Abstract
In this study, a 96-h laboratory reduction test was conducted with strain BDHSH06 (GenBank accession no. EF011103) as the test strain for Bdellovibrio and like organisms (BALOs) and 20 susceptible marine bacterial strains forming microcosms as the targets. The results showed that BDHSH06 reduced the levels of approximately 50% of prey bacterial strains within 96 h in the seawater microcosms. An 85-day black tiger shrimp (Penaeus monodon) rearing experiment was performed. The shrimp survival rate, body length, and weight in the test tanks were 48.1% ± 1.2%, 99.8 ± 10.0 mm, and 6.36 ± 1.50 g, respectively, which were values significantly (P < 0.05) higher than those for the control, viz., 31.0% ± 2.1%, 86.0 ± 11.1 mm, and 4.21 ± 1.56 g, respectively. With the addition of BDHSH06, total bacterial and Vibrio numbers were significantly reduced (P < 0.05) by 1.3 to 4.5 log CFU · ml−1 and CFU · g−1 in both water and shrimp intestines, respectively, compared to those in the control. The effect of BDHSH06 on bacterial community structures in the rearing water was also examined using PCR amplification of the 16S rRNA gene and denaturing gradient gel electrophoresis (DGGE). The DGGE profiles of rearing water samples from the control and test tanks revealed that the amounts of 44% of the bacterial species were reduced when BDHSH06 was added to the rearing water over the 85-day rearing period, and among these, approximately 57.1% were nonculturable. The results of this study demonstrated that BDHSH06 can be used as a biocontrol/probiotic agent in P. monodon culture.
INTRODUCTION
Black tiger shrimp (Penaeus monodon) are one of the most widely cultured prawn species and are an important aquatic protein source. However, aquatic diseases have significantly compromised shrimp aquaculture worldwide since 1993 (1), and shrimp aquaculture in China has also suffered from diseases caused by opportunistic and pathogenic bacteria (2).
Bacterial diseases, mainly caused by Vibrio species, are very common in shrimp aquaculture. Disease outbreaks attributed to Vibrio species, such as Vibrio harveyi, V. alginolyticus, V. parahaemolyticus, V. vulnificus, and V. penaeicida, have been documented to be a cause of larval mortality and to occur in P. monodon nurseries and grow-out ponds (3–7). The natural abundance, ubiquity, and fast multiplication rate of vibrios and the ability of vibrios to adapt to environmental changes in shrimp culture ecosystems increase these dangers (8). A reduction in either their quantity or quality could pave the way for the healthy growth of shrimp in shrimp aquaculture.
Bdellovibrio and like organisms (BALOs) comprise a group of Gram-negative predatory bacteria that require prey cells in order to invade, grow, and reproduce (9). They hunt other pathogenic and/or potentially pathogenic bacteria in water and biofilms (10), and the spectrum of bacteria on which they prey is very nonspecific (9, 11). BALOs have also been shown to be unable to prey on eukaryotic cells and as such pose no direct risk to human or animal health (12–15). These features make BALOs possible candidates as biocontrol agents (10, 11, 16, 17).
In aquaculture or related fields, BALOs are attracting attention as possible alternative biocontrol agents for farmed animals (18, 19). However, the potential of BALOs in aquaculture at the production level or near the production level has rarely been studied. No data on the effect of BALOs on P. monodon rearing and/or aquatic microbial communities are available.
In this study, we examined the efficacy of BALO strain BDHSH06 (GenBank accession no. EF011103) in improving black tiger shrimp (P. monodon) survival and growth, controlling potentially harmful and/or pathogenic bacteria and their numbers, and altering bacterial community structures in rearing waters. The results of this study should indicate whether BDHSH06 could be used as a biocontrol agent in mariculture.
MATERIALS AND METHODS
Bacterial strain, media, and growth conditions.
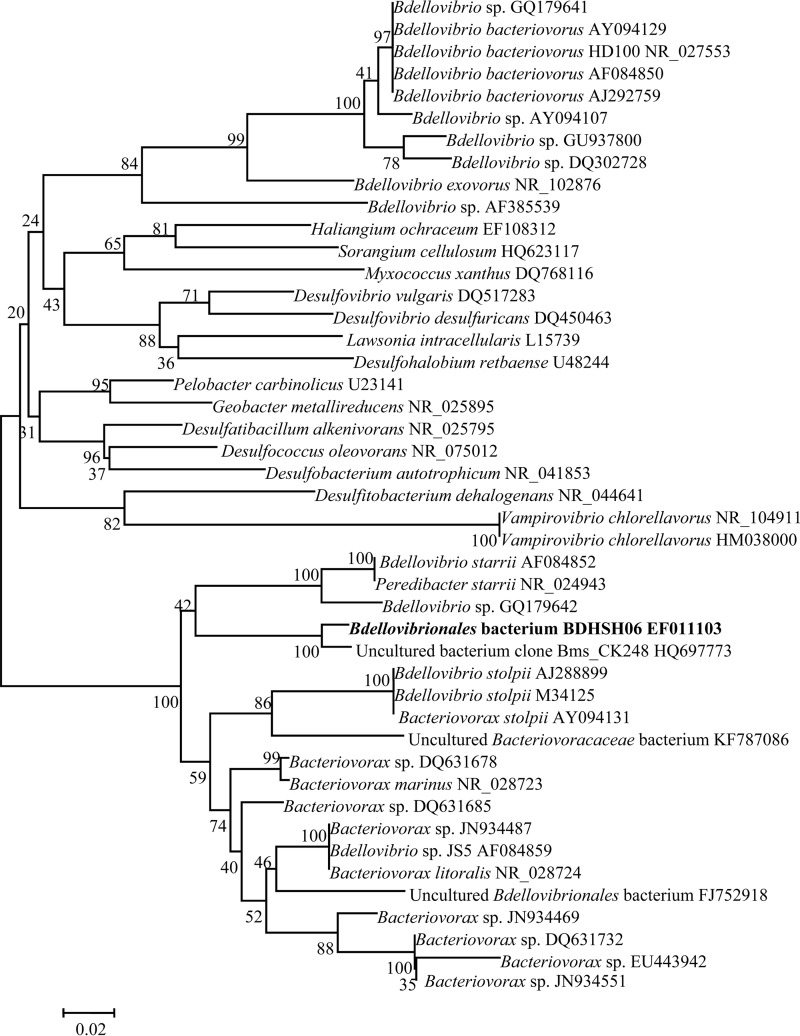
The prey strain used for growing BALOs in this study was Vibrio parahaemolyticus strain SH06, which was isolated from the guts of abalone from an abalone farm in Shanwei, Guangdong Province, China. The organism was identified by 16S rRNA gene sequencing in our laboratory (College of Light Industry and Food Sciences) and stored at 4°C (20). V. parahaemolyticus strain SH06 contains no thermostable direct hemolysin (TDH) and no TDH-related hemolysin (TRH) gene (20). The predatory bacterium used here was BALO strain BDHSH06 (GenBank accession no. EF011103), which was isolated from Daya Bay in Shenzhen, China, with V. parahaemolyticus strain SH06 as the prey using the double-layer agar technique (21). Partial 16S rRNA gene sequencing analysis revealed that BDHSH06 displays 97% sequence similarity with its closest relative, the uncultured bacterial clone Bms_CK248 (GenBank accession number HQ697773). Phylogenetic tree analysis showed that BDHSH06 and uncultured bacterial clone Bms_CK248 form an independent branch with no close relationships with members of any of the known genera Bdellovibrio, Bacteriovorax, Vampirovibrio, and Peredibacter (Fig. 1). BDHSH06 could lyse 48 strains of 57 prey tested, including Vibrio alginolyticus, V. parahaemolyticus, V. cholerae, V. fluvialis, V. minicus, Shewanella putrefaciens, and Pseudomonas aeruginosa (21). Biological characterizations revealed that BDHSH06 had an optimal growth temperature of 30°C (21).
