Abstract
The antimicrobial action of the curing agent sodium nitrite (NaNO2), which is added as a preservative to raw meat products, depends on its conversion to nitric oxide and other reactive nitrogen species under acidic conditions. In this study, we used RNA sequencing to analyze the acidified-NaNO2 shock and adaptive responses of Salmonella enterica serovar Typhimurium, a frequent contaminant in raw meat, considering parameters relevant for the production of raw-cured sausages. Upon a 10-min exposure to 150 mg/liter NaNO2 in LB (pH 5.5) acidified with lactic acid, genes involved in nitrosative-stress protection, together with several other stress-related genes, were induced. In contrast, genes involved in translation, transcription, replication, and motility were downregulated. The induction of stress tolerance and the reduction of cell proliferation obviously promote survival under harsh acidified-NaNO2 stress. The subsequent adaptive response was characterized by upregulation of NsrR-regulated genes and iron uptake systems and by downregulation of genes involved in anaerobic respiratory pathways. Strikingly, amino acid decarboxylase systems, which contribute to acid tolerance, displayed increased transcript levels in response to acidified NaNO2. The induction of systems known to be involved in acid resistance indicates a nitrite-mediated increase in the level of acid stress. Deletion of cadA, which encodes lysine decarboxylase, resulted in increased sensitivity to acidified NaNO2. Intracellular pH measurements using a pH-sensitive green fluorescent protein (GFP) variant showed that the cytoplasmic pH of S. Typhimurium in LB medium (pH 5.5) is decreased upon the addition of NaNO2. This study provides the first evidence that intracellular acidification is an additional antibacterial mode of action of acidified NaNO2.
INTRODUCTION
Nontyphoidal Salmonella gastroenteritis, with an estimated 93.8 million cases worldwide each year, is a major health burden (1). In the vast majority of these cases (an estimated 80.3 million), the organism is food borne (1). Salmonella bacteria naturally reside in the intestinal tracts of animals, which explains their high prevalence in animal produce, especially in meat (2). Pig and bovine meats, which are traditionally used in the production of raw fermented sausages in Germany (3), are often associated with Salmonella enterica subsp. enterica serovar Typhimurium (2). To prevent or control the growth of Salmonella and other pathogenic bacteria in foodstuffs, different measures, such as acidification, low temperatures, and food additives, are combined in the manufacturing process, a concept known as hurdle technology (4). An important hurdle during the production of raw fermented sausages is the curing agent sodium nitrite (NaNO2), which is especially critical at the early stages of ripening (5). According to European Union legislation (directive 2006/52/EC), 150 mg NaNO2 per kg meat is the maximum ingoing amount permitted in cured meat products (6). Under the mildly acidic conditions (pH ∼5.5) prevalent in raw meat and influenced by additives such as ascorbate and salt, NO2− is converted to diverse reactive nitrogen species (RNS), such as nitrous acid (HNO2), dinitrogen trioxide (N2O3), and, most importantly, nitric oxide (NO) (7, 8). NO, in turn, modifies pigments and proteins in the meat, leading to the typical color and flavor of cured meat products, and further acts as a scavenger of lipid- or protein-derived radicals (8, 9). Moreover, NO and its congeners exhibit antimicrobial activity. RNS are also crucial players in the host's immune response to Salmonella infection (reviewed by Henard and Vázquez-Torres [10]). NO has been shown to modify multiple cellular targets and interferes with crucial metabolic processes, including key enzymes of the tricarboxylic acid cycle (11), DNA (12), respiration (13), and the acid tolerance response (14). To avoid the deleterious effects of NO, S. Typhimurium is able to detoxify NO aerobically or anaerobically via the flavohemoglobin HmpA (15), the flavorubredoxin NorV (16), and the periplasmic cytochrome c nitrite reductase NrfA (17). Moreover, genes under the control of the NO-responsive regulator NsrR have been suggested to be active in nitrosative-stress protection (18). Whereas several studies using different sources of nitrosative stress, including acidified nitrite and S-nitrosoglutathione (GSNO), have been performed to study the stress response in Escherichia coli (19–22), only a few transcriptional studies (11, 14), all of which used NO delivered by donor compounds, have been conducted for S. Typhimurium. However, in food matrices such as NaNO2-cured raw sausages, a variety of reactive intermediates apart from NO are produced upon acidification of NO2− (8) and might additionally influence the growth of Salmonella. Nitrous acid (HNO2), for example, has been suggested to be an important player in the antifungal action of acidified nitrite by causing intracellular acidification (23). It is not known whether this mechanism is also active in bacteria. Furthermore, weak organic acids, such as lactic acid, account for the mildly acidic pH in meat, in contrast to the strong inorganic acid HCl in the stomach. The type of acidulant might also influence the reactivity of NO2− and thereby that of the transcriptome, but no studies using NO2− acidified by lactic acid have been conducted.
In this study, we performed RNA sequencing (RNA-seq) of S. Typhimurium treated with 150 mg/liter NaNO2 acidified with lactic acid at an ambient temperature of 24°C in order to gain insight into both the shock and adaptive responses of this pathogen to acidified-nitrite stress under conditions relevant for food processing. Moreover, we identified the lysine decarboxylase CadA as an important player in the acidified-nitrite stress tolerance of S. Typhimurium, and we provide evidence that intracellular acidification may constitute an additional antibacterial mode of action of acidified nitrite.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are summarized in Table 1. Bacterial cells were routinely grown in LB broth (Lennox) (neutral LB; pH ∼7) or agar (1.5%, wt/vol) (Oxoid, Wesel, Germany). To test the impact of acidified sodium nitrite (NaNO2) on S. Typhimurium, LB broth adjusted to pH 5.5 with lactic acid (90%; Merck, Darmstadt, Germany) (LB [pH 5.5]) prior to autoclaving was used. If necessary, the following antibiotics were added: ampicillin at 150 μg/ml (USB, Cleveland, OH, USA); tetracycline at 17.5 μg/ml (Sigma-Aldrich, Taufkirchen, Germany). All strains were stored at −80°C in LB broth containing 20% glycerol.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| WT | Salmonella enterica subsp. enterica serovar Typhimurium 14028; wild-type strain | DSM 19587 |
| WT pBR322 | WT strain carrying empty plasmid pBR322 | This study |
| ΔcadA pBR322 | Strain with in-frame deletion of the WT STM14_3138 DNA sequence encoding amino acids 7 to 703 of CadA (ΔcadA), carrying empty plasmid pBR322 | This study |
| ΔcadA-comp | ΔcadA strain carrying complementation plasmid pBR322-cadA | This study |
| WT pEGFP | WT strain carrying pEGFP for intracellular pH measurements | This study |
| Plasmids | ||
| pBR322 | pMB1 replicon cloning vector; Ampr Tetr | 24 |
| pBR322-cadA | Complementation plasmid containing the cadA coding sequence under the control of its native promoter, PcadBA; Ampr | This study |
| pEGFP | EGFP expression vector; Ampr | Clontech, Germany |
Ampr, ampicillin resistance; Tetr, tetracycline resistance.
Construction and complementation of a cadA deletion mutant.
