Abstract
This study intends to establish and apply methods evaluating both viral capsid and genome integrities of human noroviruses (NoVs), which thus far remain nonculturable. Murine norovirus 1 (MNV-1) and human NoV GII.4 in phosphate-buffered saline suspensions were treated with heat, UV light, or ethanol and detected by reverse transcription-quantitative PCR (RT-qPCR), long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCR. For MNV-1 heated at 60°C for 2 and 30 min, limited reductions of genomic copies (<0.3-log) were obtained by RT-qPCR and long-range RT-qPCR, while the cell-binding pretreatments obtained higher reductions (>1.89-log reduction after 60°C for 30 min by binding long-range RT-qPCR). The human NoV GII.4 was found to be more heat resistant than MNV-1. For both MNV-1 and human NoV GII.4 after UV treatments of 20 and 200 mJ/cm2, no significant difference (P > 0.05) was observed between the dose-dependent reductions obtained by the four detection methodologies. Treatment of 70% ethanol for 1 min was shown to be more effective for inactivation of both MNV-1 and human NoV GII.4 than the heat and UV treatments used in this study. Subsequently, eight raspberry and four shellfish samples previously shown to be naturally contaminated with human NoVs by RT-qPCR (GI and GII; thus, 24 RT-qPCR signals) were subjected to comparison by this method. RT-qPCR, long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCR detected 20/24, 14/24, 24/24, and 23/24 positive signals, respectively, indicating the abundant presence of intact NoV particles.
INTRODUCTION
Noroviruses (NoVs) are a group of important food-borne viruses and the leading cause of human gastroenteritis worldwide (1, 2). Methods for the detection of NoVs have been progressing over a number of years. Due to the inability to culture NoVs in vitro (3, 4), reverse transcription-quantitative PCR (RT-qPCR) is still recognized as the gold standard for virus detection, although this method cannot differentiate between infectious and noninfectious viruses (5, 6). Thus far, prediction of NoV infectivity has been attempted from the integrities and/or functions of viral RNA molecules and capsid proteins (5), which are the two essential parts for an intact and infectious virus particle.
As for the genome integrity evaluation, although it is possible to amplify nearly full-length human NoV genomes (7), the amplification efficiency decreases with fragment size, thus making amplification of full-length genomic RNA relatively insensitive. Wolf et al. (8) suggested that one important factor for assessing the genomic integrities of RNA viruses is the dependency of the reverse transcription (RT) reaction on high-quality, nonfragmented template RNA. This fact can be exploited by separating the PCR amplification site and RT priming site within the virus genome. This approach has been used to examine the integrity of the human NoV genome after high-temperature (72°C) and UV treatment (8).
Multiple studies have been conducted to assess the number of infectious NoVs based on the capsid integrity or function (9–11). Recently, binding-based RT-PCRs were developed in our laboratory and were able to decrease the detection of noninfectious NoVs by approximately 1 to 3 log, whereas all infectious viral particles were detected (12). For murine norovirus 1 (MNV-1), the cell line RAW 264.7 and the ganglioside GD1a were used as binding receptors and, for human NoVs, differentiated Caco-2 cells and pig gastric mucin were tested as the binding receptors (12).
Our aim here was to apply these methods (RT-qPCR, long-range RT-qPCR, and binding RT-qPCR) and the combination method (binding long-range RT-qPCR) in order to indicate the viral integrities of NoVs. First, murine norovirus 1 (MNV-1, a surrogate of human NoVs) and human NoV GII.4 were treated in phosphate-buffered saline (PBS) suspensions by heat, UV light, and ethanol and detected by RT-qPCR, long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCR. Second, raspberry and shellfish samples prior shown by RT-qPCR to be naturally contaminated with human NoVs were also subjected to a comparison test for these methods.
MATERIALS AND METHODS
Viruses and cells.
Cells of the murine macrophage cell line RAW 264.7 (ATCC TIB-71; kindly provided by H. W. Virgin, Washington University School of Medicine, St. Louis, MO) were maintained in complete Dulbecco modified Eagle medium (DMEM) and grown at 37°C under a 5% CO2 atmosphere. Complete DMEM consisted of DMEM (Lonza, Walkersville, MD) containing 10% low-endotoxin fetal bovine serum (HyClone, Logan, UT), 100 U/ml penicillin, 100 μg/ml streptomycin (Lonza), 10 mM HEPES (Lonza), and 2 mM l-glutamine (Lonza).
RAW 264.7 cells were infected with MNV-1.CW1, passage 7, at a multiplicity of infection of 0.05 (MNV-1:cells) for 2 days. After two freeze-thaw cycles, low-speed centrifugation was used to remove cellular debris from the virus lysate, as described by Wobus et al. (13). The lysate was stored in aliquots at −75°C.
Cells of the human enterocytic cell line Caco-2 (ECACC 86010202) were cultured in Eagle minimum essential medium with Earle's salts (EMEM; Lonza) supplemented with 10% low-endotoxin fetal bovine serum (HyClone), 100 U/ml penicillin, 100 μg/ml streptomycin (Lonza), and 2 mM l-glutamine (Lonza) and grown at 37°C under a 5% CO2 atmosphere.
Human NoV GII.4 sample was kindly provided by the Netherlands National Institute for Public Health and the Environment (RIVM). The virus genome was partially sequenced with the primers G2SKF/G2SKR as described by Nishida et al. (14), and 99% identification was obtained with the norovirus GII.4 strain (GenBank accession number KF475965.1). The sample was vortexed vigorously, and low-speed centrifugation was used to remove solid material.
