Abstract
Shiga toxin-producing Escherichia coli (STEC) infections are a critical public health concern because they can cause severe clinical outcomes, such as hemolytic uremic syndrome, in humans. Determining the presence or absence of virulence genes is essential in assessing the potential pathogenicity of STEC strains. Currently, there is limited information about the virulence genes carried by swine STEC strains; therefore, this study was conducted to examine the presence and absence of 69 virulence genes in STEC strains recovered previously from finishing swine in a longitudinal study. A subset of STEC strains was analyzed by pulsed-field gel electrophoresis (PFGE) to examine their genetic relatedness. Swine STEC strains (n = 150) were analyzed by the use of a high-throughput real-time PCR array system, which included 69 virulence gene targets. Three major pathotypes consisted of 16 different combinations of virulence gene profiles, and serotypes were determined in the swine STEC strains. The majority of the swine STEC strains (n = 120) belonged to serotype O59:H21 and carried the same virulence gene profile, which consisted of 9 virulence genes: stx2e, iha, ecs1763, lpfAO113, estIa (STa), ehaA, paa, terE, and ureD. The eae, nleF, and nleH1-2 genes were detected in one swine STEC strain (O49:H21). Other genes encoding adhesins, including iha, were identified (n = 149). The PFGE results demonstrated that swine STEC strains from pigs raised in the same finishing barn were closely related. Our results revealed diverse virulence gene contents among the members of the swine STEC population and enhance understanding of the dynamics of transmission of STEC strains among pigs housed in the same barn.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) infections are a critical public health concern, leading to 170,000 cases of human illness (1) and an economic burden of 280 million dollars (2) annually in the United States. STEC strains represent a subset of E. coli strains that produce one or more bacteriophage-encoded cytotoxins known as Shiga toxins (Stx1 and Stx2) (3, 4). Enterohemorrhagic E. coli (EHEC) strains represent a subgroup of STEC strains defined by the presence of an Stx-encoding bacteriophage and the presence of the locus of enterocyte effacement (LEE) pathogenicity island, which is important for the development of attaching and effacing lesions (5). The LEE island is not present in all STEC strains. Prior studies have found that EHEC is more commonly associated with severe clinical cases (6); however, LEE-negative STEC strains have been linked to severe clinical cases as well as to outbreaks (7). STEC is often acquired by consuming contaminated food or water (8), and cattle are viewed as the most important animal reservoirs (9). Food of bovine origin has been implicated in many STEC infections and outbreaks, though a number of other food products, including pork products, have been confirmed as the source of STEC in a number of outbreaks (10–16). For example, a recent STEC O157:H7 outbreak was associated with consuming large cuts of pork from a whole roasted pig (16).
Although the way in which the pork products became contaminated in these outbreaks was uncertain (10–16), the likelihood that on-farm pigs were the source of STEC contamination cannot be overlooked. Unlike cattle, which do not usually present clinical symptoms due to STEC infection, pigs, specifically postweaning and young finishing pigs, can suffer from edema disease caused by STEC strains carrying the stx2e variant (17). Epidemiological studies conducted in different regions of the world have reported a wide range of STEC prevalences in swine populations (18–20). However, the epidemiology and virulence characteristics of STEC carried by on-farm pigs remain largely unknown, as does whether swine-derived STEC strains are similar to human-derived STEC strains and have the potential to contribute to human infections.
Human STEC infections are associated with a range of clinical symptoms: diarrhea, hemorrhagic colitis (HC), and the life-threatening hemolytic uremic syndrome (HUS) (6, 21). The pathogenesis of STEC in human patients has been reviewed elsewhere (9, 22–26). Although Shiga toxins are critical in STEC pathogenesis because they inhibit host cell protein synthesis (27) and induce apoptosis (reviewed in reference 28), virulence factors other than Stx are also important (29). For example, following initial attachment to host intestinal cells, EHEC strains that express intimin, which is encoded by eae on the LEE pathogenicity island, can intimately attach to host cells (23, 30). The N-terminal region of intimin is highly conserved, whereas the C-terminal region is variable and accounts for the definition of eae subtypes. For STEC strains that lack eae (LEE-negative STEC) and cause diseases in humans, other adherence structures have been suggested to be important. For example, the STEC autoagglutinating adhesin encoded by saa increases adherence of the pathogen to human epithelial cells (31). Other structures contributing to STEC colonization, such as the long polar fimbriae encoded by lpf, which has many variants, have also been found in both EHEC strains and LEE-negative STEC strains (23, 32, 33). A number of other virulence factors have been proposed to contribute to STEC pathogenesis, for instance, catalase peroxidase encoded by katP, which protects STEC from peroxide-mediated oxidative damage (34). Some non-LEE-carried effector (nle) genes, which encode proteins having various functions, such as inhibiting phagocytosis, have been detected in EHEC strains isolated from human patients with severe clinical disease, namely, HC or HUS (29, 35, 36).
Knowing that various combinations of virulence factors contribute to STEC pathogenesis, it is essential to determine the presence or absence of specific virulence genes to better assess the potential pathogenicity of STEC strains (29, 32, 37). A few studies have examined the virulence gene profiles of swine STEC strains (38–42). However, every study selected different panels of virulence genes, which makes comparison of results across different studies highly challenging. The presence of many virulence genes, for example, nle genes and allelic variants, has seldom been examined in swine STEC strains. In general, little is known about the virulence characteristics of STEC originating from swine that may contribute to disease in humans and swine. To fill in the current knowledge gap and better evaluate the potential pathogenicity of swine-derived STEC strains, we utilized a PCR microarray method to examine the presence of 69 virulence genes in STEC strains recovered previously from finishing pigs in a longitudinal study (43). Moreover, the genetic relatedness of these strains was also examined to better understand STEC transmission dynamics in swine throughout the finishing period.