The cadA in-frame deletion mutant (ΔcadA strain) was constructed in the genetic background of S. Typhimurium 14028 (the wild-type [WT] strain) using the lambda Red recombinase method (25) as described previously (26). The specific oligonucleotides used for the construction of the ΔcadA strain, del_cadA_F, del_cadA_R, test_cadA_F, and test_cadA_R, are listed in Table 2. For the complementation of the ΔcadA mutant, a PCR product corresponding to the coding sequence of cadA under the control of its own promoter was introduced at the HindIII and BamHI cloning sites of pBR322. Since cadA is the second gene of the cadBA operon, it was fused to its promoter via an artificially generated 84-bp “scar” sequence of pKD4 that usually remains after FLP-mediated excision of the antibiotic cassette (25). This cloning procedure is based on a previously described system of complementation of the ΔcadA mutation (27). This was done by 3′ overhangs on primers C_cadA_B and C_cadA_C corresponding to the scar sequence. Briefly, the cadBA promoter region and the cadA coding sequence were amplified using primer combinations C_cadA_A/C_cadA_B and C_cadA_C/C_cadA_D (Table 2), respectively. The PCR products were ligated via a natural XbaI restriction site in the “scar” sequence, and the corresponding fragment was amplified using primers C_cadA_A and C_cadA_D. The product was cloned into vector pBR322, resulting in the complementation vector pBR322-cadA. Finally, for the construction of the complementation mutant (ΔcadA-comp), pBR322-cadA was introduced into S. Typhimurium ΔcadA. As controls, plasmid pBR322 was transformed into the ΔcadA as well as the WT strain, resulting in the ΔcadA pBR322 and WT pBR322 strains, respectively.
TABLE 2.
Oligonucleotides used for DNA manipulationsa
| Primer | Sequence (5′–3′)b |
|---|---|
| del_cadA_F | CGGGAGGGGCCCACTTTACCAGGAACAAGACTATGAACGTTATTGCTATCgtgtaggctggagctgcttc |
| del_cadA_R | CTTCCCTTTGGTACTTATTTCGTATTTTCTTTCAGCACCTTAACGGTGTAcatatgaatatcctcctta |
| test_cadA_F | CTTCGAACTCTCCGGCAC |
| test_cadA_R | GTAAGGCACGCATGCCGT |
| C_cadA_A (HindIII)c | AATAAGCTTATTTAACGCTGAACCATGAC |
| C_cadA_B (XbaI) | ttctctagaaagtataggaacttcgaagcagctccagcctacacGTTCATTTCTCCTGAGCTGT |
| C_cadA_C (XbaI) | ctttctagagaataggaacttcggaataggaactaaggaggatattcatatgCCGCTAACTCCTTTTTCTCA |
| C_cadA_D (BamHI)c | AATGGATCCCGCCACGATGTAAAAAATCG |
The oligonucleotides were purchased from Eurofins MWG Operon (Ebersberg, Germany).
Priming sites for, or sequence parts corresponding to, pKD4 are in lowercase. Restriction enzyme sites are underlined.
The primer sequence binding to the S. Typhimurium 14028 genome is taken from reference (27).
Growth analysis using Bioscreen C.
In vitro growth analysis of bacterial strains in a Bioscreen C system was performed by following the protocol for aerobic cultures as described previously (26). The time needed to reach an optical density at 600 nm (OD600) of 0.6, corresponding to the half-maximum OD600, was used as the parameter for displaying growth differences. Mean values and standard deviations were calculated from the results of three independent biological experiments, each including technical duplicates.
Screening of a S. Typhimurium insertion mutant library for phenotypes sensitive to acidified NaNO2.
A S. Typhimurium insertion mutant library was screened for phenotypes sensitive to acidified nitrite. This random library was constructed by Knuth et al. (28) and was generously provided by Thilo M. Fuchs. The library comprises insertion mutants derived from homologous recombination of the temperature-sensitive vector pIDM1, containing small chromosomal fragments, with the respective chromosomal site at a nonpermissive temperature (37°C). For the purpose of screening, growth curves of single insertion mutants in LB (pH 5.5) without NaNO2 or with 150 mg/liter NaNO2 at 37°C were recorded and analyzed in a Bioscreen C system. In mutants showing a nitrite-sensitive phenotype, the site of insertion was determined via amplification and partial sequencing of the chromosomal fragments cloned into pIDM1.
Cell harvesting and RNA extraction.
A shaken (160 rpm) overnight culture in LB broth was diluted 1:100 in fresh LB broth (pH 5.5) and was grown at 24°C with shaking (160 rpm). Growth was monitored by measuring the OD600 (Ultrospec 2000 UV/visible spectrophotometer; Pharmacia Biotech, Freiburg, Germany). To analyze the shock response to acidified nitrite, a 150-ml culture in a 500-ml baffled flask at an OD600 of 0.80 to 0.85 was split into two 50-ml cultures in 200-ml nonbaffled flasks. NaNO2 at 150 mg/liter was added to one of these cultures, while the other was left untreated to serve as a control. After further incubation for 10 min at 24°C with shaking (160 rpm), cells from both cultures were harvested (see Fig. S1A in the supplemental material). To analyze the adaptive response to acidified nitrite, a 50-ml reference culture and a 50-ml culture to which 150 mg/liter NaNO2 was added at an OD600 of 0.80 to 0.85 were grown in 200-ml nonbaffled flasks until they reached an OD600 of 1.50 ± 0.05 (see Fig. S1B in the supplemental material). After centrifugation (8 min, 4,186 × g, room temperature), the supernatant was discarded, and the pellets were snap-frozen in liquid nitrogen and were stored at −80°C.
For RNA isolation, the pellets were resuspended in TRI reagent (Sigma-Aldrich). Total RNA was extracted using TRI reagent and the RNeasy minikit (Qiagen, Hilden, Germany) as described previously (26).
Transcriptome library preparation and SOLiD sequencing.
For RNA-seq, 90 μg of TRI reagent-extracted RNA was subjected to the column-based purification steps of the RNeasy minikit without prior DNase digestion. Also, the on-column DNase treatment was omitted. 16S rRNA and 23S rRNA were then removed from 5 μg total RNA using the MICROBExpress kit (Ambion, Life Technologies, Darmstadt, Germany). In addition to the capture oligonucleotide mixture supplied with the kit, two additional oligonucleotides targeting fragments of the S. Typhimurium 23S rRNA (5′-xCCTCGGGGTACTTAGATGTTTCA-3′ and 5′-xGTCGGTTCGGTCCTCCAGTTAGT-3′ [where x represents the sequence needed for hybridization to Oligo MagBeads]) were added (2 μl of a 10 μM mixture, corresponding to 20 pmol of each probe), and annealing was performed for 30 min. The mRNA-enriched sample was then treated with the Turbo DNA-free kit (Ambion) to remove residual DNA and was concentrated by ethanol precipitation. The sequencing library was constructed with the SOLiD Total RNA-Seq kit and the SOLiD transcriptome multiplexing kit (Applied Biosystems, Foster City, CA, USA) as described previously (29), but cDNA was purified and was size-selected by two rounds of bead capture using the Agencourt AMPure XP reagent (Beckman Coulter, Krefeld, Germany) according to the SOLiD manual. The size distribution and yield of the purified libraries were assessed on an Agilent 2100 Bioanalyzer system with a DNA 1000 or High Sensitivity DNA chip kit (Agilent Technologies, Santa Clara, CA, USA) and with a Qubit 2.0 fluorometer and the Qubit dsDNA HS assay kit (Life Technologies, Darmstadt, Germany). SOLiD system templated bead preparation and sequencing on the 5500xl SOLiD system were conducted by CeGaT GmbH (Tübingen, Germany). Samples for the adaptive and shock responses were sequenced in independent runs. Differentially bar-coded libraries derived from acidified-NaNO2-treated and control samples were pooled and were sequenced on three (shock response) or six (adaptive response) lanes of one SOLiD slide, along with four additional libraries that do not fall within the scope of this study. For each library, the SOLiD output files (.csfasta, .qual) from the different lanes were merged into single files for further analysis.
