spectrophotometer (ND-100; NanoDrop Technologies, Willington, DE) and evaluated for purity with a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Checks for genomic DNA contamination of RNA were performed using nifH primers (mentioned below), 16S rRNA gene 27F (26), and 1492R primers (27).
PCR conditions for nifH amplification.
A nested-PCR approach was used to amplify nifH gene fragments as described previously (28). Two degenerate primers, nifH 3 (5′-ATR TTR TTN GCN GCR TA-3′) and nifH 4 (5′-TTY TAY GGN AAR GGN GG-3′), were used to amplify an ∼460-bp region. One microliter of the resultant PCR product was used as the template to amplify an ∼362-bp region using degenerate primers nifH 1 (5′-TGY GAY CCN AAR GCN GA-3′) and nifH 2 (5′-AND GCC ATC ATY TCN CC-3′). All sample manipulations were performed under laminar flow to avoid airborne contamination. PCRs were initiated with denaturation at 95°C (5 min); 30 cycles of denaturation at 95°C (30 s), annealing at 55°C (1 min), and extension at 72°C (1 min); and a final extension at 72°C (10 min). Reverse transcription-PCR (RT-PCR) was done as described by Zani et al. (28), and cDNA synthesis was carried out using 1 μg of DNase-treated total RNA with primer nifH 3 and SuperScript III reverse transcriptase according to the manufacturer's instructions (Invitrogen, USA). Nested PCR was performed with the nifH 3 and nifH 4 primer combination, followed by the nifH 1 and nifH 2 combination. No-RT (i.e., no cDNA) controls were included for all RNA samples analyzed to check for genomic DNA contamination.
nifH amplification products were separated by electrophoresis through 2.0% agarose gels and visualized with Sybr green. Amplified fragments were excised from gels and purified using a Promega gel extraction kit (Promega, USA) and then cloned into the pGEM-T Easy vector (Promega, USA). Bacterial transformation was done using Z-competent cells (Zymo Research, USA). Forty-eight white colonies per sample were picked, and colonies were grown in 96-well plates.
nifH sequencing and phylogenetic analysis.
Recombinant pGEM-T plasmids containing nifH fragments were sequenced using an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, USA). DNA sequences were edited to remove vector sequences using GeneDoc (29), and identical sequences were grouped into operational taxonomic units (OTUs), using the FastGroupII tool (30) with the method of percent sequence identity with gaps of 100% identity. DNA sequences were translated into protein amino acid sequences using BioEdit software (31). Protein sequences were subjected to BLAST analysis (32) against GenBank, and nifH protein sequences from cultivated bacteria that showed the closest match to query sequences were included for phylogenetic analysis. OTUs representing 4 or more sequences were subjected to phylogenetic analysis. The MAFFT web interface was used for alignment (33).
Phylogenetic trees were constructed using MEGA (ver. 5.0) with the maximum likelihood method based on a Poisson correction model (34). The bootstrap consensus tree was produced from 1,000 replicates, and Methanothermococcus okinawensis NifH (accession no. NZ_AEDA01000001) was used as an outgroup.
Data analysis.
nifH sequences from root DNA, root RNA (cDNA), and shoot DNA of individual plants were subjected to rarefaction analysis, and Shannon-Weaver and Simpson diversity indices were calculated using PAST statistical software (35). The percentage of total sequence diversity captured in sequenced clones of each organ for each plant was estimated by Good's method using the formula [1 − (n/N)] × 100, where n is the number of clones appearing only once in a library and N is the total number of clones sequenced (36). Venn diagrams were drawn using Excel (Microsoft).
Accession numbers.
Representative sequences from each OTU were deposited in GenBank under accession numbers KF541058 to KF541088, and the protein accession numbers are AIE38835 to AIE38865.
RESULTS
Diversity of nifH sequences.
A total of 28 clone libraries were produced from nifH PCR amplicons derived from DNA of roots and shoots separately from 10 independent plants and RNA from roots of 8 of these plants. Up to 48 clones were sequenced from each library, yielding a total of 1,087 high-quality nifH sequences. Based upon shared sequence identity, these sequences formed 52 distinct OTUs, which were tentatively associated with known species based on the highest sequence similarity (Table 1).
TABLE 1.
Phylotypes of nifH associated with switchgrass
| Phylotype | Closest relative | Accession no. | Similarity (%) | No. of sequences per sample type |
Taxonomic description | |||
|---|---|---|---|---|---|---|---|---|
| Root |
Shoot DNA | Total | ||||||
| DNA | RNA | |||||||
| OTU-01 | Bradyrhizobium sp. strain BTAi 1 | BAC07281 | 98–100 | 35 | 0 | 15 | 50 | Alphaproteobacteria |
| OTU-02 | Bradyrhizobium sp. strain MAFF 210318 | BAC07283 | 94–98 | 46 | 2 | 71 | 119 | Alphaproteobacteria |
| OTU-03 | Mesorhizobium loti | BAF95636 | 100 | 0 | 0 | 4 | 4 | Alphaproteobacteria |
| OTU-04 | Methylocystis sp. strain LW2 | AAK97418 | 97–98 | 24 | 2 | 8 | 34 | Alphaproteobacteria |
| OTU-05 | Burkholderia sp. strain WSM3937 | ABX80637 | 93–98 | 58 | 17 | 1 | 76 | Betaproteobacteria |
| OTU-06 | Burkholderia sp. strain JPY105 | AFM28513 | 97 | 7 | 7 | 3 | 17 | Betaproteobacteria |
| OTU-07 | Burkholderia sp. strain PTK47 | AAU85620 | 96–100 | 3 | 0 | 20 | 23 | Betaproteobacteria |