FIG 1.
Phylogenetic interrelationships between strain BDHSH06 from this study and other published BALOs. The tree was constructed using the neighbor-joining method and is based on partial 16S rRNA gene sequence data available from the GenBank and EBML databases. Bootstrap confidence was calculated from 1,000 replicates. GenBank accession numbers are given after the strain names.
V. parahaemolyticus strain SH06 was cultured in 50 ml sterile nutrient broth (NB; salinity, 30‰) with shaking at 200 rpm at 28°C to late exponential phase (ca. 18 h). Then, it was harvested by centrifugation at 5,000 × g for 20 min at 4°C, washed, and resuspended with 2 ml dilute nutrient broth (DNB; 1/10 NB, pH 7.4; salinity, 30‰). The resulting pellet was then resuspended and adjusted to 1 × 1011 CFU · ml−1 with DNB after enumeration by a series of 10-fold dilutions and plating on marine 2216E agar medium (22). It was then stored at 4°C.
To prepare BALOs, a BDHSH06 plaque was picked with a sterile inoculation loop from a freshly grown double-layer agar plate and inoculated into an Erlenmeyer flask that contained 50 ml DNB medium and 0.5 ml freshly prepared prey strain. Incubation was performed at 30°C with shaking at 200 rpm for 12 h. BDHSH06 reached 1 × 104 PFU · ml−1, as determined by a standard double-layer agar plating technique with a series of 10-fold dilutions and V. parahaemolyticus strain SH06 as the prey. Then, it was used as a seed culture and stored at 4°C. To expand the BDHSH06 culture, a 25-ml seed culture was added to 1,000 ml DNB with a suspension of 10 ml prepared prey (1 × 1011 CFU · ml−1) to coculture with shaking at 200 rpm at 30°C for 72 h. At such time, the concentration of BDHSH06 reached 1 × 108 PFU · ml−1, as determined by the standard double-layer agar plating technique. The culture was twice filtered through a 0.45-μm-pore-size membrane to remove the prey bacteria, and the filtrate was stored at 4°C before use.
Ninety-six-hour laboratory reduction test.
To investigate the reduction effect of BDHSH06 in a marine environment, a laboratory test was conducted for 96 h at 28°C with the establishment of marine microcosms in six 20-liter plastic aquaria (23). In this test, 20 marine bacteria which were susceptible to BDHSH06 (21) (V. alginolyticus 2, V. alginolyticus 13, V. parahaemolyticus 8, V. parahaemolyticus 9, V. parahaemolyticus Sh06, V. cholerae 6, V. fluvialis Bh02, V. fluvialis Bh03, V. fluvialis Bh05, V. minicus Bh10, V. minicus Bh12, V. minicus Bh15, Grimontia hollisae Be08, S. putrefaciens 12, S. putrefaciens 27, S. putrefaciens 28, P. aeruginosa 17, P. aeruginosa 35, P. aeruginosa 29, and Aeromonas salmonicida 33) were used to establish marine microcosms. The strains were identified by both analysis with an API 20E system and 16S rRNA gene sequencing (24). They were individually inoculated into an Erlenmeyer flask with 50 ml alkaline peptone water (APW) and incubated at 30°C with shaking at 160 rpm for 16 h. Individual cultures were then centrifuged at 5,000 × g for 20 min at 4°C. The resulting pellets were resuspended and adjusted to a concentration of 5 × 109 CFU · ml−1 with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) after the bacterial concentration was determined on marine 2216E agar medium by the spread plate technique. They were then stored at 4°C before use.
Twenty liters of sterile artificial seawater with a salinity of 20‰ (pH 7.5) was dispensed into six plastic aquaria which had been disinfected with 6 mM potassium permanganate. Marine microcosms were established with the addition of 10 ml of each of the prepared marine bacteria (final concentration of each bacterium, ca. 2.5 × 106 CFU · ml−1, determined by the spread plate technique). Three aquaria were treated as the test aquaria with the addition of 20 ml of BALO strain BDHSH06 (final concentration, about 1 × 105 PFU · ml−1, determined by a standard double-layer agar plating technique; total bacterial concentration in each aquarium, ca. 5.00 × 107 CFU · ml−1, determined by the spread plate technique). The other three aquaria were treated as controls with no BDHSH06 addition (total bacterial concentration, ca. 4.99 ×107 CFU · ml−1, determined by the spread plate technique). Then, the aquaria were incubated for 96 h at room temperature (28°C) with nonstop but mild aeration (the dissolved oxygen concentration was maintained above 6.0 mg · liter−1), and the air was sterilized by fitting a 0.22-μm-pore-size filter (Millipore Corp., USA) to the aeration tube. Five-hundred-milliliter water samples from the control and test tanks were filtered through a membrane filter (pore size, 0.22 μm) to collect microorganisms at 12 h, 24 h, 48 h, 72 h, and 96 h. After filtering, microorganisms retained on the membrane were washed into a sterile 10-ml centrifuge tube with 8 ml filtered seawater, and then 1 ml 1× TE buffer (20 mM Tris-HCl, pH 7.8, 50 mM EDTA, 20 mM NaCl) was added. These samples were temporarily stored at −20°C and then analyzed by PCR amplification of the 16S rRNA gene and denaturing gradient gel electrophoresis (DGGE).
P. monodon rearing experiment.
The shrimp rearing experiment was conducted for 85 days in an aquafarm in Yangjing City, Guangdong Province, China. Six 1,000-liter plastic tanks (1.1 m in diameter by 1.1 m in height) were prepared and disinfected with 6 mM potassium permanganate. Then, sea mud was loaded into each tank to a thickness of about 5 cm to simulate the shrimp aquatic environment. Seawater with a salinity of 17 g · liter−1 was filtered through 113-μm-pore-size gauze to remove debris and larger organisms and was used to fill each tank (volume, 1 m3). The dissolved oxygen concentration in each tank was maintained at or above 6.0 mg · liter−1 by aeration. The post-larval stage P. monodon shrimp used in this experiment were 1.1 to 1.3 cm in body length and purchased from a local company. One thousand obviously healthy post-larval stage shrimp were randomly chosen and stocked into each of the six tanks.
The shrimp were fed three times a day with a formulated shrimp diet (Yue-hai Feed Company, Guangdong Province, China) composed of 40.50% crude protein, 8.61% crude lipid, 10.73% ash, and 9.44% moisture (energy content, 17.02 kJ · g−1 dry matter). Every 7 days, to simulate shrimp pond water exchange, water 30 cm in height (approximately 33% of the total tank volume) was removed from all tanks and the same amount of freshly filtered (through a 0.22-μm-pore-size membrane filter) seawater with the same salinity (17 g · liter−1) was added.
Three tanks were treated as test tanks with the addition of BDHSH06 to each tank to a final concentration of 1 × 105 PFU · ml−1 by dilution of the appropriate amount of the BDHSH06 stock. The rest were used as controls with no BDHSH06 addition. When rearing water was partially exchanged, 1 liter BDHSH06 was added to each of the test tanks at a concentration of 1 × 108 PFU · ml−1.
Sample collection.
Water and shrimp intestinal samples from both controls and test tanks were collected at 7-day intervals both before the partial exchange of rearing water (BPERW) and at 4 h after BDHSH06 addition (4HA) (mainly to monitor the initial presence of BDHSH06). The rearing water (500 ml) was filtered through a membrane filter (pore size, 0.22 μm) to collect microorganisms. After filtering, microorganisms retained on the membrane were washed into a sterile 10-ml centrifuge tube with 8 ml filtered seawater and processed on site for plate counting for determination of the total number of bacteria (TNB), the total number of vibrios (TNV), and the total number of BALOs.
