Abstract
The management and control of mosquito vectors of human disease currently rely primarily on chemical insecticides. However, larvicidal treatments can be effective, and if based on biological insecticides, they can also ameliorate the risk posed to human health by chemical insecticides. The aerobic bacteria Bacillus thuringiensis and Lysinibacillus sphaericus have been used for vector control for a number of decades. But a more cost-effective use would be an anaerobic bacterium because of the ease with which these can be cultured. More recently, the anaerobic bacterium Clostridium bifermentans subsp. malaysia has been reported to have high mosquitocidal activity, and a number of proteins were identified as potentially mosquitocidal. However, the cloned proteins showed no mosquitocidal activity. We show here that four toxins encoded by the Cry operon, Cry16A, Cry17A, Cbm17.1, and Cbm17.2, are all required for toxicity, and these toxins collectively show remarkable selectivity for Aedes rather than Anopheles mosquitoes, even though C. bifermentans subsp. malaysia is more toxic to Anopheles. Hence, toxins that target Anopheles are different from those expressed by the Cry operon.
INTRODUCTION
Vector-borne diseases, specifically those transmitted by mosquitoes, continue to persist with constant threats of reemergence. Aggressive insecticide usage over the years contained mosquito-borne diseases, such as dengue fever, chikungunya, and yellow fever, filariasis, and malaria, primarily to a number of tropical areas. However, these diseases began to resurface in the 1970s, and previous levels of progress in the control of these diseases could no longer be sustained (1). Several factors can contribute to the reemergence of mosquito-borne diseases, including vector insecticide and pathogen drug resistance and genetic changes in both the vector and the vectored pathogens (2). Major global demographic and societal changes also contribute significantly. Moreover, with global warming and growth in the world's population, especially in the tropical regions, the incidence of mosquito-borne diseases will rise.
Large numbers of deaths worldwide are caused by vector-borne diseases. For example, yellow fever causes thousands of deaths annually in sub-Saharan Africa and tropical South America and can cause hemorrhagic fever, which is fatal 20% to 50% of the time (3). In addition, dengue fever is one of the major causes of morbidity and mortality in many Asian and South American countries (4). Similarly, resurging epidemics of malaria are seen in areas where the disease was previously thought to have been constrained (5). Consequently, mortality due to malaria is still responsible for close to a million deaths per year (6).
The discovery of species-specific biological insecticides for mosquito control produced by Bacillus thuringiensis subsp. israelensis and Lysinibacillus sphaericus has opened up new prospects for a safer and targeted mosquito control. Even though Bacillus thuringiensis subsp. israelensis formulations have been used for more than 4 decades, no substantial resistance has been observed in the field, although resistance to individual toxins has been observed in the laboratory (7). However, resistance to L. sphaericus toxins has been observed in the field in a number of countries (8–10). This ability of organisms to adapt and develop resistance to new insecticides indicates a need for the development or discovery of new biological insecticides.
Clostridium bifermentans subsp. malaysia is an anaerobic Clostridium strain possessing larvicidal properties and was isolated in Malaysia (11, 12). Clostridium is a large and diverse genus that is characterized by Gram-positive bacteria capable of forming endospores. Most species are well known for their metabolic properties and distinctive industrial applications. However, a small number of clostridial species are responsible for causing human and animal diseases. But only C. bifermentans strains to date have shown toxicity to mosquitoes, and subsp. malaysia is highly toxic to Anopheles but has lower toxicity to Aedes and Culex (13, 14). C. bifermentans subsp. malaysia cells show no toxicity to mammals and goldfish (14). Other studies have shown that C. bifermentans subsp. malaysia has no toxicity against mammals and various other nontarget organisms (15). These characteristics make C. bifermentans subsp. malaysia a promising focus for identification of a biological insecticide for management of mosquito populations.
Like B. thuringiensis subsp. israelensis and L. sphaericus, C. bifermentans subsp. malaysia toxicity is expressed during the sporulation stage but decreases significantly with cell lysis (16). But, unlike B. thuringiensis subsp. israelensis, C. bifermentans subsp. malaysia produces no parasporal inclusions that could be associated with toxicity (16). Biochemical analysis of C. bifermentans subsp. malaysia cultures showed that three proteins of 66 kDa, 18 kDa, and 16 kDa were involved in toxicity (17). In addition, it was suggested that these proteins could aggregate into a complex and are unstable when subjected to various methods of purification (17).
Subsequent efforts showed that these toxic components consisted of four major proteins in crude cultures. Those putative toxins include a doublet of 66 kDa to 68 kDa (encoded by genes cbm71 and cbm72) and two other small proteins of 18 kDa and 16 kDa (Cbm17.1 and Cbm17.2) (17). The cbm71 and cbm72 genes (accession numbers X94146 and X99478, respectively) were subsequently cloned and characterized, and the proteins were renamed Cry16A and Cry17A toxins and shown to have low levels of toxicity to Anopheles, Aedes, and Culex mosquitoes (18, 19). The gene encoding the Cbm17.1 and Cbm17.2 proteins was also identified (accession number Y10457), and these proteins have low amino acid similarity (44%) to a hemolysin from Aspergillus fumigatus (18, 19). Importantly, all four of these proteins were immunologically unrelated to toxins produced in B. thuringiensis subsp. israelensis and L. sphaericus and were determined to belong to a novel class of insecticidal toxins (20). Although early reports (18, 19) indicated that the Cry16A and Cry17A toxins had mosquitocidal activity, a subsequent report (21) indicated that the Cry16A and Cry17A toxins are not mosquitocidal and that the Cbm17.1 and Cbm17.2 proteins are not hemolytic.