| OTU-08 | Sphingomonas azotifigens | BAE71134 | 95–99 | 81 | 0 | 55 | 136 | Alphaproteobacteria |
| OTU-09 | Novosphingobium sp. strain Rr 2-17 | ZP_10360790 | 98–99 | 29 | 0 | 0 | 29 | Alphaproteobacteria |
| OTU-10 | Bradyrhizobium japonicum | ACT67985 | 92–98 | 71 | 0 | 2 | 73 | Alphaproteobacteria |
| OTU-11 | Rubrivivax gelatinosus | BAE15985 | 90–94 | 0 | 5 | 0 | 5 | Betaproteobacteria |
| OTU-12 | Methylobacterium nodulans | AAQ82902 | 94–97 | 6 | 38 | 4 | 48 | Alphaproteobacteria |
| OTU-13 | Amorphomonas oryzae | BAF48338 | 96 | 0 | 1 | 7 | 8 | Alphaproteobacteria |
| OTU-14 | Rhizobium helanshanense | ADP37388 | 95–100 | 0 | 88 | 40 | 128 | Alphaproteobacteria |
| OTU-15 | Sinorhizobium sp. strain SCAU224 | AFH96094 | 98 | 3 | 0 | 1 | 4 | Alphaproteobacteria |
| OTU-16 | Azospirillum lipoferum | ABG88868 | 96–100 | 3 | 2 | 22 | 27 | Alphaproteobacteria |
| OTU-17 | Methylomonas sp. strain MG30 | CCH22595 | 93 | 5 | 0 | 0 | 5 | Gammaproteobacteria |
| OTU-18 | Pseudomonas stutzeri | CAC03734 | 97 | 7 | 4 | 0 | 11 | Gammaproteobacteria |
| OTU-19 | Klebsiella sp. strain AL060224_04 | ACM68399 | 99–100 | 0 | 0 | 18 | 18 | Gammaproteobacteria |
| OTU-20 | Azoarcus sp. strain BH72 | YP_932042 | 96–98 | 1 | 21 | 0 | 22 | Betaproteobacteria |
| OTU-21 | Desulfuromonas acetoxidans | ZP_01312343 | 94–99 | 0 | 32 | 31 | 63 | Deltaproteobacteria |
| OTU-22 | Anaeromyxobacter sp. strain Fw109-5 | YP_001380211 | 93–97 | 12 | 0 | 15 | 27 | Deltaproteobacteria |
| OTU-23 | Geobacter uraniireducens Rf4 | YP_001229952 | 92 | 1 | 0 | 5 | 6 | Deltaproteobacteria |
| OTU-24 | Geobacter sp. strain M21 | YP_003021955 | 93–96 | 4 | 3 | 0 | 7 | Deltaproteobacteria |
| OTU-25 | Geobacter sp. strain M21 | YP_003021955 | 97–98 | 35 | 15 | 5 | 55 | Deltaproteobacteria |
| OTU-26 | Geobacter uraniireducens Rf4 | YP_001229952 | 95–97 | 3 | 0 | 6 | 9 | Deltaproteobacteria |
| OTU-27 | Anaeromyxobacter sp. strain Fw109-5 | YP_001380211 | 88–91 | 0 | 5 | 2 | 7 | Deltaproteobacteria |
| OTU-28 | Desulfotomaculum gibsoniae | ZP_09101754 | 93–95 | 0 | 0 | 15 | 15 | Firmicutes |
| OTU-29 | Geobacter sp. strain M21 | YP_003021955 | 89–92 | 13 | 0 | 0 | 13 | Deltaproteobacteria |
| OTU-30 | Desulfovibrio magneticus RS-1 | YP_002953433 | 89 | 4 | 0 | 0 | 4 | Deltaproteobacteria |
| OTU-31 | Syntrophobacter fumaroxidans | YP_845148 | 90–93 | 0 | 0 | 16 | 16 | Deltaproteobacteria |
| OTU-32 | Methylocystis sp. strain LW5 | AAK97419 | 98 | 2 | 0 | 0 | 2 | Alphaproteobacteria |
| OTU-33 | Coraliomargarita akajimensis | YP_003550022 | 89 | 1 | 0 | 1 | 2 | Verrucomicrobia |
| OTU-34 | Cupriavidus sp. strain pp2.75 | ADM25241 | 92 | 1 | 0 | 0 | 1 | Betaproteobacteria |
| OTU-35 | Rhizobium sp. strain SMF 466_6 | CCN27451 | 98 | 0 | 0 | 2 | 2 | Alphaproteobacteria |
| OTU-36 | Paenibacillus graminis | BAH23271 | 94 | 0 | 0 | 2 | 2 | Firmicutes |
| OTU-37 | Rhizobium sp. strain SMF 466_6 | CCN27451 | 92 | 0 | 2 | 0 | 2 | Alphaproteobacteria |
| OTU-38 | Nostoc sp. strain PCC 7120 | NP_484917 | 99 | 0 | 0 | 2 | 2 | Cyanobacteria |
| OTU-39 | Calothrix sp. strain LEGE 06100 | AGG40738 | 97 | 0 | 0 | 1 | 1 | Cyanobacteria |
| OTU-40 | Ectothiorhodospira haloalkaliphila | ABN10975 | 88 | 1 | 0 | 0 | 1 | Gammaproteobacteria |
| OTU-41 | Desulfatibacillum alkenivorans | YP_002430688 | 93 | 1 | 0 | 0 | 1 | Deltaproteobacteria |
| OTU-42 | Bradyrhizobium sp. strain cmy11 | AEP33459 | 96 | 1 | 0 | 0 | 1 | Alphaproteobacteria |
| OTU-43 | Rhizobium sp. strain SMF 466_6 | CCN27451 | 97 | 0 | 0 | 1 | 1 | Alphaproteobacteria |
| OTU-44 | Cupriavidus sp. strain pp2.75 | ADM25241 | 98 | 1 | 0 | 0 | 1 | Betaproteobacteria |
| OTU-45 | Dechloromonas aromatica | YP_284634 | 98 | 1 | 0 | 0 | 1 | Betaproteobacteria |
| OTU-46 | Ectothiorhodospira haloalkaliphila | ABN10975 | 90 | 1 | 0 | 0 | 1 | Gammaproteobacteria |
| OTU-47 | Heliorestis baculata | BAD80875 | 88 | 0 | 1 | 0 | 1 | Firmicutes |
| OTU-48 | Mesorhizobium tianshanense | CAR57837 | 96 | 0 | 0 | 1 | 1 | Alphaproteobacteria |
| OTU-49 | Methylocystis echinoides | AAO49390 | 89 | 1 | 0 | 0 | 1 | Alphaproteobacteria |
| OTU-50 | Pelobacter carbinolicus | YP_006717849 | 93 | 0 | 0 | 1 | 1 | Deltaproteobacteria |
| OTU-51 | Geobacter bemidjiensis | YP_002138883 | 95 | 1 | 0 | 0 | 1 | Deltaproteobacteria |
| OTU-52 | Halorhodospira halophila | ABN10970 | 94 | 0 | 1 | 0 | 1 | Gammaproteobacteria |
Estimations of species coverage, diversity, and dominance were calculated for each library separately (Table 2). Good's coverage averaged over 90% for root DNA libraries, over 95% for root RNA, and over 98% for shoot DNA libraries, indicating that the majority of nifH-containing and nifH-expressing bacteria associated with switchgrass plants were represented in the sequences obtained. Based on the number of OTUs represented by sequences in each library, bacteria containing nifH were significantly more diverse in roots than in shoots (P = 0.0145) of switchgrass plants, with an average of approximately 10 and 6 OTUs per plant, respectively (Table 2). About one-third of nifH-containing bacteria were found to express the gene in association with switchgrass roots (Table 1). Simpson's and Shannon's indices supported these conclusions about nifH bacterial diversity in the different sample types (Table 2). Overall dominance of one or a few OTUs was highest in root RNA, followed by shoot DNA and root DNA (Table 2). In three of the eight plants analyzed for RNA, only a single OTU was represented in the nifH sequences (i.e., dominance = 1).
