Abstract
The marine cyanobacterium Synechococcus sp. strain PCC 7002 synthesizes two alkenes, 1-nonadecene and 1,14-nonadecadiene. Whereas the genetic basis for the biosynthesis of the terminal double bond in both alkenes has been characterized, the origin of the internal double bond in 1,14-nonadecadiene has not. In this study, we demonstrate that a gene encoding an uncharacterized desaturase is involved in the formation of the internal double bond of 1,14-nonadecadiene. Further, at low temperatures, the desaturase gene is essential for growth, and in wild-type cells the levels of 1,14-nonadecadiene increase relative to that of cells grown at 38°C. These data suggest that 1,14-nonadecadiene plays a role in responding to cold stress.
INTRODUCTION
Hydrocarbon biosynthesis is a common trait among both eukaryotes and prokaryotes that has gained significant attention for use in developing sustainable alternatives to petroleum (1–3). Although it has been known for decades that cyanobacteria accumulate hydrocarbons with C17 to C20 chains (4, 5), the cellular function of these molecules and the biosynthetic pathways responsible for their production remain poorly understood. A recent survey of sequenced cyanobacteria discovered that all strains possess one, but not both, of two pathways for producing saturated or unsaturated straight-chain hydrocarbons (6), indicating that hydrocarbons play an important role in cyanobacterial physiology. The majority of strains harbor the FAR/ADO pathway that was first characterized in Synechococcus elongatus PCC 7942 (7). The pathway synthesizes saturated alkanes (heptadecane and pentadecane) from acyl-acyl carrier proteins (acyl-ACP) via the action of an acyl-ACP reductase and an aldehyde-deformylating oxygenase. Conversely, a smaller percentage of cyanobacteria produce long-chain alpha-olefins via the Ols pathway, first characterized in Synechococcus sp. strain PCC 7002 (8). The Ols pathway is comprised of a single, multidomain enzyme (Ols) possessing modular organization similar to that of a polyketide synthase. Based on the protein's domain architecture and experimental feeding studies, Ols was predicted to produce alpha-olefins via elongation and decarboxylation of C18 acyl-ACP, the terminal product of fatty acid biosynthesis and substrate for phospholipid synthesis (8). Unlike other cyanobacterial strains that possess a more complex hydrocarbon profile in terms of length and degree of unsaturation (4, 5, 9), Synechococcus sp. strain PCC 7002 synthesizes only two alkenes: 1-nonadecene (C19:1) and 1,14-nonadecadiene (C19:2). The only difference between these compounds is the presence of an internal double bond at position 14 of the C19:2 hydrocarbon. Whereas the biosynthesis of the terminal double bond in both hydrocarbons was explained by the decarboxylation and dehydration reactions performed by the Ols domains, the biosynthesis of the internal double bond in 1,14-nonadecadiene was not. In this study, we linked the presence of this internal double bond to a gene predicted to encode a desaturase. Further, we demonstrated an increase in C19:2 abundance as an inverse function of temperature. This finding suggests that the hydrocarbons play a role in responding to cold stress similar to the way unsaturated fatty acid content in the cell membrane is modulated in many bacteria (10).
MATERIALS AND METHODS
Reagents, media, and growth conditions.
Enzymes and reagents were purchased from New England BioLabs or Fisher Scientific unless otherwise noted. Oligonucleotides used in this study were purchased from Integrated DNA Technologies, Inc., and are listed in Table 1. Isolation of genomic DNA and purification of DNA fragments were performed with commercial reagents (Promega, Qiagen).
TABLE 1.
Oligonucleotides used in this studya
| No. | Oligonucleotide | Sequence (5′→3′) |
|---|---|---|
| 1 | desA-a2 | CCGTTCTGCAGGTTTCTTGGCGCAAGGGTTACAGCTTCC |
