Abstract
Human norovirus is the leading cause of epidemic and sporadic acute gastroenteritis. Since no cell culture method for human norovirus exists, cultivable surrogate viruses (CSV), including feline calicivirus (FCV), murine norovirus (MNV), porcine enteric calicivirus (PEC), and Tulane virus (TuV), have been used to study responses to inactivation and disinfection methods. We compared the levels of reduction in infectivities of CSV and Aichi virus (AiV) after exposure to extreme pHs, 56°C heating, alcohols, chlorine on surfaces, and high hydrostatic pressure (HHP), using the same matrix and identical test parameters for all viruses, as well as the reduction of human norovirus RNA levels under these conditions. At pH 2, FCV was inactivated by 6 log10 units, whereas MNV, TuV, and AiV were resistant. All CSV were completely inactivated at 56°C within 20 min. MNV was inactivated 5 log10 units by alcohols, in contrast to 2 and 3 log10 units for FCV and PEC, respectively. TuV and AiV were relatively insensitive to alcohols. FCV was reduced 5 log10 units by 1,000 ppm chlorine, in contrast to 1 log10 unit for the other CSV. All CSV except FCV, when dried on stainless steel surfaces, were insensitive to 200 ppm chlorine. HHP completely inactivated FCV, MNV, and PEC at ≥300 MPa, and TuV at 600 MPa, while AiV was completely resistant to HHP up to 800 MPa. By reverse transcription-quantitative PCR (RT-qPCR), genogroup I (GI) noroviruses were more sensitive than GII noroviruses to alcohols, chlorine, and HHP. Although inactivation profiles were variable for each treatment, TuV and MNV were the most resistant CSV overall and therefore are the best candidates for studying the public health outcomes of norovirus infections.
INTRODUCTION
Noroviruses are the leading cause of epidemic and sporadic acute gastroenteritis (1, 2) and infect people of all ages worldwide. The primary mode of transmission is person to person (3, 4), but food-borne transmission also plays a significant role in outbreaks (5). Transmission of norovirus disease is promoted by several factors, including a large human reservoir, shedding of large amounts of virus (105 to 109 particles/g stool), prolonged virus shedding, environmental stability, relative resistance to disinfection, and environmental contamination (6). Over the past decade, the development of sensitive molecular techniques has provided a better understanding of the burden and molecular epidemiology of norovirus disease and has been useful in the detection of norovirus in a variety of environmental settings. Despite significant efforts, human noroviruses cannot be cultivated in cell culture (7–10), and therefore, the effects of physiochemical exposure and specific disinfectants, or the best way to inactivate infectious viruses, cannot be evaluated quantitatively. Consequently, the most common approach to understanding the response to disinfectants and the environmental stability of infectious norovirus has been the use of cultivable surrogate viruses (CSV), while detection of viral RNA by reverse transcription-quantitative PCR (RT-qPCR) has been employed to compare the effects of inactivation methods on viral RNA (11). The latter approach, however, does not provide information about virus infectivity, and only a small region of the RNA genome is amplified, which may underestimate the efficacy of treatments.
Surrogates have most commonly been used in microbiology as indicators within an environmental context, such as water or food, to represent the potential presence of pathogens (12). The use of surrogates should be considered when no methods are available to study the effects of physiochemical conditions or inactivation methods on a pathogen. An ideal surrogate should have biological, biochemical, and biophysical characteristics similar to those of the pathogen of interest. Noroviruses are nonenveloped, single-stranded RNA viruses with a diameter of 27 to 40 nm. They belong to the family Caliciviridae, which consists of five genera, Norovirus, Sapovirus, Lagovirus, Vesivirus, and Nebovirus, and three proposed genera, Recovirus, Valovirus, and Chicken calicivirus (13). In 2008, Tulane virus (TuV) was first described as the first member of the proposed genus Recovirus (14). Viruses from the family Caliciviridae are the logical surrogate choice, and because feline calicivirus (FCV) is cultivable, this virus has been used as the preferred surrogate for norovirus in numerous studies since the 1970s (15). With the discovery of murine norovirus (MNV) in 2003 (16), many researchers are now using this newly described virus, because it is genetically more closely related to human noroviruses and is resistant to low pHs (17, 18). Many studies comparing FCV and MNV with or without other surrogates, such as MS2 coliphage and hepatitis A virus, as well as human norovirus (using reverse transcription-quantitative PCR [RT-qPCR]), have been published (4, 18–24). However, because MNV is highly sensitive to alcohols (18), the focus has expanded to cultivable caliciviruses such as TuV (14, 25–28) and to porcine enteric calicivirus (PEC), a genogroup III (GIII) sapovirus (29, 30).
Although numerous studies have been published comparing two or more CSV, data are difficult to compare across studies because inactivation conditions, such as exposure time, concentrations of disinfectants, and the final matrix, have not been consistent. In this study, we compared four surrogates of human norovirus head to head using identical experimental conditions. In addition, Aichi virus (AiV) was also included because of its established environmental stability (31). AiV was first detected in humans in 1989 (32) and has since been detected in water and sewage worldwide (33). We chose to compare five virus inactivation methods, including physiochemical treatments (heat and pH), commonly used disinfectants (chlorine and alcohol), and high hydrostatic pressure (HHP), an emerging method for the inactivation of pathogens in food (34). Inactivation was measured by quantitative loss of infectivity for each of the five viruses. In addition, the effects of selected conditions on the RNAs of CSV and human norovirus were compared.
MATERIALS AND METHODS
Viruses, cells, and infectivity assays.
FCV strain F9 (ATCC VR-782; ATCC, Manassas, VA) was propagated in CRFK cells (ATCC CCL-94) in EMEM (minimal essential medium with Earle's salts; Gibco Life Technologies, Grand Island, NY) plus 10% fetal bovine serum (FBS) (HyClone, Logan, UT) as described previously (22). Confluent monolayers of CRFK cells were inoculated with stock virus into the growth medium and were harvested at 2 days postinfection (p.i.), when the cytopathic effect (CPE) was complete. The infected cells were frozen and thawed three times, and this was followed by centrifugation for 30 min at 3,000 × g and 4°C. Aliquots of FCV (1 × 109 PFU/ml) were stored at −70°C, and each aliquot was freshly thawed for each experiment and was used only once. All virus aliquots, experimental controls, and treated samples were assayed by plaque assays in 60-mm dishes as described previously (22).
MNV strain CW3 (a gift from Skip Virgin, Washington University, St. Louis, MO) was propagated in RAW 264.7 cells (ATCC TIB-71) in DMEM (Dulbecco's MEM) with no pyruvate, plus 10% low-endotoxin FBS (HyClone, Logan, UT). Infectious MNV was assayed by inoculation of 60-mm dishes of RAW monolayer cells with 500 μl of 10-fold serial dilutions, incubation for 1 h at 37°C with rotation, and subsequent addition of an overlay with a medium consisting of MEM plus 2% FBS and 0.5% agarose. At 2 days p.i., a second agarose overlay containing 66 mg/ml neutral red (Sigma, St. Louis, MO) was added, and visible plaques were counted within 8 h. Frozen aliquots of MNV had titers of 107 to 108 PFU/ml.
AiV, a gift from Pierre Poitier (Dijon University Hospital, Lyon, France), was propagated and assayed by plaque assays in Vero cells (ATCC CCl-81) by using the same protocols as those described for FCV above. AiV had a titer of 1 × 107 PFU/ml.
TuV (14) was propagated in LLC-MK2 cells cultured in Opti-MEM (Gibco Life Technologies, Grand Island, NY) plus 2% FBS. Monolayers of cells were infected for 1 h at 37°C at a multiplicity of infection (MOI) of 0.001 in 10 ml of serum-free medium in T150 flasks, followed by the addition of 40 ml Opti-MEM plus 2% FBS. The virus was harvested at 48 to 72 h, when the CPE was complete, clarified at 3,000 × g for 30 min, and frozen in aliquots at −70°C until use. TuV plaque assays were performed similarly to FCV, AiV, and MNV plaque assays except that viral dilutions were made in Opti-MEM plus 2% FBS. As described for MNV, after 2 days an agarose overlay containing neutral red was used to stain plaques. The TuV titer was 107 PFU/ml.