TABLE 2.
nifH diversity and coverage estimates for root DNA and RNA and shoot DNA
| DNA/RNA type and parameter | Value for plant no.: |
Avg | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E-01 | E-04 | E-06 | E-08 | E-13 | E-18 | E-28 | E-31 | E-36 | E-40 | ||
| Root DNA | |||||||||||
| No. of taxa | 11 | 12 | 14 | 15 | 8 | 10 | 5 | 5 | 9 | 9 | 9.8 |
| No. of clones | 48 | 48 | 48 | 44 | 48 | 46 | 46 | 44 | 44 | 48 | 46.4 |
| Good's estimator (%) | 89.58 | 83.33 | 93.75 | 79.55 | 95.83 | 93.48 | 97.83 | 93.18 | 93.18 | 95.83 | 91.55 |
| Dominance | 0.201 | 0.237 | 0.133 | 0.137 | 0.248 | 0.158 | 0.306 | 0.717 | 0.185 | 0.170 | 0.249 |
| 1 − Simpson diversity index | 0.799 | 0.763 | 0.867 | 0.863 | 0.752 | 0.842 | 0.694 | 0.283 | 0.815 | 0.829 | 0.750 |
| Shannon's diversity index | 1.897 | 1.8 | 2.311 | 2.266 | 1.647 | 2.002 | 1.333 | 0.622 | 1.86 | 1.929 | 1.766 |
| Evenness | 0.606 | 0.50411 | 0.721 | 0.643 | 0.649 | 0.740 | 0.759 | 0.372 | 0.711 | 0.765 | 0.646 |
| Root RNA | |||||||||||
| No. of taxa | 5 | 6 | 3 | 1 | 1 | 3 | 1 | 6 | 3.25 | ||
| No. of clones | 41 | 31 | 32 | 33 | 25 | 19 | 32 | 34 | 30.87 | ||
| Good's estimator (%) | 97.56 | 93.33 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 98.86 | ||
| Dominance | 0.327 | 0.611 | 0.639 | 1 | 1 | 0.424 | 1 | 0.279 | 0.659 | ||
| 1 − Simpson diversity index | 0.673 | 0.389 | 0.361 | 0 | 0 | 0.576 | 0 | 0.721 | 0.340 | ||
| Shannon's diversity index | 1.271 | 0.884 | 0.656 | 0 | 0 | 0.972 | 0 | 1.492 | 0.659 | ||
| Evenness | 0.713 | 0.404 | 0.643 | 1 | 1 | 0.882 | 1 | 0.741 | 0.798 | ||
| Shoot DNA | |||||||||||
| No. of taxa | 3 | 4 | 5 | 3 | 6 | 7 | 8 | 9 | 8 | 9 | 6.2 |
| No. of clones | 38 | 38 | 44 | 41 | 47 | 32 | 31 | 42 | 28 | 35 | 37.6 |
| Good's estimator (%) | 100.00 | 97.37 | 95.45 | 100.00 | 100.00 | 90.63 | 93.55 | 97.62 | 89.29 | 91.43 | 95.53 |
| Dominance | 0.367 | 0.540 | 0.371 | 0.411 | 0.231 | 0.320 | 0.213 | 0.194 | 0.212 | 0.211 | 0.307 |
| 1 − Simpson diversity index | 0.633 | 0.459 | 0.629 | 0.589 | 0.769 | 0.679 | 0.787 | 0.806 | 0.788 | 0.789 | 0.693 |
| Shannon's diversity index | 1.043 | 0.872 | 1.155 | 0.967 | 1.579 | 1.437 | 1.769 | 1.874 | 1.753 | 1.827 | 1.428 |
| Evenness | 0.946 | 0.598 | 0.635 | 0.877 | 0.809 | 0.601 | 0.733 | 0.724 | 0.722 | 0.691 | 0.733 |
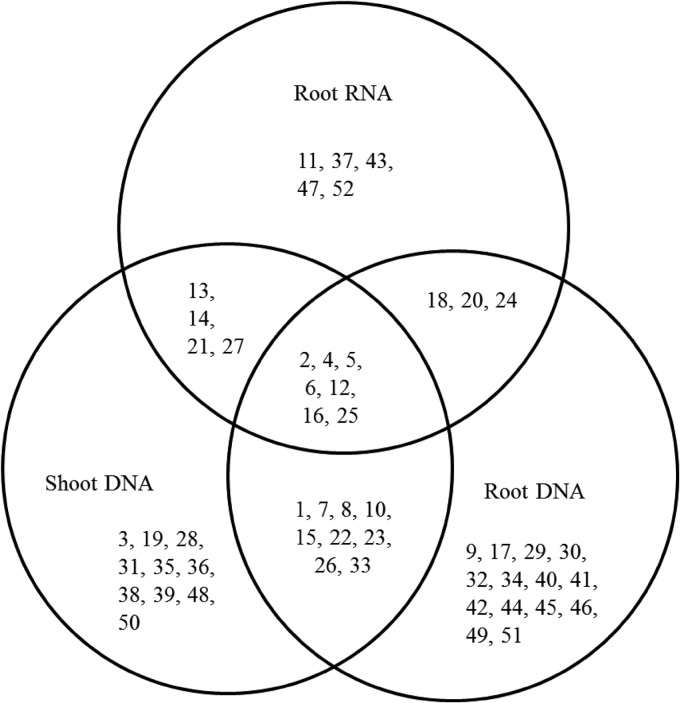
Seven OTUs were common to all sample types, while 22 were specific to roots (DNA plus RNA), and 10 OTUs were specific to shoots (Fig. 1).
