Abstract
Metformin is commonly used as the first line of medication for the treatment of metabolic syndromes, such as obesity and type 2 diabetes (T2D). Recently, metformin-induced changes in the gut microbiota have been reported; however, the relationship between metformin treatment and the gut microbiota remains unclear. In this study, the composition of the gut microbiota was investigated using a mouse model of high-fat-diet (HFD)-induced obesity with and without metformin treatment. As expected, metformin treatment improved markers of metabolic disorders, including serum glucose levels, body weight, and total cholesterol levels. Moreover, Akkermansia muciniphila (12.44% ± 5.26%) and Clostridium cocleatum (0.10% ± 0.09%) abundances increased significantly after metformin treatment of mice on the HFD. The relative abundance of A. muciniphila in the fecal microbiota was also found to increase in brain heart infusion (BHI) medium supplemented with metformin in vitro. In addition to the changes in the microbiota associated with metformin treatment, when other influences were controlled for, a total of 18 KEGG metabolic pathways (including those for sphingolipid and fatty acid metabolism) were significantly upregulated in the gut microbiota during metformin treatment of mice on an HFD. Our results demonstrate that the gut microbiota and their metabolic pathways are influenced by metformin treatment.
INTRODUCTION
Metformin is a common antidiabetic agent in the biguanide class and is known to suppress glucose production in the liver, increase insulin sensitivity, and enhance peripheral glucose uptake in hepatic and skeletal muscle (1). The metformin-induced increase in AMP-activated protein kinase (AMPK) plays an important role in energy balance and glucose metabolism; the cellular AMP/ATP ratio is maintained by increasing ATP consumption and decreasing ATP production, which is associated with AMPK activation (2, 3). Recent studies also reported that metformin regulates hepatic gluconeogenesis and improves hyperglycemia independently of the AMPK pathway (4), which suggests that metformin-induced improvement of metabolic disorders is associated with the energy state of the body.
The gut microbiota is known to play an important role in harvesting energy from food, metabolic processes, and immune modulation. The composition of the gut microbiota is significantly associated with obesity, type 2 diabetes (T2D), and metabolic syndromes (5–7). Dysbiosis of the gut microbiota is also associated with other diseases, such as inflammatory bowel disease (IBD) (8), cardiovascular disease (9), and autism (10). Recently, metformin-induced changes in the abundance of Akkermansia muciniphila were shown to be associated with metabolic improvement (11). Another study reported that treatment with A. muciniphila, which was identified from enterotype gut microbiota, improved metabolic disorders (12). However, other gut microbiota and metabolic pathways related to metformin treatment remain uncharacterized. In addition, the effect of diet and metformin treatment on the gut microbiota (including Akkermansia spp.) has not been examined. Therefore, we investigated metformin-induced changes in the composition and metabolic functions of the gut microbiota in mouse models of obesity with mice receiving a high-fat diet (HFD) and a normal diet (ND).
MATERIALS AND METHODS
Animal model.
Age-matched male and female C57BL/6 mice were purchased from Orientbio Inc., South Korea. To induce metabolic disorders (obesity and T2D), 6-week-old mice were fed an HFD (60% lipid; Harlan Laboratories Inc.) for 28 weeks. Metformin (300 mg/kg of body weight 1,1-dimethylbiguanide hydrochloride; Sigma-Aldrich) was administered every day during the HFD for 10 weeks (HFD-Met group). As negative controls, groups of mice receiving HFD without metformin treatment (HFD group), a dietary change from an HFD to an ND (5% lipid; Zeigler Bros., Inc.) (HFD-ND group), an ND without metformin treatment (ND group), and metformin treatment on an ND (ND-Met group) were included in all procedures. All experimental protocols in this study were approved by the Seoul National University Institutional Animal Care and Use Committee (SNU-111208).
Metabolic measurements for diagnosis of metabolic syndrome.
Body weight, food intake, and glucose levels were measured every week. Serum glucose levels were measured using an Accu-Chek Performa system (Roche) after starvation for 12 h. To estimate glucose tolerance, oral glucose tolerance tests (OGTTs) were performed. Mice were administered 2 g glucose/kg body weight orally in phosphate-buffered saline, and glucose levels were measured 30, 60, and 120 min after oral administration. Levels of serum total cholesterol (TC) and high-density lipoprotein (HDL) were measured using an EnzyChrom HDL and low-density lipoprotein/very-low-density lipoprotein assay kit (BioAssay System). Serum insulin was analyzed by a commercial company (Komabiotech, Seoul, South Korea) using flow cytometry (Luminex).
Tissue preparation for histological analysis.
The liver, pancreas, and small intestine were dissected from both male and female mice. Epididymal and parametrial fat pads were removed from male and female mice, respectively, and subjected to subsequent analysis. Hematoxylin-eosin (H&E) staining of the liver, pancreas, and jejunum tissues was performed by pathologists for pathological analysis of steatosis and inflammation (Reference Biolabs, Seoul, South Korea). Extended steatosis was diagnosed pathologically and given a score of from 0 to 3, as follows: 0 for no involvement, 1 for mild involvement, 2 for moderate involvement, and 3 for severe involvement (13). In addition, immunohistochemistry for analysis of the mucosa of the small intestine was performed by a commercial company (Reference Biolabs, Seoul, South Korea) using anti-MUC5AC antibody (LifeSpan BioSciences, Inc.).
Metabolic and inflammatory biomarkers for the evaluation of metabolic disorders.