In order to decrease/eliminate the interfering effect of cell culture medium (for MNV-1) or human fecal material (for human NoV GII.4), viral suspensions for inactivation studies were prepared by 1,000-fold dilution (for heat and UV treatments) or 100-fold dilution (for ethanol treatments) of MNV-1 lysate and NoV GII.4 stool supernatant in PBS (Lonza).
Inactivation treatments.
The heat treatments were performed using the GeneAmp PCR System 2400 (Perkin-Elmer, Waltham, MA). MNV-1 and human NoV GII.4 suspensions (1,000-fold dilution) were subjected to heat treatment (60°C for 2 min and 60°C for 30 min) in thin-walled PCR tubes (100 μl of each), respectively.
The UV treatments were performed in a prototype apparatus with two low-pressure mercury lamps producing mainly 254-nm UV light (TUVN4 4K P 165 lamp, 4 W each; UV-Technik Speziallampen GmbH, Germany) as described in Li et al. (15) and a CleanView UV cabinet (BiocomDirect, United Kingdom) with four low-pressure mercury lamps producing mainly 254-nm UV light (GE T8-Germicidal, 15 W each; GE Lighting, Japan). Viral suspensions (1,000-fold dilution) were exposed to the emitted UV light in a 24-well plate (200 μl of suspension per well). The UV irradiance was measured with a radiometer (model PMA2100; Solar Light Co.) and calculated as 20 and 200 mJ/cm2.
The ethanol treatments were performed by the dilution of ethanol (Sigma-Aldrich, Steinheim, Germany) with viral suspensions (100-fold dilution) to reach a final concentration of 70%. After mixing well by vortex, the mixtures were incubated at room temperature for 1 min before 10-fold dilution by complete DMEM as the neutralization.
Viral extraction from raspberry and shellfish.
Frozen raspberry puree (prescreened for NoV presence as described elsewhere [A. Keuckelaere, A. Stals, B. Deliens, and M. Uyttendaele, unpublished data]) was thawed at room temperature. Twenty grams of raspberry puree was divided into two subsamples of 10 g each. The first subsample (10 g) was spiked with MNV-1 solution as a process control to calculate the recovery efficiency of the extraction process. Each subsample was mixed with 30 ml of Tris-glycine-beef extract buffer (0.1 M Tris-HCl, 3% beef extract, 0.05 M glycine [pH 9.5] adjusted with 10 M NaOH). To prevent the formation of a gel, 150 μl of Pectinex (Sigma-Aldrich, Steinheim, Germany) was added to the elution buffer. The viruses were then concentrated by using the PEG 6000/NaCl precipitation technique. The final pellet was dissolved in 1.5 ml of PBS (Lonza), 1 ml of which was subjected to further purification by a chloroform-butanol purification step. The supernatant was stored at −75°C. Virus extraction of the second subsample (10 g) was performed in parallel with the first subsample, but during the RT step MNV-1 RNA was added to the reaction mix as an RT control to control the amplification efficiency.
Oyster digestive tissues with NoV contamination during wastewater treatment were kindly provided by Sinéad Keaveney (Marine Institute, Ireland). A 2-g portion of the digestive tissues was transferred to a 50-ml centrifuge tube and cut into fine pieces using sterile scissors. Mengo virus vMC0 solution was added as a process control to calculate the recovery efficiency and the amplification efficiency. Then, 2 ml of proteinase K (0.1 mg/ml; Sigma-Aldrich, St. Louis, MO) was added, followed by incubation at 37°C with shaking at 200 rpm for 30 min, followed in turn by incubation at 60°C for 15 min in a warm water bath. After centrifugation at 3,000 × g for 5 min at 4°C, the supernatant (ca. 2 ml) was collected and stored at −75°C.
Plaque assay.
The number of MNV-1 PFU/ml was determined by plaque assay, as described by Wobus et al. (13). Briefly, RAW 264.7 cells were seeded into six-well plates at a density of 2 × 106 viable cells per well. On the following day, 10-fold dilutions of the samples of unknown virus titer were prepared in complete DMEM, and 1 ml per dilution of the sample was plated onto two wells (0.5 ml per well). The plates were incubated for 1 h at room temperature and manually rocked every 15 min before aspirating the inoculum and overlaying the cells with 1.5% SeaPlaque agarose (Cambrex, Rockland, ME) in minimum essential Eagle medium (MEME; Lonza) supplemented with 10% low-endotoxin fetal bovine serum, 1% HEPES, 1% penicillin-streptomycin, and 2% glutamine (complete MEME) per well. The plates were incubated at 37°C and 5% CO2 for 2 days. To visualize the plaques, cells were stained with 1.5% SeaKem agarose in complete MEME containing 1% neutral red (Sigma-Aldrich, St. Louis, MO) per well for 6 h.
Cell-binding pretreatment.
The cell-binding pretreatment was performed as previously described (12). RAW 264.7 or Caco-2 cells were seeded into 24-well plates at a density of 5 × 105 viable cells per well. RAW 264.7 cells were used on the following day, while Caco-2 cells were incubated for at least 21 days postconfluence and used as differentiated Caco-2 cells. MNV-1 or human NoV samples were 10-fold diluted in complete DMEM (for MNV-1) or PBS (for human NoVs) and then 200 μl per dilution of the MNV-1 or human NoV sample was plated into two wells (100 μl per well). Inoculated cells were incubated for 1 h at 4°C and rocked manually every 15 min. The inocula were removed after 1 h of incubation, and the cells were washed three times with PBS. The first washing step was performed by adding 0.5 ml of PBS and rocking the plates manually. Second, the liquid was removed, 0.5 ml of PBS was added again, and the cells were scraped off. The suspensions were vortexed and centrifuged at 6,000 × g for 5 min (Eppendorf 5417C5417C). The supernatant was removed. Third, the pellets were resuspended in PBS, vortexed, and centrifuged again. The final pellets were resuspended in PBS (100 μl per sample) and stored at −20°C until the RNA extraction and RT-qPCR were performed.