FIG 1.
Venn diagram showing the number of shared and unique OTUs among the three sample types (root DNA, root RNA, and shoot DNA). Numbers indicated in the diagram are OTU identity numbers (details of each OTU are given in Table 1).
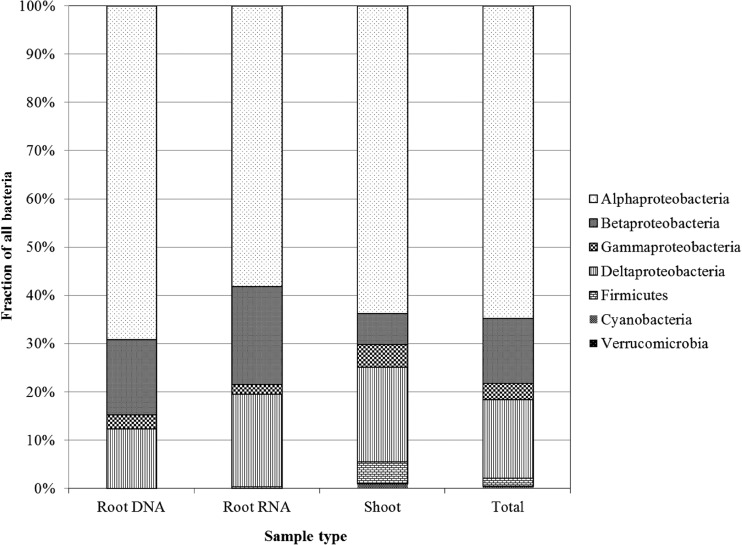
OTUs were matched to the most closely related known nitrogen-fixing species based on nifH sequence comparisons using the BLAST algorithm. A total of 23 OTUs representing 704 sequences appeared to derive from alphaproteobacteria, 9 OTUs (177 sequences) from deltaproteobacteria, 8 from betaproteobacteria (146 sequences), 6 from gammaproteobacteria (37 sequences), 2 from cyanobacteria (3 sequences), and 1 OTU each from Firmicutes (15 sequences) and Verrucomicrobia (3 sequences) (Table 1 and Fig. 2).
FIG 2.
Class-level composition of nifH clone libraries. Percent abundances per clone library from root RNA (242 sequences), root DNA (451 sequences), shoot DNA (397 sequences), and overall sequences (1,060) are shown.
nifH diversity and expression in roots.
Among the nifH sequences derived from root-associated DNA, more than 65% originated from alphaproteobacteria, 15% from betaproteobacteria, and 12% from deltaproteobacteria. The largest number of sequences in a single OTU (OTU-8; 81 sequences) were most similar (95 to 99% identity) to nifH from Sphingomonas azotifigens (accession no. BAE71134). Approximately one-third of all sequences (33%) were affiliated with various Bradyrhizobium species and distributed into 3 OTUs (1, 2, and 10) (Table 1 and Fig. 3). At the individual plant level, OTUs 1, 2, 4, 5, 8, and 10 (all alphaproteobacteria) were present in 7 to 8 out of 10 plants (see Table S2 in the supplemental material).
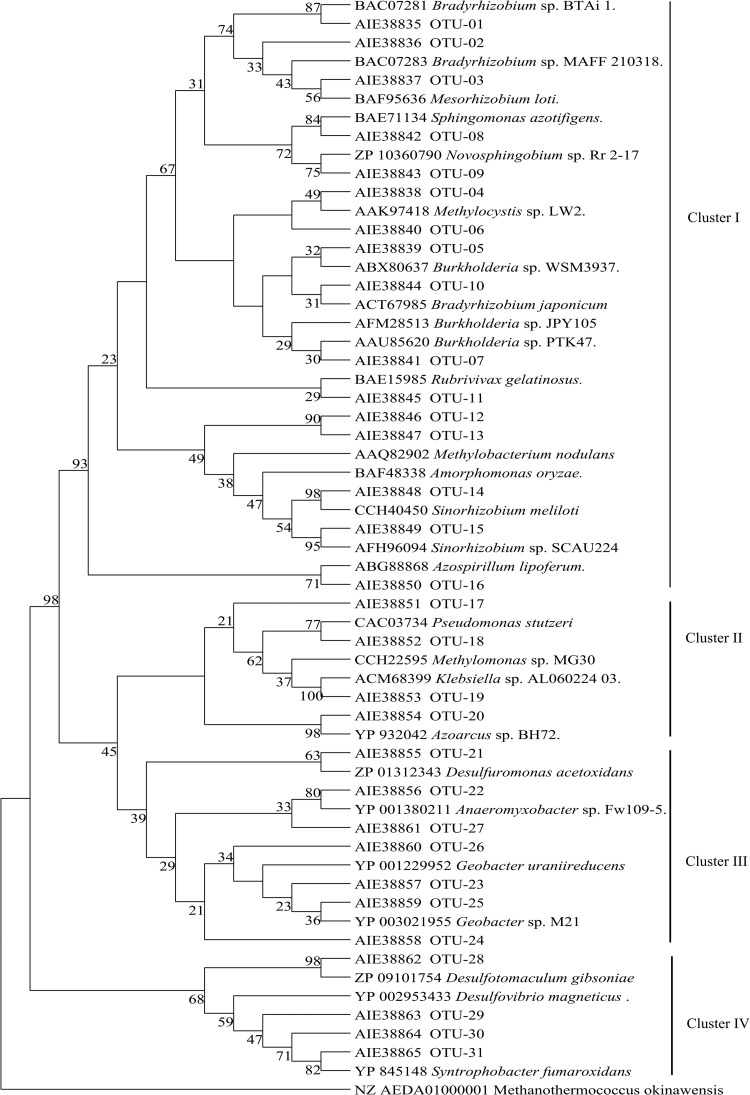
FIG 3.
