Abstract
“Candidatus Liberibacter asiaticus” is an uncultured alphaproteobacterium that systemically colonizes its insect host both inter- and intracellularly and also causes a severe, crop-destroying disease of citrus called huanglongbing, or citrus “greening.” In planta, “Ca. Liberibacter asiaticus” is also systemic but phloem limited. “Ca. Liberibacter asiaticus” strain UF506 carries two predicted prophages, SC1 and SC2. Bacteriophage particles have been observed in experimentally “Ca. Liberibacter asiaticus”-infected periwinkle but not in any other host. Comparative gene expression analysis of predicted SC1 late genes showed a much higher level of late gene expression, including holin transcripts (SC1_gp110), in “Ca. Liberibacter asiaticus”-infected periwinkle relative to “Ca. Liberibacter asiaticus”-infected citrus. To functionally characterize predicted holin and endolysin activity, SC1_gp110 and two predicted endolysins, one within SC1 (SC1_gp035) and another well outside the predicted prophage region (CLIBASIA_04790), were cloned and expressed in Escherichia coli. Both SC1 genes inhibited bacterial growth consistent with holin and endolysin function. The holin (SC1_gp110) promoter region was fused with a uidA reporter on pUFR071, a wide bacterial host range (repW) replicon, and used to transform Liberibacter crescens strain BT-1 by electroporation. BT-1 is the only liberibacter strain cultured to date and was used as a proxy for “Ca. Liberibacter asiaticus.” pUFR071 was >95% stable without selection in BT-1 for over 20 generations. The reporter construct exhibited strong constitutive glucuronidase (GUS) activity in culture-grown BT-1 cells. However, GUS reporter activity in BT-1 was suppressed in a dose-dependent manner by crude aqueous extracts from psyllids. Taken together with plant expression data, these observations indicate that “Ca. Liberibacter asiaticus” prophage activation may limit “Ca. Liberibacter asiaticus” host range and culturability.
INTRODUCTION
Huanglongbing (HLB), commonly known as citrus “greening,” is the most economically severe disease of citrus worldwide. HLB is caused by several species of the fastidious alphaproteobacterium “Candidatus Liberibacter,” the most prevalent of which is “Candidatus Liberibacter asiaticus” (1). In planta, “Ca. Liberibacter asiaticus” bacteria are systemic, intracellular, and phloem limited. “Ca. Liberibacter asiaticus” is persistently transmitted among citrus species and relatives by the psyllid vector Diaphorina citri Kuwayama (Sternorrhyncha: Psyllidae). In contrast to plant host infections, “Ca. Liberibacter asiaticus” colonizes the psyllid host systemically in multiple organs, including the alimentary canal and salivary glands, both inter- and intracellularly, but with little apparent pathogenic effect (2, 3, 4, 5). Despite numerous attempts, “Ca. Liberibacter asiaticus” has not been cultured in axenic medium, making functional genomics difficult. Indeed, of six known species of liberibacter, only a single strain from a single species of liberibacter, Liberibacter crescens, has been cultured (6). Originally isolated from the sap of mountain papaya (Carica stipulata × Carica pubescens), L. crescens has no known insect host, and attempts to reinoculate it into many plants have failed, warranting speculation that L. crescens may no longer be parasitic or pathogenic.
Some prophage sequences were found in the “Ca. Liberibacter asiaticus” strain Psy62 (for psyllid 62) chromosome (7). Subsequently, all pathogenic “Ca. Liberibacter asiaticus” strains examined were found to carry two nearly identical prophages; these were named SC1 and SC2 in “Ca. Liberibacter asiaticus” strain UF506 (8). SC1 and SC2 are largely syntenic, and large stretches are nearly 100% identical at the DNA level. UF506 prophage SC2 was found to replicate as an excision plasmid with a copy in the chromosome adjacent to prophage SC1. SC1 was not found in replicative form in psyllid hosts but was found replicating in citrus and replicating at higher copy in infected periwinkle (Catharanthus roseus) (8). The early genes of SC1 generally had homologs in SC2, and genes unique to either phage were mostly late genes. Of the two UF506 prophages, only SC1 was annotated as encoding holin (SC1_gp110) and endolysin (SC1_gp035) genes (8). Although phage particles with icosahedral heads and short tails (Podoviridae) were consistently visualized in artificially inoculated periwinkle, phage have never been documented in citrus, despite numerous electron micrographs taken from multiple samples in many countries over years (8).
The lysogenic cycle can be terminated by physiological changes (particularly stress responses) in the host cell. Treatments that are capable of inducing the lytic cycle in bacteriophages include UV irradiation, the addition of agents that damage DNA, such as mitomycin C, the addition of energy poisons, and heat treatment (9, 10, 11). The lytic cycle is characterized by activation of expression of phage structural genes, phage structural self-assembly, and expression of genes allowing cell egress, including cell wall-degrading enzymes called endolysins and cell inner membrane-permeabilizing proteins called holins (12).
To date, there is little evidence for lytic cycle activation in “Ca. Liberibacter asiaticus”-infected citrus (SC1 replicates in citrus) and none in psyllids, and the mechanism behind the activation or derepression of the lytic cycle in (artificially inoculated) periwinkles is unknown. The purpose of this research was to identify and characterize late genes and late gene promoter regions from the “Ca. Liberibacter asiaticus” prophages, to utilize L. crescens strain BT-1 as a culturable proxy for “Ca. Liberibacter asiaticus,” and to construct reporter vectors for screening potential late gene activators or repressors in Escherichia coli and L. crescens.
