Abstract
Microcystin is a common and well-known cyanobacterial toxin whose intracellular role is still under investigation. Increasing knowledge on microcystin gene expression and regulation can contribute to the understanding of its putative cellular function. In this work, reverse transcription-quantitative PCR (RT-qPCR) was used to investigate the transcriptional response of the mcyD gene to nitrogen (nitrate and ammonium) and phosphorus limitation in two toxic Microcystis strains. The existence of a direct correlation between transcripts of mcyD and ntcA genes was also identified. In previous studies, NtcA (global nitrogen regulator) has been described as a potential component in the control of microcystin biosynthesis. This research showed that stress agents linked to nutrient deprivation could lead to a significant increase of microcystin production in both strains studied. The more toxic strain proved to be more resistant to nutrient limitation. The similar outcomes of mcyD regulation observed for all nutrients suggest that this response can be linked to oxidative stress of cells undergoing adverse growth conditions.
INTRODUCTION
Bloom-forming cyanobacteria occur worldwide and produce toxins that may be harmful to humans and animals (1). Earlier studies suggested that microcystin net production depends primarily on cellular growth rate, while environmental conditions would affect microcystin production rather indirectly, via the cellular growth rate itself (2, 3). Other research papers, however, tried to verify the influence of environmental factors on microcystin production (4, 5, 6). Controversial results were generated using direct measurements of intracellular toxins, mostly because of differences in culturing techniques, growth conditions, as well as experimental design and analyses (7). Recently, some authors (8) showed a significant effect of environmental factors on microcystin production, and they observed that the effect was independent of influences on growth rate. For phosphorus and irradiance, intrinsic growth rate and microcystin production coefficient were inversely correlated.
Investigations about the role and function of microcystin are still widely discussed. Several potential roles were suggested, as, for example, iron chelator (siderophores) (2, 9), defense mechanism (10), photosynthesis or other light-related processes (11), and intercellular intraspecies communication (12). With the discovery that microcystin was not produced by ribosomes and the existence of a specific gene set, called mcy (13), there was a significant increase in studies on factors that may regulate microcystin production, contributing to the understanding of ecological questions about microcystin function (14, 15, 16, 17).
Recent studies indicated an intracellular function related to protein ligand (18) and found that several proteins of toxic and nontoxic strains were differentially expressed as a result of strain toxicity (19). It was also shown that the addition of hydrogen peroxide had less detrimental effects on toxic Microcystis strains than on nontoxic ones (20), suggesting protection by microcystin. Another report (21) demonstrated the increase of microcystin synthesis, but not of the gene transcription for toxins, in cells exposed to severe limitation of iron. Also, experiments run under iron deficiency (22) found increasing transcription levels of the mcy gene and of microcystin synthesis. When testing nitrogen limitation, Sevilla et al. (23) did not see any effect of nitrate reduction in the production of microcystin. However, Ginn and Neilan (24) showed that the microcystin synthetase gene transcription was responsive to nitrogen, increasing under nitrogen limitation. These authors also discovered that the mcyABC and mcyDEFGHIJ (hearafter called mcyA/D) promoter region has binding sites for the universal regulator of nitrogen (NtcA), indicating its importance for the regulation of microcystin production.
As primary producers, cyanobacteria need nitrogen and phosphorus as essential macronutrients, and changes in their concentration observed in the environment have several physiological effects. Phosphorus is important for the cellular synthesis of nucleic acids and membrane phospholipids, as well as for energy transfer through tri- and biphosphorylated nucleotides (25). In aquatic environments, dissolved inorganic phosphorus is biologically available as orthophosphate (26) and can easily become limiting. Nitrogen assimilation is ruled by the available nitrogen source (27). Inorganic forms like ammonium and nitrate are the most common in water and can both be assimilated by cyanobacteria following different pathways (28). As all planktonic organisms, cyanobacteria live in permanently changing environments, undergoing seasonal challenges, and may have to overcome periods of nutrient limitation. Since it triggers physiological changes, nutrient limitation can also affect the mechanisms of toxin production. A better knowledge of these mechanisms may help to evaluate the importance of cyanotoxins for cyanobacteria. In addition, it can provide tools that will improve the environmental control of these organisms.
Many studies used Microcystis aeruginosa PCC 7806 as a model organism. However, different authors already advised that the strains present in nature represent numerous ecotypes able to adapt and to survive in a specific environment (29, 30). Among others, one work (21) drew attention to the dangers of using just this strain as a model for the entire Microcystis group, since strains may present specific acclimation processes under stress conditions. Therefore, to study the mechanisms of microcystin production and their effects on metabolism, it would be meaningful to investigate new strains and to follow their response to particular surroundings.
The aim of this study was to measure changes in mcyD gene transcription in response to nitrogen and phosphorus limitation in two Microcystis strains, isolated from Brazilian water bodies. Both strains are toxic, but they produce different amounts of microcystin. We also wanted to prove the existence of a correlation between mcyD and ntcA transcripts, previously found for Microcystis PCC 7806. The 16S rRNA gene was used as the reference (endogenous control) and cpcB as the control (known response). The results of this research can contribute to the existing literature that will help with the understanding of the ecological importance of microcystins for cyanobacterial cell metabolism.
MATERIALS AND METHODS
Growth conditions and experimental design.
