Abstract
We examined gene expression patterns in the lignin-degrading fungus Phanerochaete chrysosporium when it colonizes hybrid poplar (Populus alba × tremula) and syringyl (S)-rich transgenic derivatives. A combination of microarrays and liquid chromatography-tandem mass spectrometry (LC-MS/MS) allowed detection of a total of 9,959 transcripts and 793 proteins. Comparisons of P. chrysosporium transcript abundance in medium containing poplar or glucose as a sole carbon source showed 113 regulated genes, 11 of which were significantly higher (>2-fold, P < 0.05) in transgenic line 64 relative to the parental line. Possibly related to the very large amounts of syringyl (S) units in this transgenic tree (94 mol% S), several oxidoreductases were among the upregulated genes. Peptides corresponding to a total of 18 oxidoreductases were identified in medium consisting of biomass from line 64 or 82 (85 mol% S) but not in the parental clone (65 mol% S). These results demonstrate that P. chrysosporium gene expression patterns are substantially influenced by lignin composition.
INTRODUCTION
Efficient and complete natural degradation of woody plant cell walls is generally ascribed to certain wood decay basidiomycetes. Collectively referred to as white rot fungi, these unique microbes are capable of degrading all cell wall components, including hemicelluloses, cellulose, and the more recalcitrant lignin (1). Commonly inhabiting woody debris and forest litter, these fungi play an important role in carbon cycling. To degrade and fully mineralize cell wall polymers, the model white rot fungus Phanerochaete chrysosporium employs a complex array of extracellular oxidative and hydrolytic systems (2).
Analysis of the P. chrysosporium genome revealed 177 putative glycoside hydrolases (3), most of which are likely involved in the degradation of cellulose or hemicellulose. On the basis of sequence comparisons, these enzymes were assigned to 45 glycoside hydrolase (GH) families (4; http://www.cazy.org/). Beyond GHs, considerable evidence supports a role for cellobiose dehydrogenase (CDH) and lytic polysaccharide monooxygenases (LPMOs) in the oxidative attack on cellulose and hemicellulose (5–8). Consistent with this view, elevated transcript levels and proteins corresponding to various GHs, CDH, and LPMOs have been observed in medium containing microcrystalline cellulose (9–12) and ground Populus grandidentata (13), Quercus rubra (14–16), Pinus strobus (17), or Pinus nigra (18).
Woody feedstocks are increasingly viewed as sources for high-value, low-molecular-weight products (19). However, their conversion to simple, fermentable carbohydrates remains a formidable barrier, and harsh chemical treatments are typically employed to enhance substrate accessibility and lignin removal. In recent years, lignin composition in poplar has been modified by gene misregulation, and, in one case, altered lignin improved the efficiency of saccharification and fermentation to ethanol (20). The enzymatic mechanisms underlying lignin degradation remain uncertain, but recent analysis suggests that transgenic poplar with increased ratios of syringyl to guaiacyl lignin monomers are relatively more resistant to fungal attack (21).
It is generally thought that lignin depolymerization is catalyzed by an array of oxidative enzymes, especially lignin peroxidase (LiP), manganese peroxidase (MnP), and versatile peroxidase (VP). Recent genome investigations reveal that all efficient lignin degraders possess some combination of these enzymes, all of which are class II members of the plant-fungal-prokaryotic peroxidase superfamily (22, 23). Ligninolytic peroxidases, together with auxiliary enzymes, have been classified into 10 families (24). Of these, class II peroxidases (family AA2), various glucose-methanol-choline (GMC) oxidoreductases (AA3), copper radical oxidases (CROs; AA5), benzoquinone reductases (AA6), and the aforementioned LPMOs (AA9) are expressed in P. chrysosporium cultures (reviewed in reference 25). Owing to the insolubility of the substrate, little is known of the influence of native lignin on gene expression. However, the secretomes of cultures amended with sulfonated lignin (11) or vanillin (26–28), a probable degradation product, have been reported. Aldehyde dehydrogenases (AADs), CDH, copper radical oxidases, glutathione transferases, and benzoquinone reductases, all implicated in transformations of aromatic metabolites, were identified in these submerged cultures. Characterization of proteins extracted from colonized wood blocks has been more challenging, but Ravalason and coworkers successfully identified a copper radical oxidase in P. nigra wood chips decayed by P. chrysosporium strain CIRM-BRFM41 (18).
Substrate preference among certain wood decay fungi is well established (1, 29). Numerous studies involving defined medium have demonstrated the substantial influence of substrate on P. chrysosporium gene expression (reviewed in references 30 and 31). Differential transcriptional regulation and shifting secretome patterns have also been observed in comparisons of defined-medium cultures to cultures containing a woody substrate (13, 15, 18), and comparisons of white pine (Pinus strobus) to bigtooth aspen (Populus grandidentata) also showed significant modulation of gene expression in response to highly divergent substrates (17). In this study, we have examined gene expression in hybrid poplar lines with drastically different lignin compositions, a trait of considerable importance to many industrial processes.
MATERIALS AND METHODS
Culture conditions and characterization.
RNA and protein were obtained from P. chrysosporium strain RP78 (Forest Mycology Center, Forest Products Laboratory) grown in Highley's basal salt medium (32) containing 0.5% (wt/vol) glucose (Glc) or Wiley-milled (1-mm mesh) wood. For each of the three wood samples (parental hybrid clone line P717, transgenic line 82, and transgenic line 64) multiple branches were harvested, debarked, and dried. These samples were derived from independent trees that were clonally replicated and grown in a greenhouse for 2 years. The milled material was used as is, i.e., without solvent extraction. Two-liter Erlenmeyer flasks contained 250 ml of medium and were inoculated with approximately 107 P. chrysosporium spores scraped from the surface of YMPG (yeast extract, malt extract, peptone, glucose) agar. Cultures were incubated for 5 days on a rotary shaker (150 rpm) at 37°C. Carbohydrate and lignin composition levels of wood samples have been reported previously (21). Notably, line 64, line 82, and parental line P717 had similar overall lignin contents but feature approximately 94 mol%, 85 mol%, and 65 mol% syringyl units, respectively.
