Abstract
Different lineages of fungi produce distinct classes of ergot alkaloids. Lysergic acid-derived ergot alkaloids produced by fungi in the Clavicipitaceae are particularly important in agriculture and medicine. The pathway to lysergic acid is partly elucidated, but the gene encoding the enzyme that oxidizes the intermediate agroclavine is unknown. We investigated two candidate agroclavine oxidase genes from the fungus Epichloë festucae var. lolii × Epichloë typhina isolate Lp1 (henceforth referred to as Epichloë sp. Lp1), which produces lysergic acid-derived ergot alkaloids. Candidate genes easH and cloA were expressed in a mutant strain of the mold Aspergillus fumigatus, which typically produces a subclass of ergot alkaloids not derived from agroclavine or lysergic acid. Candidate genes were coexpressed with the Epichloë sp. Lp1 allele of easA, which encodes an enzyme that catalyzed the synthesis of agroclavine from an A. fumigatus intermediate; the agroclavine then served as the substrate for the candidate agroclavine oxidases. Strains expressing easA and cloA from Epichloë sp. Lp1 produced lysergic acid from agroclavine, a process requiring a cumulative six-electron oxidation and a double-bond isomerization. Strains that accumulated excess agroclavine (as a result of Epichloë sp. Lp1 easA expression in the absence of cloA) metabolized it into two novel ergot alkaloids for which provisional structures were proposed on the basis of mass spectra and precursor feeding studies. Our data indicate that CloA catalyzes multiple reactions to produce lysergic acid from agroclavine and that combining genes from different ergot alkaloid pathways provides an effective strategy to engineer important pathway molecules and novel ergot alkaloids.
INTRODUCTION
Ergot alkaloids are agriculturally and pharmaceutically relevant secondary metabolites synthesized by several species of fungi. Historically, ergot alkaloids caused periodic mass poisonings due to infection of grain crops by the ergot fungus Claviceps purpurea (1). Agriculturally, ergot alkaloids in forage grasses colonized by endophytic Epichlë spp. (including many fungi recently realigned from the genus Neotyphodium [2]) reduce weight gain and fitness in grazing animals (3, 4). Clinically, the structural similarities of ergot alkaloids to neurotransmitters allow ergot alkaloids to treat cognitive and neurological maladies, including dementia, migraines, and Parkinson's disease, in addition to endocrine disorders such as type 2 diabetes and hyperprolactinemia (5–8). Indeed, the neurotransmitter-mimicking activities of ergot alkaloids are most infamously evident in the psychoactive drug lysergic acid diethylamide (LSD), a semisynthetic ergot alkaloid derivative.
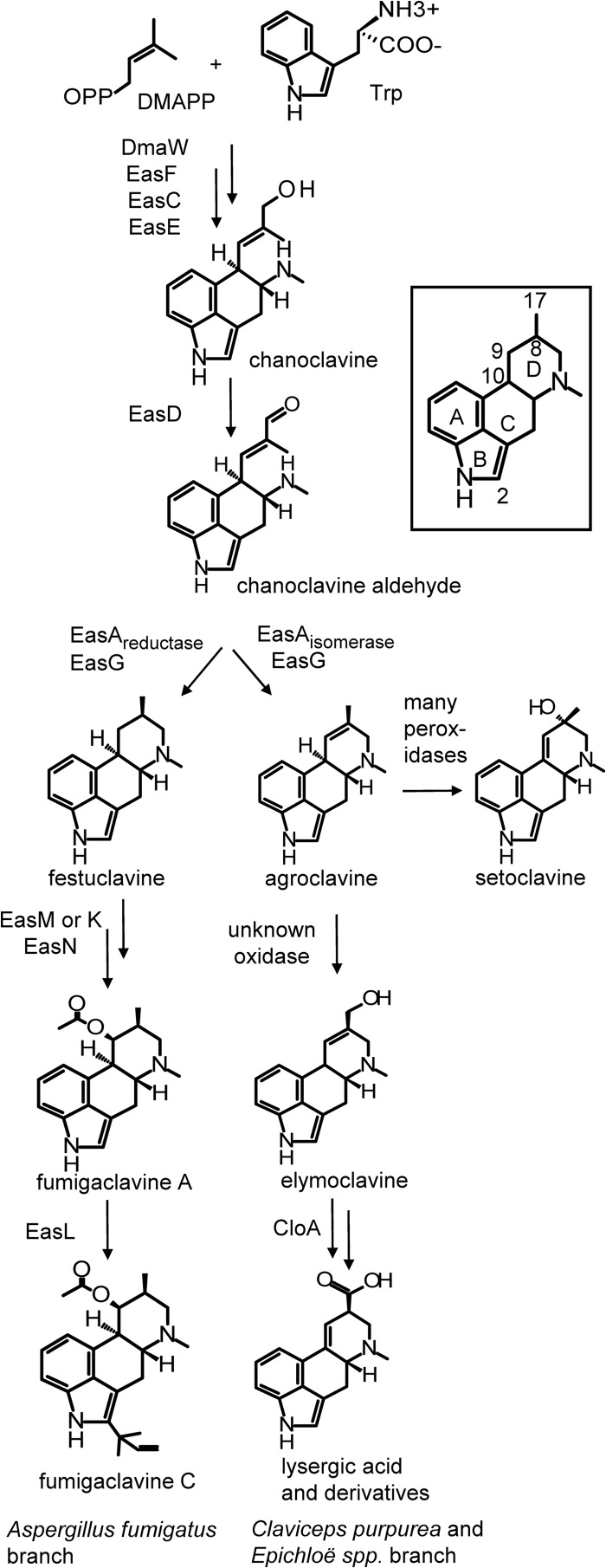
Representatives of two major families of fungi—the Clavicipitaceae and the Trichocomaceae—produce ergot alkaloids. All ergot alkaloid-producing fungi share early pathway steps before diverging to produce lineage-specific classes of ergot alkaloids (Fig. 1). Members of the Clavicipitaceae, including Claviceps purpurea and the endophytic Epichloë species such as E. festucae var. lolii × E. typhina isolate Lp1 (2, 9) (henceforth called Epichloë sp. Lp1), synthesize lysergic acid-based alkaloids in which the D ring of the ergoline nucleus is unsaturated between carbons 9 and 10 and carbon 17 is highly oxidized (Fig. 1) (4, 10, 11). Ergot alkaloid-producing fungi in the Trichocomaceae, such as the opportunistic human pathogen Aspergillus fumigatus, produce clavine-based derivatives in which the D ring is saturated and carbon 17 remains reduced as a methyl group (12, 13).
FIG 1.