Two Microcystis aeruginosa toxic strains, strains Ma19 and Ma26, were provided by the Phycology Laboratory of the Universidade Federal de Minas Gerais (Belo Horizonte, Brazil). Both strains had been isolated from Brazilian reservoirs. For these experiments, cells were grown in a WC medium (31) under batch conditions, at a temperature of 24°C, 40 μmol of photons m−2 s−1, and a 12-h light and 12-h dark photoperiod. When growing on complete WC medium, strain 26 is approximately 10 times more toxic than strain 19 (previous observations; see Results). For the nitrogen-limited experiments, strains were grown under high nitrogen supply (1 mmol liter−1 of N-NO3− or 0.2 mmol liter−1 of N-NH4+, original N concentration in WC medium) and two nitrogen-limited conditions: 0.1 and 0.01 mmol liter−1 of N-NO3− or 0.02 and 0.002 mmol liter−1 of N-NH4+ (respectively, 1/10 and 1/100 dilution). A similar procedure was performed for the phosphorous experiments. The original concentration was 0.050 mmol liter−1 of P-PO4−, and limitation was 0.005 mmol liter−1 and 0.0005 mmol liter−1 (1/10 and 1/100 of the original concentration).
Strains were inoculated in 200 ml of medium at the three different nutrient levels and grown under these conditions for about 1 week, for acclimatization. After this period, subsamples containing approximately 20,000 cells ml−1 were reinoculated in their respective medium, and the experiment started. Experiments were finalized after 6 days, during the exponential growth phase. Aliquots of 20 ml of the culture samples were harvested on glass fiber filters (GF/F), and filters were kept frozen (−80°C) until analysis. During the experiments, small aliquots (2 ml) of cultures were removed every 2 days to estimate the growth rate. Cell counting was carried out in a Fuchs-Rosenthal hemocytometer. All the experiments were done in triplicate (three independent subcultures for each treatment, growing under the same experimental conditions) and repeated at least two times. The entire experimental procedure was performed under completely axenic conditions.
Chlorophyll a and microcystin measurements.
Chlorophyll a was extracted using hot ethanol, measured according to Nusch (32), and quantified on a spectrophotometer at a 665-nm wavelength.
Microcystin was extracted with 75% (vol/vol) methanol. The analyses were performed with an Abraxis-ADDA enzyme-linked immunosorbent assay (ELISA) kit, by following the manufacturer's instructions. Plates were analyzed on a microtiter plate ELISA reader (Bio-Tek, Elx 800) at 450 nm within 15 min after the addition of the stop solution.
Isolation of mRNA and RT-qPCR.
RNA was extracted after cell lysis with TES (25% [wt/vol] sucrose, 100 mM EDTA, and 50 mM Tris-HCl, pH 8.0). Samples were left for 2 h at 4°C and then, after addition of lysozyme (5 mg/ml), for 1 h at 37°C and, after addition of proteinase K (100 μg/ml), for 1 h at 60°C; finally, TRIzol (Invitrogen) was added according to the manufacturer's recommendations. Total RNA was resuspended in 50 μl of DEPC-H2O, and RNA was treated with 1 U/μg of DNase (Promega) at 37°C for 30 min. The reaction was stopped by the addition of a stop solution and by heating for 10 min at 65°C. Removal of DNA traces was confirmed by PCR. RNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). RNA quality was verified on 1% DNase-free agarose gel.
About 500 ng of RNA was used for RT-PCR. The cDNA was generated using a High Capacity kit (Applied Biosystems) with RT random primers. Concentrations and PCR cycling conditions were established according to the manufacturer's recommendations.
Real-time PCR was performed using a StepOne system (Applied Biosystems) with 1 μl of cDNA sample, 0.3 μl of each primer (10 pmol μl−1), 5 μl of Power SYBR green I (Applied Biosystems), and sterile Milli-Q water for a final volume of 10 μl. The reactions were done in duplicate or triplicate, and specifications for the PCR cycle followed the manufacturer's guidelines. Primers used in this study are described in Table 1. The new primers were designed with the help of Primer-BLAST tools (NCBI) to amplify 141-bp products for the global nitrogen regulator gene (ntcA), 73-bp products for the phycocyanin gene (cpcB), and 103-bp products for the microcystin synthetase D gene (mcyD). The primer described by Sevilla et al. (22) for the 16S rRNA was used as a housekeeping gene. The primer amplification efficiencies for ntcA, cpcB, mcyD, and 16S rRNA genes were calculated, and they yielded E values of 1.93, 2.11, 1.95, and 1.97, respectively, where an E value of 2 indicates 100% PCR efficiency. Relative quantification of the ntcA, cpcB, and mcyD target genes was compared with the 16S rRNA reference gene and was represented as the change in transcription compared to the result observed under the control conditions (high nutrient). According to Pfaffl (33), threshold cycle (ΔCT = CT target gene – CT housekeeping gene) values were used for correlation analyses between mcyD and ntcA genes. The 16S rRNA reference gene was selected because it is known to be a good housekeeping gene in M. aeruginosa PCC7806 (14), and it was already used in several other studies.
TABLE 1.