Porcine enteric calicivirus (PEC; strain Cowden) was obtained from Linda Saif (The Ohio State University) (29). LLC-PK cells (ATCC CL-101) were passaged in MEM plus 5% FBS every 3 to 5 days, as described previously (30). Briefly, 3-day-old monolayers were rinsed twice with MEM prior to infection at an MOI of 0.01. After virus adsorption for 1 h, complete MEM plus 2% FBS containing 100 mM GCDCA (glycochenodeoxycholic acid sodium salt; Sigma) was added to the LLC-PK1 cell monolayers. Complete CPE appeared at 72 h p.i. Flasks were frozen and thawed three times, and cell debris were removed by centrifugation at 3,000 × g for 30 min to obtain serum-free virus aliquots, which were stored at −70°C. The PEC titer was 0.38 × 106 50% tissue culture infective doses (TCID50)/ml. Ninety-six-well plates (Costar; Corning Inc., Corning, NY) of LLC-PK cells were inoculated with PEC-treated or untreated samples. Infectious PEC was measured by a previously described TCID50 assay (35); the virus was detected by using a porcine anti-PEC polyclonal serum (kindly provided by Linda Saif) followed by a horseradish peroxidase (HRP)-labeled goat anti-swine antibody (KPL, Gaithersburg, MD); and color was developed with a 3-amino-9-ethylcarbazole development kit (Sigma, St. Louis, MO). Cells were observed by microscopy for reddish staining of the cytoplasm, and the results for 5 wells for each inoculum were used to calculate the TCID50 titer by the Reed and Muench method (36).
Clinical samples, preparations, and viral RNA extraction.
GI.1 norovirus-positive fecal specimens were kindly provided by Christine Moe, Emory University. GI.5 and GII.13 norovirus-positive fecal specimens were obtained from outbreak specimens submitted to the National Calicivirus Laboratory at the CDC. Clarified 10% fecal suspensions were prepared in PBS (pH 7.2) and were centrifuged at 10,000 × g for 30 min at 4°C. To prepare semipurified (SP) stool preparations, the clarified stool suspensions were buffer exchanged into MEM plus 10% FBS using an Amicon 50K Ultra-15 ultrafilter (Millipore, Billerica, MA). Viral RNA concentrations were determined by norovirus GI/GII TaqMan real-time RT-PCR as described below. The GI.1, GI.5, and GII.13 suspensions contained 4.69 × 105, 1.47 × 104, and 5.94 × 106 RNA copies/μl, respectively. The GI.5 SP preparation contained 1.95 × 105 RNA copies/μl, and the GII.13 SP preparation contained 1.23 × 107 RNA copies/μl. Norovirus preparations for experimental work were stored in aliquots at −70°C, and thawed aliquots were used only once.
RNase treatment prior to nucleic acid extraction was performed by incubation with RNase One RNase (1 U/μl) (Promega, Madison, WI) at 37°C for 2 h, and the reaction was stopped by the addition of SDS to a final concentration of 0.1%. Viral nucleic acid from control and treated samples was extracted using the KingFisher instrument and the MagMAX total-RNA isolation kit (Ambion, Austin, TX) according to the manufacturers' instructions.
TaqMan real-time RT-qPCR.
All RT-qPCR assays were TaqMan based and were performed using the AgPath One-Step RT-PCR kit (Ambion, Austin, TX) or the QuantiTect Probe RT-PCR kit (Qiagen, Valencia, CA) on an Applied Biosystems (Foster City, CA) 7500 platform. GI/GII norovirus RNA was detected as described previously (37). Two standard curves consisting of 10-fold serial dilutions of GI.4 RNA transcripts and GII.7 RNA transcripts were included in each assay as described previously (38). MNV, FCV, and AiV RT-qPCRs were performed as described previously (22, 39).
TuV RT-qPCR was performed in a 25-μl reaction volume consisting of 3 μl of template RNA, 12.5 μl of 2× RT-PCR buffer (Ag-Path One-Step RT-PCR kit; Ambion, Austin, TX), 0.6 μl of each oligonucleotide primer (400 nM), 0.3 μl (200 mM) of the TaqMan probe, 1 μl of 25× RT-PCR mixture (Ag-Path One-Step RT-PCR kit; Ambion, Austin, TX), and RNase-free water. The reaction conditions consisted of an RT step of 10 min at 45°C, followed by 10 min at 95°C to activate the Taq polymerase and then 40 cycles of amplification for 15 s at 95°C and 1 min at 60°C. The TaqMan oligonucleotide probe (6-carboxyfluorescein [FAM]-5′-TTTTCCATC[C]CATTCA[C]AAGT-3′-black hole quencher [BHQ], with the internal G-clamp indicated by [C]) and primers TuV-F (5′-TTCACCCGACCAACCCTG-3′) and TuV-R (5′-ACGCCCCAACGCACCTA-3′) were designed to target TuV open reading frame 1 (ORF-1) (GenBank accession number EU391643.1) (14).
A standard curve for each virus was generated by amplifying 10-fold serial dilutions of RNA. The threshold cycle (CT) values for each dilution were used to plot a standard curve against the corresponding PFU/ml. The concentration for each sample (treated or control) was extrapolated from this curve. The concentrations of GI and GII viral RNA (expressed in copies/μl) were calculated from standard curves of transcripts that were included in each experimental assay.
pH treatment.
Each virus aliquot (FCV, MNV, AiV, TuV) was diluted 1:10 into 100 mM citric acid buffer solutions at pH 2 or 3 and into 100 mM carbonate buffer at pH 9 or 10 in 4.0-ml polystyrene tubes. After 30 min at 37°C, reactions were stopped by diluting the solutions 1:10 into DMEM or into MEM plus 2% FBS to adjust the pH to 7.0 to 8.0. Therefore, the virus titers in the treated samples were 1:100 dilutions of stock viruses. Experimental controls were also incubated for 30 min at 37°C.
Heat treatment.
Virus aliquots were diluted 1:10 in 0.017 M PBS (pH 7.4) (HyClone, Logan, UT) and were inactivated at 56°C in a digital dry bath (Labnet International, Inc.) for 2, 5, 10, or 20 min. The treatment was stopped by transferring the 1.8-ml microcentrifuge tubes containing 1.0 ml of diluted sample to an ice bath.
Alcohol treatment.
Virus aliquots were diluted 1:10 into the test alcohol solutions for a final 1.0-ml volume and were incubated for 1 min in 1.8-ml microcentrifuge tubes, followed by 1:10 dilution into EMEM plus 10% FBS to stop inactivation. Alcohol solutions were made by adding the appropriate volume of cell culture-grade water (Life Technologies, Grand Island, NY) to molecular biology-grade absolute ethanol or isopropanol (Fisher Scientific, Fairlawn, NJ).
Chlorine surface disinfection.
We used a modification of a previously described method for the assessment of decontamination of surfaces (40). Ten microliters of each virus in EMEM, DMEM, or Opti-MEM (each containing 10% FBS) was dried on 16-mm stainless steel discs (Muzeen & Blythe Ltd., Winnipeg, Manitoba, Canada), which were placed inside 24-well plates in a laminar flow hood for 1 to 1.5 h. Fifty microliters of a chlorine solution at 200 ppm or 1,000 ppm, prepared by dilution of commercial bleach (Clorox, with 6% sodium hypochlorite) in distilled water, was added to each dried virus for 5 min. Disinfection was stopped by the addition of 450 μl of 0.1% thiosulfate in MEM plus 10% FBS. The solution was removed after vigorous repipetting onto the disc 6 to 8 times. Duplicate control discs with only MEM treatment were included in order to calculate the level of virus recovery.
HHP treatment.