Hepatic AMP-activated protein kinase alpha 1 (AMPKα1), peroxisome proliferator-activated receptor alpha (PPARα), glucose transporter 2 (GLUT2), and glucose-6-phosphatase (G6Pase) levels were determined. In addition, the levels of adiponectin, leptin, monocyte chemoattractant protein 1 (MCP-1), tumor necrosis factor alpha (TNF-α), and interleukin-6 (IL-6) in fat pads were analyzed. The pattern of expression of the MUC2 and MUC5 genes in the ileum was analyzed. Tissues (∼100 mg) were homogenized completely prior to RNA extraction. Total RNA was extracted using an Easy-Spin total RNA extraction kit (iNtRON, Sungnam, South Korea), and cDNA synthesis was performed using a high-capacity RNA-to-cDNA kit (Applied Biosystems) according to the manufacturer's instructions. To quantify the levels of mRNA for metabolic and inflammatory biomarkers, a QuantiTect SYBR green PCR kit (Qiagen) and 7300 real-time PCR system (Applied Biosystems) were used. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as the internal control.
Next-generation sequencing of gut microbiota and the bioinformatics pipeline.
Total DNA was extracted from frozen feces using a QIAamp DNA stool minikit (Qiagen) and a QIAcube system (Qiagen). For each sample, 16S rRNA genes were amplified using the 27F/534R primer set targeting the V1-V3 region (for 27F, forward primer 5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGAGAGTTTGATCCTGGCTCAG-3′; for 534R, a reverse primer containing a unique 10-nucleotide bar code for tagging each PCR product designated NNNNNNNNNN, 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGNNNNNNNNNNATTACCGCGGCTGCTGG-3′ [boldface nucleotides are specific sequences of 27F/534R primers]) (14). The amplified PCR products were purified using a QIAquick PCR purification kit (Qiagen). Bacterial 16S rRNA pyrosequencing was performed by a commercial company (Macrogen Inc., Seoul, South Korea) using a GS-FLX system (Roche). To analyze the 16S rRNA sequence, quality filtering, including determination of the sequence length (>200 bp), end trimming, and determination of the number of ambiguous bases and the minimum quality score, was performed. Sequences were then assigned to operational taxonomic units (OTUs; 97% identity), followed by the selection of representative sequences using the QIIME software package (Quantitative Insights into Microbial Ecology) (15). We next examined the alpha and beta diversity and performed a UniFrac analysis and a principal coordinate analysis (PCoA). Linear discriminant analysis of the effect size (LEfSe) was used to estimate taxonomic abundance and characterize differences between groups (16). The UniFrac distance between categorized samples was visualized using Cytoscape software (v2.8.3). In addition, phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) was performed to identify functional genes in the sampled microbial community on the basis of the data in the KEGG pathway database (17).
Cultivation of A. muciniphila in BHI with metformin.
Stool samples from the HFD-Met group were inoculated in brain heart infusion (BHI) broth supplemented with 0.1 M metformin and phenformin and incubated in an anaerobic chamber. At 3 and 6 h postincubation, total DNA was extracted using an Easy-Spin total DNA extraction kit (iNtRON, Sungnam, South Korea). Quantification of A. muciniphila and total bacteria was performed using SYBR quantitative PCR (qPCR) and TaqMan qPCR, respectively (18, 19).
Statistical analysis.
All parameters were expressed as the average and standard deviation of each group. To quantify metabolic and inflammatory biomarker levels, qPCR was performed in triplicate. The 2−ΔΔCT relative quantification method, where CT is the threshold cycle and where ΔΔCT is equal to (CT, target − CT, GAPDH)group 1 − (CT, target − CT, GAPDH)group 2, was used to assess the level of biomarker gene expression compared to the level of GAPDH expression as an internal control. Statistical significance was assessed by one-way analysis of variance, followed by Duncan's post hoc test, with P values of <0.05 considered to indicate significance. On the basis of the relative abundance analysis using LEfSe and on the basis of the results of the Kruskal-Wallis and Wilcoxon tests, a P value of <0.05 was considered to indicate statistical significance, and the threshold on the logarithmic linear discriminant analysis (LDA) score was 3.0 to 4.0. Spearman's correlation coefficient was calculated to identify correlations between metabolic/inflammatory biomarkers and bacterial abundance.
RESULTS
Metabolic biomarker levels after a dietary change or metformin treatment.
Figure 1 shows the changes in body weight and glucose, TC, and HDL levels due to diet and metformin treatment. The body weight of mice on an HFD increased significantly over 28 weeks compared with the body weight of mice on an ND. As expected, body weight significantly decreased after a dietary change from an HFD to an ND after 18 weeks (Fig. 1A). In contrast, body weight remained constant after metformin treatment, despite continuation of the high-fat diet (Fig. 1A). The serum glucose level significantly increased in both male (P < 0.001) and female (P = 0.001) mice on an HFD, and the serum glucose level decreased after either a dietary change to an ND (P = 0.032) or metformin treatment (P = 0.016), especially in female mice (Fig. 1B). Impaired glucose tolerance, which is known to be a risk factor for T2D, was measured by OGTTs and was found to decrease significantly after a dietary change from an HFD to an ND in both male (P = 0.036) and female (P = 0.008) mice but was unaffected by metformin treatment (Fig. 1B). In addition, the homeostatic model assessment-insulin resistance (HOMA-IR), estimated on the basis of glucose and insulin levels, showed a trend to reduced insulin resistance after a change from an HFD to an ND or metformin treatment, but these changes were not statistically significant (Fig. 1C). In addition, the homeostatic model assessment-beta cell function (HOMA-β) did not show significant differences after either a change from an HFD to an ND or metformin treatment in mice. TC levels decreased significantly after a dietary change from an HFD to an ND in both male (P < 0.001) and female (P < 0.001) mice. The effects of metformin treatment on the TC level differed depending on the gender of the mice. The TC level significantly decreased in female mice (P = 0.023) but not male mice (P = 0.875). HDL levels also significantly decreased after a change from an HFD to an ND in both male (P < 0.001) and female (P = 0.001) mice and decreased after metformin treatment in both male (P = 0.055) and female (P = 0.055) mice. Metformin treatment had no effect on the metabolic biomarker levels in the ND group.