MATERIALS AND METHODS
E. coli strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1, along with their relevant characteristics and source or reference. E. coli strains were grown in Luria-Bertani (LB) medium at 37°C and were used and stored according to standard protocols (13). Antibiotics were used as needed at the following concentrations, unless otherwise stated (in μg/ml): ampicillin (Amp), 100; kanamycin (Kan), 50; gentamicin (Gm), 3.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli Mach1-T1R | F− ϕ80lacZΔM15 ΔlacX74 hsdR(rK− mK+) ΔrecA1398 endA1 tonA | Invitrogen |
| E. coli DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Gibco |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| E. coli BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | Invitrogen |
| Liberibacter crescens BT-1 | Originally isolated from mountain papaya | 14 |
| Plasmids | ||
| pCR2.1-TOPO | 3.9 kb; PCR cloning vector, Apr, Knr | Invitrogen |
| pUC19 | 2.7 kb; PCR cloning vector, Apr | Invitrogen |
| pUFR071 | 9.4 kb; repW, ColE1, Mob+, LacZ+, Par+, Cmr, Gmr | 22 |
| pET27b | 5.4 kb; expression vector, T7 promoter, Knr | Novagen |
| pBI221 | 6.0 kb; transient expression vector, source of uidA, Apr | Clontech |
| pLF040 | SC1gp_110 358-bp amplicon (full-length gene) in pCR2.1-TOPO | This work |
| pLF041 | NdeI-NheI fragment of pLF040 in pET27b | This work |
| pLF042 | SC1gp_110 256-bp amplicon (N-terminal truncation) in pCR2.1-TOPO | This work |
| pLF043 | NdeI-NheI fragment of pLF042 in pET27b | This work |
| pLF044 | 511-bp fragment of SC1_gp110 promoter region fused with promoterless lacZ of pUC19 | This work |
| pLF045 | 359-bp fragment of SC1_gp110 promoter region fused with promoterless lacZ of pUC19 | This work |
| pLF046 | 223-bp fragment of SC1_gp110 promoter region fused with promoterless lacZ of pUC19 | This work |
| pLF047 | 124-bp fragment of SC1_gp110 promoter region fused with promoterless lacZ of pUC19 | This work |
| pLF049 | CLIBASIA_04790 576-bp amplicon (NdeI-NheI ends) in pCR2.1-TOPO | This work |
| pLF051 | SC1_gp035 1,452-bp amplicon (NdeI-NheI ends) in pCR2.1-TOPO | This work |
| pLF053 | NdeI-NheI fragment of pLF049 in pET27b | This work |
| pLF055 | NdeI-NheI fragment of pLF051 in pET27b | This work |
| pLF056 | PciI-HindIII fragment of pLF044 (405 bp of SC1_gp110 promoter region) in pUFR071 with lacZ promoter deleted | This work |
| pLF057 | 405-bp fragment of SC1_gp110 promoter region fused with promoterless uidA from pBI221 (hol::uidA) in pUFR071 with lacZ promoter deleted | This work |
| pLF058 | Promoterless BamHI-HindIII fragment of uidA from pBI221 fused with lacZ promoter (lacZ::uidA) of pUFR071 | This work |
L. crescens growth conditions and transformation.
L. crescens strain BT-1 was maintained in liquid BM7 medium containing 2 g of alpha-ketoglutarate, 10 g of N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) buffer, and 3.75 g of KOH in 550 ml water, pH 6.9, followed by the addition of 300 ml of filter-sterilized fetal bovine serum (HyClone Laboratories, Logan, UT, USA) and 300 ml of modified Grace's insect culture medium (TNM-FH; HyClone Laboratories) and gentle shaking at 150 rpm at 28°C (14). Transformation of BT-1 cells was done by electroporation. Five-day-old 90-ml L. crescens cultures (optical density at 600 nm [OD600] of 0.65) were chilled on ice for 30 min and harvested by centrifugation at 3,500 rpm for 15 min at 4°C. The bacterial pellet was rinsed twice in 20 ml of ice-cold sterile distilled water and resuspended in 2 ml of ice-cold 10% glycerol. Forty-microliter aliquots of competent cells were flash-frozen in liquid nitrogen and stored at −80°C. For transformation, an aliquot of competent cells was thawed on ice and added to a chilled 1-mm electroporation cuvette containing ∼500 ng of plasmid DNA. Following electroporation at 1,800 V (with the time constant usually in the range of 4.8 to 6.2 ms), the cells were immediately recovered in 900 μl of BM7 broth (without antibiotics) and transferred into 5-ml sterile tubes. The transformed cells were allowed to recover for 16 h, with gentle shaking at 29°C at 150 rpm, prior to selection on BM7 medium solidified with 1.5% agar (Difco Laboratories, Detroit, MI, USA) and supplemented with Gm (2 μg/ml).
Reverse transcription-quantitative PCR (RT-qPCR) of “Ca. Liberibacter asiaticus”-infected citrus and periwinkle.
The presence of “Ca. Liberibacter asiaticus” in specific leaves was assayed in DNA extracted from leaf discs of citrus (Citrus paradisi and Citrus sinensis) and periwinkle (Catharanthus roseus) plants using a DNeasy Plant minikit (Qiagen, Valencia, CA, USA). Conventional PCR using the primer set OI1/OI2c (15) was followed by nested PCR using the primer set CG03F/CG05R (16) for confirmation. The remaining midribs of confirmed “Ca. Liberibacter asiaticus”-infected periwinkle and citrus leaves were ground with a mortar and pestle in lysis buffer RLT (provided with a Qiagen RNeasy Plant minikit), and RNA was extracted according to the manufacturer's protocol. RNA extract yield and purity were estimated with a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Extracts were diluted with nuclease-free water to 200 ng/μl and treated with a Turbo DNA-free (DNase) kit (Ambion, Austin, TX, USA).
Reverse transcription was carried out with 0.9 to 1 μg of template using an iScript Advanced cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Quantitative PCR was carried out using a CFX96 Touch Real-Time PCR detection system (Bio-Rad). The reaction mixture contained 10 μl of 2× SsoFast Probes Supermix (Bio-Rad), 300 nM gene-specific primers (Table 2), 200 nM 6-carboxyfluorescein (FAM)-labeled ZEN double-quenched probes (Integrated DNA Technologies, Coralville, IA) (Table 2), and 90 to 100 ng of cDNA template in a total volume of 20 μl. Reaction mixtures were incubated at 95°C for 30 s and cycled 40 times at 95°C for 10 s and 56°C for 30 s. The reference gene gyrB was used for normalization. Two technical replicates were performed for each of six and eight biological replicates for citrus and periwinkle samples, respectively, along with no-template controls and no-reverse transcriptase controls.
TABLE 2.
