Abstract
How Arctic climate change might translate into alterations of biogeochemical cycles of carbon (C) and nitrogen (N) with respect to inorganic and organic N utilization is not well understood. This study combined 15N uptake rate measurements for ammonium, nitrate, and urea with 15N- and 13C-based DNA stable-isotope probing (SIP). The objective was to identify active bacterial and archeal plankton and their role in N and C uptake during the Arctic summer and winter seasons. We hypothesized that bacteria and archaea would successfully compete for nitrate and urea during the Arctic winter but not during the summer, when phytoplankton dominate the uptake of these nitrogen sources. Samples were collected at a coastal station near Barrow, AK, during August and January. During both seasons, ammonium uptake rates were greater than those for nitrate or urea, and nitrate uptake rates remained lower than those for ammonium or urea. SIP experiments indicated a strong seasonal shift of bacterial and archaeal N utilization from ammonium during the summer to urea during the winter but did not support a similar seasonal pattern of nitrate utilization. Analysis of 16S rRNA gene sequences obtained from each SIP fraction implicated marine group I Crenarchaeota (MGIC) as well as Betaproteobacteria, Firmicutes, SAR11, and SAR324 in N uptake from urea during the winter. Similarly, 13C SIP data suggested dark carbon fixation for MGIC, as well as for several proteobacterial lineages and the Firmicutes. These data are consistent with urea-fueled nitrification by polar archaea and bacteria, which may be advantageous under dark conditions.
INTRODUCTION
The Arctic is already experiencing the impacts of global climate change, which has the potential to disrupt the ecology of the Arctic Ocean, causing broad changes in the physical, chemical, and biological realms (1–5). How such changes might translate into alterations of ecosystem dynamics and of the overall balances and rates of biogeochemical cycles of carbon (C) and nitrogen (N) is not well understood (6, 7). Generally, short days and sea ice coverage during the Arctic winter limit light and primary production of phytoplankton, while summer is characterized by episodic phytoplankton blooms following sea ice melt (8). High levels of primary productivity during the summer are thought to be sustained through the buildup of NO3− in the water column during the dark winter period, as well as by inputs from ice melt and allochthonous riverine nutrient sources (9). Production could additionally be augmented by dissolved organic nitrogen (DON), which can represent between 18 and 85% of the total dissolved N pool in coastal and open ocean surface water (10, 11). The interplay between inorganic and organic N utilization, with respect to heterotrophic versus autotrophic activities, could be an important contributor to biological production but remains poorly resolved, especially in Arctic environments (12).
Among DON compounds, urea has long been recognized as an important N source in tropical, subtropical, and temperate marine environments (13–16). Urea usually occurs at nanomolar levels in the open oceans but can be found at concentrations as high as 50 μM in coastal ecosystems (17), where it can be an important N substrate that promotes large seasonal blooms of phytoplankton (18). The importance of urea at high latitudes is, however, less well understood. In addition to riverine input, other natural sources of urea in the Arctic can include excretion and sloppy feeding by zooplankton (19) and inputs from the melting of seasonal fast ice (20). Urea production has also been attributed to sediment-associated bacteria, which may mediate the release of urea into the water column via thermal or wind-driven mixing (21). In the Canadian Arctic, urea has been observed to account for >50% of total dissolved nitrogen (TDN) (22). The same study reported urea uptake rates that mimicked the distributional patterns of urea concentrations while accounting for approximately 32% of N productivity (nitrate, ammonium, and urea). A more recent study in the Beaufort Sea found that urea supplied almost half of the phytoplankton N uptake annually (23), and on a seasonal basis it was reported that urea uptake increased relative to that of other N substrates as the year progressed from winter to spring to summer (S. E. Baer, R. E. Sipler, Q. N. Roberts, P. L. Yager, M. E. Frischer, and D. A. Bronk, submitted for publication). In the northern Baffin Bay, cycloheximide and streptomycin were utilized as inhibitors, and it was found that urea was consumed primarily by phytoplankton (58 to 95%) but may also be utilized by bacteria (5 to 42%) (24). Collectively, these studies suggest that urea is likely an important source of N in Arctic systems. It remains uncertain, however, what the dynamics of competition for urea between phytoplankton and bacteria are at the community level and which populations of cells successfully compete for urea N under different conditions. Size fractionation experiments often retain significant numbers of bacteria in the “phytoplankton fraction” (traditionally collected on GF/F filters with a nominal pore size of 0.7 μm), and they provide no phylogenetic information about active microbial community members.
The ability of pelagic bacteria and archaea to fix carbon independently of light in oxygenic waters is becoming widely recognized (25–28), especially in deeper oceanic waters (29–31). However, the extent to which dark carbon fixation is occurring in the world's oceans and how important this metabolism is for the life strategies of specific taxa are still unknown. It is hypothesized that dark carbon fixation can be important in compensating for metabolic imbalances under oligotrophic conditions. Since large proportions of the world's oceans are oligotrophic, the significance of dark carbon fixation could therefore be large. This may be especially true under dark winter conditions in the Arctic, when photosynthesis rates are low. Previously, Alonso-Sáez et al. (26) found that certain taxa of bacteria (e.g., Oleispira and Pseudoalteromonas-Colwellia) collected from shelf waters in the Arctic and cultured in aged seawater had the potential to fix carbon. The authors concluded that heterotrophs were primarily responsible for the observed bicarbonate uptake and proposed that this metabolism would be advantageous for survival during periods of low nutrient availability.
In this study, we combined dissolved inorganic nitrogen (DIN) and urea uptake rate measurements with 15N- and 13C-based DNA stable-isotope probing (SIP) in order to investigate the role of bacterial and archaeal plankton in C and N cycling during the Arctic summer and winter seasons. We hypothesized that both bacteria and archaea would successfully compete for N from NO3− and urea during the Arctic winter but not during the summer, when phytoplankton dominate the absolute uptake of these N sources. Further, 13C-based SIP was used to investigate the in situ capabilities of both bacteria and archaea to incorporate carbon from bicarbonate into DNA in order to demonstrate their potential involvement in the fixation of carbon during the dark winter months.
MATERIALS AND METHODS
Field sample collection.
To capture the extreme Arctic light and physical conditions, sampling took place during the summer (August 2011) and winter (January 2012) ∼2.5 km offshore of Barrow, AK. A YSI sonde was used to measure temperature and salinity throughout the water column. During the summer, samples were collected via a low-pressure electric pump at a depth of 8 m from a 17-m water column located at 71°18.13′N, 156°43.16′W. During the winter, when the water was covered with ice, a small hole was drilled through the ice to allow access for sample collection. Due to ice conditions, winter samples were collected at a 1-m depth to minimize the intake of resuspended sediment in a shallow, 8-m water column at 71°22.12′N, 156°34.34′W. Every effort was made to reduce stress on the organisms by limiting light and temperature changes. A small tent was erected and was heated to approximately −1°C (near the temperature of the ambient seawater) to prevent the pumped seawater and sampling equipment from freezing.
15N and 13C additions.
Water was collected into a series of 2-liter acid-washed polyethylene terephthalate glycol (PETG) bottles. A subset was used for the determination of ambient nutrient concentrations. Samples for SIP and uptake rate incubations were each run in duplicate and were inoculated with unlabeled (14N) or labeled (15N) ammonium (NH4+), nitrate (NO3−), and urea (>98% 15N). Previously reported ambient concentrations were used to establish N additions for uptake rate incubations. Since DNA stable-isotope probing (SIP) requires substantial isotopic labeling, incubations for SIP samples were made with saturating additions of 2.0 μmol N liter−1 in the form of NH4+, NO3−, and urea during the summer. Winter additions were 3.25 μmol N liter−1 for NH4+ or urea and 7.7 μmol N liter−1 for NO3−. For dark carbon fixation experiments, duplicate sets of samples were incubated with either labeled (13C) or unlabeled (12C) bicarbonate at 200 mM. The bottles were then surrounded by ambient seawater, placed in insulated coolers, and brought to the laboratory within 1 h of collection to prevent freezing. Samples were incubated in a temperature-controlled chamber for 24 h at ambient water temperature (+4.7°C in the summer; −1.8°C in the winter). To mimic spectral attenuation from the field during the summer, light levels were maintained by GAMColor blue films and were confirmed using a Li-Cor PAR sensor. Winter samples were incubated in the dark. At the ends of incubations, samples were filtered separately for uptake rates and SIP analyses, and water was collected for nutrient analyses. For uptake rate determinations, samples were filtered through Whatman GF/F filters (nominal pore size, 0.7 μm). The filters were placed in cryovials and were frozen until analysis. For the determination of nutrient concentrations at the ends of incubations, the filtrate was poured into polypropylene tubes and was frozen until analysis. Samples from SIP incubation bottles were filtered onto 0.45-μm Supor filters (Pall Life Sciences) and were frozen in 750 μl STE buffer (1 M NaCl, 100 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0]).
