Abstract
This study determined the effects of specific metabolic traits of Lactobacillus reuteri on its competitiveness in sourdoughs. The competitiveness of lactobacilli in sourdough generally depends on their growth rate; acid resistance additionally contributes to competitiveness in sourdoughs with long fermentation times. Glycerol metabolism via glycerol dehydratase (gupCDE) accelerates growth by the regeneration of reduced cofactors; glutamate metabolism via glutamate decarboxylase (gadB) increases acid resistance by generating a proton motive force. Glycerol and glutamate metabolisms are lineage-specific traits in L. reuteri; therefore, this study employed glycerol dehydratase-positive sourdough isolates of human-adapted L. reuteri lineage I, glutamate decarboxylase-positive strains of rodent-adapted L. reuteri lineage II, as well as mutants with deletions in gadB or gupCDE. The competitivenesses of the strains were quantified by inoculation of wheat and sorghum sourdoughs with defined strains, followed by propagation of doughs with a 10% inoculum and 12-h or 72-h fermentation cycles. Lineage I L. reuteri strains dominated sourdoughs propagated with 12-h fermentation cycles; lineage II L. reuteri strains dominated sourdoughs propagated with 72-h fermentation cycles. L. reuteri 100-23ΔgadB was outcompeted by its wild-type strain in sourdoughs fermented with 72-h fermentation cycles; L. reuteri FUA3400ΔgupCDE was outcompeted by its wild-type strain in sourdoughs fermented with both 12-h and 72-h fermentation cycles. Competition experiments with isogenic pairs of strains resulted in a constant rate of strain displacement of the less competitive mutant strain. In conclusion, lineage-specific traits of L. reuteri determine the competitiveness of this species in sourdough fermentations.
INTRODUCTION
Sourdough fermentation with lactic acid bacteria has been used as a leavening agent in artisanal baking, and sourdough or sourdough products are increasingly being applied as a baking improver in industrial baking (1). In artisanal as well as industrial practice, sourdoughs are generally maintained by continuous back-slopping. The formation of a stable sourdough microbiota consisting of lactic acid bacteria alone or in association with yeasts depends on complex interactions between the inoculum, the cereal substrate, processing parameters, and the environment (2). Type I doughs are propagated by frequent inoculation at ambient temperatures. The use of type I sourdoughs as leavening agents requires fermentation processes that maintain continuous metabolic activity and CO2 formation by fermentation microbiota (1). Type I doughs propagated in temperate climates are generally dominated by Lactobacillus sanfranciscensis (3, 4), but type I doughs propagated in tropical climates harbor thermophilic lactobacilli, including Lactobacillus reuteri (5, 6). The frequent occurrence of L. sanfranciscensis has been attributed to the rapid growth of this organism, the efficient use of maltose as a carbon source, the use of fructose as an electron acceptor, and its small genome size (7–10). In contrast, type II doughs are used as baking improvers to modify flavor, texture, or shelf life in industrial processes in the form of active dough or dough that has been stabilized by drying or pasteurization (1). The elevated temperatures, long fermentation times, and high water content select for thermophilic and acid-resistant organisms (9, 11, 12). L. reuteri prevails in type I and type II sourdoughs fermented at elevated temperatures (2, 4, 5).
L. reuteri not only occurs in food fermentations but is known predominantly for its lifestyle as a vertebrate gut symbiont (13). L. reuteri is associated with humans, pigs, rodents, and different species of birds. Strains of L. reuteri have evolved into phylogenetically distinct and host-adapted lineages (13), with lineage-specific metabolic and genetic traits reflecting adaptation to different hosts (14). Some rodent-lineage strains are acid resistant due to the presence of the glutamate decarboxylase GadB (12, 15). Human-lineage strains convert glycerol or 1,2-propanediol to regenerate NAD+ (14, 16, 17). Sourdough isolates of L. reuteri do not represent extraintestinal lineages but can be assigned to host-adapted lineages, reflecting their long-term association with intestinal ecosystems prior to adopting an alternative lifestyle as a fermentation organism in sourdough (18). The genetics, metabolism, and phylogeny of L. reuteri have been studied extensively (14, 19, 20), making it an excellent model organism to identify strain-specific metabolic traits that affect competitiveness in sourdough.
Previous studies suggested that specific metabolic traits of lactic acid bacteria influence their growth rate in cereal substrates and determine their competitiveness in artisanal and industrial sourdough fermentations (8, 10, 12). However, experimental validation of the effects of specific metabolic traits of lactobacilli on their competitiveness in sourdough is generally lacking. It was therefore the aim of this study to determine whether lineage-specific metabolic traits contribute to the competitiveness of L. reuteri in type I and type II sourdoughs. Experiments were focused on glycerol metabolism, which may influence the growth rate in cereal substrates, and glutamate metabolism, which influences acid resistance in sourdough. Experiments evaluated the competitiveness of sourdough isolates of L. reuteri representing different host-adapted lineages that differ in glycerol and glutamate metabolism in wheat and sorghum sourdoughs. Furthermore, the competitiveness of isogenic mutants with disruptions in glycerol and glutamate metabolism was compared to that of the cognate wild-type strains.
MATERIALS AND METHODS
Sourdough fermentation.
Previously, seven strains of L. reuteri isolated from rye, wheat, and sorghum sourdoughs were assigned to three host lineages (18). To investigate the competitiveness of L. reuteri, this study used five strains: L. reuteri LTH5448 (rye/rodent-adapted lineage I), L. reuteri FUA3400 and FUA3401 (wheat/human-adapted lineage II), and L. reuteri FUA3168 and FUA3324 (sorghum/undefined lineage). Sourdough isolates of rodent-adapted lineage III were not suitable for use in competition experiments because they produce reutericyclin, an antimicrobial compound that inhibits other strains of L. reuteri (21). Strains were inoculated in modified MRS (mMRS) broth (8) and incubated at 37°C overnight. Two culture cocktails were prepared as biological repeats by mixing equal volumes of one strain from each origin to obtain approximately equal cell counts at the time of inoculation. Cocktail 1 contained L. reuteri FUA5448, FUA3400, and FUA3168, and cocktail 2 contained L. reuteri FUA5448, FUA3401, and FUA3324. Whole wheat and white sorghum sourdoughs were prepared with commercial flours and a dough yield (100 × weight [flour + water]/weight [flour]) of 200; sourdoughs were propagated at 37°C by back-slopping every 12 or 72 h with 10% of the ripe sourdoughs. Sourdough samples were taken after inoculation and at the end of each fermentation cycle for analysis, as outlined below. Two independent experiments with each cocktail were performed.