Primers and probes used in this study
| Primer or probe function and name | DNA sequence (5′–3′) | Reference or source |
|---|---|---|
| Identification of leaves positive for “Ca. Liberibacter asiaticus” | ||
| OI1 | GCG CGT ATG CAA TAC GAG CGG CA | 15 |
| OI2c | GCC TCG CGA CTT CGC AAC CCA T | 15 |
| CG03F | RGG GAA AGA TTT TAT TGG AG | 16 |
| CG05R | GAA AAT AYC ATC TCT GAT ATC GT | 16 |
| RT-qPCR | ||
| SC1_gp025F | AGC TAG ATC ATT GAC TCT TCC | This work |
| SC1_gp025R | AAA GAT GTT GGT CGT AAA CTA G | This work |
| SC2_gp095F | GGT GAA TAG CTA GAA TCC CG | This work |
| SC2_gp095R2 | TGT TGC TGG AGC CTA TAA CG | This work |
| SC2_gp100F | GTT CCA ATT CTC TAT AAG CGG | This work |
| SC2_gp100R2 | CGA ATG ATC CGA TTG CTT TAG | This work |
| SC1_gp110F2 | TCG TAC ATG CAC CCC TGA TA | This work |
| SC1_gp110R2 | AAG TGA GAC GCC AGG AAA GT | This work |
| LasprfAF | TGT CTG AAT CGC CTT CTG TC | This work |
| LasprfAR | GAT CAC CGA TGA CAG TAT GC | This work |
| LasgyrBF | TTG AAC AAG CTG TAA TTT CTG G | This work |
| LasgyrBR | ATC TGT TTG CCA ATT TAG AAG C | This work |
| SC1_gp025P | 6-FAM-ATA TTC ATC-ZEN-TTT TGT GTT AAA ACG GC-IABkFQa | This work |
| SC2_gp095P | 6-FAM-TGT TAG AGA-ZEN-AAC TAT CAC GTT CAA C-IABkFQ | This work |
| SC2_gp100P | 6-FAM-TCA AAT CCG-ZEN-TAA TAT AAA GGG CTA TT-IABkFQ | This work |
| SC1_gp110P | 6-FAM-TAT TCT ATT-ZEN-CAT CTC GTA AGC ACG T-IABkFQ | This work |
| LasprfAP | 6-FAM-TCA GTC CTA-ZEN-TAA TAT CGA AGA TTA GT-IABkFQ | This work |
| LasgyrBP | 6-FAM-TTA CAT TAA-ZEN-TTT CTT GAT CAC TTT CAC-IABkFQ | This work |
| Functional analysis of potential lysis genes | ||
| CLas endolysin NdeI F | TTA CAT ATG GTG TGT ATT ATA AAC AGA ATA ATA T | This work |
| CLas endolysin NheI R | TTA GCT AGC ATC ATA TCT ACA ATC CTC TTC TC | This work |
| SC1_gp035 NdeI F | TAT CAT ATG TAT TTC AAC GCG GTA AGC | This work |
| SC1_gp035 NheI R | AAT GCT AGC TCA TAA AGA TCC TCC TGT TCG | This work |
| Holin NdeI F | TTA CAT ATG AGT TTC TTA GAT TCG AGT G | This work |
| Holin NdeI F2 | TTA CAT ATG GGC GGA GCA GCA GGT | This work |
| Holin NheI R | AAT GCT AGC GAT CTT TGA TTC ATC GTA CAT G | This work |
| Promoter expression reporter constructs | ||
| HPromRIF | CUA CUA CUA CUA CCT AGG CCG ATA AAC TCC AAA AAA CGA G | This work |
| HPromFIR1 | GAU GAU GAU GAU CGT ACG TGA CGC AAA TAA CAC TGG TGC | This work |
| HPromFIR2 | GAU GAU GAU GAU CGT ACG AGT TTG CGA GCC TTA TCA ACC | This work |
| HPromFIR3 | GAU GAU GAU GAU CGT ACG CGG GCT TAT GTA TAC CTT TGC | This work |
| HPromFIR4 | GAU GAU GAU GAU CGT ACG GCG GAG GTT TGA ATA TGA CTC | This work |
| pUC19VF2 | UAG UAG UAG UAG GAG ACA GCT ATG ACC ATG ATT AC | This work |
| pUC19VR | AUC AUC AUC AUC CGG TTT GCG TAT TGG GCG C | This work |
| pGUSIR | GAU GAU GAU GAU CGT ACG ATA CCT AGG AGT CCC TTA TGT TAC GTC CTG | This work |
| pGUS IR2 | TTA GGA TCC AGG AGT CCC TTA TGT TAC GTC CTG | This work |
| pGUS IF2 | TTA AAG CTT CAG GAG AGT TGT TGA TTC ATT G | This work |
IABkFQ, Iowa Black FQ quencher.
The specificity of each primer set was initially assessed by conventional PCR, and the amplicons were then cloned into pCR2.1-TOPO (Invitrogen Corp., Carlsbad, CA, USA). To calculate the amplification efficiency of each primer/probe set, qPCR was performed on a dilution series of each plasmid construct across a linear dynamic range of six log10 concentrations (6.25 × 10−4 ng/μl to 6.25 × 10−9 ng/μl). Amplification efficiencies ranged from 92.7% to 101.8%, with r2 values ranging from 0.991 to 0.998 for the six primer/probe sets. Relative normalized expression levels, corrected for the various amplification efficiencies and using citrus values as calibrator controls, were calculated by the ΔΔCT (where CT is threshold cycle) method using the Bio-Rad CFX Manager, version 3.0, software package (17).
Bioinformatic and functional analyses of putative holin and endolysins.
TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) (18) and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) (19) were used to predict transmembrane α-helices from protein sequences with default settings. SignalP, version 4.0, was used to identify potential secretion signals from protein sequences (http://www.cbs.dtu.dk/services/SignalP/) (20) using both default (discrimination [D] score cutoff of 0.57) and sensitive (D score cutoff of 0.42) values. Functional analyses of SC1_gp110 (holin), SC1_gp035 (endolysin), and CLIBASIA_04790 (phage-related lysozyme) were carried out by cloning and heterologous expression in E. coli. The entire coding sequences of each of the three genes, along with a second construct comprising a truncated fragment of SC1_gp110 beginning at the second in-frame methionine codon, were PCR amplified using Accuprime Taq DNA Polymerase High Fidelity (Invitrogen) with primers designed to fuse an NdeI site to the start codon and to add an NheI site after the stop codon (Table 2) and cloned into pCR2.1-TOPO for sequence confirmation. The constructs were digested with enzymes NdeI and NheI, gel purified, and ligated into expression vector pET27b (Novagen Inc., Madison, WI, USA). These constructs were transformed into E. coli TOP10, which lacks the T7 polymerase needed to drive expression of the desired gene. Plasmids were isolated from overnight cultures and diluted to 10 ng/μl, and 1 μl was used for the transformation of E. coli BL21(DE3). Cells were plated on LB medium supplemented with Kan (30 μg/ml) under noninducing conditions. Individual colonies of BL21(DE3) containing the plasmid construct or empty vector pET27b were transferred to LB medium supplemented with Kan (30 μg/ml) and shaken at 37°C at 225 rpm overnight. For growth curves, approximately 375 μl of overnight cultures was transferred to 25 ml of prewarmed fresh LB medium supplemented with Kan (30 μg/ml) and incubated at 37°C and 225 rpm for 60 to 90 min. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.5 mM, and cultures were returned to incubate at 37°C and 225 rpm. The OD600 was measured at 30- to 60-min intervals.
Construction of promoter expression reporter vectors.
