Abstract
Rhizobial bacteria are commonly found in soil but also establish symbiotic relationships with legumes, inhabiting the root nodules, where they fix nitrogen. Endophytic rhizobia have also been reported in the roots and stems of legumes and other plants. We isolated several rhizobial strains from the nodules of noninoculated bean plants and looked for their provenance in the interiors of the seeds. Nine isolates were obtained, covering most known bean symbiont species, which belong to the Rhizobium and Sinorhizobium groups. The strains showed several large plasmids, except for a Sinorhizobium americanum isolate. Two strains, one Rhizobium phaseoli and one S. americanum strain, were thoroughly characterized. Optimal symbiotic performance was observed for both of these strains. The S. americanum strain showed biotin prototrophy when subcultured, as well as high pyruvate dehydrogenase (PDH) activity, both of which are key factors in maintaining optimal growth. The R. phaseoli strain was a biotin auxotroph, did not grow when subcultured, accumulated a large amount of poly-β-hydroxybutyrate, and exhibited low PDH activity. The physiology and genomes of these strains showed features that may have resulted from their lifestyle inside the seeds: stress sensitivity, a ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) complex, a homocitrate synthase (usually present only in free-living diazotrophs), a hydrogenase uptake cluster, and the presence of prophages. We propose that colonization by rhizobia and their presence in Phaseolus seeds may be part of a persistence mechanism that helps to retain and disperse rhizobial strains.
INTRODUCTION
Bacteria can populate diverse environments, from soil to water, even in the most extreme sites on the earth. These organisms can also be found on and inside other organisms, such as fungi, animals, and plants. The microbiome is a recently developed concept used to define the prokaryotic populations in close relationship with more-complex organisms (1, 2). Bacteria can flourish as endophytes inside plant roots, stems, leaves, and seeds (3) in crops (e.g., wheat, rice, maize, sorghum, and sugarcane), legumes (e.g., clover, common bean, and alfalfa), trees (e.g., Populus), grasses (e.g., switchgrass), and model plants (e.g., Arabidopsis) (4). Most common endophytic bacterial species have been described as members of the phyla Firmicutes and Proteobacteria (5).
Legumes are economically valuable plants in agriculture and establish symbiotic relationships with rhizobial bacteria (6). These bacteria contribute to the formation of plant root nodules, colonizing them and fixing atmospheric nitrogen. This phenomenon has been studied intensively (7). Endophytic rhizobia, in tissues other than nodules, have also been isolated from clover and pea (e.g., Rhizobium phaseoli and Rhizobium leguminosarum bv. trifolii) (8). Several nonrhizobial bacterial species have been isolated from Phaseolus tissues (roots, stems, and seeds); these include Acinetobacter, Bacillus, Enterococcus, Nocardioides, Paracoccus, Phyllobacterium, Sphingomonas, and Streptomyces spp., as well as an ineffective rhizobial species named Rhizobium endophyticum (9). Phaseolus spp. are promiscuous plants that can be effectively nodulated by several rhizobial species, albeit with diverse nitrogen fixation efficiencies (1, 10–13). Conversely, rhizobial bacteria have also been isolated from the tissues of nonleguminous plants, such as wheat (14) and rice (15) roots, maize stems (16), and Arabidopsis seeds (4).
We hypothesize that given the long-term and intimate nature of legume-rhizobium relationships, it is possible to find bacterial strains with intact nitrogen fixation abilities inside legume seeds. In this work, we report the isolation and characterization of nitrogen-fixing rhizobial strains from noninoculated Phaseolus vulgaris plants that can be propagated vertically in the interiors of the seeds.
MATERIALS AND METHODS
Bacterial strains and culture media.
The strains used are listed in Table 1. Strains were maintained either in solid PY medium, containing 0.5% peptone, 0.3% yeast extract, and 7 mM CaCl2, or in LB medium, plus antibiotics. Liquid PY medium was used at 30°C with shaking at 200 rpm (17). Minimal medium (MM) contained 1.2 mM K2HPO4, 0.8 mM MgSO4, 1.5 mM CaCl2, 10 mM NH4Cl (N source), and 10 mM succinic acid (C source). Other C and N sources were also used at 10 mM unless otherwise specified. Subcultures in MM were performed as described previously (17). Antibiotics (in micrograms per milliliter) were added as follows: nalidixic acid, 20; streptomycin, 200; rifampin, 100; neomycin, 60; tetracycline, 10; fosfomycin, 200; cycloheximide, 10.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| Strains | ||
| Rhizobium etli | ||
| CFN42 | Wild-type strain; Nalr | Laboratory collection |
| CFNX192 | CFNX89/p42d::Tn5-mob Nalr Kmr Neor | 18 |
| R. phaseoli | ||
| CIAT652 | Wild-type strain; Nalr | 43 |
| CCGM1 | Strain isolated from nodules of noninoculated P. vulgaris cv. Negro Jamapa plants grown in N-free medium; Nalr | This study |
| CCGM1.1 | CCGM1::Tn5-mob Nalr Kmr Neor | This study |
| CCGM2 | Strain isolated from nodules of noninoculated P. vulgaris cv. Negro Jamapa plants grown in N-free medium; Nalr | This study |
| CCGM8 | Strain isolated from nodules of noninoculated P. vulgaris cv. Negro Jamapa plants grown with combined nitrogen; Nalr | This study |
| CCGM9 | Strain isolated from nodules of noninoculated P. vulgaris cv. Negro Jamapa plants grown with combined nitrogen; Nalr | This study |
| R. leguminosarum | ||
| CCGM4 | Strain isolated from nodules of noninoculated P. vulgaris cv. Negro Jamapa plants grown in N-free medium; Nalr | This study |
| CCGM5 | Strain isolated from nodules of noninoculated P. vulgaris cv. Negro Jamapa plants grown in N-free medium; Nalr | This study |
| CCGM6 | Strain isolated from nodules of noninoculated P. vulgaris cv. Flor de Mayo plants grown in N-free medium; Nalr | This study |
| R. grahamii CCGM3 | Strain isolated from nodules of noninoculated P. vulgaris cv. Negro Jamapa plants grown in N-free medium; Nalr | This study |
| Agrobacterium tumefaciens | ||
| GMI9023 | C58 cured of its native plasmids | 56 |
| GMI9023/p42d | GMI9023/p42d::Tn5-mob Nalr Kmr Neor | 18 |
| S. americanum | ||
| CCGM7 | Strain isolated from nodules of noninoculated P. vulgaris cv. Negro Jamapa plants grown in N-free medium; Nalr | This study |
| CCGM7-dsRed | CCGM7 derivative with plasmid pFAJ1708::dsRed; Tcr | This study |
| Escherichia coli | ||
| HB101 | F− hsdS20-recA13 | 57 |
| S17-1 | recA1 thi pro Tra+ | 58 |
| Plasmids | ||
| pRK2013 | Helper plasmid; ColE1 mob+ Tra+ Kmr | 19 |
| pSUP5011 | Tn5-mob vector; Neor | 21 |
| pFAJ1708::dsRed | dsRed fluorescence gene from pHKT3, cloned on the SacI site of the pFAJ1708 vector; Tcr | J. Althabegoiti and G. Torres-Tejerizo, unpublished data |
Manipulation of strains.
The Agrobacterium tumefaciens GMI9023/p42d derivative was obtained by conjugation of GMI9023, CFNX192 (p42d::Tn5-mob) (18), and the helper strain HB101/pRK2013 for mobilization (19) and by selection with rifampin and neomycin. Plasmid profiles and pSym transference were analyzed by using a modified Eckhardt procedure (20). The presence of the symbiotic plasmid was determined by hybridization of blotted Eckhardt gels using a [32P]CTP-labeled Rhizobium etli nifH probe. In order to produce CCGM1.1, CCGM1 was randomly mutagenized with Tn5-mob and was mated with Escherichia coli S17.1/pSUP5011 (21). Plasmid pFAJ1708::dsRed (J. Althabegoiti and G. Torres-Tejerizo, unpublished data) was introduced into strain CCGM7 by conjugation to produce the CCGM7-dsRed derivative.
Source of bean seeds and disinfection.