To determine the roles of glycerol and glutamate metabolism in sourdough ecology, sourdough was inoculated with equal cell counts of L. reuteri FUA3400 and L. reuteri FUA3400ΔgupCDE or L. reuteri 100-23 and L. reuteri 100-23ΔgadB. L. reuteri FUA3400ΔgupCDE, which lacks the ability to use glycerol as an electron acceptor, was generated in this study (see below); the generation of L. reuteri 100-23ΔgadB with a disruption in the gene coding for glutamate decarboxylase was described previously (12). Competition experiments in sourdough were performed as described above, in triplicate independent experiments.
Generation of the L. reuteri FUA3400ΔgupCDE mutant.
The gupCDE gene (20) in L. reuteri FUA3400 was truncated by using pJRS233 (22) according to a deletion strategy described previously (12). The 5′-flanking fragment of gupCDE was amplified from genomic DNA of L. reuteri FUA3400 by using primers 5′-GGA GGT CGA CAG GCT TCA GTT GAT GCC GGA G-3′ and 5′-ACC ATG CAT TGG GGT ACC TTA AAC AAA TGT ATC TTG ATG AAT TGG-3′. The 3′-flanking fragment was amplified by using primers 5′-CTG GTA CCT ATG AAA GTC GTA AGA AGC TAA AGG GCG ATA ACT AA-3′ and 5′-CAA ATG CAT CGG ATC CCT TTC CTG TAA GAT CTG CCA TTG TTT-3′. The 5′-flanking fragment was ligated into the pGEMTeasy vector (Promega) to generate pGUP-A. Plasmid pGUP-A and the 3′-flanking fragment were digested with the restrictive enzymes KpnI and NsiI, purified, and ligated to create pGUP-AB. The DNA fragment in pGUP-AB was digested with SalI and BamHI and ligated into pJRS233. The resulting plasmid, pGUP-KO, was electrotransformed into competent L. reuteri FUA3400 cells. Transformants were grown in mMRS-erythromycin broth (5 mg/liter) at 42°C to 44°C for 80 generations to select for single-crossover mutants. L. reuteri with pGUP-KO AB integrated into the chromosome was cured by culturing in mMRS broth at 37°C for 100 generations. The culture was plated onto mMRS agar, and erythromycin-sensitive double-crossover mutants were identified by replica plating onto mMRS and mMRS-erythromycin agar. The truncation of gupCDE in L. reuteri FUA3400ΔgupCDE was confirmed by PCR and sequencing. The phenotype was confirmed by the absence of 1,3-propanediol production, as determined by high-performance liquid chromatography (HPLC) (6), and the absence of 3-hydroxypropionaldehyde (reuterin) production, as determined by a colorimetric assay (23).
Culture-dependent quantification of sourdough microbiota.
Sourdough samples were homogenized and diluted with a 0.8% (wt/vol) NaCl solution. L. reuteri cells were enumerated after surface plating onto mMRS agar. Plates were incubated anaerobically at 37°C for 24 h. When the individual strains used in the respective strain cocktails could be differentiated on the basis of their colony morphology, strains were differentially enumerated. The pH of sourdough was measured with a glass electrode; the sorghum sourdough pHs were 3.44 ± 0.03 and 3.38 ± 0.09 after 12 and 72 h of fermentation, respectively, and the wheat sourdough pHs were 3.40 ± 0.06 and 3.46 ± 0.09 after 12 and 72 h of fermentation, respectively.
Differentiation of L. reuteri strains by using a molecular beacon.
Two grams of sourdough was homogenized with 80 ml of 0.8% (wt/vol) saline. The homogenate was centrifuged at 1,500 rpm for 5 min to remove solids. The cells were harvested by centrifugation at 5,000 rpm for 15 min. DNA was extracted with a DNeasy blood and tissue kit (Qiagen, USA) according to the manufacturer's instructions.
A molecular beacon was designed to distinguish strains from different lineages based on differences in melting temperatures (24). The primers used were forward (limiting) primer 5′-AAT ATG CAG AAG CCT TAG-3′ and reverse (excess) primer 5′-TAT CAC CCA TAT CAC CAT-3′. The sequence of the molecular beacon was 5′-CGC GAT CAT GAT TAC GAA AAC AAG TTT GTG GAT GGA TCG CG-3′ (the complementary sequences of the beacon stem are underlined). Each PCR mixture contained 8 µl TaqMan Fast Universal PCR master mix (2×), No AmpErase UNG (Applied Biosystems, Canada), 2 μM excess primer, 0.2 μM limiting primer, 0.2 μM molecular beacon, and bacterial DNA in a final volume of 16 μl. PCR was done with an Applied Biosystems 7500 Fast real-time PCR system (Life Technologies, USA). The PCR conditions were as follows: denaturation for 5 min at 95°C, followed by 60 cycles of denaturation at 95°C for 10 s, annealing at 50°C for 15 s, and extension at 72°C for 20 s. At the melting-curve stage, the temperature was held at 95°C for 2 min and at 25°C for 60 min and was then increased from 25°C to 90°C at 1°C/step, with a 25-s holding time at each step. The melting temperatures were 47°C for human-lineage, 53°C for rodent-lineage, and 61°C for sorghum strains.
Relative quantification of L. reuteri wild-type and mutant bacteria by quantitative PCR.
DNA was extracted from sourdough as described above, and cells of L. reuteri wild-type and mutant strains were quantified with strain-specific primers. The primers for wild types were gad-WT-F (5′-ATC TAG ATT ATC CTG CCA TAG ATA AAA-3′), gad-WT-R (3′-TAA AGC ACG AGC ATC ATT CG-3′), gup-WT-F (5′-GCA TTC GCA ACT GTT CTT GA-3′), and gup-WT-R (5′-ACT GTC GTC CCC TTT GAT TG-3′). The primers for mutant strains were gad-M-F (5′-AAA TTA ACC TAG GAG GTT TTA TCT ATG-3′), gad-M-R (5′-CAG GAC GCA GCA AAG AAG TA-3′), gup-M-F (5′-CTG TTA TGG CTG GAC GTG AA-3′), and gup-M-R (5′-TCG CCC TTT AGC TTC TTA CG-3′). Each PCR mixture contained 1× QuantiFast SYBR green PCR master mix (Qiagen, Canada), 1 μM each primer, and 1 μl bacterial DNA in a final volume of 20 μl. PCR was done on an Applied Biosystems 7500 real-time PCR instrument (Life Technologies, Canada). The PCR conditions were as follows: denaturation for 5 min at 95°C, followed by 60 cycles of denaturation at 95°C for 10 s and annealing at 56°C for 30 s. Standard curves for absolute quantification of the strains in the sourdoughs were generated with purified PCR products obtained for the respective strains, as described previously (25).
In vitro growth rate of L. reuteri strains.
