Abstract
The haloarchaeon Natrinema sp. strain J7-2 has the ability to degrade chitin, and its genome harbors a chitin metabolism-related gene cluster that contains a halolysin gene, sptC. The sptC gene encodes a precursor composed of a signal peptide, an N-terminal propeptide consisting of a core domain (N*) and a linker peptide, a subtilisin-like catalytic domain, a polycystic kidney disease domain (PkdD), and a chitin-binding domain (ChBD). Here we report that the autocatalytic maturation of SptC is initiated by cis-processing of N* to yield an autoprocessed complex (N*-IWT), followed by trans-processing/degradation of the linker peptide, the ChBD, and N*. The resulting mature form (MWT) containing the catalytic domain and the PkdD showed optimum azocaseinolytic activity at 3 to 3.5 M NaCl, demonstrating salt-dependent stability. Deletion analysis revealed that the PkdD did not confer extra stability on the enzyme but did contribute to enzymatic activity. The ChBD exhibited salt-dependent chitin-binding capacity and mediated the binding of N*-IWT to chitin. ChBD-mediated chitin binding enhances SptC maturation by promoting activation of the autoprocessed complex. Our results also demonstrate that SptC is capable of removing proteins from shrimp shell powder (SSP) at high salt concentrations. Interestingly, N*-IWT released soluble peptides from SSP faster than did MWT. Most likely, ChBD-mediated binding of the autoprocessed complex to chitin in SSP not only accelerates enzyme activation but also facilitates the deproteinization process by increasing the local protease concentration around the substrate. By virtue of these properties, SptC is highly attractive for use in preparation of chitin from chitin-containing biomass.
INTRODUCTION
Haloarchaea thrive in hypersaline environments such as solar salterns, salt lakes, and salt deposits, and many of them secrete proteases to degrade proteins into small peptides and amino acids which are then fed into central metabolism (1). Almost all of the extracellular proteases isolated from haloarchaea are subtilisin-like serine proteases (subtilases) (2), known as halolysins (1, 3). With whole-genome sequence data for many organisms quickly becoming available, an increasing number of halolysin genes have been identified, emphasizing the physiological significance of halolysins in the haloarchaea. Because halolysins are active and stable under hypersaline conditions, they not only represent an attractive model for investigating the molecular basis of the halophilicity of enzymes but also show great potential as biocatalysts for the synthesis of oligopeptides due to the increase of their esterase/amidase activity ratios in organic mixtures of low water content (1, 4, 5). Additionally, the application of halophilic proteases at high salt concentrations can minimize the risk of microbial contamination (6).
Halolysins have been characterized from a number of haloarchaea, and these include halolysin 172P1 from Natrialba asiatica (3), R4 from Haloferax mediterranei (7), SptA from Natrinema sp. strain J7 (8), and Nep from Natrialba magadii (9). Each of these extracellular enzymes is synthesized as a precursor containing a signal peptide, an N-terminal propeptide, a subtilisin-like catalytic domain, and a C-terminal extension (CTE). Like bacterial subtilases, the signal peptide of halolysin precursors is removed after secretion by a signal peptidase to generate a proform, and then the N-terminal propeptide of the proform is autoprocessed to yield the active mature halolysin (10). The mature forms of halolysins are composed of not only a highly conserved catalytic domain (∼60 to 80% identity) but also a CTE. The CTEs of the above-mentioned halolysins also share high sequence identity (∼40 to 70%) and are reportedly important for the stability and activity of R4 (7), SptA (11), and Nep (10). The attachment of an extension to the C terminus of the catalytic domain appears to be a common strategy employed by halolysins to adapt to hypersaline environments.
The genus Natrinema comprises seven recognized species isolated from salted hide and cod (12), fish sauce (13), and salt lakes (14–17). Natrinema sp. J7 was isolated from a salt mine in Hubei Province, China, in the early 1990s (18), and it grows optimally at 3.1 to 3.8 M NaCl and at 40 to 45°C. Among the three 16S rRNA genes of Natrinema sp. J7-2 (a subculture of strain J7 that lacks the plasmid pHH205) (19), two copies show 100% identity to the 16S rRNA gene of Natrinema gari isolated from fish sauce in Thailand (13), while the third one differs from the latter at two nucleotides. This suggests that Natrinema sp. J7-2 is closely related to Natrinema gari. We previously cloned three halolysin-encoding genes from the Natrinema sp. J7 genome, named sptA, sptB (these two genes are arranged in tandem; GenBank accession number AY800382), and sptC (GenBank accession number DQ137266). The enzymatic properties of SptA and the function of its CTE have been characterized (8, 11). Interestingly, whole-genome sequencing of Natrinema sp. J7-2 (19) revealed that sptC is located in a chitin metabolism-related gene cluster, and we recently found that strain J7-2 was capable of degrading chitin. This led us to investigate the potential role of SptC in deproteinization of chitin-containing biomass.
The CTE of SptC shows no significant homology (<17% identity) to the CTEs of SptA, SptB, or other known halolysins (e.g., 172P1, R4, and Nep). In contrast to the CTEs of SptA and Nep, which are predicted to adopt a single β-jelly roll-like domain (11, 20), the CTE of SptC is predicted to contain a polycystic kidney disease domain (PkdD) and a chitin-binding domain (ChBD). Such a molecular architecture has been reported only for the chitin-binding protease AprIV from Pseudoalteromonas piscicida strain O-7 (formerly Alteromonas sp. strain O-7), wherein AprIV was found to participate in chitin degradation by the bacterium (21). The presence of the ChBD in SptC emphasizes the possibility that this enzyme is involved in degradation of chitin-containing biomass. In the present study, we investigated the maturation process and enzymatic properties of SptC. Several truncation and deletion mutants of SptC were constructed to probe possible roles for the PkdD and ChBD in enzyme stability, activity, and chitin binding. The mechanism of the chitin-accelerated activation of SptC is also discussed. In addition, we investigated the capacity of this halophilic protease to remove proteins from chitin-containing biomass at high salt concentrations.
MATERIALS AND METHODS
Strains and growth conditions.
Natrinema sp. J7 (CCTCC AB91141) was isolated from a salt mine in China (18). Natrinema sp. J7-2, a subculture of strain J7 lacking the plasmid pHH205 (19, 22), was grown in liquid modified growth medium with 18% total salts (18% MGM) as described previously (8) and used for preparation of genomic DNA. For examination of chitinolytic activity of strain J7-2, the haloarchaeon was first cultured at 37°C in 18% MGM until mid-log phase. Subsequently, cells were collected by centrifugation, washed with sterile 3 M NaCl solution, transferred to solid (1.5% [wt/vol] agar) basal medium (per liter, 230 g of NaCl, 43 g of MgSO4·7H2O, 2.5 g of KCl, 0.7 g of CaCl2·2H2O, 3.6 μg of MnCl2·4H2O, 4.4 μg of ZnSO4·7H2O, 0.5 μg of CuSO4·5H2O, 33 μg of FeSO4·7H2O, 0.27 g of NH4Cl, 2 ml of 0.5 M potassium phosphate buffer [pH 7.0], 50 ml of 1 M Tris-HCl [pH 7.2]) without or with 1 g/liter of colloidal chitin, and then incubated at 37°C for 2 weeks. Escherichia coli DH5α and E. coli BL21-CodonPlus(DE3)-RIL were used as the hosts for cloning and expression, respectively. Bacteria were grown at 37°C in Luria-Bertani (LB) medium supplemented with chloramphenicol (34 μg/ml) or kanamycin (30 μg/ml), as needed.
DNA manipulation, plasmid construction, and mutagenesis.