Sequences of the primers used in this study for mcyD, cpcB, ntcA, and 16S rRNA
| Primer | Typea | Sequence (5′ to 3′) | Reference |
|---|---|---|---|
| q-mcyD | F | GCATCTTCTAAAGAAAAGACTCC | 47 |
| q-mcyD | R | TATTCCCCAAGATTGCCATAATTT | 47 |
| QcpcB | F | GGTATCACCCCCGGCGATTG | This study |
| QcpcB | R | GCAGCAGCAGCGCGATCGAAGTA | This study |
| QntcA | F | TGCAGGGTTTGTCCTCGCGG | This study |
| QntcA | R | CCCGGATGCCATCGGTGGTG | This study |
| 16S-rRNA | F | TGCGTAGAGATTGGGAAGAACATC | 23 |
| 16S-rRNA | R | GCTTTCGTCCCTGAGTGTCA | 23 |
F, forward; R, reverse.
The analysis of the fluorescent melting curve was performed to determine the amplification melting temperature of the single PCR products in the samples, by gradually increasing the temperature from 70°C to 95°C at a rate of 0.1°C s–1. Fluorescence intensity data were collected continuously and were converted to melting peaks using the LightCycler software (StepOne Software, version 2.0).
Statistical analyses.
Statistical analyses were performed by using the general linear model (GLM) using R software (34). Analysis of variance (ANOVA) was used to test for difference among treatments under different nutrient concentrations. Significant difference was accepted when P values were ≤0.05. ANOVA was run on a Gaussian (normal) distribution for most data sets, but in a few cases, according to the data dispersion, the gamma (chi-square) distribution was preferred and shown to be more robust. Contrast analysis was used to verify differences among treatments. Correlation analyses were run between ΔCT values of mcyD and ntcA genes for each nutrient treatment.
RESULTS
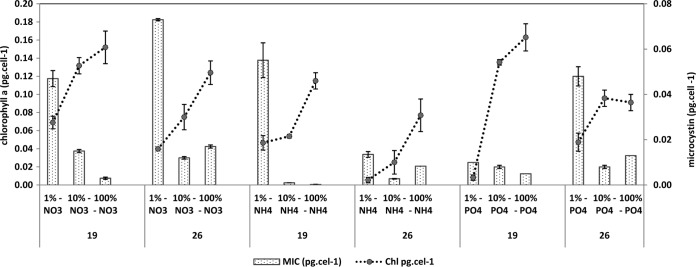
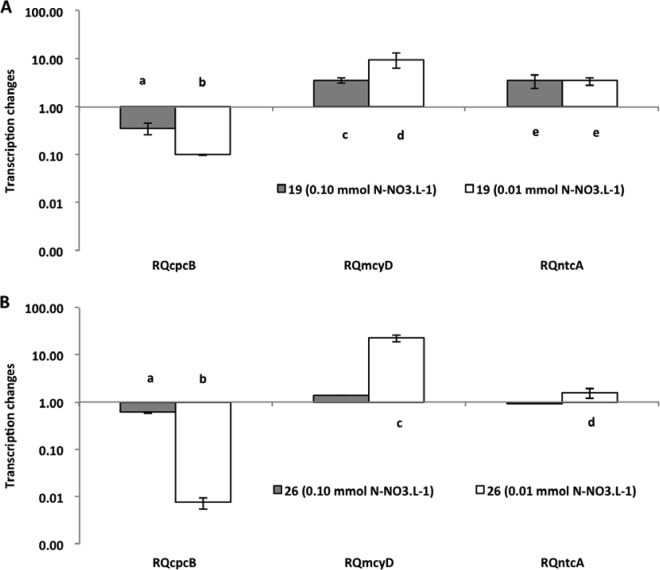
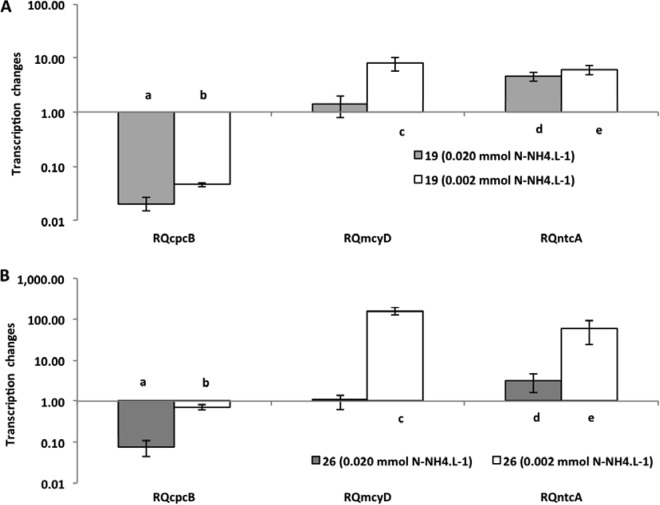
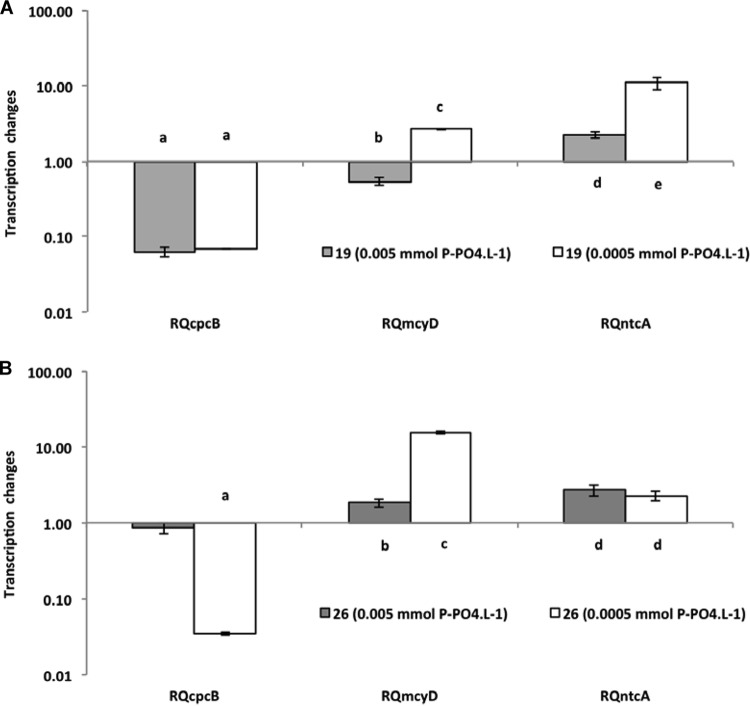
The treatment with the highest nutrient concentration was always used as a reference for the quantification of the gene expression of the treatments with less nutrients. Despite the differences in toxicity of the two strains investigated (strain 26 is approximately 10 times more toxic than strain 19 [previous observations and also Fig. 1]), they performed very similarly, and only small differences were observed between them relative to gene transcription. The mcyD gene expression showed changes under all extreme limiting conditions. In the nitrate limitation experiment (Fig. 2), strain 19 had a significant increase in expression level (P < 0.001) under both limited conditions, but strain 26 had a significant increase in expression level only under the most extreme condition (P < 0.001), when nutrient was reduced 100 times from the original concentration. Interestingly, strain 26 was the strain with 10 times higher content of microcystin and did