The main sugars and electron acceptors for wheat and rye sourdoughs are maltose and sucrose; in contrast, glucose and glycerol are the most abundant substrates for use as a carbon source and as an electron acceptor, respectively, in sorghum sourdoughs (25). Modified MRS broth, which mimics sugar compositions of wheat and sorghum with and without an electron acceptor, was prepared with the following sugars as the sole carbon source: 10 g of maltose and 2 g of glucose (W1 medium); 10 g of maltose, 2 g of glucose, and 10 g of sucrose (W2 medium); 2 g of maltose and 10 g of glucose (S1 medium); and 2 g of maltose, 10 g of glucose, and 5 g of glycerol (S2 medium). mMRS-glucose medium, containing 10 g of glucose and lacking both maltose and electron acceptors, was used to prepare the precultures. Strains of L. reuteri were subcultured twice overnight in mMRS-glucose medium and grown to an optical density (at 600 nm) of 0.5, corresponding to the exponential phase of growth. mMRS medium and W1, W2, S1, or S2 medium were inoculated with a 10% inoculum of exponentially growing cultures and incubated in 96-well microtiter plates. The cultures were overlaid with 50 μl paraffin oil to maintain an anaerobic environment. The plate was cultured at 37°C for 24 h, and the optical density was measured every 30 min, with 10 s of shaking before each measurement. Growth rates were calculated by fitting the optical density data to the logistic growth curve (26).
Quantification of 1,3-propanediol concentrations in bacterial cultures and sourdoughs.
Bacterial cultures grown overnight or sourdough samples were homogenized in equal volumes of 7% HClO4 and incubated at 4°C overnight to precipitate proteins. The sample was centrifuged at 14,000 rpm for 10 min. The supernatant was diluted three times with water and then used for HPLC. The 1,3-propanediol concentration was quantified by using an Aminex HPX-87 column (300 mm by 7.8 mm; Bio-Rad, USA) based on refractive index detection (6). Samples were eluted with 5 mmol/liter H2SO4 at 70°C with a flow rate of 0.4 ml/min.
Statistics.
Data analysis was performed with the PROC MIXED procedure (SAS v.9.2; SAS Institute, USA), using two-way analysis of variance (ANOVA). A P value of ≤0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
The sequences of gupCDE and the truncated gene were deposited in GenBank under accession numbers KJ435307 to KJ435310.
RESULTS
Microbial ecology L. reuteri strains in wheat and sorghum sourdoughs.
To determine the effects of substrate and fermentation time on sourdough microbial ecology, wheat and sorghum sourdoughs were propagated every 12 or 72 h over 12 fermentation cycles. Culture-dependent analysis and the characterization of sourdough microbiota by high-resolution melting-curve quantitative PCR (HRM-qPCR) demonstrated that the L. reuteri strains used as inoculums accounted for >99% of the bacteria in all of the sourdoughs (Fig. 1 and Table 1). The rodent-lineage strains and sorghum isolates could not be distinguished from each other by colony morphology; they were enumerated together by culture-dependent quantification (Fig. 1) but were readily differentiated by HRM-qPCR (Table 1). The total cell count after 12 h of fermentation was about 1 log higher than the cell counts after 72 h of fermentation (data not shown). After 12 fermentation cycles with a 72-h fermentation time in wheat or sorghum sourdoughs, rodent-lineage strains were identified in high cell counts in all sourdoughs; sorghum isolates were additionally identified in two of the four doughs. In wheat or sorghum sourdoughs fermented with 12-h fermentation times, human-lineage strains were identified in high cell counts after 12 fermentation cycles; sorghum isolates were additionally present in the two of the four doughs (Fig. 1 and Table 1). These results imply that different lineage-specific metabolic traits account for the competitiveness of L. reuteri in sourdoughs with short or long fermentation times.
FIG 1.
Microbial compositions of wheat sourdoughs (A and C) and sorghum sourdoughs (B and D) that were inoculated with three L. reuteri strains. Sourdoughs were propagated with 72-h fermentation cycles (A and B) or 12-h fermentation cycles (C and D). Symbols indicate human-lineage L. reuteri FUA3400 (●) and L. reuteri FUA5448 or FUA3168 (○) for cocktail 1 and human-lineage L. reuteri FUA3401 (▼) and L. reuteri FUA5448 or FUA3324 (▽) for cocktail 2. Representative data from two independent experiments are shown.
TABLE 1.
Lineage-specific detection of L. reuteri in wheat and sorghum sourdoughs propagated with 72-h and 12-h fermentation cyclesd
| Fermentation time and sourdough mixture | Strain(s) of L. reuteri detected after indicated no. of fermentation cyclesc |
||||
|---|---|---|---|---|---|
| 0 | 4 | 8 | 10 | 12 | |
| 72 h | |||||
| Wheat, cocktail 1a | HRS | RS | HRS | RS | RS |
| Wheat, cocktail 2b | HRS | HRS | HRS | RS | RS |
| Sorghum, cocktail 1 | HRS | RS | RS | R | R |
| Sorghum, cocktail 2 | HRS | R | R | R | R |
| 12 h | |||||
| Wheat, cocktail 1 | HRS | HRS | HRS | HRS | HS |
| Wheat, cocktail 2 | HRS | HRS | H | H | H |
| Sorghum, cocktail 1 | HRS | HRS | HS | HS | HS |
| Sorghum, cocktail 2 | HRS | H | H | H | H |
Cocktail 1 was composed of L. reuteri FUA3400 (human lineage II) (H), L. reuteri FUA5448 (rodent lineage I) (R), and L. reuteri FUA3168 (undefined lineage) (S).
Cocktail 2 was composed of L. reuteri FUA3401 (human lineage II) (H), L. reuteri FUA5448 (rodent lineage I) (R), and L. reuteri FUA3324 (sorghum isolate, undefined lineage) (S).
Strains detected by observation of the lineage-specific melting peaks if they accounted for 1% or more of the total fermentation microbiota. Letters denote strains of the same host lineage.
Duplicate experiments were conducted with two strain cocktails. Lineage-specific detection of L. reuteri strains was achieved with HRM-qPCR.
In vitro growth rate of L. reuteri strains.
