Abstract
A combination of 454 pyrosequencing and Sanger sequencing was used to sample and characterize the transcriptome of the entomopathogenic oomycete Lagenidium giganteum. More than 50,000 high-throughput reads were annotated through homology searches. Several selected reads served as seeds for the amplification and sequencing of full-length transcripts. Phylogenetic analyses inferred from full-length cellulose synthase alignments revealed that L giganteum is nested within the peronosporalean galaxy and as such appears to have evolved from a phytopathogenic ancestor. In agreement with the phylogeny reconstructions, full-length L. giganteum oomycete effector orthologs, corresponding to the cellulose-binding elicitor lectin (CBEL), crinkler (CRN), and elicitin proteins, were characterized by domain organizations similar to those of pathogenicity factors of plant-pathogenic oomycetes. Importantly, the L. giganteum effectors provide a basis for detailing the roles of canonical CRN, CBEL, and elicitin proteins in the infectious process of an oomycete known principally as an animal pathogen. Finally, phylogenetic analyses and genome mining identified members of glycoside hydrolase family 5 subfamily 27 (GH5_27) as putative virulence factors active on the host insect cuticle, based in part on the fact that GH5_27 genes are shared by entomopathogenic oomycetes and fungi but are underrepresented in nonentomopathogenic genomes. The genomic resources gathered from the L. giganteum transcriptome analysis strongly suggest that filamentous entomopathogens (oomycetes and fungi) exhibit convergent evolution: they have evolved independently from plant-associated microbes, have retained genes indicative of plant associations, and may share similar cores of virulence factors, such as GH5_27 enzymes, that are absent from the genomes of their plant-pathogenic relatives.
INTRODUCTION
The entomopathogenic oomycete Lagenidium giganteum is known to infect and kill mosquito larvae and therefore has been seen as a potential biological control agent against disease vector mosquitoes (1). However, little is known about the pathological process of L. giganteum in its mosquito host and the molecular basis underlying this process. The study of entomopathogenic oomycetes has yet to benefit from the tremendous advances in oomycete research, including the sequencing of several complete genomes from plant pathogens and the identification of major groups of effectors (2–4). Oomycete effectors include RXLR and crinkler (CRN) proteins, which are known to enter plant cells, as well as other molecules, such as cellulose-binding elicitor lectin (CBEL) and elicitin proteins, which have been associated with the induction of plant defense responses (5). Complementing the wealth of molecular data from plant pathogens, interest in animal-pathogenic oomycetes is increasing (6). Sequencing efforts for the fish pathogen Saprolegnia parasitica (7) and the human pathogen Pythium insidiosum (8) have been initiated. A transcriptome project for the mycoparasite Pythium oligandrum has also been reported (9). The relationship between L. giganteum and these other pathogenic oomycetes, including potential similarities at the molecular level, remains unclear. Genome analysis of the vertebrate pathogen S. parasitica indicated that oomycete effectors are absent in animal pathogens and may be restricted to plant-pathogenic oomycetes (7). However, L. giganteum is more closely related to Phytophthora spp. and other phytopathogens than to S. parasitica (6), and therefore, its genome may be hypothesized to contain similar virulence factors.
Despite a close phylogenetic relationship to Pythium and Phytophthora spp., the genus Lagenidium has virtually never been associated with plants; rather, it is associated primarily with pathogenic interactions with invertebrate hosts. Lagenidium caudatum has been described as a nematode pathogen (10); Lagenidium callinectes and Lagenidium thermophilum are pathogens of marine crustaceans, such as crabs and shrimps (11); and L. giganteum creates natural epizootics in mosquito populations (12, 13). Although some Lagenidium sp. infections have been reported in mammals, including dogs (14) and humans (15), these cases may be categorized, respectively, as examples of taxonomic misclassification (16) or rare keratitis caused by an invertebrate pathogen, similar to the cases caused by the entomopathogenic fungi Metarhizium anisopliae and Beauveria bassiana (17, 18). Because L. giganteum consistently behaves as a virulent pathogen of certain mosquito species, it has been registered with the U.S. Environmental Protection Agency and several states, including California and Florida, for use as a mosquito control tool (1). It was also briefly mass-produced and commercialized under the name Laginex (1, 19). The release of a commercial product was preceded by numerous safety studies that demonstrated the specificity of the L. giganteum interactions with a narrow range of invertebrate hosts (20–23). These studies demonstrated that plants such as corn, rice, sorghum, onions, soybeans, tomatoes, cotton, carrots, lettuce, sunflowers, and duckweed are not affected by prolonged exposure to high dosages of several developmental stages of L. giganteum (21). This oomycete remains primarily a host-specific pathogen of mosquito larvae that is not typically associated with plants, although, like many other aquatic fungi and oomycetes (24), it can also grow saprophytically on rotten vegetation (25).
As an entomopathogen, L. giganteum has traditionally been amalgamated with more-common insect-pathogenic fungi, such as Metarhizium anisopliae and Beauveria bassiana, based not only on a shared filamentous morphology but also on a common infectious strategy that involves the disruption of the insect host exoskeleton by using germinating structures and subsequent colonization of the hemocoel, the nutrient-rich primary body cavity of insects (26). In mosquito larvae, the L. giganteum infectious cycle is initiated by zoospores specifically recognizing (21) and binding to the host cuticle, where they swell and germinate to penetrate the exoskeleton and reach the hemocoel (1). Once the zoospores are in the hemocoel, mycelial growth leads to host death and terminates with the reproduction and release of infectious zoospores (1). Although genome sequence analyses have demonstrated that oomycetes are phylogenetically distant from fungi, emerging evidence gathered from phytopathogen genomes has indicated that filamentous pathogens (fungi and oomycetes) exhibit convergent evolution (27). The similarities observed at the morphological and pathological levels are reflected at the molecular level, and similar proteins, or protein motifs, are used by fungi and oomycetes during plant host infection (28–30). Therefore, the recent completion of the M. anisopliae (31) and B. bassiana (32) genome sequences provides a valuable basis for analyses aimed at identifying conserved pathogenicity factors shared by L. giganteum and fungal entomopathogens, whereas comparative analyses using the sequenced oomycete genomes may reveal infection strategies shared by L. giganteum and all other oomycetes.
In an effort to accelerate gene discovery and better characterize the molecular basis of entomopathogenicity, a pyrosequencing-based transcriptome analysis was initiated for L. giganteum. The set of full-length L. giganteum transcripts described in this study reveals that L. giganteum expresses both canonical oomycete effectors characteristic of phytopathogens and additional virulence factors shared by invertebrate pathogens. This information may ultimately be used to develop L. giganteum-based products into effective and environmentally sustainable mosquito control tools.
MATERIALS AND METHODS
Microbial culture and RNA extraction.
The oomycete Lagenidium giganteum Couch (ARSEF 373) was obtained from the U.S. Department of Agriculture–Agricultural Research Service Collection of Entomopathogenic Fungal Cultures (ARSEF, Ithaca, NY) and was grown in Sabouraud dextrose broth plus 2% yeast extract (SDY) at room temperature. Total RNA was extracted from liquid cultures using the Qiagen RNeasy Plant minikit as described previously (33).
DNA sequencing and gene annotation.