MATERIALS AND METHODS
Swine STEC strains.
A total of 150 STEC strains recovered from 95 finishing pigs in a longitudinal study were included in this study (43). Individual fecal samples were collected from three cohorts of finishing pigs (n = 50/cohort, n = 150 in total). Each cohort was raised in a separate finishing barn at two finishing sites (cohort 1 at site A, cohorts 2 and 3 at site B) within one all-in, all-out multisite production system in the midwestern United States. The samples in each cohort were collected every 2 weeks during the finishing period (8 farm visits/cohort). A sample was considered to be positive for STEC when an STEC isolate was recovered. The presence of virulence genes (stx1, stx2, stx2e, and eae) and the O:H serotypes were determined in these STEC isolates (43). At least one STEC strain was selected from each positive sample for virulence gene characterization by PCR microarray (see below). STEC strains belonging to different serotypes recovered from the same sample were also included in the study to better understand the diversity of STEC strains within the animals.
Selection of virulence gene targets.
The general rationale for the selection of virulence gene targets was based on their function, role in pathogenesis, and association with human illness and/or disease severity in human patients. According to the previous characterization results, only 1 among the 150 swine STEC strains carried the eae gene (43). Additionally, all swine STEC strains recovered in the longitudinal study were in non-O157 serogroups (43). Therefore, the 69 virulence genes targeted in the microarray were selected based on genes found in different STEC serogroups and included putative genes encoding adhesins (iha, paa, orfA, orfB, toxB, eibG, saa, eae, and allelic variants), toxins {stx [all variants, including stx2e], ent (espL2), cdtI, cdtIII, astA, estIa (STa), elt (LT), and subAB}, fimbriae (lpfAO157, lpfAO113, lpfAO26, and sfp), and others found in pathogenic STEC strains [terE, ureD, espV, espK, espN, espX7, espO1-1, nleG5, nleG6-2, Z2096, Z2098, Z2099, nleA, nleF, nleH1-2, espM1, espM2, nleB, nleE, efa1 (lifA), pagC, ecs1822, ecs1763, irp2, fyuA, ehaA, hlyA, bfp, cnf2, ecf1, ecf2, ecf3, efc4, katP, ehxA, etpD, stcE, espP, epeA, and sab]. Some of the virulence genes were also those associated with swine diseases: postweaning diarrhea [estIa (STa), elt (LT), orfA, orfB, and hlyA] and edema disease (stx2e, orfA, orfB, and hlyA). We also included fimbrial genes that are associated with swine neonatal diarrhea [fasA (F6) and fimF41a (F41)], postweaning diarrhea [fedA (F18)], and swine edema disease [fedA (F18)].
High-throughput real-time PCR microarray.
The BioMark real-time PCR system (Fluidigm) was used for high-throughput microfluidic real-time PCR amplification using 96.96 dynamic arrays (Fluidigm). Amplifications were performed using EvaGreen DNA binding dye (Biotium Inc., Hayward, CA) with TaqMan Gene Expression master mix in accordance with the recommendations of the manufacturer (Applied Biosystems, Courtaboeuf, France). The thermal profile comprised 10 min at 95°C, followed by 35 cycles of 95°C for 15 s and 60°C for 1 min, followed by a melting-curve analysis.
Besides the 69 selected virulence genes, O-group-associated genes specific for serogroups O26, O157, O145, O103, O111, O121, O45, O118, O128, O146, O91, O104, O113, and O55 were included in the PCR microarray. Moreover, the microarray chip also targeted flagellar genes for H-groups H11, H7, H21, H2, H28, H8, H19, H16, H25, H4, and H32 (44). Most of the primers used in the PCR microarray were described previously (29, 38, 44, 45), and some were designed for this study. The sequences of all primers are reported in Table S1 in the supplemental material.
Strain selection strategy for PFGE.
A subset of swine STEC strains (n = 49) was selected for pulsed-field gel electrophoresis (PFGE) analysis to determine their genetic relatedness. The selection criteria were based on serotype, virulence gene profiles, and epidemiological information related to the pigs. Within the predominant serotype, O59:H21, strains recovered in the early, middle, and late stages of the finishing period were selected. We included STEC strains of the same serotype and recovered from the same pig at different farm visits over the finishing period to examine changes over time. STEC strains of the same serotype but with different virulence gene profiles were also selected for PFGE analysis. In total, 29 O59:H21 STEC strains were selected, as well as 2 O59:H19 STEC strains. Thirteen additional STEC strains belonging to serotype O untypeable:H19, four O98:H12 STEC strains, and one O98:H19 strain were also analyzed.
PFGE.
PFGE was conducted according to the standardized Centers of Disease Control and Prevention (CDC) PulseNet protocol (46). In summary, STEC DNA was embedded in agarose and digested with 50 U of XbaI for 2 h at 37°C. A CHEF DR-III system (Bio-Rad, Munich, Germany) was used to separate the restriction fragments by electrophoresis at pulse times of 2.16 to 54.17 s in 0.5× Tris-borate-EDTA buffer with 50 μM thiourea at 14°C for 16.2 h. The H9812 Salmonella enterica serovar Braenderup strain (CDC, Atlanta, GA) was utilized as a molecular size marker. BioNumerics software package 6.6 (Applied Maths, Ghent, Belgium) was used to analyze the PFGE restriction-digested band patterns. The dendrogram was built by analyzing Dice coefficients and by using the unweighted-pair group method using average linkages (UPGMA) with 0.5% band position tolerance. The genetic relatedness of the strains was assessed by the percentages of similarity of the PFGE patterns.
RESULTS
Virulence gene profiles of swine STEC strains.
