Abstract
The vertical transmission of symbiotic microorganisms is omnipresent in insects, while the evolutionary process remains totally unclear. The oriental chinch bug, Cavelerius saccharivorus (Heteroptera: Blissidae), is a serious sugarcane pest, in which symbiotic bacteria densely populate the lumen of the numerous tubule-like midgut crypts that the chinch bug develops. Cloning and sequence analyses of the 16S rRNA genes revealed that the crypts were dominated by a specific group of bacteria belonging to the genus Burkholderia of the Betaproteobacteria. The Burkholderia sequences were distributed into three distinct clades: the Burkholderia cepacia complex (BCC), the plant-associated beneficial and environmental (PBE) group, and the stinkbug-associated beneficial and environmental group (SBE). Diagnostic PCR revealed that only one of the three groups of Burkholderia was present in ∼89% of the chinch bug field populations tested, while infections with multiple Burkholderia groups within one insect were observed in only ∼10%. Deep sequencing of the 16S rRNA gene confirmed that the Burkholderia bacteria specifically colonized the crypts and were dominated by one of three Burkholderia groups. The lack of phylogenetic congruence between the symbiont and the host population strongly suggested host-symbiont promiscuity, which is probably caused by environmental acquisition of the symbionts by some hosts. Meanwhile, inspections of eggs and hatchlings by diagnostic PCR and egg surface sterilization demonstrated that almost 30% of the hatchlings vertically acquire symbiotic Burkholderia via symbiont-contaminated egg surfaces. The mixed strategy of symbiont transmission found in the oriental chinch bug might be an intermediate stage in evolution from environmental acquisition to strict vertical transmission in insects.
INTRODUCTION
Insects that feed exclusively on nutritionally limited or persistent food sources, such as plant phloem sap, vertebrate blood, or woody materials, commonly possess symbiotic microorganisms in their guts (1–3). Symbiotic microbes are essential for host survival and reproduction and play pivotal roles in host metabolism, such as providing essential nutrients and digesting food materials (2, 4, 5). To ensure that offspring acquire these microbial partners, insects have evolved diverse mechanisms for vertical transmission of the symbionts, including ovarial transmission in aphids, egg smearing in anobiid beetles, coprophagy in termites, milk gland transmission in tsetse flies, and capsular transmission in plataspid stinkbugs (1, 6–10). In many cases, host insects and symbiotic microbes have phylogenetic congruence (5, 8, 11), strongly suggesting that these symbiotic associations have been maintained by strict vertical transmission. Despite the diversity of sophisticated vertical transmission mechanisms in insects, the evolution of this trait remains unclear.
A number of phytophagous species of the Heteroptera harbor symbiotic bacteria in the lumen of midgut crypts (1, 12, 13). Molecular phylogenetic studies have revealed that stinkbug symbionts are diverse: Gammaproteobacteria in the stinkbug superfamily Pentatomoidea, Actinobacteria in the Pyrrhocoroidea, and isolates of the Betaproteobacteria genus Burkholderia in other insects (3, 14, 15). A recent broad survey of Burkholderia infection in the heteropteran insects has demonstrated that the Burkholderia symbiosis is prevalent among the superfamilies Coreoidea and Lygaeoidea (16). The symbiotic Burkholderia isolates identified from these stinkbugs formed a cluster mixed together with soil-derived strains that is called the stinkbug-associated beneficial and environmental group (SBE) (17). In the SBE clade, the phylogeny of the stinkbug-associated Burkholderia did not reflect host species or populations (16), suggesting that host-symbiont promiscuity was caused by environmental acquisition of the symbionts. The environmental transmission of Burkholderia symbionts has been proven in a coreoid species, Riptortus pedestris (18).
Microbial symbiosis without vertical transmission is omnipresent among marine invertebrates and terrestrial plants, such as the well-known squid-Vibrio and legume-Rhizobium symbioses, respectively (19). However, a similar association is rarely found in terrestrial invertebrates, except in the stinkbug group, whiteflies (20), and thrips (21). Symbiotic associations without vertical transmission are thought to be remnants of an early evolutionary stage (or preliminary stage) of the more commonly found endosymbiosis in insects that requires the strict vertical transmission of symbionts. Hence, understanding Burkholderia symbiosis in the Coreoidea and Lygaeoidea stinkbug lineages will provide insight into the evolution of symbiotic relationships in insects.
The oriental chinch bug, Cavelerius saccharivorus (Fig. 1A) (Lygaeoidea: Blissidae), an economically important sugarcane (Saccharum officinarum) pest, is widely distributed in southeastern Asia (22, 23). In a closely related blissid species, Blissus insularis, the gut symbiotic association has been characterized in detail (24). As shown in other coreoid and lygaeoid stinkbugs (16, 25), there was no phylogenetic congruence between the symbiont and host population, and symbiont strains formed a mixed clade with soil-derived Burkholderia strains, suggesting environmental transmission of the symbionts in B. insularis (24). However, quantitative PCR (qPCR) revealed that there were also dense populations of Burkholderia associated with B. insularis eggs (24), which prompted us to hypothesize that there is vertical transmission of symbionts via eggs in the chinch bug-Burkholderia symbiotic association.
FIG 1.
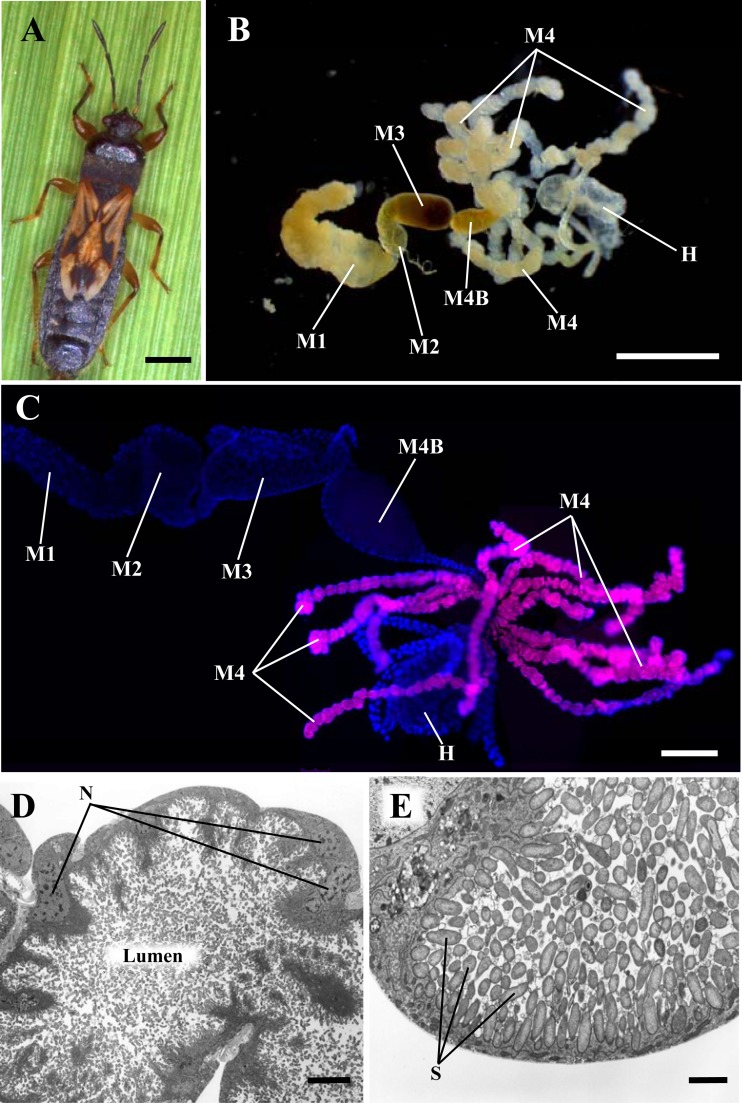
Bacterial endosymbiosis in the oriental chinch bug, C. saccharivorus. (A) A male adult. (B) A dissected alimentary tract. (C) Fluorescent in situ hybridization targeting 16S rRNA of bacteria in a dissected midgut. Red, bacteria hybridizing to an Alexa 555-conjugated EUB338 probe; blue, host insect nuclei stained with DAPI (4′,6-diamidino-2-phenylindole). Abbreviations in panels B and C: M1, first midgut section; M2, second midgut section; M3, third midgut section; M4, fourth midgut section with tubular crypts (symbiotic organ); M4B, M4 bulb; H, hindgut. (D) Transmission electron microscope image of a midgut crypt. (E) An enlarged transmission electron microscope image of a midgut crypt. Abbreviations in panels D and E: N, host nucleus; S, symbiotic bacterium. Bars, 1 mm (A and B), 0.5 mm (C), 10 μm (D), and 2 μm (E).