Double-stranded cDNA was generated using the SMARTer PCR cDNA synthesis kit (Clontech) and was processed for 454 Titanium GS-FLX pyrosequencing by the University of Florida Interdisciplinary Center for Biotechnology Research. Contig assembly was performed using CAP3, with default parameters (34). The resulting sequences were annotated in BLAST2GO (35) using BLASTX analysis with an E value cutoff of 10−3. Selected sequences were used for the design of gene-specific primers. Subsequent RACE (rapid amplification of cDNA ends) PCRs incorporated the gene-specific primers and cDNA obtained using the SMARTer RACE cDNA amplification kit (Clontech). The RACE PCR fragments were purified and sequenced commercially using Sanger technologies. Following the generation of full-length transcripts, predicted protein sequences were annotated through homology (BLAST) and motif (InterProScan) searches. Selected motifs were aligned to construct sequence logos using WebLogo, version 3 (36).
Cellulose synthase phylogenetic analyses.
The Lagenidium giganteum cellulose synthase 3 (CesA3) gene (GenBank accession number KM025055) was incorporated into a previously published oomycete CesA3 multiple-sequence alignment (TreeBASE S12300) using Clustal X (37). The alignment was optimized manually by editing the L. giganteum sequence to preserve the aligned nucleotide positions used previously to infer oomycete phylogeny (38). The final data set consisted of 3,344 characters for 26 taxa. The jModelTest program (39) was used to identify the most appropriate maximum likelihood (ML) base substitution model for this data set. The best-fit model selected by all analyses was the general time-reversible model with an inferred proportion of invariable sites and a gamma distribution for the variable sites (GTR+I+G). ML analyses that incorporated the model and parameters calculated by jModelTest were performed using PhyML, version 3.0 (40). ML bootstrap analyses were conducted using the same model and parameters in 1,000 replicates. The phylogenetic tree corresponding to the ML analyses was edited with TreeDyn (41), as implemented in the phylogeny.fr workflow (42).
GH5 phylogenetic analyses.
Glycoside hydrolase family 5 (GH5) protein sequences were selected on the basis of a recent subfamily classification (43) and were obtained from the CAZy (44), FungiDB (45), and NCBI databases (GH5 subfamily 2 [GH5_2], GH5_12, GH5_20, GH5_33, GH5_27, GH5_28, and GH5_29). Fragments corresponding to the catalytic module sequences were aligned using MUSCLE, version 3.7, as described previously (43). The resulting alignment was inspected visually and was validated using the Pfam GH5 hidden Markov model (HMM) (PF00150). Additional editing restricted the alignment to a block ranging from the conserved arginine (R) to the conserved glutamic acid (E) residue (PF00150 HMM positions 39 to 243). The final data set consisted of 493 characters for 51 taxa. The best-fit model for this data set was identified as WAG+I+G by the ProtTest program (46). Phylogenetic reconstruction, bootstrap analyses (100 replicates), and tree editing were performed using phylogeny.fr (42).
Sequencing of GH5_27 orthologs in additional invertebrate pathogens.
Genomic DNA preparations from the entomopathogenic fungi Hirsutella thompsonii strain H3 (47), Isaria fumosorosea strain Ifr AsC (48), and Paecilomyces reniformis strain IndGH96 (49) were obtained from Drion Boucias. These samples were used in PCRs in combination with the custom-designed primers FungalGH5_F (5′-AAAAGGTGAACCAGGACACG-3′) and FungalGH5_R (5′-GTCRCCMAGSCCRTCAAAGT-3′). In addition, mycelial cultures for the oomycetes Lagenidium caudatum (ARSEF 2003) and Leptolegnia chapmanii (ARSEF 2681) were produced in SDY at room temperature and were processed for genomic-DNA extraction using the Qiagen DNeasy Plant minikit. These DNA preparations were amplified by PCRs using primers GH_F3 (5′-CTGGCAGTACAAGTTTCACGAC-3′) and GH_3R (5′-TCGCTCTTCCAGTCAATCTT-3′). All PCRs were performed using the following pattern repeated for 30 cycles: 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min. Products were visualized on a 1% agarose gel, extracted, and sequenced commercially (Macrogen USA).
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the full-length transcript sequences of Lagenidium giganteum are given in Table 1. The sequences corresponding to GH5_27 orthologs from various invertebrate pathogens have been deposited in these databases under accession numbers KM025057 to KM025061. The raw L. giganteum 454 sequences identified as effector orthologs (4 CBEL homologs, 7 transglutaminase homologs, 17 elicitin homologs, and 30 crinkler homologs) are available in the NCBI Sequence Read Archive (SRA) database under accession number SRX661279 as part of BioProject PRJNA256125.
TABLE 1.
Primer sequences and accession numbers for the eight full-length L. giganteum transcripts
| Gene name (transcript length [bp]) | PCR primer |
GenBank accession no. | |
|---|---|---|---|
| Name | Sequence | ||
| Cellulose synthase 3 (3,782) | CSF | CTCTGGTGGGTACTGCATGG | KM025055 |
| CSR | TGGCTGTACAACCTCGTCAC | ||
| CSF2 | CTCAACTTCTTCCTCGGTCTGT | ||
| CSR2 | TCATCGAGTACATGATCCAAGG | ||
| Elicitin (718) | ELICITINF | TCTTGTCCAACCCAGACCTC | KF562858 |
| ELICITINR | TCGCTATTGAACGCCTTAGC | ||
| ELICITINF2 | CAGGCTTGCAAGTCGACAAC | ||
| ELICITINR2 | TGTTCAACTTGGCACCACTG | ||
| CBEL (1,120) | CBELF | GACGACTTGCAGACCGTGTA | KF562859 |
| CBELR | ACCTACGGGAGACTGGCTCT | ||
| CBELF2 | GGACTGTTGTGCCAAGTGTG | ||
| CBELR2 | CACATCATGCATCCTACCCAATCTA | ||
| CBEL (586) | CBEL2F | TGGGGGTAGTTGATGGCTAC | KF562860 |
| CBEL2R | GACCTTGCGCTTCTGGTTAC | ||
| CBEL2F2 | ACTCTGTCGACCCTGTACGGCATT | ||
| CBEL2R2 | CTTCGCTTCAGCACGCACTTT | ||
| Crinkler (DBF) (1,849) | CRINKLERF | GCTGTGGCCAAGAAGAGAAA | KF562861 |
| CRINKLERR | CGCTACCGATCTCAACCTTC | ||
| CRINKLERF2 | GTGGAAGTTGGGTGAAGTGGTTG | ||
| CRINKLERR2 | CAACCACTTCACCCAACTTCCAC | ||
| Crinkler (DXX-DAB) (1,973) | CRNF | GACGCATCGAGTCAGTTCAA | KF562862 |
| CRNR | ACTCCGACAGCATCTTCAGC | ||
| CRNF2 | ACACGTGGGGTCAATCAGTT | ||
| CRNR2 | ATAGCGTCATATCCGGGTTG | ||
| Crinkler (DXX-DAB) (1,854) | CRN2F | AATGCGTTGAATGGTTCCTTGAG | KF562863 |
| CRN2R | ACTCCGACAGCATCTTCAGC | ||
| CRN2F2 | TTCTGACTCGTGCAAGGGAGTTC | ||
| CRN2R2 | AACAGACGGTTGATGCTTGAAGA | ||
| GH5_27 (2,075) | LAGGHF | ACACCATTCGTTGACGGTCT | KM025056 |
| LAGGHR | GAGAACAGGTCCTGGTGGAA | ||
| LAGGHF2 | TGGTTCAGCAAGACTTCGACTA | ||
| LAGGHR2 | CATCAATGACTGGATGATACGG | ||
RESULTS
Transcriptome sequencing overview.
