Abstract
Bacillus thuringiensis vegetative insecticidal proteins (Vip3A) have been recently introduced in important crops as a strategy to delay the emerging resistance to the existing Cry toxins. The mode of action of Vip3A proteins has been studied in Spodoptera frugiperda with the aim of characterizing their binding to the insect midgut. Immunofluorescence histological localization of Vip3Aa in the midgut of intoxicated larvae showed that Vip3Aa bound to the brush border membrane along the entire apical surface. The presence of fluorescence in the cytoplasm of epithelial cells seems to suggest internalization of Vip3Aa or a fragment of it. Successful radiolabeling and optimization of the binding protocol for the 125I-Vip3Aa to S. frugiperda brush border membrane vesicles (BBMV) allowed the determination of binding parameters of Vip3A proteins for the first time. Heterologous competition using Vip3Ad, Vip3Ae, and Vip3Af as competitor proteins showed that they share the same binding site with Vip3Aa. In contrast, when using Cry1Ab and Cry1Ac as competitors, no competitive binding was observed, which makes them appropriate candidates to be used in combination with Vip3A proteins in transgenic crops.
INTRODUCTION
Cry proteins produced by Bacillus thuringiensis are the active components of the most widely used biopesticides in biological control. They have been used in spray formulations for more than 60 years in forestry and agriculture. The importance of Cry proteins has increased dramatically following the introduction of cry genes into a number of major crops (known as Bt crops), mainly maize and cotton, to make them resistant to insect attack. Development of insect resistance to B. thuringiensis insecticidal proteins is an important concern for the long-term use of both spray products and Bt crops. In the last decade, high levels of insect resistance raised against Bt crops have been identified in several lepidopteran pests (1–5).
A different class of insecticidal proteins from B. thuringiensis are the Vip (vegetative insecticidal proteins) proteins, which have been referred to as second generation insecticidal proteins. These proteins were initially found to be secreted into the medium during the vegetative growth phase of this bacterium, and they were discovered much later than Cry proteins (6). Vip proteins share no sequence or structural homology with the Cry proteins, and those belonging to the Vip3A class are active against a wide range of lepidopteran insects (7, 8). One interesting feature presented by the Vip3A proteins is that they extend their activity to some agronomically important pests that have little or no susceptibility to B. thuringiensis Cry proteins, such as the black cutworm, Agrotis ipsilon (6, 8). Moreover, studies so far on the mode of action of Vip3A proteins have revealed differences from that of Cry proteins: in particular, they seem to bind to different binding sites from those targeted by the Cry proteins (9–12).
The above-mentioned characteristics of Vip3A proteins make them interesting candidates to complement Cry proteins in Bt crops to broaden the insecticidal spectrum and for resistance management purposes. For this reason, several agro-biotech companies, such as Dow Agrosciences, Bayer CropScience, and Syngenta, have shown an interest in introducing the vip3A genes in plants, to combine them with the already transferred cry genes (13, 14).
Vip3A proteins are molecules of around 88 to 90 kDa that, once ingested by lepidopteran larvae, are processed by the intestinal serine peptidases to a number of proteolytic fragments. Only the 62-kDa fragment of Vip3Aa has been shown to bind to brush border membrane vesicles (BBMV) from Helicoverpa armigera (11) and is considered to be the active form of the toxin (8, 9, 15, 16). After crossing the peritrophic membrane, the activated toxin specifically binds to the brush border membrane and forms pores (11, 15). All of these steps give rise to the paralysis and complete degeneration of the gut epithelium cells and eventually to the insect's death (17, 18, 19).
In the present work, we show the in vivo binding of Vip3Aa to the brush border membrane of the midgut epithelium cells of Spodoptera frugiperda using immunofluorescence. We also set up the conditions for the in vitro binding of the radiolabeled Vip3Aa to BBMV of this insect, testing different conditions of pH, sodium chloride, chelating agents, and concentrations of divalent cations. Radiolabeling of Vip3Aa has allowed us to estimate quantitative binding parameters of a Vip3A protein for the first time and to perform heterologous competition experiments among Vip3Aa and a number of Cry and Vip3A proteins to determine whether they share binding sites.
MATERIALS AND METHODS
Source of B. thuringiensis Vip3A and Cry1 proteins.
Vip3Aa was prepared from recombinant Escherichia coli BL21 expressing the vip3Aa16 gene from the B. thuringiensis subsp. kurstaki BUPM95 strain (20). The genes encoding the Vip3Ad (NCBI accession no. CAI43276), Vip3Ae (NCBI accession no. CAI43277), and Vip3Af (NCBI accession no. CAI43275) proteins were kindly supplied by Bayer CropScience N.V. (Ghent, Belgium); these proteins were expressed in pMac5-8 in E. coli (21). Cry1Ab was obtained from B. thuringiensis recombinant strain EG7077 (Ecogen, Inc.) and Cry1Ac from recombinant E. coli strain XL-1 (kindly supplied by Ruud de Maagd).
Toxin preparation.
Conditions for bacterial culture and expression of Vip3A proteins from recombinant E. coli strains were described before for Vip3Aa (16). For Vip3Ad, Vip3Ae, and Vip3Af, the protocol by Ruiz de Escudero (22) was followed. The Vip3Aa protein used to intoxicate the larvae was purified by using a HiTrap chelating high-performance (HP) column (GE Healthcare) equilibrated with Ni2+. The lysate supernatant of the induced E. coli cells carrying the vip3Aa gene was loaded in the preequilibrated column and eluted with (50 mM phosphate buffer [pH 8.0], containing 0.3 M NaCl and 200 mM imidazole) elution buffer. Fractions (1 ml) were collected in tubes containing 5 mM EDTA, and the more concentrated ones were pooled and dialyzed against 20 mM Tris-HCl buffer (pH 9)–0.3 M NaCl.
Since we found out that the Vip3Aa protein purified with the protocol described above was not suitable for binding assays, a different purification protocol was used to prepare Vip3A proteins for radiolabeling and competition assays. Expressed Vip3A proteins were partially purified and trypsin activated as follows. After cell lysis and centrifugation, the pH of the lysate supernatant was lowered to 5.5 with 0.1 M acetic acid. The pellet was recovered by centrifugation and dissolved in 20 mM Tris-HCl–150 mM NaCl (pH 7.4). Vip3A protoxins were incubated with 1% trypsin (wt/wt) for 2 h at 37°C. Activated toxins obtained at this point were used as competitors in the binding assays. Vip3Aa to be used for labeling was further purified by anion-exchange chromatography. After overnight dialysis against 20 mM Tris-HCl, (pH 9), Vip3Aa was purified on a HiTrap Q HP (5-ml bed volume) column equilibrated in the same dialysis buffer, using an ÄKTA explorer 100 chromatography system (GE Healthcare, United Kingdom). Proteins were eluted with a 100-ml linear gradient (0 to 80%) of 1 M NaCl.