The objectives of this study were to investigate the symbiont transmission mechanism and microdiversity of symbiotic microbiota in the oriental chinch bug, C. saccharivorus. A histological approach was used to examine the in vivo distribution of microbes in C. saccharivorus. The microdiversity and potential environmental transmission of symbionts were determined using molecular phylogenetic analysis by low-throughput and deep sequencing of bacterial 16S rRNA genes. A Burkholderia group-specific diagnostic PCR comparing hatchlings from sterilized and unsterilized egg surfaces was used to determine vertical transmission.
MATERIALS AND METHODS
Insects.
The sources of the 134 C. saccharivorus chinch bugs that were collected from five different islands in Okinawa Prefecture of Japan and examined in this study are listed in Table 1. The insects were freshly dissected under a dissection microscope in a plastic petri dish filled with phosphate-buffered saline (PBS), and a pair of fine forceps was used to remove the midgut fourth section (Fig. 1B) from either 5th instar nymphs or adult insects. The dissected tissues were subjected to DNA extraction and PCR. Of the 134 insects, 12 insects (6 5th instar nymphs and 6 adults) representing different populations from five islands of Okinawa Prefecture were used for cloning and low-throughput sequence analyses, 10 adults from Minami Daito Island were used for deep sequencing, and the other 112 adult insects were used for diagnostic PCR (Table 1).
TABLE 1.
Number and sources of Cavelerius saccharivorus insects examined in this study
| Sampling islande | Sample identifier | Collection date (yr.mo.day) | Collector | No. of insects tested |
|
|---|---|---|---|---|---|
| Sequencing | Diagnostic PCRd | ||||
| Okinawa | OK | 2010.8.4 | A. Nagayama | 3c | 54 |
| Kita-Daito | KD | 2011.6.15 | A. Nagayama | 2c | 4 |
| Minami-Daito | MDa | 2010.9.15 | A. Nagayama | 3c | 45 |
| MD-Ib | 2013.6.28 | A. Nagayama | 10 | ||
| MD-IIa | 2014.5.19 | M. Aizawa | |||
| Ishigaki | IG | 2010.9.15 | H. Kodama | 2c | 4 |
| Yonaguni | YG | 2010.8.25 | A. Nagayama | 2c | 5 |
Eggs from several pairs were inspected by diagnostic PCR and subjected to the egg surface sterilization test; results are summarized in Table 4. The pairs were excluded from sequencing and diagnostic PCR analyses.
Insects used for 16S rRNA gene Illumina deep sequencing of the 16S rRNA gene; results are summarized in Table S2 in the supplemental material.
Insects used for 16S rRNA gene low-throughput Sanger sequencing; results are summarized in Table S1 in the supplemental material.
Results are summarized in Table 3.
All samples were collected from sugarcane fields.
Laboratory production of insect eggs and hatchlings.
In order to produce eggs and hatchlings, nine pairs of adult insects from Minami-Daito Island (Okinawa Prefecture, Japan) were kept together in the laboratory at 25°C under a long-day regime (16 h light and 8 h dark). A diet of fresh sugarcane shoots was regularly provided. Eggs were collected and transferred to a sterile plastic petri dish. About half the eggs (n = 107) were used for DNA extraction, and the remaining 112 were incubated under conditions similar to those used for the adults until they hatched. The hatchlings were reared under aseptic conditions in sterilized petri dishes and fed distilled water supplemented with 0.05% ascorbic acid for 4 days, after which their DNA was extracted and then subjected to diagnostic PCR for detection of the Burkholderia symbionts. The eggs and hatchlings were reared in the absence of adult insects.
Egg surface sterilization.
Egg surface sterilization was used to test whether the bacteria on egg surfaces were a potential source of gut symbionts in the hatchlings. Ninety-seven eggs were collected from laboratory-reared adult pairs of C. saccharivorus chinch bugs and treated with 70% ethanol for 10 min to sterilize the surfaces. The experimental eggs were kept in a sterile petri dish with a wet cotton ball at 25°C until they hatched. Then, hatchling feeding and subsequent DNA extraction and PCR were carried out as described above for the nonsterilized eggs. The infection status of the parent insects was confirmed by diagnostic PCR detection.
Transmission electron microscopy.
Insects collected from the main island of Okinawa were placed in 0.1 M sodium phosphate buffer (pH 7.4) containing 2.5% glutaraldehyde and dissected with fine forceps. The midgut crypts were isolated, prefixed in the fixative at 4°C overnight, and then postfixed in 2% osmium tetroxide at 4°C for 60 min. After a series of dehydration steps with ethanol, the materials were embedded in Epon 812 resin (TAAB Ltd.). Ultrathin sections were made using an ultramicrotome (EM UC7; Leica), mounted on copper mesh, stained with uranyl acetate and lead citrate, and then observed under a transmission electron microscope (H-7600; Hitachi).
FISH.