Nutrient analysis and uptake rates.
Nutrients were measured on ambient seawater and on water incubated with a labeled substrate in order to correct for isotope dilution in the uptake rate calculations. NH4+ concentrations were measured in triplicate using the phenol-hypochlorite method (32). Duplicates of NO3− and nitrite (NO2−) were measured on a Lachat QuikChem 8500 autoanalyzer (33). Urea was measured in duplicate using the manual monoxime method (34). A Europa Scientific Geo 20/20 mass spectrometer with an ANCA autosampler was used to make isotopic measurements of 15N samples. Nitrogen uptake rates were calculated according to the method of Dugdale and Goering (35) and carbon uptake rates according to that of Hama et al. (36). The NH4+ pool was isolated at the end of the incubation by solid-phase extraction (37, 38), and the 15N enrichment was determined, so that NH4+ uptake rates could be corrected for isotope dilution and NH4+ regeneration rates could be calculated (39).
Stable-isotope probing.
DNA was extracted as described by Wawrik et al. (40). Cesium chloride (CsCl) density centrifugation and fractionation were conducted as described previously (40–43) by loading 2 μg of DNA and spinning for 48 to 72 h in a Beckman VTi 65.2 rotor at 140,000 × g. Thirty 150-μl fractions were then collected from each tube with a fraction collector (Beckman) by displacing the contents with mineral oil at a constant rate using a peristaltic pump. Densities for each fraction were calculated from their refractive indices (40, 42). After ethanol reprecipitation with 1.0 μl of molecular-grade glycogen (Ambion), DNA was suspended in 30 μl sterile nuclease-free deionized (DI) water. These purified fractions served as templates for quantitative PCR (qPCR) and 16S rRNA gene PCR for Illumina sequencing. All direct comparisons made in this paper (i.e., 14N versus 15N treatment for a specific substrate) are for fractions and gradients from the same centrifuge run, which used the same batch of buffer.
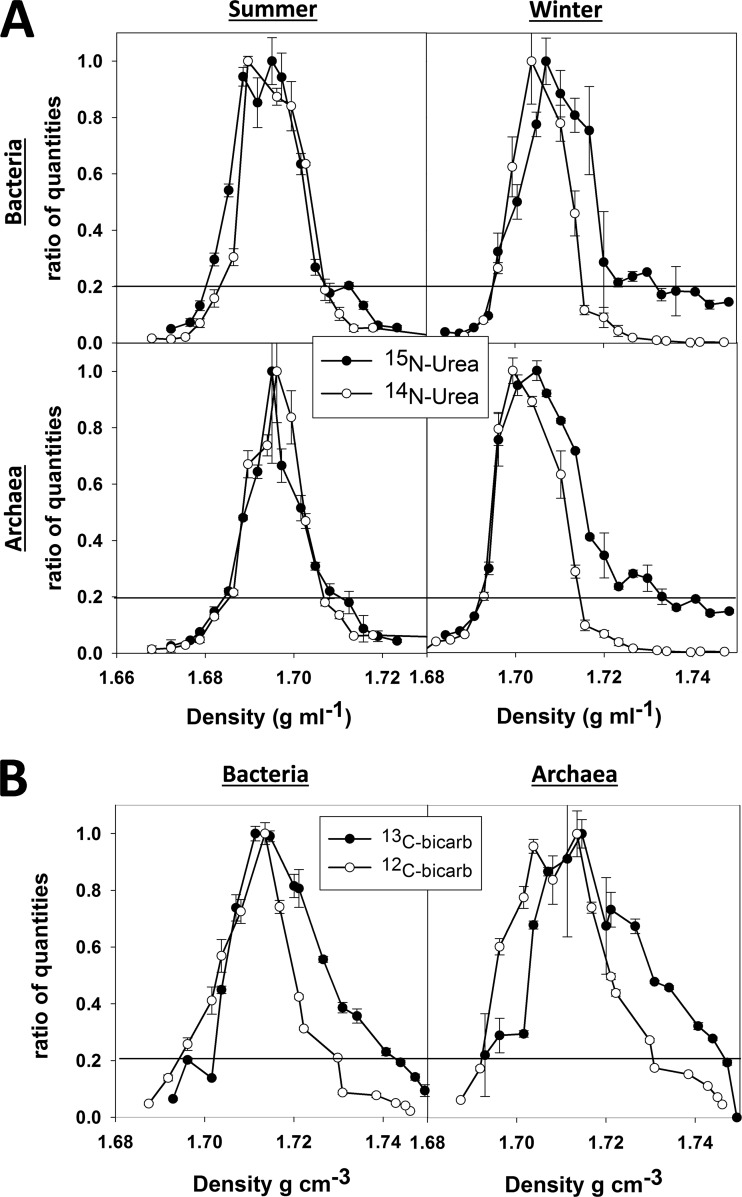
Labeling was assessed by integrating abundance peaks observed in gradients and thereby estimating their average density. For bacteria and archaea, the proportional abundance from qPCR was used. Average densities for individual phylogenetic groups and individual operational taxonomic units (OTUs) were calculated from 16S rRNA gene community OTU frequency data. To minimize biases introduced by baseline variability (i.e., because target DNA is typically found throughout gradients, and because the density ranges obtained can differ slightly among gradient runs), only the major peak of DNA was integrated. All fractions that contained >20% of the maximum observed quantity were hence integrated (see Fig. 2). The percentage of labeling was calculated by assuming that 100% labeling of DNA with 15N or 13C would yield a density shift of 0.016 g cm−3 or 0.036 g cm−3, respectively (42). Sequential gradient fractions differed, on average, by ∼0.0035 ± 0.0001 g cm−3. Shifts of >30% and >15% for 15N and 13C, respectively, were therefore interpreted as positive labeling. This corresponds to a density shift of approximately 1.5 fractions in our gradients, where complete (i.e., 100%) labeling would correspond to a shift of 4.6 or 10 fractions for 15N or 13C, respectively.
FIG 2.
Examples of qPCR analysis of SIP gradient fractions for bacterial and archaeal 16S rRNA gene copies of samples collected from the near-shore Arctic. The relative quantities detected in each fraction are shown as a function of density. All data were normalized to the highest quantities observed and are hence shown as ratios, where 1 equals the highest value observed. Error bars indicate one standard deviation calculated from three replicate qPCR measurements. The horizontal lines indicate the threshold above which quantities were integrated to calculate the average DNA density and the percentage of incorporation. (A) Comparison of winter and summer [14N]urea (○) and [15N]urea (●) treatments. (B) Dark carbon fixation SIP experiment showing [12C]bicarbonate (○) and [13C]bicarbonate (●) treatments for bacteria and archaea.
qPCR for 16S rRNA gene copy numbers of bacteria and archaea.
Bacterial and archaeal 16S rRNA gene copy numbers were determined for each SIP fraction via qPCR. The bacterial 16S rRNA gene qPCR primers were forward primer 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and reverse primer 519R (5′-GWA TTA CCG CKG CTG-3′). The archaeon-specific qPCR primers were forward primer 8AF (5′-TCC GGT TGA TCC TGC C-3′) and reverse primer A344R (5′-TCG CGC CTG CTC CIC CCC GT-3′). SYBR green qPCRs were run in 30-μl volumes using the Power SYBR green PCR master mix (Applied Biosystems), 500 nM (final concentration) each primer, and 2 μl of template DNA. Using an Applied Biosystems ABI 7300 real-time PCR system, qPCR was conducted as follows: 50°C for 2 min and 95°C for 8 min, followed by 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. Genomic DNA of Roseobacter denitrificans Och114 was used as a standard for bacteria, while a linearized plasmid containing the complete 16S rRNA gene of Methanospirillum hungatei JF-1 served as a standard for archaea.
16S rRNA gene community analysis.