Genomic DNA of strain J7-2 was isolated and prepared according to the method of Kamekura et al. (3) and was used as the template for PCR. The oligonucleotide sequences of the primers used in this study are listed in Table 1. The genes encoding wild-type SptC (SptC*), the C-terminal truncation mutants (SptCΔCTE and SptCΔChBD), the N-terminal propeptide deletion mutant (SptCΔN), and the C-terminal fragments (ChBD*, PkdD*, and CTE*) were amplified from genomic DNA by PCR using the primer pairs listed in Table 2. The PkdD deletion mutant (SptCΔPkdD) was constructed using the overlapping extension PCR method, as described previously (23). Briefly, the 5′ and 3′ ends of sptC were amplified from genomic DNA using the primer pairs sptC-F/MChBD2 and MChBD1/sptCH-R, respectively. The first-round 5′- and 3′-end PCR products were then used for the second-round PCR without adding primer. In the third-round PCR, the intact gene encoding the PkdD deletion mutant (SptCΔPkdD) was amplified using the primer pair sptC-F/sptCH-R, with the products of the second-round PCR serving as the template. The above-mentioned amplified genes were then inserted into pET26b or pET28a to generate expression plasmids for target proteins (Table 2). The QuikChange site-directed mutagenesis method (24) was employed to construct the active-site mutant SptCS230A using the primers sptCS230A-F and sptCS230A-R (Table 1). The sequences of all recombinant plasmids were confirmed by DNA sequencing.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Oligonucleotide sequencea |
|---|---|
| sptC-F | 5′-CGGAATTCATATGACAAATAACAACGACCGAGACAG-3′ |
| sptC-R | 5′-TTTAAGCTTTCAGCAGTCCGCGACGTGTTCCCA-3′ |
| sptCH-R | 5′-TTTAAGCTTTCAGTGGTGGTGGTGGTGGTGGCAGTCCGCGACGTGTTCCCA-3′ |
| sptCΔC54H-R | 5′-TTTAAGCTTTCAGTGGTGGTGGTGGTGGTGCTCGCCGGTGCCGTCGTTGTC-3′ |
| sptCΔC143H-R | 5′-TTTAAGCTTTCAGTGGTGGTGGTGGTGGTGGTCGCCGCCCTCGTCGACGAG-3′ |
| sptCΔN127-F | 5′-GGGAATTCCATATGGAACAGCGCACCCCCGAC-3′ |
| sptCC151-F | 5′-GGGAATTCCATATGGCTCTCGTCGACGAGGGCGGC-3′ |
| sptCC54-F | 5′-GGGAATTCCATATGCAACCGGGCGACTGCGGCGAC-3′ |
| MChBD1 | 5′-GCTCTCGTCGACGAGGGCGGCCAACCGGGCGACTGCGGCGAC-3′ |
| MChBD2 | 5′-GTCGCCGCAGTCGCCCGGTTGGCCGCCCTCGTCGACGAGAGC-3′ |
| sptCS230A-F | 5′-GTCGGGCACGGCGATGGCGTCGC-3′ |
| sptCS230A-R | 5′-TCGCCGTGCCCGACAGCCGCTCGTAGCCGC-3′ |
Underlined sequences indicate the restriction enzyme sites used in this study. Bold indicates the mutated nucleotides. Italicized sections indicate the His6 tag-encoding DNA sequences.
TABLE 2.
Primer pairs, templates, and restriction enzyme sites used in plasmid construction
| Plasmid | Primer pair | Template | Restriction site |
|---|---|---|---|
| pET26b-sptC* | sptC-F + sptCH-R | Genomic DNA | NdeI-HindIII |
| pET26b-sptCΔCTE | sptC-F + sptCΔC143H-R | Genomic DNA | NdeI-HindIII |
| pET26b-sptCΔChBD | sptC-F + sptCΔC54H-R | Genomic DNA | NdeI-HindIII |
| pET26b-sptCΔPkdD | sptC-F + MChBD2 (5′ end of the gene) | Genomic DNA | |
| MChBD1 + sptCH-R (3′ end of the gene) | Genomic DNA | ||
| sptC-F + sptCH-R (3rd-round PCR) | 2nd-round PCR product | NdeI-HindIII | |
| pET26b-sptCΔN | sptCΔN127-F + sptCH-R | Genomic DNA | NdeI-HindIII |
| pET26b-CTE* | sptCC151-F + sptCH-R | Genomic DNA | NdeI-HindIII |
| pET26b-PkdD* | sptCC151-F + sptCΔC54H-R | Genomic DNA | NdeI-HindIII |
| pET28a-ChBD* | sptCC54-F + sptC-R | Genomic DNA | NdeI-HindIII |
| pET26b-sptCS230A | sptCS230A-F + sptCS230A-R | pET26b-sptC |
Expression, purification, and activation.
E. coli BL21-CodonPlus(DE3)-RIL cells harboring the recombinant plasmids were cultured at 37°C until the optical density at 600 nm reached ∼0.6. Expression of recombinant proteins was induced by the addition of isopropyl-β-d-thiogalactopyranoside (0.4 mM) and continued cultivation at 37°C for 4 h. The cells were then harvested, suspended in buffer A (50 mM Tris-HCl, 10 mM CaCl2 [pH 8.0]) containing 8 M urea, and disrupted by sonication on ice. After centrifugation at 13,000 × g for 10 min (4°C), the soluble fraction was collected and subjected to affinity chromatography on a column containing Ni2+-charged chelating Sepharose Fast Flow resin (GE Healthcare) equilibrated with buffer A containing 8 M urea. After washing of the column with buffer A containing 8 M urea and 30 mM imidazole, bound His-tagged proteins were eluted with buffer A containing 8 M urea and 200 mM imidazole. The purity of isolated proteins was assessed by SDS-PAGE. Thereafter, the elution fraction was supplemented with 10 mM β-mercaptoethanol (β-ME) and kept at room temperature for 30 min. The sample was then dialyzed overnight at 4°C against buffer A containing 5 M NaCl to remove the urea and imidazole and allow the denatured protein to refold.
To prepare the mature enzymes, samples of refolded SptC*, SptCΔChBD, SptCΔPkdD, and SptCΔCTE were incubated at 37°C for 90 h in buffer A containing 5 M NaCl to activate the enzymes. The activated enzymes were then subjected to affinity chromatography on a bacitracin-Sepharose 4B (Amersham Biosciences, Sweden) column equilibrated with buffer A containing 5 M NaCl. After a washing with buffer A containing 5 M NaCl, the bound enzymes were eluted with buffer A containing 3 M NaCl and 15% isopropanol. Finally, fractions containing the purified enzymes (MWT, MΔChBD, MΔPkdD, and MΔCTE) were dialyzed overnight at 4°C against buffer A containing 5 M NaCl to remove the isopropanol. If needed, the enzyme-containing solutions were concentrated using Micron YM-3 centrifugal filters (Millipore, Bedford, MA). The concentration of each purified enzyme sample was determined using the Bradford method (25), with bovine serum albumin (BSA) as the standard.
SDS-PAGE, immunoblot analysis, and N-terminal sequencing.
SDS-PAGE was performed using the Tris-glycine system (26). To prevent self-degradation of the enzymes during sample preparation (boiling) or electrophoresis, the proteins were precipitated with 20% (wt/vol) trichloroacetic acid (TCA), washed with acetone, solubilized in loading buffer containing 8 M urea, and then subjected to electrophoresis without prior heat treatment. An anti-His tag monoclonal antibody (Novagen) was used for immunoblot analysis, as described previously (27). For N-terminal sequencing, proteins separated by SDS-PAGE were electroblotted onto a polyvinylidene difluoride membrane and then stained with Coomassie brilliant blue R-250. The target protein bands were excised and subjected to N-terminal amino acid sequence analysis using a Procise 492 cLC peptide sequencer (Applied Biosystems).
Enzymatic activity assay.
Unless otherwise indicated, the azocaseinolytic activity of the enzyme was measured at 40°C for 60 min in 200 μl of reaction mixture containing 0.25% (wt/vol) azocasein (Sigma) and 100 μl of enzyme sample in buffer A containing 3.5 M NaCl. The reaction was terminated by addition of 200 μl of 40% (wt/vol) TCA to the reaction mixture. The mixture was maintained at room temperature for 15 min and then centrifuged at 13,400 × g for 10 min, after which the absorbance of the supernatant was measured in a 1-cm cell at 366 nm (A366). One unit of azocaseinolytic activity was defined as the amount of enzyme required to increase the A366 value by 0.01 per min under the conditions described above.
Proteolytic activity against N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (suc-AAPF-pNA) (Sigma) was measured at 40°C in buffer A containing 3.5 M NaCl and 0.1 mM substrate. The initial velocity of suc-AAPF-pNA hydrolysis was monitored at 410 nm in a thermostat-controlled spectrophotometer (Cintra 10e; GBC, Australia), and the activity was calculated based on the extinction coefficient for p-nitroaniline (pNA) (8,480 M−1 cm−1 at 410 nm) (28). One unit of enzyme activity was defined as the amount of enzyme required to produce 1 μmol of pNA per min under the assay conditions described.
Chitin powder binding assay.
Prior to use, 5 mg of chitin powder (Sigma) was washed three times with deionized water, suspended in 500 μl of buffer A containing 5 M NaCl, and then kept at 0°C for 1 h. After centrifugation at 13,400 × g for 10 min, the resulting pellet was suspended in 300 μl of buffer A containing 5 M NaCl and 5 μg/ml of the target protein. Unless otherwise indicated, the mixture was incubated at 0°C to prevent activation of the enzyme (i.e., to prevent activation of the autoprocessed complex). After 1 h of incubation at 0°C, the soluble fraction was recovered by centrifugation as described above and subjected to SDS-PAGE analysis. The levels of unbound proteins were estimated from the band intensities on the SDS-PAGE gels using GeneTools gel imaging software (Syngene, Cambridge, United Kingdom).