Oligonucleotide probes EUB338 and BET940 (26, 27) whose 5′ ends were labeled with Alexa Fluor 555 were used for 16S rRNA-targeted fluorescent in situ hybridization (FISH) (Table 2). Insects from the main island of Okinawa were dissected, thoroughly washed in PBS (137 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4 [pH 7.5]), and fixed in Carnoy's solution (ethanol, chloroform, acetic acid [6:3:1]). After overnight fixation, the tissues were treated with 6% hydrogen peroxide in 80% ethanol for several days to quench the autofluorescence of the tissues (28). The tissues were washed with absolute ethanol and a buffer consisting of PBS containing 0.2% Tween 20 (PBST) and then incubated in a hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% formamide) three times for 5 min each time. Then, the samples were hybridized with hybridization buffer containing the probes (100 nM each) and SYTOX green (0.25 μM) and incubated overnight at room temperature. After thorough washing with PBST, the tissues were mounted with Slowfade antifade solution (Invitrogen) and observed under a fluorescence microscope (DMI 4000 B; Leica). To confirm the specificity of the detection, a no-probe control assay and FISH using the anti-EUB338 probe were performed.
TABLE 2.
Primers and probes used in this study
| Target group | Target gene | Primer/probe name | Nucleotide sequence (5′ → 3′) | Approximate product size (kb) | Annealing temp (°C) | Reference or source |
|---|---|---|---|---|---|---|
| Primers | ||||||
| Eubacteria | 16S rRNA | 16SA1 | AGAGTTTGATCMTGGCTCAG | 1.5 | 55 | 29 |
| 16SB1 | TACGGYTACCTTGTTACGACTT | 29 | ||||
| Eubacteria | 16S rRNA | 515F | GTGCCAGCMGCCGCGGTAA | 0.3 | 54 | 36 |
| 806R | GGACTACHVGGGTWTCTAAT | 36 | ||||
| Burkholderia | ||||||
| SBE clade | 16S rRNA | SBE160F | CGCATACGACCTAAGGGA | 1.3 | 55 | This study |
| SBE1400R | CTTGCGGTTAGGCTACCT | This study | ||||
| BCC clade | 16S rRNA | BCC370F | TTTTGGACAATGGGCGAAAG | 0.8 | 55 | This study |
| Burk16SR | GCTCTTGCGTAGCAACTAAG | 25 | ||||
| PBE clade | 16S rRNA | Burk16SF | TTTTGGACAATGGGGGCAAC | 0.5 | 55 | 25 |
| PBE822R | CTTCGTTACCAAGTCAATGAAGA | This study | ||||
| Invertebrates | COI | LCO1490 | GGTCAACAAATCATAAAGATATTGG | 0.7 | 48 | 35 |
| HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | 35 | ||||
| Probes | ||||||
| Eubacteria | 16S rRNA | EUB338 | GCTGCCTCCCGTAGGAGT | 26 | ||
| Betaproteobacteria | 16S rRNA | BET940 | TTAATCCACATCATCCACCG | 27 |
DNA extraction.
Extraction of DNA from the dissected tissues, eggs, and hatchings was performed using a QIAamp DNA minikit (Qiagen) according to the manufacturer's instructions. The quantity and quality of the extracted DNA were checked using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) and by calculating the ratio of the absorbance at 260 nm/absorbance at 280 nm.
DNA cloning and low-throughput sequencing.
Using DNA extracts from midgut tissues, a 1.5-kb region of the bacterial 16S rRNA gene was amplified by PCR using primers 16SA1 and 16SB1 (29) (Table 2) and AmpliTaq Gold DNA polymerase (Applied Biosystems). Thermal cycling conditions were 95°C for 10 min, followed by 30 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 2 min. Cloning and sequencing of the amplified products were performed as previously described (18).
Molecular phylogenetic analysis of clone library data.
Sequences obtained from clone library analyses were subjected to a BLASTn search against the sequences in the Greengenes database (30) using the BLAST (version 2.2.27+) program (31). Multiple alignments of the nucleotide sequences were generated using the MAFFT program (32). The neighbor-joining (NJ) and maximum likelihood (ML) phylogenies were inferred using MEGA (version 4.0.2) software (33). The ML tree was estimated using the Tamura-Nei substitution model (34). Bootstrap tests were performed with 1,000 replications in the NJ and ML analyses.
Diagnostic PCR.
Burkholderia-specific PCR was used to detect the presence of specific clades in total DNA extracts from the midgut crypts or whole abdomens of adult insects. It was also used to investigate the vertical transmission of the symbionts using total DNA extracted from individual eggs and hatchlings (i.e., 1st instar nymphs) (see Table 4). PCR was performed using Ampdirect Plus (Shimazu) and Burkholderia group-specific primers (Table 2) under a temperature profile of 95°C for 10 min, followed by 30 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min. The specificity of the primer sets was confirmed by a BLAST search of the primer sequences, sequence analysis of the amplified products, and PCR amplicon confirmation of target and nontarget Burkholderia strains: Burkholderia sp. strain RPE64 and Burkholderia sp. strain SFA1 for the SBE clade, Burkholderia gladioli MDT24-1 for the Burkholderia cepacia complex (BCC) clade, and Burkholderia caribensis MWAP64 (DSM13236) and Burkholderia fungorum P763-2 (DSM17061) for the plant-associated beneficial and environmental (PBE) clade. To check the quality of the DNA samples, a 0.65-kb region of the insect mitochondrial cytochrome oxidase I (COI) gene was amplified with primers LCO1490 and HCO2198 (35) (Table 2).
TABLE 4.
Diagnostic PCR of Burkholderia symbionts in eggs and hatchlings of Cavelerius saccharivorus
| Sample source | No. (%) of insects with the following infection patternb: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Single |
Double |
Triple (SBP) | None | Total | |||||
| S | B | P | SB | SP | BP | |||||
| Eggs | 107 | 50 | 3 | 3 | 0 | 0 | 0 | 0 | 64 | 56 (52.3) |
| Hatchlings | ||||||||||
| Untreated | 112 | 28 | 2 | 2 | 0 | 0 | 0 | 0 | 80 | 32 (28.6) |
| Sterilizeda | 97 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 97 | 0 (0) |
Egg surfaces were sterilized by washing in 70% ethanol (see Materials and Methods for more details).
Single, diagnostic PCR detection of only one of the three clades tested, either the SBE (S), BCC (B), or PBE clade (P); Double, PCR detection of any two of the three clades; Triple, PCR detection of all three clades; None, PCR detection of none of the three clades.
Deep sequencing of 16S rRNA gene.
DNA extracted from the midgut crypts of 10 females collected from Minami-Daito Island was individually subjected to PCR amplification of the 16S rRNA gene for Illumina deep sequencing. The V4 variable region of the bacterial 16S rRNA genes was amplified using universal primers 515F and 806R (36) (Table 2). The PCR mixture was comprised of 50 μM deoxynucleoside triphosphates, 0.4 μM primer 515F with Illumina P5 sequences attached, 0.4 μM primer 806R with 6-base indexes and Illumina P7 sequences (Illumina), Q5 high-fidelity DNA polymerase with Q5 reaction buffer (New England BioLabs), and extracted insect DNA as the template. The PCR conditions were as follows: initial denaturation at 98°C for 90 s, followed by 35 cycles of 98°C for 10 s, 54°C for 30 s, and 72°C for 30 s. The PCR products were first purified using AMPure XP beads (Agencourt Bioscience). The presence of DNA with the desired size was confirmed using a 0.7% agarose gel containing 1.15% SYNERgel (Diversified Biotech) stained with SYBR Gold nucleic acid gel stain (Invitrogen). After electrophoresis, the PCR amplicons were excised from the gels and purified using a QIAquick gel extraction kit (Qiagen). DNA libraries containing all tagged amplicons and the internal control (bacteriophage phiX) were generated for paired-end sequencing using a MiSeq reagent kit (version 2; Illumina) and sequenced using an Illumina MiSeq instrument according to the manufacturer's instruction.
