Abstract
Bifidobacteria are members of the gut microbiota, but the genetic basis for their adaptation to the human gut is poorly understood. The analysis of the 2,203,222-bp genome of Bifidobacterium adolescentis 22L revealed a nutrient acquisition strategy that targets diet/plant-derived glycans, in particular starch and starch-like carbohydrates. Starch-like carbohydrates were shown to support the growth of B. adolescentis 22L. Transcriptome profiling of 22L cultures grown under in vitro conditions or during colonization of the murine gut by RNA sequencing and quantitative real-time PCR assays revealed the expression of a set of chromosomal loci responsible for starch metabolism as well as for pilus production. Such extracellular structures include so-called sortase-dependent and type IVb pili, which may be involved in gut colonization of 22L through adhesion to extracellular matrix proteins.
INTRODUCTION
The human intestine is essentially sterile at birth, but then it rapidly becomes colonized by bacteria, which leads to the development of an intestinal microbiota (1). Bifidobacterial species display an intriguing adaptation to different stages of human life, in that there appears to be infant-specific bifidobacterial species and adult-specific bifidobacterial taxa (2, 3). However, there is no definite proof to state that certain bifidobacterial taxa are detected in either adults or infants only, and the distinction between such adult-type and infant-type bifidobacteria is based on their relative abundance in the adult or infant gut (3). Recent investigations have demonstrated that infant-associated bifidobacterial species, e.g., Bifidobacterium longum subsp. infantis ATCC 15697 and Bifidobacterium bifidum PRL2010, can metabolize host-derived glycans, in particular human milk oligosaccharides (HMOs), typically present in the gut of (breast-fed) neonates (4, 5). However, so far very little is known about how such diet-derived glycans, being recalcitrant to the enzymes of their mammalian host, have shaped the genome sequences of adult-type bifidobacteria. In this context, starch represents one of the most abundant carbohydrates in the human diet, both in adults as well as in infants after weaning (6). Starch breakdown by gut bacteria occurs through the combined action of amylases (EC 3.2.1.1, EC 3.2.1.2, and EC 3.2.1.3) and amylopullulanases (APU; EC 3.2.1.41). Starch is metabolized by different members of the gut microbiota belonging to various microbial taxa, such as Ruminococcus bromii (7), Bacteroides thetaiotaomicron (8), and Roseburia inulinivorans (9), where various amylases have been identified (7, 8). Even if bifidobacteria are nondominant members of the adult gut microbiota, their biological roles in the metabolism of dietary and host-derived glycans have only recently been proposed (5, 10, 11). At present, very limited information is available on the utilization by bifidobacteria of complex dietary glycans such as starch (12), although a class II pullulanase, encoded by Bifidobacterium breve UCC2003 and containing both an α-amylase as well as a pullulanase domain, was identified and molecularly characterized (13).
The genome sequences of relatively few members of human intestinal commensal bifidobacterial species have been reported, e.g., B. longum subsp. longum, B. longum subsp. infantis, Bifidobacterium animalis subsp. lactis, B. bifidum, and B. breve (for a review see Ventura et al. [14]), yet such genomic information is crucial to unravel the intricate interactions that occur between the host and its resident bifidobacteria. Here, we report on the genome analysis of B. adolescentis 22L, which showed that this strain possesses specific metabolic traits involved in the breakdown of diet/plant-derived complex carbohydrates but lacks the necessary genetic arsenal for the breakdown of milk-related sugars.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and chromosomal DNA extraction.
Bifidobacterial cultures were incubated in an anaerobic atmosphere [2.99% (vol/vol) H2, 17.01% (vol/vol) CO2, and 80% (vol/vol) N2] in a chamber (Concept 400; Ruskin) in de Man-Rogosa-Sharpe (MRS) medium (Scharlau Chemie) supplemented with 0.05% (wt/vol) l-cysteine hydrochloride and incubated at 37°C for 16 h. Bacterial DNA was extracted as described previously (15) and subjected to further phenol-chloroform purification using a previously described protocol (16).
Carbohydrate growth assay.
Cell growth on semisynthetic MRS medium supplemented with 1% (wt/vol) of a particular sugar was monitored by optical density at 600 nm (OD600) using a plate reader (Biotek, VT, USA). The plate reader was run in discontinuous mode, with absorbance readings performed in 12-h intervals and preceded by 30 s of shaking at medium speed. Bacteria were cultivated in the wells of a 96-well microtiter plate, with each well containing a different sugar, and incubated in an anaerobic cabinet. Growth was evaluated by monitoring the OD600 of cultures in the microtiter plates on a plate reader every 12 h. Cultures were grown in biologically independent triplicates, and the resulting growth data were expressed as the means from these replicates. Carbohydrates tested in this study include RS2-resistant starch, amylopectin, pullulan, maltotriose, maltodextrin, glycogen, FOS, GOS, inulin, xylan, mucin, arabinogalactan, N-acetyl-d-galactosamine, N-acetylglucosamine, lactose, lactulose, mannose, sucrose, maltose, mannitol, xylose, raffinose, galactose, and glucose, all of which were purchased from Sigma and Carbosynth (Berkshire, United Kingdom).
Sample preparation and AFM imaging.
Bacteria from four ml bacterial cultures were harvested by centrifugation and resuspended in 200 μl of phosphate-buffered saline (PBS; or 20 mM HEPES, pH 7.5, 1 mM EDTA). Two hundred μl of 5% glutaraldehyde was added, followed by gentle mixing and incubation for 1 min at room temperature. Thereafter, bacteria were washed four times with PBS by repeated resuspension and collection by centrifugation (4,000 rpm). The washed pellet then was resuspended in 200 μl of PBS and kept on ice until AFM imaging. To facilitate adhesion of bacteria to the mica support used for AFM imaging, mica was coated with polylysine (PL) as follows: 10 μl of a polylysine solution (10 ng/ml) was deposited onto freshly cleaved mica for 1 min. Mica then was rinsed with MilliQ water (Millipore) and dried with nitrogen. Following this, 20 μl of bacterial suspension was deposited onto PL-coated mica for 2 to 5 min depending on the particular strain or specific cultivation conditions. The mica disk then was rinsed with MilliQ water and dried under a weak gas flow of nitrogen. The quality of the sample and density of surface-bound bacteria were verified using an optical microscope.
AFM imaging was performed on dried samples with a Nanoscope III microscope (Digital Instruments) equipped with scanner J and operating in tapping mode. Commercial diving board silicon cantilevers (MikroMasch) were used. The best image quality was obtained with high driving amplitude (1 to 3V) and low scan rate (0.5 Hz). Filamentous structures at the periphery of bacteria were visible in images of 512 by 512 pixels, representing a scan size of 10 μm or less. During imaging, both height and amplitude signals were collected. Height images were flattened using Gwyddion software.
Genome sequencing and assembly.