For RNA, mycelia from triplicate cultures were collected by filtration through Miracloth (Calbiochem, EMD Biosciences, Gibbstown, NJ), squeeze dried, and snap-frozen in liquid nitrogen. Pellets were stored at −80°C until use. For mass spectroscopic analysis, culture filtrates were stored at −20°C before use.
Expression microarrays.
P. chrysosporium Roche NimbleGen array designs are available under platform GPL18011 within the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/index.cgi). The arrays feature 9,927 gene targets and differ from the previously used design platform GPL8022 (10, 13, 17, 33) by the elimination of repetitive elements and the addition of 25 open reading frames (protein target identification [ID] numbers 150002 to 150027).
Total RNA was purified from frozen mycelial pellets, converted to Cy3-labeled cDNA, hybridized to microarrays, and scanned as described previously (13). The arrays used in these experiments were scanned on an Axon 4000B scanner (Molecular Dynamics), and data were extracted using NimbleScan, version 2.4. Quantile normalization and robust multiarray averaging (RMA) (34) were applied to the raw data using DNASTAR ArrayStar, version 4 (Madison, WI). Expression levels were based on log2 signals, and significant differences in expression were determined using a moderated t test (35) with a false discovery rate (FDR) (36) threshold set at a P value of <0.05.
Mass spectrometry.
Soluble extracellular protein was precipitated from culture filtrates by direct addition of solid trichloroacetic acid (TCA) to 10% (wt/vol), and trypsin-generated peptides were analyzed by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) using an Agilent 1100 Nanoflow system (Agilent, Palo Alto, CA) connected to a hybrid linear trap quadrupole-Orbitrap mass spectrometer (LTQ-Orbitrap; Thermo Fisher Scientific, San Jose, CA) equipped with a nanoelectrospray ion source as described previously (13). Using protein databases for P. chrysosporium (BestModels_2.0 [http://jgi.doe.gov/whiterot]), the MS/MS spectra were analyzed using an in-house Mascot search engine (version 2.2.07; Matrix Science, London, United Kingdom). Mascot searches were done with a fragment ion mass tolerance of 0.6 Da, parent ion tolerance of 15 ppm, and methionine oxidation as variable modification. Scaffold (version 4; Proteome Software, Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Protein identifications were accepted if they contained at least two uniquely identified peptides and if protein probabilities exceeded 95.0% as determined by the Protein Prophet algorithm (37).
Detailed information on individual gene/protein models can be directly accessed via the Joint Genome Institute (JGI) genome portal (38). The protein pages include information from the Gene Ontology (GO) database for each InterPro domain. Function or putative function was assigned when determinations were supported by direct experimental evidence or when comparisons to known proteins revealed conserved catalytic features and/or significant alignment scores (bit scores of >150) to known proteins within the SwissProt database. All other proteins were designated hypothetical.
Microarray data accession number.
The MIAME (minimum information about a microarray experiment)-compliant (39) microarray expression data were deposited in NCBI's Gene Expression Omnibus under GEO accession number GSE52922.
RESULTS
Transcriptome analysis revealed 113 genes upregulated on poplar relative to glucose-grown cultures (Fig. 1). Seventy-nine of these were upregulated in the parental hybrid, P717, and these included a wide array of glycoside hydrolases (GHs), oxidoreductases, transporters, and hypothetical proteins. With exceptions, P. chrysosporium gene signal strength on Wiley-milled P717 was positively correlated (r = 0.96, n = 9,927) (see Data Set S1 in the supplemental material) with previously reported ball-milled Populus grandidentata (17). Notable outliers included genes encoding an endoxylanase (Phach1_7045), two lytic polysaccharide monooxygenases (LPMOs; Phach1_41563 and Phach1_41125), and a class 3 lipase (Phach1_2540), all of which exhibited high signal strength and significant upregulation on P717 relative to P. grandidentata. Possibly, these differences were influenced by wood composition, but substrate pretreatment may have also played a role. The Wiley-milled material used here is more coarsely ground and less accessible to enzyme penetration than pulverized ball-milled wood.
FIG 1.