Phaseolus vulgaris cv. Negro Jamapa seeds were cultivated in 2010 to 2012 in pots in the greenhouse, irrigated with water alternating with Fahraeus solution (22), and fertilized with nitrogen. Each year, seeds from the previous crop were propagated. Phaseolus vulgaris cv. Flor de Mayo seeds were obtained from Mexican commercial stores. The P. vulgaris cv. Negro Jamapa and P. vulgaris cv. Flor de Mayo seeds were disinfected with 0.525% and 10% sodium hypochlorite, respectively, for 10 min with shaking and were washed exhaustively until the odor disappeared (8 and 15 min). The final wash specimens were plated on PY and LB dishes (five 200-μl samples), and seeds (1 of every 4) were smeared throughout the surfaces of PY and LB plates to check for contamination. No growth was observed during these washing procedures for either cultivar.
Plant assays.
The bean seeds were germinated and were inoculated as described previously (23), but inoculant strains were washed twice, and the optical density at 540 nm (OD540) was adjusted to 0.1; 2- to 3-day-old seedlings were transferred to sterilized pots and were inoculated with 1 ml bacterial suspension. Fahraeus medium without combined nitrogen was neutralized at pH 6.8. At 17, 24, and 32 days postinoculation (dpi), the nitrogenase activities of 10 plants (2 pots) for each strain were measured using the acetylene reduction method (23); the numbers and weights of the nodules and plants were determined. For each sampling time and condition, 10 nodules from each plant were used to verify the presence or absence of a strain according to their resistance or sensitivity to the corresponding antibiotics. Control plants, grown with or without combined nitrogen, were included. The strains reported in this study were obtained by following the procedure described above except that the plants were not inoculated. Student's t test for significance was performed as described previously (24).
Assays with Leonard jars.
A. tumefaciens GMI9023/p42d::Tn5-mob was used to inoculate P. vulgaris cv. Negro Jamapa and P. vulgaris cv. Flor de Mayo in Leonard jars (25) containing vermiculite in the top container. The jars were then covered with aluminum foil. The bean seeds were treated as described above. Two plantlets of each variety were grown in each jar in the greenhouse, and 4 to 8 jars were analyzed at each sampling time (18, 25, and 32 dpi). CFNX192-treated, N-treated, and N-free plants were included as controls. Tests with no inoculation were also performed.
Extraction of rhizobial strains from P. vulgaris cv. Negro Jamapa seeds produced by plants inoculated with each strain.
Pods (12 cm long) produced by plants (12 to 15 days after flowering) were sampled under sterile conditions in the laminar flow hood. Seeds detached from 1 to 2 pods were scarified with sandpaper A-99 type 320, placed in a 50-ml Falcon tube, washed 3 times with sterile water for 1 min each time, and vortexed. To disinfect the seeds, 15 to 20 ml of 0.525% sodium hypochlorite was added for 10 min with constant vortex agitation, followed by five 1-min washes and soaking in 15 ml of water for 4 h at 29°C in the dark or incubation for 48 h; a last change of water was performed with gentle agitation. To check for contamination, the final wash specimens were plated on PY and LB dishes. The seeds were then dissected and the embryos collected in an Eppendorf tube containing 60 μl of water; they were smashed with a sterile glass or with a homogenizer (Bullet Blender; Next Advance, Averill Park, NY, USA) and were streaked onto PY medium containing 10 μg ml−1 cycloheximide to allow only the growth of bacteria. The plates were incubated at 29°C for as long as 5 days. The supernatant was used for direct isolation in agar plates (containing fosfomycin or nalidixic acid) or for PCR tests. The Rhizobium isolates were selected for their morphological appearance and growth characteristics and were further identified using the procedures described above.
PCR.
Amplifications were performed in a T100 thermocycler (Bio-Rad, Hercules, CA, USA) using Phusion High-Fidelity DNA polymerase (Thermo Scientific, Pittsburgh, PA, USA), under the conditions given in Table 2. The 25-μl reaction mixture consisted of 3.5 μl (50 ng) of genomic DNA, 12.5 μl of 2× buffer, 1.5 μl of primers (15 pmol each), and 6 μl of water. For the second round of PCR, genomic DNA was replaced with 3.5 μl of the mixture for the first round. Potential seed inhibition was assessed with a control consisting of a seed extract obtained from noninoculated plants grown with combined nitrogen and mixed with rhizobial DNA.
TABLE 2.
Conditions for PCR amplifications
| Oligonucleotide class | Bacterial group/strain specificity | Gene (size, bp) | Oligonucleotide name | Oligonucleotide type | Oligonucleotide sequence | PCR conditions |
||
|---|---|---|---|---|---|---|---|---|
| Initial step | Amplification (35 cycles) | Final step | ||||||
| Generic | Rhizobiales | nifH (95) | nifH_UN_FW | Forward | CGGGCGTCGGNTGYGCNGG | 15 s at 95°C | 2 s at 98°C, 2 s at 65°C and 2 s at 60°C, 5 s at 72°C | 25 s at 72°C |
| nifH_UN_R | Reverse | CAAAGCCACCGCANACNACRTCNCC | ||||||
| nifH (350)a | KAD | Forward | CATCTTGTGGCGACCGCGATCTC | 15 s at 95°C | 2 s at 98°C, 2 s at 58°C and 2 s at 55°C, 20 s at 72°C | 25 s at 72°C | ||
| GEM | Reverse | ATSGCCATCATYTCRCCGGA | ||||||
| Rhizobium, Sinorhizobium, and Mesorhizobium | rpoB (786) | rpoB_RSM_FW | Forward | ACCCGCGATATTCCGAAYGTHTC | 15 s at 95°C | 2 s at 98°C, 2 s at 68°C and 2 s at 65°C, 25 s at 72°C | 30 s at 72°C | |
| rpoB_RSM_RV | Reverse | GTTCAGAACGACGTCGACRTG | ||||||
| Rhizobiales | nodC (384 bp) | nodC_FW_UNi2 | Forward | GATATGGAGTACTGGCTSGCNTG | 15 s at 95°C | 2 s at 98°C, 2 s at 65°C and 2 s at 60°C, 10 s at 72°C | 30 s at 72°C | |
| nodC_RV_UNi2 | Reverse | GTTCTGTCCGACGACGTCGAGCGTVAGRAASCG | ||||||
| Alphaproteobacteria | rpoB (740–752)b | Br3200F | Forward | TGAAGATGGTCAAGGTCTTCGT | 15 s at 95°C | 2 s at 98°C, 2 s at 60°C and 2 s at 55°C, 25 s at 72°C | 30 s at 72°C | |
| Br3950R | Reverse | GTCCGACTTSACHGTCAGCAT | ||||||
| Bacteria | 16S rRNA (1,300)c | 63F | Forward | CAGGCCTAACACATGCAAGTC | 15 s at 95°C | 2 s at 98°C, 2 s at 57°C and 2 s at 54°C, 30 s at 72°C | 30 s at 72°C | |
| L1401 | Reverse | GCGTGTGTACAAGACCC | ||||||
| Specific | S. americanum CCGM7 | rpoB (264) | rpoB_CCGM7_F2 | Forward | CTCGAGGCCTACCACGAGAGC | 15 s at 95°C | 2 s at 98°C, 2 s at 65°C and 2 s at 60°C, 10 s at 72°C | 30 s at 72°C |
| rpoB_CCGM7_R2 | Reverse | CGTCGCCTGTACGTCCGTCG | ||||||
| R. phaseoli CCGM1 | rpoB (290) | rpoB_CCGM1_FW | Forward | GATGTAGCCCACCGTGACC | 15 s at 95°C | 2 s at 98°C, 2 s at 65°C and 2 s at 60°C, 10 s at 72°C | 30 s at 72°C | |
| rpoB_CCGM1_RV | Reverse | CTTACAAGGCGAATGGCAAC | ||||||
PDH activity.
Cells were cultivated in MM supplemented with succinate and ammonium and were collected after 8 h. The cells were lysed via sonication, and the pyruvate dehydrogenase (PDH) activity in desalted extracts was determined as reported previously (26). Protein determination was performed by the Lowry method.
PHB determination.
The polymer poly-β-hydroxybutyrate (PHB) was quantified by using the spectrophotometric method of Law and Slepecky (27) at 235 nm.