Viral RNA extraction.
Total RNA was extracted from 100 μl of virus suspension or food extraction using the NucliSENS easyMAG automated system (bioMérieux) according to the standard protocol of the manual (i.e., elution in 25 μl of elution buffer).
RT.
Reverse transcription (RT) was performed as described by Wolf et al. (8) with a few modifications. For the long-range RT-qPCR, The 10-μl RT reactions comprised of 200 U of SuperScript III reverse transcriptase (Invitrogen), 20 U of RNase inhibitor (Invitrogen), 2.5 μM oligo(dT) (9)-VN primer containing three locked nucleic acid (Exiqon), 0.3 mM concentrations of deoxynucleoside triphosphates (dNTPs), 1× first-strand RT buffer, and 5 μl of viral RNA. The RNA, primer, and dNTPs were initially heated to 65°C for 10 min and then cooled on ice for at least 1 min before the other reagents were added. RT was carried out at 50°C for 60 min, followed by 70°C for 15 min. cRNA was removed with 2 U/μl RNase H (Invitrogen) at 37°C for 20 min. For the conventional RT-qPCR, the respective reverse primers were used in the RT reactions. The conditions were as described above except that the primer and dNTP concentrations were 0.1 μM and 0.5 mM, respectively. Only 100 U of SuperScript III was used, and the RT reaction was carried out for 30 min without RNase H digestion.
qPCR detection.
qPCR detection of MNV-1 and human NoVs was performed as described by Baert et al. (16) and Stals et al. (17). For the singleplex qPCR detection of MNV-1, the 25-μl reaction mixture contained 4 μl of template DNA, 500 nM forward primer, 900 nM reverse primer, 250 nM probe, and 12.5 μl of TaqMan Universal PCR master mix (Applied Biosystems). The amplicon of MNV-1 was 108 bp, targeting on the ORF1/ORF2 junction of the virus genome. For duplex qPCR detection of human NoV GI and GII, the 25-μl reaction mixture contained 4 μl of template DNA, 500 nM each forward primer, 900 nM each reverse primer, 250 nM concentrations of each probe, and 12.5 μl of TaqMan Universal PCR master mix (Applied Biosystems). The amplicons of human NoV GI and GII were 85 and 88 bp, targeting the ORF1/ORF2 junctions of the virus genomes. Real-time quantification was performed on an ABI 7300 system (Applied Biosystems) with the following temperature profile: 50°C for 2 min, 95°C for 10 min, and 50 cycles of 95°C for 15 s and 60°C for 1 min. To obtain representative positive-control standards, plasmids containing primer-probe binding sites were used for the quantifications. Tenfold serial dilutions ranging from 105 to 10 copies of plasmids per reaction were used to prepare the standard curves. The plasmid quantity was determined by spectrophotometric analysis (NanoDrop; Thermo), followed by serial 10-fold dilution.
Data analysis.
Samples were tested in triplicate. Each error bar represents the data range. Statistical analyses were performed by one-way analysis of variance (Games-Howell was used as a post hoc test) with SPSS 17.0 for Windows (SPSS, Inc.). Significant differences were considered when the P value was <0.05. For data sets lower than the detection limit, the detection limit was used instead to perform statistical analyses in comparison to other data sets.
RESULTS
Effects of inactivation treatments on MNV-1 and human NoV GII.4.
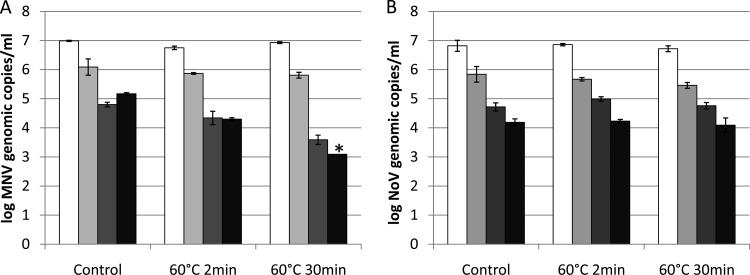
Using a plaque assay, 1.91 ± 0.13 log PFU reduction in MNV-1 infectivity was obtained by heat treatment at 60°C for 2 min, and the MNV-1 infectivity was reduced to a nondetectable level (>4-log PFU reduction) by heat at 60°C for 30 min. The numbers of MNV-1 and human NoV GII.4 genomic copies detected by RT-qPCR, long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCR before and after the heat treatments are presented in Fig. 1 (MNV-1 [Fig. 1A] and human NoV GII.4 [Fig. 1B]). For MNV-1, limited reductions (<0.3-log genomic copies) were obtained by RT-qPCR and long-range RT-qPCR after heat treatments at 60°C for both 2 and 30 min, while the cell-binding pretreatments (for binding RT-qPCR and binding long-range RT-qPCR) were found to indicate higher reductions in a dose-dependent way. The combination of binding and long-range RT-qPCR indicated reduction to nondetectable level (>1.94-log genomic copies) after 60°C treatment for 30 min, which is significantly higher than the reductions obtained by RT-qPCR and long-range RT-qPCR (P < 0.05). In contrast for human NoV GII.4, limited reductions (<0.4-log genomic copies) were obtained by all four methods after heat treatments at 60°C for both 2 and 30 min.
FIG 1.