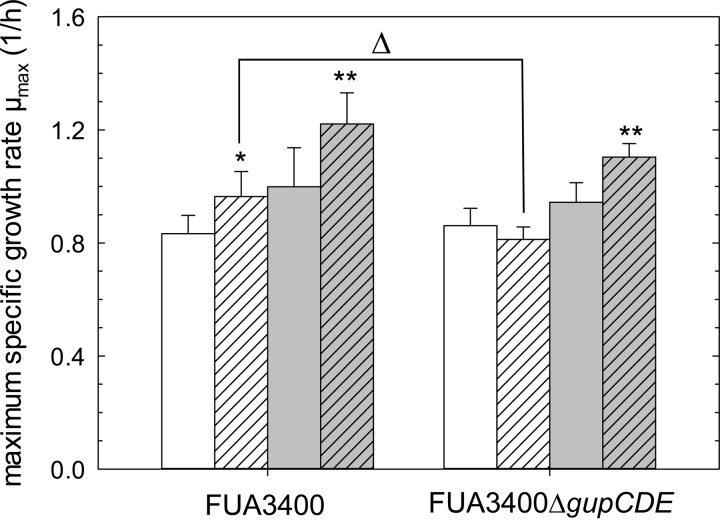
In order to determine the effect of electron acceptors on growth of L. reuteri, media were prepared to represent the main carbon sources in wheat and sorghum with sucrose and glycerol as electron acceptors (W2 and S2 media) and without electron acceptors (W1 and S1 media). For all strains, the addition of sucrose as the electron acceptor increased the growth rate (Fig. 2). In contrast, glycerol as the electron acceptor increased the growth rate of human-lineage strains only. For human-lineage strains, the increased growth rate was less pronounced irrespective of whether sugar or glycerol was the electron acceptor.
FIG 2.
Growth rates of L. reuteri strains in mMRS broth. White bars indicate glucose as a carbon source (S1 and S2 media), gray bars indicate maltose as a carbon source (W1 and W2 media), and hatched bars indicate the addition of electron acceptors (10 mmol/liter glycerol in S2 medium and 10 mmol/liter sucrose in W2 medium), between S1 and S2 media (*), between W1 and W2 media (**), and between the wild-type strain FUA3400 and the mutant strain FUA3400ΔgupCDE (Δ). Data shown are means ± standard deviations from triplicate independent experiments.
Generation of the FUA3400ΔgupCDE mutant.
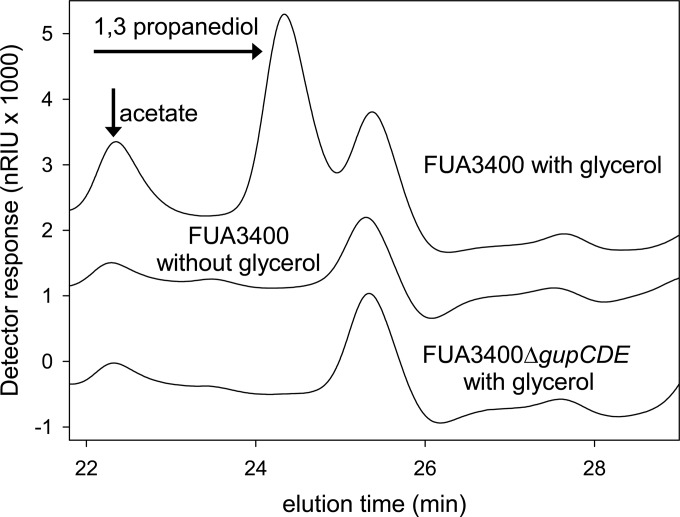
In human-lineage strains of L. reuteri, glycerol is converted to 1,3-propanediol by glycerol dehydratase and 1,3-propanediol dehydrogenase to regenerate NAD+ (16, 27). In this study, the glycerol dehydratase gene was truncated to disable glycerol metabolism in L. reuteri FUA3400 because of the possible presence of more than one 1,3-propanediol dehydrogenase in the genome (28). Truncation of the glycerol dehydratase avoids accumulation of the antimicrobially active intermediate reuterin; besides, the lack of propanediol dehydrogenase may be compensated for by other dehydrogenase enzymes with broad substrate specificity (29). The candidate genes in L. reuteri FUA3400 show high sequence similarity to their counterparts in L. reuteri JCM1112 and were thus also designated gupCDE (17). The sequences of gupCDE and the truncated gene were deposited in GenBank (accession numbers KJ435307 to KJ435310). L. reuteri FUA3400 quantitatively converted glycerol to 1,3-propanediol (Fig. 3 and data not shown). L. reuteri FUA3400ΔgupCDE did not convert glycerol to 1,3-propanediol, and glycerol supplementation of mMRS-glucose did not support acetate formation (Fig. 3). The colorimetric assay detected reuterin in cultures of L. reuteri FUA3400 but not in cultures of L. reuteri FUA3400ΔgupCDE. The growth rates of L. reuteri FUA3400 and FUA3400ΔgupCDE in media containing maltose, maltose and fructose, glucose, or glucose and glycerol (designated W1, W2, S1, and S2 media) were compared (Fig. 4). The addition of sucrose in W2 enhanced the growth rates of both strains, while the addition of glycerol in S2 increased the growth rate of L. reuteri FUA3400 but not the growth rate of L. reuteri FUA3400ΔgupCDE, indicating that glycerol metabolism had a positive effect on growth without affecting other key aspects of microbial metabolism.
FIG 3.
Separation of acetate and 1,3-propanediol in media fermented with L. reuteri FUA3400 or L. reuteri FUA3400ΔgupCDE. Where indicated, media were supplemented with 10 mmol/liter glycerol as an electron acceptor. Chromatograms were offset by 1,500 nRIU (nano-refractive index units).
FIG 4.
Growth rates of L. reuteri wild-type strain FUA3400 and L. reuteri mutant strain FUA3400ΔgupCDE in mMRS broth. White bars indicate glucose as a carbon source, matching the major carbon source in sorghum doughs (S1 and S2 media); gray bars indicate maltose as a carbon source, matching the major carbon source in wheat doughs (W1 and W2 media); and hatched bars indicate the addition of electron acceptors (10 mmol/liter glycerol in S2 medium and 10 mmol/liter sucrose in W2 medium). Symbols indicate significant differences (P < 0.05) between S1 and S2 media (*), between W1 and W2 media (**), and between the wild-type strain FUA3400 and the mutant strain FUA3400ΔgupCDE (Δ). Data shown are means ± standard deviations from triplicate independent experiments.
Role of glycerol and glutamate metabolism in short-term and long-term fermentations.