There were 11 distinct virulence gene profiles among the swine STEC strains tested in this study, with 16 different combinations based on serotype and virulence gene profiles (Table 1). The strains had between 6 and 20 genes among the 69 virulence genes examined, and most strains (82% [123/150]) carried the same virulence gene profile (virulence gene profile 1), which consisted of the following 9 genes: stx2e, iha, ecs1763, lpfAO113, estIa (STa), ehaA, paa, terE, and ureD. The second-most-prevalent virulence gene profile was profile 3, found in 10% (15/150) of the strains, and contained stx2e, iha, estIa (STa), paa, terE, and ureD. One strain, serotype O49:H21, carried virulence gene profile 11, which included eae and two nle variants (nleF and nleH1-2) as well as stx2e, katP, iha, ecs1763, lpfAO113, astA, estIa (STa), ecf1, ecf2, ecf3, ecf4, irp2, fyuA, ehaA, paa, terE, and ureD. Furthermore, the data showed that the strains can be grouped into three major pathotypes according to the virulence genes they carried (Table 1). Pathotype I was defined as the strains (12.7% [19/150]) possessing 5 core genes, including stx2e, iha, paa, terE, and ureD, along with other virulence genes. In addition to the 5 core genes, pathotype II strains (82.7% [124/150]) possessed the 4 genes ecs1763, estIa (STa), lpfAO113, and ehaA along with other virulence genes. Finally, pathotype III strains carried stx1 (2.7% [4/150]) and eae (0.7% [1/150]), as well as other virulence genes (1.3% [2/150]).
TABLE 1.
Distribution of swine STEC strains by serotype and virulence gene profiles
| Serotype | No. of swine STEC strains | No. of strains analyzed by PFGE | Virulence gene profile | Virulence gene profile code | Pathotype |
|---|---|---|---|---|---|
| O59:H21 | 118 | 27 | stx2e, iha, ecs1763, lpfAO113, estIa (STa), ehaA, paa, terE, ureD | 1 | II |
| 2 | 2 | stx2e, iha, ecs1763, lpfAO113, ehaA, paa, terE, ureD | 2 | I | |
| O untypeable:H19 | 13 | 12 | stx2e, iha, estIa (STa), paa, terE, ureD | 3 | I |
| 1 | 1 | stx2e, iha, astA, estIa (STa), terE, ureD | 4 | III | |
| O59:H19 | 2 | 2 | stx2e, iha, estIa (STa), paa, terE, ureD | 3 | I |
| O98:H12 | 2 | 2 | stx1, pag C, katP, iha, astA, ecf1, ecf2, ecf3, ecf4, paa, terE, ureD | 9 | III |
| 2 | 2 | stx1, pag C, katP, iha, ecf1, ecf2, ecf3, ecf4, paa, terE, ureD | 10 | III | |
| O untypeable:H21 | 2 | 0 | stx2e, iha, ecs1763, lpfAO113, estIa (STa), ehaA, paa, terE, ureD | 1 | II |
| O20:H21 | 1 | 0 | stx2e, iha, ecs1763, lpfAO113, estIa (STa), ehaA, paa, terE, ureD | 1 | II |
| O49:H21 | 1 | 0 | stx2e, eae, nleF, nleH1-2, katP, iha, ecs1763, lpfAO113, astA, estIa (STa), ecf1, ecf2, ecf3, ecf4, irp2, fyuA, ehaA, paa, terE, ureD | 11 | III |
| O89:H19 | 1 | 0 | stx2e, iha, lpfAO113, estIa (STa), ehaA, paa, terE, ureD | 7 | I |
| O98:H19 | 1 | 1 | stx2e, pagC, katP, iha, ecf1, ecf2, ecf3, ecf4, paa, terE, ureD | 5 | I |
| O115:H19 | 1 | 0 | stx2e, iha, ecs1763, lpfAO113, astA, estIa (STa), orfA, orfB, ehaA, paa, terE, ureD | 6 | II |
| O119:H21 | 1 | 0 | stx2e, iha, ecs1763, lpfAO113, estIa (STa), ehaA, paa, terE, ureD | 1 | II |
| O167:H21 | 1 | 0 | stx2e, iha, ecs1763, lpf AO113, estIa (STa), ehaA, paa, terE, ureD | 1 | II |
| O untypeable:H4 | 1 | 0 | stx2e, fedA (F18), hlyA, orfA, orfB, paa, terE | 8 | III |
| Total | 150 | 49 |
Although only 1 of the 150 strains had eae, other genes encoding proteins associated with attachment were present in the swine STEC strains. For example, iha, which encodes the iron-regulated gene A homolog adhesin (47), was detected in 99.3% (149/150) of the STEC strains. The lpfAO113 gene, which encodes long polar fimbriae (48), was detected in 85.3% (128/150) of the strains, and the fedA gene, which encodes fimbrial adhesin F18 and is associated with swine edema disease and postweaning diarrhea (49), was present in 0.7% (1/150) of the strains. The orfA and orfB genes, which encode adhesins involved in diffuse adherence (AIDA) (50, 51), were present in 1.3% (2/150) of the strains. Moreover, the paa gene, which encodes the porcine attaching and effacing-associated adhesin (52), was detected in 99.3% (149/150) of the strains. Among the fimbrial genes (fimF41a and fasA), which contribute to colonization in swine and are associated with swine neonatal diarrhea (17), fimF41a was detected in 0.6% (1/150) of the strains, while fasA was not detected in any of the strains.