In this study, we show that the Cry operon carries four genes, all of which are required as a complex for toxicity to mosquito larvae. The toxin complex, however, shows surprising species selectivity, particularly to Aedes mosquitoes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
C. bifermentans (ATCC) was used as the wild-type reference strain, and C. bifermentans subsp. malaysia was from the collection of the Institute for Medical Research, Malaysia (13). These strains were used for bioassays and for isolation of genomic DNA (gDNA) for cloning. Escherichia coli strain DH10 beta electrocompetent cells (New England BioLabs [NEB], Ipswich, MA) were used for cloning, and the NEB transformation protocol was used. B. thuringiensis subsp. israelensis 4Q7 (Bacillus Stock Center, Ohio State University, Columbus, Ohio) was used for expression of toxin genes using a published protocol (22). Clostridial strains were streaked for isolation on tryptone-yeast extract-glucose (TYG) agar, and liquid cultures were grown in TYG medium at 30°C under anaerobic conditions using BD GasPakEZ (Becton-Dickinson Microbiology, Cockeysville, MD). The liquid cultures for Clostridium strains were grown anaerobically at 30°C, while the Bacillus cultures were grown at 30°C with shaking. Nutrient broth medium was used for B. thuringiensis, and Luria broth medium was used for E. coli. Ampicillin (100 μg/ml) and erythromycin (25 μg/ml) were added when required.
To monitor mosquitocidal activities in C. bifermentans subsp. malaysia, the cultures after various times of growth were used in bioassays. The cultures were also centrifuged in a high-speed centrifuge to isolate pellet and supernatant fractions. C. bifermentans subsp. malaysia also underwent autolysis by 12 h; however, bacterial cell lysis was enhanced by sonication when needed.
General DNA techniques.
All DNA manipulations were performed according to standard protocols. The primers used in this study were created using the sequences from existing databases. PCRs were performed in an automated thermocycler (C 1000 Touch; Bio-Rad). Choice-Taq mastermix DNA polymerase (Denville Scientific, Metuchen, NJ) was used for all PCRs for products of <2 kb. Advantage 2× polymerase (Clontech, Mountain View, CA) or Phusion polymerase (NEB) was used for all PCRs for products of >2 kb. PCR products were separated in 1% agarose gels and subsequently cut and purified using Wizard SV gel and PCR purification kits (Promega, Madison, WI). Sequencing of purified DNA products was performed by the Genomics Core facility at the University of California, Riverside.
Construction of plasmids expressing the full and partial Cry operon and individual toxins.
The vector pHT315 (23) was used for construction of expression plasmids (Table 1). Subcloning was accomplished using a Gibson assembly mastermix (NEB). The Cyt1A promoter, a constitutive promoter from B. thuringiensis subsp. israelensis, was used for expression of all gene constructs. Hence, the 508 bp upstream of the first ATG of cyt1A was amplified from pWF45 (24) using primers 1 and 2 (Table 2). The cassettes containing the genes of interest were amplified using primers 3 and 4 for plasmid pCryO (containing the Cry operon), primers 3 and 5 for pCry16 (cry16A), and primers 4 and 6 for pCbm17.2 (cbm17.2) (Table 2). pHT315 was linearized with SmaI in the multiple cloning site. All DNA products were gel purified and purified using Wizard SV gel and PCR (Promega) purification kits to provide the desired DNA concentrations. A Gibson assembly protocol (25) was used to assemble the fragments, followed by transformation in DH10 beta electrocompetent cells (NEB). Plasmids pCryO (containing the Cry operon), pCry16 (cry16Aa), and pCbm17.2 (cbm17.2) were made using this approach. However, for plasmids pCry17 (containing cry17Aa) and pCbm17.1 (cbm17.1), the genes were synthesized (GenScript, Piscataway, NJ) and cloned in pHT315. The constructs pCry16/17 and pCryO/Δ17.1 were obtained using NcoI and PacI restriction, respectively, of pCryO followed by religation of the resulting restricted plasmid. Following confirmation of correct ligation, the plasmid constructs (Table 1) were extracted from DH10 beta cells in ample quantities. All plasmids were then used to independently transform B. thuringiensis subsp. israelensis 4Q7 cells by electroporation, and colonies were isolated for erythromycin resistance as previously described (26).
TABLE 1.
List of constructs used to assess toxicity of genes in the Cry operon
| Construct namea | C. bifermentans subsp. malaysia gene(s) used |
|---|---|
| pCryO | cry16Aa, cry17Aa, cbm17.1, cbm17.2 |
| pCry16 | cry16Aa |
| pCry17 | cry17Aa |
| pCbm17.1 | cbm17.1 |
| pCbm17.2 | cbm17.2 |
| pCry16/17 | cry16Aa, cry17Aa |
| pCryO/Δ17.1 | cry16Aa, cry17Aa, cbm17.2 |
All constructs were in pHT315 using the Cyt1A promoter from B. thuringiensis subsp. israelensis.
TABLE 2.
Primers used to create pCryO, pCry16, and pCbm17.2
| Primer no. | Description | Sequence |
|---|---|---|
| 1 | Promoter_f_overlap_pHT315 | AGTGAATTCGAGCTCGGTACCCTTTTCGATTTCAAATTTTCCAAAC |
| 2 | Promoter_r_no_overlap | AAATAAACAACTCCTTAAAGTTAATTAGAATAGTGG |
| 3 | CryO_f_overlap_promoter | CTAATTAACTTAAGGAGTTGTTTATTTATGAATACTAATATTTTTTCTACACATG |
| 4 | CryO_r_overlap_pHT315 | GGTCGACTCTAGAGGATCCCCCATCTGTATCATCTCATAAAAGATTAGGG |
| 5 | Cry16A_r_overlap_pHT315 | GGTCGACTCTAGAGGATCCCCAAGATTATCATTTGAGCTTGTGCCAG |
| 6 | 17.2_f_overlap_promoter | CTAATTAACTTAAGGAGTTGTTTATTTATGAACAATAATTGTGAAGTTAATTGTGAG |
Bioassays.
Total cultures of C. bifermentans, C. bifermentans subsp. malaysia, and recombinant B. thuringiensis strains were assayed against Aedes aegypti, Anopheles gambiae, Anopheles stephensi, and Culex quinquefasciatus larvae. Bioassays were performed at 24°C using 25 third-instar larvae in 200 ml water. Live bacterial cultures were used in bioassays. Larval mortality was determined by counting the number of larvae still alive at 24, 48, or 72 h. Bioassays were repeated 2 or 3 times, and 50% lethal concentrations (LC50s) were determined by probit analysis (USDA) and plotted using the Origin program (Origin Lab, Northampton, MA).