Two shrimp from each tank were collected when water sampling was performed (six shrimp in total for the control tanks and for the test tanks). They were first weighed, and then their body surface was washed and disinfected using 75% ethanol. Then, the whole shrimp (when they were very small) or shrimp intestinal tracts were removed with sterile forceps and scissors and placed in a sterile 1.5-ml centrifuge tube. Two shrimp or the intestinal tracts of two shrimp from the same tank were pooled in one centrifuge tube and homogenized. Homogenates were then diluted with 1 ml sterilized saline solution (0.9% [wt/vol] NaCl) and processed on site for plate counting for determination of TNB, TNV, and the total number of BALOs, as described below.
All samples, including water and shrimp/intestine samples, were collected in duplicate. One was for bacteriological analysis as specified above. The second was for PCR-DGGE analysis. These samples were suspended in 1 ml sterile 1× TE buffer and stored at −20°C before use.
Conventional plate counting.
Bacterial enumeration was performed in triplicate. TNB were counted on marine 2216E agar medium using the spread plate technique. After incubation at 28°C for 48 h, colonies were counted. TNV were determined on thiosulfate citrate bile salts (TCBS) agar plates using the method published in a compendium of microbiological methods (25), even though TCBS agar may support the growth of some other genera (26) and thus may not provide accurate counts of total vibrios. Counting of the BALOs was performed by a standard double-layer agar plating technique with V. parahaemolyticus strain SH06 as the prey.
DNA extraction.
Relevant samples from the laboratory reduction test and P. monodon rearing experiment were centrifuged at 12,000 × g for 2 min. After centrifugation, 567 μl of 1× TE buffer (pH 8.0) was added to resuspend the pellet. Then, 25 μl of lysozyme (25 mg · ml−1; TaKaRa, China) was added and the mixture was incubated for 30 min at 37°C. Thirty microliters of 10% sodium dodecyl sulfate (SDS) and 3 μl of proteinase K solution (50 μg · ml−1; TaKaRa, China) were added, and the mixture was incubated for 1 h at 37°C. One hundred microliters of 5 mol · liter−1 NaCl and 80 μl of cetyltrimethylammonium bromide (CTAB)-NaCl solution (5 g CTAB in 100 ml 0.5 M NaCl) were added, and the mixture was further incubated for 10 min at 65°C. The lysates were then purified by repeated extraction with 800 μl of a mixture of phenol-chloroform-isoamyl alcohol (25:24:1; Guangzhou Chemical Reagent Factory, China) and then centrifuged at 12,000 × g for 10 min to allow DNA extraction. The genomic DNA was precipitated with isopropanol and washed with 1 ml of 70% ethanol, centrifuged at 12,000 × g for 10 min, and air dried at room temperature. The DNA obtained was finally suspended in 25 μl of 1× TE buffer. The DNA concentration was assayed at 260 nm and 280 nm using a model MPS-2000 Shimadzu spectrophotometer and preserved at −20°C before 16S rRNA gene amplification.
16S rRNA gene amplification by PCR.
The extracted genomic DNA was used as the template DNA in the PCR amplification. PCRs based on the V3 variable region were performed. Primer 518R (5′-ATTACCGCGGCTGCTGG-3′) and primer 357F (27) incorporating a 40-GC clamp (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCCCTACGGGAGGCAGCAG-3′) were used to amplify the 16S rRNA genes sequence. PCR amplification was done in a 50-μl reaction solution, which contained 21 μl of double-distilled H2O, 25 μl of premix (containing 1.25 U Taq DNA polymerase, 0.8 mM a mixture of deoxynucleoside triphosphates, 40 mM Tris-HCl [pH 8.3], 8 mM MgCl2, 200 mM KCl; TaKaRa, China), 1 μl of primer 357F incorporating a 40-GC clamp (10 μM), 1 μl of primer 518R (10 μM), and 2 μl (approximately 100 ng) of DNA. PCR was conducted in a Mastercycler PCR apparatus (Eppendorf, Germany) with the following parameters: initial denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 45 s, and extension at 72°C for 1 min; and a final extension at 72°C for 10 min. The amplified products were separated by 1% (mass/volume) agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV light.
DGGE.
The DGGE profile was used to reveal the possible changes in bacterial communities. The 230-bp PCR fragments were separated using DGGE, performed with a Dcode universal mutation detection system (Bio-Rad Laboratories, USA) according to the manufacturer's instruction. The 10-μl PCR products (approximately 450 ng) were applied to 8% (m/v) polyacrylamide gels in 1× TAE buffer (40 mM Tris, 20 mM acetate, 1.0 mM Na2EDTA, pH 8.0; Guangzhou Chemical Reagent Factory, China) with a gradient of between 30% and 60%. Gradients were created by polyacrylamide containing 0 to 100% denaturant (7 M urea, 40% [vol/vol] formamide). Electrophoresis was performed at 200 V for 5 h at a constant temperature of 60°C. The DGGE gel was stained using silver nitrate, as described by Sambrook et al. (28), and photographed. The densities and migration patterns of the bands were calculated by applying an image analysis system to the DGGE band profiles. Then, principle component analysis, based on the densities and migration of the bands, was performed using Quantity One software (version 4.6.2; Bio-Rad Laboratories, USA). The microbial community diversity was determined by use of the Shannon index (H′), which is calculated by the formula −Σpi ln pi, where pi is the proportion of each species in lane i at each sampling time (29). The richness (R) values were calculated using the formula Ri = Li/LT, where Ri is the richness value of the bacterial community in lane i, Li is the number of bands in lane i, and LT is the total number of bands in the DGGE profile.
Individual bands were excised, reamplified, and inspected according to the protocol described by Li et al. (30). PCR products for sequencing were purified using a QIAquick gel extraction kit (Bao Bioscience and Technology Company, China). Sequencing reactions were analyzed by the Huada Biotechnology Company, Shenzhen, China. Sequences were compared with known sequences using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Measurement of shrimp growth and survival rate.
Every 7 days during the 85-day experiment, 10 shrimp from each tank were caught just prior to the partial exchange of the rearing water to measure their body weight and length. At the end of the experiment, all shrimp from both the test and control tanks were counted and measured for body weight and length. Length and weight gain, average daily gain (ADG), and survival rates were calculated using the following formulae: length gain (percent) = 100 × [(Lt − L0)/L0], weight gain (percent) = 100 × [(Wt − W0)/W0], ADG (mg · day−1) = 1,000 × [(Wt − W0)/85], and survival rate (percent) = 100 × (Nt/N0), where Nt is the sum of the number of shrimp at the termination of experiment and the number of shrimp sampled during the experiment, N0 is the number of shrimp at the start of the experiment, Lt is the final length (mm) of the shrimp, L0 is the initial length (mm) of the shrimp, Wt is the final weight (g) of the shrimp, and W0 is the initial weight (g) of the shrimp.
Statistical analysis.
In this study, mean values are given, and error bars indicated 1 standard deviation of the mean. Data were analyzed by analysis of variance (ANOVA), and least significant differences (LSDs) were calculated at the 5% significance level to compare treatment means by use of the SAS system (version 8.2; SAS Institute Inc., NC, USA).
RESULTS
Reduction in bacterial abundance by BDHSH06 in the laboratory test.
