Abstract
The diversity of bacterial species in the human oral cavity is well recognized, but a high proportion of them are presently uncultivable. Candidate division TM7 bacteria are almost always detected in metagenomic studies but have not yet been cultivated. In this paper, we identified candidate division TM7 bacterial phylotypes in mature plaque samples from around orthodontic bonds in subjects undergoing orthodontic treatment. Successive rounds of enrichment in laboratory media led to the isolation of a pure culture of one of these candidate division TM7 phylotypes. The bacteria formed filaments of 20 to 200 μm in length within agar plate colonies and in monospecies biofilms on salivary pellicle and exhibited some unusual morphological characteristics by transmission electron microscopy, including a trilaminated cell surface layer and dense cytoplasmic deposits. Proteomic analyses of cell wall protein extracts identified abundant polypeptides predicted from the TM7 partial genomic sequence. Pleiomorphic phenotypes were observed when the candidate division TM7 bacterium was grown in dual-species biofilms with representatives of six different oral bacterial genera. The TM7 bacterium formed long filaments in dual-species biofilm communities with Actinomyces oris or Fusobacterium nucleatum. However, the TM7 isolate grew as short rods or cocci in dual-species biofilms with Porphyromonas gingivalis, Prevotella intermedia, Parvimonas micra, or Streptococcus gordonii, forming notably robust biofilms with the latter two species. The ability to cultivate TM7 axenically should majorly advance understanding of the physiology, genetics, and virulence properties of this novel candidate division oral bacterium.
INTRODUCTION
The human oral cavity contains a diverse range of bacteria, with current estimates of up to 1,000 species present (1). The use of culture-independent methods for analyzing the oral microbiome has provided new insights into the complexity of oral microbial communities (2, 3). However, a significantly large proportion of microorganisms from the human oral cavity remain uncultivated (4). This has implications both for studies of human health and disease and for taxonomic classification, which requires axenic cultivation. When previously uncultivated bacteria have been cultivated (5, 6) or domesticated (4), it has been possible to then investigate their interactions with other oral microorganisms (7) or obtain accurate genomic sequences (8).
The bacteria of candidate division TM7 were first detected in a German peat bog (9) and are found in a diverse range of environments around the world (9, 10). Candidate division TM7 bacteria have also been detected at a number of human body sites, including the skin (10), distal esophagus (11), gut (12), and the oral cavity (3, 13–18). Initially, TM7 was found in relatively low abundance in oral cavity population samples (13), but in a more recent study, a high abundance of TM7 in subgingival plaque seemed to correlate with periodontal disease (3). However, it is difficult to assess the properties of these organisms in health and disease, because they have been uncultivable, with no pure-culture representatives. Phylogenetic analyses indicated that there were three subdivisions, based on 16S rRNA gene clones. The main morphotype in reactor sludge was Gram-variable nonbranching filaments up to 200 μm in length, each filament composed of closely packed cells and a cell wall with trilaminated appearance and surrounded by a sheath (9). In other studies, using fluorescent in situ hybridization (FISH) with a TM7-specific probe, long filamentous cells with striations were detected along with short rods and diplococci (10). The first genomic sequence of a TM7 organism (TM7a) was generated after single-cell isolation and whole-genome amplification, but the sequence was fragmented and incomplete (14). Metagenomic sequencing data have allowed a hybrid assembly approach to reconstruct a more complete version of a TM7 group genome (3). The final assembly is still fragmented, but 703 genes have been identified that were not present in the original assembly. Clearly, if a candidate division TM7 bacterium could be cultivated in pure culture, much more information could be obtained genetically and regarding virulence attributes.
In orthodontics, the use of fixed appliances on teeth interferes with cleaning methods and can lead to plaque accumulation and potentially increased risk for periodontal diseases (19, 20). Traditional culturing techniques have shown elevated numbers of periodontal pathogens, such as Porphyromonas gingivalis and Tannerella forsythia, present at the gingival margins during orthodontic treatment (21). In the present study, the aim was to monitor shifts of candidate division TM7 bacteria in plaque at the gingival margin associated with orthodontic treatment. From a supragingival plaque culture containing at least 12 phylotypes of TM7, we were able to domesticate a single isolate and grow this in pure culture. The organism corresponded to the previous morphological description of TM7 (9) by growing as long filaments, sometimes striated, within agar plate colonies or biofilms. However, the isolate underwent morphological changes when grown in biofilms in the presence of different oral bacterial species.
MATERIALS AND METHODS
Study design.
Twenty patients (11 to 14 years old) ready to begin orthodontic treatment at Bristol Dental Hospital participated in this study (approved by the UBHT Ethics Committee [04/Q2006/53]). Exclusion criteria included those patients with known systemic diseases and those who had taken antibiotics within 3 months before the start or during treatment. Cross-randomization to opposing contralateral quadrants of the mouth was applied (i) in the assignment of molar bands and bonded molar tubes and (ii) for self-ligating brackets (SLB) with or without elastomeric ligatures in the upper arch (19) (see Fig. S1 in the supplemental material). Samples were obtained immediately prior to (T0), 3 months into (T1), at completion of (T2), and 3 months after (T3) treatment (19). Plaque samples were placed into 50 mM Tris-EDTA buffer (pH 8.0) (TE buffer) and stored frozen at −70°C prior to processing.
Microbial culture.
Plaque samples were collected from patients in anaerobic transport medium (ATM Systems) and inoculated into fastidious anaerobic broth (FAB; LabM) containing kanamycin (100 μg/ml) to inhibit growth of Gram-negative bacteria. After 48 h of incubation at 37°C under anaerobic conditions, samples were subcultured into FAB for up to 1 week. Portions of cultures (0.1 ml) were then inoculated onto fastidious anaerobic agar (FAA) plates containing 5% defibrinated horse blood and incubated for 7 days anaerobically under N2-CO2-H2 (80:10:10). Single colonies were subsequently picked and subjected to multiple rounds of subculturing onto fresh FAA plates until pure culture isolates were obtained. All bacterial strains, including Streptococcus gordonii, Parvimonas micra, Actinomyces oris, Fusobacterium nucleatum subsp. nucleatum, Prevotella intermedia, and Porphyromonas gingivalis, were cultivated on FAA or in FAB.
DNA extraction.
DNA was extracted from plaque samples or cells harvested from broth cultures using a GeneElute bacterial genomic DNA kit (Sigma-Aldrich Company Ltd., Dorset, United Kingdom) according to the manufacturer's instructions, and DNA concentrations were estimated spectrophotometrically at 260 nm.
PCR for DGGE.