| 2 | desA-a1 | GCAACCATGGGAAACCCAACGCAAGG |
| 3 | desA-b2 | CCGTTGGATCCGTACGCTTCCATACCATGTTCACCAATATCG |
| 4 | desA-b1 | CCTCACAGGTTCGGCCTACAGTGG |
| 5 | desB-a2 | CCGTTCTGCAGCTTTACAACCCCTAATCCGCCTTTATTCATTTCC |
| 6 | desB-a1 | CAGATCGAGGGGAACCTGGTTTGCG |
| 7 | desB-b2 | CCGTTGGATCCCAACAACGCCTTGCAGAAAATCCCCAGC |
| 8 | desB-b1 | CCAGTTTTAACAGACCTTGGGTAAAGGCTTC |
| 9 | desC-a2 | CCGTTCTGCAGCCTTGTCACCTACGGCGAAGGTTGG |
| 10 | desC-a1 | GGAATACACTGACGAATACCGCGATGGG |
| 11 | desC-b2 | CCGTTGGATCCGGTAATTCCAATGCCGCCGGTAATCCAGT |
| 12 | desC-b1 | GGTTTCTGGGCGAATTGCGATCTTGAGG |
| 13 | desE-a2 | CCGTTCTGCAGATTGTTGTCCCCAAAGGGAAATATCTATTCCG |
| 14 | desE-a1 | GACCCAGAAAACCCCGTAGAATAATCACG |
| 15 | desE-b2 | CCGTTGGATCCAATGCTTTCCTAACGAGTTGAGAATATCTTCTATG |
| 16 | desE-b1 | CGTCGATTTTGCCTCATTAATTTAGTTAAAGCAGC |
| 17 | desF-a2 | CCGTTCTGCAGGAATAAATTTGCGTTTGATCATTACCGCCAATTC |
| 18 | desF-a1 | GGTTGAAACCATTTAGGGAAACCCATACTG |
| 19 | desF-b2 | CCGTTGGATCCGAATTCTCAAACAATAGAACAAGACAAAGGGGAATATC |
| 20 | desF-b1 | CGAGGCAGGTTTTGAGAGCGTCAAC |
| 21 | desE-US-Fw | TATGCACATATGCGTCGATTTTGCCTCATTAATTTAGTTAAAGCAGC |
| 22 | desE-US-Rv | GCAGTTTCATTTGATGCTCGATGAGTTTTTCTAAGCTTTCCTAACGAGTTGAGAATATCTTCTATGAAACCG |
| 23 | KM-Fw | TTAGAAAAACTCATCGAGCATCAAATGAAACTGC |
| 24 | KM-Rv | GGACTCTTCTCTACAGGTGGGTATAGATTTGTTAAGCTTTGGCAGGATCCGGCTGCTAACAAA |
| 25 | Cpc-prom-Fw | CTTAACAAATCTATACCCACCTGTAGAGAAGAGTCC |
| 26 | Cpc-prom-Rv | GGGTCAAGAACGTTGCTGTAATGCGTCATGGAATTAATCTCCTACTTGACTTTATGAGTTGGG |
| 27 | desE-DS-Fw | ACGCATTACAGCAACGTTCTTGACCC |
| 28 | desE-DS-Rv | CTGCCGCCAGGCAAATTCTGTTTTATCCATATGCTAGGGATTGGCCGCGTTTTGTAGATC |
| 29 | pBAD18-Fw | GATAAAACAGAATTTGCCTGGCGGCAG |
| 30 | pBAD18-desE-USRv | GCTGCTTTAACTAAATTAATGAGGCAAAATCGACGCATATGTGCATAGGAGAAACAGTAGAGAGTTGCGATAAAAAGCG |
| 31 | pBAD18-NdeI-Fw | CATATGGATAAAACAGAATTTGCCTGGCGGCAG |
| 32 | pBAD18-A-acsRv | TAGGAGGTTACGGGGAAAAGCCAATAGGCATATGTGCATAGGAGAAACAGTAGAGAGTTGCGATAAAAAGCG |
| 33 | A-acsA-Fw | CCTATTGGCTTTTCCCCGTAACCTCCTA |
| 34 | A-acsA-Rv | GCAGTTTCATTTGATGCTCGATGAGTTTTTCTAACCTCGGCAGCAAAGTCTGGTG |
| 35 | KM-desE-Rv | GTTTCGGTGGTGACAGTTTCTGGGCTTTGGCAGGATCCGGCTGCTAACAAA |
| 36 | desE-comp-Fw | CCCAGAAACTGTCACCACCGAAAC |
| 37 | desE-comp-Rv | GTGTCGCCCACAATTTCCTGACCCCCAGGGCATCGTTTTAGCAACG |
| 38 | B-acs-Fw | GGTCAGGAAATTGTGGGCGACAC |
| 39 | B-acs-Rv | CTGCCGCCAGGCAAATTCTGTTTTATCCATATGCCAACAAGCCTTTGCCGCTGATC |
| 40 | AAS675-a1 | TATGCACATATGCCTTCCATCACGTCGGCAGTAATTTC |
| 41 | AAS675-a2 | GCAGTTTCATTTGATGCTCGATGAGTTTTTCTAACAAGCCGAAATCATGGCTACAATCCTAC |
| 42 | KM-675-Rv | GAAGATTCCGCCATTCGGATCGCCTTTGGCAGGATCCGGCTGCTAACAAA |
| 43 | AAS675-b1 | GCGATCCGAATGGCGGAATCTTC |
| 44 | AAS675-b2 | CTGCCGCCAGGCAAATTCTGTTTTATCCATATGCACTGAGGCCACATCCGTCAAAATC |
| 45 | pBAD18-675Rv | GAAATTACTGCCGACGTGATGGAAGGCATATGTGCATAGGAGAAACAGTAGAGAGTTGCGATAAAAAGCG |
| 46 | AAS1977-a1 | TATGCACATATGGCAGTTCTGTAAGGCCCTACTAGAGG |
| 47 | AAS1977-a2 | GCAGTTTCATTTGATGCTCGATGAGTTTTTCTAAGTTTTCGCAGAATGGGTCATGGTGG |
| 48 | KM-1977-Rv | GCAATTTTCATCGCCACCCTTTAGAGGCTTTGGCAGGATCCGGCTGCTAACAAA |
| 49 | AAS1977-b1 | CCTCTAAAGGGTGGCGATGAAAATTGC |
| 50 | AAS1977-b2 | CTGCCGCCAGGCAAATTCTGTTTTATCCATATGCATTGACCCCAGGCTCAATCAGATTCC |
| 51 | pBAD18-1977Rv | CCTCTAGTAGGGCCTTACAGAACTGCCATATGTGCATAGGAGAAACAGTAGAGAGTTGCGATAAAAAGCG |
Restriction sites are underlined. Oligonucleotides 1 to 20 were used to construct integration cassettes for creating the desaturase knockouts (a1 and a2 primers were used for the upstream region of each gene, b1 and b2 for the downstream region). Oligonucleotides 21 to 30 were used to construct integration cassettes for creating the desE-up strain. Oligonucleotides 31 to 39 and 23 were used to amplify DNA fragments for subsequent Gibson assembly of an integration cassette used to create the ΔdesE+ complementation strain. Oligonocleotides 40 to 51 as well as 24 and 29 were used to construct integration cassettes for creating Δaas strains.