Analysis of SOLiD sequencing data.
Data-processing steps to convert SOLiD output files to sorted, indexed BAM files containing reads mapping to the reference genome of S. Typhimurium 14028 (NCBI RefSeq numbers NC_016856.1 [chromosome] and NC_016855.1 [plasmid]) were performed as described previously (29). The number of reads overlapping a gene on the same strand (counts) was calculated in Artemis (version 15.0.0) (30) based on the GenBank file of the reference genome. To assess the impact of acidified nitrite on global transcription in S. Typhimurium, counts of all protein-coding genes according to RefSeq .ptt files downloaded from the NCBI FTP database (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/ [accessed 14 January 2014]) were subjected to differential gene expression analysis using the Bioconductor (31) package edgeR (32). Genes with <10 counts per million (cpm) under both the acidified-nitrite treatment and the control (without nitrite) conditions were filtered, and library sizes were recomputed before TMM (trimmed mean of M-values) normalization (33) was applied to account for compositional differences between the libraries. A common dispersion value of 0.1 was used, as suggested for genetically identical model organisms in the edgeR user's guide (revised version, 4 May 2012). Differential expression analysis was performed using the exact test function. The false discovery rate (FDR) was controlled using the Benjamini-Hochberg (BH) method (34) in edgeR. Genes with log2 fold changes (FC) of >1 in either direction and BH-corrected P values of <0.05 were assigned to COGs (clusters of orthologous groups) according to the .ptt file of S. Typhimurium strain LT2 (NC_003197).
qPCR.
First-strand cDNA synthesis using the qScript cDNA SuperMix (Quanta BioSciences, Gaithersburg, MD) and quantitative real-time PCR (qPCR) assays using the PerfeCTa SYBR green FastMix (Quanta BioSciences) were performed as described previously (26). Gene-specific primers are listed in Table S1 in the supplemental material. For each growth condition, cDNAs synthesized from total RNA extracted from four independent cultures were analyzed. The comparative threshold cycle (CT) method implemented in the relative expression software tool (REST) (35) was used to evaluate relative changes in the transcript levels of NaNO2-treated versus control cultures. ampD or 16S rRNA was used as a nonregulated endogenous normalization control.
Measurement of pHi changes.
A pH-sensitive green fluorescent protein (GFP) variant (enhanced GFP [EGFP]) was used as an indicator (36) to monitor changes in the intracellular pH (pHi) of S. Typhimurium exposed to acidified NaNO2. A shaken overnight culture of the WT pEGFP strain grown for 17 h in LB broth supplemented with 150 μg/ml ampicillin at 24°C was collected (8 min, 4,186 × g, room temperature) and was washed first with 1 volume and then with 0.5 volume phosphate-buffered saline (PBS), pH 7.4. The OD600 was then adjusted to approximately 10 in PBS, pH 7.4, and the cell suspension was stored on ice. The suspension was diluted in sample buffer to an OD600 of 1.0 and was incubated for 5 min at room temperature before fluorescence was measured in a PerkinElmer (Waltham, MA, USA) LS-50B luminescence spectrophotometer. Emission spectra resulted from averaging the spectra from five subsequent scans recorded from 500 to 580 nm with excitation at 490 nm (slit, 3.5 to 4.0 nm; scan speed, 1,000 nm/min). To analyze the impact of NaNO2 addition on the intracellular pH in dependence on the pH of the growth medium, the fluorescence of the WT pEGFP strain in LB (pH 5.5) or neutral LB broth was measured before and immediately after the addition of 150 mg/liter NaNO2. To verify that a decrease in fluorescence intensity was due to NaNO2 rather than to mere photobleaching of EGFP due to repeated measurement of the same sample, a second sample, to which H2O was added instead of NaNO2, was measured. The experiment was performed three times independently.
RNA-seq data accession number.
RNA-seq data have been deposited in NCBI's Gene Expression Omnibus (37) and are accessible through GEO series accession number GSE57238.
RESULTS
Shock and adaptive responses of S. Typhimurium to acidified NaNO2.
To analyze the response of S. Typhimurium to NaNO2 acidified by lactic acid, transcriptional profiling of WT S. Typhimurium in LB (pH 5.5) treated with 150 mg/liter NaNO2 was performed via RNA-seq with two different experimental setups. In the first setup, the transcriptome of S. Typhimurium was analyzed as early as 10 min after the addition of 150 mg/liter NaNO2 (referred to below as the shock response). In the second setup, S. Typhimurium was grown further, to an OD600 of 1.5, and therefore endured acidified NaNO2 stress for a longer period (1.5 to 2 h), which allowed us to study its adaptive response. Differentially expressed genes were assessed by comparison with untreated reference cultures in the same growth medium. The up- and downregulated genes were grouped according to their COGs classes and are listed in Tables S2 (genes upregulated in the shock response), S3 (genes downregulated in the shock response), S4 (genes upregulated in the adaptive response), and S5 (genes downregulated in the adaptive response) in the supplemental material.
In total, 5,416 genes are annotated as protein coding on the S. Typhimurium chromosome and virulence plasmid. Filtering of genes with <10 cpm resulted in 3,095 (57.1%) genes expressed for the experimental setup of the shock response and 3,080 (56.9%) genes expressed for the experimental setup of the adaptive response, which were then subjected to differential gene expression analysis.
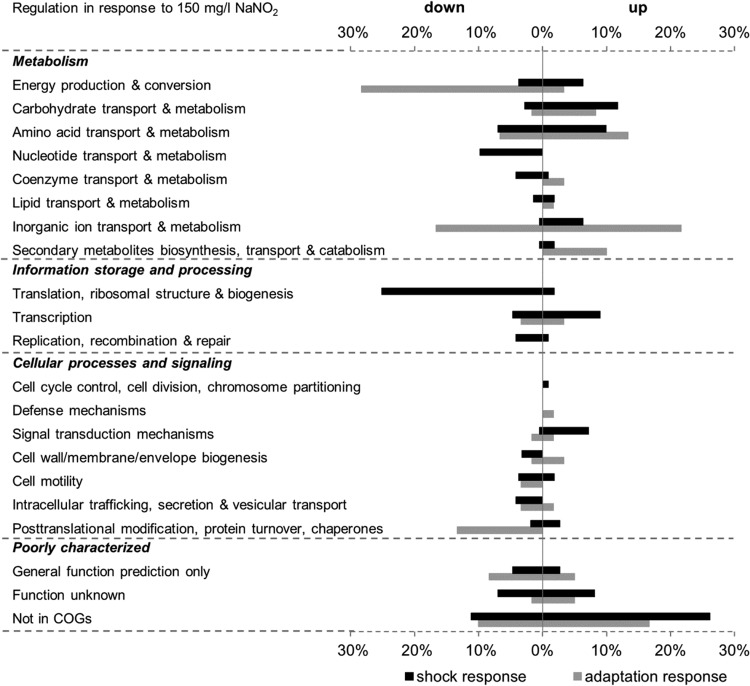
After a 10-min shock with acidified NaNO2, 102 genes (3.3%) were found to be upregulated, while 199 genes (6.4%) were downregulated, in WT S. Typhimurium. The adaptive response was characterized by increased transcription of 55 genes (1.8%) and decreased transcription of 53 genes (1.7%). These genes were functionally classified according to COGs (clusters of orthologous groups) (Fig. 1).