Environmental genomic DNA (10 ng) and 2 μl DNA purified from CsCl gradient fractions were amplified using Phusion high-fidelity DNA polymerase (Thermo Scientific) by targeting partial 16S rRNA genes with the universal (bacterial and archaeal) primers S-D-Arch-0519-a-S-15 (5′-CAG CMG CCG CGG TAA-3′) and S-D-Bact-785-a-A-21 (5′-GAC TAC HVG GGT ATC TAA TCC-3′) as described previously (44). These primers do not amplify rRNA genes from eukaryotes but cover 86.5% and 87.1% of bacterial and archaeal phyla, respectively. If one mismatch is allowed (which occurs frequently during PCR), 94.6% and 94.8% of bacterial and archaeal phyla are covered, respectively (44). Amplification of candidate divisions WS6, TM7, and OP11, as well as the phylum Nanoarchaeota, was deemed unlikely via in silico analysis (44). The forward primer was modified to include a 5′ M13 tag, used for labeling the PCR products with Illumina tags (45). Community DNA was amplified with 30 cycles. For gradient fractions, PCR was used to estimate the cycle number where PCR plateaued for each gradient. Illumina libraries were then generated from fractions by applying a cycle number (25 to 32) that maximized the amount of DNA produced but did not allow the plateauing of reaction products from individual fractions. Amplicons were checked by gel electrophoresis to confirm a single band and were then cleaned using the QIAquick PCR purification kit (Qiagen). The M13-containing amplicons were then tagged for MiSeq Illumina sequencing by including a unique 8-bp bar code in each amplicon (45). MiSeq Illumina sequencing was performed as described previously (46) with the modification of an added CC spacer between the adapter and the bar code.
Sequence classification.
Raw Illumina sequence reads were processed by first removing adapter and primer sequences and then stitching overlapping forward and reverse reads. Sequences were then clustered and assigned taxonomy using the QIIME pipeline. They were demultiplexed, clustered into OTUs using UCLUST at the 95% identity level, and checked for chimeras using USEARCH. Given the very short reads (250 bp) utilized here, the 95% level, which refers roughly to the genus level, was chosen as a compromise between a desire to retain resolution and a need for conservative interpretation of the data. This is especially true for archaea, where classification is less robust than for bacteria. A representative set of sequences was picked at random from each OTU and was aligned to the SILVA small-subunit rRNA reference alignment (www.arb-silva.de) using the PyNAST algorithm (47). Core taxa were defined as those OTUs that represented >1% of reads within a sample, and rare OTUs were defined as those representing <0.1% within a sample (48). Classification was then exported at the genus, class, and phylum levels. 16S rRNA gene frequencies at each taxonomic level were then normalized to the respective qPCR quantities (bacterial OTUs to bacterial 16S rRNA gene copy numbers by qPCR and archaeal OTUs to archaeal 16S rRNA gene copy numbers by qPCR). This was done because the frequencies of OTUs are relative to the whole 16S rRNA gene data set, which includes both bacterial and archaeal data. The qPCR normalization was also performed to account for the differential abundances and distributions of their DNA in gradients. Data were then converted to ratios of quantities in which the highest normalized frequency measured equaled 1.
Nucleotide sequence accession numbers.
Sequence data were deposited to the NCBI Sequence Read Archive under accession numbers SRR1383176 and SRR1383325.
RESULTS
The water column during late August was well mixed, with a water temperature of +4.7°C and salinity of 30.2. The chlorophyll a concentration was 0.4 μg liter−1. In winter, the water column remained well mixed, with a water temperature of −1.8°C, salinity of 33.7, and chlorophyll a concentrations of 0.01 μg liter−1. Light levels were low in the water column under the sea ice at 0.12 μmol quanta m−2 s−1 and, based on light level data from August 2010, were higher in the summer, ranging from 8 to >100 μmol quanta m−2 s−1 in the top 8 m of the water column. Summer concentrations of ambient nutrients were greatest for NH4+ (Table 1). Uptake rates were similarly dominated by NH4+; they were close to an order of magnitude greater for NH4+ than for NO3− and urea. The regeneration rate of NH4+ was more than six times greater than its uptake rate. During the winter, NO3− concentrations increased dramatically, but all of the uptake rates fell to extremely low levels, and urea uptake rates were the lowest of those measured.
TABLE 1.
Concentrations and uptake rates of ammonium, nitrate, and urea in near-shore waters of the Alaskan Arctic during January and August
| N sourcea | Concn (nmol N liter−1) (SD)b |
Uptake (nmol N liter−1 h−1) (SD)b,c |
||
|---|---|---|---|---|
| Summer | Winter | Summer | Winter | |
| NH4+ | 590 (60) | 960 (23) | 5.78 (0.34) | 0.19 (0.11) |
| NO3− | 290 (NA)d | 9,855 (1.4) | 0.82 (0.69) | 0.14 (0.04) |
| Urea | 230 (2.9) | 157 (1.9) | 0.99 (NA)d | 0.01 (0.0) |
NH4+, ammonium; NO3−, nitrate. The regeneration rate (standard deviation) for NH4+ was 38.2 (3.15) nmol N liter−1 h−1 in the summer and 10.5 (5.2) nmol N liter−1 h−1 in the winter.
Summer measurements were taken in August and winter measurements in January.
Uptake rates were determined with GF/F filters (nominal pore size, 0.7 μm).
NA, not available (n = 1).
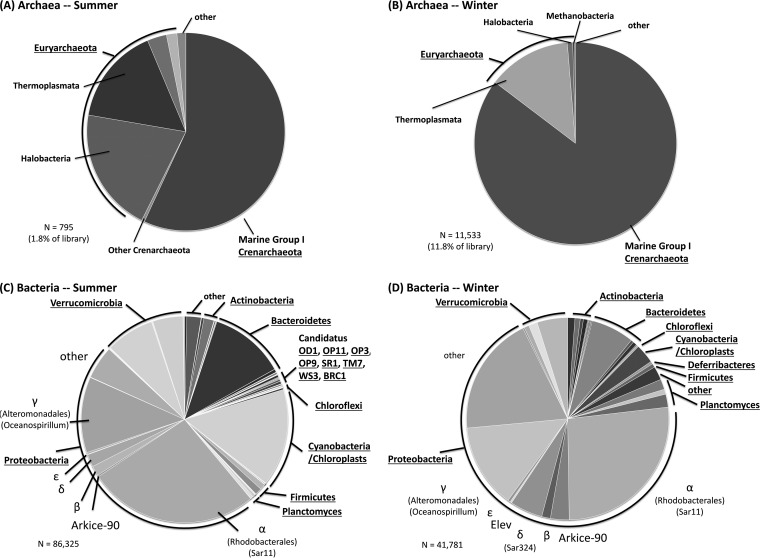
Sequencing of amplified 16S rRNA genes from ambient community DNA yielded 97,858 and 25,666 quality paired-end Illumina reads for summer and winter samples, respectively. The Shannon diversity index scores were 3.80 and 3.82, with Shannon evenness of 0.62 and 0.63 and Good's coverage of 0.99 and 0.98, for summer and winter samples, respectively. A breakdown of ambient microbial communities at the phylum/family level is shown in Fig. 1. Overall, samples contained fairly similar microbial communities during the two seasons, with some notable differences. Archaea accounted for a smaller proportion of overall reads in the summer (1.8%) than in the winter (11.8%). These data are consistent with the results from qPCR of community DNA, which detected 7.1 × 106 ± 0.41 × 106 and 2.5 × 106 ± 0.20 × 106 bacterial rRNA gene copies per ml of seawater and 8.4 × 104 ± 0.54 × 104 and 3.0 × 105 ± 0.17 × 105 archaeal rRNA gene copies per ml of seawater in summer and winter samples, respectively. Archaea were therefore 3.6-fold more abundant in winter samples than in samples collected during the summer, while bacteria were 2.8-fold more abundant during the summer. The ratio of bacterial to archaeal 16S rRNA genes was 82 in summer samples and 7.1 in winter samples, after the 2.7% and 14.9% of reads that were classified as potentially of chloroplast origin in the winter and summer, respectively, were accounted for and eliminated. Thirty-eight archaeal OTUs were shared among the summer and winter libraries, but winter communities were characterized by a greater proportion of marine group I Crenarchaeota (MGIC) and a smaller proportions of reads classified as Halobacteria (<1%) and Methanobacteria (<0.3%) (Fig. 1A and B).
FIG 1.