Phylogeny and composition of nifH phylotypes representing four or more sequences. OTUs from this study are represented by OTU number and the associated protein accession number. The evolutionary history was inferred by using the maximum likelihood method based on the Poisson correction model. The bootstrap consensus tree was inferred from 1,000 replicates. There were a total of 113 positions in the final data set, and analyses were conducted in MEGA5. Sequences from known bacteria are indicated by name and NCBI protein accession numbers. This tree was rooted with the nifH gene from the archaeon Methanothermococcus okinawensis.
Among the nifH sequences derived from root-associated RNA, 58% matched sequences from alphaproteobacteria, 20% from betaproteobacteria, and 19% from deltaproteobacteria (Table 1 and Fig. 2). Over one-third of transcribed nifH sequences (88) fell into OTU-14, which is affiliated with Rhizobium helanshanense (ADP37388; 95 to 100% identity) or Sinorhizobium meliloti (CCH40450), while 38 sequences fell into OTU-12, affiliated with Methylobacterium nodulans (AAQ82902). Five OTUs contained sequences derived from root RNA but not root DNA (OTU-11, affiliated with Rubrivivax gelatinosus; OTU-13, affiliated with Amorphomonas oryzae; OTU-14, affiliated with Rhizobium helanshanense/S. meliloti; OTU-21, affiliated with Desulfuromonas acetoxidans; and OTU-27, affiliated with Bradyrhizobium japonicum), indicating measurable nifH gene expression in several apparently low-abundance root bacteria (Table 1 and Fig. 3). Overall, 3 OTUs contained sequences derived from root DNA and RNA, 14 OTUs contained sequences from root DNA but not RNA, and 5 OTUs contained sequences from root RNA but not DNA (Table 1 and Fig. 1). At the individual plant level, OTU-4 contained nifH RNA sequences from the most plants (only 4 of 8 plants), indicating that no single nifH-expressing species was common to all switchgrass plants (see Table S2 in the supplemental material).
nifH sequences from shoot bacteria.
BLAST results indicated that 63% of sequences derived from shoot-associated DNA were affiliated with alphaproteobacteria and 20% with deltaproteobacteria. Sequences affiliated with firmicutes were associated only with shoots (OTU-28) (Fig. 2). Eighteen sequences affiliated with gammaproteobacteria (Klebsiella species accession no. ACM68399) were represented by OTU-19 (Table 1 and Fig. 3). OTU-2 contained the largest number (71 sequences) of nifH sequences from shoots, which were affiliated with an uncultured bacterium related to Bradyrhizobium sp. strain MAFF 210318 (94 to 98% identity). Ten OTUs were unique to shoots (OTUs 3, 19, 28, 31, 35, 36, 38, 39, 48, and 50; Fig. 1), among them sequences affiliated with Mesorhizobium loti (OTU-3; 100% identity) and with Syntrophobacter fumaroxidans (OTU-31; 90 to 93% identity) (Table 1 and Fig. 3). Half or more of the plants contained nifH DNA sequences affiliated with Bradyrhizobium sp. strain BTAi 1 (OTU-1), Burkholderia sp. strain PTK47 (OTU-7), and Sphingomonas azotifigens (OTU-8). A small number of nifH sequences from shoots were affiliated with cyanobacteria (Table 1 and Fig. 3).
DISCUSSION
We used a culture-independent, nifH sequence-based approach to survey the diversity of potential nitrogen-fixing bacteria associated with switchgrass. To our knowledge, this is the first study of its kind for switchgrass, and as such it lays a foundation for future work on the isolation and characterization of associative diazotrophic bacteria from switchgrass and their potential use as a nitrogen source for this biofuel crop.
Over 1,000 nifH PCR amplicons derived from root and shoot DNA and root RNA of 10 plants were cloned and sequenced in this study. Roots of switchgrass plants harbored a greater diversity of nifH-containing bacteria than did shoots (Table 2). Similar results have been obtained for maize (37). However, only a small fraction of root-associated nifH-containing bacteria expressed this gene (Table 2). Similar results have been found in sugarcane, spruce (15), and rice (14). Shannon's diversity indices for nifH transcripts associated with individual plants ranged from 0.142 to 1.245 for sugar cane, and for one spruce tree it was 0.184 (15), similar to what was found here for switchgrass.
Alphaproteobacteria were the main source of nifH sequences in both roots and shoots, accounting for 62% of all DNA and RNA sequences combined. Within this group, 58% of the sequences were related to nifH in bacteria from the order Rhizobiales and were present in all sample types. More specifically, these sequences were affiliated with the genera Bradyrhizobium, Mesorhizobium, Methylobacterium, Rhizobium, and Sinorhizobium, all of which contain species that are known to fix nitrogen in symbioses with legumes (38–45).
Based on the frequency of nifH recovered from DNA, Bradyrhizobium was the dominant genus associated with switchgrass roots and shoots, with 22% (243 sequences) of all clones distributed in 3 OTUs (Table 1). OTU-10 was closely related to Bradyrhizobium japonicum sequences. B. japonicum is a functionally diverse species (38), and some strains can fix nitrogen under free-living conditions (46). Bradyrhizobium sp. strain MAFF210318, affiliated with OTU-2, is a nonphotosynthetic bacterium that can fix nitrogen under free-living conditions (47). Bradyrhizobium sp. strain BTAi 1, affiliated with phylotype 1, is a photosynthetic rhizobium that also can fix nitrogen under free-living conditions (8, 48). Bradyrhizobium species have been found as endophytes in rice (49) and sugarcane (15, 50). Despite the abundance of Bradyrhizobium bacteria associated with switchgrass, however, the paucity of Bradyrhizobium-like nifH sequences derived from root RNA suggests that these bacteria fix little or no nitrogen in association with native switchgrass.