The genome sequence of B. adolescentis 22L was determined by GenProbio SRL (Parma, Italy) using the Ion Torrent Personal Genome Machine (PGM; Life Technologies, USA). A genomic library was generated using 10 μg of genomic DNA and an Ion Xpress Plus fragment library kit and employing the Ion Shear chemistry according to the user guide. After dilution to 2.66 × 107 molecules/μl, 4.5 × 108 molecules were used as the templates for clonal amplification on Ion Sphere particles during the emulsion PCR according to the Ion PGM template 400 kit manual. DNA quantitation was performed through the use of the library of quantitation DNA standards (Kapa Biosystems). The quality of the amplification was estimated, and the sample was loaded onto an Ion 316 chip and subsequently sequenced using 212 sequencing cycles according to the Ion PGM sequencing 400 kit user guide. This number of sequencing cycles resulted in an average reading length of approximately 400 nucleotides. The MIRA program (version 3.4.0) was used for de novo assembly of the 22L genome sequence (17). The generated sequencing output consisted of 590 Mb of DNA sequences, corresponding to an approximately 229-fold coverage of the 22L genome. In order to overcome the relatively high error rate that is sometimes detected for data obtained by the Ion Torrent sequencing platform, we applied a high-quality filter process (above 30) as well as resequencing of low-quality chromosome regions. Such quality improvement of the genome sequence involved Sanger sequencing (Macrogen, South Korea) of more than 60 PCR products across the entire genome to ensure correct assembly, double stranding, and the resolution of any remaining base conflicts.
Bioinformatic analyses.
Protein-encoding open reading frames (ORFs) were predicted using a combination of the methods Prodigal (18) and BLASTX (19). Results of the two gene finder programs were combined manually, and the predicted ORFs were used to perform a BLASTP (20) analysis against a nonredundant protein database provided by the National Centre for Biotechnology Information. Artemis (21) was employed to inspect the results of the combined gene finders and its associated BLASTP results, which was used for manual editing in order to check, or, if necessary, redefine the start of a predicted coding region or to remove or add coding regions.
The assignment of protein function to predicted coding regions of the B. adolescentis 22L genome was performed manually. Moreover, the revised gene-protein set was searched against the Swiss-Prot (http://www.expasy.ch/sprot/TrEMBL), PRIAM (http://priam.prabi.fr/), protein family (Pfam; http://pfam.sanger.ac.uk/), TIGRFAMs (http://www.jcvi.org/cms/research/projects/tigrfams/overview/), Interpro (INTERPROSCAN; http://www.ebi.ac.uk/Tools/InterProScan/), Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/), and clusters of orthologous groups (COGs; http://www.ncbi.nlm.nih.gov/COG/) databases, in addition to BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Functional assignments were defined by manual processing of the combined results. Manual corrections to automated functional assignments were completed on an individual gene-by-gene basis as needed.
rRNA genes were detected on the basis of BLASTN searches and annotated manually. tRNA genes were identified using tRNAscan-SE (22). Insertion sequence (IS) families were assigned using ISFinder (http://www-is.biotoul.fr/). Restriction/modification (R/M) systems were searched and then analyzed on the basis of the REBASE database (23). Carbohydrate-active enzymes were identified based on similarity to carbohydrate-active enzyme (CAZy) database entries (24). Transporter classification was performed according to the TC-DB scheme (25), and Enzyme Commission (EC)/Gene Onthology (GO) ORF annotation was achieved using annot8r (26). Variances in GC content were profiled by the DNA segmentation algorithm hosted at http://tubic.tju.edu.cn/GC-Profile/ (27), and atypical codon usage regions were mapped using factorial correspondence analysis with the assistance of GCUA software (28).
Proteome comparison and extraction of shared and unique genes.
Each predicted proteome of the analyzed B. adolescentis 22L, B. adolescentis ATCC 15703 (NCBI source), B. dentium Bd1 (29), B. animalis subsp. lactis Bl12 (30), B. animalis subsp. animalis ATCC 25527 (31), B. longum subsp. longum NCC2705 (32), B. longum subsp. infantis ATCC 15697 (4), B. bifidum PRL2010 (5), and B. breve UCC2003 (33) strains was searched for orthologs against the total proteome, where orthology between two proteins was defined as the best bidirectional FASTA hits (34). Identification of orthologs, paralogs, and unique genes was performed after a preliminary step consisting of the comparison of each protein to all other proteins using BLAST analysis (20) (cutoff of an E value of <1 × 10−4 and 30% identity for at least 80% of both protein sequences), after which proteins were clustered into protein families using MCL (graph theory-based Markov clustering algorithm) (35). Following this, the unique protein families for the B. adolescentis 22L genome were classified. Protein families shared between all genomes, named core gene families, were defined by selecting the families that contained at least a single protein member for each genome.
Each set of orthologous proteins was aligned using ClustalW (36), and phylogenetic trees were constructed using maximum likelihood in PhyML (37). The supertree was built using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
RNA isolation, reverse transcription, and qRT-PCR.
Total RNA was isolated from 22L cultures grown in the presence of resistant starch, starch-related carbohydrates, or glucose (reference condition) as the unique carbon source using previously described methods (38). Briefly, cell pellets were resuspended in 1 ml of QUIAZOL (Qiagen, United Kingdom) and placed in a tube containing 0.8 g of glass beads (diameter, 106 μm; Sigma). The cells were lysed by shaking the mix on a BioSpec homogenizer at 4°C for 2 min (maximum setting). The mixture then was centrifuged at 12,000 rpm for 15 min, and the RNA-containing sample was recovered from the upper phase. The RNA sample was further purified by phenol extraction and ethanol precipitation according to an established method (39). The quality and integrity of the RNA were checked by Experion (Bio-Rad) analysis. RNA concentration and purity then were determined with a Bio-Rad Smart Spec spectrophotometer, and the quality and integrity of the RNA was checked by Experion (Bio-Rad) analysis. Reverse transcription to cDNA was performed with the iScript Select cDNA synthesis kit (Bio-Rad Laboratories) using the following thermal cycle: 5 min at 25°C, 30 min at 42°C, and 5 min at 85°C. The mRNA expression levels were analyzed with SYBR green technology in quantitative real-time PCR (qRT-PCR) using SoFast EvaGreen Supermix (Bio-Rad) on a Bio-Rad CFX96 system according to the manufacturer's instructions (Bio-Rad). The primers used are indicated in Table S1 in the supplemental material. Quantitative PCR was carried out according to the following cycle: initial hold at 96°C for 30 s and then 40 cycles at 96°C for 2 s and 60°C for 5 s. Gene expression was normalized relative to a housekeeping gene as previously described by Turroni et al. (40). The amount of template cDNA used for each sample was 12.5 ng.
RNA-seq analysis performed by the Ion Torrent PGM.
For RNA sequencing (RNA-seq), we started from 10 μg of total RNA, which was treated so as to remove rRNA by the Ribo-Zero rRNA removal kit (Epicentre, Madison, WI), followed by purification of the rRNA-depleted sample by ethanol precipitation, after which the RNA was further processed according to the manufacturer's instructions. The yield of rRNA depletion was checked by Experion (Bio-Rad, United Kingdom). One hundred ng of rRNA-depleted RNAs was fragmented using RNase III (Life Technologies, USA), followed by size evaluation using Experion (Bio-Rad, United Kingdom). A whole-transcriptome library was constructed using the Ion Total-RNA Seq kit, v2 (Life Technologies, USA). Barcoded libraries were quantified by qRT-PCR, and each library template was amplified on Ion Sphere particles using a Ion One Touch 200 template kit, v2 (Life Technologies, USA). Samples were loaded into Ion 316 chips and sequenced on the PGM (Life Technologies, USA). Sequencing reads were depleted of adapters, quality filtered (with overall quality, quality window, and length filters), and aligned to the 22L reference genome through BWA (41). Counts of reads overlapping ORFs were performed using HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html), and analyses of the count data were performed using the R package DESeq (42).