FIG 1.
Overview of the differentially regulated genes according to their functional categories. Genes significantly upregulated or downregulated under conditions of acidified-NaNO2 shock (filled bars) or adaptation (shaded bars) in WT S. Typhimurium were grouped according to the NCBI COGs (clusters of orthologous groups). Each bar represents the percentage of genes with increased or decreased transcription in a given category relative to the total number of up- or downregulated genes among all COG categories (corresponding to 100%) under the respective condition. Since one gene can be classified into more than one COG class, the total number of COG assignments is greater than the number of differentially expressed genes, and relative percentages refer to the former.
More than one-third of the genes upregulated upon a 10-min acidified-NaNO2 shock are poorly characterized. Either they are characterized as genes encoding proteins with a general function based on predictions only or as proteins with an unknown function (11%) or they are not assigned to any functional category (27%). Not surprisingly, genes under the control of the dedicated NO sensors NorR (norVW) (38) and NsrR (STM14_2185, hmpA, ytfE, ygbA, hcp, yeaR-yoaG) (18) were most strongly induced in the presence of acidified NaNO2. These genes are distributed among diverse COGs. Besides these specific nitrosative-stress response regulons, several other genes with reported roles in protection against diverse stresses were also found to be upregulated. Among these, two amino acid decarboxylases and associated amino acid/polyamine antiporters for lysine (cadA, cadB) and arginine (adi, yjdE) exhibited the greatest transcriptional changes. Both amino acid decarboxylase systems have established roles in acid resistance (27, 39). Less strongly induced genes include ogt and dps, which are involved in DNA repair and protection, respectively (40, 41). Two genes, yfiA and yhbH, whose proteins mediate the inactivation of ribosomes in stationary phase (42) also showed elevated transcript levels.
Most of the downregulated genes belong to the functional category of information storage and processing, which comprises transcription, translation, and replication, processes essential for cell proliferation. The majority of these genes are involved in translation, ribosomal structure, and biogenesis. Thus, genes encoding 30S (e.g., rpsU, rpsH) and 50S (e.g., rplU, rplM) ribosomal subunits, translation initiation (infA) and termination (prfC) factors, tRNA (e.g., queA, pheS, argS, trmU, trmD, yhdG)- and rRNA (e.g., yciL, yfcB, rimM, rsmC)-modifying enzymes, and RNases (rph, rnpA) showed decreased transcript levels. Furthermore, genes coding for ATP-dependent RNA helicases (dbpA, deaD, rhlE) and GTPases (engA, era, obgE), which are involved in ribosome maturation at least in E. coli (43), were downregulated. Besides overall transcriptional decreases for genes related to translation, lower transcript abundances were observed for genes involved in transcription and in replication, recombination, and repair, such as rpoA (encoding DNA-directed RNA polymerase subunit alpha), gyrA (DNA gyrase subunit A), fis (DNA-binding protein Fis), and priB (primosomal replication protein N). Along with these, many genes required for nucleotide transport and metabolism were repressed. Several genes in the biosynthetic pathways for purines and pyrimidines were affected, and transcript levels of genes encoding transporters of uracil (uraA) and cytosine (codB) were reduced. Furthermore, the expression of several genes involved in flagellar biosynthesis (e.g., flgA, flgB, flgH, flhBA, fliE, fliFG), and thereby in cell motility, was also decreased. Noteworthy within the functional category of amino acid transport and metabolism is the downregulation of genes involved in the uptake (potAB, potC) or biosynthesis (speC, speD) of putrescine or spermidine.
When S. Typhimurium is allowed to adapt to acidified NaNO2 for a longer period, more than 60% of the upregulated genes have metabolic functions. The gene displaying the greatest fold change was hdeB, whose function is unknown and which is annotated as an acid resistance protein. Comparably to the genes involved in the shock response, genes involved in nitrosative-stress protection under the control of NsrR (hmpA, STM14_2185, ygbA, hcp, yeaR-yoaG) displayed increased transcription. Interestingly, under prolonged acidified-NaNO2 stress, the upregulated amino acid decarboxylase systems were those for ornithine (speF-potE) and arginine (adi, yjdE). The transcription of STM14_5358, STM14_5360, and STM14_5361, which have recently been shown to encode a functional arginine deiminase (ADI) pathway in S. Typhimurium (44), was also increased; these genes belong to the amino acid transport and metabolism category. The largest group of upregulated genes comprises iron uptake and transport genes, mainly in the functional categories of inorganic-ion transport and metabolism and secondary-metabolite biosynthesis, transport, and catabolism. These include genes for the synthesis of the iron-siderophore enterobactin (entCEBA, entF), the uptake of ferrous iron (feoAB-yhgG) or siderophore-bound ferric iron (fhuADB, fepA, fepB, fepC, tonB, exbD), and the release of iron from bacterioferritin or siderophores (bfd, fhuF). Most of the downregulated genes grouped into the energy production and conversion or inorganic-ion transport and metabolism subcategory (both in the metabolism category) or belonged to the posttranslational-modification, protein turnover, and chaperones subcategory (under cellular processes and signaling). Strikingly, most of the gene products are involved in anaerobic respiratory pathways. Thus, genes coding for subunits of terminal reductase complexes for dimethyl sulfoxide (DMSO) (dmsAB and two other loci putatively encoding subunits), tetrathionate (ttrBCA), nitrate (narHJI, napFDAGHBC), and nitrite (nrfA, nrfE) were downregulated. Moreover, some genes involved in the formation or maturation of hydrogenases (hypBDE) were downregulated. In conclusion, with the observed upregulation of iron import systems, transcript levels of the gene coding for the iron storage protein ftn were decreased. Another downregulated gene shown to be iron responsive (45) was yhbU, along with its downstream gene yhbV, both coding for putative proteases.
Validation of RNA-seq data via qPCR.
To validate the acidified-NaNO2-induced transcriptional changes, qPCR was performed on four biological replicates per growth condition. Genes representative of functional categories or pathways that show a differential regulation by acidified NaNO2 were selected for validation. For the shock response to acidified NaNO2, the relative transcription of six genes with increased transcript abundances (adi, cadA, hdeB, hmpA, norV, yfiA) and seven genes with decreased transcript abundances (fliF, potB, purB, pyrE, rnpA, rplM, rpsH) was analyzed. For the adaptive response, a subset of 11 differentially transcribed genes, including 7 upregulated (adi, fhuA, feoB, hdeB, hmpA, speF, STM14_5361) and 4 downregulated (nrfA, STM14_5179, ttrC, yhbU) genes, was chosen. The qPCR results showed a high correlation with the RNA-seq data for both treatments (coefficient of determination [R2], 0.92 for the shock response [Fig. 2A] and 0.96 for the adaptive response [Fig. 2B]), supporting the validity and reproducibility of the RNA-seq data.