All viruses that were not prepared in EMEM plus 10% FBS were buffer exchanged to EMEM plus 10% FBS on Amicon Ultra-15 50K centrifugal filters (Millipore, Billerica, MA) prior to shipment on dry ice to the Institute for Food Safety and Health, Illinois Institute for Technology, for high-hydrostatic-pressure (HHP) treatment. One milliliter of a virus aliquot was placed in the shaft of a Pasteur pipette, heat sealed (Heat Sealer, model 252B; Clamco, Cleveland, OH), and then vacuum sealed in 2-mil 8- by 10-in polyethylene pouches (Prime Source, St. Louis, MO). The pouches were then placed in heat-sealed high-barrier bags containing 5,000 ppm sodium hypochlorite and were transported on ice to the HHP bay. An Avure 24-liter high-pressure unit (Avure Technologies, Middleton, OH) with processing water cooled to 4°C was used. Samples were treated at 100, 200, 300, 400, 500, 600, and 800 megapascals (MPa) for 1 min at 4°C. A pressure come-up time of 55 to 65 s was not included, and release was instantaneous. HHP-treated samples were transferred to 1.7-ml cryovial tubes, immediately frozen, and shipped on dry ice to the CDC for plaque assays, TCID50 assays, or RT-qPCR. All experiments were performed in duplicate for each shipment, and two separate shipments were made for each virus, producing at least four replicates for each parameter. Virus control samples were also shipped, thawed, refrozen, and returned to the CDC to be used as the experimental controls. In some cases, as many as six replicate data points for a parameter were obtained.
Data analysis.
All experiments were performed in duplicate with at least two replicates for each experiment. The virucidal activities of the treatments were determined by calculating the reduction in the level of infectivity or RNA as the difference between log10 values for control (PFU/ml or RNA copies/μl) and treated samples. Four or more log reduction values were averaged, and the standard deviation was determined. RT-qPCR data are the means for four or more replicate samples and a triplicate assay of each sample by RT-qPCR. Statistical comparisons were made with Tukey honestly significant difference (HSD) analysis using PASW Statistics, version 18 (IBM SPSS Inc., New York, NY) (41).
RESULTS
pH stability.
We exposed the surrogate viruses to pH 2 and 3, as well as pH 9 and 10, for 30 min at 37°C (Fig. 1). Of all viruses, FCV was the most susceptible to pH extremes of pH 2, 3, and 10 (P, <0.0001). PEC infectivity was reduced by 1 log10 unit at pH 2, whereas MNV and TuV infectivities were reduced by less than 0.5 log10 unit each.
FIG 1.
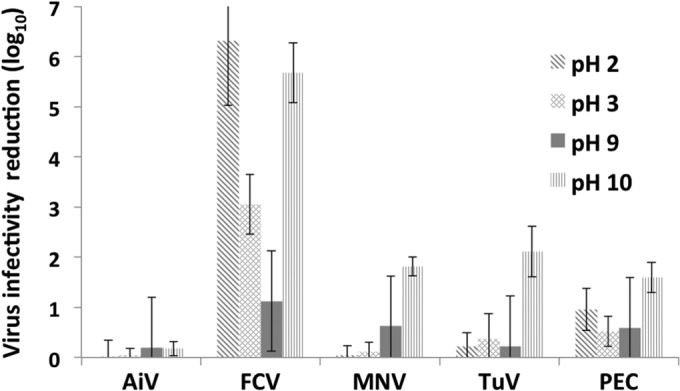
Reduction of virus infectivity after exposure to pH 2, 3, 9, or 10 for 30 min at 37°C. Virus infectivity was measured by plaque assays (AiV, FCV, MNV, and TuV) or by the TCID50 (PEC). Each bar represents the mean for 4 or more replicates. Error bars, standard deviations.
Heat treatment.
Heat treatment is commonly used to destroy viruses in food. A common treatment is pasteurization of milk at 72°C for 15 s. Inactivation at a lower temperature for longer times can reveal the kinetic differences between viruses. All viruses were inactivated rapidly at 60°C and 63°C (data not shown). Fifty-six degrees Celsius was chosen as a good temperature for observation of the kinetics of inactivation within a reasonable time. With exposure to 56°C for as long as 20 min, increasing reductions in infectivity were found for each of the viruses (Fig. 2). The maximum measurable virus reduction of 5 log10 units was obtained at 20 min for FCV, MNV, and TuV. Near-complete (4-log10-unit) reduction of AiV infectivity was achieved after 20 min of treatment. The level of reduction of PEC infectivity was lower than those of the other viruses, 2 log10 versus 4 to 5 log10 units, a difference that may be partially explained by the lower initial virus titer (0.38 × 106 TCID50/ml). Greater variation was found among replicate experiments for heat inactivation than for other test conditions.
FIG 2.
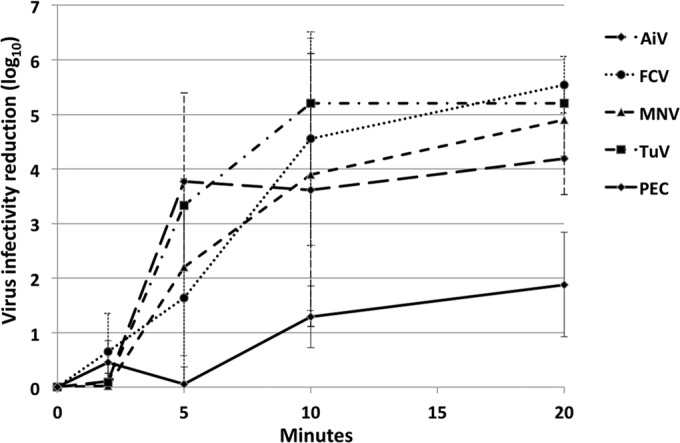
Stability of surrogate viruses at 56°C with increasing contact times. Virus infectivity was measured by plaque assay (AiV, FCV, MNV, and TuV) or by the TCID50 (PEC).
Alcohol sensitivity.
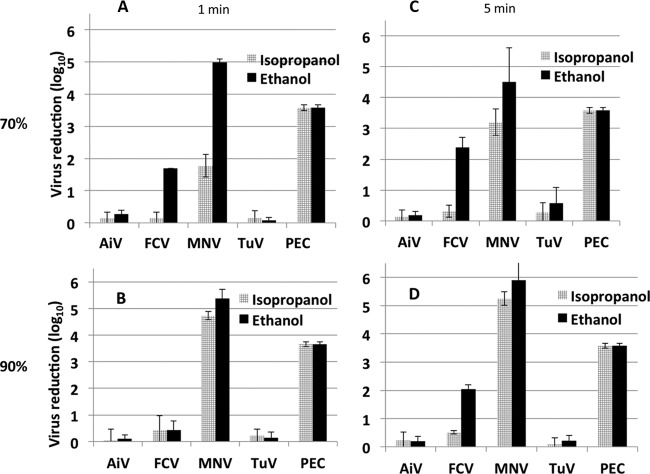
Each surrogate virus, in suspension, was exposed to four different alcohol solutions (70% or 90% isopropanol; 70% or 90% ethanol) for 1 min and 5 min (Fig. 3). MNV and PEC were the most sensitive to all test conditions, with complete reduction (6 log10 units) of MNV infectivity after 5 min of exposure to 90% ethanol. PEC infectivity was reduced completely by all treatments. The levels of reduction for each virus, except MNV, after exposure to 70% alcohol were similar at 1 min and 5 min. Exposure to 90% alcohol produced similar reductions, except for MNV, which showed a greater reduction when exposed to 90% isopropanol than when exposed to 70% isopropanol. Treatment with 70% ethanol reduced FCV infectivity more than treatment with 90% ethanol at 1 min and 5 min, as observed previously (22). AiV and TuV titers lost less than 0.5 log10 unit with all treatments.
FIG 3.
Reduction of virus infectivity after treatment with 70% (A and C) or 90% (B and D) isopropanol or ethanol for 1 min (A and B) or 5 min (C and D). Virus infectivity was measured by a plaque assay (AiV, FCV, MNV and TuV) or by the TCID50 (PEC).