Enzymatic deproteinization of SSP.
Shrimp shell powder (SSP) was prepared according to the method described by Miyamoto et al. (21). Briefly, shrimp were purchased from a local market, and the shells were stripped off and washed thoroughly with tap water. The shells were then dried at 50°C for 24 h, crushed in a mixing blender, and sieved to produce a 40-mesh powder. Prior to use, 2 mg of SSP was washed three times with buffer A containing 5 M NaCl and then pelleted by centrifugation. Unless otherwise indicated, the pellet was suspended in 200 μl of buffer A containing 5 M NaCl and 0.4 μM mature enzyme or autoprocessed complex, incubated at 40°C for various durations, and then cooled on ice. Thereafter, the supernatant was recovered by centrifugation and the amount of soluble peptides released from SSP by enzymatic hydrolysis was determined by measuring the absorbance at 280 nm (29).
Deproteinization using alkali.
SSP was mixed with 2 M NaOH at a ratio of 1:40 (wt/vol) and allowed to react at 100°C for 30 min (30). The supernatant was recovered by centrifugation and the pH was adjusted to 7.0. Next, the protein concentration in the supernatant was determined using the Bradford method (25), with BSA as the standard. To assess the degree of deproteinization of SSP by enzymatic hydrolysis, enzyme-treated SSP was washed three times with buffer A containing 5 M NaCl and then deproteinized with alkali, after which the protein concentration was determined as described above. The percentage of enzymatic deproteinization of SSP was calculated with 0% deproteinization defined as 24.4 mg of protein contained in 1 g of SSP without enzyme treatment.
RESULTS
Bioinformatic analysis of SptC.
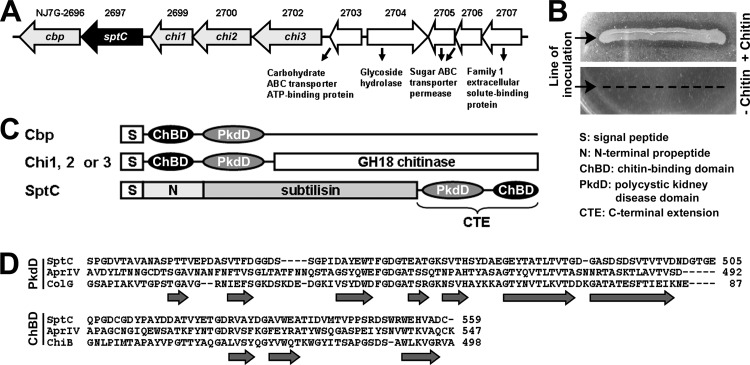
Genomic analysis of Natrinema sp. J7-2 (19) revealed the presence of a chitin metabolism-related gene cluster (NJ7G-2696 to -2707) that encodes a chitin-binding protein (Cbp), SptC, three chitinases (Chi1, Chi2, and Chi3), and five proteins involved in carbohydrate transport and metabolism (Fig. 1A). A genomic survey of sequenced haloarchaea showed that this gene cluster is highly conserved in Natrinema species (e.g., N. altunense, N. gari, and N. pallidum) and appears to be confined to the genus Natrinema. A chitin metabolism-related gene cluster has also been identified in H. mediterranei, but no protease-encoding gene was identified in that cluster (31). Strain J7-2 could grow on a plate containing colloidal chitin as the carbon source, and the chitinolytic activity of the haloarchaeon was evidenced by the formation of clear zones around the lawn (Fig. 1B). As shown in Fig. 1C, SptC, Cbp, Chi1, Chi2, and Chi3 of strain J7-2 share two domains, a PkdD and a ChBD. The PkdDs and ChBDs of these five proteins share significant sequence identity (36 to 54% for the PkdDs and 27 to 75% for the ChBDs), suggesting that an evolutionary relationship exists between these proteins. A BLAST search revealed that the homologs of SptC with a PkdD and a ChBD are present only in Natrinema species (>90% sequence identity) and Pseudoalteromonas species (∼30% sequence identity). The genes encoding the homologs in Natrinema species are all located within the chitin metabolism-related gene clusters mentioned above, while those in Pseudoalteromonas species are not clustered with chitin metabolism-related genes. Automated homology modeling of the SptC PkdD yielded a structure similar to that of the Clostridium histolyticum collagenase PkdD, which adopts a β-jelly roll-like conformation composed of seven β-strands (Fig. 1D). The ChBD of SptC shares 31 and 39% sequence identity with the ChBDs of P. piscicida AprIV (21) and Serratia marcescens chitinase B, respectively (Fig. 1D) and may adopt a conformation similar to that of the latter, which is composed of three antiparallel β-strands connected by loops (32).
FIG 1.
Organization of the chitin metabolism-related gene cluster in Natrinema sp. J7-2 and the primary structural features of SptC. (A) Schematic illustrating the organization of the chitin metabolism-related gene cluster in strain J7-2. Genes encoding chitin-binding protein (Cbp), SptC, chitinases (Chi1, Chi2, and Chi3), and proteins involved in carbohydrate transport and metabolism (NJ7G_2703-2707) are drawn to scale as arrows. The locus tag of each gene is shown above the corresponding arrow. (B) Assay of chitinolytic activity of strain J7-2. Strain J7-2 was cultured at 37°C for 2 weeks on plates containing basal medium with (+) or without (−) 1 g/liter of colloidal chitin. (C) Schematic representation of the domain organization in Cbp, Chi1, Chi2, Chi3, and SptC. (D) Amino acid sequence alignments of the PkdD and ChBD in SptC (GenBank accession number AFO57926) with those of chitin-binding protease AprIV (GenBank accession number BAB86297) from P. piscicida strain O-7, the PkdD (3JQU) of collagenase (ColG) from C. histolyticum, and the ChBD (1E15) of chitinase B (ChiB) from S. marcescens. The amino acid residues of SptC are numbered starting from the N terminus of the precursor. The β-strands in the PkdD of ColG and the ChBD of ChiB are indicated by horizontal arrows.
Autoprocessing of SptC.
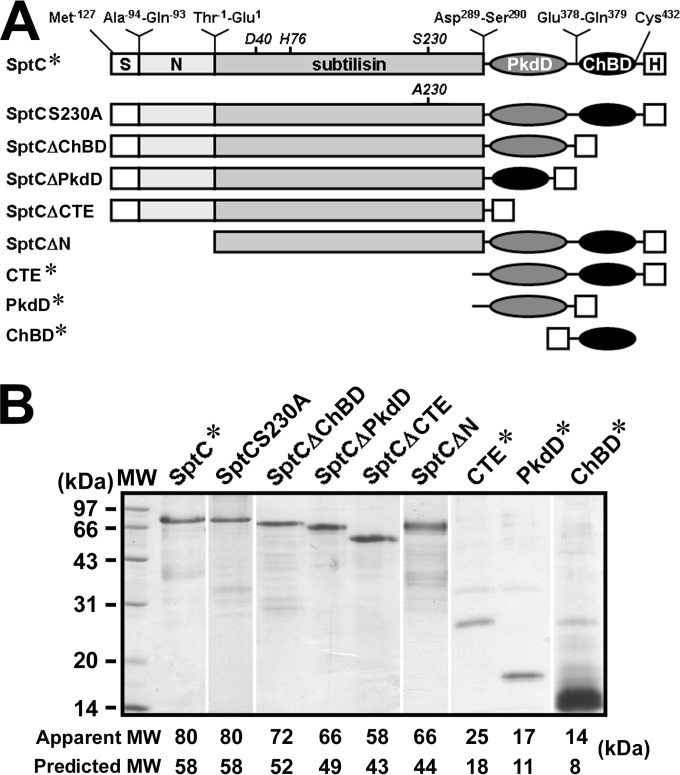
Recombinant wild-type SptC (SptC*) and its derivatives (Fig. 2A) were purified in the presence of 8 M urea by His tag affinity chromatography. It was observed that the apparent molecular masses of these recombinant proteins were higher than their predicted masses (Fig. 2B), which is a common feature of halophilic proteins with a high content of acidic residues (33). In addition to the target proteins, SDS-PAGE analysis revealed the presence of a few minor protein bands in the purified samples (Fig. 2B). Most of these minor species could be detected by anti-His tag immunoblot analysis (data not shown), indicating that they are degradation products of the target proteins.
FIG 2.
Schematic representation of the primary structures (A) and SDS-PAGE analysis of recombinant SptC and its derivatives (B). (A) The locations of the active-site residues (D40, H76, and S230) in the subtilisin-like catalytic domain and the N- and C-terminal residues of each region are shown. S, N, and H represent the signal peptide, N-terminal propeptide, and His tag, respectively. (B) Samples of purified proteins were precipitated with TCA and then subjected to SDS-PAGE analysis. The predicted molecular mass (MW) of each protein was calculated based on the amino acid sequence, and the apparent MW was determined by SDS-PAGE analysis.