Intermediates and products of the ergot alkaloid pathway (as composited from branches found in different fungi). The role of different alleles of easA (isomerase versus reductase encoding types) in controlling the branch point is indicated. Alkaloids with a 9,10 double bond (e.g., setoclavine and lysergic acid and its derivatives) often occur as diastereoisomers at position 8. Roles for enzymes discussed in the text or with genes illustrated in Fig. 2 are indicated near arrows. Double arrows indicate one or more omitted intermediates. The inset shows ring and position labeling referred to in the text. DMAPP, dimethylallylpyrophosphate.
The branch point of the pathway occurs during D ring closure. In A. fumigatus, the 8,9 double bond in chanoclavine aldehyde is reduced by the enzyme EasA reductase, allowing the aldehyde group free rotation to interact with the secondary amine to promote ring closure via Schiff base formation (14–16). The resulting iminium ion is subsequently reduced by EasG to form festuclavine (16, 17), which may be modified at carbons 9 and/or 2 to form various fumigaclavine derivatives (Fig. 1). Most ergot alkaloid-producing fungi in the Clavicipitaceae, however, synthesize the 8,9 unsaturated clavine agroclavine from chanoclavine aldehyde via the activity of an alternate version of EasA that acts as an isomerase rather than a reductase (15, 17). In C. purpurea and Epichloë spp., agroclavine is oxidized to form elymoclavine, and elymoclavine is further oxidized and isomerized to form lysergic acid (Fig. 1). Lysergic acid is then incorporated into ergopeptines and/or lysergic acid amides. Lysergic acid derivatives are the ergot alkaloids used for pharmaceutical development, but these compounds are produced exclusively in clavicipitaceous fungi and not in model organisms that would facilitate their modification and development.
The genetics of many steps in the ergot alkaloid pathway have been characterized; however, the identity of the gene encoding the oxidase that converts agroclavine to elymoclavine has remained elusive. All known genes involved in ergot alkaloid synthesis (eas genes) in both A. fumigatus and the Clavicipitaceae have been found in clusters (Fig. 2) (11, 18–24). The roles of many genes in eas clusters have been determined by gene knockout or by expression of coding sequences in Escherichia coli. Among the genes in eas clusters of lysergic acid-producing fungi, two genes stand out as candidates to encode the enzyme that oxidizes agroclavine. The gene labeled easH encodes a product with high similarity to dioxygenases (10, 19, 25); at the time this work was conducted, its role in the pathway had not been tested, but very recently EasH has been demonstrated to oxidize lysergyl-peptide lactams to facilitate their cyclolization to ergopeptines (25). This gene is present in both A. fumigatus and Epichloë spp.; however, the copy found in A. fumigatus (which lacks agroclavine and lysergic acid derivatives) is a pseudogene (10). The gene named cloA, for clavine oxidase (26), encodes a P450 monooxygenase that is a second candidate. Haarmann et al. (26) showed that CloA was required for oxidation of carbon 17 of elymoclavine during synthesis of lysergic acid and speculated that CloA also oxidized the same carbon in agroclavine. Only fungi that produce lysergic acid-derived alkaloids contain cloA in their eas clusters (18–24).
FIG 2.
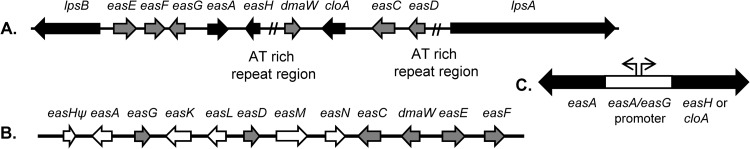
Ergot alkaloid synthesis (eas) clusters from Epichloë sp. Lp1 (A) and Aspergillus fumigatus (B) and design of transformation construct (C). (A) Epichloë sp. Lp1 eas cluster redrawn from the work of Schardl et al. (24); two AT-rich repeat regions (15 to 25 kb each) were compressed in the diagram to facilitate the presentation. (B) Aspergillus fumigatus eas cluster redrawn from the works of Unsöld and Li (22) and Coyle and Panaccione (18). Ψ, pseudogene. Genes unique to the Epichloë sp. Lp1 cluster are shown in black, and those unique to the A. fumigatus cluster are indicated in white. Genes common to both clusters are shown in gray. Although both clusters contain an allele of easA, the products of those alleles differ functionally, and so they differ in shading in their respective clusters. (C) General design of constructs generated by fusion PCR. Candidate genes were cloA or easH from Epichloë sp. Lp1. Black and white fragments correspond to Epichloë sp. Lp1 or A. fumigatus origin, as described above.
To test each candidate gene, a heterologous expression system was designed using an A. fumigatus easA knockout (easA KO) (15) as the host strain, which allowed for precise pathway control based on insertion of an agroclavine-specific allele of easA. Constructs used for transformation contained three elements: easA from Epichloë sp. Lp1, a bidirectional easA-easG promoter from A. fumigatus, and the candidate gene (either easH or cloA) amplified from Epichloë sp. Lp1 (Fig. 2). Coexpression of the Epichloë sp. Lp1 isomerase allele of easA in the easA reductase knockout background of A. fumigatus allowed accumulation of agroclavine (15, 17), which served as the substrate for the enzyme expressed from the candidate gene in the construct. This combinatorial approach allowed us to emulate the lysergic acid pathway in A. fumigatus and identify the gene encoding the agroclavine-oxidizing enzyme. Moreover, production of agroclavine in A. fumigatus in the absence of its oxidizing enzyme allowed accumulation of novel ergot alkaloids as a result of the activity of native A. fumigatus enzymes on agroclavine.
MATERIALS AND METHODS
Preparation of transformation constructs.
Each candidate oxidase gene (easH or cloA) was incorporated into a three-component construct that contained a bidirectional promoter from A. fumigatus (originating from the divergently transcribed genes easA and easG) centered between the candidate oxidase gene from Epichloë sp. Lp1 and the allele of easA from Epichloë sp. Lp1 (Fig. 2). To generate the easA-bidirectional promoter-easH construct, the following PCR amplifications were performed (Table 1). The easA portion was amplified from Epichloë sp. Lp1 DNA with primer combination 1 and Phire Hot Start II polymerase (Thermo Fisher, Waltham, MA). The initial denaturation was at 98°C for 30 s, followed by 35 cycles of 98°C (15 s), 60°C (15 s), and 72°C (30 s) and a final extension at 72°C for 60 s. The bidirectional easA-easG promoter of A. fumigatus was amplified from template DNA of A. fumigatus wild-type strain Af 293 by application of the PCR protocol described above for easA but with primer combination 2. Coding sequences and the 3′ untranslated region of easH were amplified from Epichloë sp. Lp1 DNA with the same PCR protocol as easA; however, the easH reaction was primed with primer combination 3. After each PCR, the products were cleaned with QIAquick columns (Qiagen, Gaithersburg, MD). To combine easA, the bidirectional promoter, and easH into a single construct, approximately equimolar amounts of each of the three PCR products were combined in a fusion PCR with Phire polymerase and primer combination 4. The PCR protocol had an initial denaturation at 98°C for 30 s, followed by 35 cycles of 98°C (15 s), 60°C (15 s), and 72°C (45 s) and a final elongation at 72°C for 60 s. The fusion product was purified by agarose gel electrophoresis and Qiaquick column chromatography prior to fungal transformation. The integrity of the construct was confirmed by DNA sequence analysis.