Detection of MNV-1 (A) and human NoV GII.4 (B) genomic copies by RT-qPCR (white bars), long-range RT-qPCR (light gray bars), binding RT-qPCR (dark gray bars), and binding long-range RT-qPCR (black bars) after heat treatment. Each data point is an average of triplicates, and each error bar represents the data range. *, lower than the detection limit.
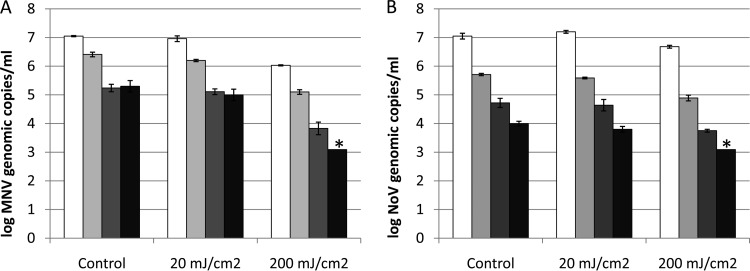
For UV treatments, MNV-1 infectivity was reduced by 1.7 ± 0.3 log PFU after 20 mJ/cm2 of UV treatment, and the MNV-1 infectivity was reduced to a nondetectable level (>4-log PFU reduction) by UV of 200 mJ/cm2. The numbers MNV-1 and human NoV GII.4 genomic copies detected by RT-qPCR, long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCR before and after the UV treatments are presented in Fig. 2 (MNV-1 [Fig. 2A] and human NoV GII.4 [Fig. 2B]). For MNV-1, although the combination of binding and long-range RT-qPCR also indicated higher reductions than the other methods (RT-qPCR, long-range RT-qPCR and binding RT-qPCR) after both 20- and 200-mJ/cm2 UV treatments, no significant difference was observed (P > 0.05). All four methods indicated dose-dependent reductions after 20-mJ/cm2 (<0.3-log genomic copies) and 200-mJ/cm2 (>1-log genomic copies) UV treatments. Similarly for human NoV GII.4, dose-dependent reductions were obtained after 20- and 200-mJ/cm2 UV treatments without significant differences between the four methods (P > 0.05).
FIG 2.
Detection of MNV-1 (A) and human NoV GII.4 (B) genomic copies by RT-qPCR (white bars), long-range RT-qPCR (light gray bars), binding RT-qPCR (dark gray bars), and binding long-range RT-qPCR (black bars) after UV treatment. Each data point is an average of triplicates, and each error bar represents the data range. *, lower than the detection limit.
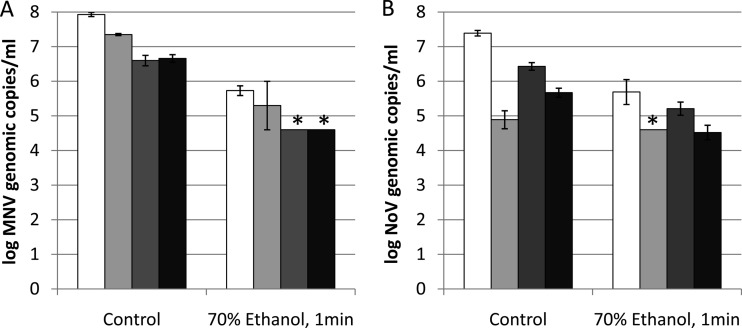
Treatment with 70% ethanol for 1 min, which reduced MNV-1 infectivity to a nondetectable level (>4-log PFU reduction), was found to be more effective in inactivating MNV-1 than the heat (60°C for 2 and 30 min) and UV (20 and 200 mJ/cm2) treatments used in the present study. Reductions of >2 log in MNV-1 genomic copies were obtained by all of the four methods (Fig. 3A). For human NoV GII.4, the treatment with 70% ethanol for 1 min also indicated higher reductions than the heat (60°C for 2 and 30 min) and UV (20 and 200 mJ/cm2) by the studied methods except long-range RT-qPCR due to its low detection limit (Fig. 3B).
FIG 3.
Detection of MNV-1 (A) and human NoV GII.4 (B) genomic copies by RT-qPCR (white bars), long-range RT-qPCR (light gray bars), binding RT-qPCR (dark gray bars), and binding long-range RT-qPCR (black bars) after ethanol treatment. Each data point is an average of triplicates, and each error bar represents the data range. *, lower than the detection limit.
Detection of human NoVs in naturally contaminated raspberry and shellfish samples.
The extraction efficiency and the amplification efficiency results for the raspberry and shellfish samples are listed in Table 1. RT-qPCR detection of NoV in raspberry samples included a process control (MNV-1) and an external control RNA (MNV-1 RNA) to check the extraction efficiency and the amplification efficiency, respectively. Raspberry samples 1 and 2 and raspberry samples 4 and 5 were the two pairs of subsamples from the same samples that had the same extraction efficiency and amplification efficiency. RT-qPCR detection of NoV in shellfish samples included a process control (Mengo virus) to calculation the extraction efficiency and the amplification efficiency together.
TABLE 1.