Hemolytic assays.
One milliliter of trypsinized sheep red blood cells (RBCs) (Colorado Serum Company) was centrifuged at 2,000 × g for 2 min. The pellet was washed three times with ice-cold 1× phosphate-buffered saline (PBS) (pH 7.4). The pellet was then resuspended in 30 ml 1× PBS (pH 7.4). To 900 μl of the RBC suspension, 100 μl of the bacterial cell culture was added, and the mixture was incubated at 37°C for 30 min. The optical density at 600 nm (OD600) was measured using a spectrophotometer (SmartSpec 3000; Bio-Rad, Hercules, CA). The RBC suspension and 1× PBS (pH 7.4) were used as negative controls, and 0.1% Triton and Cyt1A (15 ng/ml) were used as positive controls. Whole-culture samples and supernatant, obtained with the B-PER bacterial protein extraction reagent (Pierce, Rockford, IL), of recombinant B. thuringiensis strains containing pCryO, pCbm17.1, pCbm17.2, and pCryO/Δ17.1 were used as samples.
Antibodies.
Hydrophilic peptides from Cry16A and Cbm17.1 were designed based on TopPred topology and antigenicity prediction index (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html). The Cry16A peptide (CSYTDGNFEDFPKLS), comprising residues 591 to 604, and the Cbm17.1 peptide (CNKLTIDKYNTKFAI), comprising residues 115 to 128, were commercially synthesized, confirmed by mass spectral analyses (GenScript USA, Piscataway, NJ), and conjugated via the introduced cysteine to a maleimide-activated KLH carrier protein (Pierce, Rockford, IL) according to the manufacturer's protocol. This conjugate was used to immunize rabbits. (Research with animals was approved by the UC Riverside Animal Care and Use Committee and the NIH.) Cbm17.1 is comprised of 153 amino acids and shows 78% identity to Cbm17.2, which has 152 amino acids. The Cbm17.1 peptide used for antibody development is in a divergent region between the two proteins, and hence this antibody does not detect the Cbm17.2 protein.
PAGE and immunoblotting.
C. bifermentans, C. bifermentans subsp. malaysia, recombinant B. thuringiensis, and B. thuringiensis 4Q7 strains were grown in parallel for 4 to 72 h. Aliquots (100 μl) of the cultures were mixed with 2× sample buffer (Bio-Rad, Hercules, CA), and the proteins were separated by SDS-polyacrylamide (10 to 14%) gel electrophoresis. Native polyacrylamide (10%) gel electrophoresis was performed using 1× native PAGE buffer (3 g Tris, 14.5 g glycine). In parallel, the separated proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA). The membranes were treated with blocking solution (1× PBS, 5% skim milk, and 0.1% Tween 20) for 1 h at room temperature and then washed with PBST (1× PBS and 0.1% Tween 20). The blocked membrane was incubated overnight (at 4°C) with primary anti-Cry16A or Cbm17.1 antibody at a 1:1,000 dilution. A secondary antibody, enhanced chemiluminescence (ECL) anti-rabbit IgG-horseradish peroxidase-linked whole antibody (GE Healthcare, Anaheim, CA), was used at a dilution of 1:5,000. Immunoreactive bands were visualized using the ECL Western blotting kit (GE Healthcare) and exposed to an X-ray film.
Mass spectrometry.
Since there were substantial differences in mosquitocidal activity between 1- and 5-day cultures, these bacterial cultures were separated by SDS-PAGE. The protein bands that differed between the two cultures were excised from the gel, digested by trypsin, and analyzed by nano-ultraperformance liquid chromatography/tandem mass spectrometry (nano-UPLC/MS/MS) at the IIGB proteomics facility, University of California—Riverside.
RESULTS
Toxicity of Clostridium bifermentans subsp. malaysia to mosquito larvae.
In this study, we used a proteomics approach to identify the C. bifermentans subsp. malaysia toxins involved in conferring toxicity to mosquito larvae. To facilitate such analysis, we analyzed proteins that were active in the pellet and supernatant fractions obtained after centrifugation of the bacterial cultures. Thus, we monitored the toxicity of C. bifermentans subsp. malaysia live cultures to third- or fourth-instar A. stephensi mosquito larvae. At 6 to 10 h, a high level of toxicity was observed in the bacterial pellet while no toxicity was observed in the supernatant. However, at 12 to 24 h, high levels of toxicity were also observed in supernatant fractions as well as the pellet fraction. The biological activity remained high until 72 h (Fig. 1).
FIG 1.
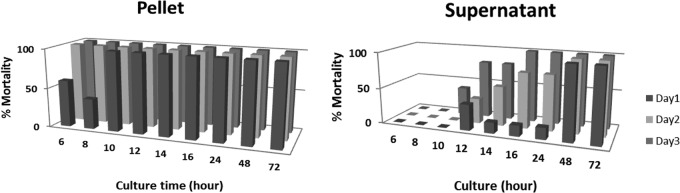
Toxicity appears in supernatants within a day of C. bifermentans subsp. malaysia cultures. To analyze the appearance of toxicity in the supernatants, both the pellet and supernatant fractions were monitored for toxicity to fourth-instar Anopheles stephensi larvae. A constant amount of the bacterial culture was assayed. A high level of toxicity in the pellet of the bacterial culture was observed within a short time of culture, while no toxicity was detected in the supernatant until 12 h, when lysis of sporangia occurs (16). Toxicity was monitored at 24, 48, and 72 h.
To determine the stability of the mosquitocidal activity, cells were lysed and tested for toxin stability after incubation at different temperatures. Under the tested conditions, it appeared that following 6 h of incubation at −80°C, 0°C, and 8°C, toxicity remained high, but it decreased at higher temperatures. However, the mosquitocidal activity was lower at all temperatures, with complete loss of toxicity at 55°C after 24 h of incubation. The data show that the overall half-life of the toxicity was about 1 day and the toxin was unstable at high temperatures. The loss of activity is likely a combination of proteolysis and low stability of C. bifermentans subsp. malaysia toxins.