A number of genes that encode toxins and hemolysins were present in the panel of swine STEC strains. For instance, 146 of the 150 swine STEC strains carried the stx2e gene, and the other four strains carried stx1. The astA gene, which encodes the enteroaggregative E. coli (EAEC) heat-stable toxin (53), was detected in 3.3% (5/150) of the strains. The estIa (STa) gene, which encodes heat-stable toxin (54), was detected in 94.7% (142/150) of the strains. Moreover, the hlyA gene, which encodes the alpha hemolysin (55), was present in 0.6% (1/150) of the strains. Interestingly, the ecs1763 gene, which encodes hypothetical proteins and was previously detected in EHEC strains (56), was present in 84.7% (127/150) of the strains. The following virulence genes, which were also targeted in the PCR microarray, were not detected in any of the 150 swine STEC strains: eae subtypes alpha, beta, gamma, epsilon, and theta, nleA, nleG5, ent (espL2), nleB, nleE, efa1 (lifA), Z2096, Z2098, Z2099, espM1, espM2, nleG6-2, espK, espN, espX7, espO1-1, espV, ecs1822, sfp, bfp, lpfAO26, lpfAO157, cdt subtypes I and III, elt (LT), cnf2, ehxA, toxB, stcE, eibG, epeA, espP, saa, subAB, and sab.
PFGE.
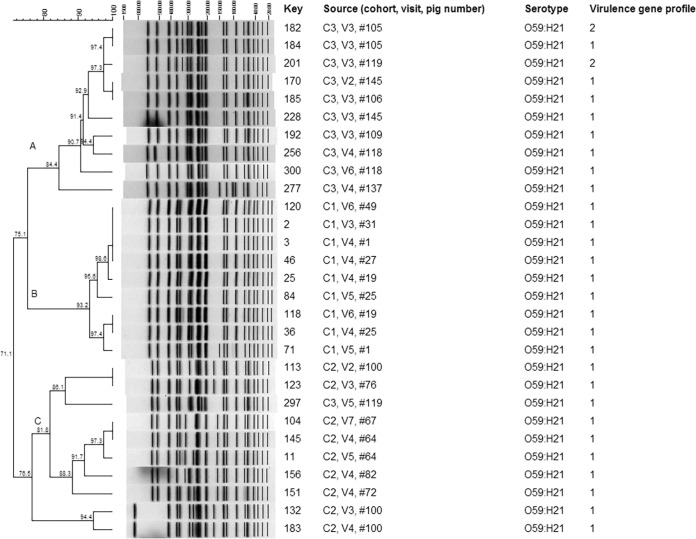
Within the O59:H21 STEC strains, three major clusters (clusters A to C) were defined at a cutoff value of 80% similarity (Fig. 1). These three major clusters were related at 71.1% similarity. The strains isolated from pigs within the same cohort were clustered. For example, cluster A contained strains from pigs in cohort 3, and cluster B contained strains from pigs in cohort 1. The only exception was that strain 297 from pig 119 in cohort 3 clustered with strains from cohort 2 (cluster C). Strains from pigs in cohort 3 (cluster A) were related to the strains from pigs in cohort 1 (cluster B) at 75.1% similarity. Within each cluster, indistinguishable PFGE patterns were observed among STEC strains recovered from samples in the same pig over time during the finishing period. For example, strains 170 and 228 with indistinguishable PFGE patterns were recovered from pig 145 at the second and third farm visits in cohort 3. In cluster A, strains carrying two different virulence gene profiles (profiles 1 and 2) clustered. The two O59:H19 strains, which carried virulence gene profile 3, were not clustered with the O59:H21 strains (24.8% similarity; data not shown).
FIG 1.
PFGE analysis of swine O59:H21 STEC strains. The “Key” column lists strain numbers. The “Source” column lists the cohort number of the pig from which each strain was collected, the number corresponding to the visit (of the eight farm visits) during which the strain was collected, and the individual pig number.
Within the O untypeable:H19 strains, 11 of the 13 strains were clustered at 83.6% similarity. One of the O untypeable:H19 strains had a PFGE pattern different from those of the other 11 strains (27.4% similarity). Moreover, strain 281, which carried a different virulence gene profile, was not clustered with the other 12 strains (see Fig. S1 in the supplemental material). Within the five strains belonging to serogroup O98, the four O98:H12 strains had indistinguishable PFGE patterns and were clustered at a similarity level of over 90%. The O98:H19 strain (n = 1) did not cluster with the four O98:H12 strains (see Fig. S2 in the supplemental material).
DISCUSSION
The objective of this study was to use molecular methods to characterize swine STEC strains recovered in a previous study (43) to determine their virulence gene profiles and genetic relatedness. This study utilized a high-throughput real-time PCR platform to examine the presence of members of a large panel of virulence gene targets in swine STEC strains. Although these swine STEC strains were recovered from samples of 95 healthy finishing pigs from three cohorts within 18 months in the same geographic area, they were composed of three major pathotypes with 16 different combinations of virulence gene profiles and serotypes. The panel of virulence genes in this study included 69 targets, and our results were in agreement with those of another study suggesting that increasing the number of virulence genes in the panel would increase the resolution of the virulence gene profiling (57). Various virulence gene profiles in swine STEC strains have also been reported elsewhere (38–42, 58–60). However, it was challenging to compare the results of this study with the results of those previous reports because all of the studies employed different panels of virulence genes. Taken together, our results and the results of previous studies of swine STEC strains have indicated that the swine STEC group consists of strains carrying diverse sets of virulence genes.
Because the swine STEC strains examined in this study represent non-O157 serotypes and because only 1 of the 150 strains carried eae, we chose to screen for the presence of many novel virulence gene targets previously reported in non-O157 and LEE-negative STEC strains. Some adhesin-encoding genes which have been detected in human-pathogenic STEC strains were present in the swine STEC strains. For example, iha and lpfAO113 were present in over 80% of the swine STEC strains. The iha gene has been detected in over 70% of the LEE-negative STEC strains associated with human clinical cases examined in studies in Germany (61) and Australia (7). Both iha and lpfAO113 have been detected in over 80% of LEE-negative STEC strains associated with human clinical cases examined in a study in Argentina (33). In addition to their potential ability to allow STEC to attach to human cells, the high prevalence of these genes in swine STEC may also suggest a role in STEC colonization of swine or enhanced transmissibility and persistence within a farm. These results warrant future research to better define the role that these attachment proteins may play in adherence to both swine and human epithelial cells.