Detection results for human NoVs in raspberry and shellfish samples obtained by RT-qPCR, long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCRa
| Sample | Log genomic copies/g sample |
Efficiency (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RT-qPCR |
Binding RT-qPCR |
Long-range RT-qPCR |
Binding long-range RT-qPCR |
Recovery | Amplification | |||||
| GI | GII | GI | GII | GI | GII | GI | GII | |||
| Raspberry | ||||||||||
| 1 | 3.5 | 2.6 | 4.1 | <LOQ | 3.3 | <LOQ | 3.2 | 2.5 | 0.47 | 46 |
| 2 | <LOD | <LOQ | 4.0 | 2.8 | <LOD | <LOD | 3.4 | <LOQ | 0.47 | 46 |
| 3 | 3.7 | <LOQ | 5.1 | 2.3 | <LOD | <LOD | 2.7 | <LOD | 1 | 33.44 |
| 4 | 3.6 | <LOQ | 4.1 | 2.5 | <LOD | <LOD | 3.6 | 2.4 | 1.41 | 58.15 |
| 5 | 2.5 | <LOD | 4.2 | 2.6 | <LOD | <LOQ | 4.1 | 2.5 | 1.41 | 58.15 |
| 6 | 4.7 | 3.0 | 4.4 | 2.3 | 3.6 | 2.7 | 3.9 | 2.3 | 1.14 | 38.12 |
| 7 | 4.5 | 2.5 | 5.2 | 2.6 | 3.9 | 2.7 | 4.2 | <LOQ | 1.88 | 50.57 |
| 8 | 3.5 | 2.4 | 3.7 | 2.3 | 3.6 | 2.4 | 3.3 | 2.4 | 0.4 | 35.57 |
| Shellfish | ||||||||||
| 1 | 5.4 | 3.3 | 6.9 | 3.7 | 5.1 | <LOQ | 4.3 | 3.4 | 3.37 | |
| 2 | 5.7 | 3.5 | 4.3 | <LOQ | 3.8 | <LOQ | 4.1 | <LOQ | 1.23 | |
| 3 | 3.4 | 3.3 | 3.2 | <LOQ | <LOD | <LOD | 3.2 | <LOQ | 0.12 | |
| 4 | <LOD | 3.6 | <LOQ | <LOQ | <LOD | <LOQ | 3.2 | <LOQ | 2.18 | |
LOD, limit of detection, 1.3-log genomic copies/g for raspberry samples and 2.1-log genomic copies/g for shellfish samples; LOQ, limit of quantification, 2.3-log genomic copies/g for raspberry samples and 3.1-log genomic copies/g for shellfish samples.
The detection results of human NoVs in raspberry and shellfish samples obtained by RT-qPCR, long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCR are shown in Table 1. Since the qPCR standard curves were generated by the detection of 106 to 10 copies of plasmids per reaction, the limit of quantification (LOQ) was 10 genomic copies/PCR, while the limit of detection (LOD) can be extrapolated to 1 genomic copy/PCR. Therefore, the LODs for raspberry and shellfish samples are 1.3- and 2.1-log genomic copies/g sample, and the LOQs for raspberry and shellfish samples are 2.3- and 3.1-log genomic copies/g sample. Within the eight raspberry and four shellfish samples (both GI and GII thus detected 24 signals for method comparison), RT-qPCR, long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCR detected 20/24, 14/24, 24/24, and 23/24 positive signals (>LOD), respectively. Eighteen of 24, 9 of 24, 19 of 24, and 18 of 24 were detected with quantitative NoV signals (>LOQ) by RT-qPCR, long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCR, respectively.
DISCUSSION
In the study of Wolf et al. (8), long-range RT-qPCR showed decreased qPCR amplification after UV exposure, which became more pronounced with increased UV light exposure time but was still significantly less than the decrease in infectivity demonstrated by the plaque assay. In one of our previous studies (18), the cell-binding RT-PCR was compared to the RNase One RT-PCR detecting NoVs after a heat treatment and revealed higher reductions for both MNV-1 and human NoV GII.4, while neither method could detect the infectivity reductions of MNV-1. In the present study, long-range and binding RT-qPCR, which demonstrate viral genome and capsid integrity, respectively, were used both separately and in combination to predict NoV infectivity in a more complete way. According to our results, nondetectable levels were reached by the use of binding long-range RT-qPCR for both MNV-1 and human NoV GII.4 after treatments that could induce the MNV-1 infectivity reduction to nondetectable levels (heat treatment at 60°C for 30 min, 200-mJ/cm2 UV treatment, and 70% ethanol treatment for 1 min [except for the heat-treated human NoV GII.4]). However, the reduction obtained using a combination of binding and long-range RT-qPCR after heat treatment was only significantly higher than reductions obtained by RT-qPCR and long-range RT-qPCR (P < 0.05) but not binding RT-qPCR (P > 0.05). No significant differences between all four methods were observed for the UV and ethanol treatments for both MNV-1 and human NoV GII.4 (P > 0.05). None of these RT-qPCR-based methods could indicate comparative reductions with plaque assay for treatments that could induce ∼2-log infectivity reductions (i.e., heat treatment at 60°C for 2 min and 20-mJ/cm2 UV treatment). Therefore, the measurement of viral integrity is still a conservative strategy. In other words, virus particles that have lost their infectivity may still maintain capsid integrity. Thus, changes in viral infectivity do not always correlate directly with changes in capsid integrity. Moreover, the cell-binding assay (coupled with either RT-qPCR method) may overestimate not only the number of infectious viruses but also the number of completely intact virus particles, since cell binding only requires partially intact viruses with an intact receptor-binding site.
It should be noted that the detection levels (genomic copies) of the starting control samples (both for MNV-1 and human NoV GII.4) by the four methods were different. The most important reason should be the presence of infective (intact) virus particles, as well as the presence of defective or noninfective virus particles, in MNV-1 lysate and human NoV clinical samples. In addition, since it is not possible to determine the proportion of intact viruses in the original samples, the absolute amplification efficiencies of long-range RT-qPCR and the viral losses caused by the handling and washing steps of binding RT-qPCR remain unknown. Despite the factors and limitations discussed above, we nevertheless demonstrated here the values of long-range and binding RT-qPCR for determining the viral integrities of NoVs inactivated by different treatments (heat, UV light, and ethanol) and in naturally contaminated food (raspberry and shellfish) samples.