A total of 58,931 reads were obtained from L. giganteum. Contig assembly resulted in 11,018 unique transcripts (including 8,967 singletons) that were subsequently processed for gene annotation through homology searches. The average length of a transcript was 240 bp, which is comparable to the lengths of the Pythium ultimum transcriptome sequences obtained using a similar pyrosequencing approach (50). The length of the assemblies (418 bp) was higher than the average length of a singleton (199 bp), and this difference was reflected in the annotation process: only 23% of the singletons resulted in significant orthology matches, while 53% of the contigs were putatively annotated on the basis of homology searches. As expected, the vast majority of the transcript sequences generated from L. giganteum were homologous to genes previously sequenced from other oomycetes (data not shown).
In agreement with previous reports (50), the length of the pyrosequencing reads remains a limiting factor for sequence assembly and annotation, and the reads may be complemented by longer sequences in order to develop more-comprehensive analyses. The 454-based L. giganteum transcriptome-sequencing effort is currently being supplemented by single-molecule, circular-consensus (CCS) reads generated using the Pacific Biosciences (PacBio) platform. The PacBio technology was recently validated for transcriptome analysis (51), and the combined sequence data will be the subject of a forthcoming article. In addition, selected 454 reads were used as seeds for RACE-PCR amplification and traditional Sanger sequencing. Eight sequences were generated (Table 1) and were used to initiate a full-length cDNA-based comparative analysis between orthologs from plant versus animal pathogens (52). In particular, the full-length transcripts provided a basis for establishing phylogenetic relationships between L. giganteum and other oomycetes and for contrasting the L. giganteum secretome with the previously characterized Phytophthora secretome (5, 52).
Cellulose synthase 3 phylogeny of oomycetes.
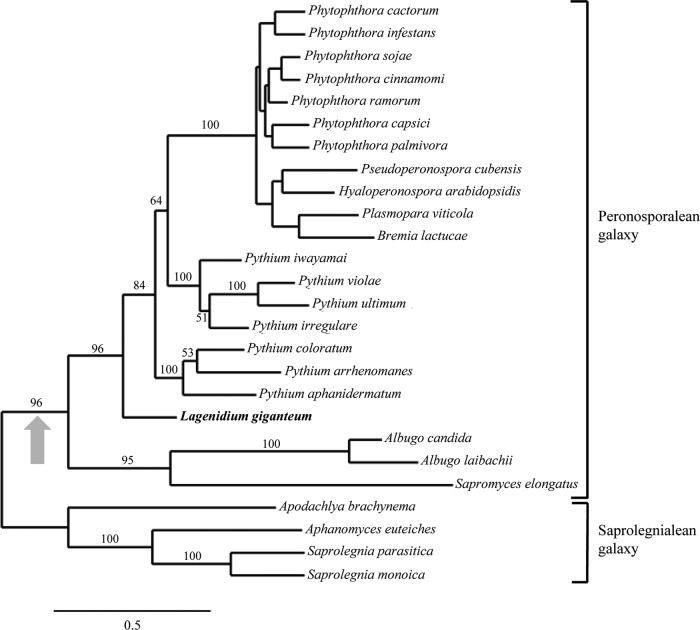
The 3,782-bp full-length L. giganteum cellulose synthase transcript was annotated as cellulose synthase 3 (CesA3) based on strong similarity to previously published sequences (38) and comparison with L. giganteum paralogs (data not shown). Fragments obtained using previously published CesA3-specific primers (38) were characterized by sequences identical to the sequences confirmed by RACE-PCR (not shown). The full-length CesA3 nucleotide sequence was used to infer the maximum likelihood phylogram presented in Fig. 1. The topology of the tree was very consistent with previously published oomycete phylogenies and depicted the well-established peronosporalean and saprolegnian galaxies (38, 53–55). Major clades corresponding to the Peronosporales (Phytophthora spp. and associated genera of downy mildews), Pythiales (Pythium spp.), Albuginales (Albugo spp.), and Saprolegniales (Saprolegnia and Aphanomyces spp.) were strongly supported by bootstrap analyses (Fig. 1). All of these clades appeared as monophyletic groups, except for Pythium spp., which have been shown to be paraphyletic (38, 53, 55). The entomopathogen L. giganteum was not grouped in any of the clades but appeared as a basal lineage to a cluster containing both Peronosporales and Pythiales. This position of the genus Lagenidium in relation to Pythium and Phytophthora spp. was consistent with some recent analyses (53, 55) but contrasted with other trees where Lagenidium was depicted as a sister taxon to a monophyletic Pythium sp. clade (6) or as a member of the Pythiales (54). Most taxon-rich oomycete phylogenies have been inferred from small-subunit (SSU) rRNA sequences ranging from 1,000 to 1,500 characters. The tree presented in Fig. 1 confirms that cellulose synthase sequences provide an unambiguously aligned >3,000-bp alternative phylogenetic signal for the reconstruction of oomycete evolutionary relationships. Based on the current analysis (Fig. 1), L. giganteum remains nested within a peronosporalean galaxy containing a large majority of plant-pathogenic oomycetes (Peronosporales, Pythiales, and Albuginales) and therefore appears to have evolved from a phytopathogenic ancestor.
FIG 1.
Maximum likelihood phylogram inferred from oomycete cellulose synthase 3 nucleotide sequences (3,344 characters). The tree is consistent with previously published oomycete phylogeny reconstructions and shows the entomopathogen L. giganteum (in boldface) as a member of the strongly supported peronosporalean galaxy clade, which contains principally plant pathogens. In accordance with the hypothesized acquisition by peronosporaleans of the ability to infect vascular plants (indicated by the shaded arrow), L. giganteum appears to have evolved from a plant-pathogenic ancestor. The numbers at the nodes represent ML bootstrap values (1,000 replicates). For purposes of clarity, bootstrap values are indicated only for the most significant nodes. The bar indicates the number of substitutions per site.
Disease-like effector families of Lagenidium giganteum.
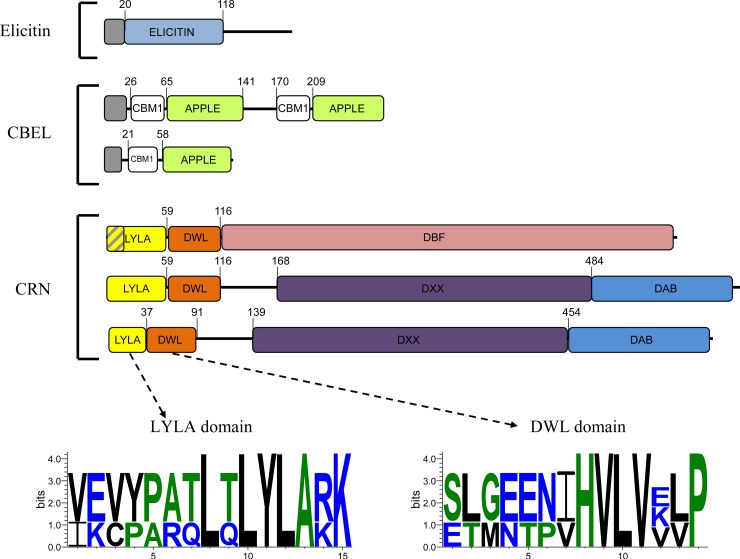
Gene annotations revealed Lagenidium giganteum transcripts orthologous to effectors catalogued for plant-pathogenic oomycetes (5). The RXLR motifs associated with Phytophthora spp. (2) and the YXSL motifs recently proposed for Pythium spp. (9) were not detected in L. giganteum sequences. Similarly, transcripts corresponding to Kazal-like serine protease inhibitors and cystatin-like cysteine protease inhibitors (5) have yet to be identified. However, a total of 58 transcript fragments showed significant similarity to cellulose-binding elicitor lectin (CBEL), elicitin, transglutaminase, and crinkler (CRN) proteins. The short pyrosequencing reads (342 ± 90 bp) were used to initiate RACE-PCR amplifications and to generate the six full-length cDNA sequences that are illustrated in Fig. 2. These six sequences include two CBEL, one elicitin, and three crinkler orthologs, ranging from 586 to 1,973 bp (Table 1; Fig. 2). Motif annotations demonstrated that the full-length L. giganteum effectors have structures similar to those of proteins described for various plant-pathogenic oomycetes. The predicted CBEL proteins are characterized by PAN/APPLE domains (InterPro IPR000177) paired with CBM1 domains (InterPro IPR000254), repeated either once or twice (Fig. 2). This organization, and the difference in motif pair numbers, has also been reported for the plant pathogen Pythium ultimum (56). Signal peptides were predicted for both CBEL sequences, suggesting that these effectors are secreted by L. giganteum (Fig. 2). A signal peptide was also identified directly preceding the elicitin domain (InterPro IPR002200). The putative crinkler proteins are longer molecules and are characterized by the modular organization previously described for Phytophthora infestans and other oomycetes (57). As shown in Fig. 2, the conserved LxLYLA(R/K) and HVLVxxP motifs previously described for Pythium spp. (9) were common to all N termini, whereas the C terminus included either the DBF (GenBank accession number KF562861) or the DXX-DAB (KF562862, KF562863) domains (57). The crinkler proteins were not associated with signal peptides, although, as previously reported for Pythium ultimum (56), the SignalP score for the N terminus of the DBF-bearing CRN protein was very close to the threshold associated with predicted secretion (Fig. 2). Significantly, the identification of Pythium-like effectors in the L. giganteum transcriptome reveals that canonical CBEL and CRN proteins are expressed by an oomycete known principally as an animal pathogen.