Phylogenetic analysis of archaeal (A and B) and bacterial (C and D) 16S rRNA gene sequences for the Arctic summer (A and C) and winter (B and D) seasons. PCR products were bar coded and sequenced using the Illumina MiSeq platform. OTUs were defined at the 95% identity level for bacteria. Representative sequences from each OTU were chosen at random, and their phylogenetic affiliations were determined using QIIME (46). Underlined taxa indicate phylum (division)-level classifications. Nonunderlined taxa are family-level assignments (not all are shown). Greek letters (α, β, γ, δ, and ε) refer to the divisions of Proteobacteria (for example, α refers to the Alphaproteobacteria).
Bacterial communities also exhibited strong similarities, with 19 core OTUs (>1.0% of reads) shared between seasons, accounting for 65% and 53% of reads during the summer and winter, respectively. The prominent difference between the two seasons consisted in greater proportions of sequences classified within the Cyanobacteria/chloroplasts and Verrucomicrobia in the summer samples (Fig. 1C and D). Winter samples, conversely, had greater proportions of reads within the Proteobacteria, Bacteroidetes, and Planctomyces than summer samples. Within the Proteobacteria, most OTUs were classified within the SAR11 clade and the Alteromonadales (Oceanospirillum) during the winter sampling. A greater diversity of taxa within the Proteobacteria was observed during the summer. Winter samples exhibited greater species richness, containing 594 unique OTUs (found only in the winter sequence libraries) compared to 144 unique taxa in summer samples. The rare biosphere was prominent, with 93% and 96% of the OTUs found in less than 0.1% of reads in libraries during the summer and winter, respectively.
CsCl DNA density gradients from 14N- and 15N-labeled NH4+, NO3−, and urea (summer and winter), as well as from 12C- and 13C-labeled bicarbonate (winter only), were fractionated. Each of the resulting fractions was assayed via qPCR for bacterial and archaeal 16S rRNA genes to estimate the degree of isotopic labeling (Fig. 2). It is assumed that minimal primer biases and minimal differences in copy numbers occurred across major taxa, given the short incubation times and the fact that no notable differences were observed between the background 16S rRNA gene distributions (shown in Fig. 1) and gradient fractions (data not shown). Keeping these caveats in mind, addition of 15NH4+ led to isotopic labeling of bacterial and archaeal DNA in summer but not winter samples, when 30% labeling for the major peak in the gradient is used as a conservative cutoff (Table 2). No evidence for the incorporation of 15NO3− into bacteria or archaea was observed in either season. Incubation with 15N-labeled urea produced no evidence of incorporation of N from urea in summer samples; however, winter samples yielded estimates of 30% and 35% isotopic labeling for bacterial and archaeal populations, respectively (Fig. 2A). Winter [14N]urea and [15N]urea treatments were therefore chosen for more-detailed analysis via high-throughput sequencing. In addition, [12C]bicarbonate- and [13C]bicarbonate-treated samples were analyzed via SIP to investigate dark (winter) carbon fixation activity by the major bacterial and archaeal populations, revealing 17 and 18% labeling of DNA with 13C for bacterial and archaeal populations in the winter, respectively.
TABLE 2.
Percentages of isotopic labeling of DNA with 15N or 13C, calculated from qPCR data of SIP fractions
| Domain | Season | Substrate | % incorporationa |
|---|---|---|---|
| Bacteria | Summer | 15NH4 | 31 |
| 15NO3− | −4 | ||
| [15N]urea | −2 | ||
| Archaea | Summer | 15NH4 | 31 |
| 15NO3− | −5 | ||
| [15N]urea | 2 | ||
| Bacteria | Winter | 15NH4 | 9 |
| 15NO3− | 6 | ||
| [15N]urea | 30 | ||
| Archaea | Winter | 15NH4 | 23 |
| 15NO3− | 17 | ||
| [15N]urea | 35 | ||
| Bacteria | Winter | [13C]bicarbonate | 17 |
| Archaea | Winter | [13C]bicarbonate | 18 |
Percentages in boldface are above the 30% and 15% thresholds used as indicators that uptake of 15N- and 13C-labeled substrates had occurred, respectively.
A total of 1.27 × 106 paired-end reads were generated from [14N]urea and [15N]urea treatments by using the Illumina MiSeq platform, yielding an average of 3.59 × 104 paired-end sequences for each fraction. The 10 most abundant divisions (proteobacteria are shown at the family level [Fig. 1]) accounted for 60% of the read data and were chosen for further analysis (Table 3). Less-abundant divisions generally did not contain sufficient read data in each fraction to resolve frequency distributions sufficiently well. Analogously, for OTU-level labeling estimates (Table 4), we restricted ourselves to OTUs that occurred at average frequencies of >0.5% of reads across gradient fractions. This cutoff was used because stochastic effects are likely to be significant at frequencies of <0.5%, given that a single read can represent a >1% difference in abundance for less-abundant taxa. For example, if a library from a fraction contains 20,000 sequences and a particular OTU is present at 0.5%, then this represents only 100 reads. Overall, labeling with N from [15N]urea appeared to be widespread among taxonomic groups and individual OTUs (Tables 3 and 4). Firmicutes and Betaproteobacteria exhibited labeling above the 30% threshold (Table 3). Alphaproteobacterial DNA displayed a shift of only 20% at the phylum level, but specific alphaproteobacterial OTUs that were classified within the SAR11 clade exhibited density shifts of >30% (Table 4). In addition, labeling of 39% and 49% was observed for a gammaproteobacterial OTU classified as Oceanospirillum and a deltaproteobacterial OTU classified within the SAR324 clade, respectively. The most abundant OTU in the data set was not classified beyond the division of Proteobacteria via QIIME but exhibited labeling for both [15N]urea and [13C]bicarbonate. MGIC exhibited labeling for both [15N]urea and [13C]bicarbonate; the dominant MGIC archaeal OTU displayed 27% labeling with both substrates.
TABLE 3.
Percentages of isotopic labeling with 15N or 13Ca derived from SIP gradients
| Domain | Division/class | Substrate | % incorporationb |
|---|---|---|---|
| Archaea | Crenarchaeota | [15N]urea | 31 |
| Bacteria | Actinobacteria | [15N]urea | 17 |
| Bacteroidetes | [15N]urea | 22 | |
| Firmicutes | [15N]urea | 31 | |
| Planctomycetes | [15N]urea | 22 | |
| Verrucomicrobia | [15N]urea | 12 | |
| Alphaproteobacteria | [15N]urea | 20 | |
| Betaproteobacteria | [15N]urea | 33 | |
| Deltaproteobacteria | [15N]urea | 25 | |
| Gammaproteobacteria | [15N]urea | 23 | |
| Archaea | Crenarchaeota | [13C]bicarbonate | 22 |
| Bacteria | Actinobacteria | [13C]bicarbonate | 14 |
| Bacteroidetes | [13C]bicarbonate | 12 | |
| Firmicutes | [13C]bicarbonate | 17 | |
| Planctomycetes | [13C]bicarbonate | 14 | |
| Verrucomicrobia | [13C]bicarbonate | 12 | |
| Alphaproteobacteria | [13C]bicarbonate | 18 | |
| Betaproteobacteria | [13C]bicarbonate | 15 | |
| Deltaproteobacteria | [13C]bicarbonate | 18 | |
| Gammaproteobacteria | [13C]bicarbonate | 15 |
Percentages were calculated from 16S rRNA gene frequency data with respect to densities of fractions.
Percentages in boldface are at or above the 30% and 15% thresholds used as indicators that uptake of 15N- and 13C-labeled substrates had occurred, respectively.
TABLE 4.