TABLE 1.
PCR primers used in this study and their products
| Primer combination | Primer sequences (5′→3′) | Product | Amplicon length(s) (bp) |
|---|---|---|---|
| 1 | TACTTGGTGGATTAGAAGCAATGTCAACTTCAAATCTTTTCAC + GCCATCATGACACCATTTGA | easA with promoter extensiona | 1,841 |
| 2 | GTGAAAAGATTTGAAGTTGACATTGCTTCTAATCCACCAAGTA + GGCTTGGATTGAACGGTCATGGTGCGGAGTGCCTACTCTA | Promoter with easA and easH extensions | 830 |
| 3 | TAGAGTAGGCACTCCGCACCATGACCGTTCAATCCAAGCC + CCTAGCTATCCATGCTCAAGC | easH with promoter extension | 1,135 |
| 4 | CGTATCACCGAGACAAAGAGG + TTGGCCATCACCTAACTATCTTG | easA-promoter-easH fusion construct | 3,250 |
| 5 | GTGAAAAGATTTGAAGTTGACATTGCTTCTAATCCACCAAGTA + GGATAACCATGGTAATATCATGGTGCGGAGTGCCTACTCTATAG | Promoter with easA and cloA extensions | 831 |
| 6 | CTATAGAGTAGGCACTCCGCACCATGATATTACCATGGTTATCC + AACACGCTAAGGGCAACAAG | cloA with promoter extension | 2,746 |
| 7 | CGTATCACCGAGACAAAGAGG + GCAACAAGCGATAAGCGTTAG | easA-promoter-cloA fusion construct | 4,905 |
| 8 | GCGAATGGATTTGATCTCGT + CCAGCGAGAGTTAGCAAGGT | easA internal sequences | 447 |
| 9 | CCAACGGTTCTCCCTTACTTC + GCACTATCTTGCCGCTCAGT | easH internal sequences | 611 |
| 10 | TTCCCGGCACGAGCTTTGCG + CTTAGAGTGCACCTCAGACGAC | cloA internal sequences | 296 (cDNA), 362 (gDNAb) |
Extension refers to incorporation of an additional 20 to 23 nucleotides at the 5′ end of a primer to add sequences that will facilitate a later fusion PCR.
gDNA, genomic DNA.
Generation of the construct containing Epichloë sp. Lp1 easA, the bidirectional promoter from A. fumigatus, and Epichloë sp. Lp1 cloA was similar to that described above for the easA-easH construct. However, the primers for each fragment were the following: combination 1 for easA, combination 5 for the promoter, and combination 6 for cloA (Table 1). Moreover, a different PCR protocol was used for the fusion reaction. This Phire reaction included primer combination 7 and had an initial denaturation temperature of 98°C for 30 s, followed by 35 cycles of 98°C (15 s), 62°C (15 s), and 72°C (65 s) and ending with an elongation period of 72°C for 60 s. The integrity of the construct was verified by DNA sequencing.
Fungal transformation.
Candidate oxidase constructs were cotransformed into the A. fumigatus easA KO strain (15), along with the selectable marker pAMD1, which contains the acetamidase gene of Aspergillus nidulans (27). Transformants capable of utilizing acetamide as a source of nitrogen were selected on acetamide medium (28). The spheroplast-polyethylene glycol (PEG)-based transformation protocol was based on previously described methods (15, 18).
mRNA analyses.
Cultures were grown in 50 ml of malt extract broth (Difco, Detroit, MI) in a 250-ml flask for 1 day with shaking at 80 rpm at 37°C to form a mat of hyphae on the surface of the broth. The mat was transferred to an empty petri dish and incubated at 37°C for an additional day to promote conidiation. RNA was extracted from approximately 100 mg of a conidiating colony with the Plant RNeasy kit (Qiagen), treated with DNase I (Qiagen), and reverse transcribed with Superscript II (Invitrogen, Carlsbad, CA). Template cDNA from each class of transformant was diluted 1:1,000 prior to amplification, and the presence of transcripts from individual genes was tested by PCR with primers (Table 1) specific for easA (combination 8), easH (combination 9), and cloA (combination 10). PCR was conducted with Phire polymerase in the following program: 98°C for 30 s, followed by 35 cycles of 98°C (10 s), 60°C (10 s), and 72°C (15 s), finishing with an elongation at 72°C for 60 s. RNA extraction and subsequent cDNA synthesis for all strains were performed concurrently to ensure consistency. The absence of genomic DNA in the easA-cloA cDNA preparation was confirmed by priming amplification with oligonucleotides that flank an intron, such that a second, larger product would be amplified from any contaminating genomic DNA. Of the three genes tested, only cloA contains introns.
Alkaloid analyses.
For quantitative analyses, six replicate colonies of one transformant of each type were grown on malt extract agar (15 g of malt extract broth medium plus 15 g of agar per liter) for 11 days. Samples of an approximately 50-mm2 surface area were collected with the broad end of a 1,000-μl pipette tip. Unless otherwise indicated, alkaloids were extracted with 98% methanol plus 2% acetic acid at 55°C for 30 min. Alternate extractions were conducted with 100% methanol or 10% aqueous ammonium acetate. Conidia in each extract were counted to provide an estimate of fungal biomass. Extracts clarified by centrifugation were then analyzed by reverse-phase high-performance liquid chromatography (HPLC) with fluorescence detection (12). Briefly, samples were separated on a 150-mm by 4.6-mm C18 column (Phenomenex Prodigy ODS3, 5-μm particle size; Torrance, CA) with a multilinear binary gradient of 5% acetonitrile plus 50 mM ammonium acetate to 75% acetonitrile plus 50 mM ammonium acetate. Lysergic acid standard was prepared by hydrolyzing 1 mg of ergotamine tartrate (Sigma-Aldrich, St. Louis, MO) in 100 μl of 1.2 M NaOH at 75°C for 6 h, followed by neutralization with a 1.2 M solution of HCl, purification on a C18 SPE column (Biotage, Charlotte, NC), and verification by liquid chromatography-mass spectrometry (LC/MS). Chanoclavine was obtained from Alfarma (Prague, Czech Republic), agroclavine was obtained from Fisher (Pittsburgh, PA), and setoclavine was prepared by oxidizing agroclavine as described previously (29, 30).