Because MNV infectivity was the most sensitive to alcohol treatments, we compared the reductions in MNV RNA levels with the reductions in GI.1, GI.5, GI.5 SP (GI.5 semipurified preparations), GII.13, and GII.13 SP RNA levels after 1 min of exposure to 70% or 90% ethanol solutions (Fig. 4). The MNV infectivity reduction was in the range of that seen in all previous experiments (Fig. 3) and was a near-complete reduction of infectious virus. The MNV RNA level was reduced by 4 to 5 log10 units in the same samples. The GI.5 SP RNA level was reduced by as much as 3.5 log10 units, whereas the GII.13 and GII.13 SP RNA levels were reduced by less than 1 log10 unit. The reductions in G1.1, G1.5, and GII.13 RNA levels, after exposure to 70% or 90% ethanol for 1 min, differed significantly from each other (P, <0.05). There was a significant difference between the reductions in the GI.5 SP and GII.13 SP RNA levels and the reductions in the RNA levels of the non-SP preparations (GI.5 and GII.13) (P, <0.05).
FIG 4.
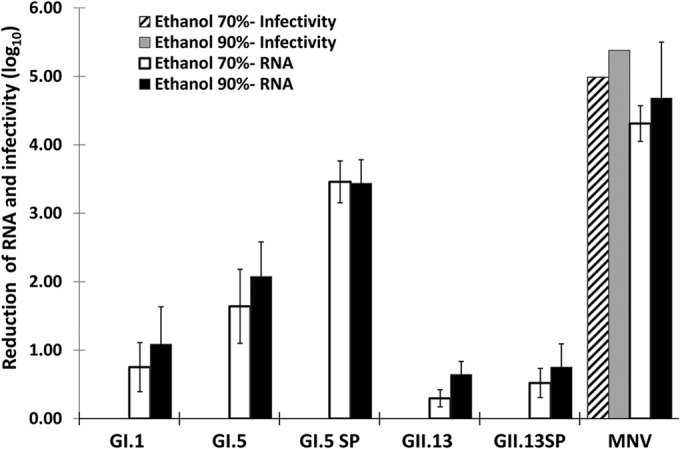
Reductions in the levels of human norovirus RNA (GI.1, GI.5, GI.5 SP, GII.13, and GII.13 SP), MNV RNA, and MNV infectivity after exposure to 70% and 90% ethanol for 1 min.
Chlorine treatment on discs.
After drying on stainless steel discs, viruses were exposed to 200 or 1,000 ppm chlorine for 5 min and were evaluated for infectivity (Table 1). After exposure to 200 ppm, <1-log10 reductions were seen for all the viruses. The FCV titer was reduced by 5 log10 units after exposure to 1,000 ppm chlorine (P, <0.0001), whereas the titers of the other viruses were reduced by about 1 log10 unit (P, <0.0001).
TABLE 1.
Chlorine treatment of surrogate viruses dried on stainless steel discs
| Chlorine concn (ppm) | Log10 reduction in infectivitya for: |
||||
|---|---|---|---|---|---|
| AiV | FCV | MNV | PEC | TuV | |
| 200 | 0.9 ± 0.2 | 0.2 ± 0.7 | 0.1 ± 0.7 | 0.4 ± 0.1 | 0.3 ± 0.1 |
| 1,000 | 1.3 ± 0.9 | 5.3 ± 0.7 | 1.4 ± 0.4 | 1.2 ± 0.5 | 1.2 ± 0.2 |
Values are means for 4 or more replicates from 2 separate experiments ± standard deviations.
Nucleic acid extracts of the same samples were tested by RT-qPCR (Fig. 5). After exposure to 1,000 ppm chlorine, the FCV RNA level was reduced by 2.5 log10 units. In contrast, TuV, MNV, and AiV RNA levels were reduced by <0.5 log10 unit following treatment with 1,000 ppm chlorine. The GI.5 SP RNA level was reduced by <1 log10 unit after treatment with 200 or 1,000 ppm chlorine. The GII.13 SP RNA level was reduced by <0.5 log10 unit.
FIG 5.
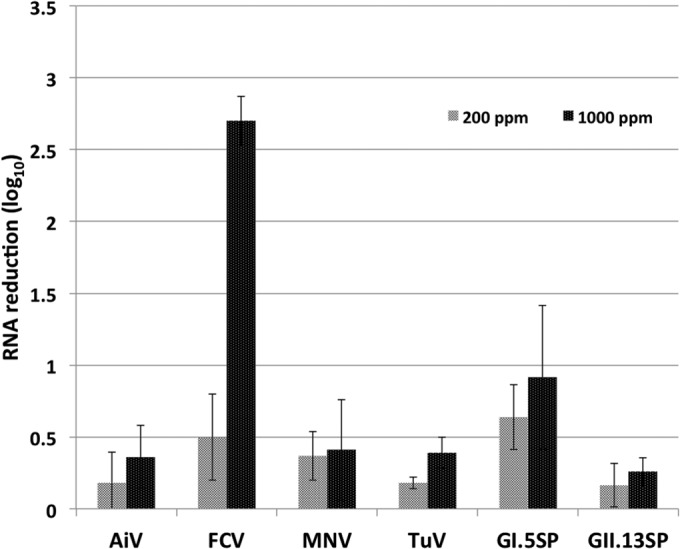
Reductions in RNA levels of surrogate viruses (AiV, FCV, MNV, TuV) and human norovirus (GI.5 SP and GII.13 SP) after treatment with 200 or 1,000 ppm chlorine. Viruses were dried on stainless steel discs as described in Materials and Methods and were then treated with chlorine for 5 min.
HHP treatment.
All CSV and AiV were tested for loss of infectivity upon treatment with increasing pressure (100 to 800 MPa) for 1 min at 4°C (Fig. 6). AiV infectivity was not reduced by pressure as high as 800 MPa, whereas FCV infectivity was reduced by 6 log10 units at 300 MPa, and MNV required 400 MPa for a 6-log10 reduction. PEC was completely inactivated (5-log10 reduction) at 400 MPa. Although TuV infectivity was reduced by 1 log10 unit at 100 MPa, in contrast to no reduction of the other viruses at 100 MPa, 600 MPa was required for complete inactivation of TuV. Thus, the inactivation kinetics of TuV appeared quite different from those of the other CSV.
FIG 6.
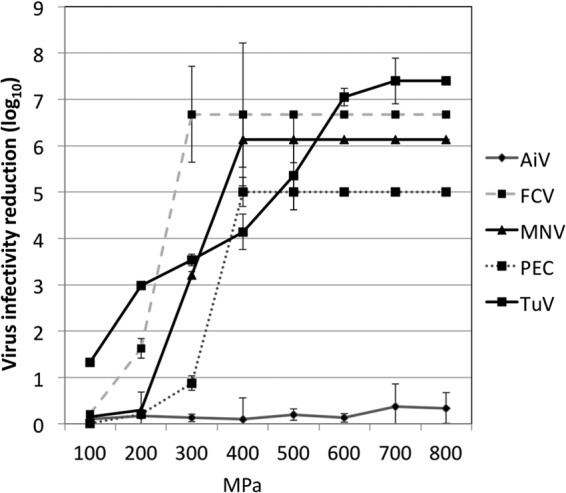
Reductions in the infectivity of surrogate viruses (AiV, FCV, MNV, PEC, and TuV) after HHP treatment at varying pressure levels (100 to 800 MPa) for 1 min at 4°C.
In another series of experiments, the reductions in the levels of RNA from human norovirus-positive semipurified stool samples (GI.5 SP and GII.13 SP) were compared with the reductions in TuV RNA levels (Fig. 7). At 200 MPa, the TuV RNA level was reduced by 1 log10 unit, with no further reduction upon HHP treatment up to 800 MPa. This was in contrast to the increasing reduction in TuV infectivity, which reached its maximum reduction level at 600 MPa. The GI.5 SP RNA level was reduced 2 log10 units with 300-MPa treatment, and no further reduction was seen at higher pressures, whereas the GII.13 SP RNA level was reduced by 1 log10 unit at 400 MPa and was not further reduced with increasing presure.