Genome sequencing, assembly, and annotation.
DNA was extracted according to standard protocols. Strain CCGM1 was sequenced at the Instituto de Biotecnología—UNAM (Cuernavaca, Mexico), with an Illumina (San Diego, CA, USA) Genome Analyzer IIx and a 400 bp-fragment paired-end library. A total of 35,521,298 paired reads were obtained, and 27,452,055 remained after trimming (genome coverage, 400×). Strain CCGM7 was sequenced at MOgene (St. Louis, MO, USA) with a 3-kb paired-end library, which was run on an Illumina MiSeq sequencer. A total of 13,317,918 paired reads were obtained; 11,646,020 remained after trimming (coverage, 104×). The reference genomes were CIAT652 and USDA257, respectively. Velvet, version 1.2.06 (28), and GapFiller, version 1.11 (29), were used for assembly, and Mauve, version 2.3.1 (30), was used for contig ordering. Circular representation graphs were obtained with GenVision (DNAStar Inc., Madison, WI, USA). Annotation was conducted with RAST, version4.0 (31).
Phylogenetic analysis.
For rpoB phylogeny, we used the rpoB sequences mentioned under “Nucleotide sequence accession numbers” below as well as those of Bradyrhizobium japonicum USDA110 (NC_004463), Bradyrhizobium sp. strain BTAi1 (NC_009485), Rhizobium grahamii CCGE502 (AEYE02000001), R. leguminosarum bv. trifolii WSM2304 (NC_011369), Rhizobium leguminosarum bv. viciae 3841 (NC_008380), and Sinorhizobium americanum CFNEI156 (synonymous with strain LMG22684) (AM295361.1). The sequences were aligned with ClustalW (32), the phylogeny was inferred with PHYLIP (33), and the tree was drawn with NJplot (34).
Comparative genomic analysis.
The completely sequenced genomes of 8 rhizobial species in the genera Rhizobium (R. etli CFN42 [NC_007761], R. phaseoli CIAT652 [NC_010994], R. phaseoli CNPAF512 [AEYZ00000000]) and Sinorhizobium (Sinorhizobium fredii HH103 [NC_016812], S. fredii NGR234 [NC_012587], S. fredii USDA257 [NC_018000], Sinorhizobium medicae WSM419 [NC_009636], and Sinorhizobium meliloti 1021 [NC_003047]) were used. To identify orthologs between CCGM1 or CCGM7 and its group, we used OrthoMCL, version 2.0 (35), with BLAST (E value, 1e−5; 30% identity and 60% overlap). Additionally, the genomes of Bradyrhizobium sp. strain ORS278 (NC_009445), Bradyrhizobium sp. strain BTAi1 (NC_009485), and Azorhizobium caulinodans ORS571 (NC_009937) were used.
Phage detection.
The presence of prophages was predicted with the PHAST database, version 1, accessed on November 2013 (36).
Nucleotide sequence accession numbers.
The whole-genome shotgun projects have been deposited in GenBank under accession numbers JFGP00000000 (CCGM1) and JFGO00000000 (CCGM7). The sequences of the rpoB genes of the following strains have been deposited in GenBank under the accession numbers given in parentheses: Rhizobium grahamii CCGM3 (KF836417), R. leguminosarum CCGM4 (KF836414), R. leguminosarum CCGM5 (KF836418), R. leguminosarum CCGM6 (KF836416), and R. phaseoli CCGM8 (KF836415).
RESULTS
In the search to assess the symbiotic phenotype of an Agrobacterium tumefaciens strain containing the symbiotic plasmid of R. etli CFN42 (GMI9023/p42d) in P. vulgaris cv. Negro Jamapa bean plants grown in sterilized Leonard jars, we observed that the inoculated strain formed white, inefficient nodules. However, sporadically, we also found pink, nitrogen-fixing nodules, formed by an unknown strain. The strain occupying these nodules was called CCGM1. It was characterized by plasmid content and presented an unknown profile.
Thinking that unintended contamination of the assay had occurred, we repeated the procedure using thoroughly surface sterilized seeds in sterilized Leonard jars that were opened only once. Strain CCGM1 appeared in effective nodules. This strain was also found when P. vulgaris cv. Flor de Mayo bean plantlets were grown under the conditions described above and was identified again by its plasmid profile. In order to investigate whether strain CCGM1 and further strains were seed borne, we performed another experiment in pots using P. vulgaris cv. Negro Jamapa without inoculation and under N-free conditions; CCGM1 was isolated again, and strains CCGM2, CCGM3, CCGM4, CCGM5, CCGM6, and CCGM7 also appeared at different times of observation. In an additional experiment with noninoculated P. vulgaris cv. Negro Jamapa plants irrigated with combined nitrogen, strains CCGM8 and CCGM9 were isolated from the nodules at later stages (55 to 90 days).
Given these results, we supposed that these strains were present in the bean seeds. To test this, CCGM1 and CCGM7 were tagged with Tn5-mob (CCGM1.1) or dsRed (CCGM7-dsRed), respectively, and were inoculated as usual into P. vulgaris cv. Negro Jamapa bean plantlets. R. phaseoli strain CIAT652 was used as a control. After successful symbiosis, we looked for the strains in the seeds produced. CCGM1.1 and CCGM7-dsRed were detected initially by PCR amplification of rpoB (Fig. 1), nodC, and nifH (see Fig. S1 in the supplemental material). The rpoB PCR products were sequenced, and they matched the rpoB sequences of the strains perfectly. Figure S2 in the supplemental material shows the PCR amplification of rpoB, nodC, and nifH from seeds from noninoculated plants grown with combined nitrogen.
FIG 1.
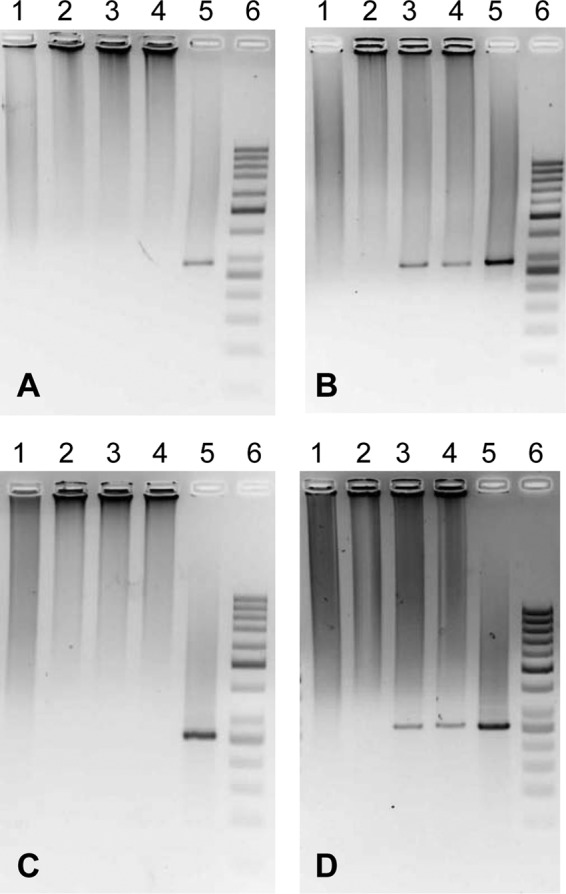
Detection of rhizobial strains in bean seeds via PCR amplification of the rpoB gene. Two sequential PCR rounds were required to obtain amplification signals. The substrate for the second round was a portion of the first reaction mixture. (A) First PCR for CCGM1.1 detection. Lanes: 1, negative control; 2, seed extract from noninoculated plants grown with combined nitrogen; 3, seed extract from plants inoculated with CCGM1.1; 4, seed extract as in lane 2 but with a CCGM1.1 lysate added (to assess inhibition); 5, CCGM1.1 lysate; 6, molecular weight marker. P. vulgaris cv. Negro Jamapa seeds (187 days old) were scarified and treated as described in Materials and Methods. (B) Second PCR for CCGM1.1 detection. Lanes are as described for panel A. (C) First PCR for CCGM7-dsRed detection. Lanes: 1, negative control; 2, seed extract from noninoculated plants grown with combined nitrogen; 3, seed extract from plants inoculated with CCGM7-dsRed; 4, seed extract as in lane 2 but with a CCGM7-dsRed lysate added; 5, CCGM7-dsRed lysate; 6, molecular weight marker. P. vulgaris cv. Negro Jamapa seeds from fresh pods (60 days old) were treated as described in Materials and Methods to obtain embryos. (D) Second PCR for CCGM7-dsRed detection. Lanes are as described for panel C.