The candidate division TM7 16S rRNA gene was PCR amplified using oligonucleotide TM7-580F (9), containing GC clamps for subsequent denaturing gradient gel electrophoresis (DGGE) and a TM7 reverse primer (5′ GTGTAGCAGTGAAATGCGTAGATAT) within a 50-μl reaction mixture containing 1.25 U GoTaq DNA polymerase (Promega), 10 μl supplied buffer (final MgCl2 concentration of 1.5 mM), 0.2 mM each deoxynucleoside triphosphates (dNTP), 1.0 mM each primer, and approximately 20 ng template DNA. The amplification cycle was performed as follows: denaturation step at 94°C for 2 min; 15 cycles at 94°C for 30 s, primer annealing steps of 30 s starting at 65°C and descending 1°C each cycle, with extension at 72°C; followed by 10 cycles of 94°C, 50°C, and 72°C for 30 s each; and a final extension step of 72°C for 10 min. Amplimers of approximately 102 bp were visualized following agarose (1.2%) gel electrophoresis.
DGGE.
PCR amplimers were separated through 10% polyacrylamide gels containing a linear gradient of urea (40 to 50%) using a CBS Scientific DGGE electrophoresis system and applied voltage of 30 V for 1 h and then 60 V overnight (16 h). The system was maintained at a constant temperature (60°C) in 22 liters of 0.5× Tris-acetate-EDTA (TAE) buffer. The gels were stained for 30 min in 1× TAE-containing SYBR green nucleic acid gel stain (104 dilution) and digitally photographed with UV light transillumination.
Real-time PCR.
Assays were performed in 96-well plates in a DNA Engine ThermoCycler Chromo4 real-time detector (Bio-Rad) with the same primer sequences that were used for DGGE-PCR but without GC clamps and with universal primers 2 and 3 for total amplifiable 16S rRNA genes (22). The total reaction volume was 25 μl per well, and each reaction mix contained 12.5 μl iTaq SYBR green supermix (Bio-Rad), 1.25 μl each primer (final concentration of 6 μM), and 10 μl (∼50 ng) template DNA. The amplification cycle was performed as follows: denaturation at 95°C for 3 min; then 30 cycles of 95°C for 15 s, 54.2°C for 45 s, 72°C for 15 s; and then a final extension step of 72°C for 10 min. The fluorescence intensity of SYBR green was measured automatically during the annealing steps. At the end of each run, a melting curve analysis was performed from 70°C to 95°C, read every 0.2°C with a 0.1-s hold. Standards were dilutions of a plasmid containing a 572-bp segment of a 16S rRNA gene closely related (99%) to the TM7a 16S rRNA gene sequence.
Amplification of 16S rRNA.
The 16S rRNA genes were amplified by PCR under standardized conditions using TM7-specific primers (forward primer, TM7-314F; reverse primer, TM7-910R) which amplify a fragment of 572 bp, as previously described (9, 13). Additional TM7-specific primers utilized were 5′GATGAACGCTGGCGGCATG and 1391R (5′GACGGGCGGTGTGTRCA). A standard protocol within a PTC-100TM thermocycler (MJ Research Inc.) was used which included denaturation at 94°C for 3 min, then 34 cycles of 94°C for 1 min, 58.4°C for 45 s, 72°C for 1 min, and a final extension step of 72°C for 10 min. The PCR products were analyzed by 1% agarose gel electrophoresis containing ethidium bromide and visualized under UV light.
Cloning procedures and sequencing.
Cloning of PCR-amplified DNA was performed with a TA cloning kit (Invitrogen) according to the instructions of the manufacturer. DNA was transformed into competent Escherichia coli DH5α cells, and transformants were selected for kanamycin resistance (50 μg/ml). Correct sizes of inserts were determined by PCR, and the 16S rRNA gene fragments were purified (QIAquick PCR purification kit) and sequenced.
16S rRNA gene sequencing and data analysis.
Numerous clones with inserts of the correct sizes (102 bp or 572 bp) were analyzed. For identification of closest relatives, the sequences of the inserts were compared to available databases by use of the Basic Local Alignment Search Tool (BLAST) to determine approximate phylogenetic affiliations and to indicate the presence of any existing sequences that might be included in the novel divisions which had not previously been identified. Phylogenetic trees were constructed by neighbor joining using the CLUSTAL W website and the PHYLIP suite of programs (23).
Probe design.
The compiled sequences were aligned using Clustal W and Clustal Omega, and the alignments were refined manually. A TM7-specific fluorescence in situ hybridization (FISH) probe (TM7-2; Cy3-GAGTTTGTGAGTTCGAATAAT) was designed based on comparative analysis of all sequences in the database. Probe sequence was subsequently confirmed for specificity using the 16S rRNA gene sequences in the Ribosomal Database Project and the GenBank databases.
FISH.
To identify the presence of TM7 in mixed-culture samples, 0.2 ml culture was centrifuged (13,000 × g, 3 min), and the pellet was fixed in 0.1 ml 4% paraformaldehyde (PFA) for 20 min on ice (2, 24). After fixation, cells were washed with 50% ethanol and mixed with 0.1 ml 0.1 M Tris-HCl (pH 7.5) containing 5 mM EDTA and 10 mg lysozyme/ml for 10 min at 37°C to permeabilize the cells. The cells were then dehydrated with a series of ethanol washes (3 min each) containing 50%, 80%, and 100% ethanol. Samples were incubated with 50 μg/ml Cy3-labeled TM7-905 oligonucleotide probe (9) or Cy3-labeled universal probe EUB-338 in hybridization buffer (0.9 mM NaCl, 20 mM Tris-HCl [pH 7.5], 25% formamide, 0.01% sodium dodecyl sulfate) at 46°C for 90 min. Samples were washed for 15 min at 48°C in buffer containing 0.16 M NaCl, 20 mM Tris-HCl (pH 7.5), 5 mM EDTA, and 0.01% sodium dodecyl sulfate. Cells were then washed in 0.4 ml ice-cold water and examined by fluorescence microscopy. Experiments were also carried out with a newly designed probe TM7-2 (see above) with the following modifications to the protocol: the initial pellet was fixed in 50% ethanol (0.15 ml) overnight at 4°C; the cells were washed with 0.4 ml phosphate-buffered saline (PBS) and centrifuged, and pellets were suspended in 0.5% Triton (0.1 ml) for 10 min at 22°C to permeabilize cells.
Preparation of saliva-coated coverslips.
Human whole saliva samples were collected from a minimum of five adult volunteers (in accordance with the Human Tissue Act and with University of Bristol ethical approval) into chilled tubes on ice, pooled, incubated for 10 min with 2.5 mM dithiothreitol (DTT), and clarified by centrifugation (10,000 × g, 10 min). The supernatant was then passed through a 0.2-μm-pore-size filter and diluted to 10% with distilled water. Portions (0.5 ml) were transferred to wells of a 24-well plate containing 13-mm-diameter glass coverslips, and plates were incubated for 16 h at 4°C. Saliva-coated coverslips were then removed from each well and utilized for biofilm experiments as described below.
Biofilm formation.