FIG 2.
Schematic representation of the six full-length Lagenidium giganteum effector orthologs. Numbers indicate predicted amino acid positions. All orthologs are characterized by the motifs and the modular organization previously described for phytopathogenic oomycete effectors (5). The signal peptides are shown in gray, whereas the InterPro domains are color-coded blue for elicitin and white and green, respectively, for the CBM1 and APPLE domains characteristic of canonical oomycete cellulose-binding elicitor lectin (CBEL) proteins. The predicted L. giganteum crinkler (CRN) proteins include the conserved N-terminal LYLA and DWL motifs (in yellow and orange, respectively), which were aligned to create the logos highlighting the conserved LxLYLA(R/K) (LYLA) and HVLVxxP (DWL) sequences previously recognized for oomycete CRN proteins (5). In contrast to the smaller proteins (CBEL and elicitins), the predicted L. giganteum CRN proteins do not include a signal peptide, although the striped area denotes a N-terminal sequence close to the threshold associated with signal peptides.
Candidate virulence factor (GH5_27) present in L. giganteum and other invertebrate pathogens.
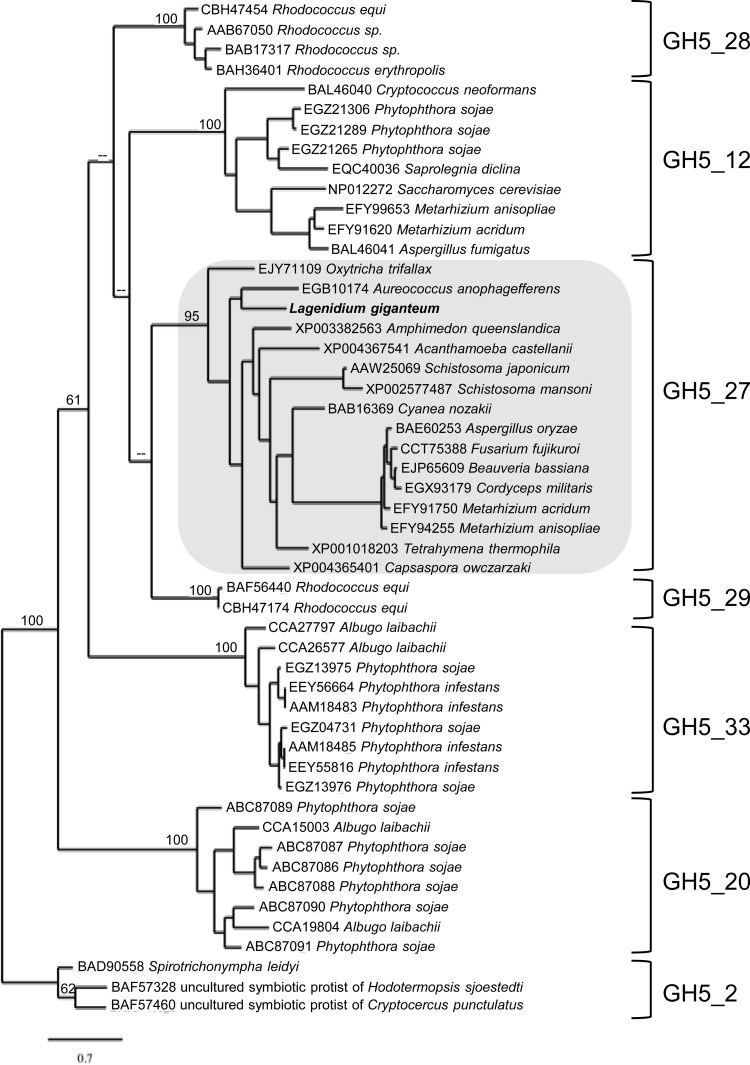
A 496-bp pyrosequencing read (singleton) was selected based on the lack of homology with any oomycete sequences. However, homology searches demonstrated that this sequence was similar to those of genes identified in the entomopathogenic fungi Metarhizium anisopliae and Beauveria bassiana (data not shown). Following RACE-PCR amplification of both 5′ and 3′ ends, a 2,075-bp transcript was obtained (Table 1) and was translated in silico. The resulting L. giganteum protein sequence was demonstrated to contain both a signal peptide and a glycoside hydrolase family 5 (GH5) domain (IPR001547). Although GH5 proteins are commonly referred to as cellulases, a recent study indicated that these enzymes can be divided into several functionally different subfamilies (43). As shown in Fig. 3, phylogenetic analyses identified the putative L. giganteum protein as a GH5_27 (GH5 subfamily 27) protein. The phylogenetic tree is rooted with true cellulases (GH5_2). It is concordant with the original GH5 classification (43) and depicts several GH5 subfamilies as strongly supported monophyletic groups (Fig. 3). Deeper nodes, indicative of relationships between the different subfamilies, are characterized by weaker support, as reported previously (43). Importantly, oomycete GH5 proteins group into three distinct clades—GH5_20, GH5_33, and GH5_12—revealing that L. giganteum GH5_27 is unique among oomycetes (Fig. 3). GH5 subfamily 20 has been referred to previously as family 5 endo-(1-4)-beta-glucanase (58) and, along with GH5 subfamily 33, appears to be specific to Stramenopiles (43). In contrast, GH5_12 enzymes are present in both fungi and oomycetes (Fig. 3). GH5 subfamily 27 also contains fungal homologs in addition to the L. giganteum sequence and other broadly distributed orthologs (Fig. 3). Genome mining using the FungiDB database indicated that GH5_27 not only is absent in plant and vertebrate pathogenic oomycetes but also appears to be underrepresented in Fungi. GH5_27 orthologs were detected in the Fusarium sp. and Aspergillus sp. genomes but were absent from the majority of the most prevalent phytopathogens, including Magnaporthe oryzae, Botrytis cinerea, Puccinia graminis, Gibberella zeae, and Ustilago maydis (59), as well as common fungal models such as Candida spp., Saccharomyces spp. and Neurospora spp. (not shown). In contrast, homology searches revealed that GH5_27 orthologs are present in all sequenced entomopathogenic fungi (Fig. 3), including M. anisopliae and Metarhizium acridum (31), B. bassiana (32), and Cordyceps militaris (60). In entomopathogenic fungi, these proteins are currently annotated as cellulases, but based on this study and the original classification of GH5 subfamilies (43), they appear to be misannotated. Biochemical characterization of GH5_27 in animals revealed that these enzymes were endoglycosylceramidases (EC 3.2.1.123) and were capable of hydrolyzing the glycosidic linkage between oligosaccharides and ceramides in various sphingolipids (61). Enzymes with similar functions have also been detected in prokaryotes and are clustered in GH5 subfamilies 28 and 29 (43). Overall, the combined data-mining and phylogenetic analyses (Fig. 3) indicate that cuticle-degrading filamentous entomopathogens secrete predicted endoglycosylceramidases (EC 3.2.1.123) and that these proteins, while not completely absent from nonentomopathogenic organisms, are predominantly represented, or preferentially retained, in entomopathogenic genomes.