not show significant changes under the less severe limitation condition. In the ammonium limitation experiment (Fig. 3), an increase of mcyD gene expression was observed only at the lowest nutrient level, for both strains (strain 19, P = 0.020; strain 26, P < 0.001). For phosphate limitation (Fig. 4), both strains had similar responses, with significantly higher expression in the extreme limited concentration (P < 0.010).
FIG 1.
Chlorophyll a and microcystin content per cell, for each strain (19 and 26) at three nutrient concentrations, in the nitrate, ammonium, and phosphate experiments. Bars represent standard errors of the means.
FIG 2.
Relative quantification of cpcB, mcyD, and ntcA gene expression in the nitrate assay for two cyanobacterial strains (19 and 26). The 1 mmol liter−1 concentration was used as a standard (control) to estimate changes in gene expression under limited conditions. Bars indicate standard errors. Different letters above the bars indicate significant differences (P ≤ 0.05), when values were compared to the control and among themselves.
FIG 3.
Relative quantification of cpcB, mcyD, and ntcA gene expression in the ammonium assay for two cyanobacterial strains (19 and 26). The 0.2 mmol liter−1 concentration was used as a standard (control). For figure explanation, see the legend to Fig. 2.
FIG 4.
Relative quantification of cpcB, mcyD, and ntcA expression genes in the phosphate assay for two cyanobacterial strains (19 and 26). The 0.05 mmol liter−1 concentration was used as a standard (control). For figure explanation, see the legend to Fig. 2.
The response of the ntcA gene showed a significant increase in its expression (P < 0.001) in all experiments under limitation (Fig. 2, 3, and 4), but individual differences were observed. For nitrate, strain 26 responded only in extreme limitation. Again, it was the more toxic strain 26 that was not affected under less severe limitation.
The analysis of the expression of the cpcB gene revealed a significant decrease in the level of transcripts in all experiments (P < 0.001) (Fig. 2, 3, and 4). However, once again, the decrease observed in strain 26 in the phosphorus experiment was significant only at the lowest concentration of P-PO4.
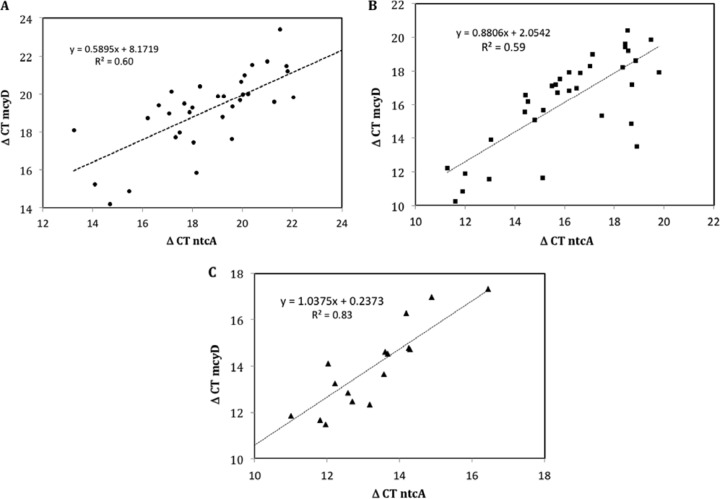
Figure 5 represents correlation analyses between ΔCT mcyD and ΔCT ntcA. All measured values for each nutrient were pooled (all replicates and experimental repetitions), in order to increase the significance of the correlation. The existence of correlation between these two variables was confirmed for all experiments that had been performed under nitrate (Fig. 5A), ammonium (Fig. 5B), and phosphorus limitation (Fig. 5C). All three correlations displayed a significant R2 value.