S. gordonii DL1 (Challis) was grown anaerobically for 24 h in fastidious anaerobic broth (FAB), while TM7 isolate UB2523, A. oris T14V, F. nucleatum subsp. nucleatum ATCC 25586, P. micra ATCC 33270, P. gingivalis ATCC 33277, and P. intermedia ATCC 25611 were grown under anaerobic conditions for up to 7 days. Cells were harvested by centrifugation, washed in FAB, and adjusted to an optical density at 600 nm (OD600) of 0.5 in the same broth. Suspensions (0.5 ml) were transferred to wells of 24-well plates containing saliva-coated coverslips and incubated for 24 h (S. gordonii, A. oris, P. micra, or P. gingivalis) or 48 h (F. nucleatum) in an anaerobic cabinet, with the well plates in a humidifying chamber. Suspensions were aspirated from the wells, and the coverslips were air dried and biofilms were stained with fluorescein isothiocyanate (FITC; 1.5 mM in 0.05 M Na2CO3 containing 0.1 M NaCl) for 15 min at 22°C in the dark or with crystal violet for 5 min. Excess stain was removed by washing the coverslips with distilled water (dH2O), and the coverslips were then air dried, inverted onto glass slides, and visualized by microscopy (transmitted light, fluorescence, or confocal scanning laser).
Biomass measurement.
To estimate biomass, biofilms grown on glass coverslips were stained with crystal violet for 5 min. Coverslips were then washed with dH2O and transferred to a fresh 24-well plate. Crystal violet was solubilized with 10% acetic acid, and absorbance at 595 nm (A595) was determined as a measure of biomass (25).
TEM.
Colonies were collected from the surface of FAA into FAB medium and centrifuged (5,000 × g, 5 min), and the cells were suspended in FAB medium and transferred into 15-ml conical centrifuge tubes. The suspension was centrifuged and to the pellet was added transmission electron microscopy (TEM) fixative (4% paraformaldehyde, 5% glutaraldehyde, 0. 1 M sodium cacodylate buffer [pH 7.2], 0.05% Tween 20; 6 ml). Tubes were shaken gently and incubated at 22°C for 15 min. The samples were centrifuged (3,000 × g, 2 min), the supernatant was carefully removed, and the pellet was suspended in TEM fixative (2% paraformaldehyde, 2.5% glutaraldehyde; 2 ml). Samples were stored at 4°C overnight. The pellets were then washed 3 times in 0.1 M sodium cacodylate buffer (pH 7.2) and set into low-melting-point 2% agarose. The agarose pellets were cut into ∼3-mm sections and incubated at 22°C for 1 h in 1% osmium tetroxide solution. The samples were rinsed in sterile H2O, dehydrated at room temperature using sequential incubations in ethanol (30%, 50%, 75%, 90%, and 100%) and then propylene oxide, and embedded in Spurr resin. The resin was cut by microtome (RMC Powertome PC) into sections of 80 to 100 nm using a diamond knife, and sections were collected on copper grids and imaged at 80 kV by TEM.
Confocal scanning laser microscopy (CSLM).
Biofilms were stained with FITC as described above and were visualized wet by using a Leica TCS-SP2 confocal imaging system attached to a Leica DMIRBE inverted microscope. Oil immersion objectives (40× and 100×) were used, and parameters were set to optimize resolution. Analysis of the data was carried out using Volocity image analysis software (Improvision).
Proteome analysis.
TM7 isolate UB2523 cells were harvested from agar plates into FAB and collected by centrifugation at 5,000 × g for 10 min. The cell pellet was suspended in 0.5 ml TE buffer and transferred to a dismembrator tube, and an equal volume of 0.5-mm glass beads (BioSpec Products Inc.) was added. The cells were disrupted using a Precellys shaker for 30 s (three times), with cooling in ice-cold water for 1 min between each step. Lysates were subjected to low-speed centrifugation to sediment the beads, transferred to a fresh tube, and centrifuged again at 13,000 × g for 10 min. The pellet, containing mainly cell wall and membrane fragments and associated cytoplasmic proteins, was then suspended in 0.5 ml 0.1 M Tris-HCl (pH 6.8) buffer containing 0.1% SDS, heated at 80°C for 10 min, and centrifuged to clarify. Portions of supernatant were electrophoresed through 5% acrylamide such that the front dye had moved 2 cm, and the gel was fixed in 50% MeOH-7% acetic acid-water for 30 min. The top 1.5-cm portions of the separating gel lanes were then excised, washed, and subjected to in-gel digestion with trypsin. The peptides were fractionated using a Dionex Ultimate 3000 nano HPLC system in line with an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific). The raw data files were processed using Proteome Discoverer software v1.2 (Thermo Scientific). These data were then searched against the Swiss-Prot Eubacteria database using the Mascot algorithm and against the TM7 partial genome sequence in the Human Oral Microbiome Database (HOMD) (26) and successively analyzed using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/).
Statistical analyses.
Results are shown as mean values and standard errors of the means (SEM) calculated for each group of samples. For normally distributed data, comparisons were tested with Student's t test. The two-tailed Mann-Whitney U test was used for comparisons between groups. A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequence for TM7 isolate UB2523 has GenBank accession number KM067151. The GenBank accession numbers for additional 16S rRNA gene clones are as follows: N1, KM251184; N2, KM251185; N3, KM251186; N4, KM251187; N7, KM251188; N8, KM251189; N9, KM251190; and N12, KM251191.
RESULTS
Phylogenetic analyses of TM7 during orthodontic treatment.
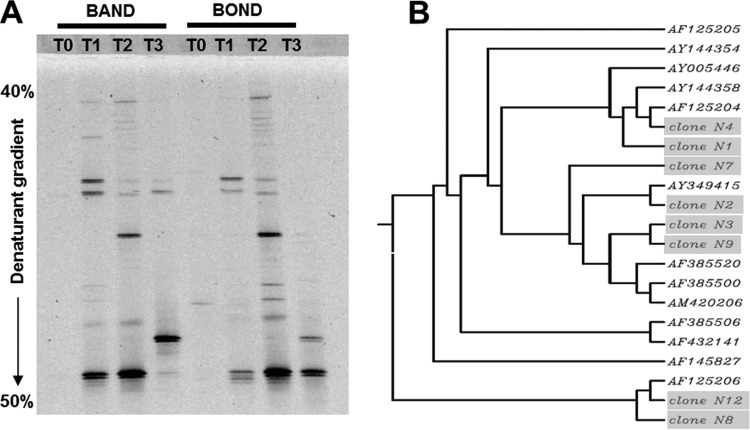
Plaque samples were collected from around appliances with orthodontic bands or bonds (see Fig. S1 in the supplemental material), and 16S rRNA gene fragments were amplified using GC clamp TM7 primers for DGGE to generate amplimers of approximately 102 bp. DGGE patterns are shown in Fig. 1A for PCR-amplified samples taken from a single patient from bands or bonds at intervals during orthodontic treatment (T0, T1, T2, and T3, as described in Materials and Methods). A single, faint TM7-associated gel band was amplified from plaque taken before commencing treatment (T0). As plaque then built up around the appliances, increased numbers of division TM7 gel bands appeared 3 months after the start of the treatment (T1) (Fig. 1A). At completion of the treatment (T2), there were at least 8 gel bands visible, but 3 months after the appliances were removed, fewer gel bands were evident (Fig. 1A). These profiles suggested that significant changes were occurring in the numbers of division TM7 bacterial phylotypes present during and after patient treatment. Each band from the gels was excised, cloned, and sequenced, and the sequences were compared with database sequences for identification. The dendrogram shown in Fig. 1B includes 8 distinct division TM7 sequences from this one patient, each described as a numbered clone, and 12 division TM7 16S rRNA sequences obtained by homology searches through the HOMD database (26). These phylogenetic analyses showed that this single individual carried at least 8 TM7 phylotypes orally.