The majority of swine STEC strains in this study carried stx2e, which is associated with edema disease in swine (9). Although stx2e was prevalent in the STEC strains in this study, none of the pigs in this study presented clinical symptoms (43). One of the potential explanations for the absence of clinical symptoms in these pigs was that only one of the strains carried the important fimbrial adhesin gene that is associated with swine edema disease (fedA) (17). Moreover, the fimbrial adhesin gene associated with swine neonatal diarrhea (fimF41a) (17) was present in only one swine STEC strain examined in this study, and fasA was not detected in any of the strains. Similarly, a previous study reported a high prevalence of stx2e (80%) and a low prevalence of fedA (4.6%) and did not detect fimF41a and fasA in STEC strains from clinically healthy pigs (38). The production of Stx alone, without an adherence factor, is deemed to be insufficient to cause severe disease. In addition, none of swine STEC strains in this study belonged to serogroups O138, O139, O141, and O147, which are associated with edema disease (17). Lastly, pigs in this study were in the finishing period (10 to 24 weeks old), which is later than the usual onset age for neonatal diarrhea (0 to 4 days old), postweaning diarrhea, and edema disease (5 weeks old) (17). In the case of edema disease, the expression of receptors for the STEC fimbrial adhesin (fedA) in pigs is associated with younger age (17). Therefore, the older age of pigs in this study may also explain why they did not develop clinical symptoms when infected with E. coli strains carrying stx2e.
In addition to swine diseases, Stx2e-producing E. coli strains are often implicated in infections of humans with mild disease or no clinical symptoms (62–64). Stx variants are known to be associated with disease severity in humans. For example, the stx2c and stx2d activatable variants are more likely to be found in STEC strains from HUS patients (64, 65), while stx1 variants are associated with STEC strains in humans with milder clinical symptoms (62–64). Therefore, the swine STEC strains in this study predominantly carrying stx2e and a few with stx1 may represent low risk to human health. Moreover, one may notice that some swine STEC strains analyzed in this study were O-untypeable and that most of those that were identified belonged to serotypes that have not previously been associated with human infections, except O59:H19 (33). Nevertheless, in some rare cases, Stx2e-producing E. coli strains have been recovered from HUS cases (66) and from humans with uncomplicated diarrhea (62, 63, 67, 68). More research is needed to characterize and examine the frequencies of different stx variant genes in swine-derived STEC strains from farms in different geographic locations.
Several of the gene targets assessed in this study have not been examined in swine STEC strains elsewhere. For example, the ecs1763 gene, which was found only in a subset of EHEC strains analyzed in a previous study (56), was prevalent in a high proportion (84.7%) of the swine strains. However, the function associated with this ecs1763 gene has not yet been determined, and the association between the presence of this gene and the clinical outcome in human cases requires more research. In addition, the combination of espK with espV, ureD, or Z2098 has been suggested to be highly prevalent in EHEC strains and can be utilized for EHEC detection purposes (45). Although ureD was present in 99.3% of our strains, the espK, espV, and Z2098 genes were absent in the panel of strains used in this study. Considering the uncertainties of the role of these putative virulence factors in causing human illness, it is difficult to determine the health risk of many of these swine STEC strains.
Our study is the first one to use PFGE to analyze STEC strains recovered from repeated samples collected from pigs, while most of the previous studies used PFGE to determine the genetic relatedness of STEC strains from swine and other species (60, 69–72). The degrees of genetic relatedness of swine STEC strains to strains from other animal species differed in those studies, and most studies focused on STEC O157:H7 (60, 69–71). Here, we found that STEC O59:H21 strains, which predominated in this swine population, were closely related among pigs in the same cohort. This suggests that the same strain disseminated throughout each cohort and provides support for the idea of a point-source outbreak at each of the three barns at two distinct finishing sites (43). Thus, the pigs may have been exposed to the same point source of infections in the finishing-site environment. A longitudinal follow-up study in cattle also reported closely related PFGE patterns among STEC strains from cattle in the same cohort on the same farm (73). More research, however, is needed to identify potential risk factors for STEC shedding in swine and the common source of infection associated with STEC strains shed by finishing swine.
It was found that swine STEC strains of the same serotype could carry different virulence gene profiles. For example, two distinct virulence gene profiles (profiles 1 and 2) were identified within O59:H21 strains; however, the strains were closely related by PFGE. This was not unexpected, as the two virulence gene profiles (profiles 1 and 2) differed by the presence or absence of estIa (STa), which is carried on a plasmid that can readily be passed between strains with similar genetic backgrounds (54). Because PFGE can determine the genetic relatedness of the swine STEC strains but cannot provide information regarding gene content (74), future studies using sequence-based molecular methods, including whole-genome sequencing, can provide insight into the genetic diversity of swine STEC strains within and across farms. Overall, these results demonstrate a high level of diversity in virulence gene contents among the members of STEC populations from a small population of swine in the same geographic location and enhance our understanding of the transmission dynamics of STEC among pigs in the same finishing barn. However, whether swine STEC strains are potentially pathogenic to humans and the role swine play in the transmission of STEC to humans require further study.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by the National Pork Board (grant number 12-069) and the U.S. Department of Agriculture, National Institute of Food and Agriculture, Agriculture and Food Research Initiative (award number 2013-67005-21189). The PCR microarray development was partially financed by the French “joint ministerial program of R&D against CBRNE risks.”