FIG 1.
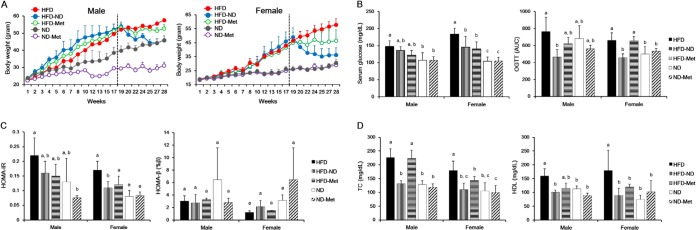
Effects of dietary changes and metformin treatment on body weight and glucose, TC, and HDL levels. Mice were induced to develop metabolic disorders while on an HFD for 18 weeks and then subjected to metformin treatment and a dietary change to an ND for the following 10 weeks (weeks 19 to 28 of the study). Several metabolic biomarkers, including body weight and glucose, TC, and HDL levels, were measured at the indicated times (n = 41). (A) Body weights of male and female mice over 28 weeks. Dotted line, time of metformin treatment and dietary changes. (B) Serum glucose level and glucose tolerance (determined by OGTTs) at 21 weeks. Glucose tolerance was expressed on the basis of the area under the curve (AUC). (C) HOMA-IR and HOMA-β were calculated from the levels of glucose and insulin measured at 21 weeks. (D) Both TC and HDL were measured at 28 weeks. Different superscript letters represent significant differences (P < 0.05) according to Duncan's post hoc test.
Effects of diet and metformin treatment on patterns of metabolic and inflammatory biomarker expression.
Figure 2 shows the relative levels of expression of various metabolic and inflammatory biomarkers in the liver and fat pads. The effects of metformin treatment and an HFD on liver tissue in mice differed according to gender. A dietary change from an HFD to an ND significantly reduced the levels of AMPKα1 (P < 0.001), PPARα (P = 0.003), and GLUT2 (P = 0.024) expression and increased those of G6Pase expression (P = 0.003) in female mice. In contrast, a dietary change from an HFD to an ND significantly reduced the level of G6Pase expression (P = 0.001) in male mice. Metformin treatment of mice on the HFD significantly decreased the levels of expression of both AMPKα1 (P = 0.003) and GLUT2 (P = 0.013) and increased the levels of expression of PPARα (P = 0.010) in female mice. In contrast, metformin treatment and an HFD significantly reduced G6Pase expression (P < 0.001) in male mice.
FIG 2.
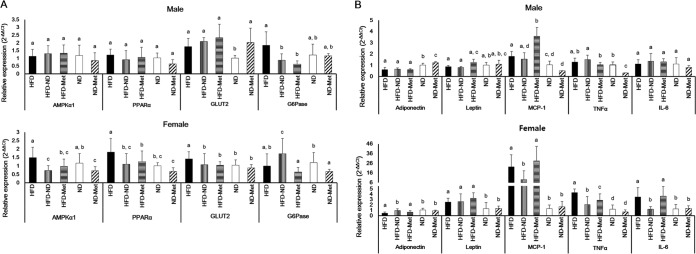
Metabolic and inflammatory biomarkers in the liver and fat pads. Relative levels of mRNA for metabolic and inflammatory biomarkers were analyzed using qPCR. (A) Expression in the livers of male and female mice; (B) expression in the epididymal fat pads of male mice and parametrial fat pads of female mice. The 2−ΔΔCT relative quantification method, described in Materials and Methods, was used for analysis of the level of biomarker expression compared to the level of GAPDH expression as an internal control. Different superscript letters indicate significant differences (P < 0.05) according to Duncan's post hoc test.
In fat pads, a dietary change from an HFD to an ND significantly reduced the levels of MCP-1 (P < 0.001), TNF-α (P < 0.001), and IL-6 (P < 0.001) expression and increased the level of adiponectin expression (P < 0.001), as shown in Fig. 2B. Metformin treatment of mice on an HFD significantly reduced the level of TNF-α expression (P < 0.001) in female mice and increased the level of MCP-1 expression (P < 0.001) in male mice.
Mucin expression and histological changes in the small intestine and liver due to metformin treatment.
Mucin levels were estimated on the basis of MUC2 and MUC5 expression in the small intestine. Expression of the genes for both proteins increased significantly after metformin treatment in female mice (for MUC2, P = 0.003; for MUC5, P = 0.010) but not in male mice (for MUC2, P = 0.110; for MUC5, P = 0.884) (Fig. 3A). These results are consistent with the histological data. Thickened intestinal mucosa was confirmed by immunohistochemical assays using an anti-MUC5AC antibody (Fig. 3B).
FIG 3.