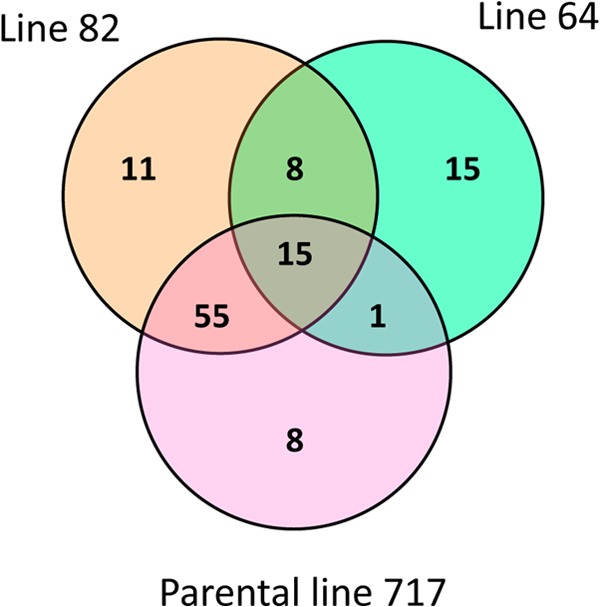
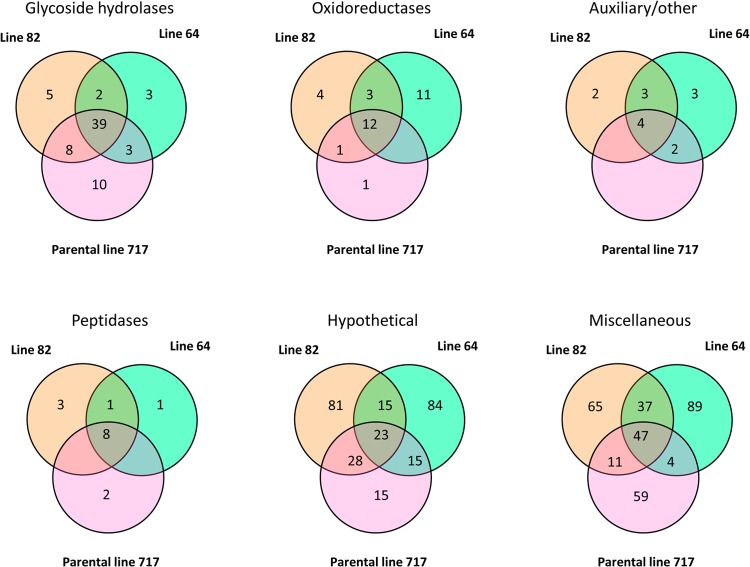
Distribution of 113 genes whose transcripts accumulate >2-fold (P < 0.05) in poplar relative to glucose medium. Of these, cultures containing high-syringyl lines 82 and 64 as the sole carbon sources had 11 and 15 genes, respectively, showing no significant increases in the parental line P717. Eight genes were significantly upregulated in both transgenic lines, but not in P717. Detailed results for all genes are available in Data Set S1 in the supplemental material.
Transcripts for 34 functionally diverse genes accumulated uniquely in line 82 and/or 64, but not in P717, relative to glucose (Fig. 1). Of genes upregulated in transgenic material relative to P717, none showed significant up- or downregulation in line 82. However, the line containing the highest syringyl lignin content, line 64, significantly (P < 0.05) induced 11 genes (Table 1). This group included a flavin-dependent oxidoreductase, formate dehydrogenase, formaldehyde dehydrogenase, phenylalanine ammonia lyase, and several hypothetical proteins. Furthermore, another flavin-dependent oxidoreductase (Phach1_140680) and a transporter (Phach1_138238) showed significant upregulation in line 64 relative to line 82.
TABLE 1.
P. chrysosporium genes upregulated in colonization of transgenic poplar line 82 or 64 relative to parental hybrid line P717
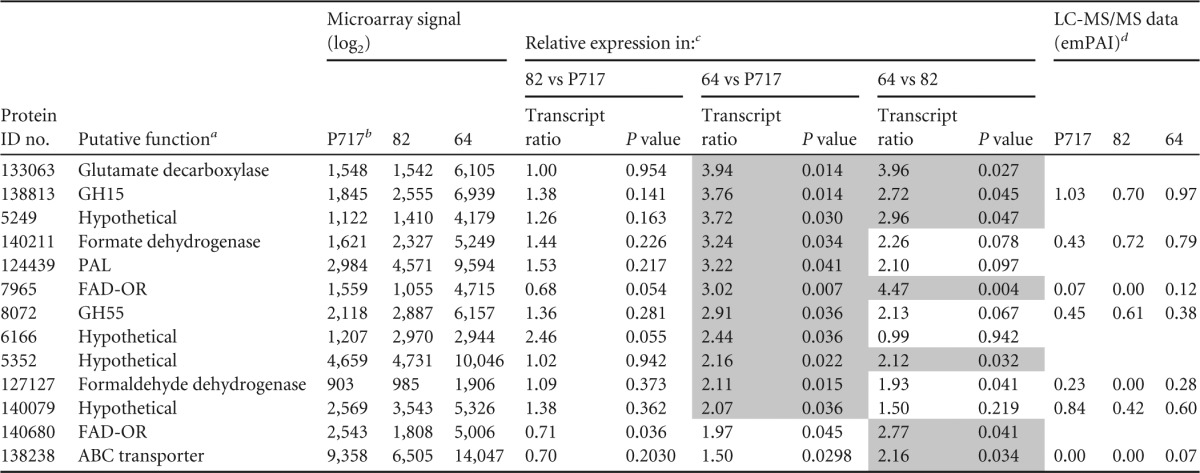
GH, glycoside hydrolase; PAL, phenylalanine ammonia lyase; FAD-OR, favin-dependent oxidoreductases similar to salicylate monooxygenase. Hypothetical proteins Phach1_5249 and Phach1_140079 were detected in previous reports (17), and the latter protein model featured a predicted secretion signal. Transmembrane helices were predicted for Phach1_5249, Phach1_6166 and Phach1_5352.
P717, wild-type parental hybrid.
Shading highlights significant upregulation (>2-fold; P < 0.05).
emPAI, exponentially modified protein abundance index.
LC-MS/MS-based secretome analysis unambiguously identified 793 proteins, each defined by ≥2 unique peptides. These proteins were broadly categorized as glycoside hydrolases (70 proteins), oxidoreductases (32), auxiliary/other proteins implicated in lignocellulose degradation (14), peptidases (15), hypotheticals (343), and miscellaneous (315) (Fig. 2). Peptides corresponding to high-oxidation-potential peroxidases were not detected, but 18 oxidoreductases were found exclusively in the transgenic lines (Table 2). All of these proteins could play a direct or indirect role in the transformation and degradation of lignin metabolites. Increased levels of glycoside hydrolases in the transgenic lines (Table 3) likely reflect broad shifts in accessibility of cellulose and hemicelluloses. Polysaccharide access might be altered by lowered molecular weight and increased proportions of β-O-4 bonds associated with lignins derived from high levels of syringyl monomer (40).