The tagged strains CCGM1 and CCGM7 were also recovered as bacterial clones from macerated seeds and extracted embryos. They were identified by exact matching of the rpoB sequence, the plasmid profile, and antibiotic resistance. Clones of the recovered strain CCGM7-dsRed were inoculated into P. vulgaris cv. Negro Jamapa bean plantlets, and nodulation and nitrogen fixation appeared normal (data not shown). CIAT652 was not detected or recovered from seeds.
Phylogenetic characterization of the endophytic strains.
We further characterized the rhizobial strains via 16S rRNA gene amplification and sequencing in order to assign the strains to the closest species. For certain strains, 16S rRNA sequences lacked sufficient resolution (data not shown); therefore, we also sequenced a segment of the DNA-directed RNA polymerase subunit β (rpoB). Thus, based on these sequences, we found the closest associations of the strains to be as follows: CCGM1 and CCGM8 with R. phaseoli CNPAF512 and CIAT652; CCGM3 with R. grahamii CCGE502; CCGM4, CCGM5, and CCGM6 with R. leguminosarum bv. trifolii WSM2304; and CCGM7 with Sinorhizobium americanum type strain CFNEI156. Figure 2 shows the rpoB phylogeny.
FIG 2.
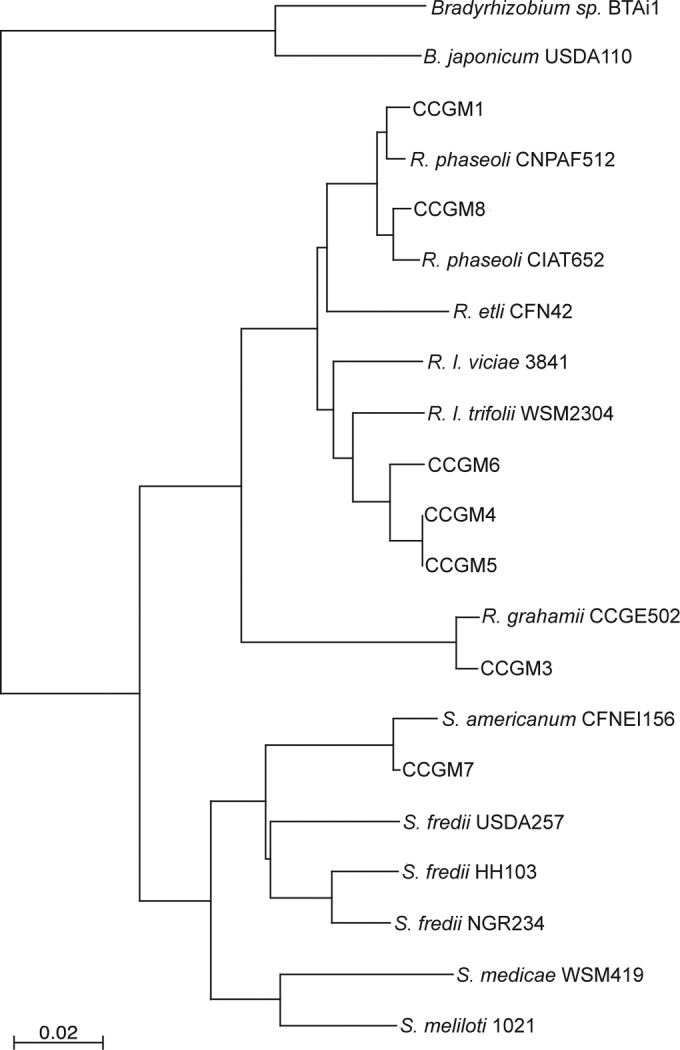
Phylogenetic tree for the rpoB genes (internal segment) of rhizobial strains isolated from bean seeds. The sequences were aligned with ClustalW; phylogeny was inferred with PHYLIP; and the tree was drawn with NJplot. The bar represents the number of substitutions per nucleotide.
Plasmid profiles of the endophytic strains.
Rhizobial strains commonly present several high-molecular-weight plasmids, and most genes related to symbiotic functions are located on the symbiotic plasmid (pSym). The plasmid profiles (Fig. 3) of the strains isolated were analyzed using Eckhardt-type gels, and the plasmids found ranged from 200 to 1,000 kb. Interestingly, strain CCGM2 presented a CCGM1-like plasmid profile, the only difference being an additional plasmid of approximately 200 kb. Additionally, strain CCGM5 exhibited a CCGM4-like profile, lacking only the smallest plasmid. Strains CCGM8 and CCGM9 showed similar profiles, differing in only one plasmid (data not shown). Unexpectedly, strain CCGM7 did not exhibit plasmids.
FIG 3.
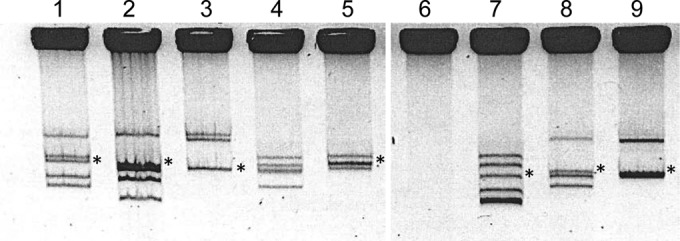
Plasmid profiles of rhizobial strains isolated from bean seeds. Lanes: 1, CCGM6; 2, CCGM2; 3, CCGM3; 4, CCGM4; 5, CCGM5; 6, CCGM7; 7, CFN42; 8, CCGM1; 9, CIAT652. Plasmids were visualized using the Eckhardt technique. Asterisks indicate the positions of plasmids hybridizing with the nifH probe, denoting the symbiotic plasmid.
Symbiotic characterization of CCGM1 strains and CCGM7.
We performed additional symbiotic, physiological, and genomic characterizations on CCGM1 and CCGM7 because they appeared consistently in seeds, were phylogenetically distant, and represent, respectively, one of the most prevalent common bean symbionts and a recently reported symbiont (37).
The symbiotic abilities of strains CCGM1 and CCGM7 were tested in P. vulgaris cv. Negro Jamapa plantlets in the greenhouse, inoculated as usual. The strain for comparison was R. phaseoli CIAT652. Strain CCGM1 showed lower nitrogenase activity than CIAT652 but promoted increased plant weight (Fig. 4). Strain CCGM7 showed sharp increases of 85% and 240% in nitrogenase activity at 24 and 32 dpi, respectively, over the control strain. The R. phaseoli strains CCGM1 and CIAT652 induced similar numbers of nodules. CCGM7 formed few but larger nodules with a lower total weight. Both strains CCGM1 and CCGM7 were efficient at inducing plant growth (Fig. 5).
FIG 4.