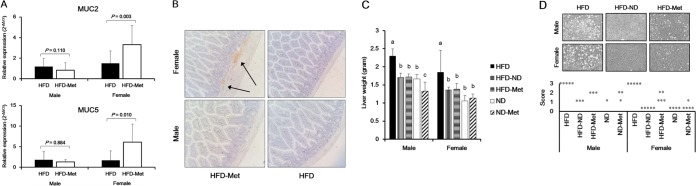
Effects of dietary change and metformin treatment on mucin expression in small intestine tissue and the histology of small intestine and liver tissue. (A) The levels of mRNA for MUC2 and MUC5 were increased after metformin treatment in female mice on an HFD. The 2−ΔΔCT relative quantification method, described in Materials and Methods, was used to analyze the level of biomarker expression compared to the level of GAPDH expression as an internal control. Statistical significance was assessed using the Mann-Whitney U test. (B) Confirmation of thickened intestinal mucosa after metformin treatment based on an immunohistochemistry assay. Arrows, MUC5AC stained by anti-MUC5AC. Magnification, ×20. (C) Weight of the liver after a dietary change to an ND and metformin treatment. (D) The extension score (0 to 3) of steatosis was evaluated by a pathologist, as follows: 0, no involvement; 1, mild involvement; 2, moderate involvement; 3, severe involvement. Each dot signifies a liver sample in which steatosis was diagnosed. Steatosis of the liver was observed using an optical microscope. Magnification, ×20. Steatosis was improved while the mice were on an ND and during metformin treatment.
The weights of the livers of the mice in the HFD-ND and HFD-Met groups differed significantly from the weights of the livers of the mice in the HFD group for both male mice (for the HFD-ND group, P = 0.036; for the HFD-Met group, P = 0.036) and female mice (for the HFD-ND group, P = 0.095; for the HFD-Met group, P = 0.095) (Fig. 3C). Severe steatosis of the liver in the HFD group was recovered by either a dietary change to an ND or metformin treatment (Fig. 3D). There was no difference in the inflammation scores between the HFD-ND and HFD-Met groups (data not shown).
Characterization of gut microbiota.
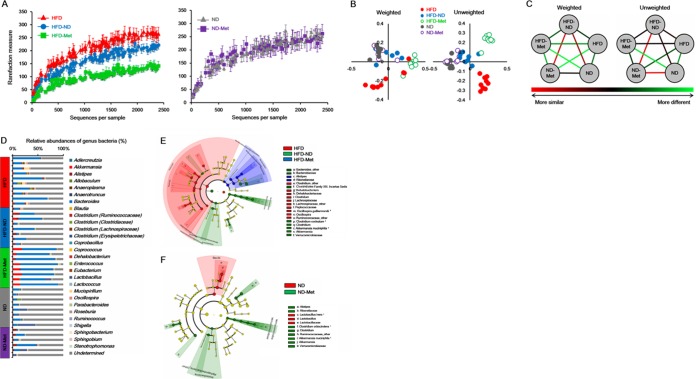
To characterize the composition of the gut microbiota, a total of 302,689 sequences were generated from 40 stool samples. Among them, 238,522 sequences were obtained after quality filtering, with an average of 5,963 ± 1,127 sequences being recovered per sample. Figure 4 shows differences in microbial diversity among the HFD, HFD-ND, and HFD-Met groups. The alpha diversities of the gut microbiota of these three groups decreased significantly in the order HFD group, HFD-ND group, and HFD-Met group (Fig. 4A). Unlike the HFD group, metformin treatment had no significant effect on the alpha diversity of the gut microbiota in the ND group (Fig. 4A). PCoA based on phylogenetic analysis indicated clustering of the gut microbiota within groups. PCoA of both unweighted and weighted UniFrac distances showed a clear separation of the HFD, HFD-Met, and HFD-ND groups, while no separation between the ND and ND-Met groups was observed (Fig. 4B). Furthermore, the UniFrac distance of the bacterial communities between the HFD and HFD-Met groups was considerably higher than that of the bacterial communities between the ND and ND-Met groups. In contrast, the distance between the bacterial communities of the HFD-Met and ND-Met groups was considerably higher than that between the bacterial communities of the HFD-ND and ND groups (Fig. 4C).
FIG 4.
Microbial diversity and difference in the bacterial community between groups categorized according to diet and metformin treatment. (A) Rarefaction curve of bacterial diversity according to dietary change and metformin treatment in mice on an HFD and ND; (B) PCoA of weighted and unweighted UniFrac distances from 40 mouse stool samples; (C) visualized UniFrac distances between groups; (D) bacterial classification at the genus level; (E) cladogram from the LEfSe results between the HFD, HFD-Met, and HFD-ND groups of mice; (F) cladogram from the LEfSe results between the ND and ND-Met groups of mice. The differences were significant (P < 0.05) both among classes (Kruskal-Wallis test) and between subclasses (Wilcoxon's test). The threshold of the logarithmic LDA score was 4.0. *, bacterial species.
Relative abundance of the gut microbiota after metformin treatment.
The composition of the phylum Bacteroidetes in the HFD group (43.79% ± 22.35%) was significantly lower than that in the ND group (79.40% ± 10.00%) (Fig. 4D). In contrast, the composition of the phylum Firmicutes was significantly higher in the HFD group (50.73% ± 19.20%) (Fig. 4D). When metformin was administered to the HFD group, the composition of the phylum Bacteroidetes increased to 77.45% ± 8.73%, which was similar to that in the ND group (Fig. 4D). Additionally, the composition of the phylum Verrucomicrobia (12.44% ± 5.26%) in the HFD-Met group significantly increased, unlike the effect of a dietary change from an HFD to an ND (Fig. 4D). In the HFD-Met group, the abundances of the families Bacteroidaceae, Verrucomicrobiaceae, and Clostridiales family XIII, incertae sedis, as well as A. muciniphila and Clostridium cocleatum changed significantly compared with those in the HFD and HFD-ND groups (Fig. 4E). Metformin treatment also affected the composition of the gut microbiota in mice fed an ND. The families Rikenellaceae, Ruminococcaceae, and Verrucomicrobiaceae, as well as Alistipes spp., Akkermansia spp., and Clostridium spp., were more abundant in the ND-Met group than the ND group (Fig. 4F).