A wild-type strain of Synechococcus sp. strain PCC 7002 obtained from the Pasteur Culture Collection was grown photoautotrophically (140 μE m−2 s−1) on solid medium A (11) (supplemented with 1 mg/ml NaNO3) agar plates or in 20-ml liquid cultures sparged with air under constant illumination from cool-white lamps at the specified temperatures. When required, cultures were supplemented with streptomycin and/or kanamycin (final concentration, 100 μg/ml).
Strain construction.
Knockout mutants were constructed by homologous recombination using a linear DNA fragment containing a resistance marker flanked by ∼1,000 bases homologous to the regions flanking the corresponding gene. For the construction of desaturase knockouts (Table 2), the upstream and downstream flanking sequences of the desaturase genes (desA, SYNPCC7002_A2756; desB, SYNPCC7002_A0159; desC, SYNPCC7002_A2198; desE, SYNPCC7002_A2833; desF, SYNPCC7002_A1989) were amplified by PCR (Phusion polymerase) from genomic DNA. PCR products were digested with BamHI and PstI and gel purified. The aadA gene (Smr) was excised from plasmid pSRA81 (12) with PstI and BamHI. The fragments were mixed in a 3:1:3 ratio (left flank:Smr:right flank) and ligated with T4 DNA ligase. Ligation products were gel extracted, reamplified, and transformed into Synechococcus sp. strain PCC 7002 as described by Frigaard et al. (12).
TABLE 2.
Cyanobacterial strains used in this study
| Strain | Phenotype | Reference or source |
|---|---|---|
| Wild type | Wild-type Synechococcus sp. strain PCC 7002 | Pasteur culture collection |
| ΔdesA | ΔdesA::aadA | This study |
| ΔdesB | ΔdesB::aadA | This study |
| ΔdesE | ΔdesE::aadA | This study |
| ΔdesF | ΔdesF::aadA | This study |
| desE-up | Φ(PcpcAB-desE) | This study |
| ΔdesE+ | ΔdesE::aadA ΔacsA::desE-aphII | This study |
| Δols | Δols::aadA | 8 |
| Δols-desE-up | Φ(PcpcAB-desE) Δols::aadA | This study |
| Δaas675 | ΔSYNPCC7002_A0675::aphII | This study |
| Δaas1977 | ΔSYNPCC7002_A1977::aphII | This study |
For the construction of the desE-up strain, the upstream region of desE was replaced with the phycocyanin (cpcBA) promoter from Synechocystis sp. strain PCC 6803, a promoter that has been reported to have high activity in Synechococcus sp. strain PCC 7002 (13). The wild-type and Δols strains (8) were transformed with a DNA cassette assembled using the in vitro enzymatic assembly method described by Gibson et al. (14). The cassette contained an upstream flanking sequence of desE, the aphII gene that confers Kmr (from pJ206 plasmid; DNA 2.0, Menlo Park, CA), the cpcBA promoter, and the 5′ end of desE.
The complementation strain (ΔdesE+) was constructed by homologous recombination using a DNA cassette containing the native desE locus (including 390 bases immediately 5′ of the desE coding sequence predicted to encode the desE promoter), the aphII gene that confers Kmr, and the upstream and downstream flanking sequences of acsA (locus SYNPCC7002_A1838). The acsA locus (target for integration of desE) encodes an acetyl coenzyme A (CoA) ligase that confers sensitivity to exogenous acrylic acid. The conditional acrylic acid sensitivity has been used as a counterselection method to facilitate the integration of heterologous genes and segregation of the resulting mutants (15). Here, the heterologous DNA was assembled from four PCR products (primers are listed in Table 2) using the in vitro enzymatic assembly method described by Gibson et al. (14). The resulting DNA cassette was transformed into the ΔdesE strain. Recombinants were selected on Km. Complete segregation of the mutants was verified by colony PCR.