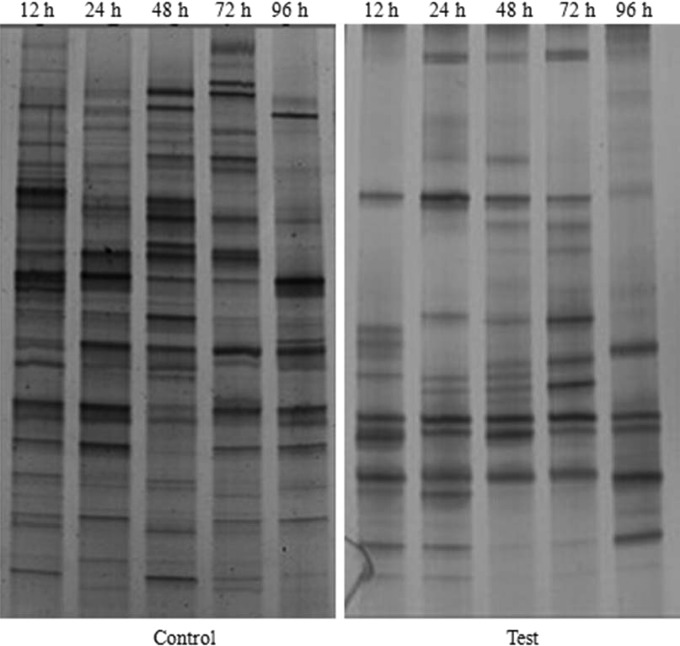
To determine the reduction in bacterial abundance by BDHSH06 in the laboratory test, DGGE analysis of the products amplified by 16S rRNA gene-based PCR was performed. As shown in Fig. 2, the numbers of discernible bands in the controls at 12 h, 24 h, 48 h, 72 h, and 96 h were 20, 17, 17, 17, and 16, respectively. Compared with the controls, test aquaria showed a trend toward fewer bands after 12 h. The numbers of discernible bands in the test aquaria were 12 and 14 at 12 h and 24 h, respectively. Afterwards, the numbers of discernible bands were reduced to 13, 11, and 8 at 48 h, 72 h, and 96 h, respectively.
FIG 2.
DGGE profiles of water samples from the 96-h laboratory reduction test. Water samples collected at 12 h, 24 h, 48 h, 72 h, and 96 h were run in the lanes indicated with those times. C, control.
Reduction of TNB and TNV in the 85-day shrimp rearing experiment.
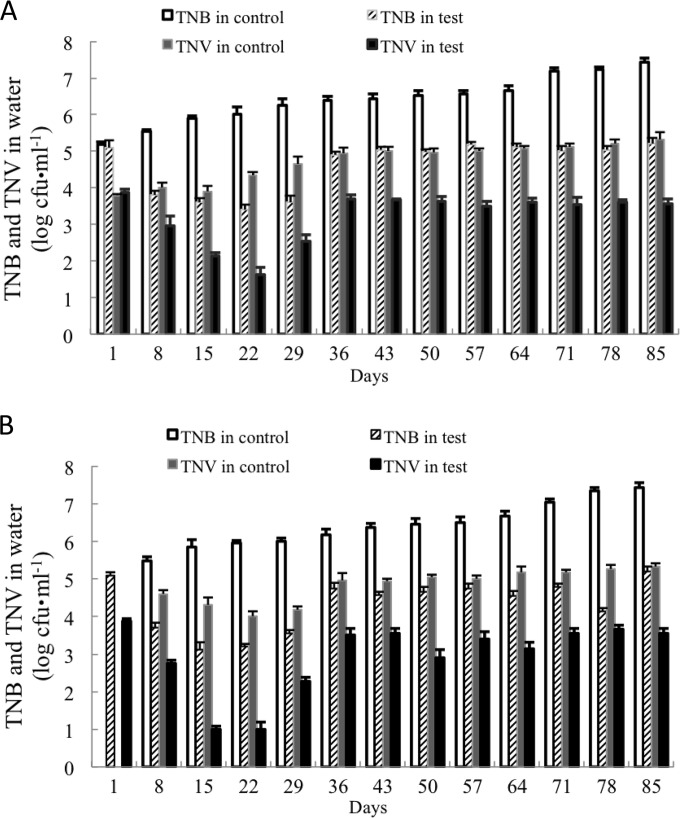
TNB and TNV in both water and shrimp intestines steadily increased for the controls over the 85-day rearing period, despite the periodic partial exchange of water, while their numbers in the test tanks remained at relatively constant levels after 36 days.
In shrimp rearing water (Fig. 3), TNB and TNV in control samples obtained before the partial exchange of rearing water (BPERW) increased from 5.18 ± 0.08 log CFU · ml−1 and 3.80 ± 0.11 log CFU · ml−1, respectively, on day 1 to 7.43 ± 0.12 log CFU · ml−1 and 5.32 ± 0.07 log CFU · ml−1, respectively, on day 85 (Fig. 3A). TNB and TNV in control water samples obtained 4 h after BDHSH06 addition (4HA) (Fig. 3B) increased from 5.47 ± 0.11 log CFU · ml−1 and 4.60 ± 0.11 log CFU · ml−1, respectively, on day 8 to 7.43 ± 0.12 log CFU · ml−1 and 5.32 ± 0.07 log CFU · ml−1, respectively, on day 85 (water was not changed on the last day).
FIG 3.
TNB and TNV in rearing water samples collected BPERW (A) and at 4HA (B) during the 85-day shrimp rearing experiment. The average of three replicates (500 ml rearing water per replicate) and standard deviation (represented by error bars; range, 0.04 to 0.21 log CFU · ml−1) are presented.
However, the test tanks showed a trend toward a significant reduction in terms of TNB (P < 0.05) and TNV (P < 0.05), regardless of sampling BPERW (Fig. 3A) or at 4HA (Fig. 3B).
TNB and TNV in water samples obtained BPERW were reduced from 5.10 ± 0.04 log CFU · ml−1 and 3.87 ± 0.09 log CFU · ml−1, respectively, on day 1 to 3.40 ± 0.21 log CFU · ml−1 and 1.63 ± 0.19 log CFU · ml−1, respectively, on day 22 and then gradually increased to 5.20 ± 0.09 log CFU · ml−1 and 3.55 ± 0.13 log CFU · ml−1, respectively, on day 85. In samples obtained at 4HA, TNB and TNV were reduced from 5.15 ± 0.16 log CFU · ml−1 and 3.85 ± 0.15 log CFU · ml−1, respectively, on day 1 to 3.11 ± 0.21 log CFU · ml−1 and 1.00 ± 0.20 log CFU · ml−1, respectively, on day 15, and then, again, they gradually increased to 5.20 ± 0.09 log CFU · ml−1 and 3.55 ± 0.13 log CFU · ml−1, respectively, on day 85.
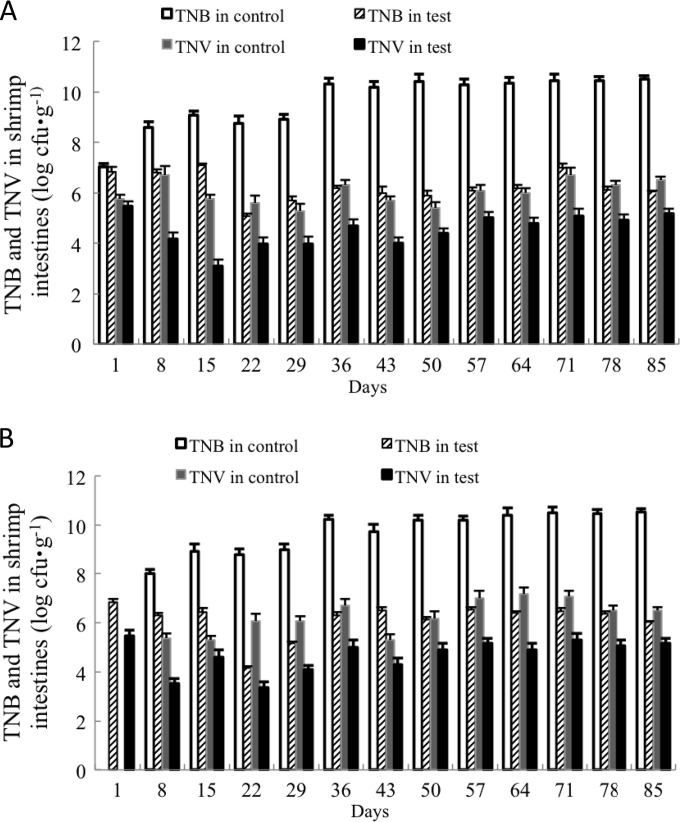
In shrimp intestinal samples from the control tanks, TNB and TNV in samples obtained BPERW (Fig. 4A) increased from 7.01 ± 0.23 log CFU · g−1 and 5.75 ± 0.19 log CFU · g−1, respectively, on day 1 to 10.52 ± 0.25 log CFU · g−1 and 6.51 ± 0.04 log CFU · g−1, respectively, on day 85. TNB and TNV in shrimp intestinal samples obtained at 4HA (Fig. 4B) increased from 8.00 ± 0.18 log CFU · g−1 and 5.38 ± 0.10 log CFU · g−1, respectively, on day 8 to 10.52 ± 0.25 log CFU · g−1 and 6.51 ± 0.04 log CFU · g−1, respectively, on day 85 (the harvest day, with no change of water).