We sincerely thank Chitrita DebRoy and the E. coli Reference Center at Pennsylvania State University for assisting with characterization of the swine STEC strains. We also acknowledge Niesa Kettler, Terrence MacManus, and the Bacteriology Branch in the Diagnostic Center for Population and Animal Health (DCPAH) at Michigan State University for assisting with the PFGE process.
Footnotes
Published ahead of print 8 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01761-14.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann S, Batz MB, Morris JG. 2012. Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J. Food Prot. 75:1292–1302. 10.4315/0362-028X.JFP-11-417 [DOI] [PubMed] [Google Scholar]
- 3.Unkmeir A, Schmidt H. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856–4864. 10.1128/IAI.68.9.4856-4864.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Brien AD, Newland JW, Miller SF, Holmes RK, Smith HW, Formal SB. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694–696. 10.1126/science.6387911 [DOI] [PubMed] [Google Scholar]
- 5.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664–1668. 10.1073/pnas.92.5.1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton HJ, Sloan J, Bulach DM, Seemann T, Allison CC, Tauschek M, Robins-Browne RM, Paton JC, Whittam TS, Paton AW, Hartland EL. 2009. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg. Infect. Dis. 15:372–380. 10.3201/eid1503.080631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 11:603–609. 10.3201/eid1104.040739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85:E45–E62. 10.2527/jas.2006-508 [DOI] [PubMed] [Google Scholar]
- 10.Paton AW, Ratcliff RM, Doyle RM, Seymour-Murray J, Davos D, Lanser JA, Paton JC. 1996. Molecular microbiological investigation of an outbreak of hemolytic-uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J. Clin. Microbiol. 34:1622–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CDC. 1995. Community outbreak of hemolytic uremic syndrome attributable to Escherichia coli O111:NM—South Australia 1995. MMWR Morb. Mortal. Wkly. Rep. 44:550–558 [PubMed] [Google Scholar]
- 12.CDC. 1995. Escherichia coli O157:H7 outbreak linked to commercially distributed dry-cured salami—Washington and California, 1994. MMWR Morb. Mortal. Wkly. Rep. 44:157–160 [PubMed] [Google Scholar]
- 13.Williams RC, Isaacs S, Decou ML, Richardson EA, Buffett MC, Slinger RW, Brodsky MH, Ciebin BW, Ellis A, Hockin J. 2000. Illness outbreak associated with Escherichia coli O157:H7 in Genoa salami. Can. Med. Assoc. J. 162:1409–1413 [PMC free article] [PubMed] [Google Scholar]
- 14.MacDonald DM, Fyfe M, Paccagnella A, Trinidad A, Louie K, Patrick D. 2004. Escherichia coli O157:H7 outbreak linked to salami, British Columbia, Canada, 1999. Epidemiol. Infect. 132:283–289. 10.1017/S0950268803001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conedera G, Mattiazzi E, Russo F, Chiesa E, Scorzato I, Grandesso S, Bessegato A, Fioravanti A, Caprioli A. 2007. A family outbreak of Escherichia coli O157 haemorrhagic colitis caused by pork meat salami. Epidemiol. Infect. 135:311–314. 10.1017/S0950268806006807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trotz-Williams LA, Mercer NJ, Walters JM, Maki AM, Johnson RP. 2012. Pork implicated in a Shiga toxin-producing Escherichia coli O157:H7 outbreak in Ontario, Canada. Can. J. Public Health 103:e322–e326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairbrother JM, Gyles CL. 2012. Colibacillosis, p 723–747 In Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW. (ed), Diseases of swine, 10th ed. John Wiley and Sons, West Sussex, United Kingdom [Google Scholar]
- 18.Fratamico PM, Bagi LK, Bush EJ, Solow BT. 2004. Prevalence and characterization of Shiga toxin-producing Escherichia coli in swine feces recovered in the National Animal Health Monitoring System's Swine 2000 study. Appl. Environ. Microbiol. 70:7173–7178. 10.1128/AEM.70.12.7173-7178.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borie C, Monreal Z, Guerrero P, Sanchez ML, Martinez J, Arellano C, Prado V. 1997. Prevalence and characterization of enterohaemorrhagic Escherichia coli isolated from healthy cattle and pigs slaughtered in Santiago, Chile. Arch. Med. Vet. 29:205–212. 10.4067/S0301-732X1997000200005 [DOI] [Google Scholar]
- 20.Oporto B, Esteban JI, Aduriz G, Juste RA, Hurtado A. 2008. Escherichia coli O157:H7 and non-O157 Shiga toxin-producing E. coli in healthy cattle, sheep and swine herds in northern Spain. Zoonoses Public Health 55:73–81. 10.1111/j.1863-2378.2007.01080.x [DOI] [PubMed] [Google Scholar]
- 21.Karmali MA, Petric M, Lim C, Fleming PC, Steele BT. 1983. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet ii:1299–1300 [DOI] [PubMed] [Google Scholar]
- 22.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 23.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8:26–38. 10.1038/nrmicro2265 [DOI] [PubMed] [Google Scholar]
- 24.Melton-Celsa A, Mohawk K, Teel L, O'Brien A. 2012. Pathogenesis of Shiga-toxin producing Escherichia coli. Curr. Top. Microbiol. Immunol. 357:67–103. 10.1007/82_2011_176 [DOI] [PubMed] [Google Scholar]
- 25.Petruzziello-Pellegrini TN, Moslemi-Naeini M, Marsden PA. 2013. New insights into Shiga toxin-mediated endothelial dysfunction in hemolytic uremic syndrome. Virulence 4:556–563. 10.4161/viru.26143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26:822–880. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, Igarashi K. 1988. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 171:45–50 [DOI] [PubMed] [Google Scholar]