FIG 2.
Distribution of proteins in broad functional categories. Miscellaneous proteins include those without secretion signals and homologous to abundant, known, intracellular proteins involved in central metabolism and protein synthesis. These are likely the result of hyphal lysis during cultivation. Detailed listings of all detected proteins are available in Data Set S1 in the supplemental material.
TABLE 2.
P. chrysosporium oxidoreductases upregulated in lines 82 and/or 64 but not P717a
| Protein distribution and ID no. | Putative function | Domainb | Comment (reference) | Microarray signal (log2) |
LC-MS/MS data (emPAI) | ||
|---|---|---|---|---|---|---|---|
| P717 | 82 | 64 | |||||
| Proteins in filtrates of line 82 | |||||||
| 6771 | Oxidoreductase | PF01408 | 1482 | 1382 | 1289 | 0.119 | |
| 8211 | GMC oxidoreductase | AA3_2 | 416 | 425 | 429 | 0.100 | |
| 10221 | Alcohol dehydrogenase | PF00248 | 3053 | 3060 | 4084 | 0.234 | |
| 9149 | GMC oxidoreductase, possible glucose oxidase | IPR001395 | Model 5′ region needs editing | 2863 | 3010 | 3117 | 0.093 |
| 1321 | Flavin-containing monooxygenasec | PF01494, PF01360 | ∼PcFMO1 (28) | 11700 | 12117 | 10235 | 0.025 |
| Proteins in filtrates of lines 82 and 64 | |||||||
| 133289 | Aldehyde dehydrogenase | PF00171 | 438 | 456 | 564 | 0.154, 0.377d | |
| 134181 | Aryl-alcohol dehydrogenase | PF00248 | 68% identity to Phach1_11055 (53) | 378 | 363 | 420 | 0.126, 0.252d |
| 3442 | Oxidoreductase | PF00107 | Model 3′ terminus needs editing | 2071 | 2018 | 2162 | 0.054, 0.050d |
| Proteins in filtrates of line 64 | |||||||
| 133757 | Formate dehydrogenase | PF00389, PF02826 | 463 | 471 | 513 | 0.365 | |
| 6283 | Flavin-containing monooxygenase | PF00743, PF13738 | 1268 | 1293 | 1557 | 0.151 | |
| 127288 | Catalase | PF00199 | 1728 | 1689 | 1755 | 0.090 | |
| 7965 | FAD-dependent oxidoreductase | PF01360 | Related to salicylate hydroxylase | 1559 | 1055 | 4715 | 0.116 |
| 7129 | Alcohol dehydrogenase | PF00107 | 529 | 586 | 717 | 0.298 | |
| 1173 | Oxidoreductase | PF01408 | 358 | 377 | 384 | 0.120 | |
| 10307 | 1,4-benzoquinone reductase | AA6 | 1214 | 1091 | 1203 | 0.249 | |
| 5793 | Alcohol dehydrogenase | PF00107 | 1287 | 1370 | 1899 | 0.369 | |
| 1056 | Oxidoreductase | PF00107 | 2382 | 2684 | 3110 | 0.133 | |
| 132896 | Flavin-containing monooxygenase | PF00743, PF13738 | 1371 | 1486 | 2073 | 0.079 | |
| 128306 | Catalase | PF00199 | 1211 | 1148 | 1210 | 0.081 | |
The Venn diagram in Fig. 2 shows the distribution of all detected proteins. Only proteins with ≥2 unique peptides are considered significant.
Pfam (http://pfam.sanger.ac.uk/), auxiliary activity families (http://www.cazy.org/Auxiliary-Activities.html), or InterPro domains (https://www.ebi.ac.uk/interpro/) are listed. See Data Set S1 in the supplemental material or individual protein pages on the JGI portal (http://genome.jgi-psf.org/Phchr1/Phchr1.home.html) for additional information.
Gene model extends beyond likely 3′ terminus; corrected gene equivalent to vanillin-induced protein PcFMO1 of P. chrysosporium strain ME446.
emPAI values for line 82 and line 64, respectively.
TABLE 3.
P. chrysosporium CAZy proteins identified in line 82 and/or 64
| Protein distribution and ID no. | Putative function | Comment (reference) | Microarray signal (log2) |
LC-MS/MS data (emPAI) | ||
|---|---|---|---|---|---|---|
| P717 | 82 | 64 | ||||
| Proteins in filtrates of line 82 | ||||||
| 31049 | AA9 lytic polysaccharide monooxygenase | C-terminal CBM1a | 11,570 | 10,399 | 3,779 | 0.252 |
| 4449 | GH28 exopolygalacturonase | Epg28B (33) | 2,548 | 2,798 | 2,738 | 0.172 |
| 135833 | GH31 alpha-glucosidase | 1,489 | 1,577 | 1,446 | 0.073 | |
| 121774 | GH5_22 endo-beta-1,4-glucanase | Probable β-1,4-glucanase (54) | 1,786 | 1,701 | 1,384 | 0.097 |
| 128446 | GH37 neutral trehalase | 1,991 | 2,194 | 1,771 | 0.059 | |
| Proteins in filtrates of lines 82 and 64 | ||||||
| 138596 | GH3 beta-glucosidase | 6,419 | 6,953 | 3,935 | 0.056, 0.052b | |
| 139732 | GH10 endo-1,4-beta-xylanase | 5′ CBM1 | 1,935 | 1,887 | 1,729 | 0.319, 0.212b |
| Proteins in filtrates of line 64 | ||||||
| 40899 | GH18 probable chitinase | 5′ incomplete | 1,266 | 1,354 | 1,083 | 0.258 |
| 126964 | GH7 exocellobiohydrolase | Cel7A | 1,024 | 1,121 | 1,090 | 0.107 |
| 35305 | GH38 alpha-mannosidase | 5′ incomplete | 2,776 | 2,948 | 2,916 | 0.040 |
CBM1, family 1 carbohydrate-binding module.
emPAI values for line 82 and line 64, respectively.