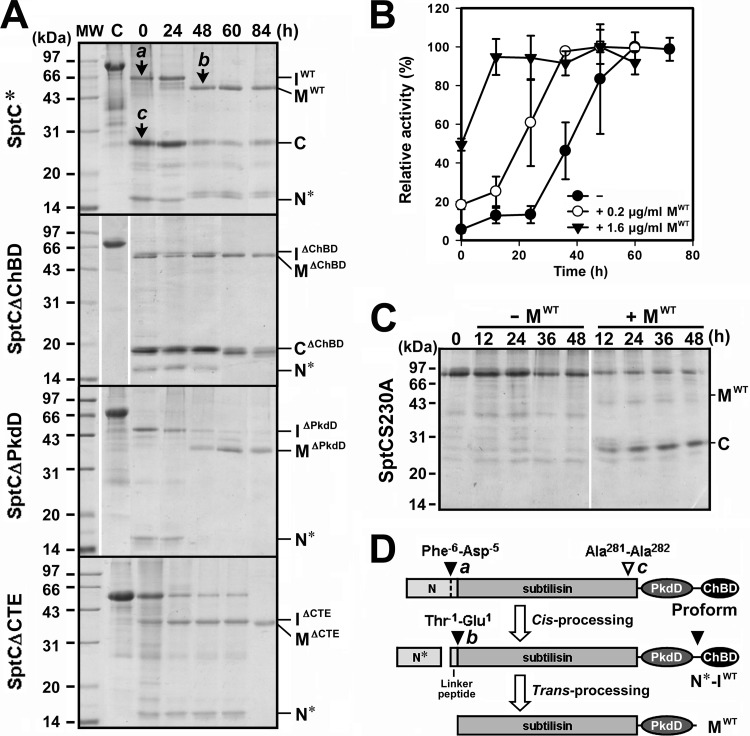
Denatured SptC* (Fig. 3A, lane C) was dialyzed at 4°C against buffer A containing 5 M NaCl to allow the protein to refold. Removal of urea by dialysis at a high NaCl concentration led to the conversion of SptC* into three major products, designated IWT, C, and N* (Fig. 3A, lane 0). During subsequent incubation of the dialyzed sample at 40°C, IWT was further processed into the mature form (MWT), and this was accompanied by the degradation of N* (Fig. 3A) and the complete release of enzymatic activity (Fig. 3B). Conversely, the active-site mutant SptCS230A remained unprocessed under the same conditions (Fig. 3C), suggesting that the maturation of SptC* proceeds autocatalytically. In addition, the N-terminal propeptide deletion mutant SptCΔN was unable to mature (data not shown), suggesting that the N-terminal propeptide acts as an intramolecular chaperone to assist enzyme folding, as in other subtilases (34).
FIG 3.
Autoprocessing of recombinant SptC and its derivatives. (A) SDS-PAGE analysis of the autoprocessing of SptC* and its derivatives. Samples of purified SptC* and its derivatives (5 μg/ml, lane C) in buffer A (50 mM Tris-HCl, 10 mM CaCl2 [pH 8.0]) containing 8 M urea and 10 mM β-ME were dialyzed overnight at 4°C against the same buffer containing 5 M NaCl (lane 0) and then incubated at 40°C. At the indicated times, aliquots were withdrawn and subjected to SDS-PAGE analysis. The bands labeled a, b, and c were subjected to N-terminal sequencing. (B) Activation of the autoprocessed complex. N*-IWT (5 μg/ml) was incubated at 40°C in buffer A containing 5 M NaCl in the absence (−) or presence (+) of 0.2 or 1.6 μg/ml of MWT. At the indicated times, aliquots were removed and subjected to azocaseinolytic-activity assay. Values are expressed as the means ± SDs (bars) of three independent experiments, and the highest mean activity value observed during the maturation process was defined as 100% to determine relative activity. (C) Processing of SptCS230A by mature SptC. SptCS230A (5 μg/ml, lane 0) in buffer A containing 5 M NaCl was incubated at 40°C in the absence (−) or presence (+) of 0.3 μg/ml of MWT. At the indicated times, aliquots were removed and subjected to SDS-PAGE analysis. (D) Schematic representation of the autoprocessing of the proform of SptC. Filled arrowheads indicate the autoprocessing sites, and the open arrowhead indicates the site of proteolytic cleavage of SptC by the active mature enzyme. The identified N termini of bands a, b, and c from panel A are indicated. The positions of the intermediates (IWT, IΔChBD, IΔPkdD, and IΔCTE), the mature forms (MWT, MΔChBD, MΔPkdD, and MΔCTE), the processed core domain of the N-terminal propeptide (N*), and the cleaved C-terminal fragments (C and CΔChBD) are also indicated to the right of each gel in panels A and C.
N-terminal sequencing identified the first five amino acid residues of IWT and MWT as DPVET and EQRTP, respectively, indicating that the N-terminal propeptide is composed of a core domain (N*) and a five-residue linker peptide that is processed in a stepwise manner at Phe−6-Asp−5 and Thr−1-Glu1, respectively (Fig. 3D). Immunoblotting against the His tag fused to the C terminus of SptC* resulted in detection of IWT but not MWT (data not shown), indicating that IWT represents a maturation intermediate derived from SptC* through autoprocessing of N* at Phe−6-Asp−5 (Fig. 3D). In addition, N* was coeluted with IWT from the Ni2+-charged resin column (data not shown), indicating that N* remains bound to IWT to form an autoprocessed complex (N*-IWT).
The inability to detect MWT by anti-His tag immunoblot analysis suggests that a C-terminal cleavage event occurred during the maturation of SptC*. To test for this possibility, we investigated the maturation processes of the C-terminal truncation mutants (SptCΔChBD, SptCΔPkdD, and SptCΔCTE). As shown in Fig. 3A, all three mutants were converted into their respective intermediates (IΔChBD, IΔPkdD, and IΔCTE) and mature forms (MΔChBD, MΔPkdD, and MΔCTE), suggesting that the CTE is not necessary for enzyme folding. As in the case of IWT, the IΔChBD, IΔPkdD, and IΔCTE intermediates formed autoprocessed complexes with N* (data not shown), which were designated N*-IΔChBD, N*-IΔPkdD, and N*-IΔCTE, respectively. Notably, MΔChBD displayed nearly the same apparent molecular mass as MWT (∼50 kDa) (Fig. 3A), indicating that the ChBD had been processed from MWT. Similarly, MΔPkdD and MΔCTE showed the same apparent molecular mass, ∼35 kDa (Fig. 3A), suggesting that the ChBD had also been cleaved from MΔPkdD. The larger apparent molecular masses of MWT and MΔChBD than of MΔPkdD and MΔCTE suggest that their PkdDs had not been processed. Therefore, the conversion of IWT to MWT must involve truncation of both the linker peptide and the ChBD (Fig. 3D). The conversion of N*-IWT to MWT and the release of enzymatic activity were accelerated at high protein concentrations (data not shown) or in the presence of supplemental active MWT (Fig. 3B), suggesting that the processing of N*-IWT to MWT occurs intermolecularly (i.e., trans-processing) (Fig. 3D).
To determine whether the SptC precursor could be trans-processed to the intermediate and/or the mature form, the active-site mutant SptCS230A was incubated with MWT. As shown in Fig. 3C, SptCS230A was not converted into the intermediate and/or the mature form but was instead degraded by MWT. Conversion of SptC* to N*-IWT, however, was independent of protein concentration (data not shown). These results suggest that conversion of SptC* to the N*-IWT complex occurs in an intramolecular manner (i.e., cis-processing) (Fig. 3D). The apparent molecular mass of the major degradation product of SptCS230A (band C in Fig. 3C) was similar to that of the degradation product derived from SptC* (band C in Fig. 3A). Both proteins could be detected upon anti-His tag immunoblot analysis (data not shown), suggesting that they are the cleaved C-terminal fragments. The five N-terminal residues of the SptC*-derived product (band c in Fig. 3A) were identified as ALVDE, indicating that cleavage of the C-terminal fragment occurs at the Ala281-Ala282 bond in the catalytic domain of SptC (Fig. 3D). Evidence showing that the C-terminal fragment (but not the catalytic domain) accumulated after degradation of SptCS230A by MWT suggests that the PkdD and ChBD are more resistant to proteolysis than the catalytic domain. Therefore, the appearance of the product (band C in Fig. 3A) during the maturation of SptC* was most likely due to degradation of the catalytic domain by the active enzyme that matured earlier.