Quantification of bacterial binding to extracellular matrix proteins.
Ninety-six-microwell plates (Maxisorp; Nunc) were coated with a solution of 500 μg/ml of extracellular matrix (ECM) protein in 100 μl PBS. ECM proteins used included fibrinogen, plasminogen, fibronectin, laminin, and collagen type IV, which were purchased from Sigma. The ECM protocol was performed as previously described by Turroni et al. (43).
Mouse experiments.
All animals used in this study were cared for in compliance with guidelines established by the Italian Ministry of Health. All procedures were approved by the University of Parma, as executed by the Institutional Animal Care and Use Committee (Dipartimento per la Sanità Pubblica Veterinaria, la Nutrizione e la Sicurezza degli Alimenti Direzione Generale della Sanità Animale e del Farmaco Veterinario). Two groups, i.e., a 22L-treated and untreated group, each containing five 3-month-old female conventional BALB/c mice, which were housed in two separate cages, were orally inoculated with bacteria. Mice were fed a standard chow diet, where starch represents approximately 45.3% of the total carbohydrate content (44). Bacterial treatment was established by means of five consecutive daily administrations whereby each animal received a dose of 109 bacterial cells using a micropipette tip placed immediately behind the incisors. The bacterial inocula consisted of B. adolescentis 22L cells harboring plasmid pNZ8048, which had been introduced in this strain by electrotransformation and selection on chloramphenicol (45).
In order to estimate the number of B. adolescentis 22L cells per gram of feces, individual fecal samples were serially diluted and cultured on selective agar (MRS) containing 3 μg/ml chloramphenicol. Following enumeration of B. adolescentis 22L in fecal samples, 100 random colonies were further tested to verify their identity using PCR primers targeting the pil4 locus.
Animals were sacrificed by cervical dislocation and their individual gastrointestinal tracts were removed, immediately treated with RNA-later, and subsequently used for RNA extraction.
The quality and integrity of the RNA was checked by Experion (Bio-Rad) analysis. cDNA was synthesized and purified using the iScript cDNA synthesis kit (Bio-Rad, CA, USA) according to the supplier's instructions. Primers used for normalization have been described previously (40). Criteria for primer design included a desired melting temperature (Tm) between 58 and 60°C and an amplicon size of 100 to 200 bp. qRT-PCR was performed using the CFX96 system (Bio-Rad, CA, USA). PCR products were detected with SYBR green fluorescent dye and amplified according to the following protocol: one cycle of 95°C for 3 min, followed by 39 cycles of 95°C for 5 s and 66°C for 20 s. The melting curve was 65°C to 95°C with increments of 0.5°C/s.
Each qRT-PCR mix contained 12.5 μl 2× SYBR supermix green (Bio-Rad, CA, USA), 1 μl of cDNA dilution, each of the forward and reverse primers at 0.5 μM, and nuclease-free water to obtain a final volume of 20 μl. In each run, a negative control (no cDNA) for each primer set was included. The expression ratio of the selected genes was calculated and analyzed using CFX Manager Expression software (Bio-Rad, CA, USA). The cutoff value applied to highlight significant change in the expression was 2.
Statistical analysis.
Statistical significance between means was analyzed using the unpaired Student's t test with a threshold P value of <0.05. Values are expressed as the means ± standard errors of the means from three experiments. Multiple comparisons are analyzed using one-way analysis of variance (ANOVA) and Bonferroni tests. Statistical calculations were performed using the software program GraphPad Prism 5 (La Jolla, CA, USA).
Nucleotide sequence accession numbers.
The sequence reported in this article has been deposited in the GenBank database (accession number CP007443). All RNA-seq raw data from this study have been submitted to the NCBI Sequence Read Archive with the corresponding BioSample (SRA) accession no. SAMN02697098 (SRS584592), SAMN02697099 (SRS584597), SAMN02697100 (SRS584604), and SAMN02697101 (SRS584606).
RESULTS AND DISCUSSION
General genome features.
Our initial interest was to understand the genetic features that support the ability of B. adolescentis 22L to colonize the human gut, and for this purpose we decided to decode the chromosome of 22L. The genome sequence consists of a single 2,203,222-bp circular chromosome with an average G+C content of 59.29%, which is similar to that of other sequenced bifidobacterial genomes, being consistent with the range of G+C mol % values previously described for Actinobacteria (46) (see Table S2 in the supplemental material). The genome of B. adolescentis 22L possesses 54 tRNAs and four rRNA operons. Identification of protein-coding sequences revealed 1,725 open reading frames (ORFs). A functional assignment was made for 71% of the predicted ORFs, while homologs with no known function from other bacterial species were identified for an additional 17.6% of the total ORFome of B. adolescentis 22L. The remaining 11.4% of ORFs specified hypothetical proteins, which appear to be unique to B. adolescentis 22L. A supertree based on the core genome sequences identified from the currently available genome sequences of bifidobacteria (http://ncbi.nlm.nih.gov), as well as comparative genomic analyses, highlight a close phylogenetic relatedness of B. adolescentis 22L with the other sequenced member of the B. adolescentis taxon (see Fig. S1 and the text of the supplemental material). The predicted B. adolescentis 22L proteome was functionally categorized according to the COG families, and the proportions in each family were compared to those of other bifidobacterial genomes. Interestingly, about 12.5% of the genes identified in the B. adolescentis 22L genome encode proteins that are predicted to be involved in carbohydrate metabolism and transport, while about 12.2% encode proteins with putative functions in amino acid metabolism and transport. These genetic features are well conserved in human-derived bifidobacterial genomes, which represent a clear sign of molecular adaptation of bifidobacteria to the mammalian gut (14).
Assessing the 22L glycome.
