Abstract
A key step in fungal l-lysine biosynthesis is catalyzed by adenylate-forming l-α-aminoadipic acid reductases, organized in domains for adenylation, thiolation, and the reduction step. However, the genomes of numerous ascomycetes and basidiomycetes contain an unexpectedly large number of additional genes encoding similar but functionally distinct enzymes. Here, we describe the functional in vitro characterization of four reductases which were heterologously produced in Escherichia coli. The Ceriporiopsis subvermispora serine reductase Nps1 features a terminal ferredoxin-NADP+ reductase (FNR) domain and thus belongs to a hitherto undescribed class of fungal multidomain enzymes. The second major class is characterized by the canonical terminal short-chain dehydrogenase/reductase domain and represented by Ceriporiopsis subvermispora Nps3 as the first biochemically characterized l-α-aminoadipic acid reductase of basidiomycete origin. Aspergillus flavus l-tyrosine reductases LnaA and LnbA are members of a distinct phylogenetic clade. Phylogenetic analysis supports the view that fungal adenylate-forming reductases are more diverse than previously recognized and belong to four distinct classes.
INTRODUCTION
In fungi, a key step in l-lysine biosynthesis is the NADPH-dependent reduction of l-α-aminoadipic acid into l-α-aminoadipate-6-semialdehyde by dedicated aminoacyl adenylate-forming reductases, such as Penicillium chrysogenum Lys2 (1, 2). They feature (i) an N-terminal adenylation (A) domain, (ii) a carrier protein (PCP) domain, also termed the thiolation (T) domain, which serves covalent binding of the substrate, and (iii) a short-chain reductase (R) domain which is located at the C terminus (Fig. 1). For functionality, T domains of all multidomain enzymes require posttranslational modification (“priming”). This is accomplished by transfer of a phosphopantetheinyl moiety from coenzyme A onto a conserved serine residue within the sequence of the T domain. This phosphopantetheinyl residue then serves as a covalent attachment site for the incoming substrate. l-α-Aminoadipic acid reductases are reminiscent of nonribosomal peptide synthetases (NRPSs). The latter also follow a multidomain setup and require A and T domains to assemble complex and often highly functionalized natural products. In addition, NRPSs also include condensation (C) domains that catalyze amide bond formation between amino acid building blocks (3). In contrast, adenylate-forming reductases lack C domains. Curiously, in numerous fungal genomes, among them those of Aspergillus flavus (4) and the basidiomycetes Ceriporiopsis subvermispora, Serpula lacrymans, and Trametes versicolor (5–7), more than one gene was identified that encodes an enzyme following the Lys2 domain architecture with a terminal R domain.
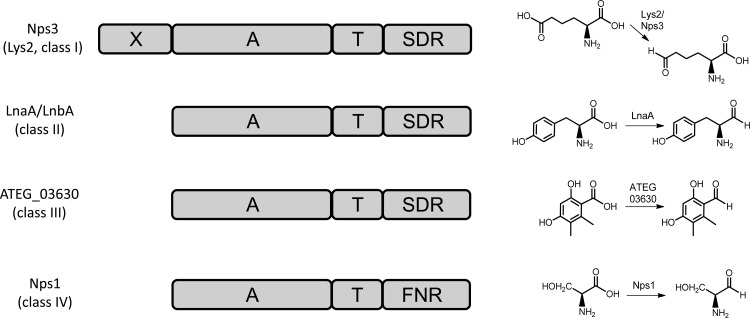
FIG 1.
Domain composition of fungal adenylate-forming reductases (left). The catalyzed reactions are shown in simplified form (right). Nps3 represents the canonical Lys2 domain structure. Domain abbreviations: A, adenylation; FNR, ferredoxin-NADP+ reductase; SDR, dehydrogenase/reductase; T, thiolation. X refers to the N-terminal portion of unknown function of l-α-aminoadipic acid reductases. ATEG_03630 functions according to reference 11.
At first glance, these additional genes seem to encode redundant l-α-aminoadipic acid (Lys2-type) reductases. Typically, they are about 1,400 amino acids (aa) in length, as shown, e.g., for Candida albicans (1,391 aa) and P. chrysogenum (1,409 aa) (8, 9). Integral to Lys2-type enzymes is an additional N-terminal portion of approximately 250 aa (X domain) (Fig. 1) which occurs with several peptide synthetases, coenzyme A (CoA) ligases, and other multifunctional proteins. Its significance has not been elucidated yet, but it may contribute to the structural integrity of the protein (9). Contrasting Lys2, other multidomain reductases lack this extra N-terminal extension and are therefore only about 1,000 to 1,050 aa in length. During a study on Aspergillus flavus secondary metabolism, first experimental evidence emerged that they do not function within l-lysine assembly: the reductases LnaA and LnbA serve the biosynthesis of piperazine natural products, which are required for proper sclerotia formation (10) and, thus, morphological development of the fungus. Further evidence stems from Aspergillus terreus, which uses an adenylate-forming reductase, encoded by the ATEG_03630 gene, to reduce a polyketide aryl acid to the respective aldehyde (11).
Here, we describe the functional characterization of Nps1 of C. subvermispora (Basidiomycota), which is a serine reductase that features a ferredoxin-NADP+ reductase (FNR)-like domain and represents a hitherto undescribed and uncharacterized class of fungal NRPS-like multidomain enzymes. We also describe Nps3 of the same species, which is a Lys2-type enzyme and the first biochemically characterized basidiomycete l-α-aminoadipic acid reductase, as well as A. flavus LnaA and LnbA (10), for which we prove tyrosine reductase activity. The latter three enzymes belong to fungal adenylate-forming reductases, which are characterized by a canonical terminal short-chain dehydrogenase/reductase (SDR) domain and which follow the standard A-T-R domain composition.
MATERIALS AND METHODS
General procedures and culture conditions.
For standard molecular biology procedures, we followed described protocols (12). For isolation of plasmid DNA from Escherichia coli and DNA restriction and ligation, we followed the instructions of the manufacturers of kits and enzymes (NEB, Fermentas, Zymo). Synthetic DNA was produced by GenScript and Life Technologies. Chemicals were from Fluka, Sigma-Aldrich, and Roth. [32P]pyrophosphate was purchased from PerkinElmer.
Cloning of reductase genes.