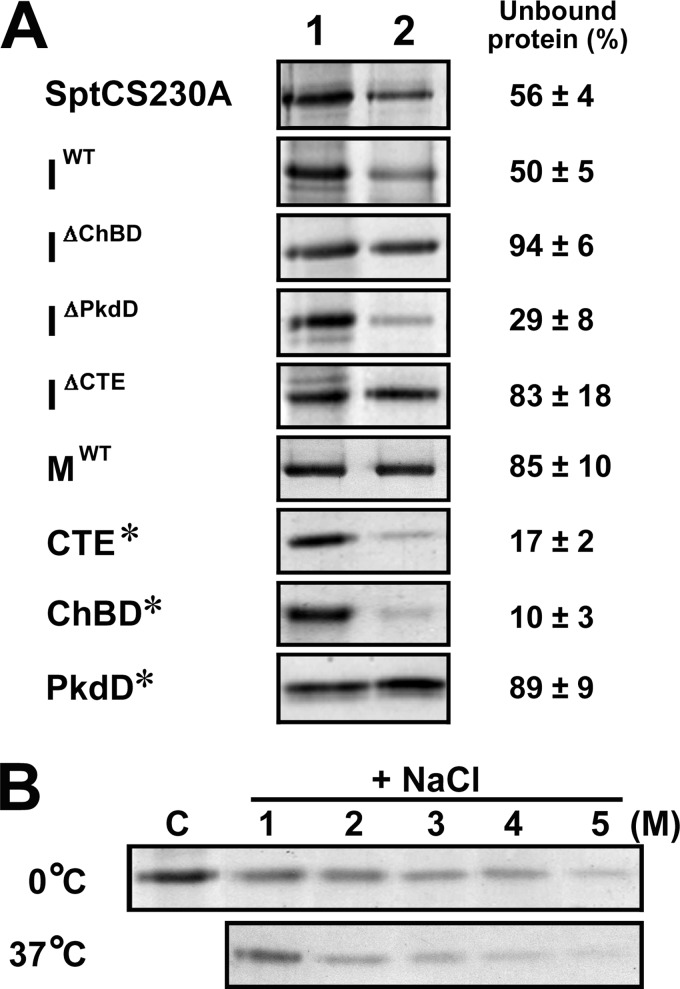
ChBD-mediated binding of SptC to chitin.
The presence of the ChBD in SptC raised the possibility that this domain mediates the binding of SptC to chitin. To examine this possibility, binding assays were carried out using insoluble chitin. As shown in Fig. 4A, proteins containing a ChBD (e.g., SptCS230A, IWT, IΔPkdD, CTE*, and ChBD*) bound to chitin to differing extents (∼50 to 90%), whereas the proteins lacking a ChBD (e.g., IΔChBD, IΔCTE, MWT, and PkdD*) showed no significant binding capacity, suggesting that the ChBD alone mediates the binding of the enzyme to chitin. We observed that about 80 to 90% of ChBD* and CTE* bound to chitin, whereas ∼50% of SptCS230A and IWT remained unbound under the same conditions (Fig. 4A), indicating that the chitin-binding capacity of the ChBD is weakened when the ChBD is covalently attached to the C terminus of the SptC catalytic domain. In addition to interacting with chitin, the ChBD probably also participates in the interdomain interaction in the proform and the autoprocessed complex of SptC, and there appears to be a certain degree of competition between the two interactions. In addition, the chitin-binding capacity of CTE* became stronger with increasing concentrations of NaCl at both 0°C and 37°C (Fig. 4B), indicating that the binding of the SptC ChBD to chitin is salt dependent.
FIG 4.
ChBD-mediated binding of SptC to chitin. (A) Chitin-binding capacities of SptCS230A, the intermediate forms (IWT, IΔChBD, IΔPkdD, and IΔCTE), the mature form (MWT), and the C-terminal fragments (ChBD*, PkdD*, and CTE*). Each protein (5 μg/ml) was incubated in 300 μl of buffer A (50 mM Tris-HCl, 10 mM CaCl2 [pH 8.0]) containing 5 M NaCl at 0°C for 30 min in the absence (lane 1) or presence (lane 2) of 5 mg of insoluble chitin. After centrifugation, 120 μl of the supernatant was subjected to SDS-PAGE analysis. Numbers given on the right of the images indicate the densitometric ratios of unbound protein (lane 2) to the control (lane 1) and are expressed as the means ± SDs of three independent experiments. (B) Salt dependence of the binding of the CTE* to chitin. CTE* (5 μg/ml) was incubated at 0 or 37°C for 30 min without chitin and NaCl (lane C) or with 5 mg of insoluble chitin in 300 μl of buffer A containing different concentrations of NaCl (1 to 5 M). After centrifugation, the supernatants were subjected to SDS-PAGE analysis.
Chitin-accelerated activation of SptC.
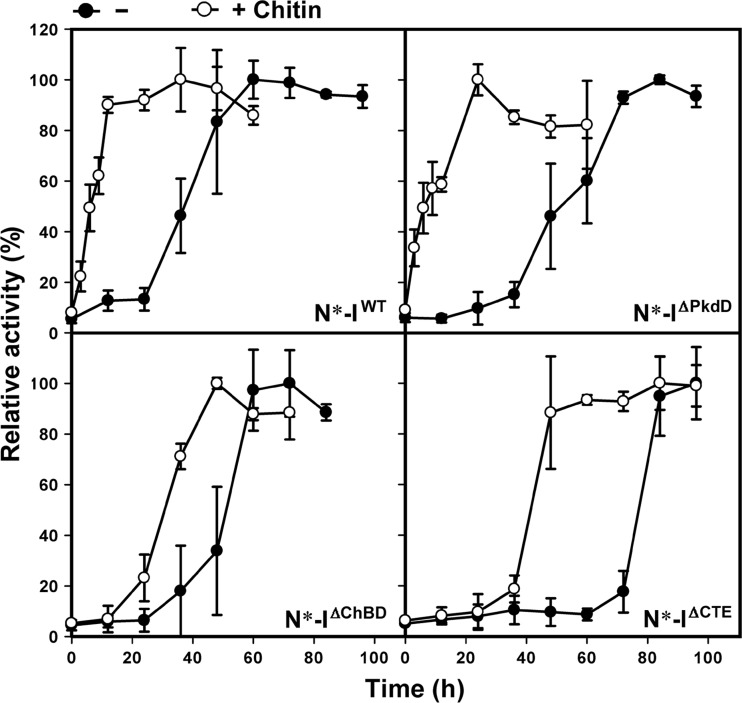
Based on the findings that the ChBD is processed from the mature form but mediates the binding of the intermediate to chitin, we hypothesized that ChBD-mediated binding of the intermediate to chitin contributes to SptC maturation. To address this issue, we investigated the effect of chitin on activation of inactive autoprocessed complexes derived from SptC*, SptCΔPkdD, SptCΔChBD, and SptCΔCTE. In the absence of chitin, N*-IWT activated faster than N*-IΔChBD, N*-IΔPkdD, and N*-IΔCTE (Fig. 5), indicating that enzyme activation is facilitated by the ChBD and the PkdD. Meanwhile, activation of the PkdD-containing autoprocessed complex (N*-IWT or N*-IΔChBD) was faster than that of the complex lacking the PkdD (N*-IΔPkdD or N*-IΔCTE), suggesting that the interaction between the PkdD and the catalytic domain is important for enzyme activation in the absence of chitin. Notably, activation of all four autoprocessed complexes was significantly accelerated in the presence of chitin (Fig. 5). The fact that chitin accelerated the activation of N*-IΔCTE suggests that the interaction of chitin with the catalytic domain and/or the N-terminal propeptide facilitates conversion of the autoprocessed complex into the active mature form. Moreover, the chitin-accelerated activation of the autoprocessed complex containing a ChBD (N*-IWT or N*-IΔPkdD) was faster than that of the autoprocessed complex lacking a ChBD (N*-IΔChBD or N*-IΔCTE) (Fig. 5). These results indicate that ChBD-mediated chitin binding aids SptC maturation by promoting activation of the autoprocessed complex.
FIG 5.
Chitin-accelerated activation of SptC. Autoprocessed complex (N*-IWT, N*-IΔChBD, N*-IΔPkdD, or N*-IΔCTE) at a concentration of 5 μg/ml was kept standing and incubated 40°C in buffer A (50 mM Tris-HCl, 10 mM CaCl2 [pH 8.0]) containing 5 M NaCl at in the absence (−) or presence (+) of 4 mg/ml of chitin. At the indicated times, aliquots were removed and subjected to azocaseinolytic-activity assay. Values are expressed as the means ± SDs (bars) of three independent experiments, and the highest mean activity value observed during the maturation process was defined as 100% to determine relative activity.