FIG 1.
Candidate division TM7 bacterial sequences in dental plaque associated with orthodontic treatment. (A) DGGE profiles, obtained by PCR with TM7 primers, of DNA extracted from plaque collected at different time points from band and bond samples in a single subject. (B) Unrooted dendrogram of candidate division TM7 phylotypes identified following PCR from pooled plaque samples collected at different time points in panel A. The dendrogram shows clonal diversity of TM7 sequences within one patient. Phylogenetic analysis of clone sequences (N, shaded in gray) and database sequences (AF, AM, and AY numbers) indicated that a single individual carried at least 8 TM7 phylotypes orally. GenBank accession numbers for clones are as follows: N1, KM251184; N2, KM251185; N3, KM251186; N4, KM251187; N7, KM251188; N8, KM251189; N9, KM251190; and N12, KM251191.
Relative TM7 numbers over the orthodontic treatment schedule.
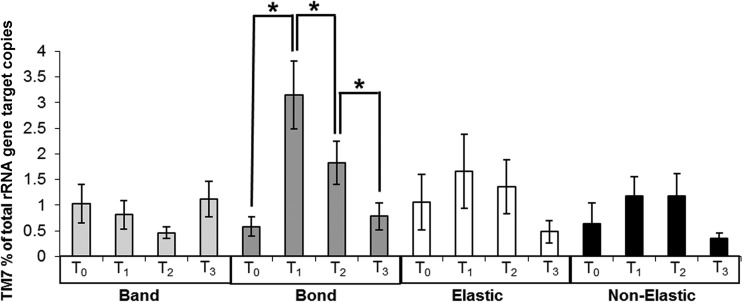
Real-time PCR analyses of plaque samples indicated a statistically significant increase in the proportion of division TM7 bacteria in gingival margin plaque samples adjacent to bonded tubes but not molar bands during treatment (Fig. 2). Three months after appliances had been removed, the proportion of division TM7 sequences relative to the total had fallen to pretreatment levels. Differences in TM7 proportions in plaque adjacent to elastic or nonelastic brackets (see Fig. S1 in the supplemental amterial) were not significantly different (Fig. 2).
FIG 2.
Quantification of candidate division TM7 bacteria in plaque samples expressed as percentage of total bacterial 16S rRNA gene copies present. Means for 20 subjects at four treatment time points (T0, T1, T2, T3) for each site of plaque collection (band, bond, elastic, and nonelastic) were calculated. Error bars are ±SEM. There were statistically significant (P < 0.05) changes in proportions of candidate division TM7 sequences associated with orthodontic bond plaque during the orthodontic treatment.
TM7 growth in mixed culture.
Successive subculturing of plaque bacteria from this single subject over several months in FAB under anaerobic conditions enabled growth of some division TM7 phylotypes in mixed cultures. Oral bacteria present were identified by 16S rRNA gene sequencing and included Actinomyces viscosus (naeslundii), Fusobacterium nucleatum, Prevotella denticola, Shuttleworthia satelles, Streptococcus gordonii, and Veillonella parvula (data not shown). DNA was extracted from samples of the mixed populations and PCR amplified with TM7-specific primers, and products were cloned into E. coli and sent for sequence analysis. Three additional division TM7 clones (designated JA1, JA2, and JA3) were identified, and these clustered with the other clones (Fig. 1B) from samples previously sequenced from plaque and with TM7 sequences available in the GenBank database, including the sequence AF125206 (clone I025) (see Fig. S2 in the supplemental material). Samples of broth were regularly inoculated onto FAA containing 5% defibrinated horse blood, and pure colony isolates were classified following 16S rRNA gene sequencing.
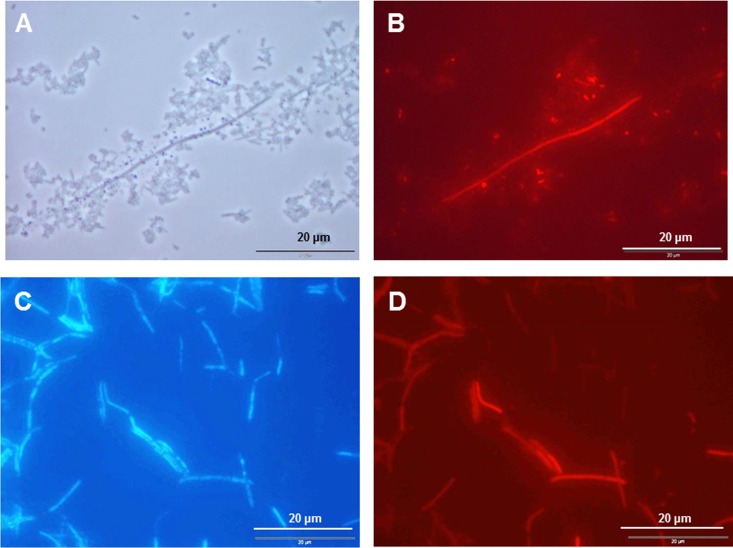
We used FISH to investigate the presence of TM7 bacteria in these mixed-broth cultures. Samples of cultures showed the presence of a range of morphotypes, often with long filamentous rods present with associated organisms (Fig. 3A). These were probed with a fluorescently labeled TM7-specific probe (see Materials and Methods), and this reacted with the long filaments (Fig. 3B) as well as with some smaller rods and cocci (Fig. 3B). Pure colony isolates of bacteria from these samples were tested for TM7 sequences by PCR. An isolated colony potentially containing TM7 bacteria was then sampled and subjected to FISH with a new probe designed for specific binding to TM7 16S rRNA genes, labeled with Cy3 fluorophore. DAPI (4′,6-diamidino-2-phenylindole) staining of cells from the colony showed the presence of filaments of various lengths (Fig. 3C), and the TM7 probe reacted with these filaments (Fig. 3D). The division TM7 bacterial isolate was designated UB2523 and formed creamy-white 0.5- to 3.0-mm-diameter colonies on FAA (see Fig. S3 in the supplemental material). There was no visible hemolysis on FAA containing blood. DNA was isolated from single colonies and subjected to PCR, with TM7 primer pairs designed to amplify fragments of 572 bp or 1,321 bp. Database homology searches with these sequences (GenBank accession number KM067151) identified a number of 99% homologous sequences from uncultured bacteria and from division TM7-designated clones.