FIG 7.
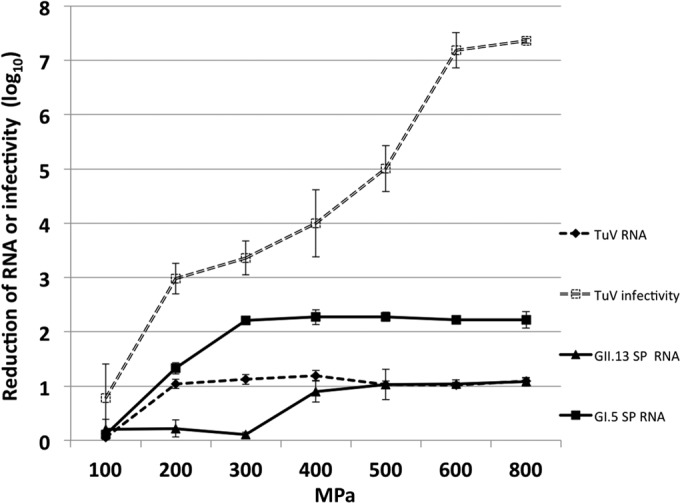
Reductions in TuV infectivity and in TuV and human norovirus (GI.5 SP and GII.13 SP) RNA levels after HHP treatment at 100 to 800 MPa for 1 min at 4°C. TuV infectivity was measured by plaque assay, and reductions in RNA levels were calculated from RT-qPCR values.
DISCUSSION
In the absence of a cell culture system for human noroviruses, cultivable surrogate viruses have been used for decades, even before the current understanding of the classification and characteristics of noroviruses. We compared the inactivation profiles of four CSV of the family Caliciviridae and AiV, which belongs to the family Picornaviridae, as surrogate viruses for the noncultivable human noroviruses, using identical experimental conditions. Although AiV is the most resistant virus to all treatments, a calicivirus surrogate would be the best indicator for assessing the efficacies of inactivation methods against human norovirus. The four calicivirus surrogates demonstrated different inactivation patterns. We confirmed results from previous studies (18, 42) showing that FCV is much more sensitive than the other caliciviruses to low pHs, which enteric viruses, such as human norovirus, need to survive in order to initiate infection. PEC, a sapovirus, is also less resistant to lower pHs than the other CSV. In addition, routine laboratory testing for PEC, in comparison with protocols for other CSV, is a challenge because of its lower titer and the need for additional incubation steps with PEC-specific antibodies for detection. Because of the importance of low-pH resistance and the need for relatively simple methods to allow the routine use of surrogates, FCV and PEC are inadequate for assessing the efficacies of methods to reduce the public health risk of norovirus infections. Based on our data and those of others, MNV and TuV are the most promising candidates for serving as surrogates for human norovirus (26). However, MNV is significantly more susceptible to alcohols than other nonenveloped viruses, as numerous studies have shown (19, 20, 22, 43). Alcohols, at concentrations of 60 to 95% (44), are important components of many hand sanitizers and are commonly used in settings where bacteria and viruses are pathogens of concern. We demonstrated that the levels of reduction of MNV and TuV infectivity by chlorine were very similar, as were their sensitivities to low pHs. The kinetics of reduction of infectivity at 56°C were slightly different, but both viruses lost almost all their infectivity after 20 min. The inactivation pattern of TuV with HHP treatment was notably different from those of the other viruses; it required 600 MPa to eliminate all infectivity, in contrast to 300 to 400 MPa for inactivation of the other norovirus surrogates. HHP is an important parameter because of growing interest in its use in food preparation (45–49). Our data demonstrated that across different inactivation procedures, TuV is the most resistant of the human norovirus surrogates. In addition, TuV consists of at least 4 different genotypes, which display diverse histo-blood group antigen (HBGA) binding patterns mimicking those of human noroviruses (50).
Like other investigators, we found that RNA quantitation after treatment of surrogate viruses with chlorine, alcohols, or HHP does not quantitate the loss of virus infectivity (22, 23, 47, 51), although in some treatments an increase in RNA reduction did parallel an increase in infectivity reduction, as was apparent for the chlorine treatments of FCV and the alcohol treatments of MNV. Interestingly, treatments of human norovirus with chlorine, alcohols, and HHP demonstrated that GII viruses are more resistant than GI viruses, as measured by reductions of RNA by RT-qPCR. In contrast to our ethanol tolerance data for a GI.1 and a GII.13 virus, which were in agreement with published work (20, 22), we found a 3 log10 reduction for a semipurified GI.5 virus. This suggests that differences in alcohol tolerance may exist between different norovirus strains and that purification of viruses from stools needs to be considered when testing additional strains of both genogroups. Such results are of particular interest because alcohols are a key component of many hand sanitizers (22, 52, 53).
Our results for the individual viruses are in general agreement with the results reported in previous studies (18, 20, 26, 27, 30, 42). However, it is difficult to make a direct comparison due to the great differences in certain test parameters (e.g., exposure time) between methods. In several instances where a 1- or 2-log reduction difference was found between our results and those of others, the methods employed did not include the exact same test conditions. PEC titers were reduced 1 to 2 log10 units at extreme pH values, which were not evaluated in a previous study (30). Both TuV and MNV were stable at lower pH values and showed similar sensitivities at pH 10. Other investigators found TuV to be more sensitive to pH 2 and pH 10 than we found, possibly due to differences in experimental parameters (26). FCV and MNV were equally sensitive to heating at 56°C in our experiments, with inactivation complete by 20 min, in agreement with a previous study (18). Our 56°C inactivation data for PEC are in the same range as those reported previously for these viruses (27, 30), whereas Tian et al. reported a 3.5 log10 reduction in TuV infectivity, in contrast to our finding of a 5 log10 reduction after 10 min of exposure (27).
Our data confirm that MNV is more sensitive to alcohols than FCV (22, 30, 54) and that FCV is more sensitive to 70% ethanol than to 90% ethanol (22). The data for PEC are in line with previously published data indicating a >2 log reduction at 30 s (30). However, in contrast to what has been reported previously (24), we found that TuV infectivity, like AiV infectivity, was very marginally reduced by ethanol and isopropanol. To compare the sensitivities of different surrogate viruses to chlorine, we chose to first dry the viruses on stainless steel coupons rather than measuring disinfection in solution so as to better mimic the environment of a food preparation area. Also, a simple dilution of commercial bleach in water was used without pH adjustment, as would be done in a food preparation environment (30, 54). In wastewater, FCV is more susceptible to chlorine than polioviruses (55). In contrast to data on the sensitivity of FCV to chlorine in solution (20), we found a <0.5 log10 reduction in FCV infectivity or decline in the FCV RNA level upon exposure to 200 ppm of chlorine, a finding similar to published data for FCV dried on a surface (24, 56). At higher chlorine concentrations (1,000 ppm), FCV was the only surrogate that was inactivated by more than 5 log10 units, whereas the infectivities of the other viruses, including TuV, were reduced by about 1 log10 unit. This finding is in contrast with data from other studies that reported several log units of virus inactivation for MNV (26, 57) and TuV (26, 27) in solution. However, our data are in agreement with another study that also evaluated chlorine disinfection on stainless steel surfaces and found that FCV was more sensitive than MNV (23). Taking these data together, it is clear that these viruses are more difficult to inactivate on surfaces than in solution, which was previously demonstrated for MS2, MNV, and human norovirus (58). Therefore, 200 ppm of chlorine (59) may not be sufficient to inactivate human norovirus on food contact surfaces.