Ergot alkaloids were quantified by comparing peak areas to standard curves prepared from external standards of ergonovine (for alkaloids fluorescing at 310 nm and 410 nm) and agroclavine (for alkaloids fluorescing at 272 nm and 372 nm) and normalizing to the number of conidia extracted. Quantities of alkaloids among strains were compared by analysis of variance (ANOVA), and when ANOVA indicated a significant effect of fungal strain on alkaloid quantity (P < 0.05), means were separated by a Tukey-Kramer test with α set at 0.05. Statistical analyses were performed with JMP (SAS, Cary, NC).
For LC/MS analysis, cultures were grown for 1 week on malt extract agar. Conidiating cultures were washed repeatedly with 4 ml of HPLC-grade methanol. After pelleting of conidia and mycelia by centrifugation, the supernatant was concentrated to 100 μl in a vacuum centrifuge, and 10 μl was analyzed by LC/MS. Samples containing novel ergot alkaloids were analyzed as described previously (31) on a Thermo Fisher LCQ DecaXP LC/MS, whereas lysergic acid samples were analyzed on a Thermo Fisher Q Exactive ultrahigh-performance liquid chromatograph-mass spectrometer (UHPLC/MS) with electrospray ionization in positive mode, a spray voltage of 3.5 kV, and a capillary temperature of 300°C. Samples were chromatographed on a 50- by 2.1-mm Hypersil Gold column with a 1.9-μm particle size (Thermo Fisher) in a linear gradient of 100% solution A (5% acetonitrile plus 0.1% formic acid) to 100% solution B (75% acetonitrile plus 0.1% formic acid) over 10 min at a flow rate of 300 μl/min. Lysergic acid was eluted at approximately 3.3 min. After an initial full scan of m/z 50 to 570, ions of m/z 269 were fragmented and analyzed.
Precursor feeding study.
The ability of strains of A. fumigatus to convert agroclavine or setoclavine into novel ergot alkaloids was tested by feeding agroclavine to A. fumigatus strains. Agroclavine is readily oxidized to setoclavine by A. fumigatus (15, 29) and many other organisms (29, 30). Strains tested included A. fumigatus NRRL 164, which lacks a functional copy of the prenyl transferase gene easL, and the A. fumigatus easA KO strain and A. fumigatus Af 293, which have functional copies of easL (32). Six replicate cultures of each strain were grown from 60,000 conidia in 200 μl of malt extract broth in a 2-ml microcentrifuge tube. Cultures were supplemented with 37 nmol of agroclavine in 1 μl of methanol or with 1 μl of methanol as a control. An additional control was malt extract broth without conidia but with 1 μl of agroclavine (37 nmol). The cultures were incubated for 1 week at 37°C and then extracted by the addition of 300 μl of methanol along with 10 3-mm-diameter glass beads, followed by bead beating in a Fastprep 120 (Bio101, Carlsbad, CA) at 6 m/s for 30 s. Alkaloids were analyzed by HPLC with fluorescence detection as described above.
RESULTS
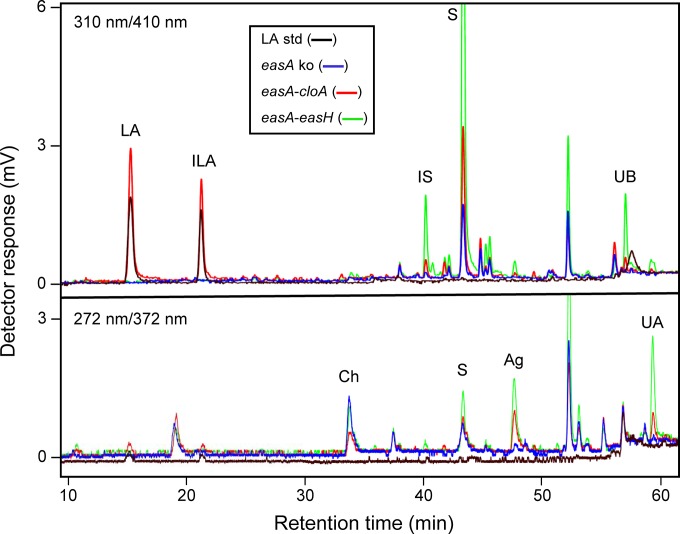
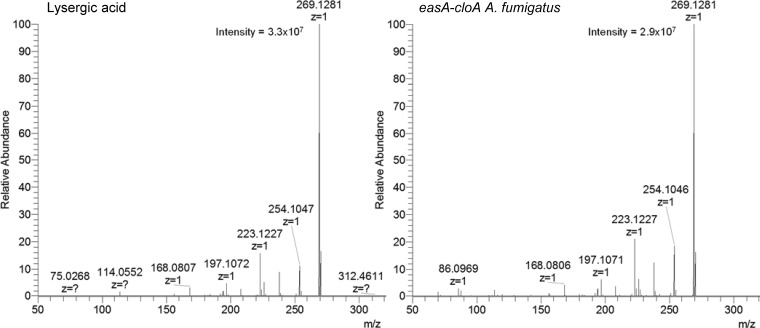
The Aspergillus fumigatus easA KO strain was successfully transformed with constructs for expressing either easA-easH or easA-cloA of Epichloë sp. Lp1. Evidence of successful transformation and expression of the Epichloë sp. Lp1 genes included accumulation of mRNA from both Epichloë sp. Lp1 genes introduced with a particular construct (Fig. 3). Further evidence of successful expression of the introduced genes was the altered ergot alkaloid profiles observed by HPLC with fluorescence detection (Fig. 4; Table 2). As described previously (15), the recipient strain, easA KO A. fumigatus, accumulated primarily chanoclavine and also small quantities of agroclavine (arising via a noncatalyzed isomerization of chanoclavine aldehyde [15, 33]) and larger quantities of its oxidation product setoclavine (Fig. 1). Transformants expressing Epichloë sp. Lp1 easA-easH accumulated chanoclavine and significantly more agroclavine and setoclavine than did the nontransformed recipient strain (P < 0.05), indicating successful expression of the easA allele of Epichloë sp. Lp1 without further modification of the ergot alkaloid profile by the product of easH. The same ergot alkaloid profile was observed in a previous study in which C. purpurea easA was expressed in the A. fumigatus easA KO strain (15). Strains that expressed the Epichloë sp. Lp1 easA-cloA construct also accumulated chanoclavine, agroclavine, and setoclavine but at levels comparable to those of the parent strain, easA KO A. fumigatus (Table 2). In addition, the easA-cloA-expressing strains accumulated a pair of polar compounds that were coeluted with lysergic acid and isolysergic acid standards (Fig. 4). The identity of the compounds as lysergic acid and its diastereoisomer was supported by UHPLC/MS analyses in which the easA-cloA strains produced parent ion and fragments consistent with those arising from the lysergic acid standard (Fig. 5). These data indicate that CloA catalyzes a cumulative six-electron oxidation of agroclavine to lysergic acid and that CloA or EasA isomerizes the 8,9 double bond in the D ring to the 9,10 position. The amount of lysergic acid extracted from easA-cloA strain cultures varied depending on the solvent used. Both 98% methanol plus 2% acetic acid and 10% (wt/vol) aqueous ammonium carbonate extracted significantly more (approximately 2.5-fold) lysergic acid than did unsupplemented methanol (P < 0.05).