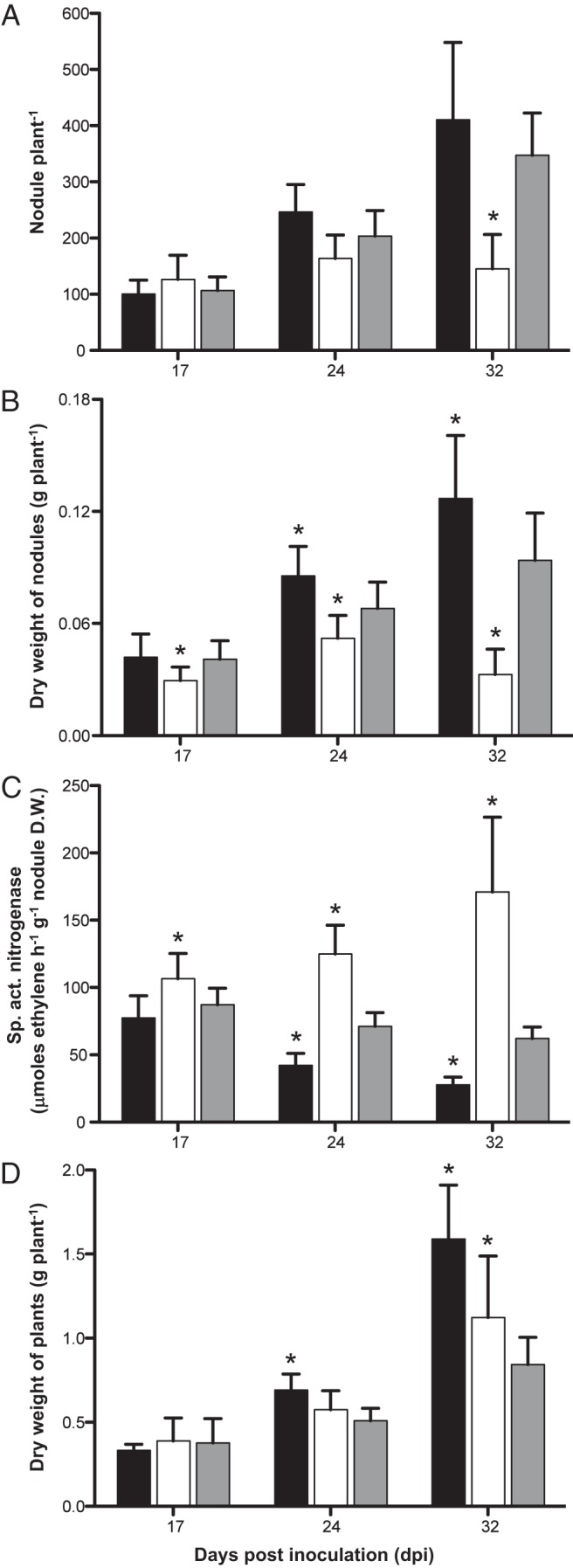
Symbiotic performance of rhizobial strains CCGM1 and CCGM7 with P. vulgaris plants. (A) Number of nodules per plant; (B) dry weight of nodules per plant; (C) specific activity of nitrogenase as measured by acetylene reduction; (D) dry weight of plants. Filled bars, strain CCGM1; open bars, strain CCGM7; shaded bars, strain CIAT652. Average values with standard deviations are shown. Ten plants per strain and date were assayed (n = 30). Asterisks indicate that the means of results for the samples are different (P, <0.05) from those for CIAT652. Results of one experiment representative of two are shown.
FIG 5.

Growth of P. vulgaris plants in symbiosis with rhizobial strains CCGM1 and CCGM7. Plants (32 days old) were maintained in the greenhouse.
Physiological characterization of strains CCGM1 and CCGM7.
To assess the growth capabilities of strains CCGM1 and CCGM7, we evaluated them in solid and liquid media with several nitrogen and carbon sources. The strains grew well in plates with MM containing C sources such as succinate, pyruvate, glucose, mannose, arabinose, galactose, or lactose, with either ammonium chloride or potassium nitrate as the N source, or with only glutamic acid or glutamine as N and C sources (see Table S1 in the supplemental material) (4). In serial subcultures of MM with succinate, strain CCGM1 exhibited reduced growth after the third subculture (Fig. 6), as did other R. phaseoli strains (17). This reduced growth effect was reversed with the addition of biotin (data not shown). Similar behavior was observed with pyruvate (data not shown). However, strain CCGM7 grew optimally in the three subcultures. It was observed that a decrease in pyruvate dehydrogenase (PDH) activity and accumulation of the polymer poly-β-hydroxybutyrate (PHB) are closely related to growth decline in subcultures (17). Therefore, we assayed both parameters with the strains grown in MM with succinate. As shown in Table 3, there was a correlation between increased PHB production and reduced PDH activity for strains CCGM1 and CIAT652. Minimal PHB production was observed for strain CCGM7; this strain also showed the highest PDH activity.
FIG 6.
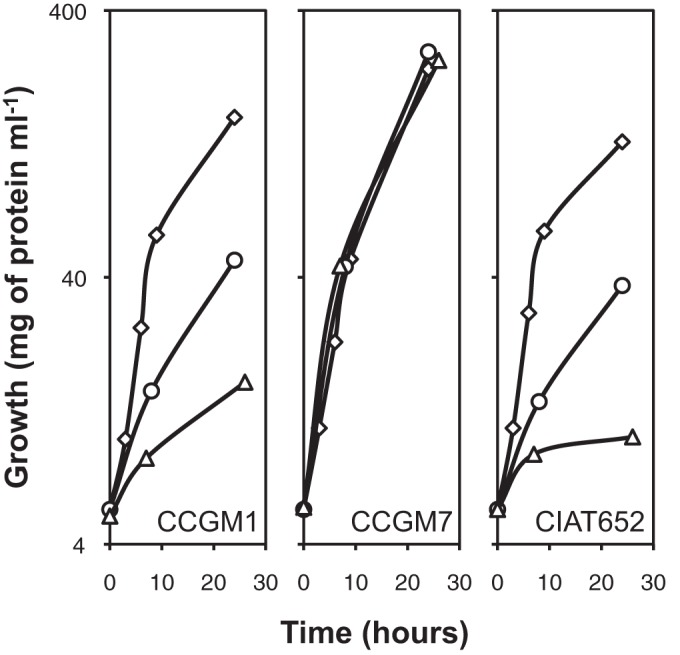
Growth of strains CCGM1 and CCGM7 in subcultures of MM with succinate and ammonium. Symbols: diamonds, 1st subculture; circles, 2nd subculture; triangles, 3rd subculture. Subcultures were performed as described previously (17). MM contained 10 mM succinic acid and 10 mM ammonium chloride. Protein content was determined using the Lowry method. The standard deviations have been omitted for clarity. Results of one experiment representative of three are shown.
TABLE 3.
Pyruvate dehydrogenase activity and poly-β-hydroxybutyrate accumulation in strains CCGM1 and CCGM7
| Strain | PDH activity (nmol NADH min−1 mg of protein−1)a,b | PHB accumulation (mg PHB mg of protein−1)a,c |
||
|---|---|---|---|---|
| 1st subculture | 2nd subculture | 3rd subculture | ||
| CCGM1 | 18.07 ± 1.15 | 0.072 ± 0.001 | 0.674 ± 0.12 | 3.464 ± 0.22 |
| CCGM7 | 68.80 ± 1.43 | 0.468 ± 0.03 | 0.376 ± 0.05 | 0.463 ± 0.05 |
| CIAT652 | 20.37 ± 0.20 | 0.061 ± 0.01 | 2.055 ± 0.03 | 4.161 ± 0.12 |
Values are averages ± standard deviations (n = 3).
The cells were grown for 8 h in minimal medium with 10 mM succinic acid and 10 mM ammonium chloride.
Measured at 24 h of growth of the 1st, 2nd, or 3rd subculture.
Strain CCGM1 exhibited unexpected stress sensitivity under normal storage conditions (PY dishes), losing viability after short periods and turning black. The latter response may be due to melanin production (data not shown). Accordingly, we were interested in comparing the phenotypes of this strain under varying stress conditions involving temperature, salt, chloride, and pH. CCGM1 was more sensitive to temperature, salt, an alkaline pH, and chloride than the closely related strain CIAT652 (see Table S2 in the supplemental material). CCGM7 showed some sensitivity to acid pH and chloride stresses.
Genomic features of strains CCGM1 and CCGM7.
We obtained and analyzed the draft genomes of strains CCGM1 and CCGM7 (Table 4; Fig. 7) and investigated specific genetic features related to symbiosis and occupancy of the seeds.
TABLE 4.
Main features of the genomes of strains CCGM1 and CCGM7
| Featurea | Value for: |
|
|---|---|---|
| CCGM1 | CCGM7 | |
| Chromosome size (Mb) | 4.48 | 6.80 |
| Plasmid size (Mb) | 2.39 | |
| G + C content (%) | 60.1 | 61.8 |
| No. of CDS: | ||
| In the chromosome | 4,243 | 6,626 |
| In the plasmids | 2,185 | |
| CDS with assigned function (%) | 68 | 68 |
| Avg CDS length (bp) | 892 | 875 |
| Coding density (%) | 93.6 | 97.4 |
| No. of: | ||
| Scaffolds in the final assembly | 55 | 99 |
| Orthologs shared with the group | 5,848 | 5,346 |
| CDS not shared with the group | 580 | 1,280 |
CDS, coding sequences.