FIG 3.
Maximum likelihood phylogram depicting various GH5 subfamilies as strongly supported monophyletic clades. The GH5_27 proteins (shaded) are putative virulence factors shared by filamentous entomopathogens (fungi and oomycetes) and underrepresented in nonentomopathogens. Accordingly, L. giganteum GH5_27 (in boldface) (GenBank accession number KM025056) is the only GH5_27 representative found in oomycetes. Sequences corresponding to GH5 from plant-pathogenic oomycetes (e.g., Phytophthora sojae) cluster in three other subfamilies: GH5_12, GH5_20, and GH5_33. ML bootstrap values of >50 (1,000 replicates) are indicated by numbers at the nodes, whereas nodes that were not supported by bootstrap analysis (values of <50) are indicated by minus signs. For purposes of clarity, the bootstrap values representative of the support for nodes within each subfamily are not shown. The bar indicates the number of substitutions per site.
Providing support for this hypothesis, genomic sequences orthologous to GH5_27 were obtained for five additional invertebrate pathogens (three fungi and two oomycetes). The fungal sequence lengths were 586 bp for Hirsutella thompsonii and Isaria fumosorosea and 600 bp for Paecilomyces reniformis. Multiple sequence alignments identified the presence of an intron at a conserved location within the predicted GH5 active site, and this intron was shown to be primarily responsible for the observed length polymorphism in the gene fragments (not shown). Homology searches demonstrated that the deduced amino acid sequences corresponded to the N termini of the predicted proteins. The sequence fragments generated for Hirsutella thompsonii and Isaria fumosorosea were 94% and 91% identical, respectively, to the published B. bassiana sequence, whereas the P. reniformis fragment was more similar to the Metarhizium sp. sequences (79% and 81% identity to M. acridum and M. anisopliae, respectively). In contrast to the polymorphic fungal sequences, the oomycete GH5_27 fragments generated from Lagenidium caudatum and Leptolegnia chapmanii were 100% identical to the L. giganteum sequence. These fragments (721 bp) corresponded to the C termini of the predicted proteins and contained an 82-bp intron at a conserved location (not shown). The direct amplification and sequencing of GH5_27 orthologs in phylogenetically diverse invertebrate pathogens contrast with their absence in nonentomopathogens (see above) and support the hypothesis that this gene may play a role during the infectious process.
DISCUSSION
The large-scale Lagenidium giganteum sequencing effort provides a strong basis for the inclusion of invertebrate pathogens in the growing field of oomycete comparative genomics. Importantly, sequences orthologous to the oomycete CBEL and CRN effectors were detected in the L. giganteum transcriptome, demonstrating that canonical effectors associated with plant pathogenicity are present in the genome of an oomycete known principally as an animal pathogen. CBEL and CRN proteins have demonstrated cytotoxic activities in plant cells (5). Therefore, these proteins represent promising candidates for investigations on the molecular basis of oomycete-mosquito interactions and suggest that the L. giganteum transcriptome may contain additional potential virulence factors, even though it was generated from in vitro cultures, with no insect interactions. The identification of L. giganteum effectors similar to the pathogenicity factors of phytopathogens is highly concordant with the phylogenetic analysis presented in Fig. 1, suggesting that L. giganteum has evolved from a plant pathogen ancestor. The putative elicitin, CBEL, and CRN proteins may reflect the evolution of L. giganteum from a plant to an insect pathogen and may indicate that L. giganteum can establish symbiotic and/or pathogenic interactions with plants. This hypothesis is remarkably similar to the recent analyses indicating that entomopathogenic fungi evolved from plant pathogens and endophytes and have retained the ability to establish endophytic relationships (62–64). Combined evidence from the filamentous entomopathogens Metarhizium anisopliae and L. giganteum suggests that entomopathogenicity has evolved from plant-associated microbes in two independent and phylogenetically distant eukaryotic lineages.
It remains unclear if the oomycete effectors identified in the L. giganteum transcriptome represent remnants from a phytopathogenic ancestor, indicate endophytic or pathogenic abilities, or play a role in mosquito infection. Tripartite interactions with both mosquitos and plants have yet to be reported for L. giganteum. This oomycete is known to grow saprophytically on rotten vegetation (25), and it has been isolated from insect larvae collected in leaf axils, suggesting close proximity to plant tissues (65). Although it is possible that L. giganteum is a plant pathogen, the recurrent observations of natural epizootics in mosquito populations, with infection rates of >85% (12, 13), and the ability to control mosquito populations with an artificial formulation (66) strongly suggest that mosquito larvae represent the main host for L. giganteum. Therefore, our primary hypothesis links the L. giganteum elicitin, CBEL, and CRN proteins with pathogenicity for the insect host. Effector motifs from Phytophthora infestans have been used to demonstrate that eukaryotic pathogens share strategies, regardless of their hosts (67). The alternating CBM1 and APPLE modules in CBEL (Fig. 2) have been associated with attachment to plant or animal host tissue through protein-carbohydrate interactions (5) and therefore may mediate the attachment of L. giganteum to the chitin-based host cuticle. Although oomycete CBM1 domains are routinely associated with binding to cellulose, a recent study demonstrated that they bind to glycoproteins through galactose or N-acetylgalactosamine residues (68). In addition, CBM1 domains are also known to bind to chitin and have been detected in M. anisopliae (associated with GH18/chitinase motifs). The hypothesis that L. giganteum CBEL proteins interact with molecules present on mosquito cuticles is under investigation. Similarly, studies have been initiated to functionally characterize the predicted elicitin and CRN proteins and to examine their roles during mosquito infection. Both elicitin and crinkler effectors have been associated with plant cell death (5). In contrast to CBEL and CRN proteins, elicitin-like proteins have been reported for animal-pathogenic oomycetes and may represent a core arsenal of secreted, active molecules that is shared among pathogens (7, 69). They are known to induce tissue necrosis, by which pathogenic oomycetes can thrive (69), and thus may play a role during oomycete-mosquito interactions. Arguments that the L. giganteum CRN proteins are crucial to the pathogenicity process include not only the strong cytotoxic activities of these proteins in plants, especially for the DBF motif (70), but also recent evidence that oomycete CRN genes have been horizontally transferred to the emerging frog pathogen Batrachochytrium dendrobatidis, suggesting that these proteins may impact animal tissues (71). Alternatively, the absence of detectable signal peptides in the CRN proteins (Fig. 2) may indicate that these effectors are no longer secreted and involved in pathogenic interactions with plant cells, supporting the hypothesized transition from phytopathogenicity to entomopathogenicity.