Cry1Ac expression, solubilization from E. coli inclusion bodies, and trypsin treatment were performed as described before (23). Cry1Ab was solubilized from B. thuringiensis parasporal crystals, trypsin activated, and chromatography purified as described elsewhere (24).
Immunohistochemical localization of Vip3Aa toxin.
Third or second instar larvae of S. frugiperda were starved overnight before being exposed to 1.2 mg/ml Vip3Aa protoxin (purified in a HiTrap chelating HP column) in a solution of Fluorella blue in 50% sucrose. After 1 h of exposure, larvae with blue-stained midgut were transferred to 4% paraformaldehyde in phosphate-buffered saline (PBS) (1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl [pH 7.4]) and incubated for 2 days at 4°C with gentle shaking, and then for 2 additional days in 30% sucrose in PBS. Whole larvae were fixed to a support and coated with Tissue Tech gel (Sakura, Japan). The gel was allowed to solidify at −30°C. Sections of 10 μm were prepared using the cryostat microtome Leica CM 1510S. Slides with the tissue sections were stored at −20°C until used.
Tissue sections were washed three times with buffer A (PBS, 0.5% bovine serum albumin [BSA], 0.3% Triton) for 10 min and blocked with the same buffer supplemented with 5% fetal bovine serum (FBS) for 30 min in a humid chamber. Sections were then coated with the anti-Vip3Aa protoxin polyclonal antibody for 2 days at 4°C in a humid chamber. Unbound antibodies were washed off with three rinses using buffer A (10 min each), and slides were subsequently incubated with a mix of fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit IgG (1:1,000 dilution) and phalloidin (1:1,000 dilution) in blocking buffer for an additional 2 h. After the slides were washed with buffer A, coverslips were mounted using Vectashield mounting medium with DAPI (4′,6-diamidino-2-phenylindole) from Vector Laboratories. The specimens were then examined using a Leica DMI2500 microscope equipped with a digital color camera (Leica DFC300 FX). Fluorescent images acquired in the red, blue, and green channels were merged using ImageJ software (25).
Radiolabeling of Vip3Aa.
Iodination was performed twice using two different batches of Vip3Aa by the chloramine T method (24, 26). Trypsin-activated and anion-exchange-purified Vip3Aa protein (25 μg) was mixed with 0.5 mCi of 125I and 1/3 (vol/vol) 18 mM chloramine T. The excess of free 125I was separated from the labeled protein using a PD10 desalting column (GE HealthCare). The purity of the 125I-labeled Vip3Aa was checked by analyzing the elution fractions by SDS-PAGE with further exposure of the dry gel to an X-ray film. The specific activity of the labeled protein was 1.1 mCi/mg in both cases. The first batch was used for the optimization of the binding parameters and the saturation experiment, and the second batch was used for the rest of experiments.
BBMV preparation.
Last instar larvae of S. frugiperda were dissected, and the midguts were frozen in liquid nitrogen and preserved at −80°C until required. Brush border membrane vesicles (BBMV) were prepared by the differential magnesium precipitation method (27), frozen in liquid nitrogen, and stored at −80°C. The protein concentration in the BBMV preparations was determined by Bradford (28) using bovine serum albumin (BSA) as a standard.
Binding assays with 125I-labeled Vip3Aa.
For all binding assays, 125I-Vip3Aa was incubated with BBMV at room temperature in a 0.1-ml final volume of PBS or Tris buffer containing 0.1% BSA. The reaction was stopped by centrifuging the tubes at 16,000 × g for 10 min at 4°C, and the pellet was washed once with 500 μl of cold buffer. The radioactivity retained in the pellet was measured in a model 2480 WIZARD2 gamma counter. An excess of unlabeled Vip3Aa toxin (300-fold) was used to estimate the nonspecific binding.
To determine the appropriate amount of BBMV to use, a fixed amount of 125I-Vip3Aa (1.2 nM) was mixed with increasing concentrations of BBMV in PBS with 0.1% BSA. For all subsequent experiments, 20 μg/ml of BBMV was chosen. For the time course experiment, BBMV were mixed with 1.2 nM 125I-Vip3Aa and incubated for 15, 30, 60, 90, and 120 min in PBS–0.1% BSA. For saturation experiments (three replicates), BBMV were incubated for 90 min with increasing amounts of 125I-Vip3Aa in PBS–0.1% BSA. To test the effect of pH, NaCl concentration, EDTA and the presence of divalent cations, two independent replicates were performed. To avoid precipitation of some divalent cations, Tris buffer was used instead of phosphate buffer.
The conditions chosen to perform the competition binding assays were 1.2 nM 125I-Vip3Aa (second labeled batch), 20 μg/ml BBMV, a 90-min incubation time, and increasing concentrations of unlabeled competitor in a 0.1-ml final volume of Tris binding buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MnCl2, 0.1% BSA). Equilibrium dissociation constants (Kd) and the concentration of binding sites (Rt) were estimated using the LIGAND software (29).
To estimate the ratio of reversible and irreversible binding within the specific binding of Vip3Aa, the procedure described by Park et al. (30) was followed. The experiment consisted of three samples, each containing 1.2 nM 125I-Vip3Aa and 2 μg of BBMV in Tris binding buffer, incubated at room temperature for 3 h. One sample was used to determine the total binding. Another sample, to which an excess of unlabeled Vip3Aa (300-fold) was added at the start of the assay, was used to determine the nonspecific binding. The third sample was used to estimate the ratio of reversible/irreversible binding by means of adding the excess of Vip3Aa 1.5 h after the start of the assay.
RESULTS
In vivo binding of Vip3Aa to S. frugiperda midgut.
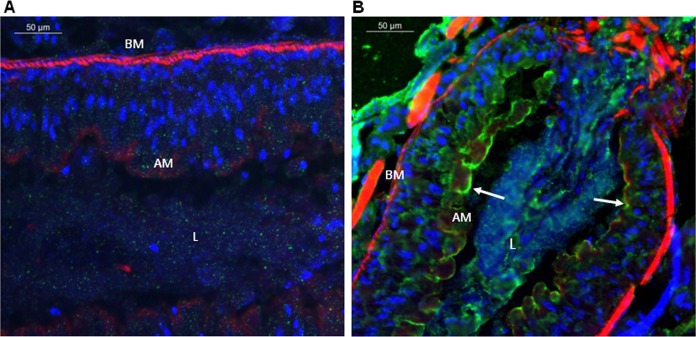
Midgut sections from S. frugiperda larvae, either exposed or nonexposed to Vip3Aa protoxin, were observed under a fluorescence microscope to investigate the fate of Vip3Aa after ingestion (Fig. 1). In the midgut of Vip3Aa-fed larvae, an intense green color, which corresponded to the toxin, was observed along the entire midgut apical surface, which was still intact (Fig. 1B). Furthermore, some green fluorescence could be observed nonhomogeneously distributed in the cytoplasm of the epithelial cells. The insect cuticle also showed intense green fluorescence, most probably due to residual Vip3Aa that adhered to the larva surface during its exposure to the protoxin. Nuclei were stained in blue and the basement membrane and the apical membrane microvilli in red. In the control sections of the nonexposed larvae, the overall midgut epithelium structure was found to be intact and well organized. No green fluorescence was detected either inside the cells or in the apical membrane of the nonexposed larvae (Fig. 1A).