Data analysis of deep sequencing.
Internal control phiX sequences were removed by use of an analysis pipeline using the Burrows-Wheeler aligner program (version 0.7.4) (37). The remaining paired sequences were joined together using the fastq-join tool in ea-utils (version 1.1.2; E. Aronesty, ea-utils: command-line tools for processing biological sequencing data [http://code.google.com/p/ea-utils]). fastq-formatted data for the combined sequences with a Q-score cutoff of >30 were converted to the fasta format using the macqiime program (version 1.6.0) (38). Chimeric and singleton sequences were removed using the Mothur program (version 1.29.2) (39). The resulting sequences were subjected to taxonomic assignment using the RDP multiclassifier (version 1.1) (40) with a 50% confidence threshold. On the basis of these assignments, Burkholderia sequences were retrieved and clustered into operational taxonomic units (OTUs), which were defined as clusters having <1% sequence differences, using the macqiime program (version 1.6.0) (38). The Burkholderia phylotypes of representative sequences of each OTU were identified by analysis against our collection of Burkholderia sequences derived from soils, plants, and stinkbugs using the BLASTn algorithm in the BLAST (version 2.2.27+) program (31).
qPCR.
Quantitative PCR (qPCR) was performed to amplify 16S rRNA genes of the domain Bacteria using a Power SYBR green PCR master mix (Applied Biosystems) and a LightCycler 96 system (Roche Applied Science). The reaction mixture was comprised of 2× SYBR green PCR master mix, 0.2 μM Bacteria group-specific primers 515F and 806R (36) (Table 2), 0.5 μg/μl bovine serum albumin, and gut tissue DNA as the template. The PCR conditions were as follows: initial denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 30 s. The total number of bacterial 16S rRNA gene copies was calculated on the basis of a standard curve constructed using a dilution series of the target PCR product of Burkholderia sp. SFA1 (DDBJ accession no. AB232333).
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA genes determined in this study have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence database under accession numbers AB916362 to AB916463 (clone libraries; see Table S1 in the supplemental material) and the MG-RAST database (http://metagenomics.anl.gov/) (41) as the Gut Symbiont of C_saccharivorus project under accession numbers 4555348.3 to 4555367.3 (Illumina sequencing libraries).
RESULTS
General observations of midgut crypts.
The midgut of the oriental chinch bug C. saccharivorus was divided into four morphologically different sections, designated M1 to M4 (Fig. 1B). White/cream-colored tubule-like crypts with branched tracheae developed in the M4 region. Light microscopy revealed large numbers of rod-shaped bacteria contained in the crypts.
Fluorescence in situ hybridization.
Symbiotic bacteria were detected in the midgut crypts of C. saccharivorus chinch bugs (Fig. 1C; see also Fig. S1 in the supplemental material) using fluorescence in situ hybridization of EUB338, a universal probe for bacteria, and BET940, a probe specific for Betaproteobacteria. No fluorescence signals were observed in the negative-control experiments (i.e., experiments with a no-probe control and FISH with the anti-EUB338 probe) (data not shown).
Transmission electron microscopy.
Transmission electron microscopic analysis of a dissected tubular crypt revealed the micromorphology of the chinch bug symbionts (Fig. 1D and E). The crypt epithelial cells were very thin, and the luminal region was filled with dense populations of rod-shaped bacteria. The rod-shaped bacteria were 2 to 3 μm in length and showed well-developed cell walls. No bacterial structures were detected inside the cytoplasm of the crypt epithelial cells.
Bacterial 16S rRNA gene sequences identified from the midgut crypts.
The top BLASTn matches of all 102 16S rRNA gene sequences cloned from 12 C. saccharivorus midgut crypt samples were to members of the genus Burkholderia (see Table S1 in the supplemental material). In each of the insects examined, sequences obtained from the same individual had greater than 99% identity, and this criterion was used to define the operative taxonomic unit (OTU) for phylogenetic analyses.
Phylogenetic placement of the gut symbiotic bacteria.
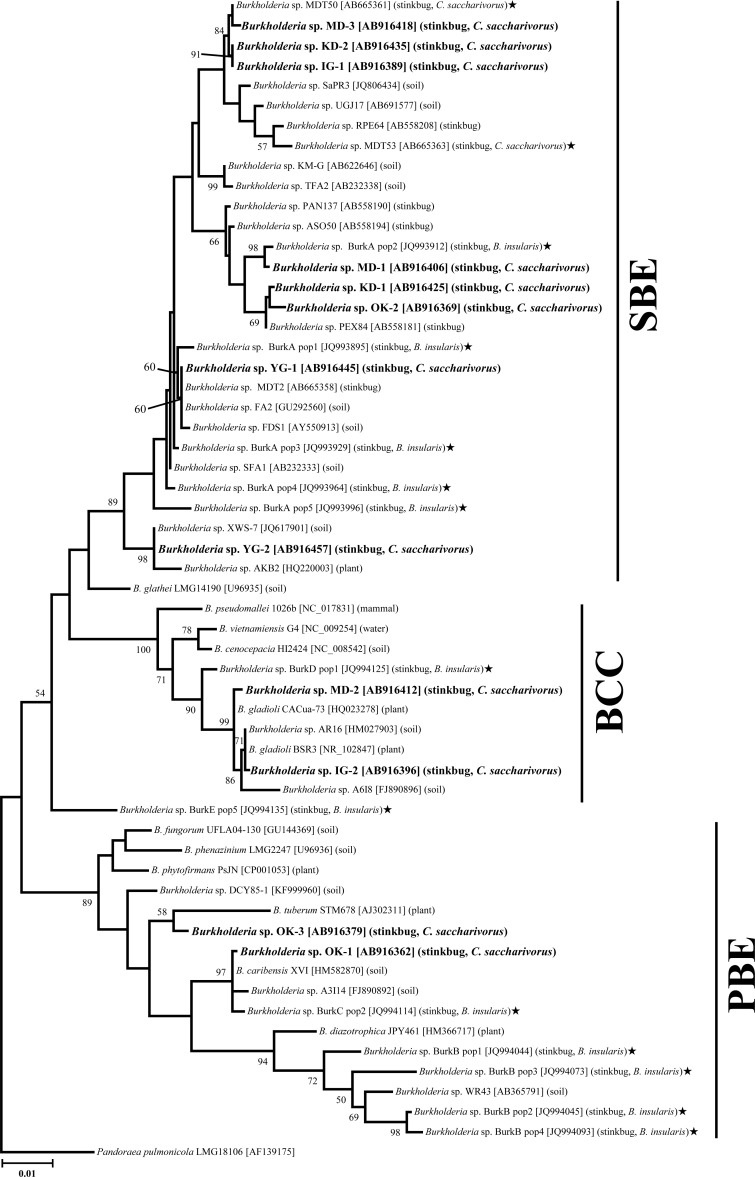
Phylogenetic analysis of the 16S rRNA gene sequences from the chinch bugs placed them into three Burkholderia clades. The majority of the sequences (67.6%) belonged to the stinkbug-associated beneficial and environmental (SBE) clade (Fig. 2) that we previously defined from diverse coreoid and lygaeoid stinkbugs (16, 17). The other Burkholderia sequences were placed into two clades previously defined by others: 15.7% in the Burkholderia cepacia complex (BCC) (42) and 16.7% in the plant-associated beneficial and environmental (PBE) group (43) (Fig. 2). The ML and NJ phylogenetic trees had similar topologies (Fig. 2; see also Fig. S2 in the supplemental material).