HHP is an attractive option for the inactivation of potential pathogens in foods for which raw character, flavor, and texture are important, such as shellfish, fruits (e.g., raspberries), and vegetable products. HHP is currently used to inactivate Vibrio vulnificus in shellfish (60), and a pressure of 275 to 300 MPa for several minutes is used for oysters (61). Inactivation of surrogates in other foods, such as milk, juice, strawberry puree, and blueberries, will expand the application of HHP pathogen inactivation to a variety of foods (47, 48, 62). A recent study on the use of HHP on contaminated oysters and clams found that 300 MPa did not inactivate GI and GII noroviruses (63). Several studies on the use of HHP to inactivate surrogate viruses seeded in a variety of nonfood matrices have been published (47, 62, 64–68). Unfortunately, a wide variety of experimental conditions and a limited range of pressures for the evaluation of each virus were used, making these results very difficult to compare across the different surrogate viruses. Because different inactivation conditions may impact virus inactivation by HHP (34), we employed the same test conditions for all viruses tested and used a range of 100 to 800 MPa for HHP. AiV is stable at 600 MPa for 5 min at 4°C to 21°C (68), and we confirmed these findings for 800 MPa at 4°C for 1 min. FCV and MNV were inactivated at roughly similar pressures, as reported previously (47, 64, 67). We found that MNV was more resistant than FCV, which was completely inactivated at 300 MPa, whereas MNV required 400 MPa for complete inactivation. Kingsley also found that MNV was more resistant than FCV (34). Using HHP, a near-4-log10 reduction has been reported for TuV and a 2 log10 reduction for MNV in a culture medium at 21°C for 2 min with 350 MPa (62), while we found a 3.5 log10 reduction in the infectivity of each virus after exposure to 300 MPa for 1 min at 4°C. These differences clearly demonstrate the challenge in comparing results from different studies. It is possible that differences in temperature (21°C versus 4°C in our study) contributed to these different findings. Use of the appropriate pressure is critical for successful inactivation of human norovirus, as was shown in a study where human volunteers were fed raw oysters that had been artificially seeded with Norwalk virus and treated with HHP. Only HHP treatment at 600 MPa, not at 400 MPa, completely inactivated the virus and resulted in no infection in any of the subjects (45), a finding comparable to the 600 MPa needed to completely inactivate Tulane virus in our study. PEC inactivation with HHP treatment has not been reported previously, but like MNV, PEC was also inactivated completely at 400 MPa.
Our study had several limitations. In order to compare the CSV with different inactivation conditions, which has not done before, we selected important test variables for each inactivation and disinfection condition. For example, we tested all viruses in cell culture medium, whereas a complex matrix, such as food or fecal matter, may provide different results. Inactivation of each surrogate virus dried on surfaces was studied only for two chlorine concentrations and one contact time, but since chlorine is commonly used at 200 ppm as a food contact sanitizer (59), and the use of 1,000 to 5,000 ppm is recommended to inactivate norovirus on nonfood surfaces (69), our data mimic the common practice in food preparation environments. Although the results are promising, additional HHP treatments of surrogate viruses, perhaps mixed with human norovirus, seeded into various food matrices are needed in order to assess the practical application of HHP. Before considering round-robin testing with several laboratories, additional data are needed with selected surrogates such as TuV. This could provide a better understanding of which inactivation methods may provide optimum treatments for the best public health outcomes. Finally, we evaluated only selected treatment conditions for RNA reduction and a limited number of human norovirus genotypes. The differences in sensitivity to alcohol between the GI.1 and GII.13 noroviruses observed in this study suggest that there may be more than one inactivation profile for human noroviruses and that therefore, additional genotypes should be tested.
Despite extensive efforts, all attempts to cultivate human norovirus in cell culture have failed to date (7–10). Consequently, the use of CSV is the approach available to most investigators for evaluating the effectiveness of control methods to prevent the spread of noroviruses and to protect public health. Our comprehensive study comparing the performance of several key norovirus control measures, such as chlorine and alcohol for surface and hand disinfection, demonstrates that depending on which treatment is evaluated, the choice of a surrogate virus for human norovirus could be narrowed down to one or two viruses. Ultimately, however, the performance of the surrogate viruses will need to be linked to reductions in the infectivity of human noroviruses.
Because of the reported differences in susceptibility to different inactivation methods between MNV and FCV, the validity of the use of surrogate viruses has been questioned, and the use of human challenge studies to determine which techniques are effective at reducing norovirus levels in foods has been recommended (70). This approach was effectively used for HHP, to determine the appropriate pressure needed for complete inactivation of norovirus in oysters (45). However, such studies are expensive and are not allowed in many countries. But since clinical trials provide us with the best possible data on which intervention methods actually work, linking well-designed surrogate studies with clinical trials in human volunteers may be the best approach to determine which methods work most effectively to reduce the number of norovirus outbreaks.
Although several molecular approaches for the detection of potentially infectious viruses have been evaluated (reviewed by Knight et al. [11]), none are fully successful. Because damage to the virus capsid is the only or primary damage inflicted by several of the inactivation methods, the methods that offer the most promise are those that rely on binding to an intact viral capsid, such as the capture of virus by HBGA, followed by detection of viral RNA by RT-qPCR (71). Ultimately, a more basic understanding of the mechanisms of disinfection is needed, including answers to questions such as which residues of the viral capsid and/or genome are involved during disinfection and whether these changes are similar for different enteric viruses (72). Such information could provide us with important insights on how to measure loss of infectivity in human noroviruses without an in vitro cell culture assay.
ACKNOWLEDGMENTS
We thank Stephen Grove, Sagar Agarwal, Eduardo Patazca, and Mathew Buenconsejo for help with the HHP work and Jianrong Li (Ohio State University) for technical advice on growing Tulane virus.
This study was supported by Agriculture and Food Research Initiative Competitive Grant 2011-68003-30395 from the U.S. Department of Agriculture, National Institute of Food and Agriculture.
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 11 July 2014
REFERENCES
- 1.Patel MM, Hall AJ, Vinjé J, Parashar UD. 2009. Noroviruses: a comprehensive review. J. Clin. Virol. 44:1–8. 10.1016/j.jcv.2008.10.009 [DOI] [PubMed] [Google Scholar]
- 2.Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinjé J, Parashar UD. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224–1231. 10.3201/eid1408.071114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee LE, Cebelinski EA, Fuller C, Keene WE, Smith K, Vinjé J, Besser JM. 2012. Sapovirus outbreaks in long-term care facilities, Oregon and Minnesota, USA, 2002–2009. Emerg. Infect. Dis. 18:873–876. 10.3201/eid1805.111843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escudero BI, Rawsthorne H, Gensel C, Jaykus LA. 2012. Persistence and transferability of noroviruses on and between common surfaces and foods. J. Food Prot. 75:927–935. 10.4315/0362-028X.JFP-11-460 [DOI] [PubMed] [Google Scholar]