A uracil DNA glycosylase (UDG)-mediated cloning strategy was used to create a series of SC1_gp110 holin (21) promoters, each fused with lacZ, as reporter constructs in pUC19. Four fragments of the promoter region of SC1_gp110 were amplified using forward primers HPromF-IR1, HPromF-IR2, HPromF-IR3, and HPromF-IR4 and reverse primer HPromR-IF to create pLF044, pLF045, pLF046, and pLF047, respectively (Table 2). The reverse primer HPromR-IF (3′ end of the promoter region) contained added AvrII and HindIII sites, and the forward primers and reverse primer had 12-bp overhangs of four dUMP-containing nucleotide repeats at their 5′ ends. The amplicons were gel purified using a Qiagen QIAquick gel extraction kit following the manufacturer's instructions. Similarly, pUC19 was amplified using pUC19-VF2 and pUC19-VR (Table 2), which sit outside the lacZ promoter region but capture the lacZ coding sequence, also containing 12-bp overhangs of four dUMP-containing nucleotide repeats at their 5′ ends. The uracil repeat overhangs of the HPromF-IR series and pUC19VR, as well as HPromR-IF and pUC19VF2, were complementary, allowing directional annealing of the promoter fragments into the amplified vector. Approximately 120 to 160 ng of the amplified promoter fragments was each mixed with 5 μl of the pUC19 amplicon, 2 μl of UDG, and 5 μl of 10× reaction buffer (New England BioLabs) and brought to a 50-μl reaction volume with sterile distilled deionized H2O. Reaction mixtures were incubated at 37°C for 30 min, and E. coli Mach1 (Invitrogen) was transformed with 3 μl of the reaction mixture. Transformed cells were plated on LB medium with appropriate antibiotics.
Since the narrow-host-range colE1 replicon of pUC19 did not function in L. crescens, the SC1_gp110 (holin) promoter was recloned into the wide-host-range (repW) vector pUFR071 (22) and fused with uidA as a reporter. First, the holin promoter region in pLF044 was directionally cloned into pUFR071 using PciI (present in the native holin promoter sequence) and HindIII, which effectively swapped the lacZ promoter consensus sequence with a putative holin promoter region. After sequences were verified in this intermediate construct (pLF056), the reporter gene uidA was amplified from pBI221 with forward primer pGUSIR, which added an AvrII site followed by a Shine-Dalgarno sequence to the 5′ end of the gene, and pGUSIF2, which added a HindIII site to the 3′ end of the gene (Table 2). The amplicon was purified and directionally cloned into the AvrII/HindIII sites of pLF056 to create pLF057. This plasmid was sequenced to confirm the insert and used to transform L. crescens.
To create a control expression vector using the lacZ promoter fused with uidA as a reporter, uidA was amplified from pBI221 with forward primer pGUSIR2 (Table 2), which added a BamHI site and Shine-Dalgarno sequence to the 5′ end of uidA, and reverse primer pGUSIF2, which added an HindIII site to the 3′ end. The amplicon was PCR purified using a Qiagen PCR Purification kit according to the manufacturer's protocol, digested with BamHI and HindIII, and directionally cloned into BamHI/HindIII-digested pUFR071 to yield pLF058.
Preparation of plant and psyllid extracts.
Crude aqueous cell-free leaf extracts of sweet orange (Citrus sinensis), periwinkle (Catharanthus roseus), eggplant (Solanum melongena), and tobacco (Nicotiana tabacum) were prepared from fully expanded young leaf tissue from greenhouse-grown plants. One gram of leaf tissue was pulverized to a fine powder and resuspended in 10 ml of deionized water. The resulting suspension was cleared by centrifugation twice at 3,220 × g for 20 min at 4°C. The cleared supernatant was filter sterilized and stored at −20°C. Likewise, sterile aqueous extracts were prepared from psyllids reared on orange jasmine (Murraya paniculata; approximately 50 insects per ml of water).
Cell viability and GUS activity assays.
Five-day-old BT-1 cultures (OD600 of 0.65) were used for cell viability and GUS activity assays following overnight incubation of 1 ml of BT-1 culture in the presence of 25 to 100 μl of sterile leaf or psyllid extract. For the cell viability assays, a 10-fold serial dilution was prepared in BM7 medium, and a 10-μl cell suspension from each dilution was spotted on triplicate BM7 agar plates. The plates were incubated at 28°C for 12 days, and colony counts were recorded.
The effect of crude aqueous leaf extracts of citrus, periwinkle, tobacco, eggplant, and psyllids on GUS activity in L. crescens cells (carrying pLF057 or pLF058) was examined using the semiquantitative histochemical GUS substrate 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc). One milliliter of 5-day-old BT-1 cell cultures was treated and incubated overnight in the presence of 50 μl of X-Gluc (40 mg ml−1 dimethyl sulfoxide [DMSO]) and checked for the development of blue colonies after incubation for 16 h. Alternatively, for the fluorimetric GUS assay, 1 ml of bacterial cultures was harvested at 2,013 × g for 15 min at 4°C. The bacterial pellets were resuspended in 30 μl of GUS extraction buffer (50 mM Na2HPO4, pH 7.0, 10 mM β-mercaptoethanol, 10 mM Na2EDTA, pH 8.0, and 0.1% Triton X-100) and incubated for 10 min at 37°C. Fluorogenic GUS substrate (4-methylumbelliferyl β-d-glucuronide [MUG]) (1 mM in GUS extraction buffer) was added to the bacterial lysate (90 μl of substrate solution per 10 μl of GUS extract) and incubated at 37°C. Reaction mixture aliquots (100 μl) were withdrawn at 10, 40, and 70 min, stopped with 50 μl of 0.2 M Na2CO3, and measured for fluorescence. The fluorescence data were normalized against a standard curve prepared using the purified GUS enzyme (type IX-A from E. coli; Sigma-Aldrich, Inc., St. Louis, MO). The data presented are mean values derived from two biological assays run in triplicate.
RESULTS
Select putative SC1 and SC2 late genes were upregulated in periwinkle relative to citrus.
To identify actively expressed late genes of SC1 and SC2, reverse transcription-quantitative PCR (RT-qPCR) was carried out in both “Ca. Liberibacter asiaticus”-infected citrus (in which phage particles have not been seen) and “Ca. Liberibacter asiaticus”-infected periwinkle plants (in which phage particles are consistently visualized in the phloem of infected plants). Comparative expression analyses of the putative phage lytic cycle genes SC1_gp025, SC2_gp095, SC2_gp100, and SC1_gp110 (holin) revealed significantly higher levels of phage transcripts in periwinkle than citrus (P values of 0.002850, 0.002338, 0.000001, and 0.000034, respectively). In contrast, “Ca. Liberibacter asiaticus” chromosomal gene prfA transcript levels were similar in both plant hosts, demonstrating plant-specific upregulation of bacteriophage late genes in periwinkle (Fig. 1).