Heat treatment is one of the most widely used techniques for food safety control and preservation (19). In recent years, there has been a greater focus on the inactivation methods of food-borne pathogens that utilize short times and low temperatures due to the demands for minimally processed foods to maintain nutrition and flavor (20, 21). The mechanism of heat inactivation of viruses is believed to be due to changes in the capsid of the virus particle, as reviewed by Hirneisen et al. (22). Accordingly, in the present study, limited reductions (<0.3-log) were obtained by RT-qPCR and long-range RT-qPCR, which solely measure the genomic integrity of MNV-1 after heat treatments at 60°C for 2 and 30 min, whereas the cell-binding pretreatments that also indicate the capsid integrity yielded higher reductions (>1.89-log reduction after 60°C for 30 min by binding long-range RT-qPCR). The human NoV GII.4 was found to be more heat resistant than MNV-1 in this study (<0.2-log reduction after 60°C for 30 min by binding long-range RT-qPCR), an observation that is consistent with the recent report by Escudero-Abarca et al. (23).
With a long history in the field of water treatment and health care facility disinfection, UV treatment (the most germicidal effect is at a wavelength of ∼254 nm) is preferred for its low cost, ease of use, and lack of toxic by-product. The UV doses used in the present study (20 and 200 mJ/cm2) were higher than those used in previous studies, since it was reported that a UV dose of 30 mJ/cm2 was able to achieve a 4-log infectivity reduction for murine norovirus, feline calicivirus, and echovirus 12 (24) and that a UV dose of 20 mJ/cm2 induced 3.3-log reduction in MNV-1 infectivity (25). It is generally believed that UV light induces damage on the viral genetic materials and only affects the capsid at higher doses. Accordingly in the present study, for MNV-1 after UV treatments of 20 and 200 mJ/cm2, although long-range RT-qPCR, binding RT-qPCR, and binding long-range RT-qPCR indicated higher reductions than RT-qPCR, no significant difference was observed between the methods based on measurement of the genomic integrity or capsid integrity. Similar trends were observed for human NoV GII.4, except that human NoV GII.4 was found to be more resistant to UV treatment than MNV-1 detected by all four methods.
Ethanol is a main component of many disinfectants. The effectiveness of ethanol against NoVs is somewhat controversial, based on findings from previous studies. For instance, a >4-log reduction in MNV-1 infectivity was observed after treatment with 60% ethanol for 0.5 min in a study by Belliot et al. (26), whereas D'Souza and Su (27) found no reduction (0.07 log) in MNV-1 infectivity after a treatment with 70% ethanol for 1 min. This discrepancy may be due to the different experimental setups (the former study was performed using suspensions, whereas the latter study was performed on surfaces), which lead to different contact situations between virus particles and the disinfectant, or to the presence of an interfering organic load, which could result in decreased viral inactivation effectiveness. Here, RT-qPCR-based methods were applied to MNV-1 and human NoV GII.4 in suspensions. The viral suspensions were prepared by 100-fold dilution of MNV-1 lysate and NoV GII.4 stool supernatant in PBS in order to decrease/eliminate the interfering effect of cell culture medium (MNV-1) or human fecal material (human NoV GII.4). Higher reductions were obtained than for the heat (60°C for 2 and 30 min) and UV (20 and 200 mJ/cm2) treatments by all four methods for both MNV-1 and human NoV GII.4.
In recent years, NoV contamination has been identified during NoV prevalence studies on food and the environment. The observed prevalence ranged from 3.9 to 76.2% in shellfish (14, 28, 29), 6.7 to 55.5% in fresh produce (30), and 15.1 to 45% in drinking water sources and surface water (31, 32). These positive NoV signals may induce recalls or rejections of food batches associated with NoV contamination, causing economic loss in international markets. However, these results obtained exclusively by the RT-qPCR method only demonstrated the presence of small segments of NoV genome since the qPCR amplicons are normally rather short (∼100 bp). In the present study, by the use of the binding and long-range RT-qPCR, which could reflect the viral capsid and genome integrity, the abundant presence of intact NoV particles has been demonstrated in NoVs naturally contaminated food samples tested.
It is notable that the cell-binding pretreatment for binding RT-qPCR and binding long-range RT-qPCR, which was expected to eliminate the false-positive results and thus indicate fewer positive samples, actually detected more positive samples, as well as higher NoV quantities, than did RT-qPCR and long-range RT-qPCR. The reason may be that during the washing step of the cell-binding treatment, most of the residual food components in the samples that could be PCR inhibitors were removed along with the unbound viruses.
Differences in the yields of viral extraction and concentration methods for noninfectious particles (i.e., genomic RNA) versus infectious particles have been reported previously (33), since some viral concentration methods can be based on the particle structure of the viruses (34) and hence favor the detection of RNA that is encapsulated over free RNA. In the present study, the use of proteinase K for virus extraction from shellfish digestive tissues may partially remove the disrupted viruses, as suggested by previous studies (35). Therefore, the selective effect of the virus extraction procedures could explain partially why long-range and binding RT-qPCRs indicated more comparative results with RT-qPCR for NoVs in food samples than in clinical samples in the inactivation study.