FIG 3.
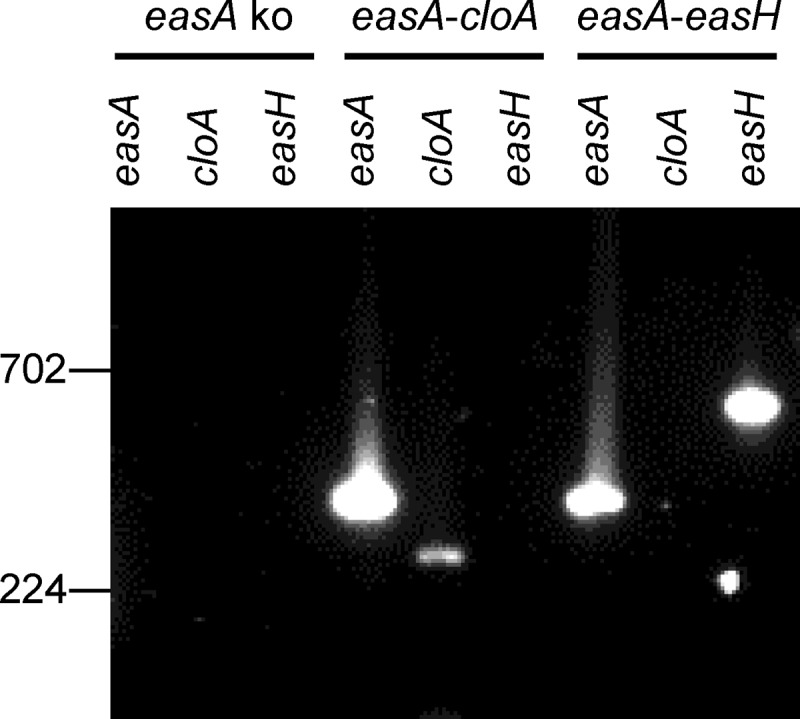
Qualitative reverse transcription-PCR (RT-PCR) demonstrating accumulation of mRNA from the indicated genes in A. fumigatus easA KO transformants. For the horizontal strain labels, easA ko refers to the nontransformed recipient strain and easA-cloA and easA-easH refer to transformants. Vertical gene labels refer to the Epichloë sp. Lp1 gene for which amplification was attempted in that lane. Primers are listed in Table 1. Each cDNA preparation was diluted 1:1,000 prior to amplification. The scale at the left indicates the relative mobility of relevant fragments from BstEII-digested bacteriophage lambda.
FIG 4.
Qualitative analysis of ergot alkaloids from transformed strains of A. fumigatus. Samples were analyzed with two fluorescence detectors; excitation and emission wavelengths are indicated. Lysergic acid and other ergot alkaloids with a 9,10 double bond fluoresce more strongly at 310 and 410 nm, whereas other ergot alkaloids fluoresce maximally with settings of 272 and 372 nm (12). Ergot alkaloids with 9,10 double bonds form diastereoisomers at carbon 8. Values for both diastereoisomers were added in quantitative analyses. Strain names and line colors are indicated in the key. Abbreviations: LA, lysergic acid; ILA, isolysergic acid; IS, isosetoclavine; S, setoclavine; UB, unknown B; Ch, chanoclavine; Ag, agroclavine; UA, unknown A. The peak eluting at 53 min is present in nontransformed A. fumigatus (15), and its fluorescence ratio indicates that it is not an ergot alkaloid (12).
TABLE 2.
Ergot alkaloid accumulation in cultures of modified strains of A. fumigatus
| Relevant genotype | Amt accumulated (amol/conidium)a |
|||||
|---|---|---|---|---|---|---|
| Chanoclavine | Agroclavine | Setoclavineb | Lysergic acidb | Unknown A | Unknown B | |
| easA KO | 0.42 ± 0.04 | 0.055 ± 0.01 B | 0.16 ± 0.01 B | NDc | ND | ND |
| easA-cloA | 0.58 ± 0.1 | 0.27 ± 0.04 B | 0.60 ± 0.07 B | 1.0 ± 0.1 | 0.16 ± 0.04 B | 0.062 ± 0.01 B |
| easA-easH | 0.59 ± 0.07 | 0.81 ± 0.1 A | 2.0 ± 0.2 A | ND | 0.38 ± 0.06 A | 0.22 ± 0.05 A |
Data are means for six cultures of the same strain ± SE; values followed by a different letter within a column differ significantly (α = 0.05) in a Tukey-Kramer test.
Values calculated from sums of both diastereoisomers.
ND, not detected; limit of detection = 0.01 amol/conidium.
FIG 5.
Mass spectra of lysergic acid standard and coeluting analyte from easA-cloA-transformed easA KO A. fumigatus. Spectra were collected with electrospray ionization in positive mode.
In addition to known ergot alkaloids described above, strains transformed with constructs containing either easA-easH or easA-cloA fragments accumulated two novel alkaloids referred to as unknown A and unknown B (Fig. 4; Table 2). The easA-easH-expressing strain, which accumulated significantly more agroclavine and setoclavine than did the easA-cloA-expressing strain, also accumulated significantly greater quantities of unknowns A and B (P < 0.05). Unknown A fluoresced more intensely at the 272- and 372-nm wavelength settings than at the 310- and 410-nm wavelength settings (Fig. 4), which is typical of ergot alkaloids lacking a double bond between positions 9 and 10 of the ergoline nucleus (12). In contrast, unknown B fluoresced more intensely at the 310- and 410-nm wavelengths than at the 272- and 372-nm wavelengths, indicating the presence of a 9,10 double bond (12). LC/MS analyses revealed that unknowns A and B had molecular ions with masses of 307.3 and 323.2, respectively (Fig. 6). The molecular ion of unknown A corresponds to the mass of [agroclavine + H]+ with an additional prenyl group, whereas the molecular ion of unknown B corresponds to the mass of [setoclavine + H]+ with an additional prenyl group.