FIG 1.
Immunolocalization of Vip3Aa in midgut tissue sections (10 μm) after in vivo ingestion by S. frugiperda larvae. Binding of Vip3Aa was revealed by Alexa Fluor-conjugated secondary antibody (green) using fluorescence microscopy. Nuclei were stained with DAPI (blue), and the apical and the basal membranes were stained with phalloidin (red). Magnification × 400. (A) Larvae not exposed to Vip3Aa; (B) larvae that ingested Vip3Aa. BM, basal membrane; AM, apical membrane; L, gut lumen. White arrows show the Vip3Aa protein bound to the midgut apical membrane.
Purification of trypsin-activated Vip3Aa for radiolabeling and binding assays.
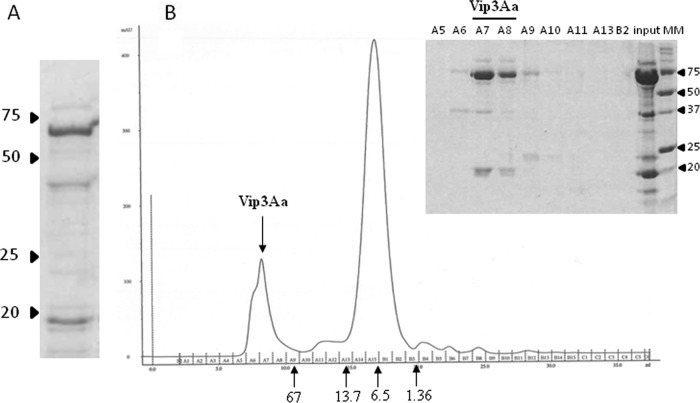
Vip3Aa was purified by isoelectric point precipitation, followed by trypsin activation and then anion-exchange chromatography (Fig. 2A). To further purify the 62-kDa fragment, the chromatographic fraction containing the highest concentration of Vip3Aa was loaded into a gel filtration column (Superdex 75 10/300 GL; GE Healthcare). The analysis of the different chromatographic fractions showed that the 62-kDa peptide and the approximately 20-kDa peptide eluted together (Fig. 2B), indicating that they remain attached after trypsinization. Since we were unable to separate these two peptides even using 1 mM dithiothreitol (DTT) (31), the preparation after anion-exchange chromatography was used for radiolabeling and competition assays.
FIG 2.
Purification of Vip3Aa. (A) SDS-PAGE of trypsin-activated Vip3Aa purified by isoelectric point precipitation and anion-exchange chromatography. (B) Gel filtration chromatography (Superdex 75 10/300 GL) of the anion-exchange-purified Vip3Aa. The inset shows the elution fractions as revealed by SDS-PAGE. The arrows below the chromatogram indicate the elution volume of molecular mass (MM) standards in kDa. The arrowheads by the electrophoresis gels indicate the molecular mass markers in kDa.
In vitro binding of 125I-Vip3Aa to S. frugiperda BBMV.
Specific binding was firstly shown by incubating a fixed amount of 125I-Vip3Aa (1.2 nM) with increasing concentrations of BBMV, in the presence or absence of excess of unlabeled Vip3Aa (Fig. 3). Around 10% of 125I-Vip3Aa used in the assay bound to the BBMV, of which approximately 50% was specific.
FIG 3.
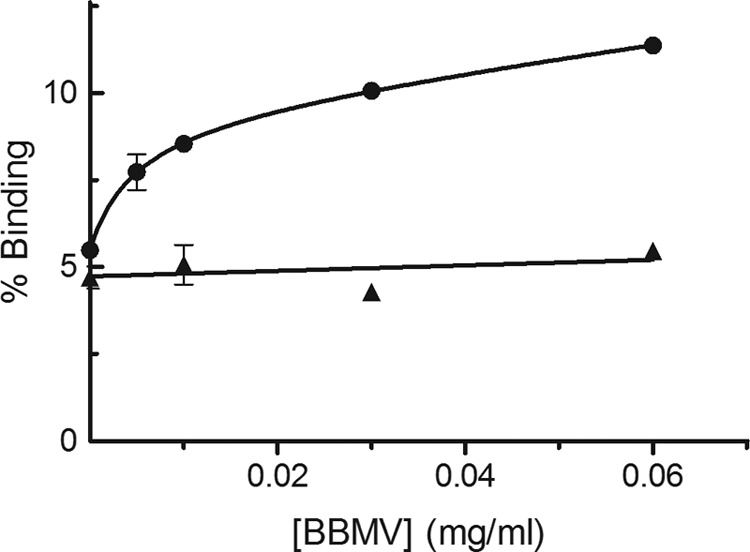
Specific binding of 125I-Vip3Aa at increasing concentrations of S. frugiperda BBMV. Data points are the mean of two replicates. The nonspecific binding was calculated using an excess of unlabeled Vip3Aa toxin. ●, total binding; ▲, nonspecific binding.
A second experiment was done, which consisted of the separation by SDS-PAGE of the proteins in the pellet after incubation of 125I-Vip3Aa with the BBMV and subsequent autoradiography (Fig. 4). Two radioactive peptides bound to the BBMV, one of 62 kDa (which corresponded to the molecular mass of the activated Vip3Aa toxin) and the second of approximately 20 kDa. Western blot analysis with a Vip3Aa-specific antibody showed that this peptide corresponded to a Vip3Aa proteolytic fragment (data not shown). As shown in Fig. 4, an excess of unlabeled Vip3Aa displaced the binding of both 125I-Vip3Aa peptides, indicating that both the activated Vip3Aa and the 20-kDa fragment bind to the BBMV specifically.
FIG 4.
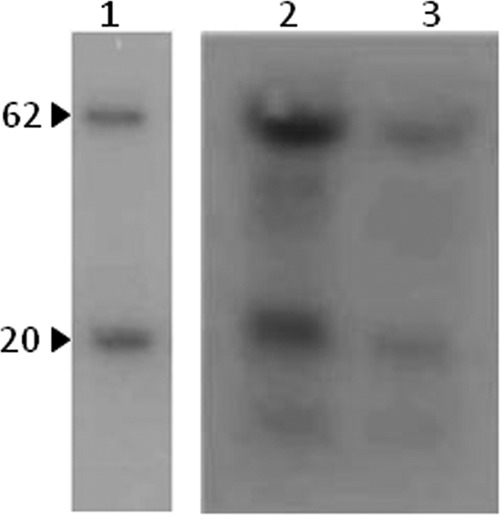
Autoradiography of 125I-Vip3Aa bound to BBMV from S. frugiperda. 125I-Vip3Aa was incubated with BBMV in the absence (lane 2) or presence (lane 3) of a 300-fold excess of unlabeled Vip3Aa. The pellet obtained after centrifugation of the reaction mixture was subjected to SDS-PAGE and exposed to X-ray film for a week. Lane 1, labeled toxin used in the assay (input). Arrowheads indicate the approximate molecular masses of the labeled peptides in kDa.