FIG 1.
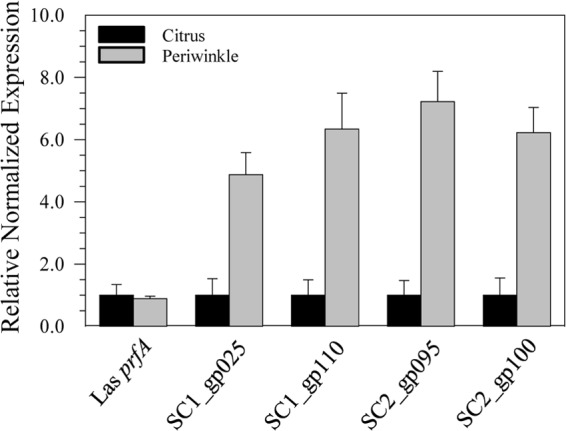
Relative expression of chromosomal gene prfA and bacteriophage genes SC1_gp025 (tail fiber), SC1_gp110 (holin), SC2_gp095 (peroxidase), and SC2_gp100 (glutathione peroxidase). Transcript expression levels were calculated by the ΔΔCT method with citrus as the calibrator sample, and the data were normalized to the chromosomal reference gene gyrB. Error bars represent the standard errors of the means.
SC1_gp035 (endolysin) and SC1_gp110 (holin) had functional activity consistent with their annotations.
As expected, within the two predicted loci annotated as endolysins (SC1_gp035 and CLIBASIA_04790), neither transmembrane helices nor secretion signals were detected. Although CLIBASIA_04790 is located well outside the prophage region of Psy62, the locus was annotated as a phage-related lysozyme, and blastp confirmed that this protein belongs to the lysozyme-like superfamily, which includes phage endolysins. Expression of CLIBASIA_04790 using pET27b in E. coli reduced growth after induction with IPTG by 12.8% (from an OD600 of 0.73 to an OD600 of 0.64) over the next 5 h of cultivation (data not shown). In contrast, expression of prophage gene SC1_gp035 in E. coli led to cessation of growth beginning 30 min after induction with IPTG and a substantial reduction in culture turbidity, with final OD600 values 33% lower than those of the controls (Fig. 2).
FIG 2.
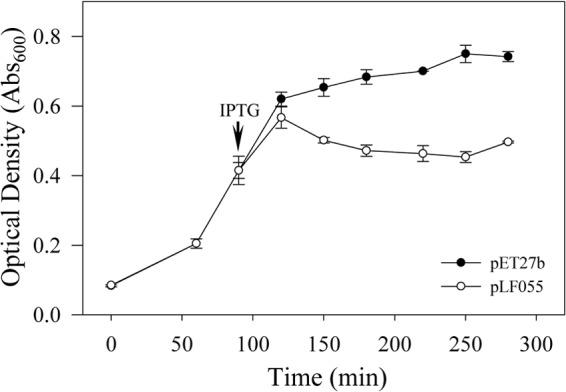
Growth kinetics of E. coli BL21(DE3)/pET27b (empty vector) or BL21(DE3)/pLF055 (expressing SC1_gp035; endolysin) in LB medium with IPTG added at the time point indicated.
TMHMM and TMpred analyses of the full-length protein sequence of the predicted holin, SC1_gp110, revealed a single transmembrane α-helix, consistent with class III holins, with the N terminus predicted in the periplasm and the C terminus in the cytoplasm (N-out, C-in topology). SC1_gp110 was cloned from the predicted start codon M1 to the predicted stop codon. In E. coli TOP10 cells, full-length SC1_gp110 in pCR2.1-TOPO reduced colony diameter when inserted in the same orientation as the lacZ promoter on the vector (data not shown). Controlled heterologous overexpression of SC1_gp110 in pET27b abruptly stopped growth of E. coli BL21(DE3) upon addition of IPTG to liquid cultures but did not cause a significant drop in OD600 values, consistent with holin activity (Fig. 3).
FIG 3.
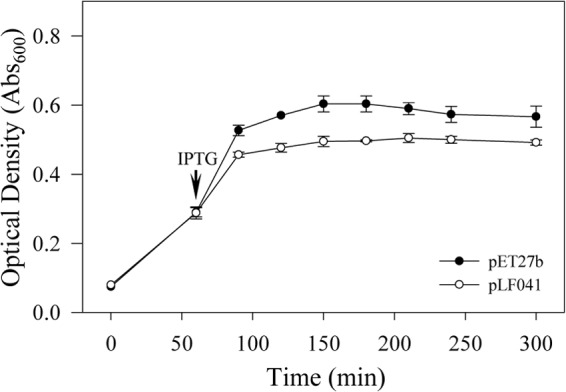
Growth kinetics of E. coli BL21(DE3)/pET27b (empty vector) or BL21(DE3)/pLF041 (expressing full-length SC1_gp110; holin) in LB medium with IPTG added at the time point indicated.
A second potential, but truncated, holin construct was created that began at M35, downstream from the first ATG translational start codon and in frame with the predicted full-length holin; this potential start codon also had a potential Shine-Dalgarno sequence (5′-AGGCG-3′) located from 6 to 10 bp upstream of the potential M35 start site. This truncated version was cloned in pET27b, but induction of expression did not affect the growth of E. coli (data not shown).
Transformation of L. crescens strain BT-1.
L. crescens strain BT-1 was consistently transformed by electroporation of the repW vector pUFR071 at high efficiencies (typically, 6 × 104 CFU/μg of DNA). Stability of pUFR071 was evaluated; this plasmid was >95% stable, without selection, when grown in BM7 medium for over 20 generations. pUFR071 was reextracted from transformed BT-1, transformed into E. coli, and appeared from restriction analysis to be unchanged (not shown).
The promoter region of SC1_gp110 (holin) was constitutively active in BT-1 but suppressible.
Four differently sized potential promoter-active regions upstream of SC1_gp110 were cloned in front of a promoterless lacZ gene to form reporter constructs pLF044, pLF045, pLF046, and pLF047. E. coli colonies carrying these plasmids were white on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) medium, indicating a lack of holin promoter activity to drive lacZ expression. However, pLF057, carrying a PciI/HindIII fragment of the holin promoter cloned directly from pLF044 and fused to a promoterless GUS reporter gene, showed constitutive and strong GUS activity in L. crescens strain BT-1 (Fig. 4A). Histochemical GUS activity was apparent beginning 6 to 7 h after incubation with X-Gluc and continued to intensify for approximately 24 h until BT-1 cells died, presumably from the relatively high incubation temperature (37°C) used for the GUS assay.