The aas knockouts were created by homologous recombination of a linear targeting cassette. Here, plasmids (pBAD18 backbone) containing a kanamycin resistance cassette (aphII gene) and the upstream and downstream regions of the corresponding genes (SYNPCC7002_A0675 and SYNPCC7002_A1977) were assembled from PCR products (primers are listed in Table 2) using the in vitro enzymatic assembly method described by Gibson et al. (14). Each plasmid was linearized before transformation. Complete segregation of the mutants was verified by colony PCR. Mutant strains were grown in air-sparged, liquid cultures supplemented with pentadecanoic acid (final concentration, 20 mg/liter) at 38°C.
Lipid analysis (GC-MS).
Cultures were grown to an optical density at 730 nm (OD730) of ∼1.0, centrifuged, resuspended in 3 ml of water, and extracted and analyzed by following previously described protocols (8, 16). Samples were analyzed using a Shimadzu GCMP QP2010S gas chromatograph mass spectrometer (GC-MS) equipped with an AOC-20i autoinjector and a Restek Rxi-5ms column (catalog no. 13423). The temperature program was a 100°C hold for 2 min, ramping up from 100°C to 150°C at 80°C per min, a hold for 4 min, ramping up from 150°C to 218°C at 4°C per min, ramping up from 218°C to 325°C at 80°C/min, and a hold at 325°C for 2.5 min. A sample injection temperature of 250°C and a volume of 1 μl was used, along with a 1:10 split ration. The MS was operated in scanning mode between 50 and 350 m/z. Quantification was achieved by comparison of integrated peaks with calibration curves of fatty acid methyl ester (FAME; Sigma) and 1-nonadecene standards (Fluka).
RESULTS AND DISCUSSION
Identification of a desaturase gene involved in alkene unsaturation.
Given the lack of functional handles on the olefins, we hypothesized that the internal bond of 1,14-nonadecadiene would be present in the substrate of the Ols-catalyzed elongation-decarboxylation mechanism, i.e., an unsaturated C18 acyl-ACP. Synechococcus sp. strain PCC 7002 synthesizes lipids that incorporate 18-carbon fatty acids with zero, one, two, or three double bonds at the Δ9, Δ12, and Δ15 (or ω3) positions at the sn-1 position of lipids and C16 fatty acids containing zero or one double bond at the Δ9 position at the sn-2 position of lipids (17–19). These unsaturated and polyunsaturated fatty acids are essential constituents of polar glycerolipids and are used to control the fluidity of membranes in response to changes in temperature (10). Three acyl-lipid desaturases, encoded by desA (Δ12), desB (Δ15), and desC (Δ9), have been shown to be involved in the biosynthesis of the unsaturated fatty acids observed in Synechococcus sp. strain PCC 7002 (20–22). Two additional genes are predicted to encode uncharacterized desaturases, desE (SYNPCC7002_A2833) and desF (SYNPCC7002_A1989). We hypothesized that one of the five desaturases was responsible for the internal double bond in 1,14-nonadecadiene.
To test this hypothesis, a disruption mutant of each desaturase was constructed by homologous recombination using a knockout cassette consisting of an antibiotic resistance gene flanked by homology targeting sequences. After multiple attempts, we were unable to obtain a ΔdesC mutant, suggesting that this mutation is lethal to the cells. The same problem was reported when trying to disrupt desC in Synechocystis sp. strain PCC 6803 (23). Conversely, fully segregated knockouts of desA, desB, desE, and desF were obtained after transformation and plating on the appropriate antibiotics. The observed fatty acid profiles of ΔdesA and ΔdesB mutants were consistent with past reports (20). Cells of the desA mutant contained 18:1(Δ9) but no 18:2(Δ9, Δ12) or 18:3(Δ9, Δ12, Δ15) fatty acids, and the cells of the desB mutant contained 18:1(Δ9) and 18:2(Δ9, Δ12) but no 18:3(Δ9, Δ12, Δ15) fatty acids. The hydrocarbon compositions of the ΔdesA, ΔdesB, and ΔdesF mutants were indistinguishable from that of wild-type Synechococcus sp. strain PCC 7002 (data not shown). Conversely, the hydrocarbon extract of the ΔdesE mutant contained no detectable C19:2 alkene (Fig. 1). When the deletion was complemented by inserting desE under its native promoter in the acsA locus (a location used previously for making chromosomal insertions [15]), 1,14-nonadecadiene production was restored. These results suggest that the desaturase encoded by desE is responsible for the internal double bond in C19:2. It should be noted that desF recently was reported to be essential for growth on plates under microoxic conditions (24), conditions that were not tested here.
FIG 1.