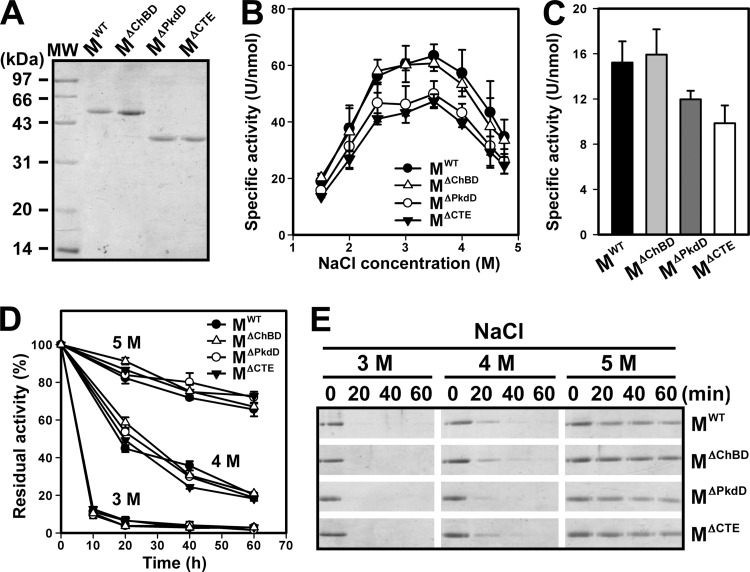
Properties of the mature enzymes.
To characterize the mature form of SptC and investigate the role of the PkdD, mature enzymes containing (MWT and MΔChBD) and lacking (MΔPkdD and MΔCTE) the PkdD were purified (Fig. 6A). Using azocasein as the substrate, the four mature forms were found to be most active at a NaCl concentration of 3 to 3.5 M (Fig. 6B). Notably, MWT and MΔChBD showed higher azocaseinolytic activity than MΔPkdD and MΔCTE over the whole range of NaCl concentrations tested (Fig. 6B). The activity of the MWT and MΔChBD forms against the small synthetic substrate suc-AAPF-pNA was higher than that of the MΔPkdD and MΔCTE forms (Fig. 6C). These results demonstrate that the PkdD enhances the enzymatic activity of SptC.
FIG 6.
Properties of the mature enzymes. (A) SDS-PAGE analysis of the purified mature enzymes. (B) Salt dependence of enzyme activity. The azocaseinolytic activity of the mature enzymes was measured at 40°C in buffer A containing different concentrations of NaCl. (C) Enzymatic hydrolysis of suc-AAPF-pNA. Enzyme activity was measured at 40°C using suc-AAPF-pNA (0.1 mM) as the substrate in buffer A containing 3.5 M NaCl. (D and E) Heat inactivation of the mature enzymes. The mature enzymes (3 μg/ml) were incubated at 60°C in buffer A (50 mM Tris-HCl, 10 mM CaCl2 [pH 8.0]) containing 3, 4, or 5 M NaCl. At the indicated times, aliquots were removed and subjected to azocaseinolytic-activity assay (D) and SDS-PAGE analysis (E). Residual activity is expressed as a percentage of the original activity of each sample (D). Values (B, C, and D) are expressed as the means ± SDs (bars) of three independent experiments.
In the presence of 3.5 M NaCl, the four mature forms were stable at 40°C and retained more than 90% of their original activities after a 1-h incubation (data not shown). To investigate whether the PkdD contributes to enzyme stability, inactivation profiles of the four mature forms were determined at elevated temperature (e.g., 60°C) and various NaCl concentrations. As shown in Fig. 6D, the four mature forms displayed similar heat inactivation profiles at NaCl concentrations of 3 to 5 M, indicating that the deletion of the PkdD has no effect on enzyme stability. As the salinity increased, the four mature forms became more resistant to heat inactivation (Fig. 6D), reflecting their halophilic nature. SDS-PAGE analysis of the heat-treated samples revealed that all four mature forms autodegraded more easily as the salinity decreased (Fig. 6E), indicating that these enzymes are more sensitive to thermal denaturation at lower NaCl concentrations and the enzymes that were denatured earlier would be degraded by those that were denatured later. These results suggest that the structure of SptC is salt dependent and that the PkdD does not confer extra stability on the enzyme.
Enzymatic deproteinization of SSP using SptC.
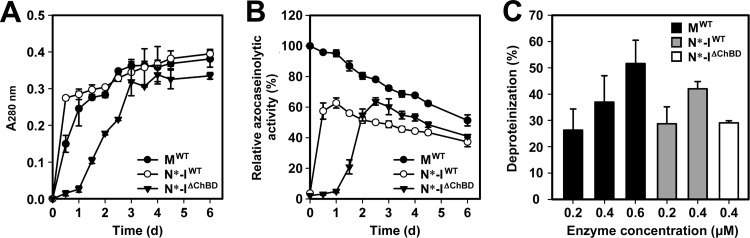
In biomass, chitin is closely associated with proteins; thus, the preparation of chitin from chitin-containing biomass involves a deproteinization process. The chitin-binding capacity of SptC prompted us to investigate whether this enzyme can remove proteins from chitin-containing biomass and whether the ChBD facilitates the deproteinization process. As shown in Fig. 7A, incubation of SSP with MWT led to the release of soluble peptides from SSP, indicating that mature SptC hydrolyzes chitin-associated proteins. A sample treated with N*-IΔChBD showed a slower release of soluble peptides than did an MWT-treated sample (Fig. 7A), probably due to the time delay in activation of the autoprocessed complex (Fig. 7B). Similar to the case of chitin-accelerated enzyme activation (Fig. 5), the ChBD-containing autoprocessed complex N*-IWT activated faster than N*-IΔChBD when incubated with SSP (Fig. 7B), suggesting that ChBD-mediated binding of N*-IWT to shrimp shell chitin facilitates enzyme maturation. It was noted that at the same enzyme concentration (0.4 μM), the N*-IWT-treated sample showed a faster release of soluble peptides than the MWT-treated sample (Fig. 7A), although removal of SSP protein in the two samples reached a plateau after 3 days and the degrees of deproteinization were comparable in the two samples (37 and 42%, respectively, Fig. 7C). One reasonable explanation for the slowdown of the deproteinization rate at the late stage of the treatment is that the accumulated soluble peptides compete with SSP-bound proteins for access to the enzyme active site. Interestingly, the amount of soluble peptides released in the N*-IWT-treated sample was ∼2-fold higher than that in the MWT-treated sample during the early stage of the treatment (e.g., 0.5 days) (Fig. 7A), but the corresponding level of azocaseinolytic activity of the former sample was significantly lower than that of the latter (Fig. 7B). It is very likely that ChBD-mediated binding of N*-IWT to SSP not only promotes enzyme activation but also increases the local concentration of active enzyme around the SSP, thus enhancing deproteinization. This is supported by the observation that the level of SSP protein removal by MWT was dependent on the enzyme concentration (Fig. 7C).
FIG 7.
Deproteinization of SSP by SptC. (A and B) Release of soluble peptides from SSP by enzymatic treatment (A) and proteolytic-activity assay of the reaction mixture (B). SSP (2 mg) was incubated with MWT, N*-IWT, or N*-IΔChBD (0.4 μM) at 40°C in 200 μl of buffer A (50 mM Tris-HCl, 10 mM CaCl2 [pH 8.0]) containing 5 M NaCl. At the indicated times, the soluble fraction was recovered from the reaction mixture by centrifugation and subjected to absorbance measurement at 280 nm (A) and azocaseinolytic-activity assay (B). Relative azocaseinolytic activity was calculated with the original activity of the MWT-treated sample, defined as 100%. (C) Percent deproteinization of SSP using different enzyme samples. After 6 days of treatment with different concentrations of MWT (0.2 to 0.6 μM), N*-IWT (0.2 to 0.4 μM), or 0.4 μM N*-IΔChBD as described above, SSP in the reaction mixture was recovered by centrifugation and the degree of deproteinization was determined. For reference purposes, 0% deproteinization was defined as 24.4 mg of protein contained in 1 g of SSP without enzyme treatment. Values are expressed as the means ± SDs (bars) of three independent experiments.