FIG 2.
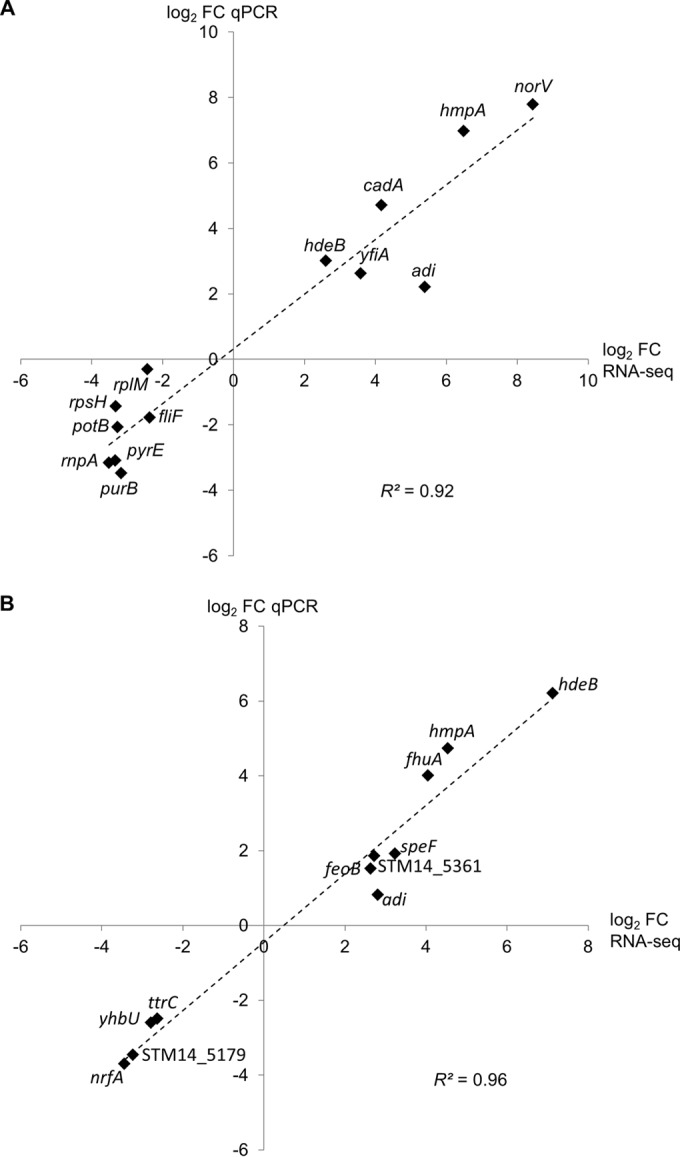
qPCR validation of RNA-seq data for selected differentially expressed genes. The relative transcription of genes found to be differentially regulated in the RNA-seq analysis of the shock response (A) or the adaptive response (B) of S. Typhimurium to acidified NaNO2 was examined by qPCR. 16S rRNA (A) or ampD (B) was used as a reference gene. Mean log2 fold changes (FC) in the transcription of genes in four independent qPCR experiments were plotted against the respective log2 FC determined by RNA-seq. The coefficient of determination (R2) was calculated in Microsoft Excel.
Impaired growth of a mutant lacking the lysine decarboxylase CadA under acidified-NaNO2 stress.
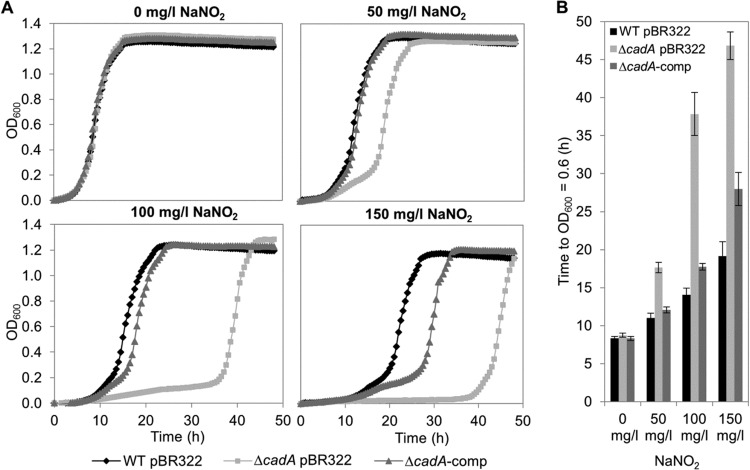
In addition to the global transcription analysis, we screened a S. Typhimurium insertion mutant library (28) for phenotypes sensitive to 150 mg/liter NaNO2 in LB (pH 5.5) in order to identify candidate genes whose products might be important for withstanding acidified-nitrite stress. Interestingly, disruption of the cadA gene resulted in a strong growth delay of the integrant in the presence of acidified NaNO2 (data not shown). cadA encodes an inducible lysine decarboxylase and constitutes an operon together with the upstream gene cadB, which codes for a lysine/cadaverine antiporter (46). Strikingly, cadA and cadB were found by RNA-seq analysis to be strongly induced upon acidified-NaNO2 shock (log2 FC, 4.17 and 4.81, respectively) (see Table S2 in the supplemental material), and cadA upregulation was verified by qPCR (log2 FC, 4.71) (Fig. 2A). Analysis of the growth of a cadA in-frame deletion mutant (ΔcadA pBR322), the WT pBR322 strain, and the respective in trans complementation mutant (ΔcadA-comp) confirmed that the phenotype observed was indeed due to lack of cadA (Fig. 3). Whereas the growth curves for the WT pBR322, ΔcadA pBR322, and ΔcadA-comp strains in LB (pH 5.5) plus 150 mg/liter ampicillin without NaNO2 are quite similar, the ΔcadA pBR322 mutant displayed an increasing growth delay with increasing concentrations of NaNO2 (50, 100, and 150 mg/liter).
FIG 3.
Impact of acidified NaNO2 on the growth of S. Typhimurium WT pBR322, the ΔcadA pBR322 mutant, and the complemented strain (ΔcadA-comp). (A) Representative growth curves (recorded in a Bioscreen C system at 24°C) of S. Typhimurium WT pBR322 (filled diamonds), the ΔcadA pBR322 mutant (shaded squares), and the ΔcadA-comp strain (shaded triangles) in LB (pH 5.5) plus 150 mg/liter ampicillin in the absence or presence of NaNO2 at 50 mg/liter, 100 mg/liter, or 150 mg/liter. (B) Time required for each strain to reach an OD600 of 0.6 (half-maximum OD600) plotted against the NaNO2 concentration. The data are means and standard deviations for three independent experiments including duplicates.
Influence of NaNO2 on the pHi of S. Typhimurium at an acidic pH.