FIG 2.
Phylogenetic placement of the gut symbiotic bacteria of C. saccharivorus on the basis of 16S rRNA gene sequences. A maximum likelihood tree inferred from aligned 1,372-bp sequences of the 16S rRNA gene is shown. Sequences detected in this study are shown in bold, and sequence identifiers, such as OK-1 and MD-1, correspond to the sample accession numbers in Table 1 and Table S1 in the supplemental material. Accession numbers in the DNA database (DDBJ/EMBL/GenBank) are shown in brackets. The origins or sources of isolation of the Burkholderia strains/sequences are represented in parentheses. Stars indicate gut symbionts detected from the southern chinch bug, Blissus insularis (24), and from C. saccharivorus in our previous study (17). The clades SBE, BCC, and PBE, as described in references 17, 42, and 43, respectively, are shown on the right. Bootstrap values of >50% are depicted on the nodes. The phylogeny estimated by neighbor-joining analysis has a similar topology (see Fig. S2 in the supplemental material).
Prevalence of the bacterial symbionts in natural populations of the chinch bug.
Diagnostic PCR surveys with clade-specific primer sets indicated that Burkholderia symbionts belonging to the three different clades, the SBE, BCC, and PBE clades, were present in the field populations of C. saccharivorus (Table 3). In the field populations, 89.3% (number of insects in which infection was detected/total number = 100/112) of the insects were infected with only one of the three clades, and the Burkholderia SBE clade was the most prevalent (Table 3). Multiple infections with different clades of Burkholderia was detected in only 9.8% (11/112) of the insects; double and triple infections were detected in 8.9% (10/112) and 0.9% (1/112) of the insects, respectively (Table 3).
TABLE 3.
Prevalence of Burkholderia symbionts in field populations of Cavelerius saccharivorus
| Collection site | No. (%) of insects with the following infection patterna: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Single |
Double |
Triple (SBP) | None | |||||
| S | B | P | SB | SP | BP | ||||
| Okinawa Island | 54 | 23 | 23 | 5 | 0 | 1 | 2 | 0 | 0 |
| Kita-Daito Island | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Minami-Daito Island | 45 | 20 | 13 | 4 | 3 | 1 | 3 | 1 | 0 |
| Ishigaki Island | 4 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Yonaguni Island | 5 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total | 112 | 53 (47.3) | 38 (33.9) | 9 (8.0) | 3 (2.7) | 2 (1.8) | 5 (4.5) | 1 (0.9) | 1 (0.9) |
Single, diagnostic PCR detection of only one of the three clades tested, either the SBE (S), BCC (B), or PBE clade (P); Double, PCR detection of any two of the three clades; Triple, PCR detection of all three clades; None, PCR detection of none of the three clades.
Diagnostic PCR of eggs and hatchlings from eggs with and without surface sterilization.
Burkholderia-specific PCR using the clade-specific primer sets indicated that 52% of eggs (number of eggs in which infection was detected/total number = 56/107) and 29% of hatchlings (number of hatchlings in which infection was detected/total number = 32/112) reared under aseptic conditions were infected with Burkholderia symbionts (Table 4). In the PCR-positive samples, the Burkholderia SBE clade was the most frequently detected, with a 47% infection rate in eggs and a 25% infection rate in hatchlings. After egg surface sterilization, no Burkholderia symbiont infection of hatchlings was found (Table 4). These results strongly suggest that in the oriental chinch bug, Burkholderia symbionts are in part transmitted from mother to offspring via eggs.
Deep sequencing and qPCR of the microbiota associated with the midgut crypts.
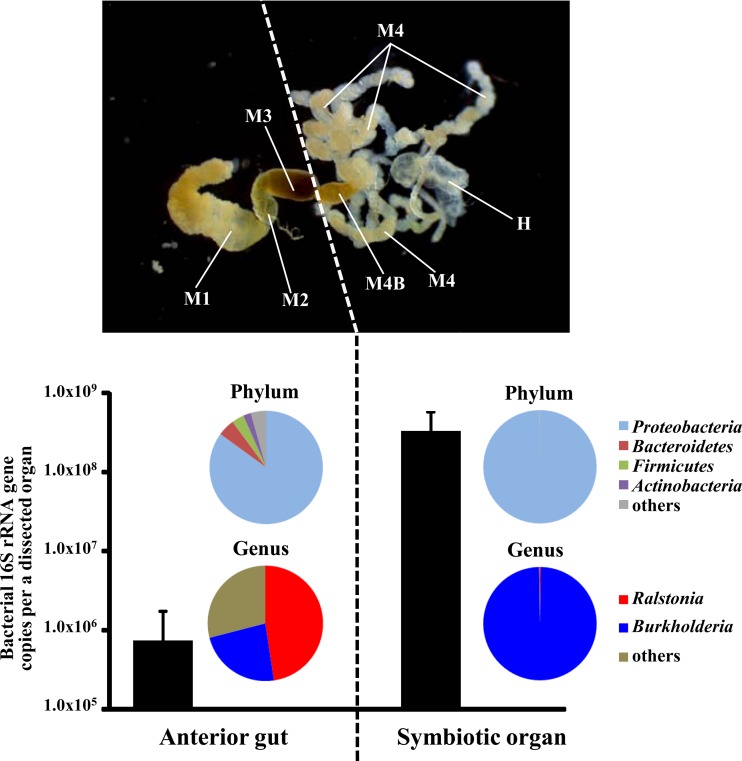
In C. saccharivorus collected from Minami-Daito Island, qPCR analysis showed that the number of copies of the bacterial 16S rRNA gene averaged 6.9 × 105 ± 9.2 × 105 in the anterior part of the midgut (i.e., M1 to M3) and 3.1 × 108 ± 2.3 × 108 in the symbiotic organ (i.e., M4B and M4) (mean ± standard deviation [SD], n = 10) (Fig. 3).
FIG 3.