DISCUSSION
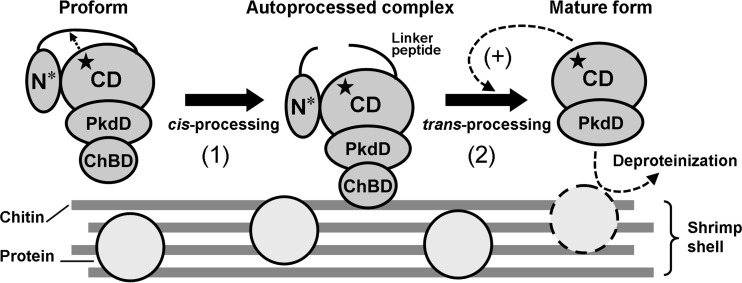
The results presented here demonstrate that the autocatalytic maturation of SptC occurs in a stepwise manner (Fig. 8). First, the core domain of the N-terminal propeptide (N*) is cis-processed to yield an autoprocessed complex (N*-IWT). Next, the five-residue linker peptide and the C-terminal ChBD are trans-processed from the intermediate (IWT) and N* is degraded, leading to generation of the active mature form composed of the catalytic domain and the PkdD. A recent report demonstrated that the maturation of halolysin Nep from N. magadii proceeds autocatalytically (10). The major difference between the maturations of the two halolysins is that the Nep precursor can be trans-processed into its mature form by active Nep, but the SptC precursor is degraded by active mature SptC. Generation of the mature enzyme via trans-processing of the N-terminal propeptide of the precursor or the proform, either auto- or heterocatalytically, has also been described for other subtilases (27, 35–38). In the precursor of the WF146 protease from thermophilic Bacillus sp. WF146, a 12-residue linker peptide between the core domain of the N-terminal propeptide and the catalytic domain comprises a proteolytically sensitive region, but the catalytic domain adopts a stable conformation that is resistant to proteolysis, allowing the trans-processing reaction to occur auto- and heterocatalytically (38). Although the SptC precursor contains a five-residue linker peptide, its catalytic domain is susceptible to proteolysis. Therefore, the presence of the active mature enzyme leads to degradation of the SptC precursor rather than trans-processing of the precursor to the mature form. In contrast to the precursor, the autoprocessed complex of SptC can be trans-processed to the mature form by the active mature enzyme, indicating that after cis-processing of N*, the catalytic domain of the autoprocessed complex adopts a proteolysis-resistant conformation. The structural change induced by the cis-processing reaction appears to be an indispensable step for proper maturation of SptC.
FIG 8.
Proposed model for autocatalytic maturation of SptC. The core domain (N*) of the N-terminal propeptide in the proform is cis-processed by the active site of the catalytic domain (CD) to yield the autoprocessed complex (step 1). The active site is indicated by a star. Next, the linker peptide and the ChBD are trans-processed and N* is degraded, generating the active mature form containing the CD and the PkdD (step 2). Active enzyme that matured earlier can catalyze (+) the step 2 reaction to accelerate the maturation process. ChBD-mediated binding of the autoprocessed complex to chitin not only accelerates conversion of the complex into the mature form but also increases the local enzyme concentration to promote enzyme activation and improve the deproteinization of chitin-containing biomass, such as shrimp shell (see Discussion for details). Note that minerals, lipids, and pigments in chitin-containing biomass are not shown.
The PkdD of SptC does not confer extra stability on the enzyme but instead contributes to the enzymatic activity of the mature form. Although the PkdD of SptC and the CTEs of other halolysins (e.g., SptA and Nep) show no significant homology, they are predicted to adopt a similar β-jelly roll-like structure. The importance of the CTE to enzymatic activity has been reported for halolysins R4 (7), SptA (11), and Nep (10). The CTE of SptA assists enzymatic activity toward protein substrates rather than small-peptide substrates by facilitating the binding of protein substrates for catalysis (11). In contrast, the presence of the PkdD increases the activity of mature SptC against both protein and smaller-peptide substrates. Whether the effect of the PkdD on enzymatic activity is due to a direct steric influence or to an indirect influence (e.g., induction of structural changes in the active site) remains to be determined. Our results also indicate that the PkdD is beneficial for the maturation of SptC, as evidenced by the finding that PkdD-containing autoprocessed complexes (N*-IWT and N*-IΔChBD) activate faster than those without a PkdD (N*-IΔPkdD and N*-IΔCTE). Apparently, the increased enzymatic activity afforded by the PkdD enables active enzyme that matures earlier to trans-process the autoprocessed complex more efficiently during the maturation process. In addition, the possibility that the PkdD interacts with the catalytic domain to promote the activation of the autoprocessed complex cannot be excluded.
In comparison with other halolysins, a unique feature of SptC is that its CTE contains an additional ChBD. The absence of a ChBD in the mature form suggests that this domain is dispensable for enzymatic activity. However, the ChBD mediates chitin binding of the autoprocessed complex to accelerate SptC maturation. Notably, activation of the autoprocessed complex of the mutant lacking both PkdD and ChBD (N*-IΔCTE) is also accelerated in the presence of chitin, demonstrating that the interaction between chitin and the catalytic domain and/or the N-terminal propeptide enhances enzyme maturation. Most likely, interaction with chitin results in a structural adjustment of the autoprocessed complex that facilitates its activation. Therefore, ChBD-mediated binding of the autoprocessed complex to chitin increases the chance of this interaction, thus further accelerating SptC maturation. Moreover, binding of the autoprocessed complex to chitin would lead to an increase in the local enzyme concentration, thus facilitating the trans-processing of the autoprocessed complex to the mature form (Fig. 8).
It has been reported that the chitin-binding protease AprIV plays an important role in the chitinolytic system of P. piscicida strain O-7 and that deproteinization of native chitin (prawn shell cuticle powder) using AprIV significantly enhances chitinase activity, probably by facilitating the approach of chitinase to chitin molecules (21). Our results showed that Natrinema sp. J7-2 possesses a chitin metabolism-related gene cluster and has the ability to degrade chitin. Although it remains to be determined whether SptC participates in chitin degradation by the haloarchaeon, the results presented here demonstrate that SptC is capable of removing proteins from chitin-containing biomass such as shrimp shell. Interestingly, ChBD-mediated binding of the SptC autoprocessed complex to chitin accelerates protein removal, probably by increasing the local protease concentration around the substrate. Meanwhile, the cleavage of the ChBD after enzyme maturation allows the mature SptC to freely access proteins that surround chitin in biomass and thus facilitates the deproteinization process. Moreover, the deproteinization reaction catalyzed by halophilic SptC can be carried out at high salt concentrations to minimize the risk of microbial contamination. By virtue of these properties, SptC is highly attractive for use in preparing chitin from chitin-containing biomass. Nevertheless, the susceptibility of SptC to proteolysis appears to be one major drawback that may limit its application. Improving the resistance of the enzyme to proteolytic attack through protein engineering is expected to overcome this problem.
ACKNOWLEDGMENTS
This work was supported in part by the National Natural Science Foundation of China (grant numbers 30870063 and 30970071) and the National Infrastructure of Natural Resources for Science and Technology Program of China (grant number NIMR-2014-8).