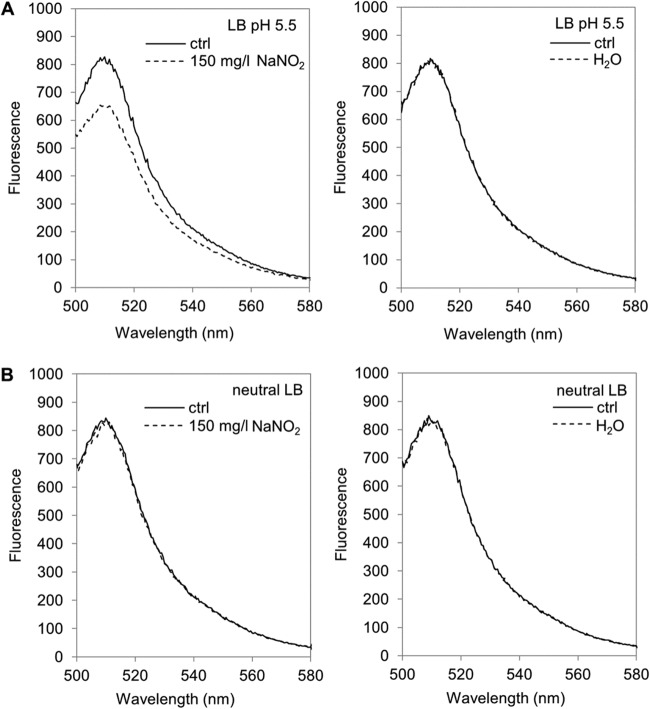
Based on the transcriptional data, we speculated that acidified nitrite activated the transcription of the cadBA operon by somehow lowering the intracellular pH (pHi). The influence of NaNO2 on the pHi of S. Typhimurium in relation to the pH of the medium was analyzed. For pHi measurements, the WT pEGFP strain, which constitutively expresses the pH-sensitive GFP derivative EGFP from a plasmid (see Materials and Methods), was used. The spectral intensity of EGFP decreases with lowered pHs, rendering it suitable for the noninvasive measurement of pHi changes (42). Fluorescence emission scans (from 500 to 580 nm) of the WT pEGFP strain in LB (pH 5.5) or neutral LB broth were recorded before and directly after the addition of 150 mg/liter NaNO2. Without added NaNO2, the fluorescence spectra of the WT pEGFP strain at the two medium pH values were similar, with the expected peak at about 510 nm, but the intensity was slightly lower at pH 5.5 (Fig. 4A) than at the neutral pH (Fig. 4B). However, the addition of NaNO2 to LB (pH 5.5) resulted in a marked decrease in fluorescence intensity around the EGFP emission peak, whereas it had no influence at the neutral pH. The addition of the same volume of H2O as a control did not alter the fluorescence spectra at either pH. Furthermore, the addition of 150 mg/liter NaNO2 did not change the external pH of the medium (data not shown). These data indicate that NaNO2, when added to LB broth acidified to pH 5.5 with lactic acid, elicits a decrease in the pHi of S. Typhimurium.
FIG 4.
Effect of acidified NaNO2 on the intracellular pH of S. Typhimurium. Shown are fluorescence emission spectra of the WT pEGFP strain in LB (pH 5.5) (A) or neutral LB (B) before (ctrl) and immediately after the addition of 150 mg/liter NaNO2 (left) or H2O (right). Representative spectra for three independent experiments are shown.
DISCUSSION
The curing agent NaNO2 is added as a preservative to control the growth of pathogenic microorganisms in raw meat products (7). The antimicrobial action depends on the conversion of NaNO2 to NO and related reactive nitrogen species (7) under the mildly acidic conditions in the meat, which are due to lactic acid. In this study, we analyzed the shock and adaptive responses of S. Typhimurium, a pathogen that is often associated with contaminated raw meat products, to NaNO2 acidified by lactic acid at an ambient temperature of 24°C. We sought to identify critical determinants in the protective response of this organism and to gain further insight into the antimicrobial action of NaNO2 under conditions relevant for food.
The NsrR regulon, implicated in nitrosative-stress protection (18), was found to be strongly induced in both the immediate and the continuous response to acidified-NaNO2 stress. This finding is consistent with previous studies of S. Typhimurium using NO donor compounds (11, 14) and underlines the importance of NO arising from acidified NaNO2 and inactivating NsrR, thereby relieving the transcriptional repression of target genes (47), including the gene encoding the NO-detoxifying protein HmpA (18). In contrast, transcriptional activation of norV, encoding the NO-reducing flavorubredoxin (16), was observed after 10 min but not after prolonged exposure. This might be due to oscillations in norV mRNA levels under aerobic conditions, as reported previously for E. coli (20). Besides this direct response to nitrosative stress, several other stress-related genes were induced, including acid resistance genes (cadBA, adi, yjdE [48]) and genes related to DNA damage (ogt [40], dps [41]). The shock response was further characterized by downregulation of the translational machinery and of genes involved in transcription and replication, which comprise crucial physiological processes. This trend was also observed in previous studies investigating the NO stress response of S. Typhimurium (11, 14) and might be a nonspecific consequence of the reduced growth rate following the addition of 150 mg/liter NaNO2 (see Fig. S1 in the supplemental material). Obviously, inducing stress tolerance and reducing cell growth promotes the survival of S. Typhimurium subjected to harsh acidified-NaNO2 stress.
The transcriptional changes observed in the adaptive response mainly affect genes involved in iron homeostasis and anaerobic respiration. The decreased transcription of the latter group of genes is consistent with the findings of previous studies investigating the response to NO stress in S. Typhimurium (11) and also in E. coli, though under anaerobic conditions (19, 21). Differential regulation was ascribed mainly to inactivation of the regulator FNR, which regulates many genes in response to oxygen availability (49, 50), and whose iron-sulfur cluster is nitrosylated by NO (51). We speculate that under the higher culture density we investigated (OD600, 1.5) for the adaptive response than for the shock response, cells might have experienced some oxygen shortage that was sufficient to result in an effect of NO on FNR regulation, as noted by Richardson et al. (11). The other large group of genes found to be deregulated under prolonged acidified-NaNO2 exposure consisted of iron-responsive genes, which are subjected to regulation by Fur (ferric uptake regulator) (45, 52). Under iron-replete conditions, dimeric Fe2+-bound Fur binds to consensus DNA sequences and represses the transcription of iron uptake systems (53). Upon nitrosylation, Fe-Fur loses its DNA-binding activity (54), resulting in the derepression of target genes involved in iron acquisition, as observed in our RNA-seq data, as well as in other studies (11, 20, 21). The transcriptional changes observed might therefore be merely a coincidental consequence of the inactivation of FNR and Fur by NO arising from acidified NaNO2. In contrast, derepression of NsrR-regulated genes may provide a physiological benefit by alleviating the nitrosative stress on the cells.