FIG 3.
FISH images of candidate division TM7 bacterial cells. (A and B) Sample of culture in FAB medium of plaque from a single subject (Fig. 1) viewed by phase-contrast microscopy (A) or probed with Cy3 fluorophore-labeled TM7-specific sequence probe TM7–905 (9) (B). The light microscopic image shows long and segmented filaments surrounded by other bacterial forms, predominantly cocci. The probe highlights the long filaments and few smaller rods and cocci. (C and D) Putative candidate division TM7 bacterial cells from a single FAA colony isolate, designated UB2523, stained with DAPI (C) or probed with TM7 probe 2 (D). DAPI staining showed the presence of variable-length filaments that were simultaneously labeled with TM7-specific probe 2.
TM7 isolate proteome.
Tryptic peptides generated from proteins extracted from UB2523 cells (as described in Materials and Methods) were interrogated against sequences in GenBank with BLASTP. Peptides were also compared directly with amino acid sequences of predicted open reading frames (ORFs) from the partial TM7a genomic sequence (3, 14). Some of the peptides matched well-conserved sequences within high-abundance proteins, such as enolase, elongation factor Tu, and ribosomal proteins (see Table S1 in the supplemental material), and matched other predicted peptide sequences from the unfinished TM7 genome through HOMD (see Table S1). Each identified TM7 protein was given a score that was the sum of the scores of the individual peptides that were matched to the protein. Very few peptides matched to each protein, because of the low sequence coverage for TM7 in the databases. For similar reasons, only 10 to 20% of the obtained peptides matched with the TM7 sequence. However, these data demonstrated unequivocally that the cultivated organism corresponded to an isolate of candidate division TM7 bacterium.
Morphological characteristics.
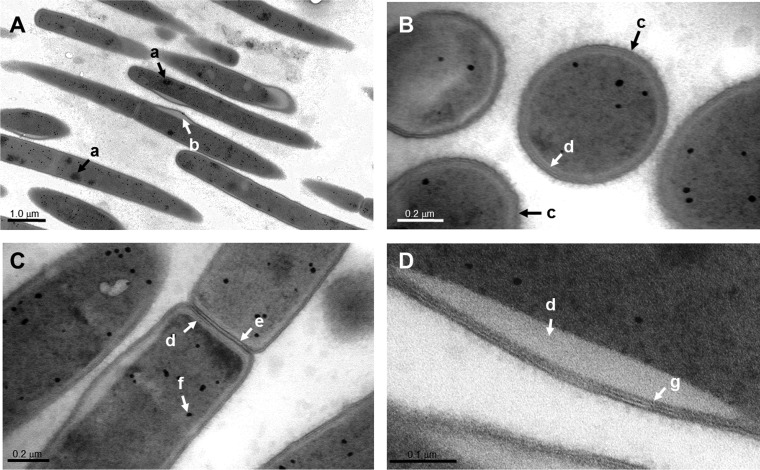
Samples from colonies of UB2523 were processed for transmission electron microscopy. Because of the lengths of the filaments, we were unable to obtain a transverse section of a complete filament. The sections shown in Fig. 4A represent partial filaments but were sufficient to demonstrate that the true poles of the cells are slightly rounded. Two features are striking: the first is that the cytoplasm of the cells contains darkly stained irregular-surfaced depositions that could be inorganic deposits or storage granules; the second is that most of the cells show a bulge at some point on their cell surface that appears to be a thickening of the underlying material between surface layers and cytoplasm. However, this could be the visual effect caused by a twist in the filament such that the plane of section is slightly changed. Vertical sections of the filaments (Fig. 4B) suggested a surface fuzzy layer, reminiscent of the Gram-positive streptococcal cell surface, and an underlying cell wall similar to that seen for many Gram-positive bacteria. In sections showing dividing cells, the septum contained at least three densely stained layers separating the cells (Fig. 4C), and the electron-dense deposits within the cytoplasm did seem to localize more toward the cell poles. Lastly, it was evident at higher magnification (Fig. 4D) that the cell surface layers formed a trilaminated structure, noted previously in electron micrographs of TM7 filaments in mixed environmental samples (9).
FIG 4.
TEM images of candidate division TM7 isolate UB2523. (A) The cells were rod shaped and of variable lengths, with the ends of the cells slightly rounded. The cytoplasm contained clusters of darkly stained material (a), and most of the cells showed one or more surface bumps (b) or swellings. These were contiguous with the surface layers and appeared to show thickening of the underlying material. (B) Vertical cross sections of the filaments revealed an outer fibrillar surface (c) somewhat like that of Gram-positive streptococci (32), a darkly stained trilaminated layer, and an underlying layer that was less well stained (d) and clearly delineated from the cytoplasm. (C) Dividing filaments showed a conventional septum (e) and densely stained cytoplasmic deposits (f), possibly accumulating to form the material marked “a” in panel A. (D) At higher magnification, it was possible to see clearly a trilaminar cell wall layer (g) overlying a surface swelling that seemed to represent a thickening of the less-electron-dense underlying material (d). Scale bars are as indicated.
Biofilm formation.
Since biofilm formation is crucial to bacterial survival in the oral cavity, we investigated the ability of the TM7 isolate to form biofilms on salivary pellicle. UB2523 TM7 formed a mat of intertwining filaments after 16 h of incubation that stained Gram positive (Fig. 5A). At 24 h, biofilms examined by CSLM consisted of elongated filaments ranging in length from 20 μm to over 200 μm (Fig. 5B), and some of the filaments were segmented and twisted (Fig. 5C). Overall thickness of the biofilms was 20 to 30 μm, and the CSLM images of the xz plane indicated that the bacteria formed a matted structure as opposed to a brush.
FIG 5.
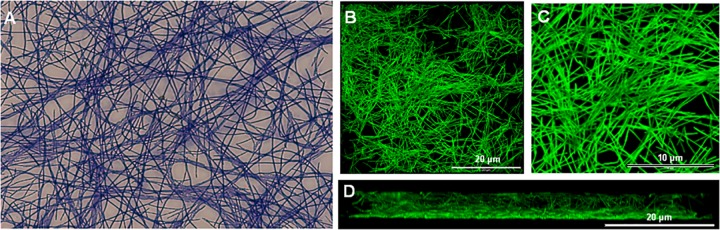
Candidate division TM7 bacterial adherence and biofilm formation. Isolate UB2523 cells in FAB medium were incubated with saliva-coated coverslips anaerobically for 24 h at 37°C. (A) Biofilm stained with crystal violet and visualized by light microscopy. (B to D) Biofilm stained with FITC and visualized by (CSLM) with a ×40 (B) or ×100 (C) objective lens, and corresponding vertical section (D). Scale bars are as indicated.
Dual-species biofilm formation.