- 28.Johannes L, Romer W. 2010. Shiga toxins—from cell biology to biomedical applications. Nat. Rev. Microbiol. 8:105–116. 10.1038/nrmicro2279 [DOI] [PubMed] [Google Scholar]
- 29.Bugarel M, Martin A, Fach P, Beutin L. 2011. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol. 11:142. 10.1186/1471-2180-11-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frankel G, Phillips AD. 2008. Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cell. Microbiol. 10:549–556. 10.1111/j.1462-5822.2007.01103.x [DOI] [PubMed] [Google Scholar]
- 31.Paton AW, Srimanote P, Woodrow MC, Paton JC. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999–7009. 10.1128/IAI.69.11.6999-7009.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Low AS, Holden N, Rosser T, Roe AJ, Constantinidou C, Hobman JL, Smith DG, Low JC, Gally DL. 2006. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ. Microbiol. 8:1033–1047. 10.1111/j.1462-2920.2006.00995.x [DOI] [PubMed] [Google Scholar]
- 33.Galli L, Miliwebsky E, Irino K, Leotta G, Rivas M. 2010. Virulence profile comparison between LEE-negative Shiga toxin-producing Escherichia coli (STEC) strains isolated from cattle and humans. Vet. Microbiol. 143:307–313. 10.1016/j.vetmic.2009.11.028 [DOI] [PubMed] [Google Scholar]
- 34.Brunder W, Schmidt H, Karch H. 1996. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:3305–3315. 10.1099/13500872-142-11-3305 [DOI] [PubMed] [Google Scholar]
- 35.Coombes BK, Wickham ME, Mascarenhas M, Gruenheid S, Finlay BB, Karmali MA. 2008. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 74:2153–2160. 10.1128/AEM.02566-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolton DJ. 2011. Verocytotoxigenic (Shiga toxin-producing) Escherichia coli: virulence factors and pathogenicity in the farm to fork paradigm. Foodborne Pathog. Dis. 8:357–365. 10.1089/fpd.2010.0699 [DOI] [PubMed] [Google Scholar]
- 37.Slanec T, Fruth A, Creuzburg K, Schmidt H. 2009. Molecular analysis of virulence profiles and Shiga toxin genes in food-borne Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 75:6187–6197. 10.1128/AEM.00874-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fratamico PM, Bhagwat AA, Injaian L, Fedorka-Cray PJ. 2008. Characterization of Shiga toxin-producing Escherichia coli strains isolated from swine feces. Foodborne Pathog. Dis. 5:827–838. 10.1089/fpd.2008.0147 [DOI] [PubMed] [Google Scholar]
- 39.Meng Q, Bai S, Zhao A, Lan R, Du H, Wang T, Shi C, Yuan S, Ji S, Jin D, Yu B, Wang Y, Sun H, Liu K, Xu J, Xiong Y. 2014. Characterization of Shiga toxin-producing Escherichia coli isolated from healthy pigs in China. BMC Microbiol. 14:5. 10.1186/1471-2180-14-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapman TA, Wu XY, Barchia I, Bettelheim KA, Driesen S, Trott D, Wilson M, Chin JJ. 2006. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 72:4782–4795. 10.1128/AEM.02885-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zweifel C, Schumacher S, Beutin L, Blanco J, Stephan R. 2006. Virulence profiles of Shiga toxin 2e-producing Escherichia coli isolated from healthy pig at slaughter. Vet. Microbiol. 117:328–332. 10.1016/j.vetmic.2006.06.017 [DOI] [PubMed] [Google Scholar]
- 42.Sonntag AK, Bielaszewska M, Mellmann A, Dierksen N, Schierack P, Wieler LH, Schmidt MA, Karch H. 2005. Shiga toxin 2e-producing Escherichia coli isolates from humans and pigs differ in their virulence profiles and interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 71:8855–8863. 10.1128/AEM.71.12.8855-8863.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tseng M, Fratamico PM, Bagi L, Manzinger D, Funk JA. 8 May 2014. Shiga toxin-producing E. coli (STEC) in swine: prevalence over the finishing period and characteristics of the STEC isolates. Epidemiol. Infect. 10.1017/S0950268814001095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bugarel M, Beutin L, Martin A, Gill A, Fach P. 2010. Micro-array for the identification of Shiga toxin-producing Escherichia coli (STEC) seropathotypes associated with hemorrhagic colitis and hemolytic uremic syndrome in humans. Int. J. Food Microbiol. 142:318–329. 10.1016/j.ijfoodmicro.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 45.Delannoy S, Beutin L, Fach P. 2013. Discrimination of enterohemorrhagic Escherichia coli (EHEC) from non-EHEC strains based on detection of various combinations of type III effector genes. J. Clin. Microbiol. 51:3257–3262. 10.1128/JCM.01471-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 47.Tarr PI, Bilge SS, Vary JC, Jelacic S, Jr, Habeeb RL, Ward TR, Baylor MR, Besser TE. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400–1407. 10.1128/IAI.68.3.1400-1407.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doughty S, Sloan J, Bennett-Wood V, Robertson M, Robins-Browne RM, Hartland EL. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761–6769. 10.1128/IAI.70.12.6761-6769.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frydendahl K. 2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 85:169–182. 10.1016/S0378-1135(01)00504-1 [DOI] [PubMed] [Google Scholar]
- 50.Benz I, Schmidt MA. 1992. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787 (O126:H27). Infect. Immun. 60:13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benz I, Schmidt MA. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An H, Fairbrother JM, Desautels C, Harel J. 1999. Distribution of a novel locus called Paa (porcine attaching and effacing associated) among enteric Escherichia coli. Adv. Exp. Med. Biol. 473:179–184. 10.1007/978-1-4615-4143-1_17 [DOI] [PubMed] [Google Scholar]