FIG 4.
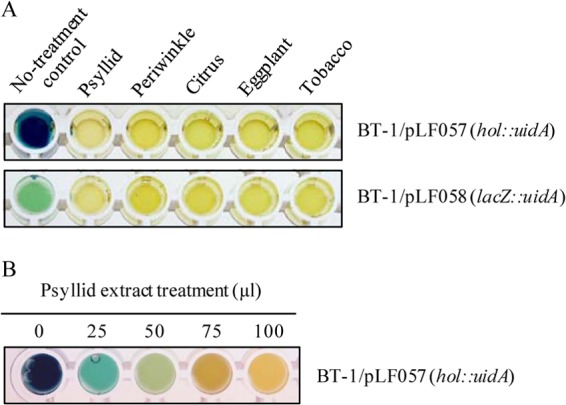
Qualitative GUS activity assays. (A) Histochemical GUS activity in BT-1 cells after treatment with 100-μl extracts, as indicated. (B) Histochemical GUS activity in BT-1/pLF057 (hol::uidA) cells after treatment with 25, 50, 75, and 100 μl of psyllid extract. Photos were taken after 16 h.
Significant inhibition of holin promoter-driven GUS activity in BT-1 cells was observed when the cells were treated with 100 μl of crude aqueous extracts from psyllids as well as from periwinkle, citrus, eggplant, and tobacco leaves (Fig. 4A, upper wells). Dose-dependent inhibition of visible GUS activity was observed in BT-1/pLF057 (hol::uidA) cells treated with 25-, 50-, 75-, and 100-μl volumes of crude psyllid extracts (Fig. 4B) as well as from plant sources (Fig. 5A). These results appeared to indicate that crude aqueous extracts from all host sources served to suppress holin promoter activity in a dose-dependent manner. However, a strong reduction in GUS activity after treatment with 75- and 100-μl volumes of crude aqueous extracts from all five sources was also seen in GUS assays using the constitutive lacZ promoter in pLF058 (lacZ::uidA) Fig. 4A, lower wells, and 5B), indicating that nonspecific inhibition of GUS activity occurred at the higher concentrations of aqueous extracts used. Notably, the observed decline in enzymatic activity, even at the highest concentrations, could not be attributed to any significant loss of cell viability due to overnight treatment of BT-1 cultures with 100 μl of psyllid or plant leaf extracts (Fig. 6).
FIG 5.
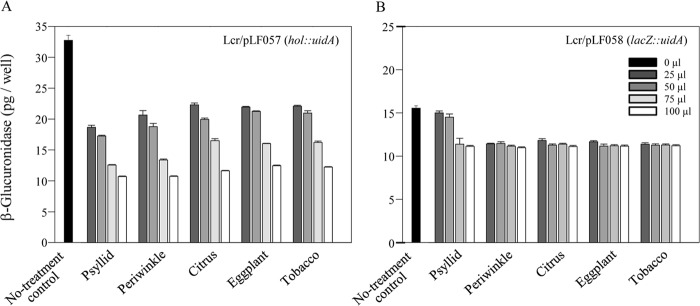
Quantitative (fluorimetric) GUS activity assays. (A) Quantification of GUS activity in BT-1/pLF057 (hol::uidA) cells treated with the aqueous crude extracts prepared from the indicated “Ca. Liberibacter asiaticus” hosts. (B) Quantification of GUS activity in BT-1/pLF058 (lacZ::uidA) cells following treatment with the “Ca. Liberibacter asiaticus” host extracts as described for panel A.
FIG 6.
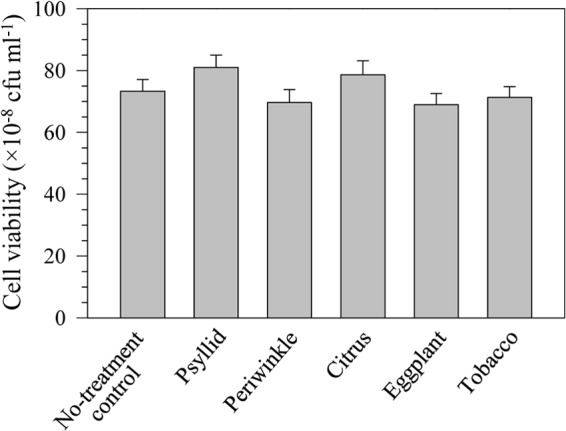
Viability of BT-1 cells following treatment with 100 μl of crude aqueous extracts prepared from the indicated “Ca. Liberibacter asiaticus” hosts after 16 h of incubation, relative to a water-treated control.
Quantitative MUG-based assays revealed that at the lower treatment levels (25 μl and 50 μl per 1 ml of culture), psyllid extracts inhibited GUS activity expressed by the holin promoter by 43% and 47%, respectively, compared to the untreated control BT-1/pLF057 cultures (Fig. 5A). In the same activity assays, psyllid extracts at these lower treatment levels failed to inhibit lacZ promoter-driven GUS activity to any significant level (Fig. 5B). The holin promoter displayed significantly stronger transcriptional activity in BT-1 cells [nearly 2.1-fold higher GUS activity in BT-1/pLF057 (hol::uidA) cells than the lacZ promoter driving the same GUS gene in BT-1/pLF058 (lacZ::uidA) cells] (Fig. 5).
DISCUSSION
“Ca. Liberibacter” species are an emerging group of phytopathogens that cause significant crop losses worldwide. The increased phage copy number and presence of phage particles in “Ca. Liberibacter asiaticus”-infected periwinkle, an artificial host of the bacterium, suggests that SC1 and/or SC2 is inducible and potentially useful for controlling HLB in citrus (8). This work shows that transcriptional levels of SC1 and SC2 late genes are significantly higher in “Ca. Liberibacter asiaticus”-infected periwinkle than in “Ca. Liberibacter asiaticus”-infected citrus, indicating lytic cycle induction. This is consistent with previous findings that phage particles were formed only in periwinkle but not in citrus. Phage SC1, which carries both the holin and endolysin genes, generally is found at a higher copy number than SC2 in both citrus and periwinkle (8).