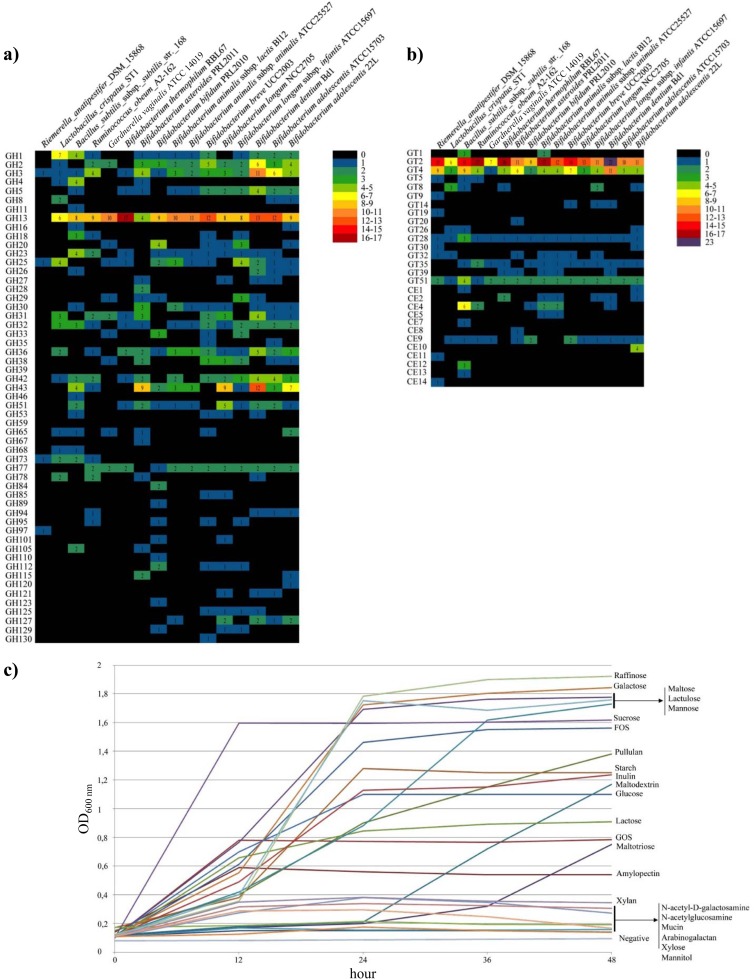
Combined genomic data suggest that B. adolescentis 22L is capable of metabolizing a broad range of carbohydrates (Fig. 1a), many of which are plant-derived carbohydrates, such as starch, maltodextrin, maltotriose, and amylopectin, as well as animal-based glycans like glycogen. These glycans are known to escape from digestion in the upper compartments of the gastrointestinal tract, and they are presumed to be abundant in the human gut, where they represent substrates for growth of enteric bacteria like B. adolescentis 22L. Classification in accordance to the CAZy system of Coutinho and Henrissat (24) revealed that the B. adolescentis 22L genome is predicted to encode 89 carbohydrate active proteins, including 55 glycoside hydrolases (GH), 24 glycosyltransferases (GT), seven glycosyl esterases (CE9), and three carbohydrate-binding module (CBM)-containing proteins (Fig. 1a and b). It is worth mentioning that the genome of strain 22L is predicted to encode a glycosyl hydrolase belonging to the GH16 family, as well as xyloglucanases (EC 3.2.1.151), endo-1,3-β-galactanases (EC 3.2.1.181), and an endo-1,3-β-glucanase (EC 3.2.1.39), enzymes that cleave β-1,4 or β-1,3 glycosidic bonds of various glucans and galactans. Notably, the genetic locus encoding the GH16-type glycoside hydrolase is absent from other publicly available bifidobacterial genomes (Fig. 1a). Furthermore, the genome of 22L encodes nine GH, predicted to belong to GH13, which encompass enzymes with amylase, pullulanase, and cyclomaltodextrinase activities. Thus, the 22L genome appears to encode the largest set of GH13 enzymes of the human gut bifidobacteria so far published (4, 5, 32, 33, 47) (see Fig. 3), suggesting superior growth performance of 22L on particular plant-derived carbohydrates.
FIG 1.
Glycome of B. adolescentis 22L. (a and b) Predicted glycoside-hydrolase (GH), glycoside-transferase (GT), and carbohydrate esterase (CE) families encoded by the genome of B. adolescentis 22L according to the CAZy database (24). The glycome identified in the genome of 22L is compared to all of the GH, GT, and CE families identified so far in currently available bifidobacterial genome sequences. (c) Growth curves of B. adolescentis 22L in a growth medium containing different carbohydrates as the sole carbon source. Growth was measured as the optical density of the medium at 600 nm. Cultures were grown in biologically independent triplicates.
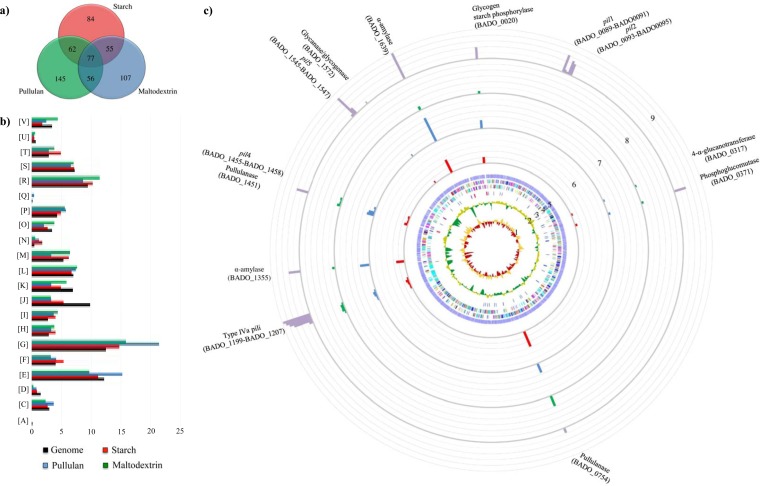
FIG 3.
Identification of B. adolescentis 22L differentially expressed genes by transcriptome analysis in response to growth on starch and starch-like carbohydrates. (a) Venn diagram showing the number of differently expressed genes during the different conditions: starch (shown in red), pullulan (represented in blue), and maltodextrin (depicted in green). The data displayed are based on RNA preparations from two independent culture experiments. (b) Functional annotation of the expressed genes of B. adolescentis 22L cultivated on starch and its derivatives according to their COG categories. Each COG family is identified by a one-letter abbreviation (National Center for Biotechnology Information database). For each category, the black bar represents the percentage of genes in that category as detected in the sequenced genome of 22L. The other bar shows the percentages of genes transcribed during growth of 22L on starch (shown in red), pullulan (represented in blue), and maltodextrin (depicted in green). The percentage was calculated as the percentage of transcribed genes belonging to the indicated COG category with respect to all transcribed genes. (c) Circular genome of B. adolescentis 22L mapped with the RNA-seq reads under in vitro and in vivo conditions by principal component analysis. From the inner circle, circle 1 shows the B. adolescentis 22L GC skew (G-C/G+C); circle 2 illustrates B. adolescentis 22L G+C% deviation; circle 3 indicates rRNAs (depicted in red) and tRNAs (depicted in blue); circle 4 shows coding regions by strand, with the color corresponding to the COG functional assignment; circle 5 denotes the ORF distribution; circles 6, 7, and 8 show the transcription level of genes encoding the different components of the starch degradation V pathway (52), when 22L cells were cultivated on starch, pullulan, and maltodextrin, respectively, and using 22L growth on glucose as a reference condition. Finally, circle 9 displays the expression of a starch metabolism-related gene upon colonization of mice using qRT-PCR. The data shown represent mean qRT-PCR values achieved for the five animals used.
The genome of B. adolescentis 22L encompasses 149 genes that are predicted to encode components of ABC-type transporters. On the basis of the TC database (25), 35 of these proteins are predicted to be involved in the internalization of carbohydrates with a preference for arabinose, maltodextrin, maltotriose, arabinose, and xylobiose, which may be derived from the breakdown of plant polysaccharides like starch. Furthermore, of the identified ABC-type carrier protein components, 34 are predicted to act in the uptake of amino acids/peptides, 10 in the uptake of vitamins, 28 in the internalization of metals, and 20 in conferring various types of resistance, while 22 have no well-defined function. Such a large arsenal of carriers for the uptake of carbohydrates is another piece of in silico proof of 22L's specialization to carbohydrate utilization. Furthermore, the predicted transporter arsenal of 22L includes two putative phosphoenolpyruvate-phosphotransferase systems (PEP-PTS) and 20 proteins that are predicted to encode the major facilitator superfamily (MFS family), of which, as based on the TC database (25), three are assigned as carbohydrate transporters, four as type 1 drug antiporters, five as type 2 drug antiporters, one as a metabolite symporter, one as a cyanate porter, and six as transporters with unknown functions.