FIG 5.
Correlation analyses between ΔCT values of the mcyD and ntcA gene expression for all experimental data obtained from the nitrate (A; dots), ammonium (B; squares), and phosphorous (C; triangles) experiments.
As expected, chlorophyll a concentration in each culture decreased as the amount of nitrogen or phosphorus supplied to the medium was reduced (P < 0.001) (Fig. 1). However, we observed a clear and opposite trend for microcystin content per cell (Fig. 1) for all treatments. For example, at a lower nitrate concentration, a significantly higher concentration of microcystin was measured (P < 0.001), showing a direct relationship between nutrient limitation and increase in microcystin.
DISCUSSION
Typically, nitrogen starvation induces physiological responses in the cell, like chlorosis, increased uptake of alternative nitrogen sources, and, once these reach complete depletion, onset of a dormant state (35). In our experiments, we detected the decrease of cpcB gene expression (phycocyanin-related gene), of chlorophyll a concentration, and of cell growth, and we observed a loss of the culture pigmentation. Chlorosis can induce cells to acquire nitrogen via degradation of the phycobiliproteins, which normally represent a large part of the protein pool in the cells (36) and may comprise up to 60% of total cellular proteins in cyanobacteria (37). In addition, during nutrient starvation, reactive oxygen species (ROS) can be formed, due to the existence of several cross-regulatory reactions between photosynthesis, redox control, and nutrient acquisition, establishing a tight control over the C/N balance in the cell (37). Furthermore, according to Dagnino et al. (38), the loss of pigmentation by discoloration, as we observed for limiting nutrient conditions, is an important sign of the occurrence of oxidative stress. Chlorophyll a bleaching was also used by Zilliges et al. (18) as an evidence of oxidative stress, in their case caused by high light conditions and later proved by the addition of hydrogen peroxide. Thus, although no direct measurement of oxidative stress was performed in this study, as previously stated by numerous authors, phenotypic changes, like the observed loss of pigmentation under nutrient limitation, provide strong evidence that the cultures were experiencing oxidative stress.
Stress conditions caused by nutrient deprivation clearly affected mcyD transcription and microcystin production in our experiments. Similar trends were found in a recent study, where higher values of microcystin and nodularin cellular content were measured in cells growing under limiting conditions of phosphate and low light (39). Ginn and Neilan (24) also observed an increase in the transcription of the mcy gene under conditions of nitrate starvation. However, Sevilla et al. (23) showed that while a decrease of nitrate caused lower cell growth, it did not directly affect mcy transcription or toxin production. Nevertheless, the lowest nitrate concentration used by these authors (0.2 mmol liter−1) was not as low as in our experiments or in experiments by Ginn and Neilan, and this is probably the reason for the difference in their results. For example, in this study, we used a nitrate concentration about 20 times lower (0.01 mmol liter−1) than that used by Sevilla et al. (23), and Ginn and Neilan (24), who found results similar to ours, used an entirely N-depleted medium (no nitrate addition). Conflicting results occur mainly because investigations on regulatory mechanisms of toxin production have not yet been standardized and experimental designs and growth controls are frequently different; therefore, outputs tend to be controversial (7).
In recent years, there was a progress in the understanding of the biological role of microcystin, and several studies pointed to potential intracellular functions of this compound. For example, studies done with mcy mutants showed differences in pigmentation (40), and toxic and nontoxic strains altered their response to low inorganic carbon concentrations because of the limited ability of the mutant to adapt to low-C conditions (41). The differences we observed between our two strains give further support to the idea of a potential microcystin role under nutrient stress. In fact, the strain with higher toxicity (Ma26) responded with significant mcyD increase only at the lowest nutrient concentration, while the less toxic strain (Ma19) showed an increase of transcripts at both limiting conditions. This may indicate that strain 26 is more resilient to environmental nutrient variations. Investigating the potential relationship of microcystin to increasing fitness, Zilliges et al. (18) also found that a microcystin-deficient mutant was more sensitive to oxidative stress conditions than the wild type. In another study, run under iron limitation and starvation, microcystin-producing cells showed a less pronounced change in iron stress-induced protein A transcript (isiA) than nonproducing ones, a finding that was also correlated with the reduced bleaching of the toxic cells under iron-starving conditions (19). Comparing toxic and nontoxic Planktothrix strains, Briand et al. (42) observed that microcystin-producing strains had clear advantages against the nontoxic strains under limiting environmental conditions. The lower toxicity of strain 19 seems to increase sensitivity to changes in nutrient supply that was detected by higher cell beaching (data not shown), as well as earlier responses of mcyD transcription and microcystin production. Therefore, different toxin content among strains may be relevant to indicate paths of fitness in toxin-producing organisms and may help explain changes in the toxicity of a natural population under shifting environmental conditions, when strains would be selected according to their ability to overcome temporary limitation.