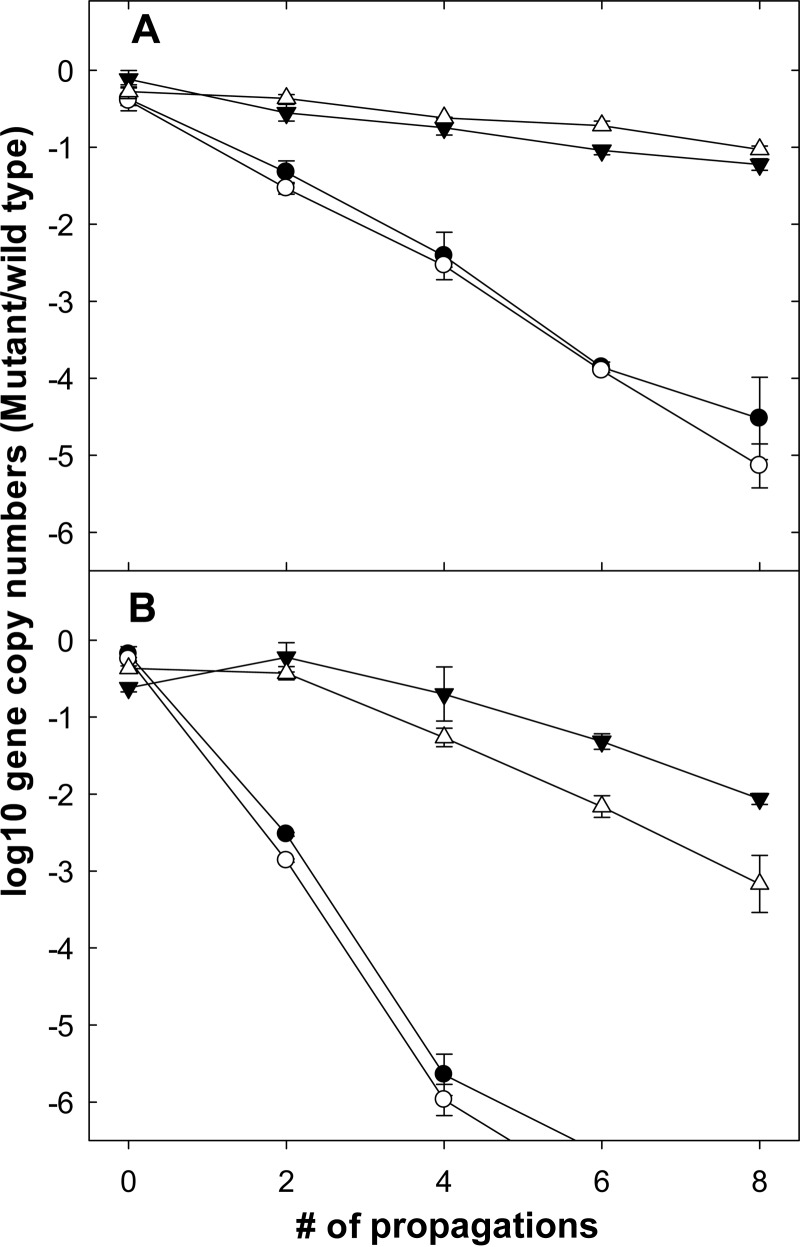
To investigate the role of glycerol and glutamate metabolisms during sourdough fermentation, wheat and sorghum sourdoughs were fermented by using two pairs of wild-type strains and their mutants. As described above, the total cell counts of all doughs fermented with 72-h fermentation cycles were about 10-fold lower than those of the doughs fermented for 12 h (data not shown), indicating cell death during extended incubation under acidic conditions (30). Disruption of the gupCDE genes impaired the growth of the mutant in both 12-h and 72-h cycles, but the strain disappeared faster in doughs fermented with 72-h cycles (Fig. 5). Disruption of gadB in L. reuteri 100-23 resulted in reduced competitiveness in doughs propagated with 72-h fermentation cycles. The wild type outcompeted the GadB mutant faster in wheat than in sorghum; however, the competitiveness of the mutant was not affected in doughs propagated with 12-h fermentation cycles.
FIG 5.
Microbial compositions in sorghum and wheat sourdough fermentations with 12-h and 72-h propagations. Symbols indicate log ratio of L. reuteri FUA3400/L. reuteri FUA3400ΔgupCDE in sorghum (●) and wheat (○) sourdoughs and the log ratio of L. reuteri 100-23/L. reuteri 100-23ΔgadB in sorghum (▼) and wheat (△) sourdoughs. The abundance of L. reuteri FUA3400ΔgupCDE was below the detection limit after 6 propagations. Data shown are means ± standard deviations from triplicate independent experiments.
Quantification of 1,3-propanediol in wheat and sorghum.
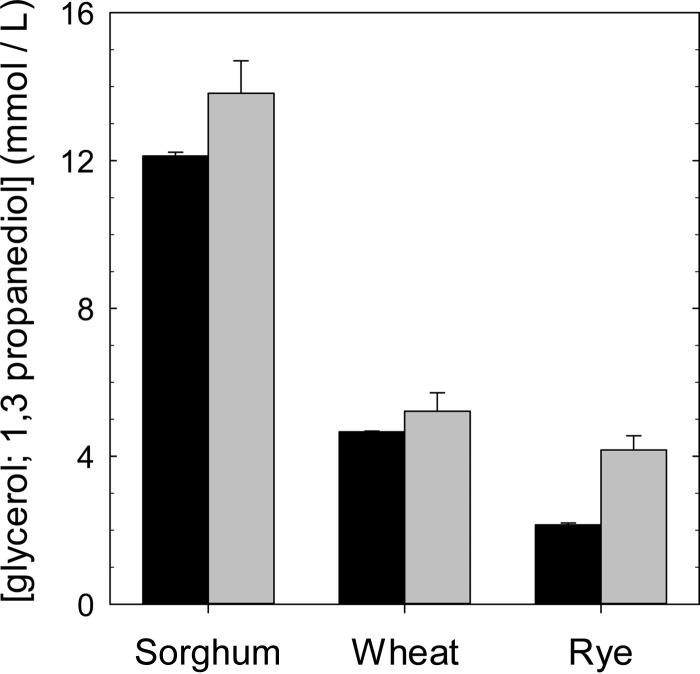
Sorghum contains glycerol in the form of phenolic esters (31). Lacking any indication that glycerol is present as an electron acceptor in wheat sourdoughs, we expected that L. reuteri FUA3400 would outcompete FUA3400ΔgupCDE in sorghum but not in wheat sourdoughs. Surprisingly, no difference between wheat and sorghum doughs was observed (Fig. 5). To determine the level of glycerol conversion in wheat, rye, and sorghum, glycerol and 1,3-propanediol were quantified in unfermented sourdoughs and in sourdoughs fermented with L. reuteri FUA3400. The concentration of 1,3-propanediol in all sourdoughs was equivalent to the glycerol concentration in unfermented sourdoughs, indicating quantitative conversion by bacterial metabolism (Fig. 6). Remarkably, glycerol and 1,3-propanediol concentrations in wheat demonstrate glycerol metabolism by L. reuteri in wheat and rye sourdoughs as well.
FIG 6.
Concentration of glycerol (black bars) in unfermented wheat, rye, and sorghum sourdoughs and concentration of 1,3-propanediol in wheat, rye, and sorghum sourdoughs after 24 h of fermentation with L. reuteri FUA3400.