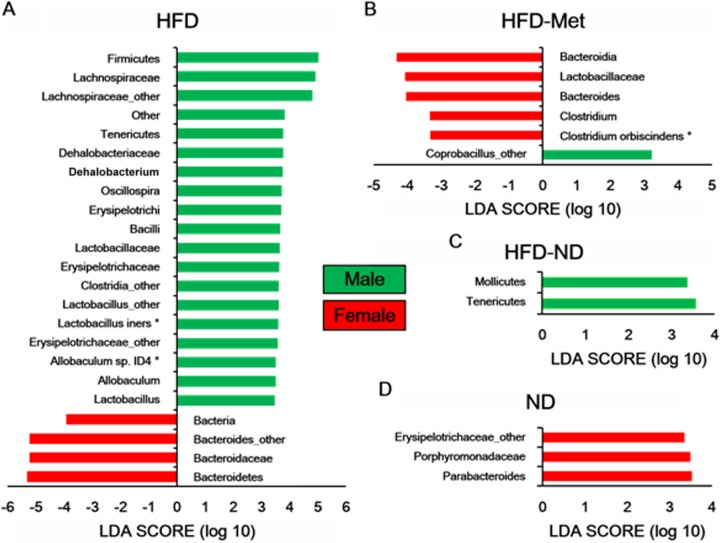
Differences in the gut microbiota between male and female mice were also examined. In the HFD group, the phylum Bacteroidetes was more abundant in female mice than male mice (Fig. 5A). In addition, the phylum Tenericutes was abundant in males of the HFD-ND group and Parabacteroides spp. were abundant in females of the ND group (Fig. 5C and D). In the HFD-Met group, Coprobacillus spp. were more abundant in males, while Clostridium spp., Bacteroides spp., and members of the family Lactobacillaceae and the class Bacteroidia were more abundant in female mice (Fig. 5B). No significant difference between male and female mice in the ND-Met group was observed.
FIG 5.
Differences in the bacterial communities between males and females. A taxonomic comparison of the bacterial communities in male and female mice in the HFD (A), HFD-Met (B), HFD-ND (C), and ND (D) groups was performed. Significant differences in LDA scores (P < 0.05) were produced among classes (Kruskal-Wallis test) and between subclasses (Wilcoxon's test). There was no significant difference between males and females in the ND-Met group. The threshold of the logarithmic LDA score was 3.0. *, bacterial species.
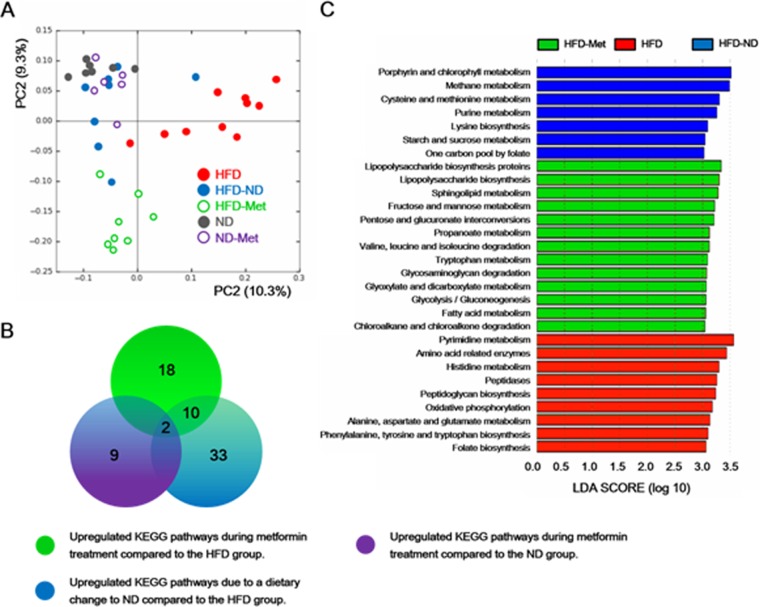
Changes in the metabolic pathways of the gut microbiota during metformin treatment.
A total of 245 KEGG pathways were generated using the composition of the gut microbiota based on PICRUSt. PCoA plots clearly showed clustering of five groups according to diet and metformin treatment (Fig. 6A). Thirty KEGG pathways were significantly upregulated in the HFD-Met group compared with their levels of regulation in the HFD group. Among them, we excluded KEGG pathways related to a dietary change only (HFD-ND) and metformin treatment on a normal diet (ND-Met). Therefore, a total of 18 KEGG pathways were exclusively associated with the specific effects of metformin in mice receiving an HFD (Fig. 6B; see Table S1 in the supplemental material). Statistically significant KEGG pathways for each group were additionally identified using LEfSe (Fig. 6C). After metformin treatment on an HFD, metabolic pathways, such as lipopolysaccharide biosynthesis, sphingolipid metabolism, fructose and mannose metabolism, pentose and glucuronate interconversions, and propanoate metabolism, were enriched significantly.
FIG 6.
Comparison of KEGG pathways predicted using PICRUSt according to diet and metformin treatment. Groups categorized according to metformin treatment and diet were clearly clustered on the basis of the KEGG pathways predicted using PICRUSt, as well as the bacterial diversity. A total of 245 KEGG pathways were generated, and the KEGG pathways that were significantly increased during metformin treatment were further analyzed using PCoA and LEfSe. (A) Clustering of five groups by KEGG pathways using PCoA. (B) Predicted KEGG pathways during metformin treatment and a dietary change to an ND. A total of 30 KEGG pathways were increased during metformin treatment of mice on an HFD. Among them, 2 and 12 KEGG pathways that were increased overlapped those that were increased in the HFD-ND and ND-Met groups, respectively. Finally, 18 unique KEGG pathways were predicted to be increased during metformin treatment of mice on an HFD. (C) LEfSe results showed a statistically significant increase in the abundance of KEGG pathways in the HFD-ND, HFD-Met, and HFD groups. LEfSe results showed a sequentially significant ranking (P < 0.05) among classes (Kruskal-Wallis test) and between subclasses (Wilcoxon's test). The threshold for the logarithmic LDA score was 3.0.