We found that nutrient stress affected mcyD transcription, and it was directly correlated with nctA gene expression, presenting significant results in all experiments. This correlation can be a strong indication of an intracellular function of microcystin. NtcA activates nitrogen-responsive genes and therefore has been used as a good marker in studies on nitrogen deprivation (43). The autoregulated ntcA gene is transcribed at a basal level in the presence of ammonium and increases under conditions of nitrogen stress (24). Ginn et al. (24) have identified regions with similarity to the consensus motifs bound by NtcA in the internal mcyA/D promoter region in M. aeruginosa PCC 7806 and suggested that the microcystin synthetase gene would be responsive to nitrogen. Similar investigation confirmed the affinity of NtcA to promoter regions of mcyA/D as well as to the regions mcyE and mcyH (44). These authors observed that the presence of oxoglutarate (2-OG) increased this affinity 2.5-fold. In cyanobacteria, 2-OG is key indicator of the C/N cell balance (27). These results suggest that NtcA activity and microcystin synthesis respond to the equilibrium between C and N metabolism (44). Some authors (45) described the pkn22 operon, which consists of genes related to iron deficiency and oxidative stress, as also being under the control of NtcA. They suggested that nitrate removal would lead to an electron transfer from ferredoxin to oxygen, generating ROS. Since ntcA expression is affected by the cell redox state and NtcA can bind to the mcy promoter region, it seems that microcystin may have an important role in the carbon and nitrogen metabolism and in the redox control and perception of redox changes (7).
Changes in the cell redox state are processes that can be intimately linked to the generation of reactive oxygen species and oxidative stress in cyanobacteria (21). Therefore, we suggest that the increase of microcystin observed in our experiments is linked to oxidative stress caused by severe nutrient limitation. By adding H2O2 to a culture in logarithmic growth phase, Schatz et al. (12) induced mcyB accumulation. Other reports showed that H2O2 stimulated the transcription of the mcyA gene and increased the activity of microcystin peptide synthetase (46). Microcystin was shown to bind to proteins under oxidative stress conditions as a possible mechanism to increase fitness of toxic Microcystis strains (19). By using high-light conditions and inducing oxidative stress, the researchers observed a specific and covalent interaction of microcystin with several proteins, resulting in their accumulation, suggesting another possible function of this metabolite.
In summary, the results reported here of increasing mcy transcription and microcystin production under nitrogen and phosphorus stress conditions are in agreement with some recent reports that point to an intracellular function of microcystin related to oxidative stress. Therefore, in our experiments, higher levels of transcript and metabolite were possibly produced for potential protection of cells under stress. Further studies may help to confirm this putative role of microcystin in the cellular metabolism and to partially elucidate the toxin dynamic observed in the field.
ACKNOWLEDGMENTS
We are grateful to Alvaro C. Nunes for assistance in the RNA procedures.
This work was supported by a scholarship to J.S.M.P. from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and by funds provided by Fundação de Amparo a Pesquisa de Minas Gerias (FAPEMIG) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) to A.G.
Footnotes
Published ahead of print 18 July 2014
REFERENCES
- 1.Chorus I. 2001. Cyanotoxin occurrence in freshwaters—a summary of survey results from different countries, p 75–82 In Chorus I. (ed), Cyanotoxins: occurrence, causes, consequences. Springer, Berlin, Germany [Google Scholar]
- 2.Orr PT, Jones GJ. 1998. Relationship between microcystin production and cell division rates in nitrogen-limited Microcystis aeruginosa cultures. Limnol. Oceanogr. 43:1604–1614. 10.4319/lo.1998.43.7.1604 [DOI] [Google Scholar]
- 3.Long BM, Jones GJ, Orr PT. 2001. Cellular microcystin content in N-limited Microcystis aeruginosa can be predicted from growth rate. Appl. Environ. Microbiol. 67:278–283. 10.1128/AEM.67.1.278-283.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivonen K, Niemelä SI, Niemi RM, Lepistö L, Luoma TH, Räsänen LA. 1990. Toxic cyanobacteria (blue-green algae) in Finnish fresh and coastal waters. Hydrobiology 190:267–275. 10.1007/BF00008195 [DOI] [Google Scholar]
- 5.Lukac M, Aegerter R. 1993. Influence of trace metals on growth and toxin production of Microcystis aeruginosa. Toxicon 31:293–305. 10.1016/0041-0101(93)90147-B [DOI] [PubMed] [Google Scholar]
- 6.Song L, Sano T, Li R, Watanabe MM, Liu Y, Kaya K. 1998. Microcystin production of Microcystis viridis (cyanobacteria) under different culture conditions. Phycol. Res. 46:19–23. 10.1046/j.1440-1835.1998.00120.x [DOI] [Google Scholar]