FIG 6.
Mass spectra of two unknown alkaloids with hypothesized structures. Spectra were collected from LC/MS analyses with electrospray ionization in positive mode. The position and reverse mode of prenylation are hypothesized based on previously characterized activities of EasL (FgaPT1) (36) and on several closely related alkaloids from A. fumigatus (36, 37). Genetic evidence of prenylation is presented in Table 3.
To test the hypothesis that unknowns A and B correspond, respectively, to prenylated versions of agroclavine and setoclavine, agroclavine was fed to three isolates of A. fumigatus: (i) easA KO A. fumigatus, the transformation recipient, which was derived from A. fumigatus isolate FGSC A1141 and contains a functional copy of the ergot alkaloid prenyl transferase gene easL (32); (ii) A. fumigatus Af 293, a wild-type isolate that also has a functional copy of easL (32); and (iii) A. fumigatus NRRL 164, an isolate unable to produce the prenylated ergot alkaloid fumigaclavine C due to a mutation resulting in a premature stop codon in easL (32). Agroclavine-fed cultures of all isolates contained some nonmetabolized agroclavine and its oxidation product setoclavine. However, only easA KO A. fumigatus and A. fumigatus Af 293, which contain functional copies of the prenyl transferase gene easL, accumulated unknowns A and B (Table 3). These data demonstrate a dependency on agroclavine for synthesis of the unknowns and are consistent with a role for the prenyl transferase encoded by easL in their biosynthesis.
TABLE 3.
Ergot alkaloids in A. fumigatus strains fed 37 nmol of agroclavine and controls
| Strain/treatment | Amt (nmol/culture) of alkaloida |
|||
|---|---|---|---|---|
| Agroclavine | Setoclavineb | Unknown A | Unknown B | |
| Af 293/agroclavine | 22 ± 1 B | 2.5 ± 0.1 A | 0.56 ± 0.04 A | 0.040 ± 0.001 A |
| NRRL 164/agroclavine | 24 ± 2 B | 2.1 ± 0.2 AB | NDc | ND |
| easA KO strain/agroclavine | 21 ± 0.3 B | 1.9 ± 0.08 B | 0.22 ± 0.03 B | 0.0078 ± 0.0007 B |
| Medium/agroclavine | 31 ± 0.5 A | 0.89 ± 0.05 C | ND | ND |
| Af 293/methanol | ND | ND | ND | ND |
| NRRL 164/methanol | ND | ND | ND | ND |
| easA KO strain/methanol | 0.041 ± 0.002 C | 0.0023 ± 0.0003 D | ND | ND |
Data are means for six samples ± SE; values followed by a different letter within a column differ significantly (α = 0.05) in a Tukey-Kramer test.
Values calculated from sums of both diastereoisomers.
ND, not detected; limit of detection = 0.01 amol/conidium.
DISCUSSION
By expressing CloA along with the isomerase form of EasA of Epichloë sp. Lp1 in an easA reductase knockout mutant of A. fumigatus, we have reprogrammed A. fumigatus to emulate the ergot alkaloid pathway of lysergic acid-producing fungi. Consistent with the data of Coyle et al. (15), both easA-easH-expressing and easA-cloA-expressing mutants produced agroclavine, as a result of expressing the isomerase form of Epichloë sp. Lp1 easA. In addition, both types of transformants accumulated setoclavine and isosetoclavine, diastereoisomers formed by the oxidation of agroclavine by endogenous peroxidases in A. fumigatus and dozens of other fungi and plants (15, 29, 30). However, comparison of the easA-cloA-expressing strain with the easA-easH-expressing strain provides evidence of an extended, multistep role for CloA in biosynthesis of lysergic acid. The data indicate that CloA performs multiple catalytic steps: a two-electron oxidation of agroclavine to elymoclavine and then a pair of two-electron oxidations to convert elymoclavine to lysergic acid, presumably via an undetected aldehyde intermediate (34). The role of CloA in catalyzing multiple oxidations to form paspalic acid or lysergic acid was previously hypothesized by Haarmann et al. (26). Our data also suggest that CloA or the coexpressed EasA catalyzes the double-bond isomerization, from position 8,9 to 9,10.
The lack of detectable elymoclavine, or any other intermediates in the oxidation series from agroclavine to lysergic acid, in our positive transformants indicates that CloA may bind agroclavine and execute successive oxidations and an isomerization before releasing lysergic acid. The lack of detectable paspalic acid (which is the 8,9 double-bond isomer of lysergic acid) in our lysergic acid-positive transformants indicates that the double-bond isomerization, from position 8,9 (as in agroclavine and elymoclavine) to position 9,10 (as in lysergic acid and derivatives thereof), occurs while substrate is bound to CloA. Moreover, young cultures (<3 days old) yielded lesser quantities of lysergic acid when extracted with methanol but significantly more when extracted with acetic acid-supplemented methanol or with ammonium carbonate. One interpretation of this observation is that the acid or base helped denature CloA, releasing otherwise bound product for detection. While some researchers have hypothesized that isomerization of paspalic acid to lysergic acid happens spontaneously over time (26), the complete lack of paspalic acid in our cultures and the existence of natural variants of Claviceps paspali that accumulate large quantities of paspalic acid and lesser quantities of lysergic acid (35) indicate that in a fully functioning lysergic acid pathway the double-bond isomerization is enzymatically catalyzed.
An unexpected and important finding of this study was the accumulation of two novel alkaloids, unknowns A and B, from both transformants but in greater quantities in the easA-easH transformants, which accumulated greater concentrations of agroclavine and setoclavine. The proposed structure of each unknown, with a prenyl group added in reverse mode to carbon 2 of either agroclavine or setoclavine, comes from five observations. (i) The accumulation of the compounds was dependent on availability of agroclavine provided by either feeding or biosynthesis. (ii) The accumulation of the compounds was restricted to strains of A. fumigatus that have a functional copy of the prenyl transferase EasL (also called FgaPT1), which is typically responsible for reverse prenylating fumigaclavine A to fumigaclavine C (Fig. 1) (36). (iii) The molecular weights of the unknowns are consistent with agroclavine and setoclavine with an additional moiety of 68 atomic mass units (amu), which corresponds to the mass of a prenyl group. (iv) The fluorescence properties of each analyte and their long retention times in reverse-phase HPLC are consistent with predicted properties of prenylated forms of agroclavine and setoclavine. (v) The recent discovery of reverse-prenylated versions of festuclavine and fumigaclavine B in A. fumigatus (37) indicates that EasL (FgaPT1) accepts other ergot alkaloids as the substrates. While our precursor-feeding and LC/MS data strongly indicate that EasL (FgaPT1) prenylates agroclavine and setoclavine, our proposal that the prenyl group is attached at position 2 in reverse mode can only be hypothesized on the basis of previous mechanistic studies of this enzyme (36) and the consistency of this mode and position of prenylation with other ergot alkaloids of A. fumigatus (36, 37). Interestingly, we saw no evidence of prenylation of lysergic acid in easA-cloA-expressing A. fumigatus, indicating that the carboxylic acid at carbon 17 interfered with enzyme binding or activity. Our data demonstrate that a combinatorial approach based on expression of enzymes from a different lineage of ergot alkaloid producers in A. fumigatus can yield novel ergot alkaloids. The biological activities of the novel alkaloids are unknown and will require further study. Based on their structures, however, it is possible that unknowns A and B will have activities similar to those of fumigaclavine C, which has anti-inflammatory activity (38).