DISCUSSION
This study used microbial competition in back-slopped sourdough as a tool to study the impact of metabolic properties on the ecological competitivenesses of closely related organisms. Sourdoughs that are propagated in the bakery are continuously contaminated by organisms from ingredients, pests, bakers, and the bakery environment (3, 4, 18). Continuous cycles of contamination and selection result in the establishment of highly competitive microbiota. The congruent evolution of microbiota in type I sourdoughs worldwide demonstrates that this process is highly selective and reproducible (3, 4, 7, 9, 32, 33).
Fermentations at a laboratory scale fail to reproduce this congruent selection process (34). Sourdough fermentations in bakeries are controlled to achieve a consistent technological function (e.g., fermentation kinetics, level of acidity, and leavening power), while sourdough fermentations in the laboratory are controlled to achieve consistent fermentation conditions with respect to time and temperature (6, 11, 12, 34). Moreover, flour is the only source of bacterial contamination in laboratory-scale fermentations, excluding other sources that are more relevant for sourdough ecology (18). Third, L. sanfranciscensis, the key organism in type I sourdoughs, was isolated only in sourdoughs with a long history—months or years—of continuous back-slopping (3, 4, 33), but laboratory-scale experiments are typically not conducted on that time scale. Competition experiments with back-slopped sourdoughs are nevertheless a sensitive tool to determine the competitivenesses of different strains or species that are inoculated into the same sourdough (11, 12, 25, 34). This study additionally demonstrates that competition experiments with knockout mutant strains provide a quantitative assessment of the contribution of individual metabolic traits to overall competitiveness. In sourdoughs inoculated with pairs of wild-type strains and knockdown mutants, the ratio of wild-type to mutant bacteria changed with a constant increment per fermentation cycle. For example, the log ratio of FUA3400ΔgupCDE to FUA3400 bacteria changed by 0.30 ± 0.04 and 0.44 ± 0.05 per fermentation cycle in sorghum and wheat sourdoughs, respectively. Despite the use of different flour and slightly different fermentation conditions, the rate of strain displacement and, hence, the impact of GadB on competitiveness in wheat sourdough determined in this study were in excellent agreement with data from previous reports (12).
Short- and long-fermented sourdoughs select for different metabolic traits. Frequent propagation selects for fast growth. Long fermentation times select for acid resistance. The conversion of arginine to ornithine, which is species specific in lactobacilli, and glutamine metabolism, which is lineage specific in L. reuteri (14), contribute to acid resistance in lactobacilli (12, 15, 35). Arginine and glutamine conversions partially compensate for the lack of glutamate decarboxylation (15, 36, 37); glutamate decarboxylase-positive rodent-lineage strains of L. reuteri thus have only a modest advantage over other strains (12; this study). The difference in competitiveness between L. reuteri 100-23 and 100-23ΔgadB was greater in wheat sourdoughs than in sorghum sourdoughs, possibly reflecting the higher content of glutamine plus glutamate in wheat proteins (31% [37]) than in sorghum proteins (19% [38]). L. reuteri 100-23 expressed gadB at the stationary stage but not at the exponential stage of growth (15); accordingly, L. reuteri 100-23 and 100-23ΔgadB were equally competitive in sourdoughs maintained by 12-h fermentation cycles.
In L. reuteri, enzymes for glycerol utilization are encoded by the pdu-cbi-cob-hem cluster (16), which is conserved in human isolates but only in a small proportion of swine or rodent isolates (14). Previous studies on glycerol metabolism by L. reuteri focused on the production of reuterin, a metabolic intermediate with antimicrobial activity at millimolar concentrations. 1,3-Propanediol has no appreciable antimicrobial activity (39). The use of glycerol as an electron acceptor enhanced the growth of obligate heterofermentative lactobacilli, including L. reuteri (27, 40). This study demonstrates that glycerol conversion to 1,3-propanediol provides an ecological advantage during growth in cereal substrates. L. reuteri FUA3400ΔgupCDE was outcompeted by the wild type in both short- and long-fermented doughs. The presence of glycerol as an electron acceptor increased the growth rate by approximately 20% (Fig. 4). A 10% inoculum in sourdoughs propagated with 12-h fermentation cycles corresponds to about 3 h of exponential growth (41). Because cell counts decreased by about 90% during the stationary phase of sourdoughs propagated with 72-h fermentation cycles, the exponential phase of growth upon back-slopping of 72-h-fermented doughs is extended, enhancing the competitive advantage for the faster-growing wild-type strain. Because L. reuteri FUA3400 and FUA3400ΔgupCDE did not differ in stationary-phase survival, this extended phase of exponential growth resulted in a more rapid decrease of the proportion of L. reuteri FUA3400ΔgupCDE bacteria in long-term-fermented sourdoughs. However, in competitions of glycerol-metabolizing human-lineage wild-type strains with glutamate-decarboxylating rodent-lineage strains, the faster growth of the former was offset by the improved stationary-phase survival of the latter (Fig. 1 and Table 1) (12).
The conversion of glycerol to 1,3-propanediol was induced by 1,2-propanediol (42). Interestingly, both 1,2-propanediol- and 1,3-propanediol-producing lactobacilli have been isolated from traditional sorghum sourdoughs (6), suggesting a possible symbiotic relationship that might benefit glycerol-metabolizing organisms. Glycerol is present in sorghum flour in the form of glycerol esters of phenolic acids (31), whereas the presence of 1,3-propanediol from glycerol in wheat sourdough has not been described. However, results from this study showed that the amount of glycerol in wheat sourdough was sufficient to provide an ecological advantage for glycerol-metabolizing strains over nonmetabolizing strains. This study did not confirm the source of glycerol in wheat; wheat lipids that are concentrated in the outer layers of the grain are a potential source for the release of glycerol by esterases during fermentation (43).
In summary, glycerol metabolism enabled rapid growth and increased competitiveness of L. reuteri in type I sourdoughs, while glutamate metabolism provided acid resistance and increased competitiveness in type II sourdoughs. The use of glycerol as an electron acceptor affects bread quality through increased levels of acetate; glutamate accumulation may alter the taste of bread (19). The observation that the competitive advantage of glycerol-metabolizing L. reuteri in sourdough is dependent on the use of reuterin as an electron acceptor may also relate to intestinal ecosystems. Glutamate-mediated acid resistance may support the survival of probiotic lactobacilli during gastric transit (12, 15). In rodents, L. reuteri colonizes the forestomach, where sucrose is available to provide fructose as an electron acceptor (44). The conserved pdu-cbi-cob-hem gene cluster in human-associated L. reuteri strains indicates that glycerol metabolism contributes to the competitiveness of the organism in humans. In humans, L. reuteri colonizes the colon, where sucrose is not available but glycerol or 1,2-propanediol may be available through lipid hydrolysis or as a fermentation product of other bacteria (45, 46).