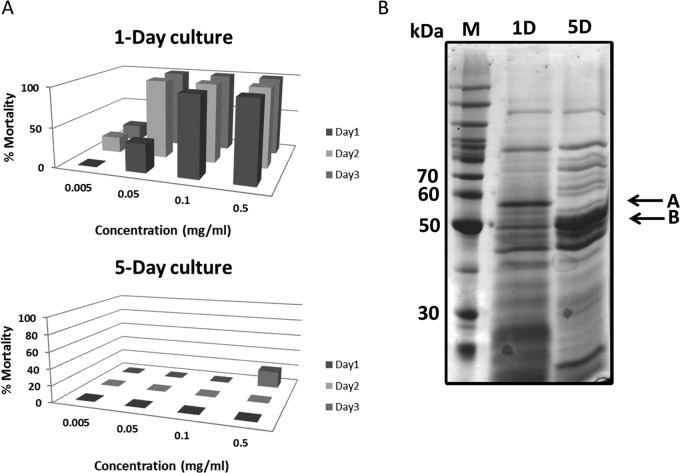
Mosquitocidal activity of supernatants obtained after cell lysis was also determined at 1 and 5 days. In the 1-day culture, a high level of toxicity was observed in the supernatant fraction, while the 5-day culture showed no toxicity, suggesting that the toxins are degraded with time after cell lysis (Fig. 2). Hence, proteins from 1- and 5-day cultures were separated immediately by SDS-PAGE, and proteins that differed were subjected to analysis by UPLC/MS/MS. Among the 21 proteins identified in day-1 culture band A, 11 of these were not present on day 5. Of these, four were DNA gyrase, one was a peptidase, one was an acetate kinase, one was a conserved protein, two were hypothetical proteins, and one was the Cry16A toxin.
FIG 2.
Toxicity is lost in C. bifermentans subsp. malaysia cultures after 5 days. (A) Toxicity in 1- and 5-day supernatants. Whole cultures were centrifuged to collect supernatants. In 1-day cultures, a high level of toxicity was observed in the supernatant. However, in 5-day cultures no toxicity was observed in 24- and 48-h bioassays. (B) Differential expressions of the protein profiles for 1- and 5-day cultures were observed in the supernatants (bands A and B). Following tryptic digestion, the peptides were sequenced using tandem mass spectrophotometry. Band B of 5-day supernatant was identified as heat shock proteins. In contrast, band A was found in 1-day but not in 5-day supernatants. One protein identified by proteomics was the Cry16A protein in the 1-day supernatant.
Expression of the Cry toxin operon.
The Cry16A toxin, as reported previously, is encoded by an operon (19). Hence, the full operon (Fig. 3A; Table 1), consisting of the cry16A, cry17A, cbm17.1, and cbm17.2 genes, was expressed using the Cyt1A promoter from B. thuringiensis subsp. israelensis (24, 26).
FIG 3.
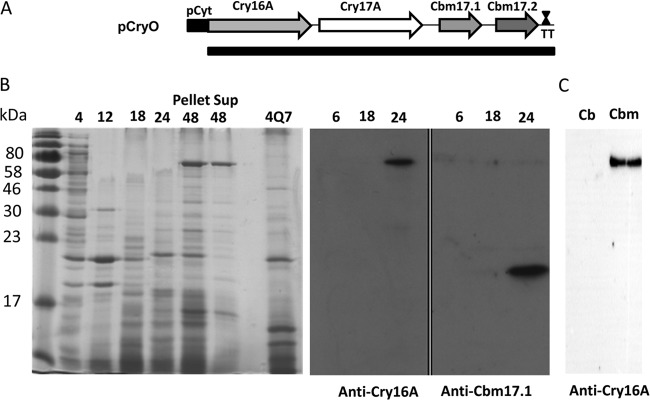
The full-length operon expresses both the Cry and hemolysin proteins. (A) Schematic of the Cry operon under the control of the Cyt1A promoter from Bacillus thuringiensis subsp. israelensis. The operon consists of the cry16A, cry17A, cbm17.1, and cbm17.2 genes, and a terminator (TT). Except for cry16A, the native Shine-Dalgarno (SD) sequences were used. For cry16A, the Cyt1A SD was used. (B) Both the Cry16A and Cbm17.1 proteins were expressed when the entire operon was used for expression. Immunoblots using anti-Cry16A and -Cbm17.1 antibodies show that these were expressed at 24 h. (C) The Cry16A protein was also observed in C. bifermentans subsp. malaysia (Cbm) cultures at 18 h but not in C. bifermentans (Cb) cultures. Sup, supernatant.
Protein expression from this transformation, recombinant B. thuringiensis pCryO, was analyzed by Coomassie blue staining and Western blotting (Fig. 3B). Cultures were grown for 4, 12, 18, 24, and 48 h to determine the optimal time for toxin production. A strong band was observed around 70 kDa for the Cry16A and Cry17A proteins, which was visible in 48-h cultures, but the approximately 20-kDa band for Cbm17.1 and Cbm17.2, which was visible at 4 h, was not visible by around 48 h of culture. These bands were not present in the control comprising of strain 4Q7 containing the pHT315 vector alone. Thus, we concluded that the entire operon is expressed under these conditions, with an optimal expression of the operon at around 24 h.
Western blotting against whole cultures with antibodies raised against the Cry16A and Cbm17.1 peptides also revealed the presence of a protein at about 70 kDa, corresponding to the predicted size of the Cry16A toxin, and one at about 20 kDa, which corresponds to the predicted size of the Cbm17.1 and Cbm17.2 proteins. Both of these bands could be detected in bacterial strains containing pCryO at 24 h. Moreover, the proteins reacting with Cry16A antibodies were observed in whole cultures of C. bifermentans subsp. malaysia (Fig. 3C), but not in C. bifermentans cultures. No protein reacting with Cry16A or Cbm17.1 antibodies was observed in whole cultures of C. bifermentans or strain 4Q7 containing the pHT315 vector alone (Fig. 3C).