- 7.Neilan BA, Pearson LA, Muenchhoff J, Moffitt MC, Dittmann E. 2013. Environmental conditions that influence toxin biosynthesis in cyanobacteria. Environ. Microbiol. 15:1239–1253. 10.1111/j.1462-2920.2012.02729.x [DOI] [PubMed] [Google Scholar]
- 8.Jahnichen S, Ihle T, Petzoldt T, Benndorf J. 2007. Impact of inorganic carbon availability on microcystin production by Microcystis aeruginosa PCC 7806. Appl. Environ. Microbiol. 73:6994–7002. 10.1128/AEM.01253-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Utkilen H, Gjølme N. 1995. Iron-stimulated toxin production in Microcystis aeruginosa. Appl. Environ. Microbiol. 61:797–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohrlack T, Dittmann E, Henning M, Borner T, Kohl JG. 1999. Role of microcystins in poisoning and food ingestion inhibition of Daphnia galeata caused by the cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 65:737–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young FM, Thomson C, Metcalf JS, Lucocq JM, Codd GA. 2005. Immunogold localisation of microcystins in cryosectioned cells of Microcystis. J. Struct. Biol. 151:208–214. 10.1016/j.jsb.2005.05.007 [DOI] [PubMed] [Google Scholar]
- 12.Schatz D, Keren Y, Vardi A, Sukenik A, Carmeli S, Börner T, Kaplan A. 2007. Towards clarification of the biological role of microcystins, a family of cyanobacterial toxins. Environ. Microbiol. 9:965–970. 10.1111/j.1462-2920.2006.01218.x [DOI] [PubMed] [Google Scholar]
- 13.Tillett D, Dittmann E, Erhard M, von Döhren H, Börner T, Neilan BA. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide–polyketidesynthetase system. Chem. Biol. 7:753–764. 10.1016/S1074-5521(00)00021-1 [DOI] [PubMed] [Google Scholar]
- 14.Kaebernick M, Neilan BA, Börner T, Dittmann E. 2000. Light and the transcriptional response of the microcystin biosynthesis gene cluster. Appl. Environ. Microbiol. 66:3387–3392. 10.1128/AEM.66.8.3387-3392.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vézie C, Rapala J, Vaitomaa J, Seitsonen J, Sivonen K. 2002. Effect of nitrogen and phosphorus on growth of toxic and nontoxic Microcystis strains and on intracellular microcystin concentrations. Microb. Ecol. 43:443–454. 10.1007/s00248-001-0041-9 [DOI] [PubMed] [Google Scholar]
- 16.Rantala A, Rajaniemi-Wacklin P, Lyra C, Lepistö L, Rintala J, Mankiewicz-Boczek J, Sivonen K. 2006. Detection of microcystin-producing cyanobacteria in Finnish lakes with genus-specific microcystin synthetase gene E (mcyE) PCR and associations with environmental factors. Appl. Environ. Microbiol. 72:6101–6110. 10.1128/AEM.01058-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida M, Yoshida T, Takashima Y, Hosoda N, Hiroishi S. 2007. Dynamics of microcystin-producing and non-microcystin-producing Microcystis populations is correlated with nitrate concentration in a Japanese lake. FEMS Microbiol. Lett. 266:49–53. 10.1111/j.1574-6968.2006.00496.x [DOI] [PubMed] [Google Scholar]
- 18.Zilliges Y, Kehr JC, Meissner S, Ishida K, Mikkat S, Hagemann M, Dittmann E. 2011. The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS One 6:e17615. 10.1371/journal.pone.0017615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexova R, Fujii M, Birch D, Cheng J, Waite TD, Ferrari BC, Neilan BA. 2011. Iron uptake and toxin synthesis in the bloom-forming Microcystis aeruginosa under iron limitation. Environ. Microbiol. 13:1064–1077. 10.1111/j.1462-2920.2010.02412.x [DOI] [PubMed] [Google Scholar]
- 20.Dziallas C, Grossart HP. 2011. Increasing oxygen radicals and water temperature select for toxic Microcystis sp. PLoS One 6:e25569. 10.1371/journal.pone.0025569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexova R, Haynes PA, Ferrari BC, Neilan BA. 2011. Comparative protein expression in different strains of the bloom-forming cyanobacterium Microcystis aeruginosa. Mol. Cell. Proteomics 10:3749–3765. 10.1074/mcp.M110.003749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sevilla E, Martin-Luna B, Vela L, Bes MT, Fillat MF, Peleato ML. 2008. Iron availability affects mcyD expression and microcystin-LR synthesis in Microcystis aeruginosa PCC7806. Environ. Microbiol. 10:2476–2483. 10.1111/j.1462-2920.2008.01663.x [DOI] [PubMed] [Google Scholar]
- 23.Sevilla E, Martin-Luna B, Vela L, Bes MT, Peleato ML, Fillat MF. 2010. Microcystin-LR synthesis as response to nitrogen: transcriptional analysis of the mcyD gene in Microcystis aeruginosa PCC7806. Ecotoxicology 19:1167–1173. 10.1007/s10646-010-0500-5 [DOI] [PubMed] [Google Scholar]
- 24.Ginn HP, Neilan BA. 2010. NtcA from Microcystis aeruginosa PCC 7806 is autoregulatory and binds to the microcystin promoter. Appl. Environ. Microbiol. 76:4362–4368. 10.1128/AEM.01862-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degerholm J, Gundersen K, Bergman B, Söderbäck E. 2006. Phosphorus-limited growth dynamics in two Baltic Sea cyanobacteria, Nodularia sp. and Aphanizomenon sp. FEMS Microbiol. Ecol. 58:323–332. 10.1111/j.1574-6941.2006.00180.x [DOI] [PubMed] [Google Scholar]