Correlations between metabolic biomarkers and bacterial abundance.
A. muciniphila was negatively correlated with serum glucose levels (Fig. 7). Clostridium orbiscindens showed a negative correlation with body weight and PPARα and GLUT2 levels and a positive correlation with TNF-α, MUC2, and MUC5 levels (Fig. 7). PPARα and GLUT2 levels were negatively correlated with Blautia producta and Allobaculum sp. strain ID4. Clostridium cocleatum was positively correlated with AMPKα1 and TC levels (Fig. 7).
FIG 7.
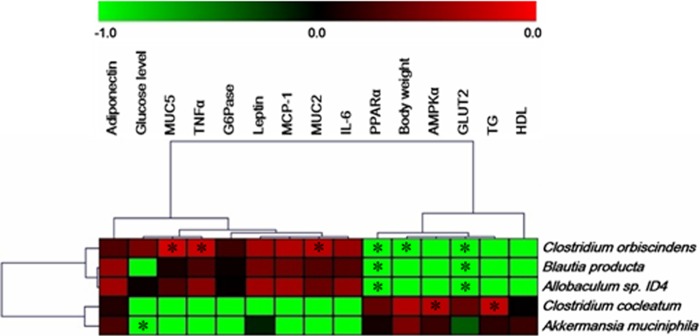
Correlation between metabolic biomarkers and bacterial abundance during metformin treatment in mice with HFD-induced obesity. *, statistical significance based on a P value of <0.05 (Spearman's correlation coefficient). The heat map was generated using MultiExperiment Viewer software (MEV; v4.8.1).
A. muciniphila enrichment by metformin or phenformin treatment in vitro.
The composition of A. muciniphila in six pooled stool samples from mice in the HFD-Met group was 14.3% ± 2.6%. At 3 and 6 h postinoculation into BHI broth, the proportions of A. muciniphila were 3.0% ± 0.5% and 3.8% ± 1.8%, respectively. In contrast, the proportion of A. muciniphila in BHI broth supplemented with 0.1 M metformin significantly increased (8.8% ± 3.7% and 6.6% ± 2.8% at 3 and 6 h postinoculation, respectively) (Fig. 8). Furthermore, when BHI was supplemented with 0.1 M phenformin (another member of the biguanide class), A. muciniphila was further enriched (19.1% ± 4.6% and 14.7% ± 2.1% at 3 and 6 h postinoculation, respectively) (Fig. 8).
FIG 8.
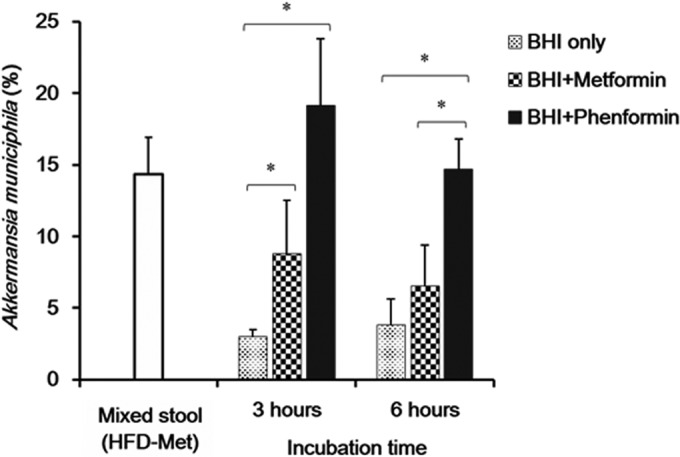
Effects of metformin and phenformin on growth of Akkermansia muciniphila as a proportion of all bacteria in BHI medium. Mixed stool, original stool samples from HFD-Met mice prior to culture. The amount of Akkermansia muciniphila bacteria as a proportion of all bacteria, considered 100%, was calculated, *, statistical significance based on a P value of <0.05 (Mann-Whitney U test).
DISCUSSION
Recent studies reported an association between the abundance of A. muciniphila, obesity, and T2D (11, 12), and changes in the abundance of A. muciniphila during metformin treatment were observed in a mouse model (11). The results of these studies were limited to an association between the gut microbiota and glucose homeostasis for a short period of time, and the association between metabolic improvement and metformin treatment due to changes in the gut microbiota remains unclear. Our results are consistent with the results of these previous studies. Additionally, we found an enrichment effect of metformin on the growth of A. muciniphila in vitro, which is consistent with the results obtained with an in vivo mouse model. Therefore, metformin appears to act as a growth factor for Akkermansia spp.
The patterns of expression of metabolic biomarkers and correlations with the composition of the gut microbiota could increase our understanding of the association between the gut microbiota and metabolic improvement. Activation of AMPK by metformin treatment is known to be the primary mechanism of improving hyperglycemia (20). Increases in cytosolic AMP by metformin were suggested to be a mechanism of AMPK activation (21). Interestingly, metabolic improvement by metformin treatment without the activation of AMPK was observed in this study (Fig. 2A). In a recent report, the effect of metformin on hepatic gluconeogenesis was determined in an AMPK-deficient mouse model (4). However, antagonism of hepatic glucagon by metformin was proposed to be another target for T2D improvement in a recent study (22). Moreover, hepatic PPARα, GLUT2, and AMPKα1 levels decreased significantly (Fig. 2A). Only the TNF-α level in the fat pad decreased during metformin treatment of mice on an HFD. A significant correlation between A. muciniphila and glucose levels was observed during metformin treatment of mice on an HFD (Fig. 7). Both PPARα and GLUT2 levels were negatively correlated with B. producta, Allobaculum sp. ID4, and C. orbiscindens, while the TNF-α level was positively correlated with C. orbiscindens during metformin treatment of mice on an HFD (Fig. 7). These results suggest that OTUs of the gut microbiota are strongly associated with specific metabolic and immunological biomarkers.