To show that binding of 125I-Vip3Aa to S. frugiperda BBMV was saturable, a fixed amount of BBMV from S. frugiperda was incubated with increasing concentrations of 125I-Vip3Aa. As shown in Fig. 5, the plot of the specific binding versus input 125I-Vip3Aa exhibited an asymptotic curve, in which saturation was observed at concentrations above 50 nM.
FIG 5.
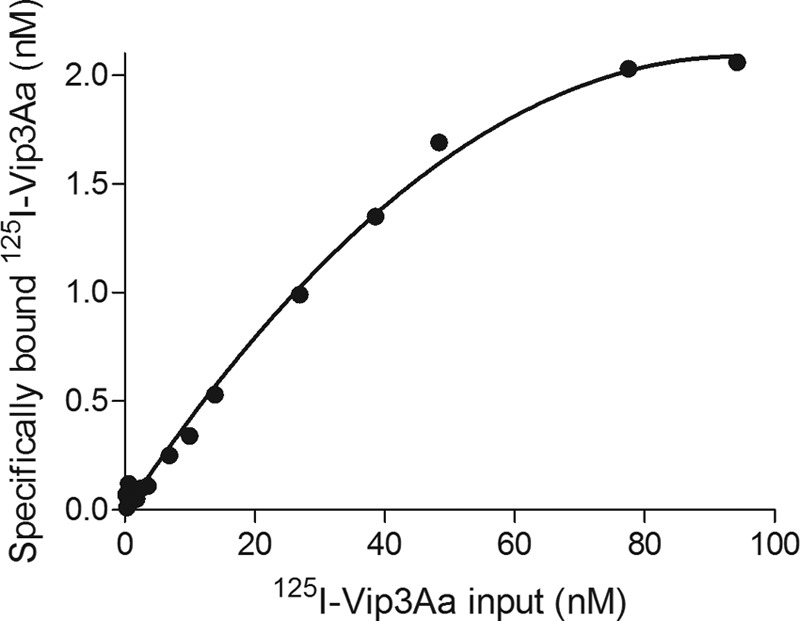
Saturation of 125I-Vip3Aa binding to S. frugiperda BBMV. A fixed amount of BBMV (2 μg) was incubated with increasing amounts of 125I-Vip3Aa for 90 min. The reaction was stopped by centrifugation, and the radioactivity retained in the pellet was measured. Specific binding was determined by subtracting nonspecific binding from the total binding.
The analysis of the irreversible and reversible components of the Vip3Aa-specific binding showed that most part of this binding was irreversible (over 87% of the specific binding) (Fig. 6).
FIG 6.
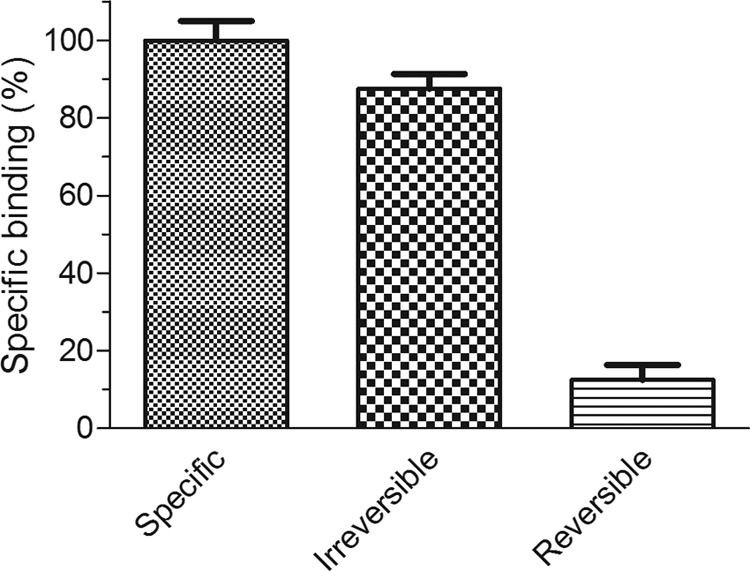
Contribution of the reversible and irreversible binding to the specific binding of 125I-Vip3Aa to S. frugiperda BBMV.
Optimization of binding conditions.
A time course experiment was performed to determine the time required for the binding reaction to reach equilibrium. Binding increased rapidly over the first 30 min, reached equilibrium at about 90 min, and remained stable for at least 2 h.
To select the best conditions to obtain the highest total binding while maintaining the nonspecific binding to a minimum, the influence of the incubation time, pH, NaCl concentration, EDTA, and the presence of divalent cations was studied (Table 1). The effect of pH was analyzed in the range of 7.4 to 9.0. The results showed that binding was pH dependent, with both the total and the specific binding decreasing as the pH increased. The concentration of NaCl also influenced the amount of total binding and the proportion of specific binding, with the optimum between 100 and 150 mM. For subsequent optimization assays, a pH of 7.4 and a concentration of NaCl of 150 mM were chosen.
TABLE 1.
Influence of pH, NaCl concentration, chelating agents, and the presence or absence of divalent cations on the binding of 125I-Vip3Aa to S. frugiperda BBMVa
| Bufferb | Binding condition |
125I-Vip3Aa binding (% of input) |
Ratio of specific to total binding | ||||
|---|---|---|---|---|---|---|---|
| pH | NaCl concn (mM) | Divalent cation | EDTA concn (mM) | Specific | Total | ||
| A | 9 | 150 | 0 | 6.3 | 0 | ||
| 8.2 | 2.5 | 7.1 | 0.35 | ||||
| B | 7.4 | 150 | 4.1 | 8.5 | 0.48 | ||
| 0 | 1.9 | 5.8 | 0.33 | ||||
| 50 | 3.5 | 7.5 | 0.47 | ||||
| 100 | 3.8 | 8.5 | 0.45 | ||||
| 300 | 2.7 | 8.1 | 0.33 | ||||
| 500 | 1.8 | 7.5 | 0.24 | ||||
| 150 | 5 | 2.8 | 7.2 | 0.39 | |||
| A | 7.4 | 150 | MnCl2 (10 mM) | 0 | 21.8 | 0 | |
| MnCl2 (1 mM) | 5.5 | 13.1 | 0.42 | ||||
| CuSO4 (1 mM) | 0 | 8.2 | 0 | ||||
| CuSO4 (0.1 mM) | 2.5 | 8.2 | 0.30 | ||||
| ZnCl2 (1 mM) | 4.2 | 10.8 | 0.39 | ||||
| MgCl2 (10 mM) | 1.2 | 6.9 | 0.17 | ||||
| CaCl2 (1 mM) | 2.4 | 7.0 | 0.34 | ||||
Values are the means of at least two replicates.