FIG 4.
TNB and TNV in shrimp intestines for samples collected BPERW (A) and at 4HA (B) during the 85-day shrimp rearing experiment. The average of three replicates (2 shrimps per replicate) and standard deviation (represented by error bars; range, 0.07 to 0.36 log CFU · g−1) are presented.
In shrimp intestinal samples from the test tanks, TNB and TNV both showed a trend toward a reduction. TNB in shrimp intestinal samples obtained BPERW (Fig. 4A) dropped from 6.82 ± 0.14 log CFU · g−1 on day 1 to 5.08 ± 0.29 log CFU · g−1 on day 22 and then gradually increased to 6.04 ± 0.13 log CFU · g−1 on day 85. TNV in shrimp intestinal samples obtained BPERW (Fig. 4A) dropped from 5.46 ± 0.21 log CFU · g−1 on day 1 to 3.10 ± 0.24 log CFU · g−1 on day 15 and then again increased to 5.18 ± 0.19 log CFU · g−1 on day 85.
TNB and TNV in shrimp samples obtained at 4HA (Fig. 4B) dropped from 6.83 ± 0.19 log CFU · g−1 and 5.18 ± 0.24 log CFU · g−1, respectively, on day 1 to 4.51 ± 0.30 log CFU · g−1 and 3.34 ± 0.23 log CFU · g−1, respectively, on day 22 and then increased to 6.04 ± 0.13 log CFU · g−1 and 5.18 ± 0.19 log CFU · g−1, respectively, on day 85.
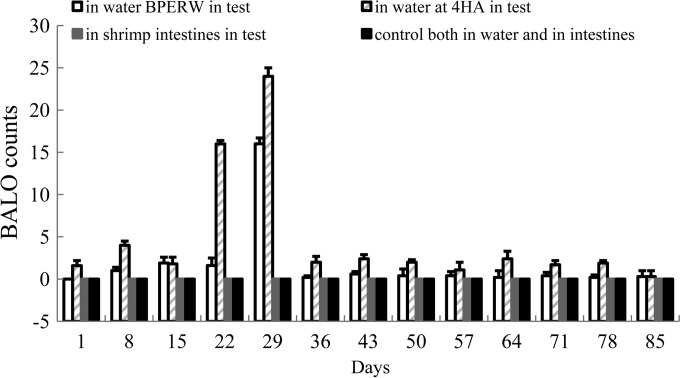
In control tanks, BALOs were not detected in any samples, either rearing water or shrimp intestines (Fig. 5).
FIG 5.
BALO counts in rearing water (PFU · ml−1) and intestines (PFU · g−1) collected BPERW and at 4HA during the 85-day rearing experiment. The average of three replicates (2 shrimp or 500 ml rearing water per replicate) and standard deviation (represented by error bars; range, 0 to 1 CFU · ml−1) are presented.
In test tanks, the BALO count in rearing water obtained BPERW (Fig. 5) went from 0 PFU · ml−1 on day 1 to 16 PFU · ml−1 on day 29 and then decreased to 0.3 ± 0.07 PFU · ml−1 on day 85. The BALO count in samples obtained at 4HA went from 0 PFU · ml−1 on day 1 to 24 PFU · ml−1 on day 29 and then down decreased to 0.3 ± 0.07 PFU · ml−1 on day 85 (the harvest day, with no change of water). Throughout the experiment, BALOs were not detected in shrimp intestines both BPERW and at 4HA.
PCR-DGGE analysis.
To understand the effect of BDHSH06 on microbial community changes over time in a shrimp aquaculture setting, a culture-independent method was employed (Fig. 6). To reduce the workload, in this experiment rearing water samples obtained BPERW were taken at intervals of every 2 weeks for DGGE analysis (Fig. 6). To avoid possible intragroup variations, the samples from replicate tanks in the same group (control or test) obtained on the same day were first mixed and then run for determination of their DGGE profiles.
FIG 6.
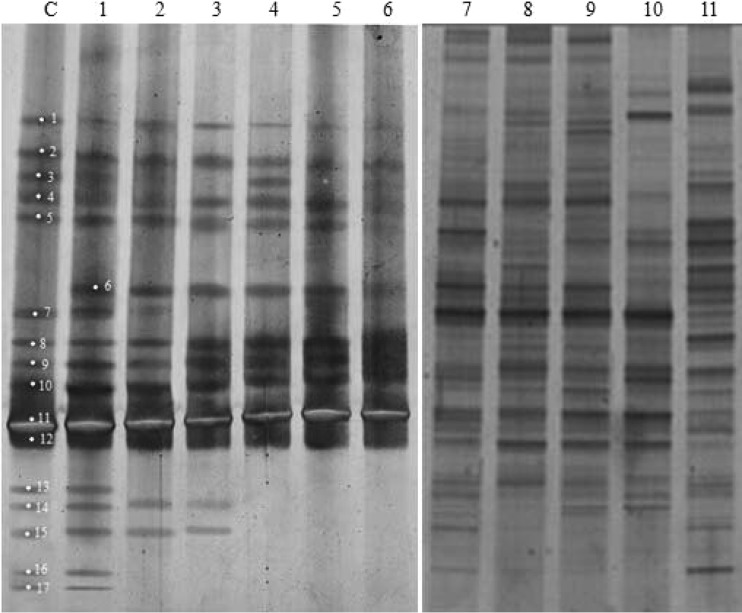
DGGE profiles of shrimp rearing water samples from the control and test tanks. Lanes C and 1, water samples collected from the control and test tanks on day 1, respectively; lanes 2 to 6, water samples collected from the test tanks on day 15, day 29, day 43, day 57, and day 85, respectively (samples were collected before the water was changed on those days); lanes 7 to 11, water samples collected from the control tanks on day 15, day 29, day 43, day 57, and day 85, respectively (samples were collected before the water was changed on those days). The excised bands are marked on the gels.
On day 1, there were 16 dominant bands in the controls (Fig. 6, lane C), which had a richness value of 0.59 (Table 1). From day 15 on, a richness value range of 0.81 to 0.96 was displayed (Table 1). According to the calculation of the relative abundance of the bands using Band Scan software, the numbers of discernible bands in the controls were 26, 24, 24, 22, and 25 on day 15, day 29, day 43, day 57, and day 85, respectively (Fig. 6, lanes 7 to 11).
TABLE 1.
H′ and R values of bacterial communities in test tanks and controls
| Day | Total no. of bandsa |
H′b |
Ric |
|||
|---|---|---|---|---|---|---|
| Control tanks | Test tanks | Control tanks | Test tanks | Control tanks | Test tanks | |
| 1 | 16 | 17 | 2.76 | 2.82 | 0.59 | 0.63 |
| 15 | 26 | 13 | 3.25 | 2.55 | 0.96 | 0.48 |
| 29 | 24 | 12 | 3.17 | 2.46 | 0.89 | 0.44 |
| 43 | 24 | 11 | 3.17 | 2.30 | 0.81 | 0.41 |
| 57 | 22 | 10 | 3.08 | 2.38 | 0.81 | 0.41 |
| 85 | 25 | 9 | 3.13 | 2.19 | 0.93 | 0.33 |
Total number of bands in a sample.
H′, Shannon diversity index, which is equal to −∑pi ln pi, where pi is the proportion of each species in lane i at each sampling time.