The genomes of the three sequenced pathogenic liberibacters (“Ca. Liberibacter asiaticus,” “Candidatus Liberibacter americanus,” and “Candidatus Liberibacter solanacearum”) each contain two prophages that appear to have the full suite of genes necessary for functionality (8, 23, 24). Not all prophages are entirely latent during lysogeny; some prophage genes can increase the fitness and/or the virulence of the bacterial host cell but have no general role in the phage life cycle (25). Examples of prophage lysogenic conversion gene products include extracellular toxins, proteins that alter antigenicity, host effector proteins, enzymes that aid in intracellular survival, and proteins that facilitate bacterial attachment to host cells (26). Both SC1 and SC2 carry genes annotated as peroxidases; these gene products could potentially aid in defense against reactive oxygen species produced by the infected plant and/or insect host. The expression levels of the SC2 peroxidases SC2_gp095 and SC2_gp100 were higher in periwinkle than in citrus, and their potential role in plant infection is currently being investigated.
The holin-endolysin lysis system was the first mechanism described for the egress of fully formed double-strand DNA bacteriophages from Gram-negative bacteria. Holins are highly diverse transmembrane proteins that are produced throughout the period of late gene expression (27). Holins can have one, two, or three α-helical transmembrane domains and are categorized as class III, II, or I holins, respectively. Upon reaching a critical concentration, the holin proteins aggregate, triggering the formation of a few large “micron-scale” holes in the cytoplasmic membrane (28). The endolysin, a muralytic enzyme, is also expressed throughout the period of late gene expression and accumulates in the cytosol. After holins trigger disruption of the inner membrane, the endolysins obtain access to the bacterial murein layer, and cell wall degradation occurs.
This work shows that SC1_gp110, annotated as a holin, and SC1_gp035, annotated as an endolysin, exhibited toxic effects on E. coli that were consistent with the annotations. The predicted presence of a single α-helical transmembrane domain of SC1_gp110 is consistent with class III holins such as t of bacteriophage T4 (29). Unlike t, the prototypical class III holin, SC1_gp110 has an N-out, C-in membrane topology similar to Gp5, a potential class III holin of mycobacteriophage Ms6 (30). Because holins often have open reading frames embedded into the primary coding sequence (31), the effect of a second, N-terminally truncated fragment of SC1_gp110 was evaluated in E. coli. This region appeared to have a Shine-Dalgarno ribosomal binding sequence but was not predicted to have α-helical secondary structure and had no detectable effect on the growth of E. coli. Controlled overexpression of the predicted endolysin (SC1_gp035) led to lysis and a significant decrease in the OD600 of E. coli cultures.
The lack of holin promoter activity in driving the LacZ reporter in E. coli may indicate the absence of a transcriptional activator necessary to drive the expression of typically repressed downstream genes. An alternative explanation is that E. coli carries a repressor that recognizes the holin promoter and silences the expression of the reporter gene. The latter explanation seems less likely due to the large phylogenetic distance between E. coli and “Ca. Liberibacter asiaticus.” Similarly, the constitutive holin promoter activity in L. crescens may be due to the presence of a transcription factor in L. crescens that recognizes the “Ca. Liberibacter asiaticus” holin promoter or to the absence of a late gene or holin-specific repressor.
The general inhibition of GUS activity by the various plant extracts and the two highest concentrations of psyllid extract used masked any potential detection of transcriptional-level activity since the presumably constitutive lacZ-driven reporter and the holin-driven reporter were both similarly affected by these extracts (Fig. 5). The GUS enzyme is quite robust; thus, the decrease in GUS activity is not likely due to direct enzyme inhibition. Based on cell viability assays (Fig. 6), the plant extracts did not have cytotoxic effects. The four plants used in this study (citrus, periwinkle, tobacco, and eggplant) are all rich in polyphenols and other secondary metabolites, and these compounds may have a negative effect on cellular metabolism and growth. (Growth rates of treated and control L. crescens cultures were not assessed largely because treatment durations of 14 to 15 h were too short to significantly impact L. crescens growth.) Although the plant extracts interfered with the GUS reporter used in these assays, the holin promoter was clearly more active in periwinkle than in citrus (Fig. 1).
At lower concentrations, crude aqueous psyllid extracts appeared to have a strong and specific inhibitory effect on expression of the GUS reporter driven by the holin promoter (43% and 47% reductions were observed using 25 and 50 μl of crude psyllid extracts, while the same treatments had no significant effect on the same reporter driven by lacZ) (Fig. 5) We hypothesize the existence of one or more ligands in the psyllid extract that is able to penetrate L. crescens cells and bind to, and interfere with, a holin promoter transcriptional activator, leading to decreased transcription. A potential model system is the MarR family of transcription factors, which can function as either an activator or repressor following ligand binding (32). Based on the increased transcriptional activity of the holin promoter in periwinkle (Fig. 1), a similarly acting ligand is also likely to occur in citrus. Further research will focus on the identification of potential transcription activators present in L. crescens as well as potential inhibitory ligands in psyllids and citrus. The elucidation of the mechanism behind lytic cycle activation of SC1 and/or SC2 may lead to a new method for HLB disease control.
The strong activity of the holin promoter in the absence of psyllid or plant extracts may help explain why liberibacters other than BT-1 have not been cultured to date. Regardless of whether phage particles form, expression of a holin alone is sufficient to stop bacterial growth. Once “Ca. Liberibacter asiaticus” bacteria are separated from a host, any host-provided ligand that may suppress transcriptional activity will become increasingly dilute, and prophage late gene expression will be derepressed. In addition, the comparative size advantage of L. crescens (1.5-Mb genome) relative to the pathogenic liberibacters, including “Ca. Liberibacter asiaticus” (1.26-Mb genome), may provide genes needed for free-living culture that are not found in other liberibacters. We speculate that factors present in hosts may suppress phage activation and may be necessary, but not alone sufficient, for free-living culture of “Ca. Liberibacter asiaticus” bacteria.
ACKNOWLEDGMENTS
We thank Patricia Rayside for excellent technical assistance and Eric Rohrig and Gloria Lotz (FDACS Division of Plant Industry) for providing psyllids.
This work was supported by the Florida Citrus Research and Development Foundation, projects 723 and 769.