DISCUSSION
Our results demonstrate the influence of lignin composition on P. chrysosporium gene expression. In particular, secretome and transcriptome profiles highlight a role for various proteins in response to an increased ratio of syringyl/guaiacyl monomers in the lignin polymer. It is unclear whether similar expression patterns would be observed in solid wood under natural conditions. Presumably, Wiley mill grinding or other commercial pretreatments enhance enzyme accessibility.
Transcript accumulation in media containing ground transgenic lines as sole carbon sources identified several regulated genes, including the flavin-binding oxidoreductases Phach1_7965 and Phach1_140680. Featuring InterPro domain IPR003042 (aromatic-ring hydroxylases), these putative monooxygenases are closely related (66% identity) and feature predicted secretion signals (Table 1; see also Data Set S1 in the supplemental material). Given their structure and relatively high transcript accumulation in the high-syringyl lignin substrate (line 64), it seems probable that these enzymes are involved in the degradation of aromatic metabolites. Upregulation of formate dehydrogenase genes encoding Phach1_140211 (Table 1) and Phach1_133757 (Table 2) may be in response to formate accumulation via the glyoxylate cycle although simultaneous increases in the expression of oxalate decarboxylase were not observed as they were in earlier studies of P. chrysosporium (17) and Dichomitus squalens (41). Nevertheless, relatively high protein levels (exponentially modified protein abundance index [emPAI] value of 1.04) (see Data Set S1 in the supplemental material) of an oxalate decarboxylase (Phach1_12177) were detected in the parental line P7171. Formate can also be generated by the successive activities of methanol oxidase (Phach1_126879) and formaldehyde dehydrogenase (Phach1_127127) (42). The latter Zn-dependent enzyme is upregulated in line 64 relative to P717 (Table 1) while the former is highly expressed in all media (see Data Set S1). Glutamate dehydrogenase Phach1_133063 and ABC transporter Phach1_138238 responses are likely induced by H2O2 and lignin-derived metabolites (43), respectively, in line 64 (Table 1). Transcript accumulation of a glucoamylase (GH15; Phach1_138813) and a 1,3-β-glucosidase (GH55; Phach1_8072) cannot be easily explained by the higher ratio of syringyl/guaiacyl monomers in line 64. Neither starch nor β-1,3-glucans are significant constituents of mature wood, but the saplings used here likely contain elevated levels. Moreover, peptides corresponding to GH15s and GH55s have been detected when various white rot fungi, including P. chrysosporium, were cultivated on mature aspen sapwood (22, 23).
In addition to their regulation, genes featuring high transcript levels undoubtedly play an important role in lignocellulose degradation. For examples, genes encoding CBH2 (GH6; Phlgi_133052), CDH (AA3_1; Phlbi_11098), an LPMO (AA9; formerly GH61; Phlgi_129325), and endoglucanase EG38 (GH5; Phlbi_6458) were among the 30 highest microarray signals observed on the parental line P717 (see Data Set S1 in the supplemental material). Along these lines, the aforementioned Phlgi_126879 accumulated at very high levels in all four media. A probable ortholog of Gloeophyllum trabeum methanol oxidase (89% identical) (44), the substrate (methanol) can be produced by lignin demethoxylation, a process common to a wide range of wood decay fungi (for example, see reference 45).
In addition to transcript analysis, protein identifications support an important role for many oxidoreductases. Among the more interesting proteins detected by LC-MS/MS, two additional flavin-containing monooxygenases, Phach1_1321 and Phach1_132896, were identified in filtrates of cultures containing line 82 and line 64, respectively (Table 2). The Phach1_1321 protein is orthologous to P. chrysosporium MO1 (PcFMO1), a flavin monooxygenase previously identified in P. chrysosporium strain ME446 cultures supplemented with vanillin (28). A closely related ME446 gene, PcFMO2, corresponds to Phach1_137230 in the RP78 genome. Although we could not detect Phach1_137230 peptides in culture filtrates, its transcripts are relatively high in line 82 relative to Glc medium (2.1-fold; P < 0.02) (see Data Set S1 in the supplemental material). Interestingly, these flavin monooxygenases were, like the aforementioned Phach1_1321 and Phach1_132896, part of a family of >34 related genes, 16 of which are located on scaffold 15. In fact, eight related genes were localized to a 32-kb region which contains Phach1_140680, Phach1_7965, Phach1_1321, and Phach1_32896. Clustering of functionally related genes in P. chrysosporium had been previously observed for LiP- and CRO-encoding genes (46, 47), but regulation is not strictly coordinated among these tightly linked sequences. Thus, a putative flavin-containing monooxygenase (Phlgi_6283) is 89% identical to Phlgi_132896 and similarly expressed on line 64 (Table 2), and yet it resides on a separate scaffold.