Gastrointestinal mucins produced by goblet cells protect the underlying epithelium from pathogens. Recently, the degradation of mucin was reported in mice on an HFD (12) and may be associated with metabolic disorders. Mucin is degraded by specific intestinal bacteria, which may comprise 1% of the colonic microbiota (23). Among them, the effect of A. muciniphila on metabolic improvement has recently been discussed. In addition, the increase in A. muciniphila abundance was associated with mucus thickness (12). In this study, the abundance of A. muciniphila increased during metformin treatment of male and female mice on an HFD. However, a significant increase in mucin gene expression and thickened intestinal mucosa were observed only in females (Fig. 3A and B). The effect of mucin on metabolic improvement may differ by gender, but the mechanisms are not fully understood. Further studies are required to explore gender differences in the role of mucin in metabolic improvement.
Bacterial diversity significantly decreased during metformin treatment and after a dietary change to an ND; metformin had a more significant effect than dietary change. The alpha diversity indicated that decreases in bacterial diversity during metformin treatment were observed only in mice on an HFD, and metformin treatment had no effect on mice fed an ND (Fig. 4A). The effects of metformin on bacterial diversity were consistent on the basis of both weighted and unweighted beta diversities and UniFrac distances (Fig. 4B and C). Therefore, metformin treatment may decrease the overall bacterial diversity, and the specific composition of the microbiota was strongly associated with metabolic improvement during metformin treatment.
The relative abundance of C. cocleatum in the HFD-Met group was 0.1% and increased significantly during metformin treatment of mice on an HFD, similar to the findings for A. muciniphila (Fig. 4E). It has been reported that C. cocleatum levels are decreased in patients with irritable bowel syndrome (IBD) (24). However, the association between C. cocleatum and metabolic syndromes (such as obesity and T2D) has not been explored. Clostridium spp. are known to degrade some animal mucin (25). Recently, an increase in Clostridium spp. was reported during metformin treatment of mice on an HFD (11). Although the relative abundance of C. cocleatum was minor compared to that of A. muciniphila, C. cocleatum may be associated with metabolic improvement during metformin treatment.
The effects of metformin treatment on metabolic biomarker expression in both liver and fat pads differed by gender (Fig. 2). In addition, differences in the gut microbiota were observed between male and female mice (Fig. 5). In previous studies, metformin treatment differentially affected body weight according to gender, and metabolic hormones—such as leptin and adiponectin—were differentially affected by metformin treatment (26, 27). The effect of the gut microbiota on metabolic disease according to gender has not been reported to date. Recently, the vaginal microbiota was shown to differ significantly on the basis of menopausal status (14), which suggests that hormones in females were highly associated with the composition of the human vaginal microbiota. It may be suggested that human gut microbiota could likewise be impacted by human hormone levels. Moreover, female hormones are involved in glucose and lipid metabolism and are known to exert a protective effect against metabolic disorders (28, 29). Therefore, differences in the gut microbiota between male and female mice during metformin treatment might be caused by differences in hormone levels, which might be associated with metabolic phenotypes.
We further predicted that the significant metabolic pathways would show differences in expression during metformin treatment of mice on an HFD, based on the bacterial abundances determined using PICRUSt. The HFD, HFD-ND, and HFD-Met groups were clearly clustered by predicted KEGG pathways and bacterial diversity (Fig. 6A). Therefore, we expected that the significant metabolic phenotypes during metformin treatment might be caused by different metabolic functions of the gut microbiota. In this study, 18 specific metabolic pathways of the gut microbiota were significantly increased by metformin alone, when other influences were controlled for (Fig. 6C). The increased metabolic activities of sphingolipid and fatty acid metabolism are closely associated with metabolic syndromes and may be important for the metabolic improvement induced by metformin treatment. This may be because specific sphingolipid signaling pathways influence the major biological processes, including insulin resistance, lipid metabolism, inflammation, and the immune response (30–32). In addition, Everard et al. have shown that the expression of lipogenic genes—such as fatty acid synthase (FASN) and acetyl coenzyme A carboxylase 1 (ACC1)—is upregulated by A. muciniphila treatment of mice on an HFD (12). Thus, the abundance of A. muciniphila may be associated with lipid metabolism by the gut microbiota.
The amelioration effect of metformin on the abundance of A. muciniphila was demonstrated using in vitro growth assays in BHI medium supplemented with metformin. Metformin or phenformin administration increased the abundance of A. muciniphila (Fig. 8). Interestingly, A. muciniphila was not detected in ND mice, and its level was slightly increased in mice on an HFD. However, after metformin treatment in mice on an HFD, the abundance of A. muciniphila increased by ∼18-fold (12.44%) compared with that in mice in the HFD group (0.68%). Therefore, the effect of A. muciniphila on metabolic disorders and improvement may be dose dependent.
In conclusion, the diversity and composition of the gut microbiota changed significantly during metformin treatment of mice on an HFD, and the changes were correlated with the levels of various metabolic biomarkers. Furthermore, specific pathways associated with lipid metabolism (including sphingolipid and fatty acid metabolism) may play a key role in the metabolic improvement induced by metformin treatment. We suggest that the specific composition of the gut microbiota during metformin treatment has therapeutic effects on metabolic diseases, including obesity and T2D.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by a grant from the National Research Foundation of Korea (NRF) funded by the South Korea government (MEST) (2012-0008692).