The C. subvermispora genes encoding Nps1 and Nps3 were synthesized as codon-optimized reading frames for expression in E. coli. Both genes were supplied in two portions (1,565 and 1,595 bp for nps1 and 2,128 and 2,160 bp for nps3), and each portion was cloned in vector pUC57. Construction of the nps1 expression plasmid was accomplished by ligating the 5′ part via the EcoRI and HindIII sites into the expression vector pET28a, to create plasmid pDK3. Subsequently, the 3′ moiety was inserted into pDK3 via the AgeI and HindIII sites to create the expression construct pDK6. Construction of the nps3 expression plasmid was as follows: the 5′ part was cloned via EcoRI and HindIII into the pMAL-c2X vector, to create plasmid pDK19. This was followed by insertion of the second part via AgeI and HindIII, to yield pDK20. The complete gene was then cloned between the EcoRI and HindIII sites of pET21a, to yield pDK21. DNA sequencing confirmed correct assembly of the respective genes. The pET15b-based plasmids to express A. flavus genes lnaA and lnbA (pSA14 and pSA15, respectively) have been described previously (10).
Gene expression and protein purification.
E. coli KRX (Promega) was transformed with expression constructs pDK6, pDK21, pSA14, and pSA15. Overnight cultures (5 ml LB medium, amended with kanamycin [50 mg/liter] for pDK6 or carbenicillin [50 mg/liter] for pDK21, pSA14, and pSA15) were used to inoculate 400 ml of LB medium. The cultures were incubated at 37°C on an orbital shaker at 180 rpm to an optical density at 600 nm (OD600) of 0.35. Then, the temperature was decreased to 16°C, for 30 min, prior to inducing expression of the respective genes by adding 0.1% l-rhamnose (wt/vol). The incubation of induced cells continued for 24 h before the biomass was collected by centrifugation (4°C, 1,200 × g, 15 min). The cells were resuspended in lysis buffer (50 mM NaH2PO4·H2O, 300 mM NaCl, 10 mM imidazole, pH 8.0) and disrupted using a Branson 450 sonifier. Subsequent centrifugation at 4°C and 14,000 × g for 15 min removed cell debris. The proteins were purified by metal affinity chromatography on Protino Ni2+-nitrilotriacetic acid (NTA) resin (Macherey-Nagel). The purified proteins were desalted on PD-10 columns (GE Healthcare), equilibrated with 100 mM phosphate buffer, pH 7.0. Purification was verified with polyacrylamide gels (12% Laemmli gels). The protein concentrations were determined by the method of Bradford (13). The phosphopantetheinyl transferase gene svp (14) was expressed as previously described (15).
In vitro characterization of adenylation domains.
All reactions were carried out in triplicate. Characterization of the Nps1 and Nps3 adenylation domains: pH and temperature optima were determined using the substrate-dependent ATP-[32P]pyrophosphate exchange assay. Reaction parameters were total assay volume of 100 μl at 25°C (varied from 4°C to 45°C to determine the temperature optimum) in 100 mM phosphate buffer (pH 7.0; varied from pH 5.0 to 8.0 to determine the optimum pH), 5 mM MgCl2, 125 nM EDTA, 5 mM ATP, 100 nM purified Nps1, 0.1 μM [32P]pyrophosphate (50 Ci/mmol), and 1 mM l-serine (Nps1) or l-α-aminoadipic acid (Nps3). The reaction proceeded for 30 min before it was stopped and further processed as described previously (15). Pyrophosphate exchange was quantified using a PerkinElmer TriCarb 2910TR scintillation counter. The substrate specificities of both enzymes were determined in a two-step process: to gain a first insight into the structural requirements of A domain substrates, pools of substrates with similar properties were tested in the ATP-[32P]pyrophosphate exchange assay. These pools contain all proteinogenic l-amino acids and glycine, dicarboxylic acids (oxalic acid, malonic acid, succinic acid, fumaric acid, maleic acid, malic acid, tartaric acid, citric acid), α-keto acids (pyruvic acid, oxaloacetic acid, α-ketoglutaric acid, phenylpyruvic acid, 4-hydroxyphenylpyruvic acid, indolyl-3-pyruvic acid), aryl acids (benzoic acid, salicylic acid, 4-hydroxybenzoic acid, 2,3-dihydroxybenzoic acid), adipic acid, l-α-aminoadipic acid, l-α-amino-4-thia-adipic acid, as well as d-alanine and d-serine, at a concentration of 1 mM each. In the second round, single-substrate tests of positive pools were performed to determine the precise substrate preference.
In vitro product formation. (i) Product formation by Nps1, LnaA, and LnbA.
Conversion of apoenzymes into holoenzymes by phosphopantetheinylation of the respective T domains was accomplished in vitro with the phosphopantetheinyl transferase Svp (14). A total of 100 nM purified enzyme (Nps1, LnaA, or LnbA) and 50 nM Svp were incubated for 30 min at 30°C in 100 mM phosphate buffer at pH 7.0 and 250 μM coenzyme A as the donor substrate. Aldehyde formation by holoenzymes was shown using a photometric assay and Brady's reagent (2,4-dinitrophenylhydrazine [2,4-DNPH]), which selectively reacts with aldehydes to yield 2,4-dinitrophenylhydrazone (16). However, 2,4-DNPH does not react with carboxy groups (i.e., the amino acid substrates in the assay). Reactions of 5 ml were set up, containing 100 mM phosphate buffer (Nps1) or 100 mM Tris-HCl buffer (LnaA/LnbA), 5 mM MgCl2, 125 nM EDTA, 5 mM ATP, 1 mM NADPH, 250 μM coenzyme A, the respective holoenzyme (Nps1, LnaA, or LnbA), and 1 mM substrate (l-serine for Nps1, l-tyrosine for LnaA and LnbA) at 25°C. For reactions with Nps1, the above-described buffer was amended with 1 mM flavin adenine dinucleotide (FAD). To record the time course, samples were taken after 5, 10, 15, 30, 60, 90, and 120 min. To stop the reactions, 500 μl ice-cold detection solution, which consisted of 0.1% (wt/vol) 2,4-DNPH (solved in 1% [vol/vol] sulfuric acid in MeOH), was dispensed into a cuvette, and an equal volume of the enzymatic reaction was added. Product formation was detected photometrically by measuring the absorption at λ = 430 nm in a ScanDrop photometer (Analytik Jena). Control reactions without amino acid substrates, without ATP, without CoA, without FAD (in the case of Nps1), or with heat-inactivated enzyme were run in parallel.
(ii) Product formation by Nps3.