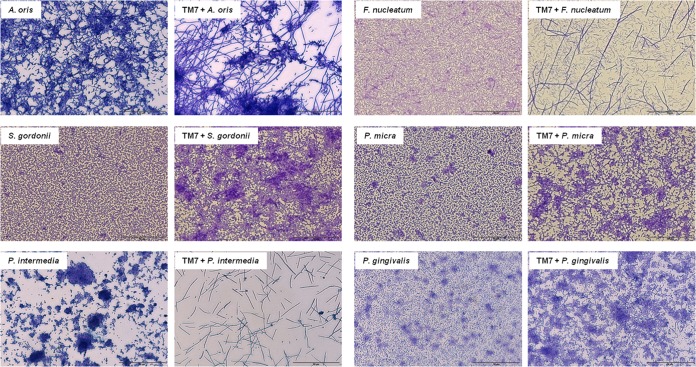
We then investigated the ability of the TM7 isolate to form biofilms with other species of oral bacteria, anticipating that the bacteria might have specific preferred partnerships since they have never before been isolated in pure culture. Consequently, strains from six different genera of oral bacteria were cultivated under biofilm-forming conditions with or without the TM7 isolate, and the effects on biofilm morphology and biomass were determined. The immediate revelation was that the TM7 isolate formed different morphologies according to the bacteria present in coculture and that each of the partnerships had unique consequences (Fig. 6). Crystal violet stain was utilized in these experiments for biofilm visualization and concomitant quantification of biomass. A. oris formed a biofilm with small clumps of cells, but with TM7 the actinomyces formed dense clumps associated with what appeared to be some form of extracellular material, and the TM7 formed long intertwined filaments. S. gordonii formed an evenly dispersed biofilm over the pellicle surface, but with TM7, larger, densely stained clumps were formed, and TM7, where visible, was present as shorter rods. P. intermedia formed a patchy biofilm with clumps of material at dispersed sites, but when TM7 was present, P. intermedia was more or less absent and TM7 formed a sparse biofilm of thin rods of up to 10 μm in length (Fig. 6). F. nucleatum formed a dual-species biofilm containing long TM7 filaments interspersed with F. nucleatum cells, while TM7 seemed to have an effect on P. micra biofilms similar to that on S. gordonii, causing matrix-like deposits to form around small TM7 filaments where they could be seen. Lastly, the presence of the TM7 isolate resulted in P. gingivalis-forming clusters in dual-species biofilm compared to a relatively evenly distributed P. gingivalis monospecies biofilm. We were unable to select for any of these different TM7 morphotypes in pure cultures over repeated laboratory transfers.
FIG 6.
Dual-species biofilm formation. Cells of six different bacterial species in FAB medium were incubated with saliva-coated coverslips anaerobically for 24 h at 37°C either as monospecies or in the presence of UB2523 (TM7). Biofilms were stained with crystal violet and visualized by light microscopy. Bacterial strains were A. oris T14V, F. nucleatum subsp. nucleatum ATCC 25586, S. gordonii DL1, P. micra ATCC 33270, P. intermedia ATCC 25611, P. gingivalis ATCC 33277. All scale bars are 70 μm.
Biomass measurements clearly demonstrated that the biofilms formed by TM7 together with F. nucleatum, P. micra, or S. gordonii were greater in biomass than the sum of the individual biomasses of the monospecies biofilms, indicating some form of synergy in growth or stimulation of matrix production (Fig. 7). In contrast, none of the other biofilms were significantly affected in biomass between monospecies and dual species. The TM7 isolate was therefore sensitive to growth modifications and to morphological changes when present in dual-species biofilms.
FIG 7.
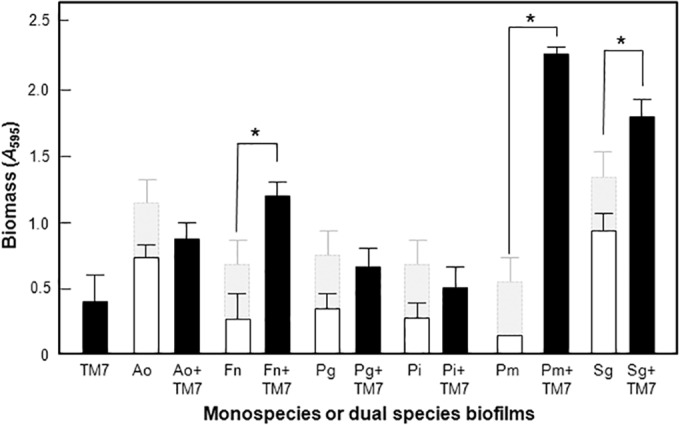
Biomass values for monospecies or dual-species biofilms as depicted in Fig. 6. Biomass values were measured by crystal violet staining. The light-gray shaded bars show the biomass of monospecies UB2523 (TM7). Error bars are ±SEM for triplicate samples from two individual experiments. Ao, A. oris; Fn, F. nucleatum subsp. nucleatum; Pg, P. gingivalis; Pi, P. intermedia; Pm, P. micra; Sg, S. gordonii. Biomass levels were significantly (P < 0.05) increased over and above the sum of the individual biomasses for dual-species biofilms of TM7 with F. nucleatum, S. gordonii, or P. micra. There were no statistically significant increases or decreases from the sum of the monospecies biomass compared to the biomasses of the dual-species biofilms for the other organisms.
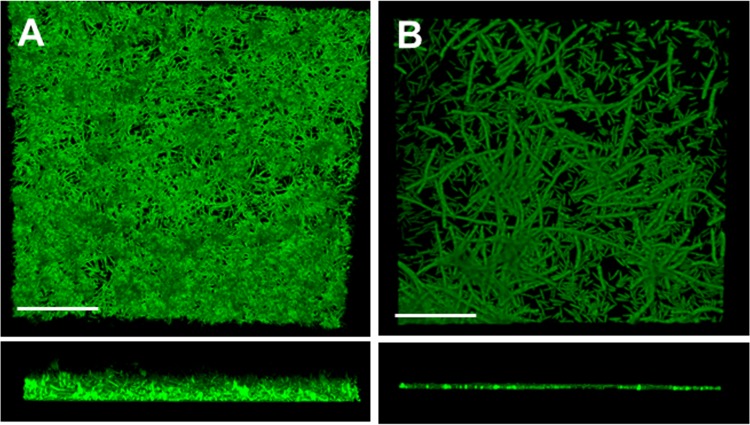
CSLM images of 24-h biofilms of A. oris, with or without TM7, showed that the biofilm growth was denser and thicker in dual species, and that a matrix-like material surrounded the actinomyces and underlying TM7 filaments (Fig. 8). F. nucleatum biofilms took longer to develop than A. oris biofilms, but in dual-species 48-h biofilms with UB2523, the TM7 filaments could be seen very clearly associated with, but not coaggregating with, the fusobacterial rods in the background. These strains of Actinomyces and Fusobacterium therefore seemed to be able to form stable dual-species biofilms with isolate UB2523, with TM7 filaments clearly visible within the biofilm images (Fig. 8).