Ri = Li/LT, where Ri is the richness value of the bacterial community in lane i, Li is the number of bands in lane i, and LT is total number of bands in the DGGE profile.
Compared with the bacterial communities in the control tanks, the bacterial communities in the test tanks shared nearly the same banding patterns except for the presence of band 6 on day 1 (Fig. 6, lane 1). However, the richness values in the test tanks showed a trend toward decreasing numbers, starting with 0.63 on day 1 and a lower range of 0.48 to 0.33 at the end (Table 1). In other words, the bacterial communities in the test tanks showed a gradual reduction in the number of bands (Fig. 6, lanes 2 to 6), from 17 discernible bands on day 1 to just 9 bands on day 85.
The Shannon index was slightly higher in the test tanks than in the control tanks at the onset of the rearing experiment but became lower from day 15 (Table 1). The difference in the Shannon index values was the largest at the end of the experiment, being 2.19 in the test tanks and 3.13 in the control tanks.
In order to determine the taxonomic position of bacteria that were predominant in and/or that disappeared from water samples from the test tanks, a total of 17 bands were excised from the DGGE gels and sequenced (Fig. 6). The strain identification/similarity obtained by a BLASTN search is shown in Table 2. Among these identified bacteria, approximately 65% (11 out of 17) were nonculturable. Sequence analysis showed that the predominant bacteria in the control tanks on day 1 were Photobacterium spp., an uncultured bacterium, an uncultured Marinobacterium, an Idiomarinaceae bacterium, an uncultured Flavobacterium sp., an uncultured Vibrio sp., and a marine sediment bacterium. Compared with the controls, predominant bacteria from the test tanks were almost the same except for the presence of a Bdellovibrionales bacterium (band 6, GenBank accession no. EF011103) on day 1. Sequence analysis showed that the Bdellovibrionales bacterium (band 6) was present in all samples from the test tanks tested by DGGE. Among the bacterial species/strains that disappeared from or went undetected in test tanks, uncultured Vibrio sp. (band 13, GenBank accession no. AB550491), an uncultured Flavobacterium sp. (band 16, GenBank accession no. FJ946537), and a marine sediment bacterium (band 17, GenBank accession no. JF750272) were gone within 15 days of the rearing test (Fig. 6). They were then followed by a Photobacterium sp. (band 3, GenBank accession no. AJ866939), uncultured bacteria (bands 7 and 14, GenBank accession no. JQ240889 and HQ891374, respectively), and an Idiomarinaceae bacterium (band 15, GenBank accession no. JX415314), which disappeared or were present at levels beneath the detectable level after 29 days (Fig. 6). Except for a marine sediment bacterium (band 17, GenBank accession no. JF750272), the rest of these bacteria were present in the controls at different times. Among these bacteria/strains that disappeared, 57.1% were nonculturable.
TABLE 2.
Species identity and sequence similarity of bands excised from DGGE gels
| DGGE band no.a | Strain/species identified | Sequence similarity (%) | GenBank accession no. |
|---|---|---|---|
| 1 | Photobacterium lipolyticum | 99 | NR025813 |
| 2 | Uncultured bacterium | 95 | FR695373 |
| 3 | Photobacterium sp. | 99 | AJ866939 |
| 4 | Uncultured bacterium | 99 | JF692373 |
| 5 | Uncultured bacterium | 99 | FN562884 |
| 6 | Bdellovibrionales bacterium | 100 | EF011103 |
| 7 | Uncultured bacterium | 100 | JQ240889 |
| 8 | Marine sediment bacterium | 100 | JF750100 |
| 9 | Uncultured bacterium | 100 | JQ245604 |
| 10 | Uncultured gamma proteobacterium | 100 | JQ753127 |
| 11 | Uncultured Marinobacterium | 100 | JQ033869 |
| 12 | Uncultured bacterium | 100 | HQ916585 |
| 13 | Uncultured Vibrio sp. | 98 | AB550491 |
| 14 | Uncultured bacterium | 99 | HQ891374 |
| 15 | Idiomarinaceae bacterium | 97 | JX415314 |
| 16 | Uncultured Flavobacterium sp. | 99 | FJ946537 |
| 17 | Marine sediment bacterium | 100 | JF750272 |
Growth and survival rate of shrimp.
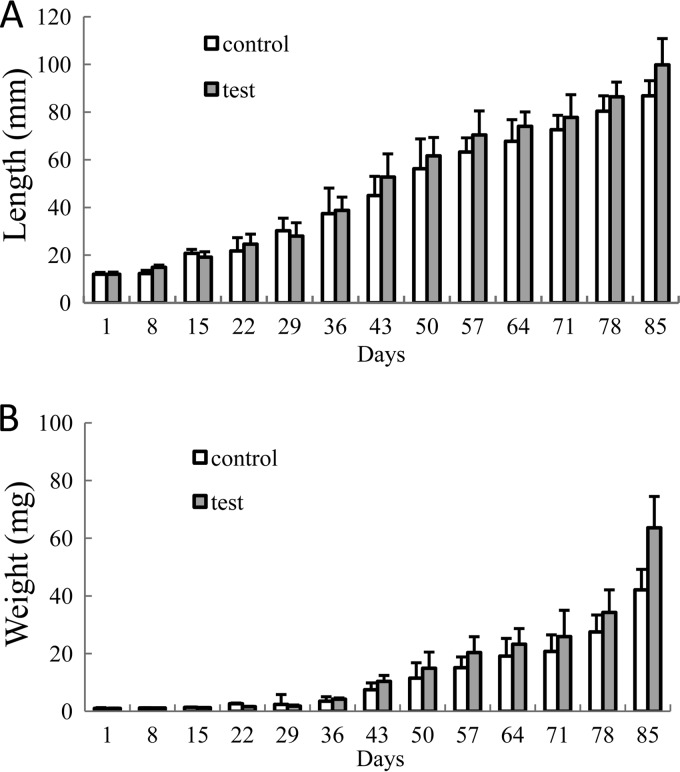
In the P. monodon rearing experiment, the initial body weight and length of the shrimp showed no significant differences (P > 0.05) between the test and the control tanks (Table 3). From day 50, their lengths and weights began to exhibit significant (P < 0.05) differences (Fig. 7).
TABLE 3.
Growth and survival rate of Penaeus monodon in test tanks and controlsa
| Group | Survival rate (%) | Length (mm) |
Length gain (%) | Wt (g) |
Weight gain (%) | ADG (mg · day−1) | ||
|---|---|---|---|---|---|---|---|---|
| Initial | Final | Initial | Final | |||||
| Control | 31.0 ± 1.2A | 12 ± 0.1A | 86.0 ± 11.1A | 616 ± 92A | 0.1 ± 0.02A | 4.21 ± 1.56A | 4,124 ± 155A | 48.4 ± 18.4A |
| Test | 48.1 ± 2.1B | 12 ± 0.1A | 99.8 ± 10.0B | 732 ± 83B | 0.1 ± 0.02A | 6.36 ± 1.50B | 6,299 ± 147B | 73.6 ± 17.6B |
Data are expressed as means ± standard deviations. Means with different superscript letters were significantly different (P < 0.05). ADG, average daily weight gain. The data were collected from the surviving shrimp in each of the six tanks at the end of the experiment, which was an average of 351 in the test tanks and an average of 180 in the control tanks. One hundred thirty shrimp in each tank were sampled during the test. The number of surviving shrimp was the number of shrimp counted at the end of the test plus the number of shrimp sampled during the test. The survival rate was calculated from the number of surviving shrimp divided by the total number of shrimp at the start of the experiment. One thousand shrimp were added to each of the six tanks at the start of the experiment.
FIG 7.
Shrimp body length (A) and weight (B) in the 85-day rearing experiment. At each sampling point, 10 shrimp were randomly assessed for weight and length. At the end of the experiment, all live shrimp (test tanks, 351 shrimp; control tanks, 180 shrimp) were measured for body weight and length. The average of three replicates (10 shrimp per replicate at each sampling point, except for the end of the experiment) and standard deviation (represented by error bars) are presented. Standard deviations of shrimp length and weight were in the range of 0.76 to 12.51 mm and 0.10 to 15.60 mg, respectively.