Buffer A is 20 mM Tris–0.1% BSA, and buffer B is 10 mM Na2HPO4/NaH2PO4-0.1% BSA.
Results in our laboratory have shown that the presence of some divalent cations, such as Ni2+, significantly reduced the toxicity of Vip3Ae and Vip3Af toward S. frugiperda (8). For this reason, the effect of the addition of the chelating agent EDTA to the binding reaction and the type and concentration of several divalent cations was tested (Table 1). The results showed that EDTA reduced both the total binding and the proportion of specific binding. The presence of some divalent cations, such as Mn2+ and Zn2+, drastically increased the total binding. However, the proportion of specific binding was strongly influenced by the concentration of the divalent cation, as could be observed with Mn2+ and Cu2+. Based on these results, 1 mM MnCl2 was included in the binding reaction in further assays with S. frugiperda BBMV.
Competition binding assays.
Homologous competition was performed with a fixed amount of 125I-Vip3Aa and increasing concentrations of unlabeled Vip3Aa (Fig. 7). The analysis of the data indicated that the curve fitted one binding-site model of high affinity (equilibrium dissociation constant [Kd] of 15 ± 2 nM) and a relatively high concentration of binding sites (Rt = 54 ± 7 pmol/mg BBMV protein). It is worth mentioning that the activated VipAa that had been purified by a Ni2+-loaded chelating column did not compete with 125I-Vip3Aa (Fig. 7).
FIG 7.
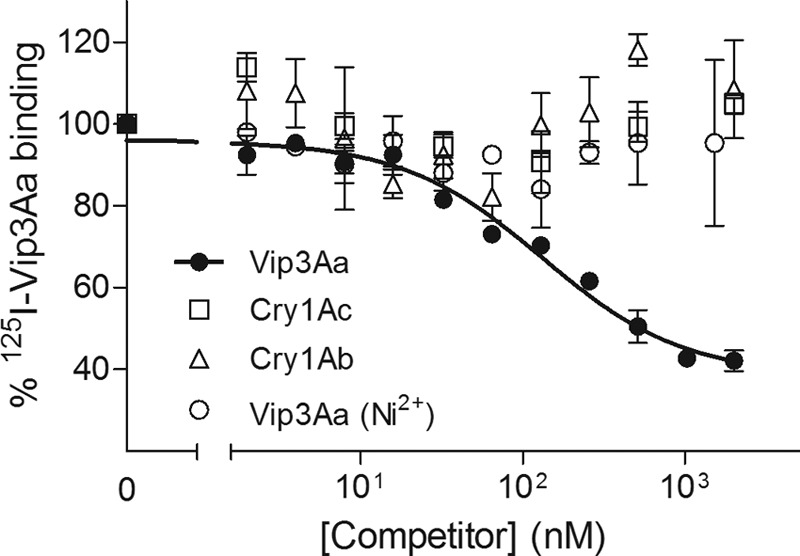
Binding of 125I-Vip3Aa to S. frugiperda BBMV at increasing concentrations of unlabeled competitor. Each data point represents the mean of two independent replicates.
To find out whether Vip3Aa shares binding sites with other B. thuringiensis toxins, binding assays with 125I-Vip3Aa and increasing concentrations of unlabeled competitors were performed (heterologous competition). No competition was observed when Cry1Ac or Cry1Ab was used as a competitor (Fig. 7). However, Vip3Ad, Vip3Ae, and Vip3Af completely displaced the specific binding of 125I-Vip3Aa (Fig. 8). These results indicate that the four Vip3A proteins share binding sites in S. frugiperda and that Cry1Ab and Cry1Ac do not bind to these sites. The estimation of Kd and Rt for the competing proteins from the heterologous data indicated that binding parameters obtained for all Vip3A proteins were comparable (Table 2).
FIG 8.
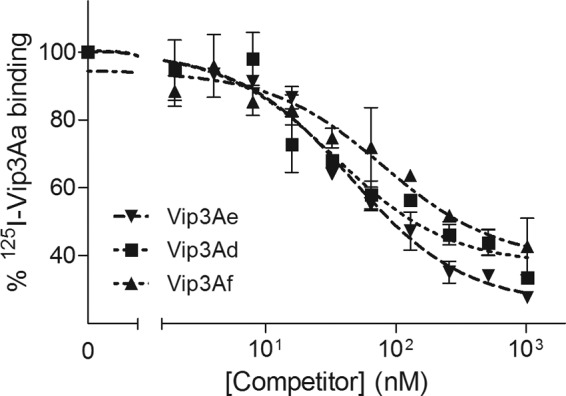
Heterologous competition of Vip3A proteins with 125I-Vip3Aa and S. frugiperda BBMV. Each data point represents the mean of two independent replicates.
TABLE 2.
Kd and Rt of Vip3A proteins with BBMV from S. frugiperdaa
| Protein | Mean ± SEM |
|
|---|---|---|
| Kd (nM) | Rt (pmol/mg) | |
| Vip3Aa | 15 ± 2 | 54 ± 7 |
| Vip3Ad | 17 ± 3 | 76 ± 17 |
| Vip3Ae | 6.1 ± 1.1 | 48 ± 8 |
| Vip3Af | 22 ± 5 | 72 ± 14 |
Equilibrium constants for Vip3Aa were estimated from homologous competition (three independent replicates), and those for Vip3Ad, Vip3Ae, and Vip3Af were estimated from heterologous competition (two independent replicates).
DISCUSSION
Vip3 proteins are considered a new generation of insecticidal proteins from B. thuringiensis as they do not share any type of sequence homology with the Cry toxins (6, 15). Vip3Aa has been recently introduced in important crops not only in an effort to widen their protection against lepidopteran pests but also as part of a strategy of resistance management in a response to the emerging resistance to the existing Cry toxins (3, 5). In addition to having different activity spectra from Cry1A and Cry2A proteins, Vip3A proteins have been shown to target distinct receptors in the insect midgut (9, 15, 32). Their discovery in 1996 was the starting point for the search for new vip3 genes, with more than 80 vip3A genes identified to date (33). Different Vip3A proteins have been shown to differ in their insecticidal spectra (8, 22). Because of the increasing interest in this new class of insecticidal proteins, it becomes important to study their mode of action in more detail.