- 53.Savarino SJ, Fasano A, Robertson DC, Levine MM. 1991. Enteroaggregative Escherichia coli elaborate a heat-stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J. Clin. Invest. 87:1450–1455. 10.1172/JCI115151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dallas WS. 1990. The heat-stable toxin I gene from Escherichia coli 18D. J. Bacteriol. 172:5490–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welch RA, Forestier C, Lobo A, Pellett S, Thomas W, Jr, Rowe G. 1992. The synthesis and function of the Escherichia coli hemolysin and related RTX exotoxins. FEMS Microbiol. Immunol. 5:29–36 [DOI] [PubMed] [Google Scholar]
- 56.Abu-Ali GS, Lacher DW, Wick LM, Qi W, Whittam TS. 2009. Genomic diversity of pathogenic Escherichia coli of the EHEC 2 clonal complex. BMC Genomics 10:296. 10.1186/1471-2164-10-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brandt SM, King N, Cornelius AJ, Premaratne A, Besser TE, On SL. 2011. Molecular risk assessment and epidemiological typing of Shiga toxin-producing Escherichia coli by using a novel PCR binary typing system. Appl. Environ. Microbiol. 77:2458–2470. 10.1128/AEM.02322-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaufmann M, Zweifel C, Blanco M, Blanco JE, Blanco J, Beutin L, Stephan R. 2006. Escherichia coli O157 and non-O157 Shiga toxin-producing Escherichia coli in fecal samples of finished pigs at slaughter in Switzerland. J. Food Prot. 69:260–266 [DOI] [PubMed] [Google Scholar]
- 59.Ateba CN, Mbewe M. 2011. Detection of Escherichia coli O157:H7 virulence genes in isolates from beef, pork, water, human and animal species in the northwest province, South Africa: public health implications. Res. Microbiol. 162:240–248. 10.1016/j.resmic.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 60.Lenahan M, O'Brien SB, Byrne C, Ryan M, Kennedy CA, McNamara EB, Fanning S, Sheridan JJ, Sweeney T. 2009. Molecular characterization of Irish E. coli O157:H7 isolates of human, bovine, ovine and porcine origin. J. Appl. Microbiol. 107:1340–1349. 10.1111/j.1365-2672.2009.04320.x [DOI] [PubMed] [Google Scholar]
- 61.Hauser E, Mellmann A, Semmler T, Stoeber H, Wieler LH, Karch H, Kuebler N, Fruth A, Harmsen D, Weniger T, Tietze E, Schmidt H. 2013. Phylogenetic and molecular analysis of food-borne Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 79:2731–2740. 10.1128/AEM.03552-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedrich AW, Bielaxzewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, Karch H. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84. 10.1086/338115 [DOI] [PubMed] [Google Scholar]
- 63.Beutin L, Krause G, Zimmermann S, Kaulfuss S, Gleier K. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099–1108. 10.1128/JCM.42.3.1099-1108.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orth D, Grif K, Khan AB, Naim A, Dierich MP, Wurzner R. 2007. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn. Microbiol. Infect. Dis. 59:235–242. 10.1016/j.diagmicrobio.2007.04.013 [DOI] [PubMed] [Google Scholar]
- 65.Bielaszewska M, Friedrich AW, Aldick T, Schurk-Bulgrin R, Karch H. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160–1167. 10.1086/508195 [DOI] [PubMed] [Google Scholar]
- 66.Thomas A, Cheasty T, Chart H, Rowe B. 1994. Isolation of Vero cytotoxin-producing Escherichia coli serotypes O9ab:H- and O101:H-carrying VT2 variant gene sequences from a patient with haemolytic uraemic syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 13:1074–1076. 10.1007/BF02111832 [DOI] [PubMed] [Google Scholar]
- 67.Pierard D, Huyghens L, Lauwers S, Lior H. 1991. Diarrhoea associated with Escherichia coli producing porcine oedema disease verotoxin. Lancet 338:762. [DOI] [PubMed] [Google Scholar]
- 68.Muniesa M, Recktenwald J, Bielaszewska M, Karch H, Schmidt H. 2000. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect. Immun. 68:4850–4855. 10.1128/IAI.68.9.4850-4855.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnsen G, Wasteson Y, Heir E, Berget OI, Herikstad H. 2001. Escherichia coli O157:H7 in faeces from cattle, sheep and pigs in the southwest part of Norway during 1998 and 1999. Int. J. Food Microbiol. 65:193–200. 10.1016/S0168-1605(00)00518-3 [DOI] [PubMed] [Google Scholar]
- 70.Franke S, Harmsen D, Caprioli A, Pierard D, Wieler LH, Karch H. 1995. Clonal relatedness of Shiga-like toxin-producing Escherichia coli O101 strains of human and porcine origin. J. Clin. Microbiol. 33:3174–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osek J, Gallien P. 2002. Molecular analysis of Escherichia coli O157 strains isolated from cattle and pigs by the use of PCR and pulsed-field gel electrophoresis methods. Vet. Med. (Praha) 47:149–158 [Google Scholar]
- 72.Rios M, Prado V, Trucksis M, Arellano C, Borie C, Alexandre M, Fica A, Levine MM. 1999. Clonal diversity of Chilean isolates of enterohemorrhagic Escherichia coli from patients with hemolytic-uremic syndrome, asymptomatic subjects, animal reservoirs, and food products. J. Clin. Microbiol. 37:778–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joris MA, Verstraete K, De Reu K, De Zutter L. 2013. Longitudinal follow-up of the persistence and dissemination of EHEC on cattle farms in Belgium. Foodborne Pathog. Dis. 10:295–301. 10.1089/fpd.2012.1277 [DOI] [PubMed] [Google Scholar]
- 74.Singer RS, Sischo WM, Carpenter TE. 2004. Exploration of biases that affect the interpretation of restriction fragment patterns produced by pulsed-field gel electrophoresis. J. Clin. Microbiol. 42:5502–5511. 10.1128/JCM.42.12.5502-5511.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.