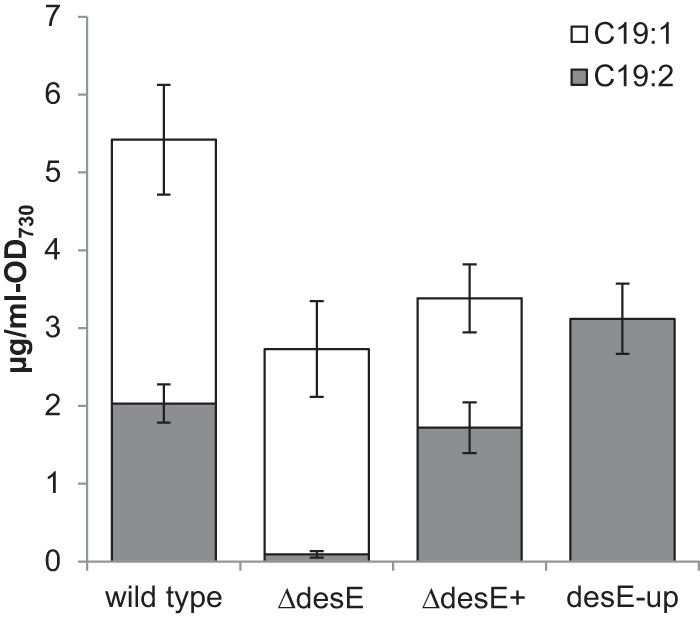
Comparison of the hydrocarbon composition from the wild-type, desE knockout (ΔdesE), ΔdesE complemented (ΔdesE+), and desE upregulated (desE-up) strains. The deletion of the desE gene eliminated only the production of the hydrocarbon with the internal double bond (C19:2). Cultures were grown autotrophically at 36°C with bubbling of air, and error bars represent the standard deviations from three biological replicates.
Precursors for 1,14-nonadecadiene biosynthesis.
The putative Ols pathway (8) calls for C18 acyl-ACP precursors to be processed via an elongation-decarboxylation mechanism catalyzed by a multimodular megasynthase. In the case of the C19:1 hydrocarbon, a fully saturated acyl-ACP (18:0) would be the precursor. However, for the C19:2 hydrocarbon to have an internal double bond at position 14, a C18 acyl-ACP with a double bond at position 13 would be required. This C18:1(Δ13) acyl-ACP could be directly synthesized by DesE acting on C18 acyl-ACP or could be the elongation product of a shorter unsaturated acyl-ACP. Potential DesE products consistent with this scheme include C16:1(Δ11), C14:1(Δ9), or C12:1(Δ7) acyl-ACPs. Compared to the ΔdesE mutant extract, the lipid profiles of the wild-type strain contained no additional fatty acids, providing no assistance in determining the DesE substrate.
The amino acid sequence of DesE is similar to the Δ9 desaturases of Rattus norvegicus (rat) and Saccharomyces cerevisiae (34 to 38%) (25). DesE contains the conserved three-histidine cluster motifs observed in Δ9 desaturases: the HXXXXH motifs (residues 79 to 84) and two HXXHH motifs (residues 116 to 120 and 242 to 246). These histidine motifs are thought to bind iron atoms and play an important role in the introduction of the double bond in the hydrocarbon chains of fatty acids (19). The high degree of similarity of DesE to Δ9 desaturases suggested that a C14 acyl-ACP was the most likely substrate to be consistent with the formation of 1,14-nonadecadiene.
If true, the C14:1(Δ9) acyl-ACP synthesized by DesE would be elongated to a C16:1(Δ11) and a C18:1(Δ13) acyl-ACP that ultimately would be the substrate for Ols (Fig. 2a). The lack of these intermediates in the lipid extracts of the wild-type strain suggested that they are not accumulating to a detectable extent. Therefore, we increased the expression of desE by replacing its promoter with the strong PcpcBA promoter from Synechocystis sp. strain PCC 6803 (13) (desE-up strain). The fatty acid profile of this strain contained no additional fatty acids compared to the wild type. Conversely, the hydrocarbon profile of desE-up was dramatically shifted from 35% C19:2 hydrocarbon in the wild type to nearly 100% C19:2 in the desE-up mutant (Fig. 1). These results suggest that the hydrocarbon profile is controlled by the relative level of Ols substrates and that the products of DesE are used exclusively in the formation of olefins. If this hypothesis is true, knocking out ols in the desE-up strain should result in the accumulation of unsaturated acyl-ACPs that ordinarily serve as intermediates in C19:2 biosynthesis. A Δols-desE-up strain was constructed, and its fatty acid profile was analyzed. Compared to the wild-type strain, the extract of the Δols-desE-up mutant contained three additional peaks in its chromatogram at retention times of 15.2, 19.6, and 19.9 min. One (15.2 min) was identified as 11-hexadecenoic acid, C16:1(Δ11), by comparison to a commercial standard (Fig. 2b). The mass spectra for the other two peaks were consistent with C18 unsaturated fatty acids, but no commercial standard for the C18:1(Δ13) fatty acid was available for structural confirmation; in order to corroborate the identity of one of these peaks as the C18:1(Δ13) fatty acid, a different approach was used.
FIG 2.