Unambiguous protein identifications coupled with high transcript levels strongly support important roles for certain genes. For example, flavin adenine dinucleotide (FAD)-dependent oxidoreductases Phach1_7965 and Phach1_132896, alcohol dehydrogenase Phach1_5793, and benzoquinone reductase Phach1_10307 are highly expressed during growth on line 64 (Table 2). The last enzyme has been implicated in quinone cycling (48), and vanillin-induced upregulation has been observed (26). In this connection, increased transcript levels of phenylalanine ammonium lyase (Table 1) could impart enhanced biogenesis of veratryl alcohol and related aromatic compounds (49). A metabolite of P. chrysosporium, veratryl alcohol, has been implicated in ligninolysis as a diffusible oxidant or LiP stabilizer (30). We also observed high expression of alcohol dehydrogenases Phach1_10221 and Phach1_5793, both of which may participate in the reductive detoxification of various lignin-derived aldehydes (42). Also consistent with an active ligninolytic system, a GMC oxidoreductase (Phach1_9149) with similarity to Aspergillus niger glucose oxidase (22% identity) was detected in cultures containing line 64. This enzyme generates glucono-1,5-lactone and H2O2 from glucose. Possibly, catalases Phach1_127288 and Phach1_128306 serve to modulate toxic accumulations of peroxide which would otherwise impact the integrity of membranes.
Impressive levels of LPMO (Phach1_31049) were detected (Table 3). This metallo-monooxygenase boosts cellulose degradation (5–7, 50), and recent investigations have demonstrated LPMO activity on hemicellulose (8). All cultures also featured substantial protein and transcript levels for CDH and aldose epimerase (ALE1). Cellobiose β-anomer, the preferred CDH substrate (51), may be generated by ALE1. CDH and CBHs were previously identified in P. chrysosporium cultures (9, 17, 33, 52), and upregulated LPMOs (Table 3; see also Data Set S1 in the supplemental material) increase oxidative degradation of cellulose and xyloglucans.
Determining the precise role of these genes in lignocellulose degradation remains a task for future research, and elucidating the roles of the many hypothetical proteins is particularly challenging. For example, 343 of the 793 proteins identified are completely uncharacterized (Fig. 2). Similarly, 39 of the 113 upregulated proteins are hypothetical. Transcripts corresponding to Phach1_5249, Phach1_6166, Phach1_5352, and Phach1_140079 accumulated in transgenic line 64 relative to line P717. These four unlinked genes had little in common (<21% identity) except for the presence of one or more transmembrane helices in Phach1_5249, Phach1_6166, and Phach1_5352. The Phach1_5249 and Phach1_5352 transcript levels were also significantly higher in line 64 than in line 82, further demonstrating a response to high syringyl lignin content. Interestingly, four Phach1_5352 homologs were clustered within a 12-kb region of scaffold 27, but all were constitutively expressed at relatively low levels (see Data Set S1). Based on transcript levels and protein identifications, these genes are potentially important in lignocellulose degradation, but the data should be cautiously interpreted. Regulated expression and secretion do not guarantee full activity. Along these lines, the absence of detectable peptides must be regarded as equivocal without information related to turnover rates, substrate binding characteristics, and the frequency of trypsin cleavage sites. Nevertheless, high transcript levels associated with extracellular peptides warrant further examination of temporal expression patterns and biochemical properties.
Conversion efficiency barriers substantially complicate enzymatic “deconstruction” of forest biomass. Much effort is currently focused on improving wood utilization, particularly through identification/improvement of enzyme systems, genetic alterations in plant cell wall composition, and development of pretreatment processes. Our results demonstrate the importance of host genotype and suggest that commercial enzyme mixtures might be improved by tailoring enzyme components to specific feedstocks. P. chrysosporium provides a uniquely suited model system because it expresses all enzymatic and nonenzymatic components necessary to disassemble wood into useful monomers. The information gained may allow the further development of enzymatic systems for biomass treatment and help guide Populus breeding programs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (grant 2007-35504-18257) and by the Agriculture and Food Research Initiative, grant number 2011-67009-20056, from the USDA National Institute of Food and Agriculture to the Forest Products Laboratory. A.A. and J.R. were funded in part by the Department of Energy Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494).
Footnotes
Published ahead of print 11 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01604-14.
REFERENCES
- 1.Eriksson K-EL, Blanchette RA, Ander P. 1990. Microbial and enzymatic degradation of wood and wood components. Springer-Verlag, Berlin, Germany [Google Scholar]
- 2.Kirk TK, Farrell RL. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465–505. 10.1146/annurev.mi.41.100187.002341 [DOI] [PubMed] [Google Scholar]
- 3.Martinez D, Larrondo LF, Putnam N, Sollewijn Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695–700. 10.1038/nbt967 [DOI] [PubMed] [Google Scholar]