ACKNOWLEDGMENTS
We acknowledge Marcia Shu-Wei Su for support with generation of the deletion mutant.
We acknowledge the National Science and Engineering Council of Canada and the Canada Research Chairs Program for financial support.
Footnotes
Published ahead of print 11 July 2014
REFERENCES
- 1.Brandt MJ. 2007. Sourdough products for convenient use in baking. Food Microbiol. 24:161–164. 10.1016/j.fm.2006.07.010 [DOI] [PubMed] [Google Scholar]
- 2.Hammes WP, Brandt MJ, Francis KL, Rosenheim J, Seitter MFH, Vogelmann SA. 2005. Microbial ecology of cereal fermentations. Trends Food Sci. Technol. 16:4–11. 10.1016/j.tifs.2004.02.010 [DOI] [Google Scholar]
- 3.Vogel RF, Knorr R, Müller MRA, Steudel U, Gänzle MG, Ehrmann MA. 1999. Non-dairy lactic fermentations: the cereal world. Antonie Van Leeuwenhoek 76:403–411. 10.1023/A:1002089515177 [DOI] [PubMed] [Google Scholar]
- 4.De Vuyst L, Vrancken G, Ravyts F, Rimaux T, Weckx S. 2009. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 26:666–675. 10.1016/j.fm.2009.07.012 [DOI] [PubMed] [Google Scholar]
- 5.Hamad SH, Dieng MC, Ehrmann MA, Vogel RF. 1997. Characterization of the bacterial flora of Sudanese sorghum flour and sorghum sourdough. J. Appl. Microbiol. 83:764–770. 10.1046/j.1365-2672.1997.00310.x [DOI] [PubMed] [Google Scholar]
- 6.Sekwati-Monang B, Gänzle MG. 2011. Microbiological and chemical characterisation of ting, a sorghum-based sourdough product from Botswana. Int. J. Food Microbiol. 150:115–121. 10.1016/j.ijfoodmicro.2011.07.021 [DOI] [PubMed] [Google Scholar]
- 7.Gänzle M, Ehmann M, Hammes W. 1998. Modeling of growth of Lactobacillus sanfranciscensis and Candida milleri in response to process parameters of sourdough fermentation. Appl. Environ. Microbiol. 64:2616–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stolz P, Böcker G, Hammes WP, Vogel RF. 1995. Utilization of electron acceptors by lactobacilli isolated from sourdough. Z. Lebensm. Unters. Forsch. 201:91–96. 10.1007/BF01193208 [DOI] [Google Scholar]
- 9.Vogel RF, Pavlovic M, Ehrmann MA, Wiezer A, Liesegang H, Offschanka S, Voget S, Angelov A, Böcker G, Liebl W. 2011. Genomic analysis reveals Lactobacillus sanfranciscensis as stable element in traditional sourdoughs. Microb. Cell Fact. 10(Suppl 1):S6. 10.1186/1475-2859-10-S1-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neubauer H, Glaasker E, Hammes WP, Poolman B, Konings WN. 1994. Mechanism of maltose uptake and glucose excretion in Lactobacillus sanfrancisco. J. Bacteriol. 176:3007–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meroth CB, Walter J, Hertel C, Brandt MJ, Hammes WP. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:475–482. 10.1128/AEM.69.1.475-482.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su MS, Schlicht S, Gänzle MG. 2011. Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Fact. 10(Suppl 1):S8. 10.1186/1475-2859-10-S1-S8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. 2010. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 4:377–387. 10.1038/ismej.2009.123 [DOI] [PubMed] [Google Scholar]
- 14.Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, Oh PL, Heng NCK, Patil PB, Juge N, Mackenzie DA, Pearson BM, Lapidus A, Dalin E, Tice H, Goltsman E, Land M, Hauser L, Ivanova N, Kyrpides NC, Walter J. 2011. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 7:e1001314. 10.1371/journal.pgen.1001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teixeira JS, Seeras A, Sanchez-Maldonado AF, Zhang C, Su MS-W, Gänzle MG. 2014. Glutamine, glutamate, and arginine-based acid resistance in Lactobacillus reuteri. Food Microbiol. 42:172–180. 10.1016/j.fm.2014.03.015 [DOI] [PubMed] [Google Scholar]
- 16.Sriramulu DD, Liang M, Hernandez-Romero D, Raux-Deery E, Lünsdorf H, Parsons JB, Warren MJ, Prentice MB. 2008. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J. Bacteriol. 190:4559–4567. 10.1128/JB.01535-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morita H, Toh H, Fukuda S, Horikawa H, Oshima K, Suzuki T, Murakami M, Hisamatsu S, Kato Y, Takizawa T, Fukuoka H, Yoshimura T, Itoh K, O'Sullivan DJ, McKay LL, Ohno H, Kikuchi J, Masaoka T, Hattori M. 2008. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 15:151–161. 10.1093/dnares/dsn009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su MS-W, Oh PL, Walter J, Gänzle MG. 2012. Intestinal origin of sourdough Lactobacillus reuteri isolates as revealed by phylogenetic, genetic, and physiological analysis. Appl. Environ. Microbiol. 78:6777–6780. 10.1128/AEM.01678-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gänzle MG, Vermeulen N, Vogel RF. 2007. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough. Food Microbiol. 24:128–138. 10.1016/j.fm.2006.07.006 [DOI] [PubMed] [Google Scholar]
- 20.Gänzle MG, Höltzel A, Walter J, Jung G, Hammes WP. 2000. Characterization of reutericyclin produced by Lactobacillus reuteri LTH2584. Appl. Environ. Microbiol. 66:4325–4333. 10.1128/AEM.66.10.4325-4333.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gänzle MG. 2003. Contribution of reutericyclin production to the stable persistence of Lactobacillus reuteri in an industrial sourdough fermentation. Int. J. Food Microbiol. 80:31–45. 10.1016/S0168-1605(02)00146-0 [DOI] [PubMed] [Google Scholar]
- 22.Perez-Casal J, Price JA, Maguin E, Scott JR. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809–819. 10.1111/j.1365-2958.1993.tb01628.x [DOI] [PubMed] [Google Scholar]
- 23.Cadieux P, Wind A, Sommer P, Schaefer L, Crowley K, Britton RA, Reid G. 2008. Evaluation of reuterin production in urogenital probiotic Lactobacillus reuteri RC-14. Appl. Environ. Microbiol. 74:4645–4649. 10.1128/AEM.00139-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin XB, Gänzle MG. 2014. Quantitative high-resolution melting PCR analysis for monitoring of fermentation microbiota in sourdough. Int. J. Food Microbiol. 186C:42–48. 10.1016/j.ijfoodmicro.2014.06.010 [DOI] [PubMed] [Google Scholar]