An unexpected finding was the upregulation of inducible amino acid decarboxylases and the respective amino acid/polyamine antiporters, which are crucial constituents of the acid stress response in enteropathogenic bacteria (48). Whereas the decarboxylation systems for lysine (cadA, cadB) and ornithine (speF, potE) were induced in response to acidified-NaNO2 shock and continuous stress, respectively, the arginine decarboxylase system (adi, yjdE) was upregulated under both conditions. The amino acid decarboxylases are known to be induced by low pHs (48), and each was shown to confer more or less acid resistance under different conditions in S. Typhimurium (27, 39). Since increased transcription of inducible amino acid decarboxylases has never been observed in bacteria exposed to NO at a neutral pH, this response is presumably specific to acidified-NaNO2 stress. The physiological role of CadA in protection against acidified-NaNO2 stress is supported by the impaired growth of the ΔcadA pBR322 deletion mutant in the presence of NaNO2. Interestingly, the CadA protein levels of a Salmonella strain missing the three major upregulated proteins (HmpA, YtfE, Hcp) were found to be elevated under RNS stress in mice (55). In uropathogenic E. coli (UPEC), the lysine decarboxylase system has been demonstrated to be involved in protection against nitrosative stress elicited by acidified NaNO2 (56). Mutations in either cadC (encoding the transcriptional activator), cadA, or cadB resulted in increased sensitivity to acidified NaNO2 (56). There are several possible mechanisms by which CadA might contribute to nitrosative-stress protection. First, the polyamine cadaverine is produced upon the decarboxylation of lysine. Bower and Mulvey (56) found that exogenous supplementation with cadaverine or other polyamines rescued the growth of the cadaverine-deficient deletion mutants, arguing that polyamines are the mediators of the protective effect. Besides this protection provided by cadaverine, the end product of lysine decarboxylation, our data indicate that the pH-homeostatic function of the lysine decarboxylase system itself (46) might account for protection against acidified-NaNO2 stress. Decarboxylation of lysine to the polyamine cadaverine consumes an intracellular proton, and the basic cadaverine is subsequently exported in exchange for extracellular lysine via the antiporter (46). Both reactions contribute to pH homeostasis and local buffering of the extracellular medium. However, this would imply that acidified NaNO2 would somehow perturb the intracellular pH of S. Typhimurium in the first place. Indeed, measurement of the pHi in S. Typhimurium via a pH-sensitive GFP derivative indicated intracellular acidification upon the addition of 150 mg/liter NaNO2 to a mildly acidic LB medium, but not to a neutral medium. The imposition of intracellular acid stress on bacteria might provide an additional mechanism for the inhibitory action of acidified nitrite, which has been reported for yeasts previously (23). The effector of intracellular acidification might be nitrous acid (HNO2), which is supposed to form upon the acidification of NO2−. As a weak acid, HNO2 might diffuse across the membrane and dissociate in the neutral cytoplasm, thereby releasing a proton (57). Lysine decarboxylase might provide a mechanism for neutralizing these protons. Furthermore, pH buffering of the surrounding environment might decrease the rate of formation of NO and RNS from NO2−, thereby contributing indirectly to nitrosative-stress protection by diminishing the growth-inhibitory effects of these species.
In conclusion, we showed that the lysine decarboxylase CadA plays an important role in protecting S. Typhimurium against acidified-NaNO2-mediated stress. Furthermore, to our knowledge, this study provides the first evidence that intracellular acidification might additionally contribute to the antibacterial action of acidified NaNO2 in foodstuffs.
Supplementary Material
ACKNOWLEDGMENTS
This research project was supported by the German Ministry of Economics and Technology (via AiF) and the FEI (Forschungskreis der Ernährungsindustrie e.V., Bonn, Germany), project AiF 16908 N, and by the Förderverein für Fleischforschung e.V., Kulmbach, Germany.
We thank Katharina Sturm, Lisa Schürch, and Jakob Schardt for excellent technical assistance and Richard Landstorfer and Svenja Simon (University of Constance, Constance, Germany) for help with analysis of the RNA-seq data. We also thank Rohtraud Pichner (Max Rubner Institute, Kulmbach, Germany) for helpful discussions. Thilo M. Fuchs is thanked for providing the Salmonella Typhimurium insertion mutant library. Matthias A. Ehrmann (Chair of Technical Microbiology, Technical University Munich, Freising, Germany) is acknowledged for providing plasmid pEGFP.
Footnotes
Published ahead of print 8 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01696-14.
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50:882–889. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 2.European Food Safety Authority, European Centre for Disease Prevention and Control. 2013. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J. 11:3129. 10.2903/j.efsa.2013.3129 [DOI] [Google Scholar]
- 3.Federal Ministry of Food and Agriculture. 15 July 2013. Deutsches Lebensmittelbuch. Leitsätze für Fleisch und Fleischerzeugnisse. http://www.bmelv.de/SharedDocs/Downloads/Ernaehrung/Lebensmittelbuch/LeitsaetzeFleisch.pdf?_blob=publicationFile
- 4.Leistner L. 2000. Basic aspects of food preservation by hurdle technology. Int. J. Food Microbiol. 55:181–186. 10.1016/S0168-1605(00)00161-6 [DOI] [PubMed] [Google Scholar]
- 5.Leistner L, Gorris LG. 1995. Food preservation by hurdle technology. Trends Food Sci. Technol. 6:41–46. 10.1016/S0924-2244(00)88941-4 [DOI] [Google Scholar]
- 6.European Parliament. 2006. Directive 2006/52/EC of the European Parliament and of the Council of 5 July 2006 amending Directive 95/2/EC on food additives other than colours and sweeteners and Directive 94/35/EC on sweeteners for use in foodstuffs. http://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32006L0052&rid=2
- 7.Cammack R, Joannou CL, Cui XY, Torres Martinez C, Maraj SR, Hughes MN. 1999. Nitrite and nitrosyl compounds in food preservation. Biochim. Biophys. Acta 1411:475–488. 10.1016/S0005-2728(99)00033-X [DOI] [PubMed] [Google Scholar]
- 8.Skibsted LH. 2011. Nitric oxide and quality and safety of muscle based foods. Nitric Oxide 24:176–183. 10.1016/j.niox.2011.03.307 [DOI] [PubMed] [Google Scholar]
- 9.Jira W. 2004. Chemische Vorgänge beim Pökeln und Räuchern. Teil 1: Pökeln. Fleischwirtschaft 84:235–239 [Google Scholar]
- 10.Henard CA, Vázquez-Torres A. 2011. Nitric oxide and salmonella pathogenesis. Front. Microbiol. 2:84. 10.3389/fmicb.2011.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, Becker LA, Navarre WW, Castor ME, Libby SJ, Fang FC. 2011. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar Typhimurium. Cell Host Microbe 10:33–43. 10.1016/j.chom.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson AR, Soliven KC, Castor ME, Barnes PD, Libby SJ, Fang FC. 2009. The base excision repair system of Salmonella enterica serovar Typhimurium counteracts DNA damage by host nitric oxide. PLoS Pathog. 5:e1000451. 10.1371/journal.ppat.1000451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevanin TM, Poole RK, Demoncheaux EAG, Read RC. 2002. Flavohemoglobin Hmp protects Salmonella enterica serovar Typhimurium from nitric oxide-related killing by human macrophages. Infect. Immun. 70:4399–4405. 10.1128/IAI.70.8.4399-4405.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourret TJ, Porwollik S, McClelland M, Zhao R, Greco T, Ischiropoulos H, Vázquez-Torres A, Aballay A. 2008. Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS One 3:e1833. 10.1371/journal.pone.0001833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford MJ, Goldberg DE. 1998. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J. Biol. Chem. 273:12543–12547. 10.1074/jbc.273.20.12543 [DOI] [PubMed] [Google Scholar]