In summation, we have reprogrammed A. fumigatus to produce lysergic acid by heterologous expression of an isomerase allele of easA in a native easA knockout strain (to produce agroclavine substrate) concomitant with the expression of the P450 monooxygenase gene cloA. This synthetic biology approach presents several intriguing possibilities. The production of lysergic acid in an experimentally tractable and fast-growing organism such as A. fumigatus is significant because lysergic acid is used as a base for modification in numerous pharmaceutical products, including the drugs nicergoline, cabergoline, and metergoline. Although A. fumigatus is an opportunistic human pathogen, it can be disarmed readily by knocking out confirmed virulence genes (39). Moreover, combinations of enzymes from different lineages gave rise to novel ergot alkaloids, such as the prenylated forms observed in our study. The apparent relaxed specificity of the prenyl transferase EasL may be exploited further for production of other alkaloids. Finally, the expression platform may be conducive to expressing alleles from lineages of fungi producing alternate ergot alkaloids, such as the pharmaceutically relevant dihydroergot alkaloids.
ACKNOWLEDGMENTS
Research with lysergic acid was conducted with licenses from the West Virginia Board of Pharmacy (TI0555042) and the U.S. Drug Enforcement Agency (RP0463353). Technology described in this article is embodied in U.S. provisional patent application no. 62/012,658. We gratefully acknowledge the use of the WVU Shared Research Facilities.
This work was supported by grant 2012-67013-19384 from USDA NIFA and Hatch funds and published with permission of the West Virginia Agriculture and Forestry Experiment Station.
Footnotes
Published ahead of print 8 August 2014
This article is scientific article number 3214 from the West Virginia Agriculture and Forestry Experiment Station.
REFERENCES
- 1.Matossian MK. 1989. Poisons of the past: molds, epidemics, and history. Yale University Press, New Haven, CT [Google Scholar]
- 2.Leuchtmann A, Bacon CW, Schardl CL, White JF, Tadych M. 2014. Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 106:202–215. 10.3852/106.2.202 [DOI] [PubMed] [Google Scholar]
- 3.Schardl CL, Young CA, Faulkner JR, Florea S, Pan J. 2012. Chemotypic diversity of epichloae, fungal symbionts of grasses. Fungal Ecol. 5:331–344. 10.1016/j.funeco.2011.04.005 [DOI] [Google Scholar]
- 4.Panaccione DG, Beaulieu WT, Cook D. 2014. Bioactive alkaloids in vertically transmitted fungal endophytes. Funct. Ecol. 28:299–314. 10.1111/1365-2435.12076 [DOI] [Google Scholar]
- 5.Baskys A, Hou AC. 2007. Vascular dementia: pharmacological treatment approaches and perspectives. Clin. Interv. Aging 2:327–335 [PMC free article] [PubMed] [Google Scholar]
- 6.Morren JA, Galvez-Jimenez N. 2010. Where is dihydroergotamine mesylate in the changing landscape of migraine therapy? Expert Opin. Pharmacother. 11:3085–3093. 10.1517/14656566.2010.533839 [DOI] [PubMed] [Google Scholar]
- 7.Perez-Lloret S, Rascol O. 2010. Dopamine receptor agonists for the treatment of early or advanced Parkinson's disease. CNS Drugs 24:941–968. 10.2165/11537810-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 8.Kerr JL, Timpe EM, Petkewicz KA. 2010. Bromocriptine mesylate for glycemic management in type 2 diabetes mellitus. Ann. Pharmacother. 44:1777–1785. 10.1345/aph.1P271 [DOI] [PubMed] [Google Scholar]
- 9.Schardl CL, Leuchtmann A, Tsai HF, Collett MA, Watt DM, Scott DB. 1994. Origin of a fungal symbiont of perennial ryegrass by interspecific hybridization of a mutualist with the ryegrass choke pathogen, Epichloë typhina. Genetics 136:1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schardl C, Panaccione DG, Tudzynski P. 2006. Ergot alkaloids—biology and molecular biology. Alkaloids Chem. Biol. 63:45–86. 10.1016/S1099-4831(06)63002-2 [DOI] [PubMed] [Google Scholar]
- 11.Lorenz N, Haarmann T, Pazoutova S, Jung M, Tudzynski P. 2009. The ergot alkaloid gene cluster: functional analyses and evolutionary aspects. Phytochemistry 70:1822–1832. 10.1016/j.phytochem.2009.05.023 [DOI] [PubMed] [Google Scholar]
- 12.Panaccione DG, Ryan KL, Schardl CL, Florea S. 2012. Analysis and modification of ergot alkaloid profiles in fungi. Methods Enzymol. 515:267–290. 10.1016/B978-0-12-394290-6.00012-4 [DOI] [PubMed] [Google Scholar]
- 13.Wallwey C, Li S-M. 2011. Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat. Prod. Rep. 28:496–510. 10.1039/c0np00060d [DOI] [PubMed] [Google Scholar]
- 14.Cheng JZ, Coyle CM, Panaccione DG, O'Connor SE. 2010. A role for old yellow enzyme in ergot alkaloid biosynthesis. J. Am. Chem. Soc. 132:1776–1777. 10.1021/ja910193p [DOI] [PubMed] [Google Scholar]
- 15.Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG. 2010. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Appl. Environ. Microbiol. 76:3898–3903. 10.1128/AEM.02914-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallwey C, Matuschek M, Li S-M. 2010. Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I to chanoclavine-I aldehyde catalyzed by a short-chain alcohol dehydrogenase FgaDH. Arch. Microbiol. 192:127–134. 10.1007/s00203-009-0536-1 [DOI] [PubMed] [Google Scholar]