As a first approach to the study of the binding of the Vip3Aa to the midgut of S. frugiperda, immunohistochemical analysis of Vip3Aa showed that binding took place in the brush border membrane of the midgut epithelial cells (Fig. 1), as had been described previously in Agrotis ipsilon and Ostrinia nubilalis larvae (17). Unlike the Cry toxins, there was no visible binding of the Vip3Aa toxin to the basal membrane of the midgut epithelial cells or to the peritrophic membrane (34, 35, 36). In addition, some green fluorescence could be observed inside the epithelial cells, which could suggest internalization of Vip3Aa or a fragment of it. Results from a previous study with Sf21 insect cells also suggested that Vip3Aa internalized after binding to the cell membrane (37). Whether internalization of Vip3Aa is actually a step in the mode of action deserves further study.
So far, all studies on the binding of Vip3A proteins to the insect midgut have been done with biotinylated Vip3A proteins, and thus, quantitative parameters of Vip3A binding were lacking. In the present work, successful radiolabeling of a Vip3 protein and its use to show saturable and specific binding to BBMV are described for the first time. Specific binding of 125I-Vip3Aa to S. frugiperda BBMV was shown by incubating a fixed amount of 125I-Vip3Aa with increasing concentrations of BBMV (Fig. 3) and confirmed by autoradiography of the bound protein after separation from BBMV by SDS-PAGE (Fig. 4). In both cases, around half of the total binding of the iodinated toxin was inhibited by the presence of an excess of unlabeled Vip3Aa. The 20-kDa fragment present in the sample of Vip3Aa used in radiolabeling also showed competition by an excess of unlabeled Vip3Aa. This is due to the fact that this fragment remains tightly linked to the 62-kDa fragment (Fig. 2B). Saturation of Vip3Aa binding sites was shown by incubating a fixed amount of BBMV and increasing concentrations of the radiolabeled protein (Fig. 5). Despite the fact that the affinity-purified Vip3Aa protoxin, after trypsin activation, showed strong toxicity against S. frugiperda (16), this toxin preparation was found to be inappropriate for binding assays: no specific binding could be obtained with the radiolabeled toxin (data not shown), and the unlabeled protein was unable to compete with radiolabeled Vip3Aa (anion-exchange purified) (Fig. 7).
For Cry1 toxins, a direct correlation between irreversible binding, pore formation, and toxicity has been described in various cases (38, 39). Vip3A proteins have been shown to form pores in different susceptible insects, such as M. sexta and H. armigera (11, 15), which indirectly indicates that binding of Vip3A to the BBMV from these insects is, at least in part, irreversible. Our study provides the first direct evidence of the irreversible binding of Vip3Aa to S. frugiperda BBMV (Fig. 6).
Since this was the first time that radiolabeled Vip3Aa was used for binding assays, it was necessary to first select the conditions under which the binding to the S. frugiperda BBMV was optimum. As in one of the first studies with radiolabeled Cry proteins (26), the influence of pH, NaCl concentration, and incubation time was tested. Furthermore, the effect of the presence of EDTA or the type and concentration of divalent cations was investigated. Since the pH of the midgut of lepidopterans is known to be alkaline, the effect of pH was tested in the range from 7.4 to 9. The binding was shown to be dependent on the pH: the higher values of specific binding were obtained at the lowest pH. The NaCl concentration also had an influence on the specific binding of 125I-Vip3Aa, most likely by stabilizing the Vip protein.
Hernández-Martínez et al. (8) showed that the purification of two different Vip3A proteins using the metal chelation columns exerted negative effect on their toxicity, and thus, EDTA was used to stabilize the toxin. However, the addition of the chelating agent EDTA in the binding reaction mixture of the 125I-Vip3Aa decreased both the total and the specific binding to the BBMV (Table 1), which indicated that the in vitro binding is sensitive to the presence of divalent cations. The concentration and type of the cations affected both the total and the specific binding of the 125I-Vip3Aa. Mn2+ at 10 mM yielded the highest total binding, although all this binding was nonspecific. However, at 1 mM Mn2+, despite that total binding decreased comparatively, a substantial amount of specific binding was obtained. Addition of Cu2+ (0.1 or 1 mM), Mg2+ (10 mM), or Ca2+ (1 mM) had relatively small effects on the total binding; however, in all cases specific binding was decreased compared with when these ions were absent. These results are in contrast with the binding of Cry1Ab to Manduca sexta BBMV, which was not affected by the presence of either 5 mM EGTA or 10 mM Mg2+ or Ca2+ (26). It is possible that some metal ions are required by some brush border membrane proteins involved in the Vip3Aa binding.
The displacement of the 125I-Vip3Aa protein observed in the homologous competition experiment confirms the occurrence of a limited number of receptors for Vip3Aa that could be saturated, adding an excess of unlabeled toxin. The heterologous competition by Vip3Ad, Vip3Ae, and Vip3Af indicates that these three proteins also bind to the same sites as Vip3Aa. However, whether Vip3Aa competes for all of the binding sites recognized by Vip3Ad, Vip3Ae, or Vip3Af (i.e., the reciprocal competition experiments) has not been tested here. Competition of Vip3Aa with biotinylated Vip3Af had been shown previously in S. frugiperda (10). The proteins Vip3Aa, Vip3Ae, and Vip3Af are known to be toxic to S. frugiperda (8, 10, 16); however, Vip3Ad is nontoxic (8). This result indicates, as occurs with Cry proteins, that binding of Vip proteins is necessary, though not sufficient, for toxicity.
The analysis of the binding parameters from the homologous and the heterologous competitions rendered Kd and Rt values similar for all four Vip3A proteins, with Kd values in the range of 6.1 to 22 nM and Rt values in the range of 48 to 76 pmol/mg of BBMV protein. These values are higher (around 10-fold) than the ones normally obtained for the Cry1A and Cry2A proteins (12, 24, 40–42), which indicates that Vip3A proteins have lower affinity but a higher number of binding sites in the BBMV than the Cry1A and Cry2A proteins. Lee et al. (15) showed that the kinetics of pore formation of activated Vip3A was more than 8-fold slower than that of Cry1Ab (at equimolar concentrations) and that the kinetics did not change after a 10-fold increase in the Vip3A concentration. Lee et al. claimed that this could be due to the fact that saturation of functional binding sites of the Vip3A proteins was hard to reach.
When Cry1Ab and Cry1Ac were used as heterologous competitors, no displacement of 125I-Vip3Aa occurred. This result, along with the competition of the Vip3A proteins for the same binding site found here and the results obtained in a previous study (10), strongly suggests that Vip3A proteins do not share binding sites with Cry1A proteins in S. frugiperda. Lack of competition between Cry1A and Cry2A proteins and Vip3Aa had already been reported in three heliothine species (9, 11, 12). The overall results suggest that these two classes of toxins (Vip3A and Cry1A/2A) use different receptors to bind to the brush border membrane of target insects.