Larvicidal activity of bacterial clone expressing the Cry operon.
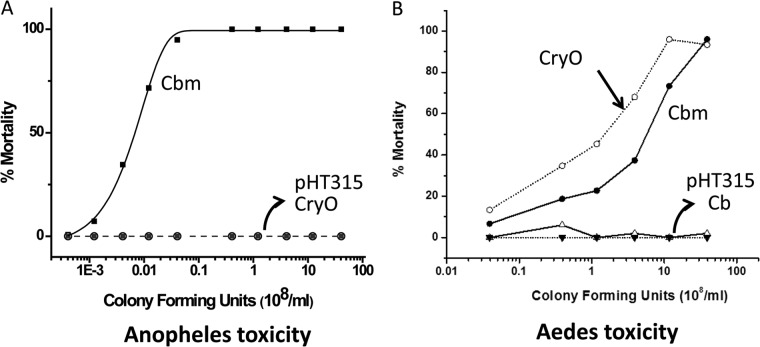
Whole-cell preparations from C. bifermentans, C. bifermentans subsp. malaysia, and recombinant B. thuringiensis transformants containing pHT315 (control) and pCryO, which expressed the entire Cry operon, were assayed for larvicidal activity using third-instar Aedes aegypti, Anopheles gambiae (Table 3), and Culex quinquefasciatus. The recombinant B. thuringiensis pHT315 control samples showed no larval toxicity against any of the three species. But the recombinant B. thuringiensis pCryO sample showed no activity against Anopheles gambiae, even though C. bifermentans subsp. malaysia is highly toxic to this species, with an LC50 of 6.4 × 105 CFU/ml (Fig. 4A; Table 3). In contrast, the recombinant B. thuringiensis pCryO showed high larvicidal activity against Aedes aegypti with a pattern comparable to that of C. bifermentans subsp. malaysia but showing higher mortality (Fig. 4B). C. bifermentans subsp. malaysia had an LC50 of 3.3 × 108 CFU/ml, whereas recombinant B. thuringiensis pCryO had an LC50 of 1.4 × 108 CFU/ml (Fig. 4B; Table 3). Recombinant B. thuringiensis pCryO also showed no toxicity to Culex quinquefasciatus larvae.
TABLE 3.
Toxicity of C. bifermentans subsp. malaysia and recombinant Bacillus thuringiensis expressing the C. bifermentans subsp. malaysia Cry operon to mosquito larvae
| Organism | LC50 (95% confidence interval) for larvae of: |
|
|---|---|---|
| Aedes aegypti (× 108 CFU/ml) | Anopheles gambiae (× 105 CFU/ml) | |
| C. bifermentans subsp. malaysia | 3.30 (1.39–10.0) | 6.39 (4.87−8.46) |
| B. thuringiensis with Cry operona | 1.39 (0.70–2.74) | Nontoxic |
Recombinant Bacillus thuringiensis expressing the C. bifermentans subsp. malaysia Cry operon.
FIG 4.
The full-length Cry operon shows high toxicity to Aedes mosquitoes but not to Anopheles mosquitoes. (A) Toxicity of C. bifermentans subsp. malaysia (Cbm) and the Cry operon to Anopheles mosquitoes. While C. bifermentans subsp. malaysia cultures show high toxicity to Anopheles gambiae mosquitoes, the recombinant B. thuringiensis containing the Cry operon is nontoxic to Anopheles mosquitoes. The toxicity curves for the recombinant B. thuringiensis containing the Cry operon with Anopheles stephensi and Culex quinquefasciatus were similar to those obtained with Anopheles gambiae. (B) Toxicity of C. bifermentans subsp. malaysia and the Cry operon to Aedes mosquitoes. Both C. bifermentans subsp. malaysia and the recombinant B. thuringiensis Cry operon were toxic to Aedes mosquitoes. However, C. bifermentans (Cb) and the recombinant B. thuringiensis pHT315 were nontoxic.
Larvicidal activity of individual Cry operon toxins.
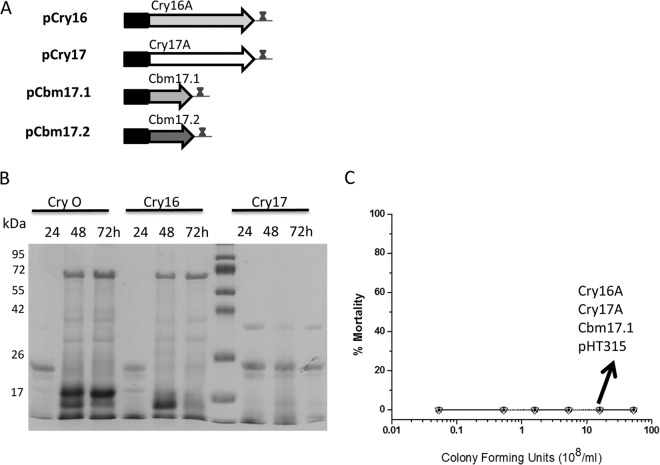
Since the entire Cry operon that expressed all four of the toxins showed high toxicity to Aedes larvae, we then analyzed the roles of the individual toxins. These toxins were cloned under the control of the Cyt1A promoter and the Cry operon terminator, giving pCry16, pCry17, pCbm17.1, and pCbm17.2 (Fig. 5A). Of these, only the pCry16 construct was observed to form a stable protein (Fig. 5B). The Cry17A protein was expressed but readily degraded in Bacillus cells even after 24 h, while the Cbm17.1 and Cbm17.2 proteins were poorly expressed on their own. Importantly, none of the bacterial strains containing these constructs showed any larval toxicity (Fig. 5C).
FIG 5.
All single genes show no toxicity to Aedes mosquitoes. (A) Each of the toxin genes in the Cry operon (CryO) was expressed under the control of the Cyt1A promoter, the Cyt1A Shine-Dalgarno sequence, and includes a 3′ terminator identical to the Cry operon construct in Fig. 3. (B) The Cry16A toxin was stably expressed, and the Cry17A toxin was expressed but readily degraded in culture even at 24 h. The Cry16A protein was not observed in SDS-PAGE at 24 h (Fig. 3) but was detected by immunoblotting at this time (Fig. 3). (C) None of the single-gene constructs expressed in B. thuringiensis showed any toxicity.