Evidence was obtained for nifH gene expression in a variety of bacterial genera associated with switchgrass, including affiliates of Rhizobium/S. meliloti (OTU-14), Methylobacterium (OTU-12), Desulfuromonas (OTU-21), Burkholderia (OTU-5 and OTU-6), Azoarcus (OTU-20), Geobacter (OTU-25), Bradyrhizobium (OTU-2, OTU-24, and OTU-27), Rubrivivax (OTU-11), Pseudomonas (OTU-18), Methylocystis (OTU-4), Azospirillum (OTU-16), and Amorphomonas (OTU-13), in descending order of prevalence based on numbers of sequenced clones. Half of these genera belong to the alphaproteobacteria, while the remainder partitioned into the beta-, gamma-, and deltaproteobacteria. Rhizobium helanshanense/S. meliloti (ADP37388/CCH40450)-affiliated sequences (alphaproteobacteria) were prominent among root nifH RNA, accounting for 36% of all root RNA-derived sequences (Table 1). However, only one sequence was detected from root DNA, and many (44 sequences) were identified from shoot DNA. OTU-14 showed over 97% identity with sequences from S. meliloti and various Rhizobium species, such as Rhizobium sp. strain CCNWSX0878 (accession no. AEB96238), R. yanglingense (AFD62623), and R. undicola (AEP04095). These results indicate active nifH gene expression in a very small population of specific rhizobia, a conclusion that warrants confirmation in future work designed to identify the best potential nitrogen-fixing bacteria for switchgrass.
Practical application of nitrogen-fixing bacteria for N supply to switchgrass will depend not only on active nitrogenase genes but also on the ability of the plant to host a sufficiently large population of the bacteria. Given the low number of rhizobial nifH DNA sequences that match nifH RNA sequences from switchgrass roots, the latter would seem to be an impediment to the development of effective nitrogen fixation in the roots of switchgrass.
A large number of nifH clones (38 in OTU-12) derived from root RNA were affiliated with Methylobacterium nodulans ORS 2060 (AAQ82902), another alphaproteobacteria able to form nitrogen-fixing nodules on legumes (51), as well as Desulfuromonas acetoxidans (32 in OTU-21) and Azoarcus sp. strain BH72 (21 in OTU-20). Here, nifH expression for these three bacteria outweighed their DNA abundance in the samples investigated.
Within the betaproteobacterial group, Burkholderia species appeared to be the source of some nifH sequences. Burkholderia species have been found to nodulate and fix nitrogen in mimosoid legumes like Mimosa pigra (52), and some are known to associate with maize and sugarcane (53, 54). Expressed nifH sequences also affiliated with the betaproteobacterium Azoarcus sp. strain BH72 (YP_932042), which was isolated from Kallar grass and can colonize rice roots and express high levels of nifH transcripts there (55, 56).
Finally, the deltaproteobacteria nifH transcripts identified in switchgrass roots were affiliated with Geobacter sp. strain M21 and D. acetoxidans (ZP_01312343). Geobacter nifH transcripts have been found previously in roots and stems of rice (22, 57).
We are aware that taxonomic inferences based on single genes, such as nifH, can be complicated by horizontal gene transfer. For this reason, the tentative assignments of bacterial genus and species associated with switchgrass made here should be substantiated by further work with isolated, cultured bacteria in the future. However, an independent, 16S rRNA gene-based metagenomic study of bacteria associated with switchgrass from the same sites in Oklahoma, harvested at different times of year, identified 14 of the 19 genera found in the present study (S. R. Chaluvadi and J. L. Bennetzen, personal communication).
In summary, we have identified some of the natural diversity of diazotrophic bacteria associated with switchgrass from the Oklahoma prairie. Evidence was presented that nifH genes of many bacterial species from the alpha-, beta-, gamma-, and deltaproteobacterial groups are expressed in switchgrass roots. Prominent among these were Rhizobium and Methylobacterium species of the alphaproteobacteria, Burkholderia and Azoarcus species of the betaproteobacteria, and Desulfuromonas and Geobacter species of the deltaproteobacteria. This work provides a basis for future work on the isolation, culture, and functional characterization of nitrogen-fixing endophytes of switchgrass and their possible use as nitrogen sources for cultivated switchgrass.
Supplementary Material
ACKNOWLEDGMENTS
Financial support for this work was provided by The Samuel Roberts Noble Foundation and the DOE BioEnergy Science Center.
We thank the Genomics/Microarray Facility at The Noble Foundation for sequencing all nifH libraries.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02091-14.