Enzyme priming was carried out with 150 nM Nps3, 150 nM Svp, and 250 μM coenzyme A. After 30 min of incubation at 30°C to produce holoenzyme, all other reagents/substrates were added. The assay was 10 mM α-aminoadipic acid, 10 mM ATP, 10 mM MgCl2·6 H2O, 1 mM reduced glutathione, and 1 mM NADPH in 100 mM Tris buffer, pH 7.5, in a total volume of 500 μl, for 120 min at 25°C. The reaction was stopped by successively adding 1 ml ethanol, 1 ml trichloroacetic acid (10%, vol/vol), and 1 ml O-aminobenzaldehyde (1% in ethanol, vol/vol), followed by incubation for 30 min. Formation of l-α-aminoadipate-6-semialdehyde in vitro was detected by a previously described spectrophotometric assay (17), also running the controls mentioned above. The samples were then lyophilized and solved in methanol, and mass spectrometric analysis was performed on a Thermo Accela instrument coupled to an Exactive Orbitrap spectrometer in negative mode and using electrospray ionization. The liquid chromatography (LC) solvents were acetonitrile (A) and 0.1% formic acid in water. After an initial hold at 5% A for 1 min, a linear gradient to 98% A within 15 min was run, at a flow rate of 1 ml/min. Detection was at λ = 450 nm.
Bioinformatic analysis.
Primary protein sequences of A domains (for sequence accession numbers, see Table S1 in the supplemental material) were aligned separately using the MAFFT algorithm (18) implemented as a plugin for the Geneious software package version 7.0 (Biomatters). Sequences of both characterized (2, 8–11) and uncharacterized reductases were chosen and retrieved from the NCBI protein database or by searching publically available genome sequences using BLAST (19). A rooted phylogenetic tree of A domains was inferred by Bayesian Inference using MrBayes 3.2 (20) using the following parameters: the mixed model of protein evolution was chosen as a prior (aamodelpr = mixed). PheA of Brevibacillus brevis was defined as the outgroup. Two heated chains were run in parallel for 1 million generations until convergence was reached (average standard deviation of splits frequency fell well below 0.01). The first 25% of generations was discarded during data analysis (burninfrac = 0.25). The model of evolution determined was the WAG model (posterior probability = 1). The consensus tree was visualized and formatted in FigTree 1.3.1 (developed by A. Rambaut, University of Edinburgh; http://tree.bio.ed.ac.uk).
RESULTS AND DISCUSSION
Phylogeny of adenylate-forming reductases.
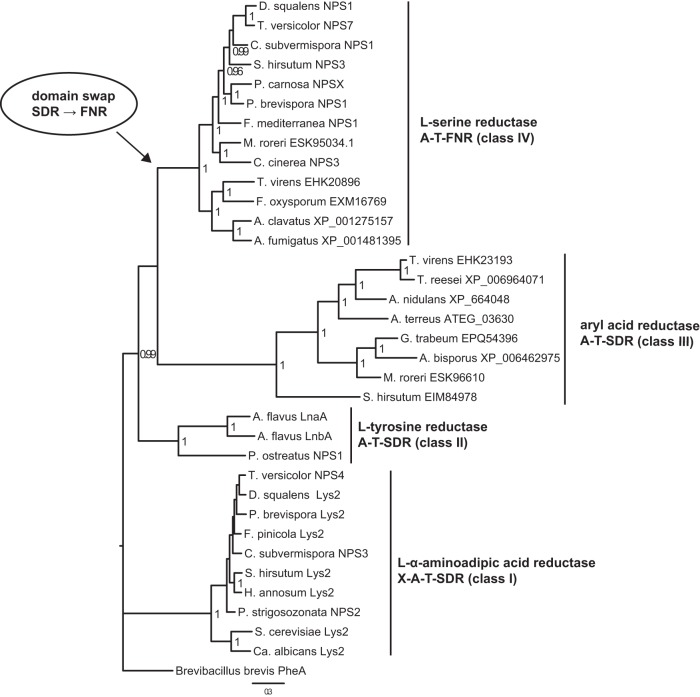
A phylogenetic tree was constructed from A domains of 34 putative adenylate-forming reductases of basidiomycete and ascomycete origin (Fig. 2), with PheA of Brevibacillus brevis as the outgroup. An earlier study on a limited set of PCR-amplified lys2 orthologs (lacking other types of adenylate-forming reductases) resolved the overall evolutionary groups of the ascomycetes (21) and concluded that a monophyletic origin of Lys2 enzymes was likely (22). We confirmed these results and demonstrate a distinct phylogenetic clustering of A domains of Lys2-type (class I), LnaA/LnbA-type (class II), and aryl acid reductase (class III) enzymes. Intriguingly, we found another phylogenetic group (class IV) that includes Nps1 of C. subvermispora. Classes II to IV appear to share an ancestor that branched off from class I prior to the evolutionary divergence of ascomycetes and basidiomycetes.
FIG 2.
Phylogenetic tree of adenylation domains taken from fungal adenylate-forming reductases. Biochemical activities, domain structures, and the proposed class are indicated on the right. Four classes (I to IV) are represented by highly supported clades. Reductase domains of class I to III enzymes belong to the short-chain dehydrogenase/reductase (SDR) superfamily, and reductase domains of class IV enzymes belong to the ferredoxin-NADP+ reductase (FNR) superfamily. This suggests a domain swap event (arrow) where an ancestral SDR domain was replaced by an FNR domain. The tree was computed using the Bayesian inference implemented in MrBayes 3.2.2. PheA of Brevibacillus brevis was used as an outgroup to root the tree. Posterior probability values of >0.95 are shown next to the nodes. The scale bar indicates the expected changes per site.
The term “nonribosomal code” (23) (Table 1) describes a specificity signature of A domains that relates to their substrate preference. To a degree, it allows for the prediction of structural features of the NRPS product. Extraction of this code of the Nps3 A domain yielded DPRHFVM(I/V)PK for l-α-aminoadipate (Table 1), which is virtually identical across basidiomycete class I enzymes (Lys2-type l-α-aminoadipate reductases) and highly similar to those of Saccharomyces cerevisiae and Ca. albicans [DPRHFVM(I/V)KK]. In addition to the standard motifs of A domains, Guo and Bhattacharjee (24) described a conserved l-α-aminoadipate binding motif [460HDP(I/V)QRD466 in Ca. albicans], designated “core C1,” in A domains of class I reductases, which does not occur outside this group of enzymes. Sequence analysis confirmed that the l-α-aminoadipate binding signature is highly conserved among class I enzymes from both ascomycetes and basidiomycetes. Notably, while the Nps1 primary sequence includes the set of typical A domain motifs, A1 to A10 (3), the core C1 motif is not present in class II to IV enzymes, which also exhibit deviating nonribosomal code signatures (Table 1).