- 4.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westereng B, Ishida T, Vaaje-Kolstad G, Wu M, Eijsink VG, Igarashi K, Samejima M, Stahlberg J, Horn SJ, Sandgren M. 2011. The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS One 6:e27807. 10.1371/journal.pone.0027807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen JC, Johansen KS, Krogh KB, Jorgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. 2011. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U. S. A. 108:15079–15084. 10.1073/pnas.1105776108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bey M, Berrin JG, Poidevin L, Sigoillot JC. 2011. Heterologous expression of Pycnoporus cinnabarinus cellobiose dehydrogenase in Pichia pastoris and involvement in saccharification processes. Microb. Cell Fact. 10:113. 10.1186/1475-2859-10-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agger JW, Isaksen T, Varnai A, Vidal-Melgosa S, Willats WG, Ludwig R, Horn SJ, Eijsink VG, Westereng B. 2014. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc. Natl. Acad. Sci. U. S. A. 111:6287–6292. 10.1073/pnas.1323629111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wymelenberg AV, Sabat G, Martinez D, Rajangam AS, Teeri TT, Gaskell J, Kersten PJ, Cullen D. 2005. The Phanerochaete chrysosporium secretome: database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. J. Biotechnol. 118:17–34. 10.1016/j.jbiotec.2005.03.010 [DOI] [PubMed] [Google Scholar]
- 10.Vanden Wymelenberg A, Gaskell J, Mozuch MD, Kersten P, Sabat G, Martinez D, Cullen D. 2009. Transcriptome and secretome analysis of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl. Environ. Microbiol. 75:4058–4068. 10.1128/AEM.00314-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manavalan A, Adav SS, Sze SK. 2011. iTRAQ-based quantitative secretome analysis of Phanerochaete chrysosporium. J. Proteomics 75:642–654. 10.1016/j.jprot.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 12.Hori C, Igarashi K, Katayama A, Samejima M. 2011. Effects of xylan and starch on secretome of the basidiomycete Phanerochaete chrysosporium grown on cellulose. FEMS Microbiol. Lett. 321:14–23. 10.1111/j.1574-6968.2011.02307.x [DOI] [PubMed] [Google Scholar]
- 13.Vanden Wymelenberg A, Gaskell J, Mozuch MD, Sabat G, Ralph J, Skyba O, Mansfield S, Blanchette RA, Martinez D, Grigoriev I, Kersten P, Cullen D. 2010. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl. Environ. Microbiol. 76:3599–3610. 10.1128/AEM.00058-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbas A, Koc H, Liu F, Tien M. 2005. Fungal degradation of wood: initial proteomic analysis of extracellular proteins of Phanerochaete chrysosporium grown on oak substrate. Curr. Genet. 47:49–56. 10.1007/s00294-004-0550-4 [DOI] [PubMed] [Google Scholar]
- 15.Sato S, Liu F, Koc H, Tien M. 2007. Expression analysis of extracellular proteins from Phanerochaete chrysosporium grown on different liquid and solid substrates. Microbiology 153:3023–3033. 10.1099/mic.0.2006/000513-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato S, Feltus FA, Iyer P, Tien M. 2009. The first genome-level transcriptome of the wood-degrading fungus Phanerochaete chrysosporium grown on red oak. Curr. Genet. 55:273–286. 10.1007/s00294-009-0243-0 [DOI] [PubMed] [Google Scholar]
- 17.Vanden Wymelenberg A, Gaskell J, Mozuch M, BonDurant SS, Sabat G, Ralph J, Skyba O, Mansfield SD, Blanchette RA, Grigoriev IV, Kersten PJ, Cullen D. 2011. Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species. Appl. Environ. Microbiol. 77:4499–4507. 10.1128/AEM.00508-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravalason H, Jan G, Molle D, Pasco M, Coutinho PM, Lapierre C, Pollet B, Bertaud F, Petit-Conil M, Grisel S, Sigoillot JC, Asther M, Herpoel-Gimbert I. 2008. Secretome analysis of Phanerochaete chrysosporium strain CIRM-BRFM41 grown on softwood. Appl. Microbiol. Biotechnol. 80:719–733. 10.1007/s00253-008-1596-x [DOI] [PubMed] [Google Scholar]
- 19.Banerjee G, Scott-Craig JS, Walton JD. 2010. Improving enzymes for biomass conversion: a basic research perspective. Bioenerg. Res. 3:82–92. 10.1007/s12155-009-9067-5 [DOI] [Google Scholar]
- 20.Van Acker R, Leple JC, Aerts D, Storme V, Goeminne G, Ivens B, Legee F, Lapierre C, Piens K, Van Montagu MC, Santoro N, Foster CE, Ralph J, Soetaert W, Pilate G, Boerjan W. 2014. Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. U. S. A. 111:845–850. 10.1073/pnas.1321673111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skyba O, Douglas CJ, Mansfield SD. 2013. Syringyl-rich lignin renders poplars more resistant to degradation by wood decay fungi. Appl. Environ. Microbiol. 79:2560–2571. 10.1128/AEM.03182-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kues U, Kumar TK, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Duenas FJ, Sabat G, Salamov A, Samejima M, Schmutz J, Slot JC, St John F, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbett DS. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. 10.1126/science.1221748 [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Fueyo E, Ruiz-Duenas FJ, Ferreira P, Floudas D, Hibbett DS, Canessa P, Larrondo LF, James TY, Seelenfreund D, Lobos S, Polanco R, Tello M, Honda Y, Watanabe T, Ryu JS, Kubicek CP, Schmoll M, Gaskell J, Hammel KE, St John FJ, Vanden Wymelenberg A, Sabat G, Splinter BonDurant S, Syed K, Yadav JS, Doddapaneni H, Subramanian V, Lavin JL, Oguiza JA, Perez G, Pisabarro AG, Ramirez L, Santoyo F, Master E, Coutinho PM, Henrissat B, Lombard V, Magnuson JK, Kues U, Hori C, Igarashi K, Samejima M, Held BW, Barry KW, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Riley R, Salamov AA, Hoffmeister D, Schwenk D, Hadar Y, Yarden O, de Vries RP, Wiebenga A, Stenlid J, Eastwood D, Grigoriev IV, Berka RM, Blanchette RA, Kersten P, Martinez AT, Vicuna R, Cullen D. 2012. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc. Natl. Acad. Sci. U. S. A. 109:5458–5463. 10.1073/pnas.1119912109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. 2013. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 6:41. 10.1186/1754-6834-6-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen D. 2013. Wood decay, p 41–62 In Martin F. (ed), Ecological genomics of fungi. Wiley-Blackwell, New York, NY [Google Scholar]
- 26.Shimizu M, Yuda N, Nakamura T, Tanaka H, Wariishi H. 2005. Metabolic regulation at the tricarboxylic acid and glyoxylate cycles of the lignin-degrading basidiomycete Phanerochaete chrysosporium against exogenous addition of vanillin. Proteomics 5:3919–3931. 10.1002/pmic.200401251 [DOI] [PubMed] [Google Scholar]