In conclusion, the successful radiolabeling of Vip3Aa in this work opens up interesting perspectives for the future of binding studies with Vip3A proteins. Using radiolabeled Vip3Aa allowed us to estimate for the first time binding parameters for this protein. Furthermore, heterologous competition has revealed that Vip3Ad, Vip3Ae, and Vip3Af competed for the Vip3Aa binding sites. The absence of competition of Cry1Ac and Cry1Ab makes them appropriate candidates to be used in combination with Vip3A proteins in transgenic crops as a strategy to delay the evolution of resistance in insects.
ACKNOWLEDGMENTS
We thank R. González-Martínez for rearing the insect colony.
This research was supported by the Spanish Ministry of Science and Innovation (grant reference no. AGL2009-13340-C02-01), by grants ACOMP/2009/313 and PROMETEO 2011/044 from the Generalitat Valenciana, and by European FEDER funds. M.C. was supported by a Santiago Grisolía Fellowship from the Generalitat Valenciana.
Juan Ferré provides consultancy services to Bayer Cropscience. This company provided plasmids for this study.
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.Tabashnik BE, Sisterson MS, Ellsworth PC, Dennehy TJ, Antilla L, Liesner L, Whitlow M, Staten RT, Fabrick JA, Unnithan GC, Yelich AJ, Ellers-Kirk C, Harpold VS, Li X, Carrière Y. 2010. Suppressing resistance to Bt cotton with sterile insect releases. Nat. Biotechnol. 28:1304–1307. 10.1038/nbt.1704 [DOI] [PubMed] [Google Scholar]
- 2.Dhurua S, Gujar GT. 2011. Field-evolved resistance to Bt toxin Cry1Ac in the pink bollworm, Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae), from India. Pest Manag. Sci. 67:898–903. 10.1002/ps.2127 [DOI] [PubMed] [Google Scholar]
- 3.Downes S, Mahon R. 2012. Evolution, ecology and management of resistance in Helicoverpa spp. to Bt cotton in Australia. J. Invertebr. Pathol. 110:281–286. 10.1016/j.jip.2012.04.005 [DOI] [PubMed] [Google Scholar]
- 4.Storer NP, Kubiszak ME, King JE, Thompson GD, Santos AC. 2012. Status of resistance to Bt maize in Spodoptera frugiperda: lessons from Puerto Rico. J. Invertebr. Pathol. 110:294–300. 10.1016/j.jip.2012.04.007 [DOI] [PubMed] [Google Scholar]
- 5.Tabashnik BE, Wu K, Wu Y. 2012. Early detection of field-evolved resistance to Bt cotton in china: cotton bollworm and pink bollworm. J. Invertebr. Pathol. 110:301–306. 10.1016/j.jip.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 6.Estruch JJ, Warren GW, Mullins MA, Nye GJ, Craig JA, Koziel MG. 1996. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. U. S. A. 93:5389–5394. 10.1073/pnas.93.11.5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milne R, Liu Y, Gauthier D, van Frankenhuyzen K. 2008. Purification of Vip3Aa from Bacillus thuringiensis HD-1 and its contribution to toxicity of HD-1 to spruce budworm (Choristoneura fumiferana) and gypsy moth (Lymantria dispar) (Lepidoptera). J. Invertebr. Pathol. 99:166–172. 10.1016/j.jip.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 8.Hernández-Martínez P, Hernández-Rodríguez CS, Van Rie J, Escriche B, Ferré J. 2013. Insecticidal activity of Vip3Aa, Vip3Ad, Vip3Ae, and Vip3Af from Bacillus thuringiensis against lepidopteran corn pests. J. Invertebr. Pathol. 113:78–81. 10.1016/j.jip.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 9.Lee MK, Miles P, Chen JS. 2006. Brush border membrane binding properties of Bacillus thuringiensis Vip3A toxin to Heliothis virescens and Helicoverpa zea midguts. Biochem. Biophys. Res. Commun. 339:1043–1047. 10.1016/j.bbrc.2005.11.112 [DOI] [PubMed] [Google Scholar]
- 10.Sena JA, Hernández-Rodríguez CS, Ferré J. 2009. Interaction of Bacillus thuringiensis Cry1 and Vip3Aa proteins with Spodoptera frugiperda midgut binding sites. Appl. Environ. Microbiol. 75:2236–2237. 10.1128/AEM.02342-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Yang A, Shen X, Hua B, Shi G. 2011. Specific binding of activated Vip3Aa10 to Helicoverpa armigera brush border membrane vesicles results in pore formation. J. Invertebr. Pathol. 108:92–97. 10.1016/j.jip.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 12.Gouffon C, Van Vliet A, Van Rie J, Jansens S, Jurat-Fuentes JL. 2011. Binding sites for Bacillus thuringiensis Cry2Ae toxin on heliothine brush border membrane vesicles are not shared with Cry1A, Cry1F, or Vip3A toxin. Appl. Environ. Microbiol. 77:3182–3188. 10.1128/AEM.02791-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anonymous. 2010. Syngenta, Dow AgroSciences sign cotton technology licensing agreements. http://www.syngenta.com/global/corporate/en/news-center/news-releases/Pages/en-100105.aspx
- 14.Anonymous. 2010. Syngenta and Bayer CropScience sign global cotton technology licensing agreement. http://www.syngenta.com/global/corporate/en/news-center/news-releases/Pages/en-100714.aspx
- 15.Lee MK, Walters FS, Hart H, Palekar N, Chen JS. 2003. The mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3Aa differs from that of Cry1Ab delta-endotoxin. Appl. Environ. Microbiol. 69:4648–4657. 10.1128/AEM.69.8.4648-4657.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakroun M, Bel Y, Caccia S, Abdelkefi-Mesrati L, Escriche B, Ferré J. 2012. Susceptibility of Spodoptera frugiperda and S. exigua to Bacillus thuringiensis Vip3Aa insecticidal protein. J. Invertebr. Pathol. 110:334–339. 10.1016/j.jip.2012.03.021 [DOI] [PubMed] [Google Scholar]
- 17.Yu CG, Mullins MA, Warren GW, Koziel MG, Estruch JJ. 1997. The Bacillus thuringiensis vegetative insecticidal protein Vip3Aa lyses midgut epithelium cells of susceptible insects. Appl. Environ. Microbiol. 63:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdelkefi-Mesrati L, Boukedi H, Dammak-Karray M, Sellami-Boudawara T, Jaoua S, Tounsi S. 2011. Study of the Bacillus thuringiensis Vip3Aa16 histopathological effects and determination of its putative binding proteins in the midgut of Spodoptera littoralis. J. Invertebr. Pathol. 106:250–254. 10.1016/j.jip.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 19.Abdelkefi-Mesrati L, Boukedi H, Chakroun M, Kamoun F, Azzouz H, Tounsi S, Rouis S, Jaoua S. 2011. Investigation of the steps involved in the difference of susceptibility of Ephestia kuehniella and Spodoptera littoralis to the Bacillus thuringiensis Vip3Aa16 toxin. J. Invertebr. Pathol. 107:198–201. 10.1016/j.jip.2011.05.014 [DOI] [PubMed] [Google Scholar]