TABLE 1.
Nonribosomal A domain specificity codes, substrate specificities, and categories of selected fungal adenylate-forming reductases
| Reductase | Nonribosomal codec | Substrate | Reductase domain type | Reductase family |
|---|---|---|---|---|
| Candida albicans Lys2 | DPRHFVMIKK | l-α-Aminoadipic acida | SDR | α-Aminoadipic acid reductase (class I) |
| Ceriporiopsis subvermispora Nps3 | DPRHFVMIPK | l-α-Aminoadipic acida | SDR | α-Aminoadipic acid reductase (class I) |
| Dichomitus squalens Lys2 | DPRHFVMIPK | l-α-Aminoadipic acidb | SDR | α-Aminoadipic acid reductase (class I) |
| Fomitopsis pinicola Lys2 | DPRHFVMLPK | l-α-Aminoadipic acidb | SDR | α-Aminoadipic acid reductase (class I) |
| Heterobasidion annosum Lys2 | DPRHFVMIPK | l-α-Aminoadipic acidb | SDR | α-Aminoadipic acid reductase (class I) |
| Phlebia brevispora Lys2 | DPRHFVMIPK | l-α-Aminoadipic acidb | SDR | α-Aminoadipic acid reductase (class I) |
| Punctularia strigosozonata Nps2 | DPRHFVMIPK | l-α-Aminoadipic acidb | SDR | α-Aminoadipic acid reductase (class I) |
| Saccharomyces cerevisiae Lys2 | DPRHFVMVKK | l-α-Aminoadipic acida | SDR | α-Aminoadipic acid reductase (class I) |
| Stereum hirsutum Lys2 | DPRHFVMIPK | l-α-Aminoadipic acidb | SDR | α-Aminoadipic acid reductase (class I) |
| Trametes versicolor Nps4 | DPRHFVMIPK | l-α-Aminoadipic acidb | SDR | α-Aminoadipic acid reductase (class I) |
| Aspergillus flavus LnaA | DVFAFGAIFK | l-Tyrosinea | SDR | Tyrosine reductase (class II) |
| Aspergillus flavus LnbA | DVFAFGAIFK | l-Tyrosinea | SDR | Tyrosine reductase (class II) |
| Pleurotus ostreatus Nps1 | DILFVGAVLK | Unknown | SDR | Tyrosine reductase (class II) |
| Trichoderma virens EHK23193 | GFLMGGL--S | Unknown | SDR | Aryl acid reductase (class III) |
| Trichoderma reesei XP_006964071 | GFLLGGL--S | Unknown | SDR | Aryl acid reductase (class III) |
| Aspergillus nidulans XP_664048 | GFVMTGH--S | Unknown | SDR | Aryl acid reductase (class III) |
| Aspergillus terreus ATEG_03630 | GFVMLGH--S | 5-Methyl orsellinic acid | SDR | Aryl acid reductase (class III) |
| Gloeophyllum trabeum EPQ54396 | GFLLAGH--S | Unknown | SDR | Aryl acid reductase (class III) |
| Agaricus bisporus XP_006462975 | GFMLVGH--S | Unknown | SDR | Aryl acid reductase (class III) |
| Moniliophthora roreri ESK96610 | GFLLAGH--S | Unknown | SDR | Aryl acid reductase (class III) |
| Stereum hirsutum EIM84978 | AMCFTAF--T | Unknown | SDR | Aryl acid reductase (class III) |
| Aspergillus clavatus XP_001275157 | DMWIAACIIK | Unknown | FNR | Serine reductase (class IV) |
| Aspergillus fumigatus XP_001481395 | DMWIAACIIK | Unknown | FNR | Serine reductase (class IV) |
| Coprinopsis cinerea Nps3 | DFWFVAAIAK | Unknown | FNR | Serine reductase (class IV) |
| Ceriporiopsis subvermispora Nps1 | DMWIAASIVK | l-Serinea | FNR | Serine reductase (class IV) |
| Dichomitus squalens Nps1 | DMWIAASIVK | Serineb | FNR | Serine reductase (class IV) |
| Fomitiporia mediterranea Nps1 | DMWIAASIVK | Serineb | FNR | Serine reductase (class IV) |
| Fusarium oxysporum EXM16769 | DMWIAACIVK | Unknown | FNR | Serine reductase (class IV) |
| Moniliophthora roreri ESK95034.1 | DMWIAACIVK | Unknown | FNR | Serine reductase (class IV) |
| Phlebia brevispora Nps1 | DMWIAASIVK | Serineb | FNR | Serine reductase (class IV) |
| Phanerochaete carnosa NpsX | DMWIAASIVK | Serineb | FNR | Serine reductase (class IV) |
| Stereum hirsutum Nps3 | DMWIAASIVK | Serineb | FNR | Serine reductase (class IV) |
| Trametes versicolor Nps7 | DMWIAACIVK | Unknown | FNR | Serine reductase (class IV) |
| Trichoderma virens EHK20896 | DMWIAACIVK | Unknown | FNR | Serine reductase (class IV) |
Experimentally confirmed.
Deduced by identity of nonribosomal codes.
Codes were determined manually by comparison of MAFFT alignments with the PheA protein of Brevibacillus brevis (23).
The R domains found typically with peptide synthetases belong to the short-chain dehydrogenase/reductase (SDR) family and contain a Rossmann fold with a GXXGXXG motif for NAD(P)H binding and a YXXX(K/R) active site motif (25). While the R domains of class I to III reductases clearly belong to the SDR family, the putative R domain of class IV reductases does not. Rather, it resembles the ferredoxin-NADP+ reductase (FNR) superfamily, with NADP+ and FAD binding sites (26). Considering that only class IV enzymes feature an FNR domain, it appears very likely that SDR represents the ancestral R domain. We therefore conclude that class IV enzymes reflect a derived domain architecture in which a terminal FNR domain replaced the SDR domain. Intriguingly, all major classes of adenylate-forming reductases include proteins of both basidiomycetes and ascomycetes, suggesting that these reductases evolved in a common ancestor of these major fungal taxa.