Taxonomic compositions of gut microbiota of C. saccharivorus at bacterial phylum- and genus-level resolutions. The 16S rRNA gene sequences obtained by Illumina deep sequencing were classified using the RDP multiclassifier with a threshold level of 50%. Insects collected from Minami-Daito Island were used. Circles on the left indicate the composition of the microbes in the anterior midgut (M1 to M3), while circles on the right indicate the composition of those in the symbiotic organ (M4B and M4, not including the hind gut). The mean proportions for 10 individuals are shown. The bar graphs indicate the numbers of copies of bacterial 16S rRNA genes (mean ± SD, n = 10).
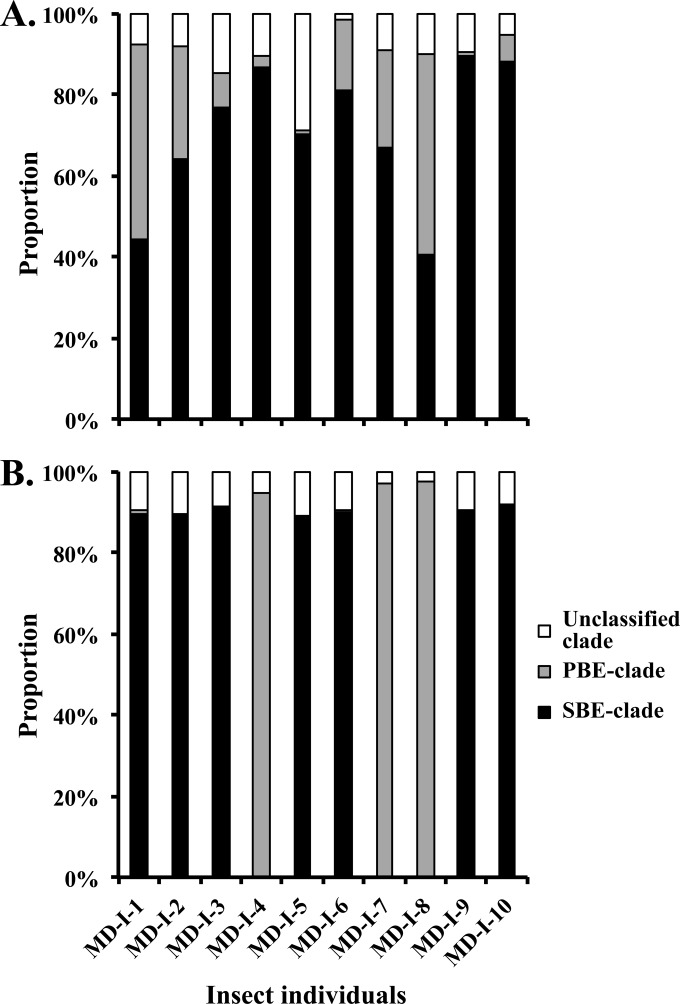
Illumina deep sequencing of the 16S rRNA genes in the anterior midgut revealed a diverse bacterial community, 329 genera belonging to 20 phyla (Fig. 3), whereas over 99% of the symbiotic organ sequences belonged to one genus, Burkholderia. The diversity of Burkholderia phylotypes classified into OTUs (defined by >99% sequence identity; see Table S2 in the supplemental material) revealed that a single OTU dominated the midgut crypts with a remarkably high (88.4% to 96.8%) relative abundance (see Table S3 in the supplemental material). The diversity of OTUs dominating the symbiotic organ was limited and clustered into either the SBE or PBE clade of Burkholderia (Fig. 4B; see Table S3 in the supplemental material). Of the 10 insects investigated, the Burkholderia SBE and PBE clades dominated in 7 and 3 individuals, respectively. The Burkholderia symbionts were also detected in the anterior part of the midgut (Fig. 4A), but their proportions were much lower than those in the midgut region (Fig. 3).
FIG 4.
Taxonomic compositions of symbiotic Burkholderia in the midgut of C. saccharivorus. (A) Anterior midgut; (B) midgut crypts. The Illumina deep sequences annotated as Burkholderia were determined using BLASTn analysis against the reference sequences used for Fig. 2. On the basis of >99% sequence identity, these sequences were categorized into either the SBE or PBE clade. The Burkholderia BCC clade was not detected in the 10 individuals examined. Note that in all of the individuals, a single sequence from either the SBE or PBE clade accounted for >88% of the sequences (see Tables S2 and S3 in the supplemental material for more detailed information).
DISCUSSION
In our previous study, a broad survey of Burkholderia infection among members of the infraorder Pentatomomorpha showed that this symbiotic bacterium is prevalent in the superfamilies Coreoidea and Lygaeoidea (16). Phylogenetic analysis indicated that Burkholderia symbionts were associated with select species of Heteroptera superfamilies, such as R. pedestris, Togo hemipterus, and Dimorphopterus pallipes. The symbionts formed a group along with some soil-derived Burkholderia strains, which is called the SBE clade (16, 17). In this study we identified two additional groups of crypt-associated Burkholderia from the oriental chinch bug that cluster in previously defined Burkholderia groups, the Burkholderia cepacia complex (BCC) and the plant-associated beneficial and environmental (PBE) group (17, 42, 43). Recently, crypt-associated symbionts of the southern chinch bug, Blissus insularis, were investigated by cloning and sequencing analyses, and diverse groups of Burkholderia, including Burkholderia of the BCC and PBE clades, were identified (24). These results suggest that these two additional groups of symbionts are commonly associated with the family Blissidae, and phylogenetically more diverse Burkholderia species might be found in coreoid and lygaeoid stinkbugs using broader and more in-depth analyses.
In the oriental chinch bug, the environmental acquisition of symbionts was indicated by three findings. First, the three Burkholderia clades (the BCC, PBE, and SBE clades) found in the oriental chinch bug did not form a monophyletic group (Fig. 2). Second, the symbiont phylogeny did not reflect the host population. Third, stinkbug-associated strains had 16S rRNA gene sequences highly similar or identical to those of soil-derived Burkholderia strains. These results strongly suggest the promiscuous nature of the symbiotic association between the chinch bug and the Burkholderia symbionts, as shown by the environmental acquisition of symbionts in the bean bug, R. pedestris (18), and suggested in other coreoid and lygaeoid species (16). The vertical transmission of symbionts in a subset of chinch bugs was supported by the fact that almost 30% of hatchlings possessed Burkholderia symbionts prior to environmental exposure (Table 4). These results demonstrate that this insect can employ both vertical and environmental mechanisms for transmitting Burkholderia symbionts.