FIG 8.
CSLM images of dual-species biofilms of UB2523 (TM7) with A. oris and F. nucleatum. (A) A. oris and UB2523 top view (xy) and side view (xz); (B) F. nucleatum and UB2523 (xy and xz stacks). In panel A, the biofilm contains matrix-like material, while in panel B, long filaments of UB2523 (TM7) may be easily differentiated from F. nucleatum cells. Scale bars = 30 μm.
DISCUSSION
The cultivable microbiota from the human oral cavity is estimated at ∼50% of species present. Concerted attempts are now being made to cultivate the uncultivable (27), recognizing experimentally that organisms may be uncultivable in the conventional way because they exist in obligate metabolic associations with other organisms. This has been suggested as a possible reason for the inability to cultivate TM7 axenically in pure culture. However, we have now successfully cultivated a division TM7 bacterial isolate based upon sequencing, proteomic, and morphological data. TM7 was first discovered using environmental sequencing data and consecutively described from soil, laboratory bioreactor, and batch reactor sludge (9, 28) and subsequently in human subgingival plaque (13). The relationships between the environmental bacteria and human oral cavity isolates are not known. The isolation of TM7 in this paper raises the possibility that this isolate might be able to complement metabolic deficiencies in other TM7 bacteria and therefore be tagged in some way and utilized in further attempts to isolate other TM7 organisms.
The clinical significance of finding TM7 associated with orthodontic bonds as opposed to bands is not clear at this time, although it is likely that the plaque that accumulates around these appliances might be more inaccessible (and remain undisturbed) in the bond situation (Fig. S1 in the supplemental material shows in vivo bonds and bands). It is interesting that we found the candidate division present in such relatively high proportions (about 3% of total sequences), which might indicate a preference for mature or undisturbed plaque. However, this is not a prerequisite for TM7 detection, since metagenomic and microbiomic studies have increasingly reported detection of TM7 at a variety of sites, healthy as well as diseased (15, 18, 29).
Some of our TM7 sequences from plaque samples were unique (e.g., clones N3 or N9) and are likely to represent new TM7 subtypes (Fig. 1B). Other sequences were very similar to those in the databases, e.g., clone N12 is likely to be derived from a TM7 bacterium very closely related to that from which the GenBank database sequence AF125206 originated. The TM7 clones obtained from broth culture clustered together with a TM7 clone previously found in the plaque used to inoculate the broth, suggesting to us that TM7 was able to grow in the mixed culture. Multiple phylotypes of other oral organisms are also being detected by molecular sequencing methods, so it is possibly unsurprising that multiple TM7 phylotypes were detected in a single human subject. Likewise, it has been reported, for example, that multiple phylotypes of Treponema can be found in a single periodontal pocket (30).
The transmission electron microscopy studies of this TM7 bacterium isolate showed a cell surface layer structure more like Gram-positive bacterial cell wall. However, unlike with environmental TM7 cell micrographs (9), we could not detect an outer sheath. This might be related to the possibility that, in the environment, more protection is required against adverse conditions, such as desiccation or temperature, than in the relatively more stable conditions of the human mouth. The trilaminated structure of the outer layers was similar to that reported for the environmental organisms (9), but it is not clear at this time what these layers represent. The thickness of the triple layer was rather less than the thickness of the outer fibrillar layer, estimated at about 50 nm. It is possible that the organism produces a surface protein array, and a detailed cell wall analysis of both carbohydrate and protein components should be able to determine the biochemical composition and molecular structure of a possible outer layer. The organism maintained long filaments under biofilm conditions, and cell units were visible within these filaments as described for the environmental organisms (9).
An interesting feature was the appearance of at least one bulge along the length of most filaments. These are unlikely to be artifacts as a result of processing, because they are too regular in appearance and not associated with any disruptions of the cell outer layers. We believe that these could represent surface twists in the filaments, not related directly to septum formation (cell division), as they are not localized specifically to the septum. As further evidence of the twist hypothesis, potential twists could be envisaged in the biofilm filaments (Fig. 5C), and some of these filaments clearly seem to contain lines of cell units.
The TM7 isolate described formed biofilms as mats. The closest associations of the bacteria were with A. oris and F. nucleatum, where biofilms of the two organisms growing together could be observed. However, A. oris did not form a synergistic relationship with TM7 UB2523 (Fig. 6). Specific physical interactions were not apparent under the conditions used, so it seemed that the organisms were coexisting and not coaggregating. We regularly detected Fusobacterium sequences during the enrichment procedures on plaque, and Fusobacterium spp. were one of the highest detectable genera present in microarray studies of orthodontic plaque (19). The TM7 did not seem to form a biofilm with P. intermedia; rather, it appeared to be inhibitory to this strain. In other dual-species biofilms, cells of TM7 could be seen present as more darkly staining entities. With P. micra, TM7 was present sparsely as short filaments, but the biofilm was hugely increased in biomass over P. micra alone, with dense clusters of bacteria surrounded by matrix. With S. gordonii, TM7 cells were present as very short rods that collected masses of S. gordonii cells with matrix production, possibly extracellular DNA (eDNA). Since sucrose was not present in the growth medium, it was unlikely that conditions would favor the production of extracellular glucans. With P. gingivalis, longer filaments of TM7 could be seen present within large clusters of P. gingivalis bacteria surrounded by matrix. Given the recurrence of the matrix observations, it would be important to determine the chemical nature of these matrix substances and which of the organisms in the dual-species biofilms are producing them. Clearly, TM7 has the ability to modulate biofilm growth of other oral bacteria, to form biofilms with a range of different oral bacterial species, and to be itself growth modulated by these species. Such interactions could promote (or inhibit) the development of a disease-associated oral microbial community.
Taken collectively, these results show that this TM7 organism from the human mouth has almost all of the morphological characteristics of the TM7 organisms that were identified within bioreactor sludge (9) but were never able to be cultivated. The presence of candidate division TM7 in dual-species interactions has multiple effects upon the biofilms produced by other bacteria. The TM7 bacteria can vary in morphology from very long filaments in monoculture (>200 μm) to shorter filaments (20 μm) or small rods or cocci (∼1 μm) in the presence of Gram-positive streptococci and P. micra. The morphological transformations must be closely related to the metabolic conditions of the mixed cultures or as a result of signaling from the partner bacteria in the biofilm. These morphological changes in the presence of different bacteria may go some way in explaining our FISH images (Fig. 3B) and the various morphological descriptions of putative TM7-like organisms in soil as cocci and filamentous cells (9) and as long and short filaments (9) in cultures of bacteria from dental samples. The ability to cultivate this isolate axenically now opens up the possibility for a more complete biochemical and genetic analysis of the organism, testing of virulence and pathogenic attributes (3, 31), and advancing understanding of this novel candidate division bacterium that has until now remained elusive.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jane Brittan, Katy Jepson, and Alan Leard for technical assistance and Graham Stafford and Nick Jakubovics for helpful discussions.