The analysis of the B. adolescentis 22L chromosome revealed the presence of conventional mobilome candidates that may have been acquired through horizontal gene transfer (HGT) (see Fig. S2 details in the text of the supplemental material) and may provide important ecological advantages while also influencing chromosome structure (46).
Starch utilization by bifidobacteria.
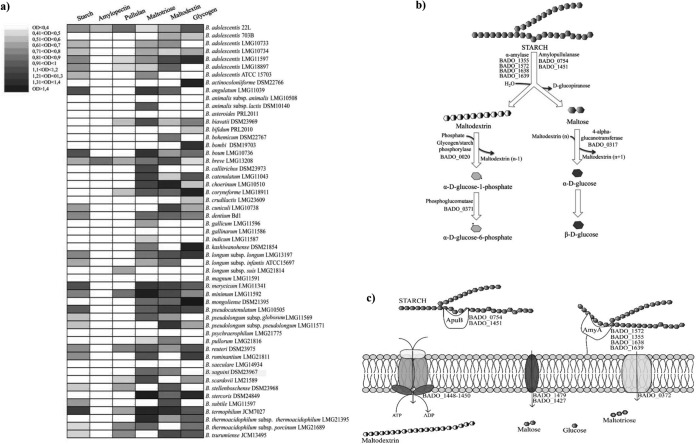
Genomic analyses of representative members of the gut microbiota suggest that the genetic capacity to metabolize complex carbohydrates that are not hydrolyzed by host enzymes can significantly affect the composition of the gut microbiome (14). As displayed in Fig. 1a, the predicted glycobiome of strain 22L coupled with its fermentation capabilities on different carbon sources (Fig. 1b) highlights that B. adolescentis strain 22L possesses fermentation abilities broader than those of the phylogenetically closely related enteric B. adolescentis ATCC 15703 strain. Notably, 22L displays a preference for the utilization of different esose-containing sugars (e.g., galactose, mannose, and glucose), as well as key plant-derived glycans present in the human diet, such as starch (Fig. 1b), fructooligosaccharides, and inulin. In contrast, 22L cells display a low level of preference for host glycans, such as mucin or host glycan derivatives (e.g., N-acetylgalactosamine and N-acetylglucosamine) (Fig. 1b). Even though the fermentation abilities of B. adolescentis 22L were shown to include a relatively large set of carbohydrate sources (Fig. 1b), we decided to focus on the genetic adaptation of 22L to starch metabolism, since this is a common and abundant carbon source available in the mammalian gut. Although a large selection of Bifidobacterium strains was used to evaluate their ability to utilize starch as a sole carbon source, involving representative strains for many species of the genus Bifidobacterium, substantial growth on starch-based medium (OD above 0.5) was noticed only for strains belonging to B. adolescentis, Bifidobacterium angulatum, Bifidobacterium boum, B. breve, Bifidobacterium cuniculi, B. dentium, B. longum subsp. longum, B. longum subsp. infantis, Bifidobacterium merycicum, Bifidobacterium minimum, Bifidobacterium reuteri, B. pseudocatenulatum, Bifidobacterium ruminantium, and Bifidobacterium thermophilum (Fig. 2a). Noticeable differences in growth on starch-based medium were evident among the B. adolescentis isolates. The strain B. adolescentis 703B did not exhibit any appreciable growth on starch, while strains B. adolescentis ATCC 15703, B. adolescentis LMG10733, B. adolescentis LMG10734, and B. adolescentis LMG 18897 exhibited some growth, although considerably less than the B. adolescentis 22L and B. adolescentis LMG11579 strains, which are considered to be superior starch-utilizing bifidobacteria (Fig. 2a). Interestingly, B. adolescentis 22L represented the only strain of this species that was capable of growth on starch and all starch derivatives (maltodextrin, maltotriose, pullulan, and glycogen). Therefore, 22L strain was selected as our model B. adolescentis strain to investigate starch metabolism.
FIG 2.
B. adolescentis 22L metabolism of starch and starch derivatives. (a) Heat map representing the growth performance of B. adolescentis 22L as well as all of the type strains of the currently recognized 47 taxa belonging to the genus Bifidobacterium on starch and starch-like carbohydrates. Cultures were grown in biologically independent triplicates. The different shading represents the optical density reached by the various cultures. (b) Schematic representation of the metabolic pathways for starch degradation followed by 22L cells. The different ORFs of B. adolescentis 22L encoding the presumed enzymes involved in the breakdown of starch are indicated. (c) Schematic overview of a cell and the location of enzymes involved in the breakdown and internalization of starch. Carbohydrates derived from starch are internalized by ABC transporters (represented in orange), PTS systems (shown in blue), and symporter (depicted in green), after which glycosyl hydrolases process such oligosaccharides to monosaccharides. The different ORFs of B. adolescentis 22L encoding the presumed enzymes involved in the breakdown and internalization of starch are indicated.
Genomics of starch utilization.
Starch functions as a glycan reservoir of plant cells and consequently is the main carbohydrate present in a plant-based diet (48). Within the human gastrointestinal tract this polysaccharide represents, as a so-called resistant starch, an important carbon source for the microbiota residing in such an environment as well as an important site for adhesion and colonization of gut bacteria (12, 49, 50). Starch consists of amylose and amylopectin moieties, with the former being a linear α-(1,4) glucose chain with a plant-specific degree of polymerization of 200 to 6,000, while the latter represents short linear α-(1,4) glucose-linked chains with α-(1,6)-linked side chains (48). Natural derivatives of starch are amylopectin, maltodextrin, maltotriose, and maltose (48). Given the diversity and complexity of starch as well as starch-derived structures found within the gut, specific strategies for deconstructing these molecules must be featured inherently in the genomes of starch-utilizing bacteria. The B. adolescentis 22L genome encodes various glycosyl hydrolases putatively implicated in degradation of starch as well as starch-derived glycans, including four proteins (BADO_1355, BADO_1572, BADO_1638, and BADO_1639) with predicted α-amylase activity (Fig. 2b), which catalyzes the hydrolysis of the α-linkage between glucose moieties and generating maltodextrin as well as maltose residues (51). Furthermore, the 22L genome encompasses two genes (BADO_0754 and BADO_1451) predicted to encode amylopullulanases, similar to the starch-degrading enzyme previously described for B. breve UCC2003 (13) (Fig. 2b). The genome of B. adolescentis 22L also encodes a glycogen/starch phosphorylase (BADO_0020) (Fig. 2b), as well as a predicted phosphoglucomutase (BADO_0371) (Fig. 2b), which release α-d-glucose-6-phosphate from maltodextrin according to the starch degradation V pathway (52). The putative phosphoglucomutase-encoding gene is located near genes encoding a phosphotransferase (PTS) system (BADO_0372) predicted to be involved in the internalization of glucose in the cell (Fig. 2c) (53). In addition, a putative 4-α-glucanotransferase (BADO_0317), involved in the generation of α-d-glucose from maltose according to starch degradation pathway V (52), was detected in the predicted proteome of B. adolescentis 22L (Fig. 2b). Finally, in silico analyses of the genome of 22L highlighted the presence of a set of genes whose products might be involved in starch utilization in a fashion similar to that previously described for B. breve UCC2003 (13, 33). This set of genes consists of two genes predicted to encode α-1,4- and α-1,6-glucosidases (BADO_1445 and BADO_1447), an ABC-type system predicted to be involved in the transport of maltose and maltodextrin (BADO_1448-BADO_1450), and a gene encoding a LacI-type regulator (BADO_1454) which flanks a gene encoding an amylopullulanase (BADO_1451) (Fig. 2c).