REFERENCES
- 1.Bouton J. 2008. Improvement of switchgrass as a bioenergy crop, p 295–308 In Vermerris W. (ed), Genetic improvement of bioenergy crops. Spinger Science and Business Media, New York, NY [Google Scholar]
- 2.Parrish DJ, Fike JH. 2005. The biology and agronomy of switchgrass for biofuels. Crit. Rev. Plant Sci. 24:423–459. 10.1080/07352680500316433 [DOI] [Google Scholar]
- 3.Parrish DJ, Fike JH. 2009. Selecting, establishing, and managing switchgrass (Panicum virgatum) for biofuels. Methods Mol. Biol. 581:27–40. 10.1007/978-1-60761-214-8_2 [DOI] [PubMed] [Google Scholar]
- 4.Ghimire SR, Charlton ND, Bell JD, Krishnamurthy YL, Craven KD. 2011. Biodiversity of fungal endophyte communities inhabiting switchgrass (Panicum virgatum L.) growing in the native tallgrass prairie of northern Oklahoma. Fungal Divers. 47:19–27. 10.1007/s13225-010-0085-6 [DOI] [Google Scholar]
- 5.Lemus R, Parrish DJ, Wolf DD. 2009. Nutrient uptake by alamo switchgrass used as an energy crop. BioEnergy Res. 2:37–50. 10.1007/s12155-009-9032-3 [DOI] [Google Scholar]
- 6.Yang JD, Worley E, Wang MY, Lahner B, Salt DE, Saha M, Udvardi M. 2009. Natural variation for nutrient use and remobilization efficiencies in switchgrass. BioEnergy Res. 2:257–266. 10.1007/s12155-009-9055-9 [DOI] [Google Scholar]
- 7.Guretzky JA, Biermacher JT, Cook BJ, Kering MK, Mosali J. 2010. Switchgrass for forage and bioenergy: harvest and nitrogen rate effects on biomass yields and nutrient composition. Plant Soil 339:69–81. 10.1007/s11104-010-0376-4 [DOI] [Google Scholar]
- 8.Spiertz JHJ. 2010. Nitrogen, sustainable agriculture and food security. A review. Agron. Sustain. Dev. 30:43–55. 10.1051/agro:2008064 [DOI] [Google Scholar]
- 9.Tjepkema JD, Burris RH. 1976. Nitrogenase activity associated with some Wisconsin prairie grasses. Plant Soil 45:81–94. 10.1007/BF00011131 [DOI] [Google Scholar]
- 10.Tjepkema J. 1975. Nitrogenase activity in the rhizosphere of Panicum virgatum. Soil Biol. Biochem. 7:179–180. 10.1016/0038-0717(75)90016-4 [DOI] [Google Scholar]
- 11.Montañez A, Abreu C, Gill PR, Hardarson G, Sicardi M. 2008. Biological nitrogen fixation in maize (Zea mays L.) by 15N isotope-dilution and identification of associated culturable diazotrophs. Biol. Fertil. Soils 45:253–263. 10.1007/s00374-008-0322-2 [DOI] [Google Scholar]
- 12.Gutiérrez-Zamora ML, Martínez-Romero E. 2001. Natural endophytic association between Rhizobium etli and maize (Zea mays L.). J. Biotechnol. 91:117–126. 10.1016/S0168-1656(01)00332-7 [DOI] [PubMed] [Google Scholar]
- 13.Rosenblueth M, Martínez-Romero E. 2004. Rhizobium etli maize populations and their competitiveness for root colonization. Arch. Microbiol. 181:337–344. 10.1007/s00203-004-0661-9 [DOI] [PubMed] [Google Scholar]
- 14.Sessitsch A, Hardoim P, Doering J, Weilharter A, Krause A, Woyke T, Mitter B, Hauberg-Lotte L, Friedrich F, Rahalkar M, Hurek T, Sarkar A, Bodrossy L, van Overbeek L, Brar D, van Elsas JD, Reinhold-Hurek B. 2012. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant Microbe Interact. 25:28–36. 10.1094/MPMI-08-11-0204 [DOI] [PubMed] [Google Scholar]
- 15.Burbano CS, Liu Y, Rosner KL, Reis VM, Caballero-Mellado J, Reinhold-Hurek B, Hurek T. 2011. Predominant nifH transcript phylotypes related to Rhizobium rosettiformans in field-grown sugarcane plants and in Norway spruce. Environ. Microbiol. Rep. 3:383–389. 10.1111/j.1758-2229.2010.00238.x [DOI] [PubMed] [Google Scholar]
- 16.Kirchhof G, Reis VM, Baldani JI, Eckert B, Dobereiner J, Hartmann A. 1997. Occurrence, physiological and molecular analysis of endophytic diazotrophic bacteria in gramineous energy plants. Plant Soil 194:45–55. 10.1023/A:1004217904546 [DOI] [Google Scholar]
- 17.Hurek T, Handley LL, Reinhold-Hurek B, Piche Y. 2002. Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Mol. Plant Microbe Interact. 15:233–242. 10.1094/MPMI.2002.15.3.233 [DOI] [PubMed] [Google Scholar]
- 18.Zehr JP, Mellon MT, Hiorns WD. 1997. Phylogeny of cyanobacterial nifH genes: evolutionary implications and potential applications to natural assemblages. Microbiology 143:1443–1450. 10.1099/00221287-143-4-1443 [DOI] [PubMed] [Google Scholar]
- 19.Hurek T, Reinhold-Hurek B. 2006. Molecular ecology of N2-fixing microbes associated with graminaceous plants: hidden activities of unknown bacteria, p 173–198 In Werner D, Newton WE. (ed), Nitrogen fixation in agriculture, forestry, ecology, and the environment. Springer, New York, NY [Google Scholar]
- 20.Zehr JP, Mellon M, Braun S, Litaker W, Steppe T, Paerl HW. 1995. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl. Environ. Microbiol. 61:2527–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lema KA, Willis BL, Bourne DG. 2012. Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78:3136–3144. 10.1128/AEM.07800-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martensson L, Diez B, Wartiainen I, Zheng W, El-Shehawy R, Rasmussen U. 2009. Diazotrophic diversity, nifH gene expression and nitrogenase activity in a rice paddy field in Fujian, China. Plant Soil 325:207–218. 10.1007/s11104-009-9970-8 [DOI] [Google Scholar]
- 23.Young JPW. 1992. Phylogenetic classification of nitrogen-fixing organisms, p 43–86 In Stacy G, Burris RH, Evans HJ. (ed), Biological nitrogen fixation. Chapman and Hall, New York, NY [Google Scholar]
- 24.Young JPW. 1993. Molecular phylogeny of rhizobia and their relatives, p 587–592 In Palacios R, Mora J, Newton W. (ed), New horizons in nitrogen fixation, vol 17 Springer, Amsterdam, Netherlands [Google Scholar]
- 25.Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321–4325. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843–7853. 10.1093/nar/17.19.7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zani S, Mellon MT, Collier JL, Zehr JP. 2000. Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl. Environ. Microbiol. 66:3119–3124. 10.1128/AEM.66.7.3119-3124.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholas KB, Nicholas HB, Jr, Deerfield DW., II 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14 [Google Scholar]