In addition to putative virulence factors that are shared by sister taxa, the entomopathogenic arsenal of L. giganteum is expected to include insect-specific molecules that are not present in phytopathogenic oomycetes. The GH5_27 genes identified in this study represent strong potential candidates for pathogenicity determinants shared by filamentous entomopathogens, suggesting that the recent genome annotations of entomopathogenic fungi may be refined by comparative genomic analysis of phylogenetically distant but morphologically similar organisms. The phylogenetic analyses (Fig. 3) demonstrated that GH5_27 proteins are shared by entomopathogenic fungi and oomycetes, and additional evidence indicated that these enzymes are likely to be active on insect carbohydrates. An expressed sequence tag (EST) homologous to the GH5_27 sequence (GenBank accession number JK742380) was reported for the entomopathogenic fungus M. acridum growing on locust wings (72), suggesting that the GH5_27 proteins may play a role in the early stage of infection, during cuticle penetration. Although they have never been formally characterized in fungi, these proteins are predicted to function as endoglycosylceramidases (EC 3.2.1.123) and may participate in disrupting the association between carbohydrates and lipidic residues in the top layer of the host cuticle. It is now well established that entomopathogenic organisms that penetrate the insect cuticle, such as the fungi M. anisopliae and B. bassiana and the oomycete L. giganteum, do not rely solely on chitinases and proteinases but also secrete active molecules that are thought to target the mixture of lipids that forms the upper exoskeleton layer (73). In agreement with the hypothesis that GH5_27 enzymes are active against the insect cuticle, no orthologous genes were found in the genome of the fungal bee pathogen Ascosphaera apis (74) or the insect-pathogenic alga Helicosporidium sp. (75), both of which are known to initiate their infectious cycles by germinating within the host digestive tract. In contrast, some orthologous gene fragments were readily amplified and sequenced from other cuticle-degrading fungi and oomycetes, including Hirsutella thompsonii, Isaria fumosorosea, Paecilomyces reniformis, and Leptolegnia chapmanii. The presence of GH5_27 extends to the nematophagous oomycete Lagenidium caudatum (this study) and the nematode-trapping fungi Drechslerella stenobrocha, Dactylellina haptotyla, and Arthrobotrys oligospora (GenBank accession numbers EWC43853, EPS43100, and EGX50094, respectively). Phylogenetic analyses inferred from full-length sequences may shed light on the evolutionary relationship between fungal and oomycete GH5_27 proteins and indicate if oomycete GH5_27 genes have been acquired by horizontal gene transfer or from a vertical ancestor. In addition, functional analyses may clarify the function of these enzymes during the pathogenicity process and explain why GH5_27 orthologs can be episodically detected in the genomes of fungi that are not normally associated with insects, such as Aspergillus oryzae or Fusarium fujikuroi (Fig. 3). The entomopathogenic ability and mosquito specificity displayed by L. giganteum likely involve more than just the presence of GH5_27. Other potential virulence factors active on the insect cuticle were recently identified through a proteomic analysis of the M. anisopliae secretome (76), providing candidate targets for further genomic exploration in L. giganteum. Interestingly, these proteomic analyses also related lipolytic activities on the cuticle with ceramidases (76).
In conclusion, the L. giganteum transcriptome provides robust evidence that the convergent evolution hypothesis proposed for entomopathogenic fungi should be extended to the phylogenetically distant oomycetes (73). A phylogenetic analysis inferred from cellulose synthase sequences (Fig. 1) and a genome content analysis revealing effectors similar to the pathogenicity factors of phytopathogens (Fig. 2) strongly suggest that L. giganteum has evolved from a plant-pathogenic ancestor. In addition, the identification of GH5_27 transcripts that are shared among cuticle-degrading organisms but are largely absent in nonentomopathogens (Fig. 3) provides additional support for a convergent evolution hypothesis for oomycetes and fungi and indicates that the two lineages may express a common core arsenal of pathogenic determinants targeting the host surface chemistry. The emergence of this convergent evolution hypothesis, combined with a deeper sequencing effort for L. giganteum, offers a strong basis for initiating comprehensive comparative genomic analyses for plant and invertebrate pathogens, and for fungi and oomycetes, in order to identify and functionally characterize additional virulence factors with potential insecticidal activities. Additional sequence information may establish L. giganteum not only as a source of bioactive compounds against mosquitoes but also as an invaluable model in which to detail the molecular events associated with the transition from plant pathogen to invertebrate pathogen.
ACKNOWLEDGMENTS
We thank Dean Fraga for critical comments on earlier drafts of this article and the anonymous reviewers for constructive comments and suggestions.
Support for next-generation sequencing technologies was provided by the University of Florida Interdisciplinary Center for Biotechnology Research. Funding for this project was provided through a Nova Southeastern University (NSU) President's Faculty Research and Development Grant (award 335482) and a grant from the U.S. Department of Agriculture (Agriculture and Food Research Initiative 2011-68004-30104).
Footnotes
Published ahead of print 8 August 2014
REFERENCES
- 1.Scholte EJ, Knols BG, Samson RA, Takken W. 2004. Entomopathogenic fungi for mosquito control: a review. J. Insect Sci. 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. 2012. Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15:483–492. 10.1016/j.pbi.2012.03.008 [DOI] [PubMed] [Google Scholar]
- 3.Stassen JH, Van den Ackerveken G. 2011. How do oomycete effectors interfere with plant life? Curr. Opin. Plant Biol. 14:407–414. 10.1016/j.pbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 4.Tyler BM. 2009. Entering and breaking: virulence effector proteins of oomycete plant pathogens. Cell. Microbiol. 11:13–20. 10.1111/j.1462-5822.2008.01240.x [DOI] [PubMed] [Google Scholar]
- 5.Kamoun S. 2006. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44:41–60. 10.1146/annurev.phyto.44.070505.143436 [DOI] [PubMed] [Google Scholar]
- 6.Phillips AJ, Anderson VL, Robertson EJ, Secombes CJ, van West P. 2008. New insights into animal pathogenic oomycetes. Trends Microbiol. 16:13–19. 10.1016/j.tim.2007.10.013 [DOI] [PubMed] [Google Scholar]