Transcriptome analyses of B. adolescentis 22L and adaptation to starch utilization.
In order to substantiate the notion that B. adolescentis 22L contains specific genes dedicated to the utilization of starch, we investigated the transcriptome of B. adolescentis 22L grown in the presence of starch and starch glycan derivatives as the unique carbon source and compared these results to data obtained when 22L was cultivated on glucose using an RNA-seq approach. In total, 278 genes displayed increased expression (more than 2-fold) when 22L was cultured in the presence of starch as the sole carbon source (Fig. 3a). Functional assignment of the starch-specific upregulated genes was performed through COG analysis. As illustrated in Fig. 3b, carbohydrate metabolism, corresponding to COG category G, is one of the COG functions of 22L most significantly affected by the presence of starch or starch derivatives, such as pullulan and maltodextrin. As expected, 22L transcriptome profiling identified starch and pullulan- and maltodextrin-induced genes that encode α-amylase (BADO_1355, BADO_1572, and BADO_1639) as well as two predicted amylopullanases (BADO_0754 and BADO_1451), which were shown to be at levels more than 4-fold higher than those in the absence of starch. Other starch/pullulan/maltodextrin-upregulated genes included those encoding glycogen/starch phosphorylase (BADO_0020), phosphoglucomutase (BADO_0371), 4-α-glucanotransferase (BADO_0317), and transporters dedicated to the uptake of starch/starch derivatives (Fig. 3c). Further genes whose transcription was induced on starch included the sortase-dependent pilus biosynthesis pil4 locus (BADO_1455-BADO_1458), which was upregulated 3- to 8-fold. Notably, although to a lesser extent, the genes encompassing the type IVa pilus biosynthesis gene cluster (BADO_1197-BADO_1207) also were shown to be upregulated (2.3-fold) when 22L was cultivated on starch. Altogether, these findings support the involvement of plant-derived glycan in the modulation of these extracellular structures, which have been implicated in host interactions (33, 43, 54).
Starch utilization by B. adolescentis 22L in the murine gut.
In order to investigate the possible upregulation of the dedicated starch utilization gene repertoire of 22L when the microorganism resides in the mammalian's gut, we performed transcription profiling of those genes predicted to be involved in starch utilization of this strain upon the presence of B. adolescentis 22L in the murine gut using a qRT-PCR approach. Conventional female BALB/c mice were administered a single daily dose of 109 CFU B. adolescentis 22L. Mice were checked a priori for the presence of bifidobacteria in fecal samples by PCR using Bifidobacterium-specific primers (55), which revealed that bifidobacteria were either absent or below the limit of detection. Animals were sacrificed 12 days later, allowing sufficient time for several cycles of turnover of the intestinal epithelium and its overlying mucus layer (56). Microbial evaluation of the murine gut showed the presence of B. adolescentis 22L at stable numbers, reminiscent of at least transient persistence over time (see Fig. S3 in the supplemental material). The mRNA levels corresponding to genes encoding putative enzymes involved in the breakdown of starch identified in the genome of 22L were shown to be variable in response to its presence in the murine gut (Fig. 3c). Interestingly, the transcription of the genes encoding the predicted α-amylase (BADO_1355 and BADO_1639), two putative amylopullanases (BADO_0754 and BADO_1451), a gene encoding glycanase/glycogenase (BADO_1572), as well as the predicted glycogen/starch phosphorylase (BADO_0020) and phosphoglucomutase (BADO_0371), 4-α-glucanotransferase (BADO_0317) were significantly enhanced (from 3- to 100-fold induction) when 22L was present in the murine gut (Fig. 3c), and this may reflect a specialization of 22L toward a particular ecological niche where starch is abundant. Furthermore, the expression of genes encoding the major and minor subunits of pil1, pil2, and pil5, as well as the genes encompassing the type IVa pilus locus, were significantly upregulated when 22L cells were retrieved from the mouse cecum. The enhancement of the expression of the genes encoding these cell appendages may be largely attributable to host factors as well as to 22L cultivated on starch.
Genetic evidence of adaptation to the human gut.
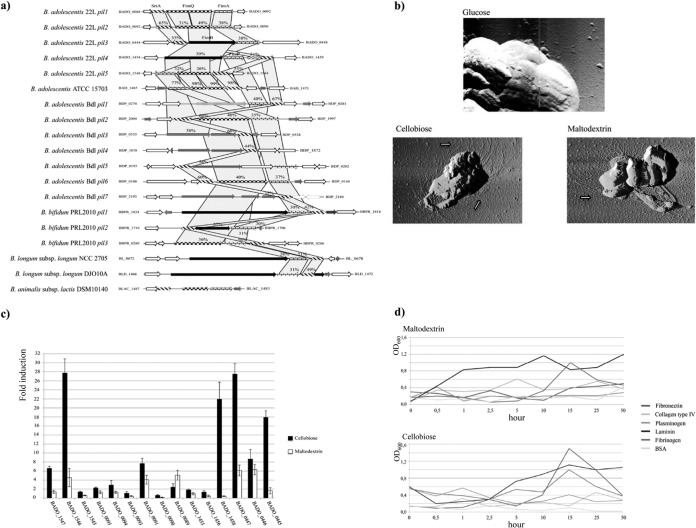
The cell envelope of gut commensals such as bifidobacteria constitutes a pivotal structural interface between gut bacteria and the environment and involves proteins and carbohydrates that facilitate bacterial attachment and colonization (14, 57). In silico analysis of B. adolescentis 22L genome sequences revealed the presence of five sortase-dependent pilus loci, i.e., pil1 (BADO_0089-BADO_0091), pil2 (BADO_0093-BADO_0095), pil3 (BADO_0445-BADO_0447), pil4 (BADO_1455-BADO_1458), and pil5 (BADO_1545-BADO_1547) (Fig. 4a). Each of these five pilus loci encodes a major subunit and a minor subunit in a fashion similar to that previously described for other sortase-dependent pili identified in bifidobacteria (43, 54). Notably, atomic force microscopy (AFM) coupled with qRT-PCR experiments targeting the major and the minor subunits provided evidence for high production of pilus-like structures on the surface of 22L cells cultivated on cellobiose or maltodextrin (Fig. 4b and c). Sortase-dependent pili encoded by B. bifidum PRL2010 have been shown to be responsible for adhesion of bifidobacterial cells to the ECM proteins (43). Thus, we examined the ability of 22L cells to adhere to various ECM proteins, such as fibrinogen, plasminogen, fibronectin, laminin, and collagen type IV. Notably, B. adolescentis 22L cells which had been cultivated under pilus-producing conditions (e.g., growth on cellobiose or maltodextrin) showed higher levels of adhesion to laminin, fibrinogen, and fibronectin than those of other tested ECM proteins (Fig. 4d).
FIG 4.