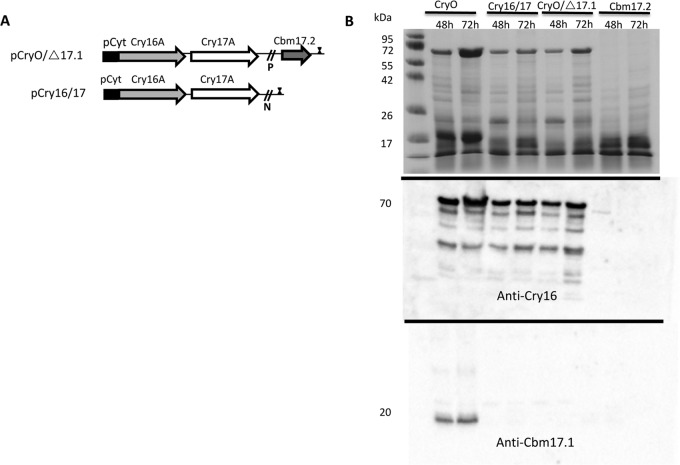
Based on these data, we concluded that some of these toxins could be acting together. We then made deletion constructs using PacI and NcoI restriction of the Cry operon. Restriction of pCryO followed by ligation of the restricted fragment led to the construction of the plasmids pCryO/Δ17.1 (deletion of PacI fragment) and pCry16/17 (deletion of NcoI fragments) (Fig. 6A). The pCryO/Δ17.1 and pCry16/17 constructs expressed protein(s) of 70 kDa (Fig. 6B), and the Cry16A protein was detected using an anti-Cry16A antibody. However, neither of these constructs was toxic to Aedes larvae.
FIG 6.
Any gene deletion in the Cry operon results in loss of toxicity. (A) Three gene deletions were made. In the first construct, pCry16, only Cry16A was expressed (Fig. 5), while in the second, CryO/Δ17.1, only cbm17.1 was deleted and in the third, pCry16/17, both cbm17 genes were deleted. The last two were derived from pCryO by restriction with PacI (P) and NcoI (N), respectively. Each of the constructs is under the control of the Cyt1A promoter and includes a 3′ terminator identical to the Cry operon construct in Fig. 3. (B) The Cry16A toxin was observed in the recombinant B. thuringiensis CryO, recombinant B. thuringiensis pCryO/Δ17.1, and recombinant B. thuringiensis pCry16/17 cultures. The recombinant B. thuringiensis CryO construct also shows expression of the Cbm17.1 protein at 48 to 72 h, as observed at 24 h (Fig. 3).
The Cry operon toxins form a complex.
Since none of the constructs showed any toxicity, other than pCryO, which includes all proteins in the operon, we hypothesized that a complex may be needed for toxicity. Indeed, analysis by native PAGE showed a complex of about 170 kDa (Fig. 7A). In contrast, recombinant B. thuringiensis containing pCry16, pCry17, or pCryO/Δ17.1 did not show the formation of a significant amount of the complex. The complex formed cross-reacted with the anti-Cbm17.1 antibody (Fig. 7B). Less distinct and larger complexes are also visible in both native PAGE and immunoblots of bacterial cultures expressing the entire Cry operon (Fig. 7A and B).
FIG 7.
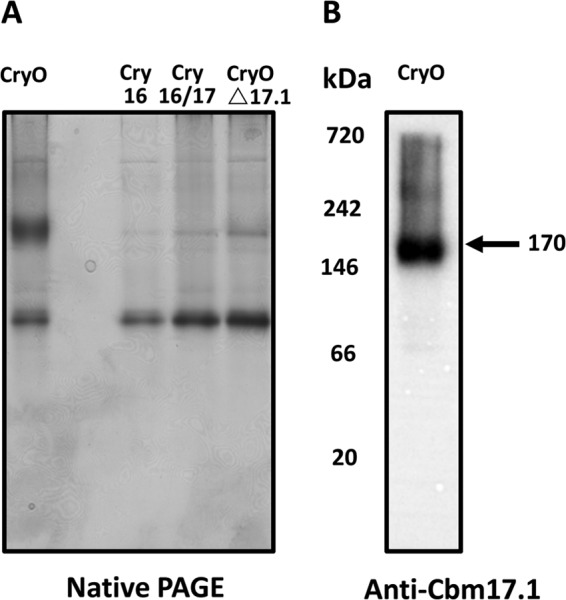
Native PAGE shows that the Cry operon forms a complex. (A) The entire Cry operon (CryO) forms a complex of about 170 kDa in 24-h recombinant B. thuringiensis cultures. This complex included the Cbm17.1 protein as shown in the immunoblot (B). None of the other three constructs (pCry16, pCry16/17, and pCryO/Δ17.1) showed the presence of the complex in recombinant B. thuringiensis cultures.
The Cry operon lacks hemolytic activity.
Hemolytic activity in recombinant B. thuringiensis strains containing pCryO, pCbm17.1, pCbm17.2, and pCryO/Δ17.1 was analyzed using sheep RBCs. None of these strains showed any hemolytic activity in either the pellet or B-Per bacterial extracts. In contrast, both 0.1% Triton and Cyt1A toxin (15 ng/ml) effectively lysed the RBCs.
DISCUSSION
The novel bacterial strain C. bifermentans subsp. malaysia is highly active against a number of mosquito genera but particularly against anopheline species (11, 13). However, the genes responsible for its toxicity have remained elusive. Prior efforts characterized the bacterium and identified proteins that had mosquitocidal activity (16, 17, 20), and the genes encoding these proteins were subsequently identified (18, 19). All of the identified proteins (16, 17, 20) are encoded by a single operon, the Cry operon (19).