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.De Castro RE, Maupin-Furlow JA, Gimenez MI, Herrera Seitz MK, Sanchez JJ. 2006. Haloarchaeal proteases and proteolytic systems. FEMS Microbiol. Rev. 30:17–35. 10.1111/j.1574-6976.2005.00003.x [DOI] [PubMed] [Google Scholar]
- 2.Siezen RJ, Leunissen JAM. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6:501–523. 10.1002/pro.5560060301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamekura M, Seno Y, Holmes ML, Dyall-Smith ML. 1992. Molecular cloning and sequencing of the gene for a halophilic alkaline serine protease (halolysin) from an unidentified halophilic archaea strain (172P1) and expression of the gene in Haloferax volcanii. J. Bacteriol. 174:736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu K, Kim J, Dordick JS. 1994. Catalytic properties and potential of an extracellular protease from an extreme halophile. Enzyme Microb. Technol. 16:266–275. 10.1016/0141-0229(94)90165-1 [DOI] [PubMed] [Google Scholar]
- 5.Ruiz DM, Iannuci NB, Cascone O, De Castro RE. 2010. Peptide synthesis catalysed by a haloalkaliphilic serine protease from the archaeon Natrialba magadii (Nep). Lett. Appl. Microbiol. 51:691–696. 10.1111/j.1472-765X.2010.02955.x [DOI] [PubMed] [Google Scholar]
- 6.Margesin R, Schinner F. 2001. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5:73–83. 10.1007/s007920100184 [DOI] [PubMed] [Google Scholar]
- 7.Kamekura M, Seno Y, Dyall-Smith M. 1996. Halolysin R4, a serine proteinase from the halophilic archaeon Haloferax mediterranei; gene cloning, expression and structural studies. Biochim. Biophys. Acta 1294:159–167. 10.1016/0167-4838(96)00016-7 [DOI] [PubMed] [Google Scholar]
- 8.Shi W, Tang X-F, Huang Y, Gan F, Tang B, Shen P. 2006. An extracellular halophilic protease SptA from a halophilic archaeon Natrinema sp. J7: gene cloning, expression and characterization. Extremophiles 10:599–606. 10.1007/s00792-006-0003-8 [DOI] [PubMed] [Google Scholar]
- 9.De Castro RE, Ruiz DM, Gimenez MI, Silveyra MX, Paggi RA, Maupin-Furlow JA. 2008. Gene cloning and heterologous synthesis of a haloalkaliphilic extracellular protease of Natrialba magadii (Nep). Extremophiles 12:677–687. 10.1007/s00792-008-0174-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz DM, Paggi RA, Gimenez MI, De Castro RE. 2012. Autocatalytic maturation of the Tat-dependent halophilic subtilase Nep produced by the archaeon Natrialba magadii. J. Bacteriol. 194:3700–3707. 10.1128/JB.06792-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Z, Du X, Li T, Gan F, Tang B, Tang X-F. 2011. Functional insight into the C-terminal extension of halolysin SptA from haloarchaeon Natrinema sp. J7. PLoS One 6:e23562. 10.1371/journal.pone.0023562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGenity TJ, Gemmell RT, Grant WD. 1998. Proposal of a new halobacterial genus Natrinema gen. nov., with two species Natrinema pellirubrum nom. nov. and Natrinema pallidum nom. nov. Int. J. Syst. Evol. Microbiol. 48:1187–1196. 10.1099/00207713-48-4-1187 [DOI] [PubMed] [Google Scholar]
- 13.Tapingkae W, Tanasupawat S, Itoh T, Parkin KL, Benjakul S, Visessanguan W, Valyasevi R. 2008. Natrinema gari sp. nov., a halophilic archaeon isolated from fish sauce in Thailand. Int. J. Syst. Evol. Microbiol. 58:2378–2383. 10.1099/ijs.0.65644-0 [DOI] [PubMed] [Google Scholar]
- 14.Xin H, Itoh T, Zhou P, Suzuki K, Kamekura M, Nakase T. 2000. Natrinema versiforme sp. nov., an extremely halophilic archaeon from Aibi salt lake, Xinjiang, China. Int. J. Syst. Evol. Microbiol. 50:1297–1303. 10.1099/00207713-50-3-1297 [DOI] [PubMed] [Google Scholar]
- 15.Xu XW, Ren PG, Liu SJ, Wu M, Zhou PJ. 2005. Natrinema altunense sp. nov., an extremely halophilic archaeon isolated from a salt lake in Altun Mountain in Xinjiang, China. Int. J. Syst. Evol. Microbiol. 55:1311–1314. 10.1099/ijs.0.63622-0 [DOI] [PubMed] [Google Scholar]
- 16.Castillo AM, Gutierrez MC, Kamekura M, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A. 2006. Natrinema ejinorense sp. nov., isolated from a saline lake in Inner Mongolia, China. Int. J. Syst. Evol. Microbiol. 56:2683–2687. 10.1099/ijs.0.64421-0 [DOI] [PubMed] [Google Scholar]
- 17.Albuquerque L, Taborda M, La Cono V, Yakimov M, da Costa MS. 2012. Natrinema salaciae sp. nov., a halophilic archaeon isolated from the deep, hypersaline anoxic Lake Medee in the Eastern Mediterranean Sea. Syst. Appl. Microbiol. 35:368–373. 10.1016/j.syapm.2012.06.005 [DOI] [PubMed] [Google Scholar]
- 18.Shen P, Chen Y. 1994. Plasmid from Halobacterium halobium and its restriction map. Yi Chuan Xue Bao 21:409–416 [PubMed] [Google Scholar]
- 19.Feng J, Liu B, Zhang Z, Ren Y, Li Y, Gan F, Huang Y, Chen X, Shen P, Wang L, Tang B, Tang X-F. 2012. The complete genome sequence of Natrinema sp. J7-2, a haloarchaeon capable of growth on synthetic media without amino acid supplements. PLoS One 7:e41621. 10.1371/journal.pone.0041621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souza TA, Okamoto DN, Ruiz DM, Oliveira LC, Kondo MY, Tersario IL, Juliano L, De Castro RE, Gouvea IE, Murakami MT. 2012. Correlation between catalysis and tertiary structure arrangement in an archaeal halophilic subtilase. Biochimie 94:798–805. 10.1016/j.biochi.2011.11.011 [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto K, Nukui E, Itoh H, Sato T, Kobayashi T, Imada C, Watanabe E, Inamori Y, Tsujibo H. 2002. Molecular analysis of the gene encoding a novel chitin-binding protease from Alteromonas sp. strain O-7 and its role in the chitinolytic system. J. Bacteriol. 184:1865–1872. 10.1128/JB.184.7.1865-1872.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye X, Ou J, Ni L, Shi W, Shen P. 2003. Characterization of a novel plasmid from extremely halophilic Archaea: nucleotide sequence and function analysis. FEMS Microbiol. Lett. 221:53–57. 10.1016/S0378-1097(03)00175-7 [DOI] [PubMed] [Google Scholar]
- 23.Bian Y, Liang X, Fang N, Tang X-F, Tang B, Shen P, Peng Z. 2006. The roles of surface loop insertions and disulfide bond in the stabilization of thermophilic WF146 protease. FEBS Lett. 580:6007–6014. 10.1016/j.febslet.2006.09.068 [DOI] [PubMed] [Google Scholar]
- 24.An Y, Lv A, Wu W. 2010. A QuikChange-like method to realize efficient blunt-ended DNA directional cloning and site-directed mutagenesis simultaneously. Biochem. Biophys. Res. Commun. 397:136–139. 10.1016/j.bbrc.2010.05.042 [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 26.King J, Laemmli UK. 1971. Polypeptides of the tail fibres of bacteriophage T4. J. Mol. Biol. 62:465–477. 10.1016/0022-2836(71)90148-3 [DOI] [PubMed] [Google Scholar]
- 27.Cheng G, Zhao P, Tang X-F, Tang B. 2009. Identification and characterization of a novel spore-associated subtilase from Thermoactinomyces sp. CDF. Microbiology 155:3661–3672. 10.1099/mic.0.031336-0 [DOI] [PubMed] [Google Scholar]
- 28.DelMar EG, Largman C, Brodrick JW, Geokas MC. 1979. A sensitive new substrate for chymotrypsin. Anal. Biochem. 99:316–320. 10.1016/S0003-2697(79)80013-5 [DOI] [PubMed] [Google Scholar]
- 29.Percot A, Viton C, Domard A. 2003. Optimization of chitin extraction from shrimp shells. Biomacromolecules 4:12–18. 10.1021/bm025602k [DOI] [PubMed] [Google Scholar]
- 30.Oh Y, Shih II, Tzeng Y, Wang S. 2000. Protease produced by Pseudomonas aeruginosa K-187 and its application in the deproteinization of shrimp and crab shell wastes. Enzyme Microb. Technol. 27:3–10. 10.1016/S0141-0229(99)00172-6 [DOI] [PubMed] [Google Scholar]
- 31.Hou J, Han J, Cai L, Zhou J, Lu Y, Jin C, Liu J, Xiang H. 2013. Characterization of genes for chitin catabolism in Haloferax mediterranei. Appl. Microbiol. Biotechnol. 2013:1–10. 10.1007/s00253-013-4969-8 [DOI] [PubMed] [Google Scholar]
- 32.van Aalten DM, Synstad B, Brurberg MB, Hough E, Riise BW, Eijsink VG, Wierenga RK. 2000. Structure of a two-domain chitotriosidase from Serratia marcescens at 1.9 Å resolution. Proc. Natl. Acad. Sci. U. S. A. 97:5842–5847. 10.1073/pnas.97.11.5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madern D, Ebel C, Zaccai G. 2000. Halophilic adaptation of enzymes. Extremophiles 4:91–98. 10.1007/s007920050142 [DOI] [PubMed] [Google Scholar]
- 34.Shinde U, Thomas G. 2011. Insights from bacterial subtilases into the mechanisms of intramolecular chaperone-mediated activation of furin. Methods Mol. Biol. 768:59–106. 10.1007/978-1-61779-204-5_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Power SD, Adams RM, Wells JA. 1986. Secretion and autoproteolytic maturation of subtilisin. Proc. Natl. Acad. Sci. U. S. A. 83:3096–3100. 10.1073/pnas.83.10.3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egnell P, Flock JI. 1992. The autocatalytic processing of the subtilisin Carlsberg pro-region is independent of the primary structure of the cleavage site. Mol. Microbiol. 6:1115–1119. 10.1111/j.1365-2958.1992.tb01549.x [DOI] [PubMed] [Google Scholar]
- 37.Han X, Kennan RM, Steer DL, Smith AI, Whisstock JC, Rood JI. 2012. The AprV5 subtilase is required for the optimal processing of all three extracellular serine proteases from Dichelobacter nodosus. PLoS One 7:e47932. 10.1371/journal.pone.0047932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu H, Xu BL, Liang X, Yang YR, Tang X-F, Tang B. 2013. Molecular basis for auto- and hetero-catalytic maturation of a thermostable subtilase from thermophilic Bacillus sp. WF146. J. Biol. Chem. 288:34826–34838. 10.1074/jbc.M113.498774 [DOI] [PMC free article] [PubMed] [Google Scholar]