Gene clusters encoding sortase-dependent pili identified in the genome of B. adolescentis 22L. (a) Schematic comparative representation of pilus-encoding loci identified in the genome of B. adolescentis 22L with the corresponding loci identified in different bifidobacterial genomes. (b) AFM images of 22L cells cultivated on glucose, maltodextrin, or cellobiose, providing evidence of the presence of pilus-like structures. Arrows highlight pilus structures on the cell surface. Scale bar, 1 μm. (c) qRT-PCR relative transcription levels of pilus-encoding genes from B. adolescentis 22L upon cultivation in a medium supplemented with maltodextrin or cellobiose, as unique carbon sources, versus growth in medium supplemented with glucose as the unique carbon source. Cultures were grown in biologically independent triplicates. The histograms indicate the relative amounts of the pilin/sortase mRNAs for the specific samples. The data displayed are based on RNA preparations from two independent culture experiments. (d) Putative involvement of pili produced by B. adolescentis 22L in adhesion to ECM proteins in the presence of different carbohydrates (maltodextrin and cellobiose). For each of these experiments, adhesion of microbial cells to BSA was used as a negative control.
Recently, a member of the so-called type IVb or Tad (tight-adherence) pilus family was shown to be expressed specifically by B. breve UCC2003 under in vivo conditions in a murine gut, where it plays a key role in the establishment of UCC2003 cells (33). Notably, the genome sequences of B. adolescentis 22L contains a complete tad locus consisting of six genes (BADO_0139 to BADO_0144) displaying high levels of identity (ranging from 38% to 77% at the protein level) to putative tad loci in other bifidobacterial genomes. In addition, the B. adolescentis 22L chromosome contains a putative type IVa pilus-encoding gene cluster (BADO_1197 to BADO_1207), which has not been detected previously in bifidobacterial genome sequences. The genes encompassing the latter locus are highly similar (ranging from 25% to 56% at the amino acid level) to genes identified in Firmicutes members, e.g., Cellulomonas fimi ATCC 484, Clostridium difficile 630, and Clostridium perfringens ATCC 13124 (see Fig. S4 in the supplemental material). Remarkably, type IVa pili have been reported to be involved in various cellular processes, such as motility, conjugation, adherence, and DNA uptake (58). qRT-PCR targeting the genes of this locus showed a high level of expression using RNA samples obtained from 22L cultures growth on cellobiose (see Fig. S4), suggesting that the production of type IVa pilus of 22L is mediated by the presence of cellobiose. Other extracellular structures that have a reported role in gut colonization by enteric bifidobacteria are represented by exopolysaccharides (EPS) (59). The genome sequence of B. adolescentis 22L contains three gene clusters (eps1 [BADO_0389 to BADO_0407], eps2 [BADO_1353 to BADO_1384] and eps3 [BADO_1638 to BADO_1647]) predicted to be involved in the production of cell surface-associated EPSs. The putative EPS gene clusters of 22L each exhibit a unique genetic structure, although all clusters contained a variable number of genes coding for putative glycosyltransferases, genes involved in the transport of carbohydrate, as well as ABC-transport genes and an MFS system (see Fig. S5). Notably, the eps2 locus encompasses two genes (BADO_1356 and BADO_1357) predicted to be involved in the biosynthesis of rhamnose (60). Another crucial protein found in the predicted proteome of B. adolescentis 22L sustaining the microbe-host interaction is represented by a serpin homolog (22L_0506), which in B. breve 210B has been shown to counter the adverse effects caused by host proteases and to have a role in modulating inflammation in the human gastrointestinal tract (38).
Conclusions.
Bifidobacteria are dominant in the infant before weaning, after which their (relative) abundance decreases during subsequent stages of life, even though they never disappear (3, 55). There are strong indications of their capabilities to adapt to the human gut thanks to the utilization of different complex carbohydrates that are not metabolized by the digestive enzymes of the host. Such a sugar utilization repertoire is influenced by diet, which changes at different stages of life. In this context, it is dominated by human milk oligosaccharides at the infant level, and after weaning these complex carbohydrates are replaced by a diet that includes plant-derived sugars, such as starch and plant cell wall carbohydrates. Here, the analysis of the genome sequence of B. adolescentis 22L presents this strain as a bifidobacterial prototype for a gut commensal with the ability to thrive on starch as its sole carbon and energy source. The 22L strain originally was isolated from a human milk sample, which suggests that infants are inoculated with adult-type bifidobacteria like B. adolescentis 22L directly from maternal milk. This finding also suggests that human milk does not act as a reservoir of infant-type bifidobacteria only but also of bifidobacterial strains like B. adolescentis 22L, which displays genetic features consistent with adaptation to the adult gut (61). During weaning, infants switch to a solid-food diet, being exposed for the first time to different (i.e., as opposed to those in mother's milk) complex carbohydrates. In infants, the lack of chewing ability, the very small amount of salivary amylase activity, and the immaturity of the infant's gut allow a large amount of such sugars, including starch, to escape digestion from the upper compartments of the gastrointestinal tract, thereby arriving in an essentially intact form in the large intestine of neonates (62). The human gut harbors various amylolytic bacteria, such as B. breve, B. longum subsp. infantis, B. longum subsp. longum, B. pseudocatenulatum, B. angulatum, B. thetaiotaomicron, Ruminococcus bromii, and Eubacterium rectale (50), and the proven ability of B. adolescentis 22L as an active degrader of resistant starch adds to this list of diet-polysaccharide users (7).
Supplementary Material
ACKNOWLEDGMENTS
We thank GenProbio SRL for financial support of the Laboratory of Probiogenomics. This work was supported financially by a FEMS Jensen Award to F.T. and by a Ph.D. fellowship (Spinner 2013, Regione Emilia Romagna) to S.D.
D.V.S. and F.T. are members of The Alimentary Pharmabiotic Centre, and D.V.S. is also a member of the Alimentary Glycoscience Research Cluster, both funded by Science Foundation Ireland (SFI) through the Irish Government's National Development Plan (grant numbers 07/CE/B1368, SFI/12/RC/2273, and 08/SRC/B1393).
Footnotes
Published ahead of print 25 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01993-14.