Characterization of Ceriporiopsis subvermispora Nps1.
The nps1 gene is disrupted by 16 introns and encodes a 1,056-aa-residue tridomain protein (GenBank identifier EMD40260.1). Contrasting the cluster paradigm of fungal natural product genes for individual or even intertwined pathways (27, 28), the vicinity up- and downstream of nps1 does not encode genes with any obvious role for secondary or amino acid metabolism.
The Nps1 apoprotein was heterologously produced in E. coli KRX, transformed with expression plasmid pDK6, which encodes the N-terminal Nps1-hexahistidine fusion protein. Purification via metal affinity chromatography on an Ni-NTA column resulted in the expected 118-kDa protein (see Fig. S1 in the supplemental material).
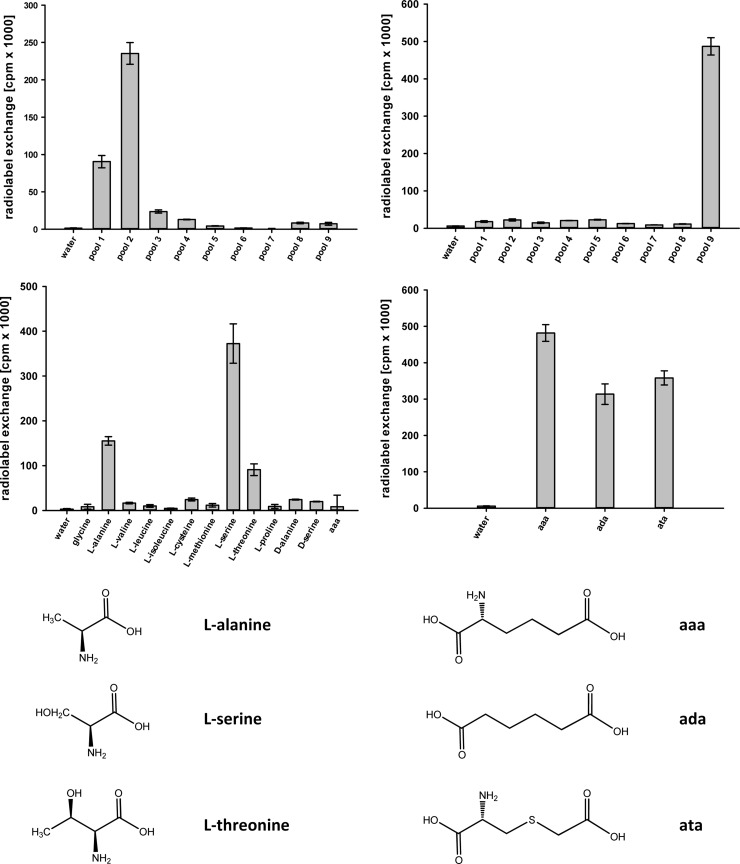
Nps1 is distinct by (i) the absent core C1 fingerprint motif [HDP(I/V)QRD] that is typical for all A domains of class I enzymes, (ii) the NRPS code (DMWIAASIVK) that deviates from the l-α-aminoadipic acid motif but is invariable among numerous other putative basidiomycete reductases and very similar to Aspergillus reductases (Table 1), and (iii) the separate position in the phylogenetic tree (Fig. 2). Therefore, we hypothesized that Nps1 may not reduce l-α-aminoadipic acid. However, as the first position of the NRPS code is an aspartic acid residue, which is indicative for α-amino acid substrates (23, 29), they still represented the most plausible substrate group. The substrate-dependent ATP-[32P]pyrophosphate exchange assay is used to detect aminoacyl adenylate formation, i.e., the reaction catalyzed by the A domain. The assay makes use of the domain's reverse reaction when aminoacyl adenylate is converted back to the free amino acid while radiolabeled ATP is formed using [32P]pyrophosphate. The specificity of the Nps1 A domain was investigated in two steps. First, pools of substrates were tested, followed by the individual substrates included in those pools that showed a positive reaction. Additionally, α-keto and aryl acids were also tested, covering a total of 41 potential substrates. The Nps1 A domain displayed a marked preference for l-serine (413,500 cpm) (Fig. 3) and, at a lower level, for l-alanine and l-threonine (193,600 and 122,400 cpm, respectively). All other l-amino acids, d-configured amino acids, and tested substrates did not result in a significant radiolabel exchange. Particularly poor adenylation was observed for l-α-aminoadipic acid (4,900 cpm), essentially at a level that represents the background of the negative control with water instead of the substrate. These results, and the unambiguous rejection of l-α-aminoadipic acid, disprove a function in catalyzing the reductive step during l-lysine biosynthesis and point to a distinct metabolic role involving l-serine. Optimal pyrophosphate exchange, i.e., maximum substrate activation, was found at 25°C and at neutral pH. The temperature optimum is consistent with the normal growth temperature of C. subvermispora.
FIG 3.
Substrate specificity of C. subvermispora reductases Nps1 (left) and Nps3 (right) based on the substrate-dependent ATP-[32P]pyrophosphate exchange assay. Diagrams represent assays with pools of substrates (top) and individual compounds (bottom). Error bars indicate the standard deviations. Composition of pools was as follows: pool 1, Gly, l-Ala, l-Val, l-Leu, l-Ile; pool 2, l-Cys, l-Met, l-Ser, l-Thr, l-Pro; pool 3, l-His, l-Phe, l-Tyr, l-Trp; pool 4, l-Asp, l-Asn, l-Glu, l-Gln; pool 5, l-Lys, l-Arg, l-Orn, oxalic acid; pool 6, pyruvic acid, α-ketoglutaric acid, phenylpyruvic acid, 4-hydroxyphenylpyruvic acid, indole-3-pyruvic acid; pool 7, malonic acid, succinic acid, fumaric acid, maleic acid, malic acid, tartaric acid, citric acid; pool 8, benzoic acid, salicylic acid, 4-hydroxybenzoic acid, 2,3-dihydroxybenzoic acid. aaa, l-α-aminoadipic acid; ada, adipic acid; ata, l-α-amino-4-thia-adipic acid.