- 17.Cheng JZ, Coyle CM, Panaccione DG, O'Connor SE. 2010. Controlling a structural branch point in ergot alkaloid biosynthesis. J. Am. Chem. Soc. 132:12835–12837. 10.1021/ja105785p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coyle CM, Panaccione DG. 2005. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl. Environ. Microbiol. 71:3112–3118. 10.1128/AEM.71.6.3112-3118.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haarmann T, Machado C, Lübbe Y, Correia T, Schardl CL, Panaccione DG, Tudzynski P. 2005. The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution. Phytochemistry 66:1312–1320. 10.1016/j.phytochem.2005.04.011 [DOI] [PubMed] [Google Scholar]
- 20.Fleetwood DJ, Scott B, Lane GA, Tanaka A, Johnson RD. 2007. A complex ergovaline gene cluster in Epichloe endophytes of grasses. Appl. Environ. Microbiol. 73:2571–2579. 10.1128/AEM.00257-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenz N, Wilson EV, Machado C, Schardl CL, Tudzynski P. 2007. Comparison of ergot alkaloid biosynthesis gene clusters in Claviceps species indicates loss of late pathway steps in evolution of C. fusiformis. Appl. Environ. Microbiol. 73:7185–7191. 10.1128/AEM.01040-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unsöld IA, Li SM. 2005. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 151:1499–1505. 10.1099/mic.0.27759-0 [DOI] [PubMed] [Google Scholar]
- 23.Schardl CL, Young CA, Hesse U, Amyotte SG, Andreeva K, Calie PJ, Fleetwood DJ, Haws DC, Moore N, Oeser B, Panaccione DG, Schweri KK, Voisey CR, Farman ML, Jaromczyk JW, Roe BA, O'Sullivan DM, Scott B, Tudzynski P, An Z, Arnaoudova EG, Bullock CT, Charlton ND, Chen L, Cox M, Dinkins RD, Florea S, Glenn AE, Gordon A, Güldener U, Harris DR, Hollin W, Jaromczyk J, Johnson RD, Khan AK, Leistner E, Leuchtmann A, Li C, Liu JG, Liu J, Liu M, Mace W, Machado C, Nagabhyru P, Pan J, Schmid J, Sugawara K, Steiner U, Takach JE, Tanaka E, et al. 2013. Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 9:e1003323. 10.1371/journal.pgen.1003323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schardl CL, Young CA, Pan J, Florea S, Takach JE, Panaccione DG, Farman ML, Webb JS, Jaromczyk J, Charlton ND, Nagabhyru P, Chen L, Shi C, Leuchtmann A. 2013. Currencies of mutualisms: sources of alkaloid genes in vertically transmitted epichloae. Toxins 5:1064–1088. 10.3390/toxins5061064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Havemann J, Vogel D, Loll B, Keller U. 2014. Cyclolization of D-lysergic acid alkaloid peptides. Chem. Biol. 21:146–155. 10.1016/j.chembiol.2013.11.008 [DOI] [PubMed] [Google Scholar]
- 26.Haarmann T, Ortel I, Tudzynski P, Keller U. 2006. Identification of the cytochrome P450 monooxygenase that bridges the clavine and ergoline alkaloid pathways. Chembiochem 7:645–652. 10.1002/cbic.200500487 [DOI] [PubMed] [Google Scholar]
- 27.Hynes MJ, Corrick CM, King JA. 1983. Isolation of genomic clones containing the amdS gene of Aspergillus nidulans and their use in the analysis of structural and regulatory mutations. Mol. Cell. Biol. 3:1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panaccione DG, Scott-Craig JS, Pocard JA, Walton JD. 1992. A cyclic peptide synthetase gene required for pathogenicity of the fungus Cochliobolus carbonum on maize. Proc. Natl. Acad. Sci. U. S. A. 89:6590–6594. 10.1073/pnas.89.14.6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Béliveau J, Ramstad E. 1967. 8-Hydroxylation of agroclavine and elymoclavine by fungi. Llyodia 29:234–238 [Google Scholar]
- 30.Panaccione DG, Tapper BA, Lane GA, Davies E, Fraser K. 2003. Biochemical outcome of blocking the ergot alkaloid pathway of a grass endophyte. J. Agric. Food Chem. 51:6429–6437. 10.1021/jf0346859 [DOI] [PubMed] [Google Scholar]
- 31.Ryan KL, Moore CT, Panaccione DG. 2013. Partial reconstruction of the ergot alkaloid pathway by heterologous gene expression in Aspergillus nidulans. Toxins 5:445–455. 10.3390/toxins5020445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson SL, Panaccione DG. 2012. Chemotypic and genotypic diversity in the ergot alkaloid pathway of Aspergillus fumigatus. Mycologia 104:804–812. 10.3852/11-310 [DOI] [PubMed] [Google Scholar]
- 33.Matuschek M, Wallwey C, Xie X, Li SM. 2011. New insights into ergot alkaloid biosynthesis in Claviceps purpurea: an agroclavine synthase EasG catalyses, via a non-enzymatic adduct with reduced glutathione, the conversion of chanoclavine-I aldehyde to agroclavine. Org. Biomol. Chem. 9:4328–4335. 10.1039/c0ob01215g [DOI] [PubMed] [Google Scholar]
- 34.Lin CL, Blair GE, Cassady JM, Gröger D, Maier W, Floss HG. 1973. Biosynthesis of ergot alkaloids: synthesis of 6-methyl-8-acetoxymethylene-9-ergolene and its incorporation into ergotoxine by Claviceps. J. Org. Chem. 38:2249–2251. 10.1021/jo00952a035 [DOI] [PubMed] [Google Scholar]
- 35.Rutschmann J, Kobel H, Schreier E. 1967. Heterocyclic carboxylic acids and their production. US patent 3,314,961
- 36.Unsöld IA, Li S-M. 2006. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: gene expression, purification, and characterization of fumigaclavine C synthase FgaPT1. Chembiochem 7:158–164. 10.1002/cbic.200500318 [DOI] [PubMed] [Google Scholar]
- 37.Ge HM, Yu ZG, Zhang J, Wu JH, Tan RX. 2009. Bioactive alkaloids from endophytic Aspergillus fumigatus. J. Nat. Prod. 72:753–755. 10.1021/np800700e [DOI] [PubMed] [Google Scholar]
- 38.Du RH, Li EG, Cao Y, Song YC, Tan RX. 2011. Fumigaclavine C inhibits tumor necrosis factor α production via suppression of Toll-like receptor 4 and nuclear factor κB activation in macrophages. Life Sci. 89:235–240. 10.1016/j.lfs.2011.06.015 [DOI] [PubMed] [Google Scholar]
- 39.Tsai H-F, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]