- 5.Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinjé J. 2014. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J. Clin. Microbiol. 52:147–155. 10.1128/JCM.02680-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopman B, Gastanaduy P, Park GW, Hall AJ, Parashar UD, Vinjé J. 2012. Environmental transmission of norovirus gastroenteritis. Curr. Opin. Virol. 2:96–102. 10.1016/j.coviro.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 7.Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MPG, Estes MK. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79–87. 10.1099/vir.0.19478-0 [DOI] [PubMed] [Google Scholar]
- 8.Papafragkou E, Hewitt J, Park GW, Greening G, Vinjé J. 2013. Challenges of culturing human norovirus in three-dimensional organoid intestinal cell culture models. PLoS One 8:e63485. 10.1371/journal.pone.0063485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbst-Kralovetz M, Radtke A, Lay M, Hjelm B, Bolick A, Sarker S, Atmar R, Kingsley D, Arntzen C, Estes M, Nickerson C. 2013. Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells. Emerg. Infect. Dis. 19:431–438. 10.3201/eid1903.121029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takanashi S, Saif LJ, Hughes JH, Meulia T, Jung K, Scheuer KA, Wang Q. 2014. Failure of propagation of human norovirus in intestinal epithelial cells with microvilli grown in three-dimensional cultures. Arch. Virol. 159:257–266. 10.1007/s00705-013-1806-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight A, Li D, Uyttendaele M, Jaykus LA. 2013. A critical review of methods for detecting human noroviruses and predicting their infectivity. Crit. Rev. Microbiol. 39:295–309. 10.3109/1040841X.2012.709820 [DOI] [PubMed] [Google Scholar]
- 12.Sinclair RG, Rose JB, Hashsham SA, Gerba CP, Haas CN. 2012. Criteria for selection of surrogates used to study the fate and control of pathogens in the environment. Appl. Environ. Microbiol. 78:1969–1977. 10.1128/AEM.06582-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke IN, Estes MK, Green KY, Hansman GS, Knowles NJ, Koopmans MK, Matson DO, Meyers G, Neill JD, Radford A, Smith AW, Studdert MJ, Thiel H-J, Vinjé J. 2012. Caliciviridae, virus taxonomy: the classification and nomenclature of viruses, p 977–986 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, Waltham, MA [Google Scholar]
- 14.Farkas T, Sestak K, Wei C, Jiang X. 2008. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 82:5408–5416. 10.1128/JVI.00070-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoover EA, Kahn DE. 1975. Experimentally induced feline calicivirus infection: clinical signs and lesions. J. Am. Vet. Med. Assoc. 166:463–468 [PubMed] [Google Scholar]
- 16.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW., IV 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578. 10.1126/science.1077905 [DOI] [PubMed] [Google Scholar]
- 17.Wobus CE, Thackray LB, Virgin HW., IV 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80:5104–5112. 10.1128/JVI.02346-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cannon JL, Papafragkou E, Park GW, Osborne J, Jaykus LA, Vinjé J. 2006. Surrogates for the study of norovirus stability and inactivation in the environment: a comparison of murine norovirus and feline calicivirus. J. Food Prot. 69:2761–2765 [DOI] [PubMed] [Google Scholar]
- 19.Belliot G, Lavaux A, Souihel D, Agnello D, Pothier P. 2008. Use of murine norovirus as a surrogate to evaluate resistance of human norovirus to disinfectants. Appl. Environ. Microbiol. 74:3315–3318. 10.1128/AEM.02148-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tung G, Macinga D, Arbogast J, Jaykus LA. 2013. Efficacy of commonly used disinfectants for inactivation of human noroviruses and their surrogates. J. Food Prot. 76:1210–1217. 10.4315/0362-028X.JFP-12-532 [DOI] [PubMed] [Google Scholar]
- 21.Bozkurt H, D'Souza DH, Davidson PM. 2013. Determination of the thermal inactivation kinetics of the human norovirus surrogates, murine norovirus and feline calicivirus. J. Food Prot. 76:79–84. 10.4315/0362-028X.JFP-12-327 [DOI] [PubMed] [Google Scholar]
- 22.Park GW, Barclay L, Macinga D, Charbonneau D, Pettigrew CA, Vinjé J. 2010. Comparative efficacy of seven hand sanitizers against murine norovirus, feline calicivirus, and GII.4 norovirus. J. Food Prot. 73:2232–2238 [DOI] [PubMed] [Google Scholar]
- 23.Park GW, Sobsey MD. 2011. Simultaneous comparison of murine norovirus, feline calicivirus, coliphage MS2, and GII.4 norovirus to evaluate the efficacy of sodium hypochlorite against human norovirus on a fecally soiled stainless steel surface. Foodborne Pathog. Dis. 8:1005–1010. 10.1089/fpd.2010.0782 [DOI] [PubMed] [Google Scholar]
- 24.Kim SW, Baek SB, Ha JH, Lee MH, Choi C, Ha SD. 2012. Chlorine treatment to inactivate norovirus on food contact surfaces. J. Food Prot. 75:184–188. 10.4315/0362-028X.JFP-11-243 [DOI] [PubMed] [Google Scholar]
- 25.Esseili MA, Wang Q, Zhang Z, Saif LJ. 2012. Internalization of sapovirus, a surrogate for norovirus, in romaine lettuce and the effect of lettuce latex on virus infectivity. Appl. Environ. Microbiol. 78:6271–6279. 10.1128/AEM.01295-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirneisen KA, Kniel KE. 2013. Comparing human norovirus surrogates: murine norovirus and Tulane virus. J. Food Prot. 76:139–143. 10.4315/0362-028X.JFP-12-216 [DOI] [PubMed] [Google Scholar]
- 27.Tian P, Yang D, Quigley C, Chou M, Jiang X. 2013. Inactivation of the Tulane virus, a novel surrogate for the human norovirus. J. Food Prot. 76:712–718. 10.4315/0362-028X.JFP-12-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Hirneisen KA, Markland SM, Kniel KE. 2013. Survival of murine norovirus, Tulane virus, and hepatitis A virus on alfalfa seeds and sprouts during storage and germination. Appl. Environ. Microbiol. 79:7021–7027. 10.1128/AEM.01704-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parwani AV, Flynn WT, Gadfield KL, Saif LJ. 1991. Serial propagation of porcine enteric calicivirus in a continuous cell line. Effect of medium supplementation with intestinal contents or enzymes. Arch. Virol. 120:115–122 [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Zhang Z, Saif LJ. 2012. Stability of and attachment to lettuce by a culturable porcine sapovirus surrogate for human caliciviruses. Appl. Environ. Microbiol. 78:3932–3940. 10.1128/AEM.06600-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallansch M, Oberste MS, Whitton JL. 2013. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses, chapter 17, p 490–530 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 32.Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. 1991. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J. Infect. Dis. 164:954–957. 10.1093/infdis/164.5.954 [DOI] [PubMed] [Google Scholar]
- 33.Lodder WJ, Rutjes SA, Takumi K, de Roda Husman AM. 2013. Aichi virus in sewage and surface water, the Netherlands. Emerg. Infect. Dis. 19:1222–1230. 10.3201/eid1908.130312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kingsley DH. 2013. High pressure processing and its application to the challenge of virus-contaminated foods. Food Environ. Virol. 5:1–12. 10.1007/s12560-012-9094-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esseili MA, Wang Q, Saif LJ. 2012. Binding of human GII.4 norovirus virus-like particles to carbohydrates of romaine lettuce leaf cell wall materials. Appl. Environ. Microbiol. 78:786–794. 10.1128/AEM.07081-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 37.Vega E, Barclay L, Gregoricus N, Williams K, Lee D, Vinjé J. 2011. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerg. Infect. Dis. 17:1389–1395. 10.3201/eid1708.101837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gentry J, Vinjé J, Guadagnoli D, Lipp EK. 2009. Norovirus distribution within an estuarine environment. Appl. Environ. Microbiol. 75:5474–5480. 10.1128/AEM.00111-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chhabra P, Payne DC, Szilagyi PG, Edwards KM, Staat MA, Shirley SH, Wikswo M, Nix WA, Lu X, Parashar UD, Vinjé J. 2013. Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. J. Infect. Dis. 208:790–800. 10.1093/infdis/jit254 [DOI] [PubMed] [Google Scholar]