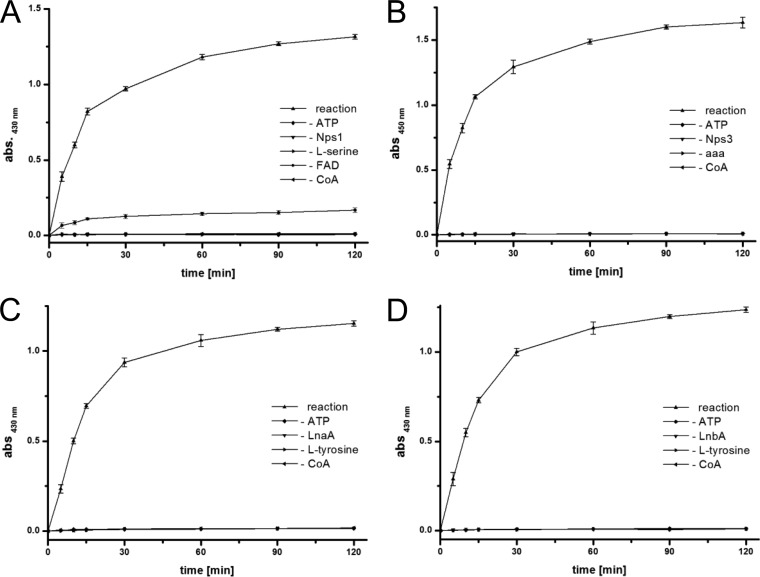
To verify its function, product formation by holo-Nps1 was shown with an established photometric assay. It is based on the selective reaction of Brady's reagent (2,4-dinitrophenylhydrazine [2,4-DNPH]) with carbonyl groups, but not with carboxylic acids, into the corresponding hydrazone, which can be detected by measuring the absorption at λ = 430 nm. A time course over 120 min showed a strong increase of the OD430, i.e., hydrazone formation, as a consequence of the present serine aldehyde, within minutes after the reaction was initiated (Fig. 4A). Controls without amino acid substrate, without ATP, without CoA, or without enzyme did not show a photometrically measurable turnover. Low but detectable turnover was found in the absence of FAD, probably due to a protein-bound cofactor carried over from the protein production host E. coli. Taken together, the above-given results demonstrate that C. subvermispora Nps1 does not possess α-aminoadipic acid reductase activity but functions as l-serine reductase.
FIG 4.
Aldehyde formation by reductases Nps1 (class IV), Nps3 (class I), LnaA and LnbA (class II). (A) Photometric detection of hydrazone formation from Nps1-produced l-serine aldehyde and 2,4-dinitrophenylhydrazine (2,4-DNPH). (B) Photometric detection of the dihydroquinazolinium cation, which is the reaction product of O-aminobenzaldehyde with Δ1-piperidine-6-carboxylic acid (the cyclic anhydride of the Nps3 product l-α-aminoadipate-6-semialdehyde). (C) Photometric detection of hydrazone formation from LnaA-produced l-tyrosine aldehyde and 2,4-DNPH. (D) Hydrazone formation from LnbA-produced l-tyrosine aldehyde and 2,4-DNPH. For clarity, error bars are not shown for negative controls. Abbreviation: aaa, l-α-aminoadipic acid.
Ceriporiopsis subvermispora Nps3 is an l-α-aminoadipic acid reductase.
The complete C. subvermispora nps3 gene includes five introns. The cDNA encodes a 1,417-aa protein (GenBank identifier EMD34789.1) with a theoretical mass of 155.1 kDa. The Nps3 A domain included the standard motifs (A1 to A10, according to Schwarzer et al. [3]), but also the class I (Lys2)-specific so-called core motif C1 (506HDPVQRD512) (24). The carrier protein and reductase domain showed typical sequences for the phosphopantetheinyl-accepting site (908LGGHSI913) and the NADP binding region (1022TGATGFLGA1030 and 1303GYGVRSA1309), respectively, which follow the consensus shown for Lys2 and other reductases (30).
Heterologous expression of the nps3 gene was accomplished in E. coli KRX, transformed with plasmid pDK21, which encodes Nps3 with a C-terminally added hexahistidine tag. Nps3 apoprotein was used to determine the substrate using the ATP-[32P]pyrophosphate exchange assay. A total of 41 substrates (see Materials and Methods), including adipic acid, l-α-amino-4-thia-adipic acid, and l-α-aminoadipic acid, were tested. Maximum radiolabel exchange occurred in the presence of l-α-aminoadipic acid (481,500 cpm) (Fig. 3). Adipic acid and l-α-amino-4-thia-adipic acid led to lower turnover (313,500 and 358,000 cpm, respectively), while all other tested compounds failed as substrates (<5% exchange, compared to l-α-aminoadipic acid). Maximum pyrophosphate exchange occurred at 25°C and at pH 7.5, i.e., under very similar conditions as those found for C. subvermispora Nps1.
Holo-Nps3 was then tested in vitro for enzymatic reduction, i.e., l-α-aminoadipate-6-semialdehyde formation. We used O-aminobenzaldehyde to detect the expected product, essentially following a described procedure (17, 31): l-α-aminoadipate-6-semialdehyde stands in chemical equilibrium with its cyclic anhydride, Δ1-piperidine-6-carboxylic acid. O-aminobenzaldehyde reacts with the latter to give a dihydroquinazolinium cation that can be measured photometrically at λ = 450 nm (Fig. 4B). High-resolution mass spectrometry supports this finding, as the expected mass matched the mass of the chromatographically detected product peak (m/z 231.1128 [M]+, expected for C13H15O2N2 231.1123) at a retention time (tR) of 11.8 min (see Fig. S2 in the supplemental material). Product formation was not observed in the control reactions without substrates or enzyme.
Prototypical l-α-aminoadipate reductases are represented by S. cerevisiae Lys2 (31, 32) and by P. chrysogenum Lys2 (2). In the latter case, a metabolic dichotomy precedes the Lys2-catalyzed step, as l-α-aminoadipic acid also serves as the substrate for δ-(l-α-aminoadipoyl)-l-cysteinyl-d-valine (ACV) synthetase, the gateway enzyme for penicillin biosynthesis. Our in vitro biochemical and chromatographic data demonstrate that C. subvermispora Nps3 is an α-aminoadipic acid reductase. It is the first characterized basidiomycete representative and, along with, e.g., Lys2 of S. cerevisiae and Ca. albicans (8, 31, 33), is one out of only very few examples that have been characterized biochemically.
l-Tyrosine aldehyde formation by Aspergillus flavus reductases LnaA and LnbA.