At the end of the 85-day experiment, the body length (99.8 ± 10.0 mm) and weight (6.36 ± 1.50 g) of the shrimp in the test tanks were significantly longer and higher than those of the controls (body length, 86.0 ± 11.1 mm; weight, 4.21 ± 1.56 g) (Table 3). The survival rate of the shrimp in the test tanks was 48.1% ± 2.1%, which was significantly higher than the rate of 31.0% ± 1.2% for the controls (Table 3). The percentages of weight and length gain and average daily growth (ADG) in test tanks were 732% ± 83%, 6,299% ± 147%, and 73.6 ± 17.6 mg · day−1, respectively, values significantly higher (P < 0.05) than those for the controls, viz., 616% ± 92%, 4,124% ± 155%, and 48.4 ± 18.4 mg · day−1, respectively (Table 3).
DISCUSSION
As a group of Gram-negative predatory bacteria that hunt other pathogenic and/or potentially pathogenic bacteria for growth, BALOs had once been and are now again being considered possible bacterial control agents. The BALOs' ubiquity in nature (31, 32), abundance in the human gut (33), inability to prey on eukaryotic cells (12–15), and ability to attack biofilms (10), as well as their very nonspecific predatory spectrum (9, 11), make them very promising candidates as biocontrol agents.
In the microcosm study carried out in the laboratory, the addition of BDHSH06 reduced about 50% of bacterial species/strains within 96 h compared with the numbers present in the controls (Fig. 2). Considering that all species/strains used were susceptible to BDHSH06 (21), it is evident that BDHSH06 also has clear prey preferences, a characteristic that is shared by other BALOs (32, 34).
In the 85-day P. monodon rearing study, TNB and TNV in the water and the intestines of shrimp in the test tanks were significantly reduced compared with the values for the controls (Fig. 3 and 4). It is clear that BDHSH06 was effective in controlling the total number of bacteria and vibrios in terms of both quantity and quality. With the significant reduction in the total number of bacteria and vibrios and their maintenance at low levels, the growth and survival rate of the P. monodon shrimp in the test tanks were significantly enhanced (Table 3). These enhancements were also supported by the fact that up to 40.6% more feed was used in the test tanks than in the control tanks over the 85-day P. monodon rearing duration; that is, if the amount of feed used by the controls was considered 100%, the shrimp in the test tanks consumed 140.6% of the amount consumed by the controls.
Surprisingly, BALO densities were not high when it is considered that an initial BALO concentration of 1 × 105 PFU · ml−1 was added to the rearing water, even though it could be detected by both the double-layer agar plating and DGGE techniques (Fig. 6). The lower than expected concentration of BALOs in the rearing water was most obvious in the water samples collected at 4HA (4 h after BALO addition). For this, we can only suggest that the majority of them were bound to the dirt and sank to the tank bottom, as the shrimp stirred up a large quantity of the sea mud on the tank bottom and made the tank water very dirty, especially when the shrimp were larger. Further, dirty particles in the water column made filtration extremely difficult. On each sampling date, each 500-ml rearing water sample took over 1 h to filter. This time-consuming process at high temperature (above 29°C) might have damaged the temperature-sensitive attack-phase/swimming-stage BALOs and reduced their viabilities and, hence, resulted in very low counts on the double-layer agar plates. We also noticed that as prey-dependent bacteria, the number of BALOs was rapidly reduced after the reduction of the prey bacteria. Then, the TNB in the water gradually recovered until the next round of addition of BALOs (Fig. 3). This means that to maintain predatory pressure on aquatic bacteria, BALOs need to be added on a regular basis. It could also imply that without further addition of BALOs, a mature bacterial community and BALOs may well reach a balance in a tit-for-tat war in nature.
The ability of BALOs to hunt other bacteria and their adaptability to many environments support their roles in bacterial reduction in the environment (10). Until now, few examples of this group of bacterial predators have been known, and knowledge of their effect on mortality in the microbial community within a particular environment has also been limited (22). To explore the role of BALOs in bacterial mortality and the shaping of community structure, the diversity of the bacterial population in an environment must be taken into account (10). We used DGGE profiling to evaluate the changes in bacterial community structures following BDHSH06 administration in the P. monodon aquaculture system. Our results showed that approximately 44% of the bacterial species/strains in the bacterial communities disappeared when BDHSH06 was added to the rearing water (Fig. 6). This was reflected by both the Shannon index and bacterial community richness values (Table 1). Our results contradict those of Purwandari and Chen (29), who found that the probiotic species Bacillus subtilis increased intestinal microbial diversity in the orange spotted grouper (Epinephelus coioides). This difference may be caused by the different mechanisms behind these two different types of bacteria: by inhibition and/or spatial/nutritional competition, as in Bacillus subtilis, and by lysis, as in BALOs.
Among the bacteria that disappeared from the test group, all of them except for a marine sediment bacterium (band 17, GenBank accession no. JF750272) were present in the controls during the experiment (Fig. 6). Considering that the only difference between the test and the control tanks was the addition of BDHSH06 to the test tanks and not to the control tanks, it is reasonable to suggest that these bacteria that disappeared most likely disappeared due to lysis by BDHSH06 rather than wild-type BALOs or phages. Even if wild-type BALOs were present in the system, they should have been present at levels below the level of detection by both the double-layer agar plating technique (Fig. 5) and the DGGE technique (Fig. 6). Phages should have exerted the same pressure on species in the test and the control tanks, as the conditions in both types of tanks were nearly the same.
Of all the species/strains that disappeared, 57.1% were nonculturable. Considering that only BDHSH06 and seawater filtered through a 0.22-μm-pore-size membrane were added weekly during the experiment, these unculturable bacteria were most likely from one or all of the following: the sea mud, post-larval stage shrimp, and/or the seawater added to the tanks.
It is known that BALOs attack Gram-negative bacteria from very distinct and different genera. However, nearly all previous studies were based on culture-dependent techniques. Little information is known about the predation of BALOs on unculturable bacteria in an aquatic/aquaculture ecosystem. Our preliminary investigation has revealed for the first time that nonculturable bacteria within an aquaculture environment are reduced/suppressed by BALOs. We also noted a nearly 50/50 disappearance rate for the culturable and nonculturable bacteria (42.9% versus 57.1%). While the mechanism behind this is not yet clear, the impact of BALOs on altering bacterial communities through predation in a mixed bacterial population is beginning to be uncovered (23).
In conclusion, the present work demonstrated the potential for the application of BALOs as biocontrol agents. Further studies will be directed toward evaluation of the role of BALOs in shaping microbial community structures and their functions in a variety of aquaculture environments.
ACKNOWLEDGMENTS
This study was supported by NSFC-40776091 from the Natural Sciences Fund of China, A200899H02 from the Ocean and Fisheries Bureau of Guangdong Province of China, 2009B090300270 from the Science and Technology Department of Guangdong Province of China, and SCUT-2008010 from the Graduate School, South China University of Technology.
We also thank Edward C. Mignot of Shandong University for linguistic advice.