Percentages of isotopic labeling of OTUs in winter samplesa with [15N]urea or [13C]bicarbonate
| Domain | Division/class | Family/genus | % labelingb with: |
|
|---|---|---|---|---|
| 15N | 13C | |||
| Bacteria | Proteobacteria | Unclassified | 35 | 18 |
| Alphaproteobacteria | SAR11 (Surface 1) | 30 | 14 | |
| Archaea | Crenarchaeota | Marine group I | 27 | 27 |
| Bacteria | Gammaproteobacteria | Oceanospirillales | 39 | 12 |
| Alphaproteobacteria | SAR11 (Surface 2) | 31 | 22 | |
| Verrucomicrobia | Verrucomicrobiaceae | 13 | 3 | |
| Alphaproteobacteria | Rhodobacteraceae | 19 | 15 | |
| Gammaproteobacteria | Alteromonadaceae | 28 | 17 | |
| Deltaproteobacteria | SAR324 (marine group B) | 49 | 13 | |
| Alphaproteobacteria | SAR11 (Chesapeake Delaware Bay) | 32 | 17 | |
| Verrucomicrobia | Verrucomicrobiaceae | 13 | 6 | |
| Deltaproteobacteria | Nitrospinaceae | 16 | 9 | |
| Alphaproteobacteria | Rhodobacteraceae | 13 | 7 | |
| Bacteroidetes | Cryomorphaceae | 42 | −9 | |
| Alphaproteobacteria | SAR11 (Surface 4) | 32 | 16 | |
| Alphaproteobacteria | SAR11 (Deep 1) | 37 | 14 | |
| Alphaproteobacteria | Rhodospirillaceae | 29 | 20 | |
| Actinobacteria | Acidimicrobiales (SVA0996) | 32 | 5 | |
Only OTUs that accounted for more than 0.5% of reads across libraries from gradient fractions are shown. OTUs are shown in rank order, with the first being the most abundant.
Percentages were calculated from 16S rRNA gene OTU frequencies in Illumina libraries with respect to densities of fractions. Percentages in boldface are at or above the 30% and 15% thresholds used as indicators that uptake of 15N- and 13C-labeled substrates occurred, respectively.
Both the bacterial and archaeal qPCR analyses of SIP fractions provided evidence of [13C]bicarbonate uptake during the winter (Fig. 2B), exhibiting 17% and 18% labeling, respectively. Sequencing of fractions from [12C]bicarbonate- and [13C]bicarbonate-treated samples produced 1.40 × 106 reads, which yielded an average of 2.38 × 104 paired-end sequences for each fraction. No appreciable difference in overall phylogenetic composition was observed between these data and the 15N-labeled fractions (data not shown). Evidence of labeling was observed for all four major proteobacterial families detected here, as well as for the Firmicutes and the Crenarchaeota.
DISCUSSION
Marine bacterial and archaeal plankton play a critical role in the biogeochemical cycling of C and N in the world's oceans (49, 50). Identification of the phylogenetic groups that are responsible for specific C or N cycling activities can provide insight into the forces that drive marine productivity and community function, as well as the spatial and temporal dynamics of individual C and N transformation processes themselves. The work presented here aimed to identify the dominant groups of bacteria and archaea that incorporate DIN and urea into DNA and to quantify the rates of uptake of these N substrates. In addition, we explored dark carbon fixation by bacteria and archaea, which can be coupled to ammonium oxidation via urea deamination.
SIP data suggest an important seasonal transition of microbial N incorporation from NH4+ during the summer to urea during the winter (Table 2). In addition, SIP implicated a broad range of microbial taxa in urea utilization during the winter, including Proteobacteria, Firmicutes, and MGIC. Utilization of [15N]urea was not necessarily expected, given that other sources of N were available, but it is not inconsistent with the phylogenetic distribution of urease genes among bacteria and archaea. Although it has been argued that urease genes are rare in the domain Archaea (17), and it has been noted that urease genes are absent from the genomes of Nitrosopumilus maritimus and “Candidatus Nitrosoarchaeum limnia” (51), it has also been observed that ureC genes are abundant in polar archaea, with an average ureC/16S rRNA gene ratio close to or greater than 1 (51). Likewise, urease-related genes were identified in “Candidatus Nitrosopumilus sediminis” AR2, a species cultured from Arctic sediments (52). A search of genome sequences available in the DOE's Joint Genome Institute IMG database (as of August 2013) identified urease genes within the genomes of several Halobacteria and Crenarchaeota; the latter are relevant to our samples. Similarly, the IMG search identified urease genes in the genomes of Alpha-, Beta-, Gamma-, and Epsilonproteobacteria, Actinobacteria, and Firmicutes, which were the main bacterial groups in our samples. In contrast to the SIP observations, urea uptake rates were very low in both summer and winter samples. This discrepancy likely reflects our choice of filters. Filters with a nominal pore size of 0.7 μm (GF/F filters) were used to determine rates, while 0.45-μm filters were used for DNA extractions for SIP. Flow cytometric analysis of seawater indicated that most bacterial cells in the winter were <0.7 μm (data not shown). Thus, the urea uptake rates reported here likely underestimate actual in situ activity, particularly in the winter. In the future, therefore, investigators conducting rate and SIP studies in the Arctic should consider filters with smaller pore sizes (e.g., 0.1 μm) in order to capture a greater fraction of active bacteria.
Summer and winter samples exhibited good incorporation of 15N from NH4+ and urea, respectively, indicating that incubation times (24 h) were sufficiently long. A lack of labeling with 15N from NH4+ or urea might indicate incubation times inconsistent with uptake rates. Conversely, incubation times that are too long can lead to cross-feeding and eliminate the ability to resolve substrate-specific uptake in SIP experiments (43). Cross-feeding results when a substrate is added in one form but is converted to another form during incubation. Nitrification, for example, can occur at high rates during the Arctic winter (53; Baer et al., submitted), and therefore, additions of 15NH4+ could result in the production of 15NO3−, which could subsequently be incorporated. Additionally, urea is likely converted to NH4+, but there is no current information on the rate of that process in the environment. Given those caveats, our relatively short incubation times, and low ambient rates of uptake (Table 1), cross-feeding was not likely to be an important factor in our results.
Incorporation of N from 15NO3− by bacterial or archaeal populations was not supported by SIP for either season (Table 2). This is in contrast to the findings of prior studies indicating that NO3− can serve as an important N source for bacterioplankton in the Arctic and sub-Arctic, accounting for 16 to 40% of NO3− uptake (24, 53, 54). A meta-analysis of these and other data suggests a pattern of greater NO3− utilization by heterotrophic bacterioplankton under low chlorophyll conditions and relatively lower contributions when phytoplankton are more prevalent (24). Although NO3− incorporation, as determined via SIP, was not observed in this study, we note that background NO3− levels (∼10 μM [Table 1]) in winter samples were substantially above the levels of our additions. SIP experiments, as performed here, require an excess of 15N-labeled substrate in order to result in >30% labeling of cellular DNA. Additions of 15NO3− to winter samples were therefore likely below the limit of detection. Summer additions, in contrast, were sufficiently high, and a lack of 15N labeling of DNA is consistent with the domination of NO3− uptake by phytoplankton (54). It has also been noted that NO3− is likely the least preferred N source for supporting bacterial growth (55); i.e., it may be the N source of last resort, especially during times other than the spring bloom, when energetically more favorable sources of N are abundant (Baer et al., submitted).
Community analysis of 16S rRNA genes revealed that summer and winter communities were largely similar, while displaying some notable differences. Minor seasonal differences in microbial community composition are in line with previous reports for Arctic Ocean microbial communities (56–58). For example, analysis of denaturing gradient gel electrophoresis (DGGE) fingerprints from Arctic and Antarctic samples revealed no seasonal variation in archaeal community structure but suggested greater richness of archaea in water from greater depths (58). The dominant contributors to the archaeal community in the study by Bano et al. (58) and in a study of five distinct water masses in the Arctic Ocean (59) were MGIC, as in the populations described here. We also observed a greater proportion of archaea during the winter, as in prior observations (51). The abundance of MGIC in the southeast Beaufort Sea has been observed to reach 18% in winter, decreasing to ca. 5% of cells in the spring of the same year (51). This change was attributed to the growth of the archaeal populations during the winter and not due to mixing with deeper water masses, which often contain greater proportions of these organisms. Overall, these data suggest that archaea are a salient and stable feature of Arctic marine planktonic communities and that their relative importance increases during the dark, cold months of the Arctic winter. Seasonal differences in bacterial communities are equally constrained. A study using samples collected from the western Arctic reported no significant seasonal differences in bacterial communities, as determined via pyrotag sequencing, despite large differences in biogeochemical parameters (56). The dominant bacterial taxa in that study were the Alpha- and Gammaproteobacteria, as well as the Bacteroidetes (Flavobacteria) (56). Our findings match these observations, although Kirchman and Wheeler did not observe a shift toward greater proportions of chloroplast-like sequences during the summer. This discrepancy likely arises from the use of 0.8-μm-prefiltered samples to remove much of the eukaryotic phytoplankton, including their chloroplasts.