- 26.Shan Y, McKelvie ID, Hart BT. 1994. Determination of alkaline phosphatase-hydrolyzable phosphorus in natural water systems by enzymatic flow injection. Limnol. Oceanogr. 39:1993–2000. 10.4319/lo.1994.39.8.1993 [DOI] [Google Scholar]
- 27.Luque I, Forchhammer K. 2008. Nitrogen assimilation and C/N balance sensing, p 335–382 In Herrero A, Flores E. (ed), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, VA [Google Scholar]
- 28.Flores E, Frías JE, Rubio LM, Herrero A. 2005. Photosynthetic nitrate assimilation in cyanobacteria. Rev. Photosynth. Res. 83:117–133. 10.1007/s11120-004-5830-9 [DOI] [PubMed] [Google Scholar]
- 29.Welker M, Brunke M, Preussel K, Lippert I, von Dohren H. 2004. Diversity and distribution of Microcystis (cyanobacteria) oligopeptides chemotypes from natural communities studied by single mass spectrometry. Microbiology 150:1785–1796. 10.1099/mic.0.26947-0 [DOI] [PubMed] [Google Scholar]
- 30.Pereira DA, Pimenta AMC, Giani A. 2012. Profiles of toxic and non-toxic oligopeptides of Radiocystis fernandoii (Cyanobacteria) exposed to three different light intensities. Microbiol. Res. 167:413-421. 10.1016/j.micres.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 31.Guillard RRL, Lorenzen CJ. 1972. Yellow-green algae with chlorophyllide C1,2. J. Phycol. 8:10–14. 10.1111/j.1529-8817.1972.tb03995.x [DOI] [Google Scholar]
- 32.Nusch EA. 1980. Comparison of different methods for chlorophyll and phaepigment determination. Arch. Hydrobiol. Beih. Ergebn. Limnol. 14:14–36 [Google Scholar]
- 33.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/ [Google Scholar]
- 35.Schwarz R, Forchhammer K. 2005. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology 151:2503–2514. 10.1099/mic.0.27883-0 [DOI] [PubMed] [Google Scholar]
- 36.Anderson DC, Campbell EL, Meeks JC. 2006. A soluble 3D LC/MS/MS proteome of the filamentous cyanobacterium Nostoc punctiforme. J. Proteome Res. 5:3096–3104. 10.1021/pr060272m [DOI] [PubMed] [Google Scholar]
- 37.Sawaki H, Sugiyama T, Omata T. 1998. Promoters of the phycocyanin gene clusters of the cyanobacterium Synechococcus sp. strain PCC 7942. Plant Cell Physiol. 39:756–761. 10.1093/oxfordjournals.pcp.a029431 [DOI] [PubMed] [Google Scholar]
- 38.Dagnino D, Meireles AD, Almeida AJC. 2006. Growth of nutrient-replete Microcystis PCC 7806 cultures is inhibited by an extracellular signal produced by chlorotic cultures. Environ. Microbiol. 8:30–36. 10.1111/j.1462-2920.2005.00866.x [DOI] [PubMed] [Google Scholar]
- 39.Kurmayer R. 2011. The toxic cyanobacterium Nostoc sp. strain 152 produces highest amounts of microcystin and nostophycin under stress conditions. J. Phycol. 47:200–207. 10.1111/j.1529-8817.2010.00931.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hesse K, Kohl JG. 2001. Effects of light and nutrient supply on growth and microcystin content of different strains of Microcystis aeruginosa, p 152–158 In Chorus I. (ed), Cyanotoxins: occurrence, causes, consequences. Springer, Berlin, Germany [Google Scholar]
- 41.Jahnichen S, Long BM, Petzoldt Y. 2011. Microcystin production by Microcystis aeruginosa: direct regulation by multiple environmental factors. Harmful Algae 12:95–104. 10.1016/j.hal.2011.09.002 [DOI] [Google Scholar]
- 42.Briand E, Yéprémian C, Humbert JF, Quiblier C. 2008. Competition between microcystin- and non-microcystin-producing Planktothrix agardhii (cyanobacteria) strains under different environmental conditions. Environ. Microbiol. 10:3337–3348. 10.1111/j.1462-2920.2008.01730.x [DOI] [PubMed] [Google Scholar]
- 43.Lindell D, Post AF. 2001. Ecological aspects of ntcA gene expression and its use as an indicator of the nitrogen status of marine Synechococcus spp. Appl. Environ. Microbiol. 67:3340–3349. 10.1128/AEM.67.8.3340-3349.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuniyoshi TM, Gonzalez A, Lopez-Gomollon S, Valladares A, Bes MT, Fillat MF, Peleato ML. 2011. 2-Oxoglutarate enhances NtcA binding activity to promoter regions of the microcystin synthesis gene cluster. FEBS Lett. 585:3921–3926. 10.1016/j.febslet.2011.10.034 [DOI] [PubMed] [Google Scholar]
- 45.Lafiti A, Ruiz M, Zhang CC. 2009. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33:258–278. 10.1111/j.1574-6976.2008.00134.x [DOI] [PubMed] [Google Scholar]
- 46.Qian H, Yu S, Sun Z, Xie X, Liu W, Fu Z. 2010. Effects of copper sulfate, hydrogen peroxide and N-phenyl-2-naphthylamine on oxidative stress and the expression of genes involved photosynthesis and microcystin disposition in Microcystis aeruginosa. Aquat. Toxicol. 99:405–412. 10.1016/j.aquatox.2010.05.018 [DOI] [PubMed] [Google Scholar]
- 47.Pimentel JS, Giani A. 2013. Estimating toxic cyanobacteria in a Brazilian reservoir by quantitative real-time PCR, based on the microcystin synthetase D gene. J. Appl. Phycol. 25:1545–1554. 10.1007/s10811-013-9996-4 [DOI] [Google Scholar]