In summary, using binding and long-range RT-qPCR, we have demonstrated here changes in NoV integrity in response to different inactivation treatments. We confirmed that heat treatment targets mainly the viral capsid, whereas UV light damages viral genetic material. Treatment with 70% ethanol for 1 min was more effective in damaging MNV-1 and human NoV GII.4 integrity than heat treatment at 60°C for 30 min or UV treatment at 200 mJ/cm2. Human NoV GII.4 was more resistant than MNV-1 to the heat, UV, and ethanol treatments used in this study. In addition, we demonstrated the abundant presence of NoV particles with intact receptor-binding sites and a large part of complete genome in the naturally contaminated food samples.
ACKNOWLEDGMENTS
This study was supported by a postdoctoral grant (D.L.) from the Research Foundation–Flanders (Fonds voor Wetenschappelijk Onderzoek–Vlaanderen) and the European Community's Seventh Framework Programme (FP7) under grant agreement 244994 (project VEG-i-TRADE).
We thank Sinéad Keaveney (Marine Environment and Food Safety, Marine Institute, Dublin, Ireland) for supplying the shellfish samples. We also thank Geert Van Cleemput (ELSCOLAB NV, Kruibeke, Belgium) for the help in measuring the UV irradiance.
Footnotes
Published ahead of print 8 August 2014
REFERENCES
- 1.European Food Safety Authority, European Centre for Disease Prevention and Control. 2012. The European Union summary report on trends and sources of zoonoses, zoonotic agents, and food-borne outbreaks in 2010. EFSA J. 2012 10:2597. 10.2903/j.efsa.2012.2597 [DOI] [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the united states-major pathogens. Emerg. Infect. Dis. 17:7–22. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MPG, Estes MK. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79–87. 10.1099/vir.0.19478-0 [DOI] [PubMed] [Google Scholar]
- 4.Herbst-Kralovetz MM, Radtke AL, Lay MK, Hjelm BE, Bolick AN, Sarker SS, Atmar RL, Kingsley DH, Arntzen CJ, Estes MK. 2013. Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells. Emerg. Infect. Dis. 19:431–438. 10.3201/eid1903.121029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight A, Li D, Uyttendaele M, Jaykus LA. 2012. A critical review of methods for detecting human noroviruses and predicting their infectivity. Crit. Rev. Microbiol. 39:295–309. 10.3109/1040841X.2012.709820 [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez RA, Pepper IL, Gerba CP. 2009. Application of PCR-based methods to assess the infectivity of enteric viruses in environmental samples. Appl. Environ. Microbiol. 75:297–307. 10.1128/AEM.01150-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostela J, Ayers M, Nishikawa J, McIntyre L, Petric M, Tellier R. 2008. Amplification by long RT-PCR of near full-length norovirus genomes. J. Virol. Methods 149:226–230. 10.1016/j.jviromet.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 8.Wolf S, Rivera-Aban M, Greening GG. 2009. Long-range reverse transcription as a useful tool to assess the genomic integrity of norovirus. Food. Environ. Virol. 1:129–136. 10.1007/s12560-009-9016-7 [DOI] [Google Scholar]
- 9.Parshionikar S, Laseke I, Fout GS. 2010. Use of propidium monoazide in reverse transcriptase PCR to distinguish between infectious and noninfectious enteric viruses in water samples. Appl. Environ. Microbiol. 76:4318–4326. 10.1128/AEM.02800-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sano D, Pinto RM, Omura T, Bosch A. 2010. Detection of oxidative damages on viral capsid protein for evaluating structural integrity and infectivity of human noroviruses. Environ. Sci. Technol. 44:808–812. 10.1021/es9018964 [DOI] [PubMed] [Google Scholar]
- 11.Topping J, Schnerr H, Haines J, Scott M, Carter M, Willcocks M, Bellamy K, Brown D, Gray J, Gallimore C. 2009. Temperature inactivation of feline calicivirus vaccine strain FCV F-9 in comparison with human noroviruses using an RNA exposure assay and reverse transcribed quantitative real-time polymerase chain reaction-a novel method for predicting virus infectivity. J. Virol. Methods 156:89–95. 10.1016/j.jviromet.2008.10.024 [DOI] [PubMed] [Google Scholar]
- 12.Li D, Baert L, Van Coillie E, Uyttendaele M. 2011. Critical studies on binding based RT-PCR detection of infectious noroviruses. J. Virol. Methods 177:153–159. 10.1016/j.jviromet.2011.07.013 [DOI] [PubMed] [Google Scholar]
- 13.Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW., IV 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:2076–2084. 10.1371/journal.pbio.0020432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishida T, Kimura H, Saitoh M, Shinohara M, Kato M, Fukuda S, Munemura T, Mikami T, Kawamoto A, Akiyama M. 2003. Detection, quantitation, and phylogenetic analysis of noroviruses in Japanese oysters. Appl. Environ. Microbiol. 69:5782–5786. 10.1128/AEM.69.10.5782-5786.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li D, Baert L, De Jonghe M, Van Coillie E, Ryckeboer J, Devlieghere F, Uyttendaele M. 2011. Inactivation of murine norovirus 1, coliphage ΦX174, and B. fragilis phage B40-8 on surfaces and fresh-cut iceberg lettuce by hydrogen peroxide and UV light. Appl. Environ. Microbiol. 77:1399–1404. 10.1128/AEM.02131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baert L, Wobus CE, Van Coillie E, Thackray LB, Debevere J, Uyttendaele M. 2008. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat treatment. Appl. Environ. Microbiol. 74:543–546. 10.1128/AEM.01039-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stals A, Baert L, Botteldoorn N, Werbrouck H, Herman L, Uyttendaele M, Van Coillie E. 2009. Multiplex real-time RT-PCR for simultaneous detection of GI/GII noroviruses and murine norovirus 1. J. Virol. Methods 161:247–553. 10.1016/j.jviromet.2009.06.019 [DOI] [PubMed] [Google Scholar]