Urea is present in relatively high concentrations in polar seawater (22, 23, 51), and its role as an important component of the Arctic N cycle has been recognized. Urea can account for as much as 30 to 50% of the N assimilated by phytoplankton annually (22, 23) and as much as 80% of the regenerated production during the spring bloom (23). Urea has also been reported to have half-saturation constants similar to those of NH4+ but greater maximum uptake rates (59). A more recent study has investigated the role of urea in nitrification by polar marine archaea (51). In that study, metagenomic analysis of Arctic winter samples revealed an abundance of urea transport and degradation genes. Quantitative PCR assays resulted in good correlation between the numbers of MGIC and ureC genes, suggesting that ureC genes were abundant in polar archaea. Experiments with 14C-labeled urea demonstrated C uptake from urea (carbon dioxide is generated from urea by urease activity) by the 0.2- to 0.6-μm fraction and showed that this activity was greater under dark conditions (51). The implication is that urea may fuel nitrification and autotrophic growth by polar archaea via the release of ammonium from urea. This may be advantageous under dark conditions, when urea can be a more reliable source of energy for ammonium oxidizers (2). Urea may also be produced continually during the Arctic winter via microbial and zooplankton (20) activities. The notion of urea-fueled nitrification is not without precedent. It has been argued that urea hydrolysis may serve to generate ammonium and carbon dioxide by ammonium oxidizers (60), allowing these bacteria to generate energy to fuel dark carbon fixation.
Studies using microautoradiography-catalyzed reporter deposition fluorescence in situ hybridization (MAR-CARDFISH) have shown that bacteria and archaea from the Beaufort and Chukchi Seas actively incorporated bicarbonate (61). The study in the Beaufort Sea targeted specific bacterial taxa and found that Gamma- and Betaproteobacteria, specifically organisms of the gammaproteobacterial genera Oleispira and Pseudoalteromonas-Colwellia, were actively assimilating bicarbonate when seawater cultures were grown on a resource-deplete medium (26). Our results demonstrate that dark carbon fixation is not limited to dilution cultures grown under enhanced substrate depletion conditions but may be a viable metabolic strategy for diverse taxa under in situ conditions. Moreover, our work expands on the previously reported diversity of bacteria and archaea active in assimilating dissolved inorganic carbon (DIC) in the Arctic during the winter.
The relevance of chemoautotrophy, such as ammonia oxidation by bacteria and archaea, in Arctic waters may increase from summer to winter, when light levels and photosynthetic primary production are low (53). The important role of ammonia-oxidizing Crenarchaeota to carbon fixation and nitrogen cycling in deeper waters of the world's oceans is becoming well established (29, 30, 62). The significance of ammonia oxidation in surface waters of the ocean has, however, been argued to be minor due to inhibition from light and competition with phytoplankton for ammonia (63, 64). Arctic winter surface waters are comparable to deeper waters in the ocean due to limited light as a consequence of sea ice coverage, short days, and low inputs of organic matter from photosynthesis. Further, ammonium concentrations in the Arctic are higher in winter than in summer (53; this study). Thus, the role of chemoautotrophy from ammonia oxidation may be notable in Arctic surface waters during winter, as it is for deeper waters in the world's oceans. In agreement with this proposition, Christman et al. (53) found that ammonia monooxygenase (amoA) genes from archaea and bacteria in surface waters near Barrow, AK, were almost 2 orders of magnitude more abundant in the winter than in the summer, with most or all Crenarchaeota likely capable of ammonia oxidation. Metagenomic analysis of winter (August) surface communities near the Antarctic Peninsula were consistent with bacterial and archaeal autotrophy, with 18 to 37% of the community belonging to known or putative chemolithoautotrophs (65). Similarly, metaproteomic analysis of coastal winter samples taken from the same region in Antarctica supported the utilization of chemolithoautotrophic metabolism by ammonia-oxidizing archaea and nitrite-oxidizing bacteria (66).
In addition to chemoautotrophic contributions to DIC uptake, all heterotrophic bacteria are thought to assimilate bicarbonate via pathways involved in anaplerotic reactions of the tricarboxylic acid cycle. However, carbon fixation by heterotrophs via anaplerotic reactions is assumed to account for only 1 to 8% of bacterial biomass production (67, 68) and thus would play a minor role in DIC uptake. In contrast, ammonium additions failed to stimulate bicarbonate assimilation, leading Alonso-Sáez et al. (26) to conclude that the high rates of bicarbonate assimilation they observed in their seawater cultures resulted from dark carbon fixation by heterotrophs. Although we cannot differentiate between chemoautotrophic and heterotrophic assimilation with [13C]bicarbonate SIP data, both metabolisms likely contributed to our results. Thus, future research should consider that dark carbon fixation may be an essential survival strategy for heterotrophs, chemoautotrophs, mixotrophs, and other organisms in the microbial loop that rely on archaeal and bacterial production during the Arctic winter.
To determine the extent to which dark carbon fixation is a significant aspect of community ecology during the Arctic winter, quantification of in situ rates of bicarbonate uptake by bacteria and archaea in the dark is essential (as in reference 69). Preliminary data from our study area in January suggest a bicarbonate uptake rate of 0.09 μg C liter−1 day−1 (S. E. Baer and D. A. Bronk, unpublished data), which is similar to rates observed by L. Alonso-Sáez (personal communication) in the Beaufort Sea and is within the range for heterotrophic production rates in the Arctic in the winter and the spring (70, 71). These scarce uptake measurements and the phylogenetic diversity of labeled populations in [15N]urea and [13C]bicarbonate SIP experiments suggest that dark carbon fixation may be notable for marine Arctic ecosystems during the winter, highlighting the need for additional observations.
The current study, in conjunction with discoveries made by others, continues to call our attention to several unresolved questions, such as the following. Is a seasonal shift in NH4+ and urea utilization widespread? What regulates N utilization in the Arctic? How does dark carbon fixation influence other members of the ecosystem, such as bacterivores, especially in oligotrophic environments such as the Arctic in winter? What are the capacity and rate of dark DIC assimilation by heterotrophic bacteria? And what is the role of urea uptake and dark carbon fixation in the N and C budgets of the world's oceans? These questions remain poorly addressed for aerobic chemoautotrophs, mixotrophs, and heterotrophs. Recent methodological advances with DNA-SIP, RNA-SIP, lipid-SIP, MAR-CARDFISH, and metagenomics hold promise for helping us resolve the importance of these metabolisms on a seasonal basis in the Arctic Ocean or on a global scale and for deepening our understanding of the life history strategies adapted for survival in resource-limited environments.
ACKNOWLEDGMENTS
We thank Q. N. Roberts, K. Sines, and R. E. Sipler for field work. We also thank M. P. Sanderson for running mass spectrometry samples.
This work would not have been possible without the guidance and support of the UMIAQ support team, led by B. France, and support by U.S. National Science Foundation grants OCE 0961900, ARC 0910252, and ARC 0909839.
Footnotes
Published ahead of print 25 July 2014
This article is contribution 3386 from the Virginia Institute of Marine Science, College of William and Mary.