This operon consists of four genes, cry16Aa, cry17Aa, cbm17.1, and cbm17.2. The derived sequences of Cry16A and Cry17A suggest that these proteins are part of the Cry-like toxin family and are similar in size, about 70 kDa (19). Cry17A has about 30% amino acid identity to the Cry27A and Cry29A proteins, while Cry16A has about 30% identity to the Cry19A protein (http://www.btnomenclature.info). In contrast, the Cbm17.1 and Cbm17.2 proteins belong to the aegerolysin family of proteins (27). These two proteins are 78% similar in amino acid sequence and are 17 kDa in size (18, 19) but run as 20-kDa proteins in SDS-PAGE.
Expression of the Cry operon showed low levels of activity to Anopheles, Aedes, and Culex larvae in a previous study (18). Importantly, in that study expression of only the Cry16A protein and part of the Cry17A protein also showed toxicity to all three mosquito species. However, subsequent efforts from the same group suggested that all these proteins are neither mosquitocidal nor hemolytic (19, 21).
In our efforts to identify novel mosquitocidal genes, we reinvestigated toxins in this bacterial strain. Proteomics of SDS-PAGE bands of this strain again identified the Cry16A and Cbm17 proteins as potential toxins, since these were observed in 1-day cultures, which were toxic, but not in 5-day cultures, which were inactive (Fig. 2B). The Cry operon encodes four proteins, but the individual proteins did not appear to have toxicity (21). The presence of a single promoter upstream of four genes, each with intact translation sites, suggested (19) that these genes were expressed simultaneously. Based on this premise, we created and expressed a clone of the entire operon under the control of a strong Bacillus promoter, cyt1A (24, 26).
Expression of the 70-kDa Cry proteins is clearly visible at 48 and 72 h. However, Western blots show that the protein can be observed at least by 24 h, and likely earlier. This may be because as previously observed, very small amounts of Cry16A are secreted before sporulation (19). The Cbm17.1 and Cbm17.2 proteins can be observed in as early as 12-h cultures. Since both the Cry16A and Cbm17 proteins are expressed at 24 h, we routinely used such cultures for bioassays.
We show here that when the C. bifermentans subsp. malaysia Cry operon was expressed in its entirety, high levels of larvicidal activity toward Aedes aegypti are observed and that the toxicity of Bacillus strain 4Q7, expressing the Cry operon, recombinant B. thuringiensis CryO, is higher than that observed with wild-type C. bifermentans subsp. malaysia (Fig. 4B). However, to our surprise this Bacillus strain 4Q7 expressing the Cry operon showed no toxicity to A. gambiae and C. quinquefasciatus larvae. Our observations differ from those reported in reference 18, since we observed much higher levels of toxicity of the Cry operon to Aedes mosquitoes. Potentially, the use of a strong Bacillus thuringiensis promoter, like cyt1A, results in the production of the high toxin levels observed here. Further, we did not observe any toxicity to Anopheles or Culex mosquitoes, nor was any of the truncated Cry operon constructs active as previously observed (18).
Since the Cry operon was larvicidal, the four genes were individually expressed under the control of the Cyt1A promoter. None of the proteins encoded by these genes were toxic, although we were able to confirm the expression of the full-length protein for only one toxin, Cry16A. Consequently, constructs that had deletion of the cbm17.1 gene or both the cbm17 genes were made, but both were also inactive. Thus, our systematic deletion of genes from the original active clone, pCryO, supports the original hypothesis that all genes within the Cry operon are required for larvicidal activity. Further, we observed the formation of a 170-kDa complex, in addition to other minor larger complexes, when the entire Cry operon is expressed. Our results are therefore consistent with previous observations (17) that a toxin complex is required for toxicity.
C. bifermentans subsp. malaysia is the first known anaerobic bacterium possessing larvicidal activity. The Cry larvicidal genes are similar to those found in B. thuringiensis. Despite the similarities to other Cry toxins, Cry16A did not cross-react with antibodies raised against any of the B. thuringiensis subsp. israelensis or other B. thuringiensis toxins (17). Further, the hemolysin-like proteins Cbm17.1 and Cbm17.2 are similar to proteins found in various fungi, including Aspergillus, but are not identical. However, the Cry operon does not show any hemolytic activity. Potentially, this is because the toxins are indeed nonhemolytic, or alternatively, the Cbm17 toxins were not obtained in an active conformation when isolated. Nevertheless, it is clear that the Cry operon in C. bifermentans subsp. malaysia contains a unique combination of toxins that act cooperatively. It is a likely possibility that C. bifermentans subsp. malaysia acquired this operon with horizontal gene transfer.
The Cry operon shows high selectivity toward Aedes mosquitoes. In contrast, wild-type C. bifermentans subsp. malaysia shows much higher toxicity to Anopheles gambiae than to Aedes aegypti. Consequently, the C. bifermentans subsp. malaysia proteins that are responsible for Anopheles toxicity are likely to be different and are unknown, and investigations are under way to identify these. In addition, in future studies, we plan to investigate the mechanisms of larvicidal activity of the Cry toxins and the roles that these proteins play in the toxicity toward Aedes larvae.
ACKNOWLEDGMENTS
The technical assistance of Maria Ramirez is gratefully acknowledged. We thank Anthony James (UC Irvine), Marcelo Jacobs-Lorena (Johns Hopkins University), William Walton (UC Riverside), and Brad White (UC Riverside) for providing mosquito larvae.
This research was funded in part through grants from the National Institutes of Health (1R21AI070873 and 1R01AI066014) and the University of California Agricultural Experiment Station.