- 7.Jiang RH, de Bruijn I, Haas BJ, Belmonte R, Lobach L, Christie J, van den Ackerveken G, Bottin A, Bulone V, Diaz-Moreno SM, Dumas B, Fan L, Gaulin E, Govers F, Grenville-Briggs LJ, Horner NR, Levin JZ, Mammella M, Meijer HJ, Morris P, Nusbaum C, Oome S, Phillips AJ, van Rooyen D, Rzeszutek E, Saraiva M, Secombes CJ, Seidl MF, Snel B, Stassen JH, Sykes S, Tripathy S, van den Berg H, Vega-Arreguin JC, Wawra S, Young SK, Zeng Q, Dieguez-Uribeondo J, Russ C, Tyler BM, van West P. 2013. Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. PLoS Genet. 9:e1003272. 10.1371/journal.pgen.1003272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krajaejun T, Khositnithikul R, Lerksuthirat T, Lowhnoo T, Rujirawat T, Petchthong T, Yingyong W, Suriyaphol P, Smittipat N, Juthayothin T, Phuntumart V, Sullivan TD. 2011. Expressed sequence tags reveal genetic diversity and putative virulence factors of the pathogenic oomycete Pythium insidiosum. Fungal Biol. 115:683–696. 10.1016/j.funbio.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 9.Horner NR, Grenville-Briggs LJ, van West P. 2012. The oomycete Pythium oligandrum expresses putative effectors during mycoparasitism of Phytophthora infestans and is amenable to transformation. Fungal Biol. 116:24–41. 10.1016/j.funbio.2011.09.004 [DOI] [PubMed] [Google Scholar]
- 10.Barron G. 1976. Nematophagous fungi: new species of the Lagenidiales endoparasitic on Rhabditis. Antonie Van Leeuwenhoek 42:131–139. 10.1007/BF00399457 [DOI] [PubMed] [Google Scholar]
- 11.Nakamura K, Nakamura M, Hatai K. 1995. Lagenidium infection in eggs and larvae of mangrove crab (Scylla serrata) produced in Indonesia. Mycoscience 36:399–404. 10.1007/BF02268623 [DOI] [Google Scholar]
- 12.Glenn F, Chapman H. 1978. Natural epizootic of the aquatic fungus Lagenidium giganteum in the mosquito Culex territans. Mosq. News 38:522–524 [Google Scholar]
- 13.Kerwin JL, Washino RK. 1988. Field evaluation of Lagenidium giganteum (Oömycetes: Lagenidiales) and description of a natural epizoötic involving a new isolate of the fungus. J. Med. Entomol. 25:452–460 [DOI] [PubMed] [Google Scholar]
- 14.Grooters AM, Hodgin EC, Bauer RW, Detrisac CJ, Znajda NR, Thomas RC. 2003. Clinicopathologic findings associated with Lagenidium sp. infection in 6 dogs: initial description of an emerging oomycosis. J. Vet. Intern. Med. 17:637–646. 10.1111/j.1939-1676.2003.tb02494.x [DOI] [PubMed] [Google Scholar]
- 15.Reinprayoon U, Permpalung N, Kasetsuwan N, Plongla R, Mendoza L, Chindamporn A. 2013. Lagenidium sp. ocular infection mimicking ocular pythiosis. J. Clin. Microbiol. 51:2778–2780. 10.1128/JCM.00783-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza L, Schurko A, Newton JC. 2009. Are strains identified as Lagenidium sp from dogs actually cryptic isolates of Pythium insidiosum? Am. J. Vet. Res. 70:163. 10.2460/ajvr.70.2.163 [DOI] [PubMed] [Google Scholar]
- 17.Motley WW, Melson AT, Mortensen JE. 2011. Pediatric Metarhizium anisopliae keratitis. J. AAPOS 15:101–103. 10.1016/j.jaapos.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 18.Figueira L, Pinheiro D, Moreira R, Pinto E, Simões J, Camisa E, Torrão L, Palmares J, Falcão-Reis F. 2011. Beauveria bassiana keratitis in bullous keratopathy: antifungal sensitivity testing and management. Eur. J. Ophthalmol. 22:814–818. 10.5301/ejo.5000152 [DOI] [PubMed] [Google Scholar]
- 19.Hallmon CF, Schreiber ET, Vo T, Bloomquist A. 2000. Field trials of three concentrations of Laginex as biological larvicide compared to Vectobac-12AS as a biocontrol agent for Culex quinquefasciatus. J. Am. Mosq. Control Assoc. 16:5–8 [PubMed] [Google Scholar]
- 20.Siegel JP, Shadduck JA. 1987. Safety of the entomopathogenic fungus Lagenidium giganteum (Oomycetes: Lagenidiales) to mammals. J. Econ. Entomol. 80:994–997 [DOI] [PubMed] [Google Scholar]
- 21.Kerwin JL, Drit DA, Washino RK. 1988. Nonmammalian safety tests for Lagenidium giganteum (Oomycetes: Lagenidiales). J. Econ. Entomol. 81:158–171 [DOI] [PubMed] [Google Scholar]
- 22.Kerwin JL, Drit DA, Washino RK. 1990. Confirmation of the safety of Lagenidium giganteum (Oomycetes: Lagenidiales) to mammals. J. Econ. Entomol. 83:374–376 [DOI] [PubMed] [Google Scholar]
- 23.Nestrud LB, Anderson RL. 1994. Aquatic safety of Lagenidium giganteum: effects on freshwater fish and invertebrates. J. Invertebr. Pathol. 64:228–233. 10.1016/S0022-2011(94)90275-5 [DOI] [PubMed] [Google Scholar]
- 24.Czeczuga B, Mazalska B, Godlewska A, Muszyńska E. 2005. Aquatic fungi growing on dead fragments of submerged plants. Limnologica 35:283–297. 10.1016/j.limno.2005.07.002 [DOI] [Google Scholar]
- 25.Kerwin JL. 2007. Oomycetes: Lagenidium giganteum. J. Am. Mosq. Control Assoc. 23:50–57. 10.2987/8756-971X(2007)23[50:OLG]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 26.Vega FE, Kaya HK. 2012. Insect pathology, 2nd ed. Academic Press, London, United Kingdom [Google Scholar]
- 27.Latijnhouwers M, De Wit PJ, Govers F. 2003. Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 11:462–469. 10.1016/j.tim.2003.08.002 [DOI] [PubMed] [Google Scholar]
- 28.Luis P, Gauthier A, Trouvelot S, Poinssot B, Frettinger P. 2013. Identification of Plasmopara viticola genes potentially involved in pathogenesis on grapevine suggests new similarities between oomycetes and true fungi. Phytopathology 103:1035–1044. 10.1094/PHYTO-06-12-0121-R [DOI] [PubMed] [Google Scholar]
- 29.Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. 2006. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr. Biol. 16:1857–1864. 10.1016/j.cub.2006.07.052 [DOI] [PubMed] [Google Scholar]
- 30.Petre B, Kamoun S. 2014. How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 12:e1001801. 10.1371/journal.pbio.1001801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Q, Jin K, Ying S-H, Zhang Y, Xiao G, Shang Y, Duan Z, Hu X, Xie X-Q, Zhou G. 2011. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 7:e1001264. 10.1371/journal.pgen.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao G, Ying S-H, Zheng P, Wang Z-L, Zhang S, Xie X-Q, Shang Y, St Leger RJ, Zhao G-P, Wang C, Feng MG. 2012. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2:483. 10.1038/srep00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson DL, Tartar A. 2006. Evolution of glutamine synthetase in heterokonts: evidence for endosymbiotic gene transfer and the early evolution of photosynthesis. Mol. Biol. Evol. 23:1048–1055. 10.1093/molbev/msj110 [DOI] [PubMed] [Google Scholar]
- 34.Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877. 10.1101/gr.9.9.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 36.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 38.Blum M, Gamper HA, Waldner M, Sierotzki H, Gisi U. 2012. The cellulose synthase 3 (CesA3) gene of oomycetes: structure, phylogeny and influence on sensitivity to carboxylic acid amide (CAA) fungicides. Fungal Biol. 116:529–542. 10.1016/j.funbio.2012.02.003 [DOI] [PubMed] [Google Scholar]
- 39.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 41.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. 10.1186/1471-2105-7-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aspeborg H, Coutinho PM, Wang Y, Brumer H, III, Henrissat B. 2012. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12:186. 10.1186/1471-2148-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF. 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 40:D675–D681. 10.1093/nar/gkr918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. 10.1093/bioinformatics/bti263 [DOI] [PubMed] [Google Scholar]