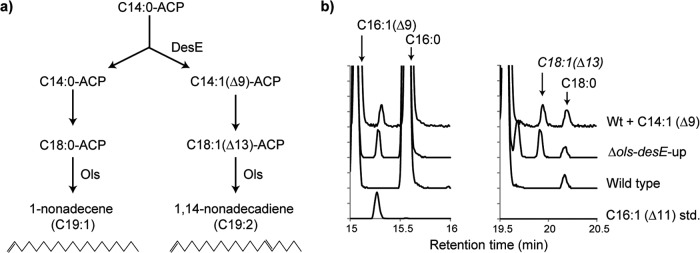
(a) Proposed route for alkene biosynthesis in Synechococcus sp. strain PCC 7002. DesE is required to place a double bond at the Δ9 position of the C14:0 acyl-ACP substrate. The product subsequently is elongated to a C18:1(Δ13) acyl-ACP that would serve as the precursor for 1,14-nonadecadiene biosynthesis by Ols. (b) Comparison of the fatty acid profiles of Synechococcus sp. strain PCC 7002 and mutants. Feeding of the C14:1(Δ9) fatty acid to the wild-type strain and overexpression of desE in the Δols strain resulted in the formation of the C16:1(Δ11) fatty acid (retention time, 15.2 min) and the C18:1(Δ13) fatty acid (retention time, 19.9 min). Attempts to chemically complement the ΔdesE mutant with C14:1(Δ9) fatty acid failed, as did attempts to chemically complement the Δols mutant with 1-nonadecene.
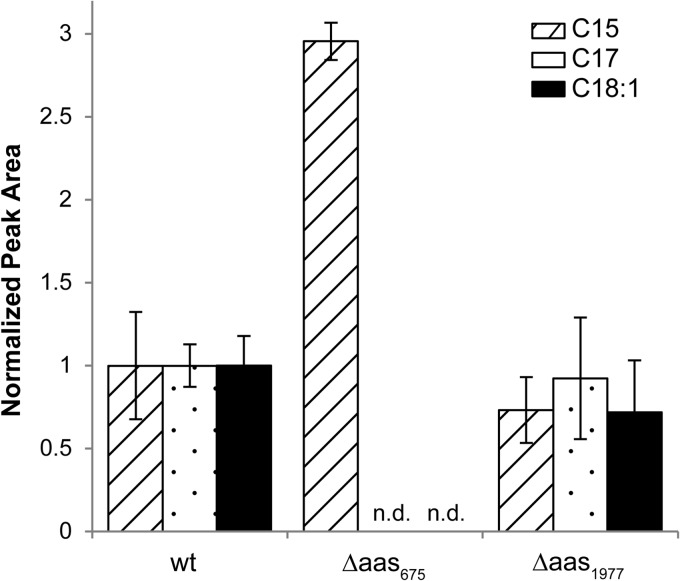
Exogenous fatty acids that are transported across the outer membrane can be incorporated into cyanobacterial lipid metabolism after being activated to the acyl-ACP form by an acyl-ACP synthetase (Aas). The Aas involved in this activation have been identified in Synechocystis sp. strain PCC 6803 and Synechococcus elongatus PCC 7942 (26). Although no Aas has been characterized in Synechococcus sp. strain PCC 7002, when pentadecanoic acid (C15) was fed to this strain, we detected the formation of heptadecenoic acid (C17) and 1-octadecene, showing that Synechococcus sp. strain PCC 7002 has the ability to elongate and incorporate exogenous fatty acids into lipid metabolism (8). Two enzymes with homology to the Aas from Synechocystis sp. strain PCC 6803 and Synechococcus elongatus PCC 7942 were identified in the genome of Synechococcus sp. strain PCC 7002 (SYNPCC7002_A0675 and SYNPCC7002_A1977). Knockout mutants were constructed for each gene (Δaas675 and Δaas1977 strains). After feeding C15 free fatty acid to each mutant, C17 fatty acid and 1-octadecene were not detected in the Δaas675 strain (Fig. 3) but were in the Δaas1977 strain, suggesting that the Aas encoded by SYNPCC7002_A0675 is responsible for the activation of exogenous fatty acids to acyl-ACPs, and its activity is required for the formation of the alkenes from exogenous fatty acids.
FIG 3.
Heptadecanoic acid (C17) and 1-octadecene (C18:1) formation after the addition of pentadecanoic acid (C15). The formation of C17 and C18:1 was observed for the wild-type (wt) and Δaa1977 strains but not for the Δaas675 strain. Peak areas obtained from GC-MS analysis were normalized to the volume, OD730, and the area of an internal standard; they are presented as a fraction of the wild-type-normalized area for the corresponding compound. Error bars represent the standard deviations from three biological replicates. n.d., not detected.
Therefore, to corroborate the identity of one of the peaks observed in the Δols-desE-up strain as C18:1(Δ13) fatty acid, 9-tetradecenoic acid, C14:1(Δ9), was fed to wild-type Synechococcus sp. strain PCC 7002 (Fig. 2b). Once activated by the Aas, the elongation of the C14:1(Δ9) fatty acid should result in the formation of the C16:1(Δ11) and C18:1(Δ13) acyl-ACPs. After analysis, feeding Synechococcus sp. strain PCC7002 with C14:1(Δ9) resulted in the formation of two additional peaks with retention times of 15.2 min, corresponding to C16:1(Δ11), and 19.9 min (Fig. 2b); these peaks were not observed when C14:1(Δ9) was fed to the Δaas675 strain. These data suggest that the peak at 19.9 min observed in the Δols-desE-up strain is indeed the C18:1(Δ13) fatty acid and that DesE is acting on C14 acyl-ACP.