Most phytophagous species of the superfamilies Pentatomoidea and Pyrrhocoroidea are associated with gammaproteobacterial and actinobacterial gut symbionts, respectively, and vertically transmit these symbionts from mother to offspring (3, 8, 14, 15, 44–51). Various mechanisms for vertical transmission have been reported for stinkbug groups. In the families Pentatomidae, Scuttelleridae, Acanthosomatidae, and Pyrrhocoridae, the mother insect superficially contaminates eggs (called “egg smearing”), and hatchlings acquire the symbionts by probing egg surfaces (14, 44, 46–50). In the families Cydnidae and Parastrachiidae, hatchlings acquire the symbionts by feeding on the excrement of their mother (51, 52). In the family Plataspidae, the mother insects provide a symbiont-containing capsule under the eggs, and hatchlings are infected with symbionts by sucking the capsule (8). In C. saccharivorus, infection through capsular transmission and coprophagy can be rejected because capsule-like materials have not been observed (data not shown) and hatchlings acquired the Burkholderia symbionts even when they were reared without their parents (Table 4). Egg surface sterilization resulted in no infection with Burkholderia symbionts (Table 4), suggesting that the Burkholderia symbionts are most likely transmitted by egg smearing.
In order to ensure the acquisition of essential microbes by offspring, insects have evolved elaborate mechanisms for vertical symbiont transmission: ovarial infection, egg smearing, coprophagy, and capsule transmission (reviewed in reference 3). The transmission mechanisms that have been studied are highly developed, and intermediate stages have not been found, making the evolutionary process of the transmission mechanisms broadly found in insects unclear. A number of insect symbionts, whether intracellular or extracellular, belong to the Enterobacteriaceae family of the Gammaproteobacteria (reviewed in reference 3), implying that symbiotic relationships may have evolved from gut bacteria that insects occasionally acquired from surrounding environments (or bacteria that contaminated insects) and that provided benefits to the host (53, 54). In this context, the symbiotic association found in C. saccharivorus may represent an intermediate stage from the evolutionarily primitive gut symbiosis from environmental transmission to the sophisticated association maintained by vertical transmission. It would be of great interest to investigate the structural and molecular basis of vertical transmission in the oriental chinch bug to understand the evolutionary process of vertical symbiont transmission in diverse insects.
Diagnostic PCR revealed only three different groups of Burkholderia, and multiple infections were rarely detected in field populations of the insect (Table 3). Illumina deep sequencing of the 16S rRNA gene confirmed that the crypts of each insect were dominated by a single Burkholderia strain, with relative proportions being >88% (Fig. 4), although the analysis was based on partial 255-bp sequences. Considering that most chinch bug hatchlings acquire symbionts from their surrounding environment and that millions of bacterial species inhabit soils (55, 56), the extraordinary simplicity of their gut symbiont community is noteworthy. Such a simple gut microbiota has been reported in the medicinal leech, Hirudo verbana, in which only two bacterial species predominate (57). The simplistic symbiotic association likely occurred through selective colonization, indicated by the high level of microbial diversity found in the anterior midgut compared to that found in midgut crypts (Fig. 3 and 4). Generally, the community composition of gut microbiota is thought to be determined by symbiont-symbiont and/or symbiont-host interactions (58). Since space and nutrients in midgut crypts are limited, severe competition between symbiont strains is inevitable, unless some mutualistic cooperation between strains occurs (59). From the host side, a simple microbial community would be favorable because symbiont-symbiont competition could cause excessive exploitation of host resources, the so-called tragedy of the commons (60). This could lead to the evolution of cheaters and the eventual collapse of the symbiotic association. To prevent exploitation by symbionts, host species have evolved sophisticated mechanisms for policing the microbial community in endosymbiotic systems without vertical transmission. For instance, leguminous plants punish non-nitrogen-fixing nodules by suppressing supplementation of oxygen (61). In the squid-Vibrio luminescent symbiosis, the light organ produces a poisonous concentration of peroxidase, which might specifically harm nonluminescent symbionts because the luciferase that they produce has a high affinity for oxygen (62). These symbiont- and host-controlled mechanisms may synergistically contribute to establishment of the remarkably simple crypt microbiota community in the oriental chinch bug.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Matsuura for supporting the FISH experiments, M. Aizawa and H. Kodama for collecting insect samples, N. Nakamura for technical assistance, T. Hoshino and T. Takushi for helpful comments, and C. H. Nakatsu for helpful comments and correction of the English.
This study was supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI grant number 24117525 and by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-Oriented Industry.
Footnotes
Published ahead of print 18 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01087-14.
REFERENCES
- 1.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, New York, NY [Google Scholar]
- 2.Bourtzis K, Miller TA. 2003. Insect symbiosis. CRC Press LLC, Boca Raton, FL [Google Scholar]
- 3.Kikuchi Y. 2009. Endosymbiotic bacteria in insects: their diversity and culturability. Microbes Environ. 24:195–204. 10.1264/jsme2.ME09140S [DOI] [PubMed] [Google Scholar]
- 4.Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17–37. 10.1146/annurev.ento.43.1.17 [DOI] [PubMed] [Google Scholar]
- 5.Baumann P. 2005. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155–189. 10.1146/annurev.micro.59.030804.121041 [DOI] [PubMed] [Google Scholar]
- 6.Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, Aksoy S. 2008. Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J. Insect Physiol. 54:1236–1242. 10.1016/j.jinsphys.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue T, Kitade O, Yoshimura T, Yamaoka I. 2000. Symbiotic association with protists. In Abe T, Bignell DE, Higashi M. (ed), Termites: evolution, sociality, symbioses, ecology. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 8.Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4:e337. 10.1371/journal.pbio.0040337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miura T, Braendle C, Shingleton A, Sisk G, Kambhampati S, Stern DL. 2003. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). J. Exp. Zool. B Mol. Dev. Evol. 295:59-81. 10.1002/jez.b.3 [DOI] [PubMed] [Google Scholar]
- 10.Pant NC, Fraenkel G. 1954. Studies on the symbiotic yeasts of two insect species, Lasioderma serricorne F. and Stegobium paniceum L. Biol. Bull. 107:420–432. 10.2307/1538590 [DOI] [PubMed] [Google Scholar]
- 11.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190. 10.1146/annurev.genet.41.110306.130119 [DOI] [PubMed] [Google Scholar]
- 12.Glasgow H. 1914. The gastric caeca and the caecal bacteria of the Heteroptera. Biol. Bull. 3:101–171 [Google Scholar]