- 27.Nakamura T, Ichinose H, Wariishi H. 2010. Cloning and heterologous expression of two aryl-aldehyde dehydrogenases from the white-rot basidiomycete Phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 394:470–475. 10.1016/j.bbrc.2010.01.131 [DOI] [PubMed] [Google Scholar]
- 28.Nakamura T, Ichinose H, Wariishi H. 2012. Flavin-containing monooxygenases from Phanerochaete chrysosporium responsible for fungal metabolism of phenolic compounds. Biodegradation 23:343–350. 10.1007/s10532-011-9521-x [DOI] [PubMed] [Google Scholar]
- 29.Worrall JJ, Anagnost SE, Zabel RA. 1997. Comparison of wood decay among diverse lignicolous fungi. Mycologia 89:199–219. 10.2307/3761073 [DOI] [Google Scholar]
- 30.Hammel KE, Cullen D. 2008. Role of fungal peroxidases in biological ligninolysis. Curr. Opin. Plant Biol. 11:349–355. 10.1016/j.pbi.2008.02.003 [DOI] [PubMed] [Google Scholar]
- 31.Kersten P, Cullen D. 2007. Extracellular oxidative systems of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Fungal Genet. Biol. 44:77–87. 10.1016/j.fgb.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 32.Highley TL. 1973. Influence of carbon source on cellulase activity of white rot and brown rot fungi. Wood Fiber 5:50–58 [Google Scholar]
- 33.Vanden Wymelenberg A, Minges P, Sabat G, Martinez D, Aerts A, Salamov A, Grigoriev I, Shapiro H, Putnam N, Belinky P, Dosoretz C, Gaskell J, Kersten P, Cullen D. 2006. Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveals complex mixtures of secreted proteins. Fungal Genet. Biol. 43:343–356. 10.1016/j.fgb.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 34.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 35.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:1544–6115. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300 [Google Scholar]
- 37.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 38.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, Otillar R, Poliakov A, Ratnere I, Riley R, Smirnova T, Rokhsar D, Dubchak I. 2012. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 40:D26–D32. 10.1093/nar/gkr947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29:365–371. 10.1038/ng1201-365 [DOI] [PubMed] [Google Scholar]
- 40.Stewart JJ, Akiyama T, Chapple C, Ralph J, Mansfield SD. 2009. The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol. 150:621–635. 10.1104/pp.109.137059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makela MR, Sietio OM, de Vries RP, Timonen S, Hilden K. 2014. Oxalate-metabolising genes of the white-rot fungus Dichomitus squalens are differentially induced on wood and at high proton concentration. PLoS One 9:e87959. 10.1371/journal.pone.0087959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma M, Wang X, Zhang X, Zhao X. 2013. Alcohol dehydrogenases from Scheffersomyces stipitis involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Appl. Microbiol. Biotechnol. 97:8411–8425. 10.1007/s00253-013-5110-8 [DOI] [PubMed] [Google Scholar]
- 43.Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye-Rowley WS. 2001. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 276:244–250. 10.1074/jbc.M007103200 [DOI] [PubMed] [Google Scholar]
- 44.Daniel G, Volc J, Filonova L, Plihal O, Kubatova E, Halada P. 2007. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl. Environ. Microbiol. 73:6241–6253. 10.1128/AEM.00977-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niemenmaa O, Uusi-Rauva A, Hatakka A. 2008. Demethoxylation of [O14CH3]-labelled lignin model compounds by the brown-rot fungi Gloeophyllum trabeum and Poria (Postia) placenta. Biodegradation 19:555–565. 10.1007/s10532-007-9161-3 [DOI] [PubMed] [Google Scholar]
- 46.Stewart P, Kersten P, Vanden Wymelenberg A, Gaskell J, Cullen D. 1992. The lignin peroxidase gene family of Phanerochaete chrysosporium: complex regulation by carbon and nitrogen limitation, and the identification of a second dimorphic chromosome. J. Bacteriol. 174:5036–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanden Wymelenberg A, Sabat G, Mozuch MD, Kersten P, Cullen D, Blanchette RA. 2006. Structure, organization, and transcriptional regulation of a family of copper radical oxidase genes in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 72:4871–4877. 10.1128/AEM.00375-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomez-Toribio V, Garcia-Martin AB, Martinez MJ, Martinez AT, Guillen F. 2009. Induction of extracellular hydroxyl radical production by white-rot fungi through quinone redox cycling. Appl. Environ. Microbiol. 75:3944–3953. 10.1128/AEM.02137-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jensen KA, Evans KM, Kirk TK, Hammel KE. 1994. Biosynthetic pathway for veratryl alcohol in the ligninolytic fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 60:709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, Xu F, Cherry J, Larsen S, Lo Leggio L. 2010. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49:3305–3316. 10.1021/bi100009p [DOI] [PubMed] [Google Scholar]
- 51.Higham CW, Gordon-Smith D, Dempsey CE, Wood PM. 1994. Direct 1H NMR evidence for conversion of β-d-cellobiose to cellobionolactone by cellobiose dehydrogenase from Phanerochaete chrysosporium. FEBS Lett. 351:128–132. 10.1016/0014-5793(94)00847-7 [DOI] [PubMed] [Google Scholar]
- 52.Mansfield SD, deJong E, Saddler JN. 1997. Cellobiose dehydrogenase, an active agent in cellulose depolymerization. Appl. Environ. Microbiol. 63:3804–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reiser J, Muheim A, Hardegger M, Frank G, Fiechter A. 1994. Aryl-alcohol dehydrogenase from the white-rot fungus Phanerochaete chrysosporium: gene cloning, sequence analysis, expression and purification of recombinant protein. J. Biol. Chem. 269:28152–28159 [PubMed] [Google Scholar]
- 54.Aspeborg H, Coutinho PM, Wang Y, Brumer H, III, Henrissat B. 2012. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12:186. 10.1186/1471-2148-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.