FIG 7.
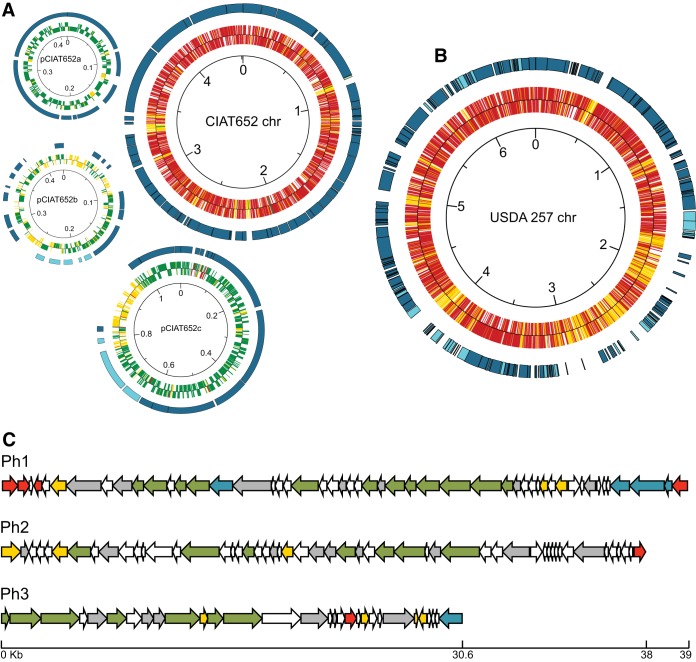
Schematic representations of the genome and prophages of strain CCGM1 and the genome of strain CCGM7. (A) Assembled genome of strain CCGM1 based on the CIAT652 backbone (chromosome and plasmids). For the chromosome (chr), the inner circle shows the prediction of open reading frames for the plus (upper) and minus (lower) strands of CCGM1 (red, homologs with the CIAT652 chromosome; green, homologs with CIAT652 plasmids; yellow, nonhomologous open reading frames), and the outer circle shows segments of homology with direct orientation (dark blue). For plasmids, the inner circles show open reading frame predictions for CCGM1 (green, homologs with the CIAT652 plasmid pCIAT652a, pCIAT652b, or pCIAT652c; red, homologs with the CIAT652 chromosome; yellow, nonhomologous open reading frames), and the outer circles show segments of homology with direct (dark blue) or inverse (light blue) orientation. The scale is in megabases. (B) Assembled genome of strain CCGM7 based on the USDA257 chromosome. The inner circle shows open reading frame predictions for CCGM7 in the plus (upper) and minus (lower) strands (red, homologs with the USDA257 chromosome; yellow, nonhomologous open reading frames). The outer circle shows segments of homology with direct (dark blue) or inverse (light blue) orientation. The scale is in megabases. Both panels A and B were drawn with GenVision. (C) Structures of predicted CCGM1 prophages. Shown are open reading frame functional predictions for prophage structure (green arrows), lysis (yellow arrows), mobilization (blue arrows), regulation of transposition (red arrows), and other functions (gray arrows), as well as open reading frames with hypothetical functions (white arrows). The scale is in kilobases.
Thus, of 6,428 potential genes in CCGM1, 580 were not shared with close strains (see Table S3 in the supplemental material). Among the unshared genes were those for the synthesis of phenazine (an antibiotic with redox activity, involved in signaling) (38), queuosine (a cytoskeletal modifier for successful symbiosis) (39), a cellulose synthase-cellulosome anchoring system, a retron transcriptase, and several pairs of toxin-antitoxin products, such as vapBC and doc-phd. We found three prophages: CCGM1 prophage 1 (CCGM1-Ph1) qualified as intact and belongs to the FluMu type, while CCGM1-Ph2 qualified as incomplete and CCGM-Ph3 as questionable (Fig. 7C; see also Table S4 in the supplemental material). Interestingly, when lawns of strain CCGM1 were mixed on petri dishes with the PY medium for CIAT652 growth (6 h, without cells), lysis plaques appeared (data not shown). This phenotype was not observed when CFN42 dishes were used.
The CCGM7 chromosome showed synteny with that of Sinorhizobium fredii strain USDA257, but some stretches showed no homology (Fig. 7B). Instead, these stretches showed homology to segments of plasmids pNGR234b and pNGR234a (data not shown). Three repABC gene copies, responsible for plasmid replication, were found. CCGM7 showed several copies of fixN (n = 2), fixK (n = 2), nodA (n = 3), and nodD (n = 5) (see Table S5 in the supplemental material). However, compared with NGR234, noeEIJKL and nolBTUVWX, which are important for a wide nodulation range, were absent. Other features of strain CCGM7 were a nifV gene (for homocitrate synthase), present only in free-living diazotrophs; a hydrogenase cluster; a complete RubisCO cluster (cbbAEFLPRSTX), also present in the photosynthetic strains Bradyrhizobium sp. strain ORS278 and Bradyrhizobium sp. strain BTAi1; and a large number of genes for secretion systems: 24 genes belonging to type II, 12 to type III, 21 to type IV, and 7 to the newly identified secretion system type VI, which contains a spike-like protein homolog to the T4 phage tail and is involved in pathogenesis or interactions with eukaryotes (40).
Other bacteria isolated from seeds and nodules of noninoculated plants.
Other types of bacteria were also recovered from the seeds, nodules, and xylem of noninoculated P. vulgaris plants. These strains, which were identified using 16S rRNA sequencing (data not shown), belonged to diverse species, such as Acinetobacter, Brevundimonas (formerly Pseudomonas), Burkholderia, Curtobacterium, Microbacterium, Paracoccus, Pseudomonas, and Stenotrophomonas spp.
DISCUSSION
Several studies have addressed the presence of bacteria inhabiting diverse plant parts, such as nodules, roots, stems, leaves, flowers, and seeds (for a recent review, see reference 41). We report the isolation of efficient, nitrogen-fixing rhizobia from the interiors of legume seeds. This finding reveals the potential for self-inoculated crops and has evolutionary implications given the vertical transmission of rhizobia in seeds.
We first found that seeds carry rhizobial strains after careful management of noninoculated bean plants grown in N-free medium in order to avoid external contamination. Then the main focus of the work was to identify and analyze these strains. All these strains were isolated from nodules (with plasmid profiles not seen previously) and were not introduced by experimentation in the greenhouse. We tested two strains and found that they were able to establish adequate symbiosis with the plant: they fixed nitrogen and promoted plant growth. Also, we assessed the phylogenetic distribution of the strains and found that they belonged to diverse phylogenetic branches that are known to establish symbiosis with common beans, such as Rhizobium leguminosarum, R. grahamii, and Sinorhizobium americanum (Fig. 2); however, we did not find R. etli strains, which are the most common symbiont of common beans in Mexican fields (42). This may have been due to insufficient sampling, or that species may be sensitive to the conditions of passage through the plant and seeds.
The plasmid profiles revealed that despite the common presence of high-molecular-size plasmids in rhizobia (43), S. americanum strain CCGM7 did not present plasmids (Fig. 3). Yet, by genomic analysis, several DNA stretches of plasmid origin were found in its chromosome; the main homology corresponded to the plasmids of S. fredii NGR234. This revealed the complex evolutionary history of these segments. For the other strains, the plasmid arrangement was unusual, because they appeared as pairs differing in just one plasmid: CCGM1-CCGM6, CCGM4-CCGM5, and CCGM8-CCGM9 (Fig. 3). Possibly this is related to global resemblance between the strains of each pair. We are analyzing their genomic characteristics, and the findings will be published elsewhere.