Footnotes
Published ahead of print 18 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01357-14.
REFERENCES
- 1.Rotella CM, Monami M, Mannucci E. 2006. Metformin beyond diabetes: new life for an old drug. Curr. Diabetes Rev. 2:307–315. 10.2174/157339906777950651 [DOI] [PubMed] [Google Scholar]
- 2.Hardie DG, Ross FA, Hawley SA. 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13:251–262. 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viollet B, Guigas B, Leclerc J, Hebrard S, Lantier L, Mounier R, Andreelli F, Foretz M. 2009. AMP-activated protein kinase in the regulation of hepatic energy metabolism: from physiology to therapeutic perspectives. Acta Physiol. 196:81–98. 10.1111/j.1748-1716.2009.01970.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foretz M, Hebrard S, Leclerc J, Zarrinpashneh E, Soty M, Mithieux G, Sakamoto K, Andreelli F, Viollet B. 2010. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Invest. 120:2355–2369. 10.1172/JCI40671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 6.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102:11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, et al. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 8.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780–13785. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ordovas JM, Mooser V. 2006. Metagenomics: the role of the microbiome in cardiovascular diseases. Curr. Opin. Lipidol. 17:157–161. 10.1097/01.mol.0000217897.75068.ba [DOI] [PubMed] [Google Scholar]
- 10.Malkki H. 2014. Neurodevelopmental disorders: human gut microbiota alleviate behavioural symptoms in a mouse model of autism spectrum disorder. Nat. Rev. Neurol. 10:60. 10.1038/nrneurol.2014.1 [DOI] [PubMed] [Google Scholar]
- 11.Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, Bae JW. 2013. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63:727–735. 10.1136/gutjnl-2012-303839 [DOI] [PubMed] [Google Scholar]
- 12.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 110:9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gove ME, Rhodes DH, Pini M, van Baal JW, Sennello JA, Fayad R, Cabay RJ, Myers MG, Jr, Fantuzzi G. 2009. Role of leptin receptor-induced STAT3 signaling in modulation of intestinal and hepatic inflammation in mice. J. Leukoc. Biol. 85:491–496. 10.1189/jlb.0808508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JE, Lee S, Lee H, Song YM, Lee K, Han MJ, Sung J, Ko G. 2013. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 8:e63514. 10.1371/journal.pone.0063514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31:814–821. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SY, Ko G. 2012. Using propidium monoazide to distinguish between viable and nonviable bacteria, MS2 and murine norovirus. Lett. Appl. Microbiol. 55:182–188. 10.1111/j.1472-765X.2012.03276.x [DOI] [PubMed] [Google Scholar]
- 19.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. 2007. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 73:7767–7770. 10.1128/AEM.01477-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirpichnikov D, McFarlane SI, Sowers JR. 2002. Metformin: an update. Ann. Intern. Med. 137:25–33. 10.7326/0003-4819-137-1-200207020-00009 [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, He H, Balschi JA. 2007. Metformin and phenformin activate AMP-activated protein kinase in the heart by increasing cytosolic AMP concentration. Am. J. Physiol. Heart Circ. Physiol. 293:H457–H466. 10.1152/ajpheart.00002.2007 [DOI] [PubMed] [Google Scholar]
- 22.Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ. 2013. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 494:256–260. 10.1038/nature11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoskins LC, Boulding ET. 1981. Mucin degradation in human colon ecosystems. Evidence for the existence and role of bacterial subpopulations producing glycosidases as extracellular enzymes. J. Clin. Invest. 67:163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malinen E, Krogius-Kurikka L, Lyra A, Nikkila J, Jaaskelainen A, Rinttila T, Vilpponen-Salmela T, von Wright AJ, Palva A. 2010. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. 16:4532–4540. 10.3748/wjg.v16.i36.4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salyers AA, West SE, Vercellotti JR, Wilkins TD. 1977. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl. Environ. Microbiol. 34:529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gui Y, Silha JV, Murphy LJ. 2004. Sexual dimorphism and regulation of resistin, adiponectin, and leptin expression in the mouse. Obesity Res. 12:1481–1491. 10.1038/oby.2004.185 [DOI] [PubMed] [Google Scholar]
- 27.Anisimov VN, Piskunova TS, Popovich IG, Zabezhinski MA, Tyndyk ML, Egormin PA, Yurova MV, Rosenfeld SV, Semenchenko AV, Kovalenko IG, Poroshina TE, Berstein LM. 2010. Gender differences in metformin effect on aging, life span and spontaneous tumorigenesis in 129/Sv mice. Aging 2:945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geer EB, Shen W. 2009. Gender differences in insulin resistance, body composition, and energy balance. Gender Med. 6(Suppl 1):S60–S75. 10.1016/j.genm.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu BN, O'Sullivan AJ. 2011. Sex differences in energy metabolism need to be considered with lifestyle modifications in humans. J. Nutr. Metab. 2011:391809. 10.1155/2011/391809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hla T, Dannenberg AJ. 2012. Sphingolipid signaling in metabolic disorders. Cell Metab. 16:420–434. 10.1016/j.cmet.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bikman BT, Summers SA. 2011. Ceramides as modulators of cellular and whole-body metabolism. J. Clin. Invest. 121:4222–4230. 10.1172/JCI57144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chavez JA, Holland WL, Bar J, Sandhoff K, Summers SA. 2005. Acid ceramidase overexpression prevents the inhibitory effects of saturated fatty acids on insulin signaling. J. Biol. Chem. 280:20148–20153. 10.1074/jbc.M412769200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.