- 30.Yu Y, Breitbart M, McNairnie P, Rohwer F. 2006. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics 7:57. 10.1186/1471-2105-7-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 33.Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26:1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammer Ø, Harper DAT, Ryan PD. 2001. Past: paleontological statistics software package for education and data analysis. Palaeontol. Electronica 4:9 [Google Scholar]
- 36.Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 37.Roesch LFW, Camargo FAO, Bento FM, Triplett EW. 2008. Biodiversity of diazotrophic bacteria within the soil, root and stem of field-grown maize. Plant Soil 302:91–104. 10.1007/s11104-007-9458-3 [DOI] [Google Scholar]
- 38.Sachs JL, Kembel SW, Lau AH, Simms EL. 2009. In situ phylogenetic structure and diversity of wild Bradyrhizobium communities. Appl. Environ. Microbiol. 75:4727–4735. 10.1128/AEM.00667-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmad MJ, Muhammad K, Amer J. 2011. Short communication: screening of Mesorhizobium ciceri isolates from rainfed area for improvement in growth and nodulation of chickpea seedlings under controlled conditions. Soil Environ. 30:160–165 [Google Scholar]
- 40.Batista JS, Hungria M, Barcellos FG, Ferreira MC, Mendes IC. 2007. Variability in Bradyrhizobium japonicum and B. elkanii seven years after introduction of both the exotic microsymbiont and the soybean host in a cerrados soil. Microb. Ecol. 53:270–284. 10.1007/s00248-006-9149-2 [DOI] [PubMed] [Google Scholar]
- 41.Dakora FD, Joseph CM, Phillips DA. 1993. Alfalfa (Medicago sativa L.) root exudates contain isoflavonoids in the presence of Rhizobium meliloti. Plant Physiol. 101:819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gangwar S, Dubey M. 2012. Chickpea (Cicer arietinum L.) root nodulation and yield as affected by micronutrients application and Rhizobium inoculation. Crop. Res. 44:37–41 [Google Scholar]
- 43.Guimaraes AP, Morais RF, de Urquiaga S, Boddey RM, Alves BJR. 2008. Bradyrhizobium strain and the 15N natural abundance quantification of biological N2 fixation in soybean. Sci. Agric. 65:516–524. 10.1590/S0103-90162008000500011 [DOI] [Google Scholar]
- 44.Laranjo M, Young JPW, Oliveira S. 2012. Multilocus sequence analysis reveals multiple symbiovars within Mesorhizobium species. Syst. Appl. Microbiol. 35:359–367. 10.1016/j.syapm.2012.06.002 [DOI] [PubMed] [Google Scholar]
- 45.Jourand P, Renier A, Rapior S, de Faria SM, Prin Y, Galiana A, Giraud E, Dreyfus B. 2005. Role of methylotrophy during symbiosis between Methylobacterium nodulans and Crotalaria podocarpa. Mol. Plant Microbe Interact. 18:1061–1068. 10.1094/MPMI-18-1061 [DOI] [PubMed] [Google Scholar]
- 46.Chaintreuil C, Giraud E, Prin Y, Lorquin J, Bâ A, Gillis M, de Lajudie P, Dreyfus B. 2000. Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl. Environ. Microbiol. 66:5437–5447. 10.1128/AEM.66.12.5437-5447.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantera JJL, Kawasaki H, Seki T. 2004. The nitrogen-fixing gene (nifH) of Rhodopseudomonas palustris: a case of lateral gene transfer? Microbiology 150:2237–2246. 10.1099/mic.0.26940-0 [DOI] [PubMed] [Google Scholar]
- 48.Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, de Lajudie P, Masson-Boivin C. 1999. Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl. Environ. Microbiol. 65:3084–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan Z, Hurek T, Vinuesa P, Muller P, Ladha JK, Reinhold-Hurek B. 2001. Specific detection of Bradyrhizobium and Rhizobium strains colonizing rice (Oryza sativa) roots by 16S-23S ribosomal DNA intergenic spacer-targeted PCR. Appl. Environ. Microbiol. 67:3655–3664. 10.1128/AEM.67.8.3655-3664.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thaweenut N, Hachisuka Y, Ando S, Yanagisawa S, Yoneyama T. 2011. Two seasons' study on nifH gene expression and nitrogen fixation by diazotrophic endophytes in sugarcane (Saccharum spp. hybrids): expression of nifH genes similar to those of rhizobia. Plant Soil 338:435–449. 10.1007/s11104-010-0557-1 [DOI] [Google Scholar]
- 51.Jourand P, Giraud E, Béna G, Sy A, Willems A, Gillis M, Dreyfus B, de Lajudie P. 2004. Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria. Int. J. Syst. Evol. Microbiol. 54:2269–2273. 10.1099/ijs.0.02902-0 [DOI] [PubMed] [Google Scholar]
- 52.Chen W-M, James EK, Chou J-H, Sheu S-Y, Yang S-Z, Sprent JI. 2005. β-Rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol. 168:661–675. 10.1111/j.1469-8137.2005.01533.x [DOI] [PubMed] [Google Scholar]
- 53.Perin L, Martínez-Aguilar L, Castro-González R, Estrada-De Los Santos P, Cabellos-Avelar T, Guedes HV, Reis VM, Caballero-Mellado J. 2006. Diazotrophic Burkholderia species associated with field-grown maize and sugarcane. Appl. Environ. Microbiol. 72:3103–3110. 10.1128/AEM.72.5.3103-3110.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Estrada P, Mavingui P, Cournoyer B, Fontaine F, Balandreau J, Caballero-Mellado J. 2002. A N2-fixing endophytic Burkholderia sp. associated with maize plants cultivated in Mexico. Can. J. Microbiol. 48:285–294. 10.1139/w02-023 [DOI] [PubMed] [Google Scholar]
- 55.Hurek T, Reinhold-Hurek B, Van Montagu M, Kellenberger E. 1994. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J. Bacteriol. 176:1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Egener T, Hurek T, Reinhold-Hurek B. 1999. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol. Plant Microbe Interact. 12:813–819. 10.1094/MPMI.1999.12.9.813 [DOI] [PubMed] [Google Scholar]
- 57.Elbeltagy A, Ando Y. 2008. Expression of nitrogenase gene (nifH) in roots and stems of rice, Oryza sativa, by endophytic nitrogen-fixing communities. Afr. J. Biotechnol. 7:1950–1957 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.