Serial growth determination in minimal medium (MM) is based on our experience with other rhizobial strains. We have studied the growth phenotypes and know that biotin synthesis and thiamine synthesis are important determinants for the maintenance of an active tricarboxylic acid (TCA) cycle and carbon assimilation (17, 44, 45). In MM serial growth, most of the rhizobia, which are biotin auxotrophs, synthesize the polymer PHB and, in doing so, limit growth and determine the capability for nitrogen fixation in symbiosis. Strains of S. meliloti, R. etli, and R. phaseoli are biotin auxotrophs, because several genes of this pathway are lacking, and show growth decline (46, 47). The R. phaseoli strain CCGM1 is also a biotin auxotroph that showed similar growth behavior and accumulated PHB. It is important that an R. etli mutant unable to synthesize PHB accumulated reducing power and organic acids that resulted in the inhibition of PDH activity (48). However, the S. americanum strain CCGM7 grew optimally with maximum yields, presented high PDH activity, and did not accumulate PHB (Fig. 6; Table 3). Moreover, CCGM7 has a complete set of genes for biotin synthesis (bioABDFMNSY). It is a matter of speculation whether the optimal flow of carbon and the accumulation of reducing power in strain CCGM7 are responsible for the optimal efficiency of the symbiotic nitrogen fixation process, even when few but larger nodules are formed. By the same token, the presence of the genes for the RubisCO complex in this strain may provide a new way to balance the generation of energy excess through the spillover of reducing power, as was proposed to occur in the Bradyrhizobium sp. strain ORS278–Aechynomene symbiosis, in which a mutant lacking RubisCO showed impaired symbiotic nitrogen fixation (49).
The synthesis of homocitrate, a function known to be present only in free-living diazotrophs (50), is a unique trait of the strain and could be responsible for a potential ability to fix nitrogen under free-living conditions. It would be important to assess the ability of CCGM7 to persist in the soil, given that it has been isolated only from seeds. A close relative, the S. americanum type strain CFNEI156, which was isolated from soil by E. Martínez-Romero and colleagues, nodulated Acacia, Leucaena, and Phaseolus plants and presented two high-molecular-size plasmids (37). We are obtaining its genomic sequence in order to make comparisons with CCGM7. A main genomic characteristic of strain CCGM1 was the presence of prophages. Possibly, in this strain, the presence of prophages has played a role in the integration of new genes and functions. Other traits, such as the queuosine or the cellulosome anchoring system, possibly are important in the interaction with Phaseolus plants.
Other bacteria isolated from the seeds and nodules of noninoculated plants included species of Curtobacterium, the agent of wilt disease in bean seeds (51, 52); Microbacterium, which has been reported to have important effects on bacterial signaling, such as quorum quenching (53); and Brevundimonas, a common endophyte and environmental bacterium (54).
Nitrogen-fixing endophytes in seed legumes, such as those reported here for the common bean, represent a potential biotechnological tool for crop improvement. We consider that increasing the number of cells within the seeds could perhaps enable their use as autoinoculated seeds. The vertical transmission of symbiotic bacteria in plant and animal models is an important research field (55). It is necessary to know the determinants and the mechanism and genes responsible for this capability, in order to increase the rhizobial population in seeds and to reach a typical nitrogen-fixing symbiosis. Also, testing of mutant strains unable to reach the seeds will provide information about the necessary functions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sandra Contreras and Oliver Castillo for technical help, Jadaú Sánchez and José Luis Zitlalpopoca for plant assays, Rosa María Ocampo and Paz Salas for Eckhardt profiles, and Luis Lozano for help in bioinformatics.
This work was partially supported by DGAPA-UNAM grant IN205113 and CONACYT grants 213606 and 152776.
We declare that no conflict of interest exists.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01491-14.
REFERENCES
- 1.Aserse AA, Räsänen LA, Assef F, Hailemariam A, Lindström K. 2012. Phylogeny and genetic diversity of native rhizobia nodulating common bean (Phaseolus vulgaris L.) in Ethiopia. Syst. Appl. Microbiol. 35:120–131. 10.1016/j.syapm.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 2.Pflughoeft KJ, Versalovic J. 2012. Human microbiome in health and disease. Annu. Rev. Pathol. 7:99–122. 10.1146/annurev-pathol-011811-132421 [DOI] [PubMed] [Google Scholar]
- 3.Hardoim PR, van Overbeek LS, van Elsas JD. 2008. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16:463-471. 10.1016/j.tim.2008.07.008 [DOI] [PubMed] [Google Scholar]
- 4.Truyens S, Weyens N, Cuypers A, Vangronsveld J. 2012. Changes in the population of seed bacteria of transgenerationally Cd-exposed Arabidopsis thaliana. Plant Biol. (Stuttg.) 15:971–981. 10.1111/j.1438-8677.2012.00711.x [DOI] [PubMed] [Google Scholar]
- 5.Mundt JO, Hinkle NF. 1976. Bacteria within ovules and seeds. Appl. Environ. Microbiol. 32:694–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herridge DF, Peoples MB, Boddey RM. 2008. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311:1–18. 10.1007/s11104-008-9668-3 [DOI] [Google Scholar]
- 7.Guan SH, Gris C, Cruveiller S, Pouzet C, Tasse L, Leru A, Maillard A, Médigue C, Batut J, Masson-Boivin C, Capela D. 2013. Experimental evolution of nodule intracellular infection in legume symbionts. ISME J. 7:1367–1377. 10.1038/ismej.2013.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sturz AV, Christie BR, Matheson BG, Nowak J. 1997. Biodiversity of endophytic bacteria which colonize red clover nodules, roots, stems and foliage and their influence on host growth. Biol. Fertil. Soils 25:13–19. 10.1007/s003740050273 [DOI] [Google Scholar]
- 9.López-López A, Rogel MA, Ormeño-Orrillo E, Martínez-Romero J, Martínez-Romero E. 2010. Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst. Appl. Microbiol. 33:322–327. 10.1016/j.syapm.2010.07.005 [DOI] [PubMed] [Google Scholar]
- 10.Michiels J, Dombrecht B, Vermeiren N, Xi C, Luyten E, Vanderleyden J. 1998. Phaseolus vulgaris is a non-selective host for nodulation. FEMS Microbiol. Ecol. 26:193–205. 10.1111/j.1574-6941.1998.tb00505.x [DOI] [Google Scholar]
- 11.Herrera-Cervera JA, Caballero-Mellado J, Laguerre G, Tichy HV, Requena N, Amarger N, Martínez-Romero E, Olivares J, Sanjuan J. 1999. At least five rhizobial species nodulate Phaseolus vulgaris in a Spanish soil. FEMS Microbiol. Ecol. 30:87–97. 10.1111/j.1574-6941.1999.tb00638.x [DOI] [Google Scholar]
- 12.Bromfield ESP, Barran LR. 1990. Promiscuous nodulation of Phaseolus vulgaris, Macroptilium atropurpureum and Leucaena leucocephala by indigenous Rhizobium meliloti. Can. J. Microbiol. 36:369–372. 10.1139/m90-065 [DOI] [Google Scholar]
- 13.Sadowsky MJ, Cregan PB, Keyser HH. 1988. Nodulation and nitrogen fixation efficacy of Rhizobium fredii with Phaseolus vulgaris genotypes. Appl. Environ. Microbiol. 54:1907–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma PK, Sarita S, Prell J. 2005. Isolation and characterization of an endophytic bacterium related to Rhizobium/Agrobacterium from wheat (Triticum aestivum L.) roots. Curr. Sci. 89:608–610 [Google Scholar]
- 15.Stoltzfus JR, So R, Malarvithi PP, Ladha JK, de Bruijn FJ. 1997. Isolation of endophytic bacteria from rice and assessment of their potential for supplying rice with biologically fixed nitrogen. Plant Soil 194:25–36. 10.1023/A:1004298921641 [DOI] [Google Scholar]
- 16.Gutiérrez-Zamora ML, Martínez-Romero E. 2001. Natural endophytic association between Rhizobium etli and maize (Zea mays L.). J. Biotechnol. 91:117–126. 10.1016/S0168-1656(01)00332-7 [DOI] [PubMed] [Google Scholar]