Efforts to heterologously express DesE in Escherichia coli (Mistic tagged [27] and His tagged under various conditions) failed to generate soluble protein or changes in the fatty acid profile (data not shown). The hydropathy profile of DesE shows two highly hydrophobic regions, suggesting that it is an integral membrane protein (25), and further optimization of the expression construct will be needed to confirm the biochemical activity of DesE.
Effect of temperature in alkene unsaturation.
It is known that upon a downshift in temperature, expression of some desaturases is upregulated and the mRNA stability of desaturase genes is altered (22), resulting in an increase of the unsaturated lipids that are used to maintain membrane fluidity at low temperatures. In the case of desE, global transcriptome profiling via RNA-seq showed that its transcript levels increased approximately 2-fold in cells grown at 22°C compared to cells grown at 38°C (28). Also, it has been shown that desE transcripts had an estimated half-life of only 1 min at 38°C and 21 min at 22°C (25), suggesting that desE is regulated to respond to changes in temperature. To test if desE upregulation at low temperatures would affect the hydrocarbon composition of Synechococcus sp. strain PCC 7002, the wild-type strain was grown at different temperatures. At 38°C, the C19:2 hydrocarbon represented only 11% of the total hydrocarbon pool, whereas at 22°C it was 96% (Fig. 4a). Except for 38°C, most of these differences were generated by a decrease in C19:1 hydrocarbon rather than an increase of C19:2. For the ΔdesE strain, there was a slight decrease in C19:1 at low temperatures; however, on average, hydrocarbon levels remained constant across all temperatures (Fig. 4b). Interestingly, no C18:1(Δ13) fatty acid was detected at any of the temperatures tested, suggesting that this intermediate is not incorporated into lipids but rather is exclusively converted to hydrocarbon by Ols. Given that Synechococcus sp. strain PCC7002 does not contain a pathway for fatty acid catabolism (e.g., beta-oxidation), other uses of the C18:1(Δ13) acyl chain are not obvious.
FIG 4.
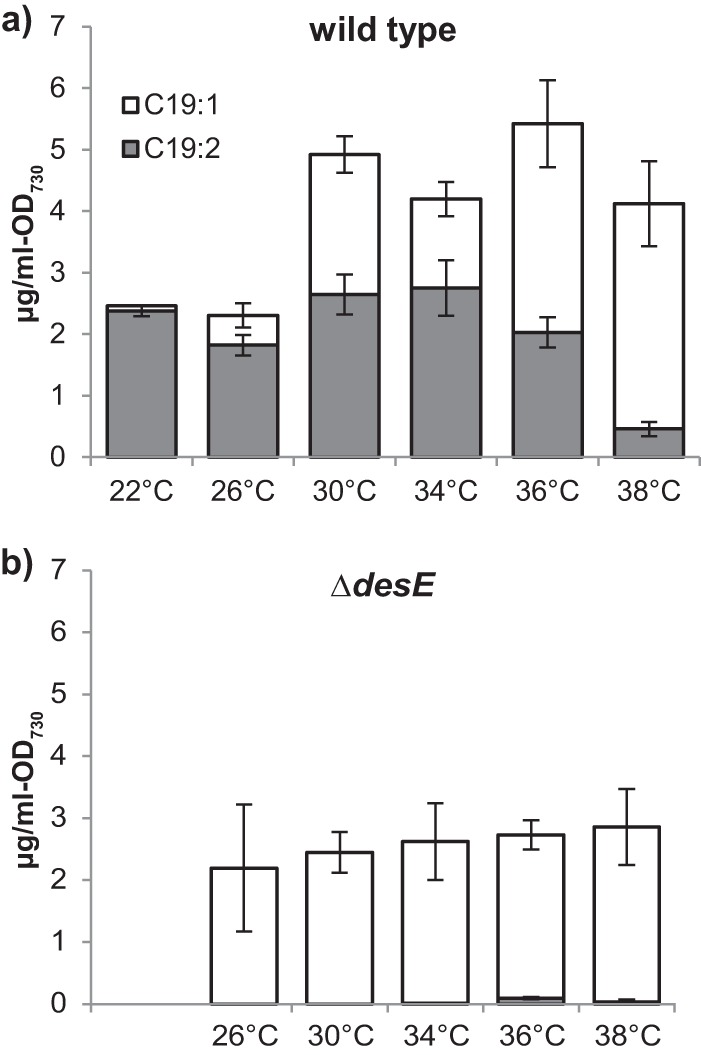
Comparison of the hydrocarbon composition from cultures grown at different temperatures. 1-Nonadecene (C19:1) levels decrease at low temperatures for the wild-type strain (a) but not for the ΔdesE strain (b). Error bars represent the standard deviations from three biological replicates.