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.Najera JA. 2001. Malaria control: achievements, problems and strategies. Parassitologia 43:1–89 [PubMed] [Google Scholar]
- 2.Lederberg J. 1992. The interface of science and medicine. Mt. Sinai J. Med. 59:380–383 [PubMed] [Google Scholar]
- 3.Staples JE, Gershman M, Fischer M. 2010. Yellow fever vaccine: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm. Rep. 59(RR-7):1–27 [PubMed] [Google Scholar]
- 4.Murray NE, Quam MB, Wilder-Smith A. 2013. Epidemiology of dengue: past, present and future prospects. Clin. Epidemiol. 5:299–309. 10.2147/CLEP.S34440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts DR, Manguin S, Mouchet J. 2000. DDT house spraying and re-emerging malaria. Lancet 356:330–332. 10.1016/S0140-6736(00)02516-2 [DOI] [PubMed] [Google Scholar]
- 6.Murray CJ, Rosenfeld LC, Lim SS, Andrews KG, Foreman KJ, Haring D, Fullman N, Naghavi M, Lozano R, Lopez AD. 2012. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet 379:413–431. 10.1016/S0140-6736(12)60034-8 [DOI] [PubMed] [Google Scholar]
- 7.Georghiou GP, Wirth MC. 1997. Influence of exposure to single versus multiple toxins of Bacillus thuringiensis subsp. israelensis on development of resistance in the mosquito Culex quinquefasciatus (Diptera: Culicidae). Appl. Environ. Microbiol. 63:1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao DR, Mani TR, Rajendran R, Joseph AS, Gajanana A, Reuben R. 1995. Development of a high level of resistance to Bacillus sphaericus in a field population of Culex quinquefasciatus from Kochi, India. J. Am. Mosq. Control Assoc. 11:1–5 [PubMed] [Google Scholar]
- 9.Charles J-F, Nielsen-LeRoux C. 2000. Mosquitocidal bacterial toxins: diversity, mode of action and resistance phenomena. Mem. Inst. Oswaldo Cruz 95(Suppl 1):201–206. 10.1590/S0074-02762000000700034 [DOI] [PubMed] [Google Scholar]
- 10.Nielsen-Leroux C, Charles JF, Thiery I, Georghiou GP. 1995. Resistance in a laboratory population of Culex quinquefasciatus (Diptera: Culicidae) to Bacillus sphaericus binary toxin is due to a change in the receptor on midgut brush-border membranes. Eur. J. Biochem. 228:206–210. 10.1111/j.1432-1033.1995.tb20251.x [DOI] [PubMed] [Google Scholar]
- 11.de Barjac H, Sebald M, Charles JF, Cheong WH, Lee HL. 1990. Clostridium bifermentans serovar malaysia, a new anaerobic bacterium pathogen to mosquito and blackfly larvae. C. R. Acad. Sci. III 310:383–387 (In French.) [PubMed] [Google Scholar]
- 12.Lee HL, Seleena P. 1990. Isolation of indigenous larvicidal microbial control agents of mosquitos: the Malaysian experience. Southeast Asian J. Trop. Med. Public Health 21:281–287 [PubMed] [Google Scholar]
- 13.Lee HL, Seleena P. 1990. Isolation and evaluation of larvicidal Clostridium bifermentans against mosquitoes of public health importance. Trop. Biomed. 7:103–106 [Google Scholar]
- 14.Thiery I, Hamon S, Gaven B, De Barjac H. 1992. Host range of Clostridium bifermentans serovar malaysia, a mosquitocidal anaerobic bacterium. J. Am. Mosq. Control Assoc. 8:272–277 [PubMed] [Google Scholar]
- 15.Yiallouros M, Storch V, Thiery I, Becker N. 1994. Efficacy of Clostridium bifermentans serovar malaysia on target and nontarget organisms. J. Am. Mosq. Control Assoc. 10:51–55 [PubMed] [Google Scholar]
- 16.Charles JF, Nicolas L, Sebald M, de Barjac H. 1990. Clostridium bifermentans serovar malaysia: sporulation, biogenesis of inclusion bodies and larvicidal effect on mosquito. Res. Microbiol. 141:721–733. 10.1016/0923-2508(90)90066-Y [DOI] [PubMed] [Google Scholar]
- 17.Nicolas L, Charles JF, de Barjac H. 1993. Clostridium bifermentans serovar malaysia: characterization of putative mosquito larvicidal proteins. FEMS Microbiol. Lett. 113:23–28. 10.1111/j.1574-6968.1993.tb06482.x [DOI] [PubMed] [Google Scholar]
- 18.Barloy F, Delecluse A, Nicolas L, Lecadet MM. 1996. Cloning and expression of the first anaerobic toxin gene from Clostridium bifermentans subsp. malaysia, encoding a new mosquitocidal protein with homologies to Bacillus thuringiensis delta-endotoxins. J. Bacteriol. 178:3099–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barloy F, Lecadet MM, Delecluse A. 1998. Cloning and sequencing of three new putative toxin genes from Clostridium bifermentans CH18. Gene 211:293–299. 10.1016/S0378-1119(98)00122-X [DOI] [PubMed] [Google Scholar]
- 20.Nicolas L, Hamon S, Frachon E, Sebald M, Barjac H. 1990. Partial inactivation of the mosquitocidal activity of Clostridium bifermentans serovar malaysia by extracellular proteinases. Appl. Microbiol. Biotechnol. 34:36–41 [Google Scholar]
- 21.Juárez-Pérez V, Delécluse A. 2001. The Cry toxins and the putative hemolysins of Clostridium bifermentans ser. malaysia are not involved in mosquitocidal activity. J. Invertebr. Pathol. 78:57–58. 10.1006/jipa.2001.5042 [DOI] [PubMed] [Google Scholar]
- 22.Macaluso A, Mettus AM. 1991. Efficient transformation of Bacillus thuringiensis requires nonmethylated plasmid DNA. J. Bacteriol. 173:1353–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arantes O, Lereclus D. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115–119. 10.1016/0378-1119(91)90495-W [DOI] [PubMed] [Google Scholar]
- 24.Wu D, Federici BA. 1993. A 20-kilodalton protein preserves cell viability and promotes CytA crystal formation during sporulation in Bacillus thuringiensis. J. Bacteriol. 175:5276–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 26.Chang C, Yu YM, Dai SM, Law SK, Gill SS. 1993. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl. Environ. Microbiol. 59:815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berne S, Lah L, Sepcic K. 2009. Aegerolysins: structure, function, and putative biological role. Protein Sci. 18:694–706. 10.1002/pro.85 [DOI] [PMC free article] [PubMed] [Google Scholar]