REFERENCES
- 1.Duarte CM, Lenton TM, Wadhams P, Wassmann P. 2012. Abrupt climate change in the Arctic. Nat. Clim. Change 2:60–62. 10.1038/nclimate1386 [DOI] [Google Scholar]
- 2.Parmentier F-JW, Christensen TR, Sorensen LL, Rysgaard S, McGuire AD, Miller PA, Walker DA. 2013. The impact of lower sea-ice extent on Arctic greenhouse-gas exchange. Nat. Clim. Change 3:195–202. 10.1038/nclimate1784 [DOI] [Google Scholar]
- 3.Holland MM, Bitz CM. 2003. Polar amplification of climate change in coupled models. Clim. Dynam. 21:221–232. 10.1007/s00382-003-0332-6 [DOI] [Google Scholar]
- 4.Stroeve JC, Serreze MC, Holland MM, Kay JE, Maslanik J, Barrett AP. 2012. The Arctic's rapidly shrinking sea ice cover: a research synthesis. Clim. Change 110:1005–1027. 10.1007/s10584-011-0101-1 [DOI] [Google Scholar]
- 5.Serreze MC, Francis JA. 2006. The arctic amplification debate. Clim. Change 76:214–264. 10.1007/s10584-005-9017-y [DOI] [Google Scholar]
- 6.Doney SC, Ruckelshaus M, Emmett Duffy J, Barry JP, Chan F, English CA, Galindo HM, Grebmeier JM, Hollowed AB, Knowlton N, Polovina J, Rabalais NN, Sydeman WJ, Talley LD. 2012. Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 4:11–37. 10.1146/annurev-marine-041911-111611 [DOI] [PubMed] [Google Scholar]
- 7.Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F. 2012. Impacts of climate change on the future of biodiversity. Ecol. Lett. 15:365–377. 10.1111/j.1461-0248.2011.01736.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perrette M, Yool A, Quartly GD, Popova EE. 2011. Near-ubiquity of ice-edge blooms in the Arctic. Biogeosciences 8:515–524. 10.5194/bg-8-515-2011 [DOI] [Google Scholar]
- 9.Sherr EB, Sherr BF, Wheeler PA, Thompson K. 2003. Temporal and spatial variation in stocks of autotrophic and heterotrophic microbes in the upper water column of the central Arctic Ocean. Deep Sea Res. Part I Oceanogr. Res. Pap. 50:557–571. 10.1016/S0967-0637(03)00031-1 [DOI] [Google Scholar]
- 10.Antia NJ, Harrison PJ, Oliveira L. 1991. The role of dissolved organic nitrogen in phytoplankton nutrition, cell biology and ecology. Phycologia 30:1–89. 10.2216/i0031-8884-30-1-1.1 [DOI] [Google Scholar]
- 11.Aluwihare LI, Meador T. 2008. Chemical composition of marine dissolved organic nitrogen, p 95–140 In Capone DG, Bronk DA, Mulholland M, Carpenter EJ. (ed), Nitrogen in the marine environment, 2nd ed. Elsevier, San Diego, CA [Google Scholar]
- 12.Letscher RT, Hansell DA, Kadko D, Bates NR. 2013. Dissolved organic nitrogen dynamics in the Arctic Ocean. Mar. Chem. 148:1–9. 10.1016/j.marchem.2012.10.002 [DOI] [Google Scholar]
- 13.L'Helguen S, Slawyk G, Le Corre P. 2005. Seasonal patterns of urea regeneration by size-fractionated microheterotrophs in well-mixed temperate coastal waters. J. Plankton Res. 27:263–270. 10.1093/plankt/fbh174 [DOI] [Google Scholar]
- 14.Remsen CC. 1971. The distribution of urea in coastal and oceanic waters. Limnol. Oceanogr. 16:732–740. 10.4319/lo.1971.16.5.0732 [DOI] [Google Scholar]
- 15.Remsen CC, Carpenter EJ, Schroeder BW. 1974. The role of urea in marine microbial ecology, p 286–304 In Colwell RR, Morita RY. (ed), Effect of the ocean environment on microbial activities. University Park Press, Baltimore, MD [Google Scholar]
- 16.McCarthy JJ. 1972. The uptake of urea by natural populations of marine phytoplankton. Limnol. Oceanogr. 17:738–748. 10.4319/lo.1972.17.5.0738 [DOI] [Google Scholar]
- 17.Solomon CM, Collier JL, Berg GM, Glibert PM. 2010. Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquat. Microb. Ecol. 59:67–88. 10.3354/ame01390 [DOI] [Google Scholar]
- 18.Glibert P, Magnien R, Lomas M, Alexander J, Tan C, Haramoto E, Trice M, Kana T. 2001. Harmful algal blooms in the Chesapeake and Coastal Bays of Maryland, USA: comparison of 1997, 1998, and 1999 events. Estuaries 24:875–883. 10.2307/1353178 [DOI] [Google Scholar]
- 19.Conover RJ, Gustavson KR. 1999. Sources of urea in arctic seas: zooplankton metabolism. Mar. Ecol. Prog. Ser. 179:41–54. 10.3354/meps179041 [DOI] [Google Scholar]
- 20.Conover RJ, Mumm N, Bruecker P, MacKenzie S. 1999. Sources of urea in arctic seas: seasonal fast ice. Mar. Ecol. Prog. Ser. 179:55–69. 10.3354/meps179055 [DOI] [Google Scholar]
- 21.Pedersen H, Lomstein BA, T. Henry B. 1993. Evidence for bacterial urea production in marine sediments. FEMS Microbiol. Ecol. 12:51–59. 10.1111/j.1574-6941.1993.tb00016.x [DOI] [Google Scholar]
- 22.Harrison WG, Head EJH, Conover RJ, Longhurst AR, Sameoto DD. 1985. The distribution and metabolism of urea in the eastern Canadian Arctic. Deep Sea Res. Part A Oceanogr. Res. Pap. 32:23–42. 10.1016/0198-0149(85)90015-9 [DOI] [Google Scholar]
- 23.Simpson KG, Tremblay JE, Brugel S, Price NM. 2013. Nutrient dynamics in the western Canadian Arctic. II. Estimates of new and regenerated production over the Mackenzie Shelf and Cape Bathurst Polynya. Mar. Ecol. Prog. Ser. 484:47–62. 10.3354/meps10298 [DOI] [Google Scholar]
- 24.Fouilland E, Gosselin M, Rivkin RB, Vasseur C, Mostajir B. 2007. Nitrogen uptake by heterotrophic bacteria and phytoplankton in Arctic surface waters. J. Plankton Res. 29:369–376. 10.1093/plankt/fbm022 [DOI] [Google Scholar]
- 25.Wuchter C, Schouten S, Boschker HTS, Damste JSS. 2003. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol. Lett. 219:203–207. 10.1016/S0378-1097(03)00060-0 [DOI] [PubMed] [Google Scholar]
- 26.Alonso-Sáez L, Galand PE, Casamayor EO, Pedros-Alio C, Bertilsson S. 2010. High bicarbonate assimilation in the dark by Arctic bacteria. ISME J. 4:1581–1590. 10.1038/ismej.2010.69 [DOI] [PubMed] [Google Scholar]
- 27.Hügler M, Sievert SM. 2011. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu. Rev. Mar. Sci. 3:261–289. 10.1146/annurev-marine-120709-142712 [DOI] [PubMed] [Google Scholar]
- 28.DeLorenzo S, Bräuer SL, Edgmont CA, Herfort L, Tebo BM, Zuber P. 2012. Ubiquitous dissolved inorganic carbon assimilation by marine bacteria in the Pacific Northwest coastal ocean as determined by stable isotope probing. PLoS One 7:e46695. 10.1371/journal.pone.0046695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, Pernthaler J. 2005. Contribution of archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71:2303–2309. 10.1128/AEM.71.5.2303-2309.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ERM, Pearson A. 2006. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc. Natl. Acad. Sci. U. S. A. 103:6442–6447. 10.1073/pnas.0510157103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yakimov MM, La Cono V, Smedile F, DeLuca TH, Juarez S, Ciordia S, Fernandez M, Albar JP, Ferrer M, Golyshin PN, Giuliano L. 2011. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea). ISME J. 5:945–961. 10.1038/ismej.2010.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koroleff F. 1983. Determination of nutrients, p 125–187 In Grasshoff K, Ehrhardt M, Kremling K. (ed), Methods of seawater analysis. Verlag Chemie, New York, NY [Google Scholar]
- 33.Parsons TR, Maita Y, Lalli CM. 1984. A manual of chemical and biological methods for seawater analysis. Pergamon, Oxford, United Kingdom [Google Scholar]
- 34.Price NM, Harrison PJ. 1987. Comparison of methods for the analysis of dissolved urea in seawater. Mar. Biol. 94:307–317. 10.1007/BF00392945 [DOI] [Google Scholar]
- 35.Dugdale RC, Goering JJ. 1967. Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol. Oceanogr. 12:196–206. 10.4319/lo.1967.12.2.0196 [DOI] [Google Scholar]
- 36.Hama T, Miyazaki T, Ogawa Y, Iwakuma T, Takahashi M, Otsuki A, Ichimura S. 1983. Measurement of photosynthetic production of a marine-phytoplankton population using a stable C-13 isotope. Mar. Biol. 73:31–36. 10.1007/BF00396282 [DOI] [Google Scholar]