- 18.Li D, Baert L, Xia M, Zhong W, Van CE, Jiang X, Uyttendaele M. 2012. Evaluation of methods measuring the capsid integrity and/or functions of noroviruses by heat inactivation. J. Virol. Methods 181:1–5. 10.1016/j.jviromet.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 19.Rahman S. 2007. Handbook of food preservation, p 571–634, 2nd ed. CRC Press, Inc, Boca Raton, FL [Google Scholar]
- 20.Baert L, Uyttendaele M, Van Coillie E, Debevere J. 2008. The reduction of murine norovirus 1, B. fragilis HSP40 infecting phage B40-8 and E. coli after a mild thermal pasteurization process of raspberry puree. Food Microbiol. 25:871–874. 10.1016/j.fm.2008.06.002 [DOI] [PubMed] [Google Scholar]
- 21.Uyttendaele M, Rajkovic A, Van Houteghem N, Boon N, Thas O, Debevere J, Devlieghere F. 2008. Multimethod approach indicates no presence of sublethally injured Listeria monocytogenes cells after mild heat treatment. Int. J. Food Microbiol. 123:262–268. 10.1016/j.ijfoodmicro.2008.02.015 [DOI] [PubMed] [Google Scholar]
- 22.Hirneisen KA, Black EP, Cascarino JL, Fino VR, Hoover DG, Kniel KE. 2010. Viral inactivation in foods: a review of traditional and novel food-processing technologies. Compr. Rev. Food Sci. Food Safety 9:3–20. 10.1111/j.1541-4337.2009.00092.x [DOI] [PubMed] [Google Scholar]
- 23.Escudero-Abarca B, Rawsthorne H, Goulter R, Suh S, Jaykus L. 2014. Molecular methods used to estimate thermal inactivation of a prototype human norovirus: more heat resistant than previously believed? Food Microbiol. 41:91–95. 10.1016/j.fm.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 24.Park G, Linden K, Sobsey M. 2011. Inactivation of murine norovirus, feline calicivirus and echovirus 12 as surrogates for human norovirus (NoV) and coliphage (F+) MS2 by ultraviolet light (254 nm) and the effect of cell association on UV inactivation. Lett. Appl. Microbiol. 52:162–167. 10.1111/j.1472-765X.2010.02982.x [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Zoh K, Ko G. 2008. Inactivation and UV disinfection of murine norovirus with TiO2 under various environmental conditions. Appl. Environ. Microbiol. 74:2111–2117. 10.1128/AEM.02442-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belliot G, Lavaux A, Souihel D, Agnello D, Pothier P. 2008. Use of murine norovirus as a surrogate to evaluate resistance of human norovirus to disinfectants. Appl. Environ. Microbiol. 74:3315–3318. 10.1128/AEM.02148-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Souza DH, Su X. 2010. Efficacy of chemical treatments against murine norovirus, feline calicivirus, and MS2 bacteriophage. Foodborne Pathog. Dis. 7:319–326. 10.1089/fpd.2009.0426 [DOI] [PubMed] [Google Scholar]
- 28.Lowther JA, Gustar NE, Powell AL, Hartnell RE, Lees DN. 2012. Two-year systematic study to assess norovirus contamination in oysters from commercial harvesting areas in the United Kingdom. Appl. Environ. Microbiol. 78:5812–5817. 10.1128/AEM.01046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woods JW, Burkhardt W., III 2010. Occurrence of norovirus and hepatitis A virus in U.S. oysters. Food Environ. Virol. 2:176–182. 10.1007/s12560-010-9040-7 [DOI] [Google Scholar]
- 30.Baert L, Mattison K, Loisy-Hamon F, Harlow J, Martyres A, Lebeau B, Stals A, Van Coillie E, Herman L, Uyttendaele M. 2011. Review: norovirus prevalence in Belgian, Canadian, and French fresh produce: a threat to human health? Int. J. Food Microbiol. 151:261–269. 10.1016/j.ijfoodmicro.2011.09.013 [DOI] [PubMed] [Google Scholar]
- 31.Lodder WJ, van den Berg H, Rutjes SA, de Roda Husman AM. 2010. Presence of enteric viruses in source waters for drinking water production in the Netherlands. Appl. Environ. Microbiol. 76:5965–5971. 10.1128/AEM.00245-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steyer A, Torkar KG, Gutiérrez-Aguirre I, Poljsak-Prijatelj M. 2011. High prevalence of enteric viruses in untreated individual drinking water sources and surface water in Slovenia. Int. J. Hyg. Environ. Health 214:392–398. 10.1016/j.ijheh.2011.05.006 [DOI] [PubMed] [Google Scholar]
- 33.Gassilloud B, Duval M, Schwartzbrod L, Gantzer C. 2003. Recovery of feline calicivirus infectious particles and genome from water: comparison of two concentration techniques. Water Sci. Technol. 47:97–101 [PubMed] [Google Scholar]
- 34.Haramoto E, Katayama H, Oguma K, Ohgaki S. 2007. Recovery of naked viral genomes in water by virus concentration methods. J. Virol. Methods 142:169–173. 10.1016/j.jviromet.2007.01.024 [DOI] [PubMed] [Google Scholar]
- 35.Nuanualsuwan S, Cliver DO. 2002. Pretreatment to avoid positive RT-PCR results with inactivated viruses. J. Virol. Methods 104:217–225. 10.1016/S0166-0934(02)00089-7 [DOI] [PubMed] [Google Scholar]