LnaA and LnbA belong to class II reductases and are two functionally redundant and highly similar enzymes (A-T-R layout, 58% identical amino acids) which are implicated in assembly of piperazine natural products from l-tyrosine in A. flavus (10). Their length is similar to that of Nps1 (1,042 and 1,029 aa, respectively), and they lack the extra N-terminal extension, whose presence is typical for class I enzymes. Both Nps1 and LnaA/LnbA use proteinogenic amino acids as substrates. Despite these similarities to Nps1, LnaA and LnbA are intriguing, as their A domains are phylogenetically separated from the other clades (Fig. 2). R domains of LnaA/LnbA belong to the SDR family. In a previous study on LnaA and LnbA (10), l-tyrosine had been identified as the preferred substrate of their A domains. The exact function of this pair of enzymes remained unclear, as two biosynthetic routes toward the piperazine products appear possible, i.e., dimerization of reductively released free amino aldehydes and, alternatively, the formation of a dimeric imine via an enzyme-tethered intermediate. To test if l-tyrosine aldehyde was liberated from the enzymes, we investigated LnaA and LnbA separately in their holo forms, again using the 2,4-DNPH-based photometric assay and 1 mM l-tyrosine as the amino acid substrate. The reactions were monitored in 1-min intervals, over six min. A strong increase of the OD430 was detectable (Fig. 4C and D), which proves hydrazone formation due to free aldehyde. Controls without substrates or enzyme did not result in a photometrically detectable change. We therefore show that both LnaA and LnbA represent l-tyrosine reductases, that l-tyrosine aldehyde is released, and that this occurs with individual enzymes and without LnaA/LnbA heterodimer formation.
Conclusion.
Our results show that fungal peptide synthetase-like reductases are phylogenetically more diverse than previously thought. Several classes of fungal adenylate-forming reductases emerged. Class I represents the well-studied l-α-aminoadipate reductases (Lys2 type) involved in l-lysine biosynthesis. Here, we show the biochemical activity of a basidiomycete class I enzyme in vitro and confirm previous hypotheses that class I has a monophyletic origin in a common ancestor of basidiomycetes and ascomycetes. Classes II to IV share an ancient common ancestor as well and have evolved distinct biochemical activities. We demonstrate that the class II enzymes LnaA and LnbA, which are involved in piperazine biosynthesis (10), act as l-tyrosine reductases and release free aldehydes in vitro. The l-serine reductase Nps1 of C. subvermispora is the first member of class IV reductases to be characterized. In class IV enzymes, a C-terminal FNR domain replaced the original SDR domain for substrate reduction. The selective advantage resulting from this domain swap remains obscure. Still, we show that l-serine is a substrate of Nps1 and that free aldehyde is formed in vitro. Considering that class II and class IV enzymes are involved in secondary metabolism, we assume that the product of the orphan biochemical pathway involving class IV adenylate-forming reductases is a promising target for future metabolomics studies. To a degree, the functional divergence among adenylate-forming reductases is reminiscent of the synthetases AtrA and MicA (34, 35), i.e., members of another class of NRPS-type fungal biosynthesis enzymes that lack a C domain. Even though AtrA and MicA share an identical domain setup (adenylation, thiolation, terminal thioesterase), these synthetases catalyze quinone and furanone formation, respectively. Stimulated by the wealth of available genomic data, more biochemical work is warranted to further explore the realm of fungal NRPS-related biosynthetic enzymes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (DFG grant HO2515/6-1 to D.H.).
We thank Andrea Perner (Leibniz Institute for Natural Product Research and Infection Biology—Hans-Knöll-Institute, Jena, Germany) for recording mass spectra.
Footnotes
Published ahead of print 1 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01767-14.
REFERENCES
- 1.Bhattacharjee JK. 1985. α-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes. Crit. Rev. Microbiol. 12:131–151. 10.3109/10408418509104427 [DOI] [PubMed] [Google Scholar]
- 2.Casqueiro J, Gutierrez S, Banuelos O, Fierro F, Velasco J, Martin JF. 1998. Characterization of the lys2 gene of Penicillium chrysogenum encoding alpha-aminoadipic acid reductase. Mol. Gen. Genet. 259:549–556. 10.1007/s004380050847 [DOI] [PubMed] [Google Scholar]
- 3.Schwarzer D, Finking R, Marahiel MA. 2003. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20:275–287. 10.1039/b111145k [DOI] [PubMed] [Google Scholar]
- 4.Cleveland TE, Yu J, Fedorova N, Bhatnagar D, Payne GA, Nierman WC, Bennett JW. 2009. Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 27:151–157. 10.1016/j.tibtech.2008.11.008 [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Fueyo E, Ruiz-Duenas FJ, Ferreira P, Floudas D, Hibbett DS, Canessa P, Larrondo LF, James TY, Seelenfreund D, Lobos S, Polanco R, Tello M, Honda Y, Watanabe T, San RJ, Kubicek CP, Schmoll M, Gaskell J, Hammel KE, St. John FJ, Vanden Wymelenberg A, Sabat G, Splinter Bondurant S, Syed K, Yadav JS, Doddapaneni H, Subramanian V, Lavin JL, Oguiza JA, Perez G, Pisabarro AG, Ramirez L, Santoyo F, Master E, Coutinho PM, Henrissat B, Lombard V, Magnuson JK, Kües U, Hori C, Igarashi K, Samejima M, Held BW, Barry KW, Labutti KM, Lapidus A, Lindquist EA, Lucas SM, Riley R, Salamov AA, Hoffmeister D, Schwenk D, Hadar Y, Yarden O, de Vries RP, Wiebenga A, Stenlid J, Eastwood D, Grigoriev IV, Berka RM, Blanchette RA, Kersten P, Martinez AT, Vicuna R, Cullen D. 2012. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc. Natl. Acad. Sci. U. S. A. 109:5458–5463. 10.1073/pnas.1119912109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H, Kohler A, LaButti K, Lapidus A, Lavin JL, Lee YH, Lindquist E, Lilly W, Lucas S, Morin E, Murat C, Oguiza JA, Park J, Pisabarro AG, Riley R, Rosling A, Salamov A, Schmidt O, Schmutz J, Skrede I, Stenlid J, Wiebenga A, Xie X, Kües U, Hibbett DS, Hoffmeister D, Hogberg N, Martin F, Grigoriev IV, Watkinson SC. 2011. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333:762–765. 10.1126/science.1205411 [DOI] [PubMed] [Google Scholar]