- 47.Maimala S, Tartar A, Boucias D, Chandrapatya A. 2002. Detection of the toxin Hirsutellin A from Hirsutella thompsonii. J. Invertebr. Pathol. 80:112–126. 10.1016/S0022-2011(02)00123-4 [DOI] [PubMed] [Google Scholar]
- 48.Meyer JM, Hoy MA, Boucias DG. 2008. Isolation and characterization of an Isaria fumosorosea isolate infecting the Asian citrus psyllid in Florida. J. Invertebr. Pathol. 99:96–102. 10.1016/j.jip.2008.03.007 [DOI] [PubMed] [Google Scholar]
- 49.Kalkar Ö, Carner G, Scharf D, Boucias D. 2006. Characterization of an Indonesian isolate of Paecilomyces reniformis. Mycopathologia 161:109–118. 10.1007/s11046-005-0133-z [DOI] [PubMed] [Google Scholar]
- 50.Cheung F, Win J, Lang JM, Hamilton J, Vuong H, Leach JE, Kamoun S, André Lévesque C, Tisserat N, Buell CR. 2008. Analysis of the Pythium ultimum transcriptome using Sanger and pyrosequencing approaches. BMC Genomics 9:542. 10.1186/1471-2164-9-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharon D, Tilgner H, Grubert F, Snyder M. 2013. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 31:1009–1014. 10.1038/nbt.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Win J, Kanneganti TD, Torto-Alalibo T, Kamoun S. 2006. Computational and comparative analyses of 150 full-length cDNA sequences from the oomycete plant pathogen Phytophthora infestans. Fungal Genet. Biol. 43:20–33. 10.1016/j.fgb.2005.10.003 [DOI] [PubMed] [Google Scholar]
- 53.Uzuhashi S, Kakishima M, Tojo M. 2010. Phylogeny of the genus Pythium and description of new genera. Mycoscience 51:337–365. 10.1007/S10267-010-0046-7 [DOI] [Google Scholar]
- 54.Beakes GW, Glockling SL, Sekimoto S. 2012. The evolutionary phylogeny of the oomycete “fungi.” Protoplasma 249:3–19. 10.1007/s00709-011-0269-2 [DOI] [PubMed] [Google Scholar]
- 55.Lara E, Belbahri L. 2011. SSU rRNA reveals major trends in oomycete evolution. Fungal Diversity 49:93–100. 10.1007/s13225-011-0098-9 [DOI] [Google Scholar]
- 56.Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J. 2010. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11:R73. 10.1186/gb-2010-11-7-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stam R, Jupe J, Howden AJ, Morris JA, Boevink PC, Hedley PE, Huitema E. 2013. Identification and characterisation CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS One 8:e59517. 10.1371/journal.pone.0059517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costanzo S, Ospina-Giraldo MD, Deahl KL, Baker CJ, Jones RW. 2007. Alternate intron processing of family 5 endoglucanase transcripts from the genus Phytophthora. Curr. Genet. 52:115–123. 10.1007/s00294-007-0144-z [DOI] [PubMed] [Google Scholar]
- 59.Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J. 2012. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13:414–430. 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng P, Xia Y, Xiao G, Xiong C, Hu X, Zhang S, Zheng H, Huang Y, Zhou Y, Wang S. 2011. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 12:R116. 10.1186/gb-2011-12-11-r116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horibata Y, Okino N, Ichinose S, Omori A, Ito M. 2000. Purification, characterization, and cDNA cloning of a novel acidic endoglycoceramidase from the jellyfish, Cyanea nozakii. J. Biol. Chem. 275:31297–31304. 10.1074/jbc.M003575200 [DOI] [PubMed] [Google Scholar]
- 62.Wyrebek M, Bidochka MJ. 2013. Variability in the insect and plant adhesins, Mad1 and Mad2, within the fungal genus Metarhizium suggest plant adaptation as an evolutionary force. PLoS One 8:e59357. 10.1371/journal.pone.0059357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.St Leger RJ, Wang C, Fang W. 2011. New perspectives on insect pathogens. Fungal Biol. Rev. 25:84–88. 10.1016/j.fbr.2011.04.005 [DOI] [Google Scholar]
- 64.Zheng P, Xia Y, Zhang S, Wang C. 2013. Genetics of Cordyceps and related fungi. Appl. Microbiol. Biotechnol. 97:2797–2804. 10.1007/s00253-013-4771-7 [DOI] [PubMed] [Google Scholar]
- 65.Frances S, Sweeney A, Humber R. 1989. Crypticola clavulifera gen. et sp. nov. and Lagenidium giganteum: oomycetes pathogenic for dipterans infesting leaf axils in an Australian rain forest. J. Invertebr. Pathol. 54:103–111. 10.1016/0022-2011(89)90146-8 [DOI] [PubMed] [Google Scholar]
- 66.Kerwin JL, Dritz D, Washino RK. 1994. Pilot scale production and application in wildlife ponds of Lagenidium giganteum (Oomycetes: Lagenidiales). J. Am. Mosq. Control Assoc. 10:451–455 [PubMed] [Google Scholar]
- 67.Haldar K, Kamoun S, Hiller NL, Bhattacharje S, van Ooij C. 2006. Common infection strategies of pathogenic eukaryotes. Nat. Rev. Microbiol. 4:922–931. 10.1038/nrmicro1549 [DOI] [PubMed] [Google Scholar]
- 68.Larroque M, Barriot R, Bottin A, Barre A, Rougé P, Dumas B, Gaulin E. 2012. The unique architecture and function of cellulose-interacting proteins in oomycetes revealed by genomic and structural analyses. BMC Genomics 13:605. 10.1186/1471-2164-13-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang RH, Tyler BM, Whisson SC, Hardham AR, Govers F. 2006. Ancient origin of elicitin gene clusters in Phytophthora genomes. Mol. Biol. Evol. 23:338–351. 10.1093/molbev/msj039 [DOI] [PubMed] [Google Scholar]
- 70.Schornack S, Van Damme M, Bozkurt TO, Cano LM, Smoker M, Thines M, Gaulin E, Kamoun S, Huitema E. 2010. Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. U. S. A. 107:17421–17426. 10.1073/pnas.1008491107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun G, Yang Z, Kosch T, Summers K, Huang J. 2011. Evidence for acquisition of virulence effectors in pathogenic chytrids. BMC Evol. Biol. 11:195. 10.1186/1471-2148-11-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He M, Hu J, Xia Y. 2012. Large scale expressed sequence tag (EST) analysis of Metarhizium acridum infecting Locusta migratoria reveals multiple strategies for fungal adaptation to the host cuticle. Curr. Genet. 58:265–279. 10.1007/s00294-012-0382-6 [DOI] [PubMed] [Google Scholar]
- 73.Ortiz-Urquiza A, Keyhani NO. 2013. Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects 4:357–374. 10.3390/insects4030357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qin X, Evans JD, Aronstein K, Murray KD, Weinstock GM. 2006. Genome sequences of the honey bee pathogens Paenibacillus larvae and Ascosphaera apis. Insect Mol. Biol. 15:715–718. 10.1111/j.1365-2583.2006.00694.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pombert J-F, Blouin NA, Lane C, Boucias D, Keeling PJ. 2014. A lack of parasitic reduction in the obligate parasitic green alga Helicosporidium. PLoS Genet. 10:e1004355. 10.1371/journal.pgen.1004355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beys-da-Silva WO, Santi L, Berger M, Calzolari D, Passos DO, Guimarães JA, Moresco JJ, Yates JR. 2014. Secretome of the biocontrol agent Metarhizium anisopliae induced by the cuticle of the cotton pest Dysdercus peruvianus reveals new insights into infection. J. Proteome Res. 13:2282–2296. 10.1021/pr401204y [DOI] [PMC free article] [PubMed] [Google Scholar]