- 13.Goodchild AJP. 1963. Studies on the functional anatomy of the intestines of Heteroptera. Proc. Zool. Soc. Lond. 141:851–910 [Google Scholar]
- 14.Kaltenpoth M, Winter SA, Kleinhammer A. 2009. Localization and transmission route of Coriobacterium glomerans, the endosymbiont of pyrrhocorid bugs. FEMS Microbiol. Ecol. 69:373–383. 10.1111/j.1574-6941.2009.00722.x [DOI] [PubMed] [Google Scholar]
- 15.Prado SS, Almeida RP. 2009. Phylogenetic placement of pentatomid stink bug gut symbionts. Curr. Microbiol. 58:64–69. 10.1007/s00284-008-9267-9 [DOI] [PubMed] [Google Scholar]
- 16.Kikuchi Y, Hosokawa T, Fukatsu T. 2011. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 5:446–460. 10.1038/ismej.2010.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. 2012. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. U. S. A. 109:8618–8622. 10.1073/pnas.1200231109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kikuchi Y, Hosokawa T, Fukatsu T. 2007. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 73:4308–4316. 10.1128/AEM.00067-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bright M, Bulgheresi S. 2010. A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol. 8:218–230. 10.1038/nrmicro2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caspi-Fluger A, Inbar M, Mozes-Daube N, Katzir N, Portnoy V, Belausov E, Hunter MS, Zchori-Fein E. 2012. Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc. Biol. Sci. 279:1791–1796. 10.1098/rspb.2011.2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Vries EJ, Jacobs G, Sabelis MW, Menken SB, Breeuwer JA. 2004. Diet-dependent effects of gut bacteria on their insect host: the symbiosis of Erwinia sp. and western flower thrips. Proc. Biol. Sci. 271:2171–2178. 10.1098/rspb.2004.2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomokuni M, Yasunaga T, Takai M, Yamashita I, Kawamura M, Kawasawa T. 1993. A field guide to Japanese bugs. Zenkoku Noson Kyoiku Kyoukai, Tokyo, Japan [Google Scholar]
- 23.Schaefer CW, Panizzi AR. 2000. Heteroptera of economic importance. CRC Press LLC, Boca Raton, FL [Google Scholar]
- 24.Boucias DG, Garcia-Maruniak A, Cherry R, Lu H, Maruniak JE, Lietze V-U. 2012. Detection and characterization of bacterial symbionts in the Heteropteran, Blissus insularis. FEMS Microbiol. Ecol. 82:629–641. 10.1111/j.1574-6941.2012.01433.x [DOI] [PubMed] [Google Scholar]
- 25.Kikuchi Y, Meng XY, Fukatsu T. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71:4035–4043. 10.1128/AEM.71.7.4035-4043.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demaneche S, Sanguin H, Pote J, Navarro E, Bernillon D, Mavingui P, Wildi W, Vogel TM, Simonet P. 2008. Antibiotic-resistant soil bacteria in transgenic plant fields. Proc. Natl. Acad. Sci. U. S. A. 105:3957–3962. 10.1073/pnas.0800072105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koga R, Tsuchida T, Fukatsu T. 2009. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl. Entomol. Zool. 44:281–291. 10.1303/aez.2009.281 [DOI] [Google Scholar]
- 29.Fukatsu T, Nikoh N. 1998. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl. Environ. Microbiol. 64:3599–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics 9:286–298. 10.1093/bib/bbn013 [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526 [DOI] [PubMed] [Google Scholar]
- 35.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3:294–299 [PubMed] [Google Scholar]
- 36.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. 10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coenye T, Vandamme P, Govan JR, LiPuma JJ. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427–3436. 10.1128/JCM.39.10.3427-3436.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suárez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonca-Previato L, James EK, Venturi V. 2012. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 63:249–266. 10.1007/s00248-011-9929-1 [DOI] [PubMed] [Google Scholar]
- 44.Abe Y, Mishiro K, Takanashi M. 1995. Symbiont of brown-winged green bug, Plautia stali Scott. Jpn. J. Appl. Entomol. Zool. 39:109–115. 10.1303/jjaez.39.109 [DOI] [Google Scholar]
- 45.Hosokawa T, Kikuchi Y, Nikoh N, Fukatsu T. 2012. Polyphyly of gut symbionts in stinkbugs of the family Cydnidae. Appl. Environ. Microbiol. 78:4758–4761. 10.1128/AEM.00867-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, Ito M, Fukatsu T. 2010. Primary gut symbiont and secondary, Sodalis-allied symbiont of the scutellerid stinkbug Cantao ocellatus. Appl. Environ. Microbiol. 76:3486–3494. 10.1128/AEM.00421-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, Ito M, Fukatsu T. 2011. Bacterial symbionts of the giant jewel stinkbug Eucorysses grandis (Hemiptera: Scutelleridae). Zool. Sci. 28:169–174. 10.2108/zsj.28.169 [DOI] [PubMed] [Google Scholar]
- 48.Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T. 2009. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 7:2. 10.1186/1741-7007-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tada A, Kikuchi Y, Hosokawa T, Musolin D, Fujisaki K, Fukatsu T. 2011. Obligate association with gut bacterial symbiont in Japanese populations of the southern green stinkbug Nezara viridula (Heteroptera: Pentatomidae). Appl. Entomol. Zool. 46:483–488. 10.1007/s13355-011-0066-6 [DOI] [Google Scholar]
- 50.Kikuchi Y, Hosokawa T, Nikoh N, Fukatsu T. 2012. Gut symbiotic bacteria in the cabbage bugs Eurydema rugosa and Eurydema dominulus (Heteroptera: Pentatomidae). Appl. Entomol. Zool. 47:1–8. 10.1007/s13355-011-0081-7 [DOI] [Google Scholar]
- 51.Hosokawa T, Hironaka M, Mukai H, Inadomi K, Suzuki N, Fukatsu T. 2012. Mothers never miss the moment: a fine-tuned mechanism for vertical symbiont transmission in a subsocial insect. Anim. Behav. 83:293–300. 10.1016/j.anbehav.2011.11.006 [DOI] [Google Scholar]
- 52.Schorr H. 1957. Zur Verhaltensbiologie und Symbiose von Brachypelta aterrima Först (Cydnidae, Heteroptera). Z. Morph. Ökol. Tiere 45:561–602. 10.1007/BF00399596 [DOI] [Google Scholar]
- 53.Harada H, Oyaizu H, Ishikawa H. 1996. A consideration about the origin of aphid intracellular symbiont in connection with gut bacterial flora. J. Gen. Appl. Microbiol. 42:17–26. 10.2323/jgam.42.17 [DOI] [Google Scholar]
- 54.Husník F, Chrudimský T, Hypša V. 2011. Multiple origins of endosymbiosis within the Enterobacteriaceae (gamma-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 9:87. 10.1186/1741-7007-9-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290. 10.1038/ismej.2007.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unno Y, Shinano T. 2013. Metagenomic analysis of the rhizosphere soil microbiome with respect to phytic acid utilization. Microbes Environ. 28:120–127. 10.1264/jsme2.ME12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graf J, Kikuchi Y, Rio RV. 2006. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 14:365–371. 10.1016/j.tim.2006.06.009 [DOI] [PubMed] [Google Scholar]
- 58.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 59.Frank SA. 1996. Host-symbiont conflict over the mixing of symbiotic lineages. Proc. Biol. Sci. 263:339–344. 10.1098/rspb.1996.0052 [DOI] [PubMed] [Google Scholar]
- 60.Hardin G. 1968. The tragedy of the commons. Science 162:1243–1248. 10.1126/science.162.3859.1243 [DOI] [PubMed] [Google Scholar]
- 61.Kiers ET, Rousseau RA, West SA, Denison RF. 2003. Host sanctions and the legume-rhizobium mutualism. Nature 425:78–81. 10.1038/nature01931 [DOI] [PubMed] [Google Scholar]
- 62.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. 10.1128/JB.182.16.4578-4586.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.