This research was funded in part by the British Orthodontic Foundation. Support of the University of Bristol Wolfson Bioimaging Facility by the Medical Research Council (United Kingdom) is gratefully acknowledged.
Footnotes
Published ahead of print 8 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01827-14.
REFERENCES
- 1.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017. 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diaz PI, Dupuy AK, Abusleme L, Reese B, Obergfell C, Choquette L, Dongari-Bagtzoglou A, Peterson DE, Terzi E, Strausbaugh LD. 2012. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol. Oral Microbiol. 27:182–201. 10.1111/j.2041-1014.2012.00642.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu B, Faller LL, Kiltgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang Y-C, Li S, Stine OC, Hasturk H, Kasif S, Segrè D, Pop M, Amar S. 2012. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One 7:e37919. 10.1371/journal.pone.0037919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sizova MV, Hohmann T, Hazen A, Paster BJ, Halem SR, Murcphy CM, Panikov NS, Epstein SS. 2012. New approaches for isolation of previously uncultivated oral bacteria. Appl. Environ. Microbiol. 78:194–203. 10.1128/AEM.06813-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Downes J, Munson M, Wade WG. 2003. Dialister invisus sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 53:1937–1940. 10.1099/ijs.0.02640-0 [DOI] [PubMed] [Google Scholar]
- 6.Vartoukian SR, Palmer RM, Wade WG. 2010. Cultivation of a Synergistetes strain representing a previously uncultivated lineage. Environ. Microbiol. 12:916–928. 10.1111/j.1462-2920.2009.02135.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Wright CJ, Dingming H, Uriarte SM, Lamont RJ. 2013. Oral community interactions of Filifactor alocis in vitro. PLoS One 10:e76271. 10.1371/journal.pone.0076271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Göker M, Held B, Lucas S, Nolan M, Yasawong M, Glavina Del Rio T, Tice H, Cheng JF, Bruce D, Detter JC, Tapia R, Han C, Goodwin L, Pitluck S, Liolios K, Ivanova N, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Rohde M, Sikorski J, Pukall R, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Lapidus A. 2010. Complete genome sequence of Olsenella uli type strain (VPI D76D-27C). Stand. Genomic Sci. 3:76–84. 10.4056/sigs.1082860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture relatives. Appl. Environ. Microbiol. 67:411–419. 10.1128/AEM.67.1.411-419.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinis JM, Barton DE, Ghadiri J, Surendar D, Reddy K, Velasquez F, Chaffee CL, Lee M-CW, Gavrilova H, Ozuna H, Smits SA, Ouverney CC. 2011. In search of an uncultured human-associated TM7 bacterium in the environment. PLoS One 6:e21280. 10.1371/journal.pone.0021280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. 2004. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. U. S. A. 101:4250–4255. 10.1073/pnas.0306398101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuehbacher T, Rehman A, Lepage P, Hellmig S, Fölsch UR, Schreiber S, Ott SJ. 2008. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J. Med. Microbiol. 57:1569–1576. 10.1099/jmm.0.47719-0 [DOI] [PubMed] [Google Scholar]
- 13.Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. 2003. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl. Environ. Microbiol. 69:1687–1694. 10.1128/AEM.69.3.1687-1694.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcy Y, Ouverney C, Bik EM, Lösekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, Quake SR. 2007. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. U. S. A. 104:11889–11894. 10.1073/pnas.0704662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. 2009. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J. Periodontol. 80:1421–1432. 10.1902/jop.2009.090185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rego RO, Oliverira CA, dos Santos-Pinto A, Jordan SF, Zambon JJ, Cirelli JA, Haraszthy VI. 2010. Clinical and microbiological studies of children and adolescents receiving orthodontic treatment. Am. J. Dent. 23:317–323 [PubMed] [Google Scholar]
- 17.Teles FR, Teles RP, Siegelin Y, Paster B, Haffajee AD, Socransky SS. 2011. RNA-oligonucleotide quantification technique (ROQT) for the enumeration of uncultivated bacterial species in subgingival biofilms. Mol. Oral Microbiol. 26:127–139. 10.1111/j.2041-1014.2010.00603.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, Krishnan K, Frias-Lopez J. 2014. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 8:1659–1672. 10.1038/ismej.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ireland AJ, Soro V, Sprague SV, Harradine NW, Day C, Al-Anezi S, Jenkinson HF, Sherriff M, Dymock D, Sandy JR. 2014. The effects of different orthodontic appliances upon microbial communities. Orthod. Craniofac. Res. 17:115–123. 10.1111/ocr.12037 [DOI] [PubMed] [Google Scholar]
- 20.Leung NM, Chen R, Rudney JD. 2006. Oral bacteria in plaque and invading buccal cells of young orthodontic patients. Am. J. Orthod. Dentofacial Orthop. 130:698.e11–698.e18. 10.1016/j.ajodo.2006.05.028 [DOI] [PubMed] [Google Scholar]
- 21.Naranjo AA, Triviño ML, Jaramillo A, Betancourth M, Botero JE. 2006. Changes in the subgingival microbiota and periodontal parameters before and 3 months after bracket placement. Am. J. Orthod. Dentofacial Orthop. 130:275.e17–275.e22. 10.1016/j.ajodo.2005.10.022 [DOI] [PubMed] [Google Scholar]
- 22.Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsenstein J. 1993. PHYLIP (Phylogeny Inference Package) version 3.6a2. Department of Genetics, University of Washington, Seattle, WA [Google Scholar]
- 24.Foster JS, Kolenbrander PE. 2004. Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl. Environ. Microbiol. 70:4340–4348. 10.1128/AEM.70.7.4340-4348.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakubovics NS, Strömberg N, van Dolleweerd CJ, Kelly CG, Jenkinson HF. 2005. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 55:1591–1605. 10.1111/j.1365-2958.2005.04495.x [DOI] [PubMed] [Google Scholar]
- 26.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a Web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010:baq013. 10.1093/database/baq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vartoukian SR, Palmer RM, Wade WG. 2010. Strategies for culture of ‘unculturable' bacteria. FEMS Microbiol. Lett. 309:1–7. 10.1111/j.1574-6968.2010.02000.x [DOI] [PubMed] [Google Scholar]
- 28.Rheims H, Rainey FA, Stackebrandt E. 1996. A molecular approach to search for diversity among bacteria in the environment. J. Indust. Microbiol. 17:159–169 [Google Scholar]
- 29.Lazarevic V, Whiteson K, Hernandez D, François P, Schrenzel J. 2010. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 11:523. 10.1186/1471-2164-11-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi BK, Paster BJ, Dewhirst FE, Gobel UB. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 62:1889–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338–344. 10.1177/154405910308200503 [DOI] [PubMed] [Google Scholar]
- 32.Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407–450. 10.1128/MMBR.00014-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.