- 37.Dudek N, Brzezinski MA, Wheeler PA. 1986. Recovery of ammonium nitrogen by solvent extraction for the determination of relative 15N abundance in regeneration experiments. Mar. Chem. 18:59–69. 10.1016/0304-4203(86)90076-9 [DOI] [Google Scholar]
- 38.Brzezinski MA. 1987. Colorimetric determination of nanomolar concentrations of ammonium in seawater using solvent extraction. Mar. Chem. 20:277–288. 10.1016/0304-4203(87)90078-8 [DOI] [Google Scholar]
- 39.Glibert PM, Lispschultz F, McCarthy JJ, Altabet MA. 1982. Isotope dilution models of uptake and remineralization of ammonium by marine plankton. Limnol. Oceanogr. 27:639–650. 10.4319/lo.1982.27.4.0639 [DOI] [Google Scholar]
- 40.Wawrik B, Callaghan AV, Bronk DA. 2009. Use of inorganic and organic nitrogen use by Synechococcus spp. and diatoms on the West Florida Shelf as measured using stable isotope probing. Appl. Environ. Microbiol. 75:6662–6670. 10.1128/AEM.01002-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buckley DH, Huangyutitham V, Hsu S-F, Nelson TA. 2007. Stable isotope probing with 15N2 reveals novel noncultivated diazotrophs in soil. Appl. Environ. Microbiol. 73:3196–3204. 10.1128/AEM.02610-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buckley DH, Huangyutitham V, Hsu S-F, Nelson TA. 2007. Stable isotope probing with 15N achieved by disentangling the effects of genome G+C content and isotope enrichment on DNA density. Appl. Environ. Microbiol. 73:3189–3195. 10.1128/AEM.02609-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wawrik B, Boling WB, Van Nostrand JD, Xie JP, Zhou JZ, Bronk DA. 2012. Assimilatory nitrate utilization by bacteria on the West Florida Shelf as determined by stable isotope probing and functional microarray analysis. FEMS Microbiol. Ecol. 79:400–411. 10.1111/j.1574-6941.2011.01226.x [DOI] [PubMed] [Google Scholar]
- 44.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wawrik B, Mendivelso M, Parisi VA, Suflita JM, Davidova IA, Marks CR, Van Nostrand JD, Liang Y, Zhou J, Huizinga BJ, Strapoc D, Callaghan AV. 2012. Field and laboratory studies on the bioconversion of coal to methane in the San Juan Basin. FEMS Microbiol. Ecol. 81:26–42. 10.1111/j.1574-6941.2011.01272.x [DOI] [PubMed] [Google Scholar]
- 46.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedrós-Alió C. 2006. Marine microbial diversity: can it be determined? Trends Microbiol. 14:257–263. 10.1016/j.tim.2006.04.007 [DOI] [PubMed] [Google Scholar]
- 49.Kirchman DL, Moran XA, Ducklow H. 2009. Microbial growth in the polar oceans: role of temperature and potential impact of climate change. Nat. Rev. Microbiol. 7:451–459. 10.1038/nrmicro2115 [DOI] [PubMed] [Google Scholar]
- 50.Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5:782–791. 10.1038/nrmicro1747 [DOI] [PubMed] [Google Scholar]
- 51.Alonso-Sáez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, Lovejoy C, Tremblay J-É, Potvin M, Heinrich F, Estrada M, Riemann L, Bork P, Pedrós-Alió C, Bertilsson S. 2012. Role for urea in nitrification by polar marine Archaea. Proc. Natl. Acad. Sci. U. S. A. 109:17989–17994. 10.1073/pnas.1201914109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park SJ, Kim JG, Jung MY, Kim SJ, Cha IT, Ghai R, Martin-Cuadrado AB, Rodriguez-Valera F, Rhee SK. 2012. Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus sediminis” AR2, from Svalbard in the Arctic Circle. J. Bacteriol. 194:6948–6949. 10.1128/JB.01869-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christman GD, Cottrell MT, Popp BN, Gier E, Kirchman DL. 2011. Abundance, diversity, and activity of ammonia-oxidizing prokaryotes in the coastal Arctic Ocean in summer and winter. Appl. Environ. Microbiol. 77:2026–2034. 10.1128/AEM.01907-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simpson KG, Tremblay J-É, Price NM. 2013. Nutrient dynamics in the western Canadian Arctic. I. New production in spring inferred from nutrient draw-down in the Cape Bathurst Polynya. Mar. Ecol. Prog. Ser. 484:33–45. 10.3354/meps10275 [DOI] [Google Scholar]
- 55.Kirchman DL, Wheeler PA. 1998. Uptake of ammonium and nitrate by heterotrophic bacteria and phytoplankton in the sub-Arctic Pacific. Deep Sea Res. Part I Oceanogr. Res. Pap. 45:347–365. 10.1016/S0967-0637(97)00075-7 [DOI] [Google Scholar]
- 56.Kirchman DL, Cottrell MT, Lovejoy C. 2010. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 12:1132–1143. 10.1111/j.1462-2920.2010.02154.x [DOI] [PubMed] [Google Scholar]
- 57.Nikrad MP, Cottrell MT, Kirchman DL. 2012. Abundance and single-cell activity of heterotrophic bacterial groups in the western Arctic Ocean in summer and winter. Appl. Environ. Microbiol. 78:2402–2409. 10.1128/AEM.07130-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bano N, Ruffin S, Ransom B, Hollibaugh JT. 2004. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl. Environ. Microbiol. 70:781–789. 10.1128/AEM.70.2.781-789.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith WO, Harrison WG. 1991. New production in polar regions: the role of environmental controls. Deep Sea Res. Part A Oceanogr. Res. Pap. 38:1463–1479. 10.1016/0198-0149(91)90085-T [DOI] [Google Scholar]
- 60.Koper TE, El-Sheikh AF, Norton JM, Klotz MG. 2004. Urease-encoding genes in ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 70:2342–2348. 10.1128/AEM.70.4.2342-2348.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirchman DL, Elifantz H, Dittel AI, Malmstrom RR, Cottrell MT. 2007. Standing stocks and activity of Archaea and Bacteria in the western Arctic Ocean. Limnol. Oceanogr. 52:495–507. 10.4319/lo.2007.52.2.0495 [DOI] [Google Scholar]
- 62.Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Sinninghe Damsté JS. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317–12322. 10.1073/pnas.0600756103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward BB. 2008. Nitrification in marine systems, p 199–262 In Capone DG, Bronk DA, Mulholland MR, Carpenter EJ. (ed), Nitrogen in the marine environment, 2nd ed. Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 64.Middelburg JJ. 2011. Chemoautotrophy in the ocean. Geophys. Res. Lett. 38:L24604. 10.1029/2011GL049725 [DOI] [Google Scholar]
- 65.Grzymski JJ, Riesenfeld CS, Williams TJ, Dussaq AM, Ducklow H, Erickson M, Cavicchioli R, Murray AE. 2012. A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J. 6:1901–1915. 10.1038/ismej.2012.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams TJ, Long E, Evans F, DeMaere MZ, Lauro FM, Raftery MJ, Ducklow H, Grzymski JJ, Murray AE, Cavicchioli R. 2012. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 6:1883–1900. 10.1038/ismej.2012.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li WKW. 1982. Estimating heterotrophic bacterial productivity by inorganic radiocarbon uptake: importance of establishing time courses of uptake. Mar. Ecol. Prog. Ser. 8:167–172. 10.3354/meps008167 [DOI] [Google Scholar]
- 68.Roslev P, Larsen MB, Jorgensen D, Hesselsoe M. 2004. Use of heterotrophic CO2 assimilation as a measure of metabolic activity in planktonic and sessile bacteria. J. Microbiol. Methods 59:381–393. 10.1016/j.mimet.2004.08.002 [DOI] [PubMed] [Google Scholar]
- 69.Reinthaler T, van Aken HM, Herndl GJ. 2010. Major contribution of autotrophy to microbial carbon cycling in the deep North Atlantic's interior. Deep Sea Res. Part II Top. Stud. Oceanogr. 57:1572–1580. 10.1016/j.dsr2.2010.02.023 [DOI] [Google Scholar]
- 70.Garneau ME, Roy S, Lovejoy C, Gratton Y, Vincent WF. 2008. Seasonal dynamics of bacterial biomass and production in a coastal arctic ecosystem: Franklin Bay, western Canadian Arctic. J. Geophys. Res. Oceans 113(C7). 10.1029/2007JC004281 [DOI] [Google Scholar]
- 71.Nguyen D, Maranger R, Tremblay JE, Gosselin M. 2012. Respiration and bacterial carbon dynamics in the Amundsen Gulf, western Canadian Arctic. J. Geophys. Res. Oceans 117(C9). 10.1029/2011JC007343 [DOI] [Google Scholar]