REFERENCES
- 1.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219–226. 10.1128/AEM.68.1.219-226.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ventura M, van Sinderen D, Fitzgerald GF, Zink R. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Van Leeuwenhoek 86:205–223. 10.1023/B:ANTO.0000047930.11029.ec [DOI] [PubMed] [Google Scholar]
- 3.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. 10.1371/journal.pone.0036957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 105:18964–18969. 10.1073/pnas.0809584105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U. S. A. 107:19514–19519. 10.1073/pnas.1011100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levitt MD. 1987. Only the nose knows. Gastroenterology 93:1437–1438 [DOI] [PubMed] [Google Scholar]
- 7.Ze X, Duncan SH, Louis P, Flint HJ. 2012. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6:1535–1543. 10.1038/ismej.2012.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Elia JN, Salyers AA. 1996. Contribution of a neopullulanase, a pullulanase, and an alpha-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J. Bacteriol. 178:7173–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott KP, Martin JC, Chassard C, Clerget M, Potrykus J, Campbell G, Mayer CD, Young P, Rucklidge G, Ramsay AG, Flint HJ. 2011. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):S4672–S4679. 10.1073/pnas.1000091107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turroni F, van Sinderen D, Ventura M. 2010. Genomics and ecological overview of the genus Bifidobacterium. Int. J. Food Microbiol. 149:37–44. 10.1016/j.ijfoodmicro.2010.12.010 [DOI] [PubMed] [Google Scholar]
- 11.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 5:220–230. 10.1038/ismej.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Conway PL, Brown IL, Evans AJ. 1999. In vitro utilization of amylopectin and high-amylose maize (amylomaize) starch granules by human colonic bacteria. Appl. Environ. Microbiol. 65:4848–4854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connell Motherway M, Fitzgerald GF, Neirynck S, Ryan S, Steidler L, van Sinderen D. 2008. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 74:6271–6279. 10.1128/AEM.01169-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventura M, Turroni F, O'Connell Motherway M, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 20:467–476. 10.1016/j.tim.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 15.Ventura M, Elli M, Reniero R, Zink R. 2001. Molecular microbial analysis of Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microbiol. Ecol. 36:113–121. 10.1111/j.1574-6941.2001.tb00831.x [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17.Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159. 10.1101/gr.1917404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gish W, States DJ. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266–272 [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 21.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 22.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts RJ, Vincze T, Posfai J, Macelis D. 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38:D234–D236. 10.1093/nar/gkp874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coutinho PM, Henrissat B. 1999. Life with no sugars? J. Mol. Microbiol. Biotechnol. 1:307–308 [PubMed] [Google Scholar]
- 25.Busch W, Saier MH., Jr 2002. The transporter classification (TC) system, 2002. Crit. Rev. Biochem. Mol. Biol. 37:287–337 [DOI] [PubMed] [Google Scholar]
- 26.Schmid R, Blaxter ML. 2008. annot8r: GO, EC and KEGG annotation of EST datasets. BMC Bioinformatics 9:180. 10.1186/1471-2105-9-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao F, Zhang CT. 2006. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 34:W686–W691. 10.1093/nar/gkl040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McInerney J, Kooij D. 1997. Economic analysis of alternative AD control programmes. Vet. Microbiol. 55:113–121 [DOI] [PubMed] [Google Scholar]
- 29.Ventura M, Turroni F, Zomer A, Foroni E, Giubellini V, Bottacini F, Canchaya C, Claesson MJ, He F, Mantzourani M, Mulas L, Ferrarini A, Gao B, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Gupta RS, Zhang Z, Beighton D, Fitzgerald GF, O'Toole PW, van Sinderen D. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 5:e1000785. 10.1371/journal.pgen.1000785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milani C, Duranti S, Lugli GA, Bottacini F, Strati F, Arioli S, Foroni E, Turroni F, van Sinderen D, Ventura M. 2013. Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl. Environ. Microbiol. 79:4304–4315. 10.1128/AEM.00984-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loquasto JR, Barrangou R, Dudley EG, Roberts RF. 2011. Short communication: the complete genome sequence of Bifidobacterium animalis subspecies animalis ATCC 25527T and comparative analysis of growth in milk with B. animalis subspecies lactis DSM 10140T. J. Dairy Sci. 94:5864–5870. 10.3168/jds.2011-4499 [DOI] [PubMed] [Google Scholar]
- 32.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422–14427. 10.1073/pnas.212527599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U. S. A. 108:11217–11222. 10.1073/pnas.1105380108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson WR. 2000. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 132:185–219 [DOI] [PubMed] [Google Scholar]
- 35.van Dongen S. 2002. Graph clustering by flow simulation. Ph.D. thesis University of Utrecht, Utrecht, The Netherlands [Google Scholar]
- 36.Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics Chapter 2:Unit 2.3. 10.1002/0471250953.bi0203s00 [DOI] [PubMed] [Google Scholar]
- 37.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 38.Turroni F, Foroni E, O'Connell Motherway M, Bottacini F, Giubellini V, Zomer A, Ferrarini A, Delledonne M, Zhang Z, van Sinderen D, Ventura M. 2010. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl. Environ. Microbiol. 76:3206–3219. 10.1128/AEM.02938-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40.Turroni F, Foroni E, Montanini B, Viappiani A, Strati F, Duranti S, Ferrarini A, Delledonne M, van Sinderen D, Ventura M. 2011. Global genome transcription profiling of Bifidobacterium bifidum PRL2010 under in vitro conditions and identification of reference genes for quantitative real-time PCR. Appl. Environ. Microbiol. 77:8578–8587. 10.1128/AEM.06352-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turroni F, Serafini F, Foroni E, Duranti S, O'Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. U. S. A. 110:11151–11156. 10.1073/pnas.1303897110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jurgonski A, Juskiewicz J, Zdunczyk Z. 2014. A high-fat diet differentially affects the gut metabolism and blood lipids of rats depending on the type of dietary fat and carbohydrate. Nutrients 6:616–626. 10.3390/nu6020616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Ruyter PG, Kuipers OP, de Vos WM. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495–548. 10.1128/MMBR.00005-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JH, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, Weimer B, O'Sullivan DJ. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. 10.1186/1471-2164-9-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Maarel MJ, van der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L. 2002. Properties and applications of starch-converting enzymes of the alpha-amylase family. J. Biotechnol. 94:137–155. 10.1016/S0168-1656(01)00407-2 [DOI] [PubMed] [Google Scholar]
- 49.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. 10.4161/gmic.19897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan SM, Fitzgerald GF, van Sinderen D. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 72:5289–5296. 10.1128/AEM.00257-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganzle MG, Follador R. 2012. Metabolism of oligosaccharides and starch in lactobacilli: a review. Front. Microbiol. 3:340. 10.3389/fmicb.2012.00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caspi R, Foerster H, Fulcher CA, Hopkinson R, Ingraham J, Kaipa P, Krummenacker M, Paley S, Pick J, Rhee SY, Tissier C, Zhang P, Karp PD. 2006. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 34:D511–D516. 10.1093/nar/gkj128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turroni F, Strati F, Foroni E, Serafini F, Duranti S, van Sinderen D, Ventura M. 2012. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl. Environ. Microbiol. 78:5002–5012. 10.1128/AEM.00629-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, O'Connell Motherway M, Viappiani A, Zhang Z, Rivetti C, van Sinderen D, Ventura M. 2011. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb. Cell Fact. 10(Suppl 1):S16. 10.1186/1475-2859-10-S1-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, Cagnasso P, Bizzarri B, de'Angelis GL, Shanahan F, van Sinderen D, Ventura M. 2009. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 75:1534–1545. 10.1128/AEM.02216-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]
- 57.Turroni F, Ventura M, Butto LF, Duranti S, O'Toole PW, O'Connell Motherway M, van Sinderen D. 2014. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life Sci. 71:183–203. 10.1007/s00018-013-1318-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imam S, Chen Z, Roos DS, Pohlschroder M. 2011. Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria. PLoS One 6:e28919. 10.1371/journal.pone.0028919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, O'Connell Motherway M, Shanahan F, Nally K, Dougan G, van Sinderen D. 2012. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. U. S. A. 109:2108–2113. 10.1073/pnas.1115621109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hidalgo-Cantabrana C, Sanchez B, Milani C, Ventura M, Margolles A, Ruas-Madiedo P. 2014. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl. Environ. Microbiol. 80:9–18. 10.1128/AEM.02977-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turroni F, Foroni E, Serafini F, Viappiani A, Montanini B, Bottacini F, Ferrarini A, Bacchini PL, Rota C, Delledonne M, Ottonello S, van Sinderen D, Ventura M. 2011. Ability of Bifidobacterium breve to grow on different types of milk: exploring the metabolism of milk through genome analysis. Appl. Environ. Microbiol. 77:7408–7417. 10.1128/AEM.05336-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bullen CL, Tearle PV. 1976. Bifidobacteria in the intestinal tract of infants: an in-vitro study. J. Med. Microbiol. 9:335–344. 10.1099/00222615-9-3-335 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.