Footnotes
Published ahead of print 8 August 2014
REFERENCES
- 1.Flegel T, Alday-Sanz V. 2007. The crisis in Asian shrimp aquaculture: current status and future needs. J. Appl. Ichthyol. 14:269–273. 10.1111/j.1439-0426.1998.tb00654.x [DOI] [Google Scholar]
- 2.Liu HD, Wang L, Liu M, Wang BJ, Jiang KY, Ma SS, Li QF. 2011. The intestinal microbial diversity in Chinese shrimp (Fenneropenaeus chinensis) as determined by PCR-DGGE and clone library analyses. Aquaculture 317:32–36. 10.1016/j.aquaculture.2011.04.008 [DOI] [Google Scholar]
- 3.Alapide-Tendencia EV, Dureza LA. 1997. Isolation of Vibrio spp. from Penaeus monodon (Fabricius) with red disease syndrome. Aquaculture 154:107–114. 10.1016/S0044-8486(97)00045-8 [DOI] [Google Scholar]
- 4.Alvarez JD, Austin B, Alvarez AM, Reyes H. 1998. Vibrio harveyi: a pathogen of penaeid shrimps and fish in Venezuela. J. Fish Dis. 21:313–316. 10.1046/j.1365-2761.1998.00101.x [DOI] [PubMed] [Google Scholar]
- 5.Goarant C, Merien F, Berthe F, Mermoud I, Perolat P. 1999. Arbitrarily primed PCR to type Vibrio spp. pathogenic for shrimp. Appl. Environ. Microbiol. 65:1145–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee KK, Yu SR, Chen FR, Yang TI, Liu PC. 1996. Virulence of Vibrio alginolyticus isolated from diseased tiger prawn, Penaeus monodon. Curr. Microbiol. 32:229–231. 10.1007/s002849900041 [DOI] [PubMed] [Google Scholar]
- 7.Lightner DV. 1996. A handbook of pathology and diagnostic procedures for diseases of penaeid shrimp, p 236 World Aquaculture Society, Baton Rouge, LA [Google Scholar]
- 8.Saulnier D, Haffner P, Goarant C, Levy P, Ansquer D. 2000. Experimental infection models for shrimp vibriosis studies: a review. Aquaculture 191:133–144. 10.1016/S0044-8486(00)00423-3 [DOI] [Google Scholar]
- 9.Martin M. 2002. Predatory prokaryotes: an emerging research opportunity. J. Microbiol. Biotechnol. 4:467–477 [PubMed] [Google Scholar]
- 10.Williams HN, Pineiro S. 2006. Ecology of the predatory Bdellovibrio and like organisms. Microbiol. Monogr. 4:213–248. 10.1007/7171_2006_058 [DOI] [Google Scholar]
- 11.Dwidar M, Monnappa AK, Mitchell RJ. 2012. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 10.5483/BMBRep.2012.45.2.71 [DOI] [PubMed] [Google Scholar]
- 12.Atterbury RJ, Hobley L, Till R, Lambert C, Capeness MJ, Lerner TR, Andrew KF, Barrow P, Sockett RE. 2011. Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl. Environ. Microbiol. 77:5794–5803. 10.1128/AEM.00426-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dashiff A, Kadouri DE. 2011. Predation of oral pathogens by Bdellovibrio bacteriovorus 109J. Mol. Oral Microbiol. 26:19–34. 10.1111/j.2041-1014.2010.00592.x [DOI] [PubMed] [Google Scholar]
- 14.Dashiff A, Junka RA, Libera M, Kadouri DE. 2011. Predation of human pathogen by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J. Appl. Microbiol. 110:431–444. 10.1111/j.1365-2672.2010.04900.x [DOI] [PubMed] [Google Scholar]
- 15.Van Essche M, Quirynen M, Sliepen I, Loozen G, Boon N, Van Eldere J, Teughels W. 2011. Killing of anaerobic pathogens by predatory bacteria. Mol. Oral Microbial. 26:52–61. 10.1111/j.2041-1014.2010.00595.x [DOI] [PubMed] [Google Scholar]
- 16.Sockett RE, Lambert C. 2004. Bdellovibrio as therapeutic agents: a predatory renaissance. Nat. Rev. Microbiol. 2:669–675. 10.1038/nrmicro959 [DOI] [PubMed] [Google Scholar]
- 17.Yair S, Yaakov D, Susan K, Jurkevitch E. 2003. Small eats big: ecology and diversity of Bdellovibrio and like organisms, and their dynamics in predator-prey interactions. Agronomie 23:433–439. 10.1051/agro:2003026 [DOI] [Google Scholar]
- 18.Cao HP, He S, Wang HC, Hou SL, Lu LQ, Yang XL. 2012. Bdellovibrios, potential bio-control bacteria against pathogenic Aeromonas hydrophila. Vet. Microbiol. 154:413–418. 10.1016/j.vetmic.2011.07.032 [DOI] [PubMed] [Google Scholar]
- 19.Lu F, Cai J. 2010. The protective effect of Bdellovibrio-and-like organisms (BALO) on tilapia fish fillets against Salmonella enterica ssp. enterica serovar Typhimurium. Lett. Appl. Microbiol. 51:625–631. 10.1111/j.1472-765X.2010.02943.x [DOI] [PubMed] [Google Scholar]
- 20.Cai C, Zhou Y, Cai J, Yang H. 2005. Bacteriological studies in a digestive tract of abalone (Haliotis diversicolor supertexta) and in the waters. Fish Sci. 24:1–5 (In Chinese.) [Google Scholar]
- 21.Li H, Liu C, Chen L, Zhang X, Cai J. 2011. Biological characterization of two marine Bdellovibrio-and-like organisms isolated from Daya Bay of Shenzhen, China and their application in the elimination of Vibrio parahaemolyticus in oyster. Int. J. Food Microbiol. 151:36–43. 10.1016/j.ijfoodmicro.2011.07.036 [DOI] [PubMed] [Google Scholar]
- 22.Zobell CE. 1941. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J. Mar. Res. 4:42–75 [Google Scholar]
- 23.Chen H, Young S, Berhane T, Williams HN. 2012. Predatory Bacteriovorax communities ordered by various prey species. PLoS One 7:e34174. 10.1371/journal.pone.0034174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai J, Han Y, Wang Z, Song Z. 2006. Lysis study of Bdellovibrios on seafood-borne potentially pathogenic vibrios. Food Sci. 27:75–78 (In Chinese.) [Google Scholar]
- 25.Downes MP, Ito K. 2001. Compendium of methods for the microbiological examination of foods, 4th ed, p 676 APHA, Washington, DC [Google Scholar]
- 26.Pfeffer C, Oliver JD. 2003. A comparison of thiosulphate-citrate-bile-salts-sucrose (TCBS) agar and thiosulphate-chloride-iodide (TCI) agar for the isolation of Vibrio species from estuarine environments. Lett. Appl. Microbiol. 36:150–151. 10.1046/j.1472-765X.2003.01280.x [DOI] [PubMed] [Google Scholar]
- 27.Muyzer G, Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Purwandari AR, Chen HY. 2013. Effects of probiotic Bacillus subtilis on intestinal microbial diversity and immunity of orange spotted grouper (Epinephelus coioides). J. Appl. Biotechnol. 1:25–36. 10.5296/jab.v1i1.3714 [DOI] [Google Scholar]
- 30.Li ZY, He LM, Wu J, Jiang Q. 2006. Bacterial community diversity associated with four marine sponges from the South China Sea based on 16S rDNA-DGGE fingerprinting. J. Exp. Mar. Biol. Ecol. 329:75–85. 10.1016/j.jembe.2005.08.014 [DOI] [Google Scholar]
- 31.Davidov Y, Friedjung A, Jurkevitch E. 2006. Structure analysis of a soil community of predatory bacteria using culture-dependent and culture-independent methods reveals a hitherto undetected diversity of Bdellovibrio-and-like organisms. Environ. Microbiol. 8:1667–1673. 10.1111/j.1462-2920.2006.01052.x [DOI] [PubMed] [Google Scholar]
- 32.Jurkevitch E, Minz D, Ramati B, Barel G. 2000. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl. Environ. Microbiol. 66:2365–2371. 10.1128/AEM.66.6.2365-2371.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iebba V, Santangelo F, Totino V, Nicoletti M, Gagliardi A. 2013. Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS One 8:e61608. 10.1371/journal.pone.0061608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogosky AM, Moak PL, Emmert EA. 2006. Differential predation by Bdellovibrio bacteriovorus 109J. Curr. Microbiol. 52:81–85. 10.1007/s00284-005-0038-6 [DOI] [PubMed] [Google Scholar]