- 17.Encarnación S, Dunn M, Willms K, Mora J. 1995. Fermentative and aerobic metabolism in Rhizobium etli. J. Bacteriol. 177:3058–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brom S, García de los Santos A, Stepkowsky T, Flores M, Dávila G, Romero D, Palacios P. 1992. Different plasmids of Rhizobium leguminosarum bv. phaseoli are required for optimal symbiotic performance. J. Bacteriol. 174:5183–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652. 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hynes MF, McGregor NF. 1990. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol. Microbiol. 4:567–574. 10.1111/j.1365-2958.1990.tb00625.x [DOI] [PubMed] [Google Scholar]
- 21.Simon R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol. Gen. Genet. 196:413–420. 10.1007/BF00436188 [DOI] [PubMed] [Google Scholar]
- 22.Fahraeus G. 1957. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16:374–381. 10.1099/00221287-16-2-374 [DOI] [PubMed] [Google Scholar]
- 23.Peralta H, Mora Y, Salazar E, Encarnación S, Palacios R, Mora J. 2004. Engineering the nifH promoter region and abolishing poly-β-hydroxybutyrate accumulation in Rhizobium etli enhance nitrogen fixation in symbiosis with Phaseolus vulgaris. Appl. Environ. Microbiol. 70:3272–3281. 10.1128/AEM.70.6.3272-3281.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steel RG, Torrie JH. 1980. Principles and procedures for statistics: a biometrical approach, 2nd ed. McGraw-Hill, New York, NY [Google Scholar]
- 25.Vincent JM. 1970. A manual for the practical study of root-nodule bacteria, p 73–97 International Biological Programme Handbook, no. 15. Blackwell Scientific Publications, Ltd, Oxford, United Kingdom [Google Scholar]
- 26.Karr DB, Waters JK, Suzuki F, Emerich DW. 1984. Enzymes of the poly-β-hydroxybutyrate and citric acid cycles of Rhizobium japonicum bacteroids. Plant Physiol. 75:1158–1162. 10.1104/pp.75.4.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law JH, Slepecky RA. 1961. Assay of poly-β-hydroxybutyric acid. J. Bacteriol. 82:33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13:R56. 10.1186/gb-2012-13-6-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felsenstein J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 34.Perrière G, Gouy M. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369. 10.1016/0300-9084(96)84768-7 [DOI] [PubMed] [Google Scholar]
- 35.Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39:W347–W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toledo I, Lloret L, Martínez-Romero E. 2003. Sinorhizobium americanus sp. nov., a new Sinorhizobium species nodulating native Acacia spp. in Mexico. Syst. Appl. Microbiol. 26:54–64. 10.1078/072320203322337317 [DOI] [PubMed] [Google Scholar]
- 38.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. 2008. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321:1203–1206. 10.1126/science.1160619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchetti M, Capela D, Poincloux R, Benmeradi N, Auriac MC, Le Ru A, Maridonneau-Parini I, Batut J, Masson-Boivin C. 2013. Queuosine biosynthesis is required for Sinorhizobium meliloti-induced cytoskeletal modifications on HeLa cells and symbiosis with Medicago truncatula. PLoS One 8:e56043. 10.1371/journal.pone.0056043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 104:15508–15513. 10.1073/pnas.0706532104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64:807–838. 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- 42.Hernández-Lucas I, Segovia L, Martínez-Romero E, Pueppke SG. 1995. Phylogenetic relationships and host range of Rhizobium spp. that nodulate Phaseolus vulgaris L. Appl. Environ. Microbiol. 61:2775–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.González V, Acosta JL, Santamaría RI, Bustos P, Fernández JL, Hernández-González IL, Díaz R, Flores M, Palacios R, Mora J, Dávila G. 2010. Conserved symbiotic plasmid DNA sequences in the multireplicon pangenomic structure of Rhizobium etli. Appl. Environ. Microbiol. 76:1604–1614. 10.1128/AEM.02039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunn MF, Encarnación S, Araíza G, Vargas MC, Dávalos A, Peralta H, Mora Y, Mora J. 1996. Pyruvate carboxylase from Rhizobium etli: mutant characterization, nucleotide sequence, and physiological role. J. Bacteriol. 178:5960–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Encarnación S, Vargas MDC, Dunn CM, Dávalos A, Mendoza G, Mora Y, Mora J. 2002. AniA regulates reserve polymer accumulation and global protein expression in Rhizobium etli. J. Bacteriol. 184:2287–2295. 10.1128/JB.184.8.2287-2295.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Streit WR, Joseph CM, Phillips DA. 1996. Biotin and other water-soluble vitamins are key growth factors for alfalfa root colonization by Rhizobium meliloti 1021. Mol. Plant Microbe Interact. 9:330–338. 10.1094/MPMI-9-0330 [DOI] [PubMed] [Google Scholar]
- 47.Guillén-Navarro K, Encarnación S, Dunn MF. 2005. Biotin biosynthesis, transport and utilization in rhizobia. FEMS Microbiol. Lett. 246:159–165. 10.1016/j.femsle.2005.04.020 [DOI] [PubMed] [Google Scholar]
- 48.Cevallos MA, Encarnación S, Leija A, Mora Y, Mora J. 1996. Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-β-hydroxybutyrate. J. Bacteriol. 178:1646–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gourion B, Delmotte N, Bonaldi K, Nouwen N, Vorholt JA, Giraud E. 2011. Bacterial RuBisCO is required for efficient Bradyrhizobium/Aeschynomene symbiosis. PLoS One 6:e21900. 10.1371/journal.pone.0021900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terpolilli JJ, Hood GA, Poole PS. 2012. What determines the efficiency of N2-fixing Rhizobium-legume symbioses? Adv. Microb. Physiol. 60:325–389. 10.1016/B978-0-12-398264-3.00005-X [DOI] [PubMed] [Google Scholar]
- 51.Collins MD, Jones D. 1983. Reclassification of Corynebacterium flaccumfaciens, Corynebacterium betae, Corynebacterium oortii and Corynebacterium poinsettiae in the genus Curtobacterium, as Curtobacterium flaccumfaciens comb. nov. J. Gen. Microbiol. 129:3545–3548 [Google Scholar]
- 52.de Souza VL, Maringoni AC, Morais-Carbonell SA, Ito MF. 2006. Resistência genética em genótipos de feijoeiro a Curtobacterium flaccumfaciens pv. flaccumfaciens. Summa Phytopathol. 32:339–344. 10.1590/S0100-54052006000400004 [DOI] [Google Scholar]
- 53.Morohoshi T, Someya N, Ikeda T. 2009. Novel N-acylhomoserine lactone-degrading bacteria isolated from the leaf surface of Solanum tuberosum and their quorum-quenching properties. Biosci. Biotechnol. Biochem. 73:2124–2127. 10.1271/bbb.90283 [DOI] [PubMed] [Google Scholar]
- 54.Thomas P, Kumari S, Swarna GK, Gowda TK. 2007. Papaya shoot tip associated endophytic bacteria isolated from in vitro cultures and host-endophyte interaction in vitro and in vivo. Can. J. Microbiol. 53:380–390. 10.1139/W06-141 [DOI] [PubMed] [Google Scholar]
- 55.Watanabe K, Yukuhiro F, Matsuura Y, Fukatsu T, Noda H. 2014. Intrasperm vertical symbiont transmission. Proc. Natl. Acad. Sci. U. S. A. 111:7433–7437. 10.1073/pnas.1402476111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenberg C, Hughet T. 1984. The pAtC58 plasmid of Agrobacterium tumefaciens is not essential for tumor induction. Mol. Gen. Genet. 196:533–536. 10.1007/BF00436205 [DOI] [Google Scholar]
- 57.Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459–472. 10.1016/0022-2836(69)90288-5 [DOI] [PubMed] [Google Scholar]
- 58.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 59.Ohkuma M, Noda S, Usami R, Horikoshi K, Kudo T. 1996. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:2747–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khamis A, Colson P, Raoult D, Scola BL. 2003. Usefulness of rpoB gene sequencing for identification of Afipia and Bosea species, including a strategy for choosing discriminative partial sequences. Appl. Environ. Microbiol. 69:6740–6749. 10.1128/AEM.69.11.6740-6749.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.López I, Ruiz-Larrea F, Cocolin L, Orr E, Phister T, Marshall M, VanderGheynst J, Mills DA. 2003. Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:6801–6807. 10.1128/AEM.69.11.6801-6807.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.