- 25.Sekwati-Monang B, Valcheva R, Gänzle MG. 2012. Microbial ecology of sorghum sourdoughs: effect of substrate supply and phenolic compounds on composition of fermentation microbiota. Int. J. Food Microbiol. 159:240–246. 10.1016/j.ijfoodmicro.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 26.Zwietering MH, Jongenburger I, Rombouts FM, van't Riet K. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talarico TL, Axelsson LT, Novotny J, Fiuzat M, Dobrogosz WJ. 1990. Utilization of glycerol as a hydrogen acceptor by Lactobacillus reuteri: purification of 1,3-propanediol:NAD oxidoreductase. Appl. Environ. Microbiol. 56:943–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevens MJA, Vollenweider S, Meile L, Lacroix C. 2011. 1,3-Propanediol dehydrogenases in Lactobacillus reuteri: impact on central metabolism and 3-hydroxypropionaldehyde production. Microb. Cell Fact. 10:61. 10.1186/1475-2859-10-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elleuche S, Fodor K, Klippel B, von der Heyde A, Wilmanns M, Antranikian G. 2013. Structural and biochemical characterisation of a NAD+-dependent alcohol dehydrogenase from Oenococcus oeni as a new model molecule for industrial biotechnology applications. Appl. Microbiol. Biotechnol. 97:8963–8975. 10.1007/s00253-013-4725-0 [DOI] [PubMed] [Google Scholar]
- 30.Stromeck A, Hu Y, Chen L, Gänzle MG. 2011. Proteolysis and bioconversion of cereal proteins to glutamate and γ-aminobutyrate (GABA) in rye malt sourdoughs. J. Agric. Food Chem. 59:1392–1399. 10.1021/jf103546t [DOI] [PubMed] [Google Scholar]
- 31.Svensson L, Sekwati-Monang B, Lutz DL, Schieber A, Gänzle MG. 2010. Phenolic acids and flavonoids in nonfermented and fermented red sorghum (Sorghum bicolor (L.) Moench). J. Agric. Food Chem. 58:9214–9220. 10.1021/jf101504v [DOI] [PubMed] [Google Scholar]
- 32.Siragusa S, Di Cagno R, Ercolini D, Minervini F, Gobbetti M, De Angelis M. 2009. Taxonomic structure and monitoring of the dominant population of lactic acid bacteria during wheat flour sourdough type I propagation using Lactobacillus sanfranciscensis starters. Appl. Environ. Microbiol. 75:1099–1109. 10.1128/AEM.01524-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minervini F, Di Cagno R, Lattanzi A, De Angelis M, Antonielli L, Cardinali G, Cappelle S, Gobbetti M. 2012. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical Italian breads: interactions between ingredients and microbial species diversity. Appl. Environ. Microbiol. 78:1251–1264. 10.1128/AEM.07721-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minervini F, Lattanzi A, De Angelis M, Di Cagno R, Gobbetti M. 2012. Influence of artisan bakery- or laboratory-propagated sourdoughs on the diversity of lactic acid bacterium and yeast microbiotas. Appl. Environ. Microbiol. 78:5328–5340. 10.1128/AEM.00572-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rollan G, Lorca GL, Font de Valdez G. 2003. Arginine catabolism and acid tolerance response in Lactobacillus reuteri isolated from sourdough. Food Microbiol. 20:313–319. 10.1016/S0740-0020(02)00139-9 [DOI] [Google Scholar]
- 36.Arena ME, Manca de Nadra MC. 2001. Biogenic amine production by Lactobacillus. J. Appl. Microbiol. 90:158–162. 10.1046/j.1365-2672.2001.01223.x [DOI] [PubMed] [Google Scholar]
- 37.Vermeulen N, Gänzle MG, Vogel RF. 2007. Glutamine deamidation by cereal-associated lactic acid bacteria. J. Appl. Microbiol. 103:1197–1205. 10.1111/j.1365-2672.2007.03333.x [DOI] [PubMed] [Google Scholar]
- 38.Wieser H, Koehler P. 2008. The biochemical basis of celiac disease. Cereal Chem. 85:1–13. 10.1094/CCHEM-85-1-0001 [DOI] [Google Scholar]
- 39.Vollenweider S, Evers S, Zurbriggen K, Lacroix C. 2010. Unraveling the hydroxypropionaldehyde (HPA) system: an active antimicrobial agent against human pathogens. J. Agric. Food Chem. 58:10315–10322. 10.1021/jf1010897 [DOI] [PubMed] [Google Scholar]
- 40.Schütz H, Radler F. 1984. Anaerobic reduction of glycerol to propanediol-1.3 by Lactobacillus brevis and Lactobacillus buchneri. Syst. Appl. Microbiol. 5:169–178. 10.1016/S0723-2020(84)80018-1 [DOI] [Google Scholar]
- 41.Brandt MJ, Hammes WP, Gänzle MG. 2004. Effects of process parameters on growth and metabolism of Lactobacillus sanfranciscensis and Candida humilis during rye sourdough fermentation. Eur. Food Res. Technol. 218:333–338. 10.1007/s00217-003-0867-0 [DOI] [Google Scholar]
- 42.Amin HM, Hashem AM, Ashour MS, Hatti-Kaul R. 2013. 1,2 Propanediol utilization by Lactobacillus reuteri DSM 20016, role in bioconversion of glycerol to 1,3 propanediol, 3-hydroxypropionaldehyde and 3-hydroxypropionic acid. J. Genet. Eng. Biotechnol. 11:53–59. 10.1016/j.jgeb.2012.12.002 [DOI] [Google Scholar]
- 43.Morrison WR. 1994. Wheat lipids: structure and functionality, p 128–142 In Bushuk W, Rasper VF. (ed), Wheat. Springer, Boston, MA [Google Scholar]
- 44.Sims IM, Frese SA, Walter J, Loach D, Wilson M, Appleyard K, Eason J, Livingston M, Baird M, Cook G, Tannock GW. 2011. Structure and functions of exopolysaccharide produced by gut commensal Lactobacillus reuteri 100-23. ISME J. 5:1115–1124. 10.1038/ismej.2010.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Badía J, Ros J, Aguilar J. 1985. Fermentation mechanism of fucose and rhamnose in Salmonella typhimurium and Klebsiella pneumoniae. J. Bacteriol. 161:435–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott KP, Martin JC, Campbell G, Mayer C-D, Flint HJ. 2006. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans.” J. Bacteriol. 188:4340–4349. 10.1128/JB.00137-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathieson M, Haikerwal AR. 1971. The protein content and amino acid composition of sorghum grain. Cereal Chem. 48:690–699 [Google Scholar]