- 20.Mesrati AL, Tounsi S, Jaoua S. 2005. Characterization of a novel vip3-type gene from Bacillus thuringiensis and evidence of its presence on a large plasmid. FEMS Microbiol. Lett. 244:353–358. 10.1016/j.femsle.2005.02.007 [DOI] [PubMed] [Google Scholar]
- 21.Stanssens P, Opsomer C, McKeown YM, Kramer W, Zabeau M, Fritz HJ. 1989. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acids Res. 17:4441–4454. 10.1093/nar/17.12.4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz de Escudero I, Banyuls N, Bel Y, Maeztu M, Escriche B, Muñoz D, Caballero P, Ferré J. 2014. A screening of five Bacillus thuringiensis Vip3A proteins for their activity against lepidopteran pests. J. Invertebr. Pathol. 117:51–55. 10.1016/j.jip.2014.01.006 [DOI] [PubMed] [Google Scholar]
- 23.Bosch D, Schipper B, van der Kleij H, de Maagd RA, Stiekema WJ. 1994. Recombinant Bacillus thuringiensis crystal proteins with new properties: possibilities for resistance management. Nat. Biotechnol. 12:915–918. 10.1038/nbt0994-915 [DOI] [PubMed] [Google Scholar]
- 24.Hernández-Rodríguez CS, Van Vliet A, Bautsoens N, Van Rie J, Ferré J. 2008. Specific binding of Bacillus thuringiensis Cry2A insecticidal proteins to a common site in the midgut of Helicoverpa species. Appl. Environ. Microbiol. 74:7654–7659. 10.1128/AEM.01373-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. 1989. Specificity of Bacillus thuringiensis delta-endotoxins. Importance of specific receptors on the brush border membrane of the mid-gut of target insects. Eur. J. Biochem. 186:239–247 [DOI] [PubMed] [Google Scholar]
- 27.Wolfersberger MG, Luthy P, Maurer P, Parenti P, Sacchi VF, Giordana B, Hanozet GM. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86A:301–308 [Google Scholar]
- 28.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 29.Munson P, Rodbard D. 1980. LIGAND: a versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 107:220–239. 10.1016/0003-2697(80)90515-1 [DOI] [PubMed] [Google Scholar]
- 30.Park Y, González-Martínez RM, Navarro-Cerrillo G, Chakroun M, Kim Y, Ziarsolo P, Blanca J, Cañizares J, Ferré J, Herrero S. 2014. ABCC transporters as receptors for Bacillus thuringiensis toxins revealed by bulk segregant analysis of a Bt-resistant Spodoptera exigua colony. BMC Biol. 12:46. 10.1186/1741-7007-12-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao JZ, Collins HL, Tang JD, Cao J, Earle ED, Roush RT, Herrero S, Escriche B, Ferré J, Shelton AM. 2000. Development and characterization of diamondback moth resistance to transgenic broccoli expressing high levels of Cry1C. Appl. Environ. Microbiol. 66:3784–3789. 10.1128/AEM.66.9.3784-3789.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdelkefi-Mesrati L, Rouis S, Sellami S, Jaoua S. 2009. Prays oleae midgut putative receptor of Bacillus thuringiensis vegetative insecticidal protein Vip3LB differs from that of Cry1Ac toxin. Mol. Biotechnol. 43:15–19. 10.1007/s12033-009-9178-4 [DOI] [PubMed] [Google Scholar]
- 33.Crickmore N, Baum J, Bravo A, Lereclus D, Narva K, Sampson K, Schnepf E, Sun M, Zeigler DR. 2013. Bacillus thuringiensis toxin nomenclature. http://www.btnomenclature.info/
- 34.Bravo A, Jansens S, Peferoen M. 1992. Immunocytochemical localization of Bacillus thuringiensis isinsecticidal crystal proteins in intoxicated insects. J. Invertebr. Pathol. 60:237–246. 10.1016/0022-2011(92)90004-N [DOI] [Google Scholar]
- 35.Rodrigo-Simón A, de Maagd RA, Avilla C, Bakker PL, Molthoff J, González-Zamora JE, Ferré J. 2006. Lack of detrimental effects of Bacillus thuringiensis Cry toxins on the insect predator Chrysoperla carnea: a toxicological, histopathological, and biochemical analysis. Appl. Environ. Microbiol. 72:1595–1603. 10.1128/AEM.72.2.1595-1603.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rouis S, Chakroun M, Jaoua S. 2008. Comparative study of Bacillus thuringiensis Cry1Aa and Cry1Ac delta-endotoxin activation, inactivation and in situ histopathological effect in Ephestia kuehniella (Lepidoptera: Pyralidae). Mol. Biotechnol. 38:233–239. 10.1007/s12033-007-9021-8 [DOI] [PubMed] [Google Scholar]
- 37.Singh G, Sachdev B, Sharma N, Seth R, Bhatnagar RK. 2010. Interaction of Bacillus thuringiensis vegetative insecticidal protein with ribosomal S2 protein triggers larvicidal activity in Spodoptera frugiperda. Appl. Environ. Microbiol. 76:7202–7209. 10.1128/AEM.01552-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ihara H, Kuroda E, Wadano A, Himeno M. 1993. Specific toxicity of δ-endotoxin from Bacillus thuringiensis to Bombyx mori. Biosci. Biotechnol. Biochem. 57:200–204. 10.1271/bbb.57.200 [DOI] [PubMed] [Google Scholar]
- 39.Liang Y, Patel SS, Dean DH. 1995. Irreversible binding kinetics of Bacillus thuringiensis CryIA δ-endotoxins to gypsy moth brush border membrane vesicles is directly correlated to toxicity. J. Biol. Chem. 270:24719–24724. 10.1074/jbc.270.42.24719 [DOI] [PubMed] [Google Scholar]
- 40.Garczynski SF, Crim JW, Adang MJ. 1991. Identification of putative insect brush border membrane-binding molecules specific to Bacillus thuringiensis delta-endotoxin by protein blot analysis. Appl. Environ. Microbiol. 57:2816–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rang C, Bergvingson D, Bohorova N, Hoisington D, Frutos R. 2004. Competition of Bacillus thuringiensis Cry1 toxins for midgut binding sites: a basis for the development and management of transgenic tropical maize resistant to several stemborers. Curr. Microbiol. 49:22–27. 10.1007/s00284-003-4258-3 [DOI] [PubMed] [Google Scholar]
- 42.Caccia S, Hernández-Rodríguez CS, Mahon RJ, Downes S, James W, Bautsoens N, Van Rie J, Ferré J. 2010. Binding site alteration is responsible for field-isolated resistance to Bacillus thuringiensis Cry2A insecticidal proteins in two Helicoverpa species. PLoS One 5:e9975. 10.1371/journal.pone.0009975 [DOI] [PMC free article] [PubMed] [Google Scholar]