Footnotes
Published ahead of print 25 July 2014
REFERENCES
- 1.Planet P, Jagoueix S, Bové JM, Garnier M. 1995. Detection and characterization of the African citrus greening Liberobacter by amplification, cloning, and sequencing of the rplKAJL-rpoBC operon. Curr. Microbiol. 30:137–141. 10.1007/BF00296198 [DOI] [PubMed] [Google Scholar]
- 2.Ammar E-D, Shatters RG, Jr, Lynch C, Hall DG. 2011. Detection and relative titer of Candidatus Liberibacter asiaticus in the salivary glands and alimentary canal of Diaphorina citri (Hemiptera: Psyllidae) vector of citrus huanglongbing disease. Ann. Entomol. Soc. Am. 104:526–533. 10.1603/AN10134 [DOI] [Google Scholar]
- 3.Ammar E-D, Shatters RG, Jr, Hall DG. 2011. Localization of Candidatus Liberibacter asiaticus, associated with citrus huanglongbing disease, in its psyllid vector using fluorescence in situ hybridization. J. Phytopathol. 159:726–734. 10.1111/j.1439-0434.2011.01836.x [DOI] [Google Scholar]
- 4.Inoue H, Ohnishi J, Ito T, Tomimura K, Miyata S, Iwanami T, Ashihara W. 2009. Enhanced proliferation and efficient transmission of Candidatus Liberibacter asiaticus by adult Diaphorina citri after acquisition feeding in the nymphal stage. Ann. Appl. Biol. 155:29–36. 10.1111/j.1744-7348.2009.00317.x [DOI] [Google Scholar]
- 5.Gottwald TR. 2010. Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48:119–139. 10.1146/annurev-phyto-073009-114418 [DOI] [PubMed] [Google Scholar]
- 6.Leonard MT, Fagen JR, Davis-Richardson AG, Davis MJ, Triplett EW. 2012. Complete genome sequence of Liberibacter crescens BT-1. Stand. Genomic Sci. 7:271–283. 10.4056/sigs.3326772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan Y, Zhou L, Hall DG, Li W, Doddapaneni H, Lin H, Liu L, Vahling CM, Gabriel DW, Williams KP, Dickerman A, Sun Y, Gottwald T. 2009. Complete genome sequence of citrus huanglongbing bacterium, “Candidatus Liberibacter asiaticus” obtained through metagenomics. Mol. Plant Microbe Interact. 22:1011–1020. 10.1094/MPMI-22-8-1011 [DOI] [PubMed] [Google Scholar]
- 8.Zhang S, Flores-Cruz Z, Zhou L, Kang B-H, Fleites LA, Gooch MD, Wulff NA, Davis MJ, Duan Y-P, Gabriel DW. 2011. “Ca. Liberibacter asiaticus” carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant infections. Mol. Plant Microbe Interact. 24:458–468. 10.1094/MPMI-11-10-0256 [DOI] [PubMed] [Google Scholar]
- 9.Lieb M. 1964. Ultraviolet sensitivity of Escherichia coli containing heat-inducible λ prophages. Science 145:175–176. 10.1126/science.145.3628.175 [DOI] [PubMed] [Google Scholar]
- 10.Otsuji N, Sekiguchi M, Iijima T, Takagi Y. 1959. Induction of phage formation in the lysogenic Escherichia coli K-12 by mitomycin C. Nature 184(Suppl 14):1079–1080. 10.1038/1841079b0 [DOI] [PubMed] [Google Scholar]
- 11.Young R. 2013. Phage lysis: do we have the hole story yet? Curr. Opin. Microbiol. 16:790–797. 10.1016/j.mib.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young R, Wang I-N, Roof W. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8:120–128. 10.1016/S0966-842X(00)01705-4 [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch EF, Maniatis TA. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 14.Fagen JR, Leonard MT, McCullough CM, Edirisinghe JN, Henry CS, Davis MJ, Triplett EW. 2014. Comparative genomics of cultured and uncultured strains suggests genes essential for free-living growth of Liberibacter. PLoS One 9:e84469. 10.1371/journal.pone.0084469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jagoueix S, Bové JM, Garnier M. 1996. PCR detection of the two “Candidatus” liberobacter species associated with greening disease of citrus. Mol. Cell. Probes 10:43–50. 10.1006/mcpr.1996.0006 [DOI] [PubMed] [Google Scholar]
- 16.Zhou LJ, Gabriel DW, Duan YP, Halbert SE, Dixon WN. 2007. First report of dodder transmission of huanglongbing from naturally infected Murraya paniculata to citrus. Plant Dis. 91:227. 10.1094/PDIS-91-2-0227B [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 18.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 19.Hofmann K, Stoffel W. 1993. TMbase: a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166 [Google Scholar]
- 20.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 21.Rashtchian A, Buchman GW, Schuster DM, Berninger MS. 1992. Uracil DNA glycosylase-mediated cloning of polymerase chain reaction-amplified DNA: application to genomic and cDNA cloning. Anal. Biochem. 206:91–97. 10.1016/S0003-2697(05)80015-6 [DOI] [PubMed] [Google Scholar]
- 22.De Feyter R, Gabriel DW. 1991. Use of cloned DNA methylase genes to increase the frequency of transfer of foreign genes into Xanthomonas campestris pv. malvacearum. J. Bacteriol. 173:6421–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin H, Lou B, Glynn JM, Doddapaneni H, Civerolo EL, Chen C, Duan Y, Zhou L. 2011. The complete genome sequence of “Candidatus Liberibacter solanacearum,” the bacterium associated with potato zebra chip disease. PLoS One 6:e19135. 10.1371/journal.pone.0019135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wulff NA, Zhang S, Setubal JC, Almeida NF, Martins EC, Harakava R, Kumar D, Thiberio Rangel L, Foissac X, Bové JM, Gabriel DW. 2014. The complete genome sequencer of “Candidatus Liberibacter americanus,” associated with citrus huanglongbing. Mol. Plant Microbe Interact. 27:163–176. 10.1094/MPMI-09-13-0292-R [DOI] [PubMed] [Google Scholar]
- 25.Brüssow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602. 10.1128/MMBR.68.3.560-602.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyd EF, Brüssow H. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521–529. 10.1016/S0966-842X(02)02459-9 [DOI] [PubMed] [Google Scholar]
- 27.Wang I-N, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799–825. 10.1146/annurev.micro.54.1.799 [DOI] [PubMed] [Google Scholar]
- 28.Dewey JS, Savva CG, White RL, Vitha S, Holzenburg A, Young R. 2010. Micron-scale holes terminate the phage infection cycle. Proc. Natl. Acad. Sci. U. S. A. 107:2219–2223. 10.1073/pnas.0914030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young R, Bläsi U. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 17:191–205. 10.1016/0168-6445(94)00079-4 [DOI] [PubMed] [Google Scholar]
- 30.Catalão MJ, Gil F, Moniz-Pereira J, Pimentel M. 2011. Functional analysis of the holin-like proteins of mycobacteriophage Ms6. J. Bacteriol. 193:2793–2803. 10.1128/JB.01519-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bläsi U, Young R. 1996. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol. Microbiol. 21:675–682. 10.1046/j.1365-2958.1996.331395.x [DOI] [PubMed] [Google Scholar]
- 32.Perera IC, Grove A. 2010. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell Biol. 2:243–254. 10.1093/jmcb/mjq021 [DOI] [PubMed] [Google Scholar]