- 16.Mills PC, Richardson DJ, Hinton JCD, Spiro S. 2005. Detoxification of nitric oxide by the flavorubredoxin of Salmonella enterica serovar Typhimurium. Biochem. Soc. Trans. 33:198–199. 10.1042/BST0330198 [DOI] [PubMed] [Google Scholar]
- 17.Mills PC, Rowley G, Spiro S, Hinton JCD, Richardson DJ. 2008. A combination of cytochrome c nitrite reductase (NrfA) and flavorubredoxin (NorV) protects Salmonella enterica serovar Typhimurium against killing by NO in anoxic environments. Microbiology (Reading, Engl.) 154(Part 4):1218–1228. 10.1099/mic.0.2007/014290-0 [DOI] [PubMed] [Google Scholar]
- 18.Karlinsey JE, Bang I, Becker LA, Frawley ER, Porwollik S, Robbins HF, Thomas VC, Urbano R, McClelland M, Fang FC. 2012. The NsrR regulon in nitrosative stress resistance of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 85:1179–1193. 10.1111/j.1365-2958.2012.08167.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Justino MC, Vicente JB, Teixeira M, Saraiva LM. 2005. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 280:2636–2643. 10.1074/jbc.M411070200 [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. U. S. A. 101:745–750. 10.1073/pnas.0307741100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pullan ST, Gidley MD, Jones RA, Barrett J, Stevanin TM, Read RC, Green J, Poole RK. 2007. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J. Bacteriol. 189:1845–1855. 10.1128/JB.01354-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flatley J, Barrett J, Pullan ST, Hughes MN, Green J, Poole RK. 2005. Transcriptional responses of Escherichia coli to S-nitrosoglutathione under defined chemostat conditions reveal major changes in methionine biosynthesis. J. Biol. Chem. 280:10065–10072. 10.1074/jbc.M410393200 [DOI] [PubMed] [Google Scholar]
- 23.Mortensen HD, Jacobsen T, Koch AG, Arneborg N. 2008. Intracellular pH homeostasis plays a role in the tolerance of Debaryomyces hansenii and Candida zeylanoides to acidified nitrite. Appl. Environ. Microbiol. 74:4835–4840. 10.1128/AEM.00571-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113 [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mühlig A, Kabisch J, Pichner R, Scherer S, Müller-Herbst S. 2014. Contribution of the NO-detoxifying enzymes HmpA, NorV and NrfA to nitrosative stress protection of Salmonella Typhimurium in raw sausages. Food Microbiol. 42:26–33. 10.1016/j.fm.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 27.Viala JPM, Méresse S, Pocachard B, Guilhon A, Aussel L, Barras F. 2011. Sensing and adaptation to low pH mediated by inducible amino acid decarboxylases in Salmonella. PLoS One 6:e22397. 10.1371/journal.pone.0022397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knuth K, Niesalla H, Hueck CJ, Fuchs TM. 2004. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol. Microbiol. 51:1729–1744. 10.1046/j.1365-2958.2003.03944.x [DOI] [PubMed] [Google Scholar]
- 29.Landstorfer R, Simon S, Schober S, Keim D, Scherer S, Neuhaus K. 2014. Comparison of strand-specific transcriptomes of enterohemorrhagic Escherichia coli O157:H7 EDL933 (EHEC) under eleven different environmental conditions including radish sprouts and cattle feces. BMC Genomics 15:353. 10.1186/1471-2164-15-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 31.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289–300 [Google Scholar]
- 35.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kneen M, Farinas J, Li Y, Verkman AS. 1998. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 74:1591–1599. 10.1016/S0006-3495(98)77870-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tucker NP, D'Autreaux B, Studholme DJ, Spiro S, Dixon R. 2004. DNA binding activity of the Escherichia coli nitric oxide sensor NorR suggests a conserved target sequence in diverse proteobacteria. J. Bacteriol. 186:6656–6660. 10.1128/JB.186.19.6656-6660.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alvarez-Ordóñez A, Fernández A, Bernardo A, López M. 2010. Arginine and lysine decarboxylases and the acid tolerance response of Salmonella Typhimurium. Int. J. Food Microbiol. 136:278–282. 10.1016/j.ijfoodmicro.2009.09.024 [DOI] [PubMed] [Google Scholar]
- 40.Yamada M, Sedgwick B, Sofuni T, Nohmi T. 1995. Construction and characterization of mutants of Salmonella typhimurium deficient in DNA repair of O6-methylguanine. J. Bacteriol. 177:1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calhoun LN, Kwon YM. 2011. The ferritin-like protein Dps protects Salmonella enterica serotype Enteritidis from the Fenton-mediated killing mechanism of bactericidal antibiotics. Int. J. Antimicrob. Agents 37:261–265. 10.1016/j.ijantimicag.2010.11.034 [DOI] [PubMed] [Google Scholar]
- 42.Polikanov YS, Blaha GM, Steitz TA. 2012. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336:915–918. 10.1126/science.1218538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaczanowska M, Rydén-Aulin M. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71:477–494. 10.1128/MMBR.00013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi Y, Choi J, Groisman EA, Kang D, Shin D, Ryu S. 2012. Expression of STM4467-encoded arginine deiminase controlled by the STM4463 regulator contributes to Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 80:4291–4297. 10.1128/IAI.00880-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bjarnason J, Southward CM, Surette MG. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185:4973–4982. 10.1128/JB.185.16.4973-4982.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park YK, Bearson B, Bang SH, Bang IS, Foster JW. 1996. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol. Microbiol. 20:605–611. 10.1046/j.1365-2958.1996.5441070.x [DOI] [PubMed] [Google Scholar]
- 47.Tucker NP, Hicks MG, Clarke TA, Crack JC, Chandra G, Le Brun NE, Dixon R, Hutchings MI. 2008. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One 3:e3623. 10.1371/journal.pone.0003623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao B, Houry WA. 2010. Acid stress response in enteropathogenic gammaproteobacteria: an aptitude for survival. Biochem. Cell Biol. 88:301–314. 10.1139/O09-182 [DOI] [PubMed] [Google Scholar]
- 49.Spiro S, Guest JR. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6:399–428 [DOI] [PubMed] [Google Scholar]
- 50.Fink RC, Evans MR, Porwollik S, Vazquez-Torres A, Jones-Carson J, Troxell B, Libby SJ, McClelland M, Hassan HM. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J. Bacteriol. 189:2262–2273. 10.1128/JB.00726-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crack JC, Le Brun NE, Thomson AJ, Green J, Jervis AJ. 2008. Reactions of nitric oxide and oxygen with the regulator of fumarate and nitrate reduction, a global transcriptional regulator, during anaerobic growth of Escherichia coli. Methods Enzymol. 437:191–209. 10.1016/S0076-6879(07)37011-0 [DOI] [PubMed] [Google Scholar]
- 52.Troxell B, Fink RC, Porwollik S, McClelland M, Hassan HM. 2011. The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol. 11:236. 10.1186/1471-2180-11-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Escolar L, Pérez-Martín J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D'Autreaux B, Touati D, Bersch B, Latour J, Michaud-Soret I. 2002. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. U. S. A. 99:16619–16624. 10.1073/pnas.252591299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burton NA, Schürmann N, Casse O, Steeb AK, Claudi B, Zankl J, Schmidt A, Bumann D. 2014. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe 15:72–83. 10.1016/j.chom.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 56.Bower JM, Mulvey MA. 2006. Polyamine-mediated resistance of uropathogenic Escherichia coli to nitrosative stress. J. Bacteriol. 188:928–933. 10.1128/JB.188.3.928-933.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lambert RJ, Stratford M. 1999. Weak-acid preservatives: modelling microbial inhibition and response. J. Appl. Microbiol. 86:157–164. 10.1046/j.1365-2672.1999.00646.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.