- 40.Springthorpe VS, Sattar SA. 2005. Carrier tests to assess microbicidal activities of chemical disinfectants for use on medical devices and environmental surfaces. J. AOAC Int. 88:182–201 [PubMed] [Google Scholar]
- 41.D'Agostino RB, Sullivan LM, Beiser AS. 2005. Introductory applied biostatistics. Brooks/Cole, Belmont, CA [Google Scholar]
- 42.Duizer E, Bijkerk P, Rockx B, De Groot A, Twisk F, Koopmans M. 2004. Inactivation of caliciviruses. Appl. Environ. Microbiol. 70:4538–4543. 10.1128/AEM.70.8.4538-4543.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sattar SA, Ali M, Tetro JA. 2011. In vivo comparison of two human norovirus surrogates for testing ethanol-based handrubs: the mouse chasing the cat! PLoS One. 6:e17340. 10.1371/journal.pone.0017340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.WHO. 2008. CDC/WHO Hand Hygiene Guidelines crosswalk. Jt. Comm. Perspect 28:4–7 [PubMed] [Google Scholar]
- 45.Leon JS, Kingsley DH, Montes JS, Richards GP, Lyon GM, Abdulhafid GM, Seitz SR, Fernandez ML, Teunis PF, Flick GJ, Moe CL. 2011. Randomized, double-blinded clinical trial for human norovirus inactivation in oysters by high hydrostatic pressure processing. Appl. Environ. Microbiol. 77:5476–5482. 10.1128/AEM.02801-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Chen H, Kingsley DH. 2013. The influence of temperature, pH, and water immersion on the high hydrostatic pressure inactivation of GI.1 and GII.4 human noroviruses. Int. J. Food Microbiol. 167:138–143. 10.1016/j.ijfoodmicro.2013.08.020 [DOI] [PubMed] [Google Scholar]
- 47.Kovac K, Diez-Valcarce M, Raspor P, Hernandez M, Rodriguez-Lazaro D. 2012. Effect of high hydrostatic pressure processing on norovirus infectivity and genome stability in strawberry puree and mineral water. Int. J. Food Microbiol. 152:35–39. 10.1016/j.ijfoodmicro.2011.10.005 [DOI] [PubMed] [Google Scholar]
- 48.Horm KM, Harte FM, D'Souza DH. 2012. Human norovirus surrogate reduction in milk and juice blends by high pressure homogenization. J. Food Prot. 75:1984–1990. 10.4315/0362-028X.JFP-12-003 [DOI] [PubMed] [Google Scholar]
- 49.Arcangeli G, Terregino C, De Benedictis P, Zecchin B, Manfrin A, Rossetti E, Magnabosco C, Mancin M, Brutti A. 2012. Effect of high hydrostatic pressure on murine norovirus in Manila clams. Lett. Appl. Microbiol. 54:325–329. 10.1111/j.1472-765X.2012.03211.x [DOI] [PubMed] [Google Scholar]
- 50.Farkas T, Cross RW, Hargitt E, III, Lerche NW, Morrow AL, Sestak K. 2010. Genetic diversity and histo-blood group antigen interactions of rhesus enteric caliciviruses. J. Virol. 84:8617–8625. 10.1128/JVI.00630-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diez-Valcarce M, Kovac K, Raspor P, Rodriguez-Lazaro D, Hernandez M. 2011. Virus genome quantification does not predict norovirus infectivity after application of food inactivation processing technologies. Food Environ. Virol. 3:141–146. 10.1007/s12560-011-9070-9 [DOI] [Google Scholar]
- 52.Gehrke C, Steinmann J, Goroncy-Bermes P. 2004. Inactivation of feline calicivirus, a surrogate of norovirus (formerly Norwalk-like viruses), by different types of alcohol in vitro and in vivo. J. Hosp. Infect. 56:49–55. 10.1016/j.jhin.2003.08.019 [DOI] [PubMed] [Google Scholar]
- 53.Liu P, Escudero B, Jaykus LA, Montes J, Goulter RM, Lichtenstein M, Fernandez M, Lee JC, De Nardo E, Kirby A, Arbogast JW, Moe CL. 2013. Laboratory evidence of Norwalk virus contamination on the hands of infected individuals. Appl. Environ. Microbiol. 79:7875–7881. 10.1128/AEM.02576-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D'Souza DH, Su X. 2010. Efficacy of chemical treatments against murine norovirus, feline calicivirus, and MS2 bacteriophage. Foodborne Pathog. Dis. 7:319–326. 10.1089/fpd.2009.0426 [DOI] [PubMed] [Google Scholar]
- 55.Tree JA, Adams MR, Lees DN. 2005. Disinfection of feline calicivirus (a surrogate for Norovirus) in wastewaters. J. Appl. Microbiol. 98:155–162. 10.1111/j.1365-2672.2004.02442.x [DOI] [PubMed] [Google Scholar]
- 56.Gulati BR, Allwood PB, Hedberg CW, Goyal SM. 2001. Efficacy of commonly used disinfectants for the inactivation of calicivirus on strawberry, lettuce, and a food contact surface. J. Food Prot. 64:1430–1434 [DOI] [PubMed] [Google Scholar]
- 57.Lim MY, Kim JM, Ko G. 2010. Disinfection kinetics of murine norovirus using chlorine and chlorine dioxide. Water Res. 44:3243–3251. 10.1016/j.watres.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 58.Park GW, Boston DM, Kase JA, Sampson MN, Sobsey MD. 2007. Evaluation of liquid- and fog-based application of Sterilox hypochlorous acid solution for surface inactivation of human norovirus. Appl. Environ. Microbiol. 73:4463–4468. 10.1128/AEM.02839-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.FDA. 1999. Food code 1999. U.S. Department of Health and Human Services, Public Health Service, Food and Drug Administration, College Park, MD [Google Scholar]
- 60.Iwamoto M, Ayers T, Mahon BE, Swerdlow DL. 2010. Epidemiology of seafood-associated infections in the United States. Clin. Microbiol. Rev. 23:399–411. 10.1128/CMR.00059-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cruz-Romero M, Kelly AL, Kerry JP. 2007. Effects of high-pressure and heat treatments on physical and biochemical characteristics of oysters (Crassostrea gigas). Innov. Food Sci. Emerg. 8:30–38. 10.1016/j.ifset.2006.05.002 [DOI] [Google Scholar]
- 62.Li X, Ye M, Neetoo H, Golovan S, Chen H. 2013. Pressure inactivation of Tulane virus, a candidate surrogate for human norovirus and its potential application in food industry. Int. J. Food Microbiol. 162:37–42. 10.1016/j.ijfoodmicro.2012.12.016 [DOI] [PubMed] [Google Scholar]
- 63.Ye M, Li X, Kingsley DH, Jiang X, Chen H. 2014. Inactivation of human norovirus in contaminated oysters and clams by high hydrostatic pressure. Appl. Environ. Microbiol. 80:2248–2253. 10.1128/AEM.04260-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen H, Hoover DG, Kingsley DH. 2005. Temperature and treatment time influence high hydrostatic pressure inactivation of feline calicivirus, a norovirus surrogate. J. Food Prot. 68:2389–2394 [DOI] [PubMed] [Google Scholar]
- 65.Kingsley DH, Chen H. 2008. Aqueous matrix compositions and pH influence feline calicivirus inactivation by high pressure processing. J. Food Prot. 71:1598–1603 [DOI] [PubMed] [Google Scholar]
- 66.Sanchez G, Aznar R, Martinez A, Rodrigo D. 2011. Inactivation of human and murine norovirus by high-pressure processing. Foodborne Pathog. Dis. 8:249–253. 10.1089/fpd.2010.0667 [DOI] [PubMed] [Google Scholar]
- 67.Lou F, Neetoo H, Chen H, Li J. 2011. Inactivation of a human norovirus surrogate by high-pressure processing: effectiveness, mechanism, and potential application in the fresh produce industry. Appl. Environ. Microbiol. 77:1862–1871. 10.1128/AEM.01918-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kingsley DH, Chen H, Hoover DG. 2004. Inactivation of selected picornaviruses by high hydrostatic pressure. Virus Res. 102:221–224. 10.1016/j.virusres.2004.01.030 [DOI] [PubMed] [Google Scholar]
- 69.Barclay LE, Park GW, Vega E, Hall A, Parashar U, Vinjé J, Lopman BA. 2014. Infection control for norovirus. Clin. Microbiol. Infect. 10.1111/1469-0691.12674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richards GP. 2012. Critical review of norovirus surrogates in food safety research: rationale for considering volunteer studies. Food Environ. Virol. 4:6–13. 10.1007/s12560-011-9072-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang D, Tian P. 2014. Inactivation conditions for human norovirus measured by an in situ capture-qRT-PCR method. Int. J. Food Microbiol. 172:76–82. 10.1016/j.ijfoodmicro.2013.11.027 [DOI] [PubMed] [Google Scholar]
- 72.Wigginton KR, Kohn T. 2012. Virus disinfection mechanisms: the role of virus composition, structure, and function. Curr. Opin. Virol. 2:84–89. 10.1016/j.coviro.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]