- 7.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Lombard ALV, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Duenas FJ, Sabat G, Salamov A, Samejima M, Schmutz J, Slot JC, John FS, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbett DS. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. 10.1126/science.1221748 [DOI] [PubMed] [Google Scholar]
- 8.Guo S, Evans SA, Wilkes MB, Bhattacharjee JK. 2001. Novel posttranslational activation of the LYS2-encoded alpha-aminoadipate reductase for biosynthesis of lysine and site-directed mutational analysis of conserved amino acid residues in the activation domain of Candida albicans. J. Bacteriol. 183:7120–7125. 10.1128/JB.183.24.7120-7125.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hijarrubia MJ, Aparicio JF, Martin JF. 2003. Domain structure characterization of the multifunctional alpha-aminoadipate reductase from Penicillium chrysogenum by limited proteolysis. Activation of alpha-aminoadipate does not require the peptidyl carrier protein box or the reduction domain. J. Biol. Chem. 278:8250–8256. 10.1074/jbc.M211235200 [DOI] [PubMed] [Google Scholar]
- 10.Forseth RR, Amaike S, Schwenk D, Affeldt KJ, Hoffmeister D, Schroeder FC, Keller NP. 2013. Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew. Chem. Int. Ed. Engl. 52:1590–1594. 10.1002/anie.201207456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, Beissner M, Zhao H. 2014. Aryl-aldehyde formation in fungal polyketides: discovery and characterization of a distinct biosynthetic mechanism. Chem. Biol. 21:257–263. 10.1016/j.chembiol.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Russell DW. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13.Bradford M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 14.Sanchez C, Du L, Edwards DJ, Toney MD, Shen B. 2001. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC 15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem. Biol. 8:725–738. 10.1016/S1074-5521(01)00047-3 [DOI] [PubMed] [Google Scholar]
- 15.Schneider P, Weber M, Rosenberger K, Hoffmeister D. 2007. A one-pot chemoenzymatic synthesis for the universal precursor of antidiabetes and antiviral bis-indolylquinones. Chem. Biol. 14:635–644. 10.1016/j.chembiol.2007.05.005 [DOI] [PubMed] [Google Scholar]
- 16.Brady OL, Elsmie GV. 1926. The use of 2:4-dinitrophenylhydrazine as a reagent for aldehydes and ketones. Analyst 51:77–78. 10.1039/an9265100077 [DOI] [Google Scholar]
- 17.Larson RL, Sandine WD, Broquist HP. 1963. Enzymic reduction of α-aminoadipic acid: relation to lysine biosynthesis. J. Biol. Chem. 238:275–282 [Google Scholar]
- 18.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 20.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An KD, Nishida H, Miura Y, Yokota A. 2002. Aminoadipate reductase gene: a new fungal-specific gene for comparative evolutionary analyses. BMC Evol. Biol. 2:6. 10.1186/1471-2148-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An KD, Nishida H, Miura Y, Yokota A. 2003. Molecular evolution of adenylating domain of aminoadipate reductase. BMC Evol. Biol. 3:9. 10.1186/1471-2148-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stachelhaus T, Mootz HD, Marahiel MA. 1999. The specificity conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493–505. 10.1016/S1074-5521(99)80082-9 [DOI] [PubMed] [Google Scholar]
- 24.Guo S, Bhattacharjee JK. 2003. Site-directed mutational analysis of the novel catalytic domains of alpha-aminoadipate reductase (Lys2p) from Candida albicans. Mol. Genet. Genomics 269:271–279. 10.1007/s00438-003-0833-3 [DOI] [PubMed] [Google Scholar]
- 25.Kavanagh KL, Jornvall H, Persson B, Oppermann U. 2008. Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65:3895–3906. 10.1007/s00018-008-8588-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrillo N, Ceccarelli EA. 2003. Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur. J. Biochem. 270:1900–1915. 10.1046/j.1432-1033.2003.03566.x [DOI] [PubMed] [Google Scholar]
- 27.Hoffmeister D, Keller NP. 2007. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 24:393–416. 10.1039/b603084j [DOI] [PubMed] [Google Scholar]
- 28.Wiemann P, Guo CJ, Palmer JM, Sekonyela R, Wang CC, Keller NP. 2013. Prototype of an intertwined secondary-metabolite supercluster. Proc. Natl. Acad. Sci. U. S. A. 110:17065–17070. 10.1073/pnas.1313258110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalb D, Lackner G, Hoffmeister D. 2013. Fungal peptide synthetases: an update on functions and specificity signatures. Fungal Biol. Rev. 27:43–50. 10.1016/j.fbr.2013.05.002 [DOI] [Google Scholar]
- 30.Hijarrubia MJ, Aparicio JF, Casqueiro J, Martin JF. 2001. Characterization of the lys2 gene of Acremonium chrysogenum encoding a functional alpha-aminoadipate activating and reducing enzyme. Mol. Gen. Genet. 264:755–762. 10.1007/s004380000364 [DOI] [PubMed] [Google Scholar]
- 31.Ehmann DE, Gehring AM, Walsh CT. 1999. Lysine biosynthesis in Saccharomyces cerevisiae: mechanism of alpha-aminoadipate reductase (Lys2) involves posttranslational phosphopantetheinylation by Lys5. Biochemistry 38:6171–6177. 10.1021/bi9829940 [DOI] [PubMed] [Google Scholar]
- 32.Eibel H, Philippsen P. 1983. Identification of the cloned S. cerevisiae LYS2 gene by an integrative transformation approach. Mol. Gen. Genet. 191:66–73. 10.1007/BF00330891 [DOI] [PubMed] [Google Scholar]
- 33.Sagisaka S, Shimura K. 1960. Mechanism of activation and reduction of α-aminoadipic acid by yeast enzyme. Nature 188:1189–1190. 10.1038/1881189a0 [DOI] [Google Scholar]
- 34.Schneider P, Bouhired S, Hoffmeister D. 2008. Characterization of the atromentin biosynthesis genes and enzymes in the homobasidiomycete Tapinella panuoides. Fungal Genet. Biol. 45:1487–1496. 10.1016/j.fgb.2008.08.009 [DOI] [PubMed] [Google Scholar]
- 35.Yeh HH, Chiang YM, Entwistle R, Ahuja M, Lee KH, Bruno KS, Wu TK, Oakley BR, Wang CC. 2012. Molecular genetic analysis reveals that a nonribosomal peptide synthetase-like (NRPS-like) gene in Aspergillus nidulans is responsible for microperfuranone biosynthesis. Appl. Microbiol. Biotechnol. 96:739–748. 10.1007/s00253-012-4098-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.