Although the physiological functions of hydrocarbons in cyanobacteria remain poorly understood, several hypotheses have been proposed, including intra- or interspecies chemical signaling, prevention of desiccation, enhanced buoyancy, and membrane fluidity/stability (6). The alkenes synthesized by Synechococcus sp. strain PCC 7002 are a significant portion of the total lipid content in this strain (for example, at 36°C the total alkene content is ∼5.5 μg/ml-OD730 and the total fatty acid content is ∼35 μg/ml-OD730). Furthermore, olefins are found exclusively in the cell pellet and not in the spent media, indicating that olefins are not secreted outside the cell (results not shown). Therefore, the change in olefin composition with temperature suggested that these compounds are important for growth at low temperatures. To test this hypothesis, the growth of the wild-type, ΔdesE, and Δols strains was compared at 38°C and 22°C on solid media. As can be seen in Fig. 5a, the three strains grew at 38°C after 4 days; at 22°C, however, only the wild-type strain was able to grow to comparable levels, suggesting that the alkenes are important for growth at low temperatures. Similar results were observed when the strains were grown in liquid cultures (Fig. 5b).
FIG 5.
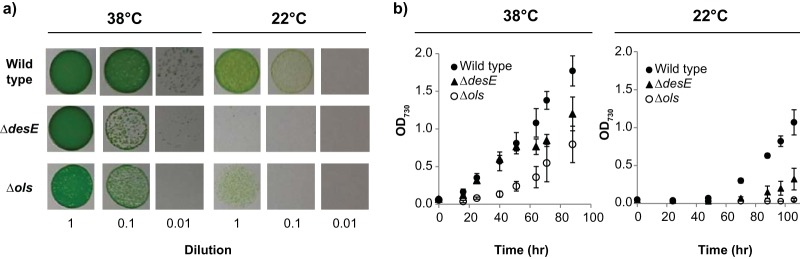
Effect of temperature on growth. (a) Cells were harvested from liquid cultures and suspended to the same initial cell concentration (OD730, ∼0.05) and then serially diluted (1:10). Seven μl of each dilution was plated on solid medium A (supplemented with 1 mg/ml NaNO3), and cells were grown for 4 days at the specified temperatures. (b) Growth curves for the wild-type (wt), ΔdesE, and Δols strains in liquid cultures.
Conclusions.
In summary, we demonstrated the involvement of a desaturase gene (desE) in the formation of the internal double bond in the 1,14-nonadecadiene synthesized by the cyanobacterium Synechococcus sp. strain PCC 7002. The amino acid sequence encoded by the desE gene shows a high degree of similarity to Δ9 desaturases, suggesting that its most likely substrate is a C14 fatty acid, which, after elongation to a C18:1(Δ13) fatty acid, would serve as the precursor for the formation of the hydrocarbon with the internal double bond at position 14. When expression of desE was increased by replacing its promoter, we observed the formation of only the C19:2 (and not the C19:1) hydrocarbon, suggesting that the products of DesE are earmarked for the hydrocarbon pathway. Moreover, since no hydrocarbons coming from the other unsaturated C18 fatty acids synthesized by Synechococcus sp. strain PCC 7002 have been detected, such as C18:1(Δ9) or C18:2(Δ9, Δ11), it seems that not only the presence of the additional unsaturation is important but also its location. Since the hydrocarbon content in Synechococcus sp. strain PCC 7002 represents about 10 to 15% of the total lipid content, the increase in C19:2 (diunsaturated hydrocarbon) production and the essentiality of DesE at low temperatures suggests that hydrocarbons play a role in responding to cold, possibly in maintaining membrane fluidity.
ACKNOWLEDGMENTS
We acknowledge the contributions of Jackie Cooper, Mick McGee, John Tyler Youngquist, Jeff Cameron, and Joe Villanueva to this project.
This work was funded by the United States Air Force Office of Scientific Research through grant FA9550-11-1-0038.
Footnotes
Published ahead of print 25 July 2014
REFERENCES
- 1.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. 2008. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19:556–563. 10.1016/j.copbio.2008.10.014 [DOI] [PubMed] [Google Scholar]
- 2.Rude MA, Schirmer A. 2009. New microbial fuels: a biotech perspective. Curr. Opin. Microbiol. 12:274–281. 10.1016/j.mib.2009.04.004 [DOI] [PubMed] [Google Scholar]
- 3.Lennen RM, Pfleger BF. 2013. Microbial production of fatty acid-derived fuels and chemicals. Curr. Opin. Biotechnol. 21:1044–1053. 10.1016/j.copbio.2013.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winters K, Parker PL, van Baalen C. 1969. Hydrocarbons of blue-green algae: geochemical significance. Science 163:467–468. 10.1126/science.163.3866.467 [DOI] [PubMed] [Google Scholar]
- 5.Han J, Calvin M. 1969. Hydrocarbon distribution of algae and bacteria, and microbiological activity in sediments. Proc. Natl. Acad. Sci. U. S. A. 64:436–443. 10.1073/pnas.64.2.436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coates RC, Podell S, Korobeynikov A, Lapidus A, Pevzner P, Sherman DH, Allen EE, Gerwick L, Gerwick WH. 2014. Characterization of cyanobacterial hydrocarbon composition and distribution of biosynthetic pathways. PLoS One 9:e85140. 10.1371/journal.pone.0085140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. 2010. Microbial biosynthesis of alkanes. Science 329:559–562. 10.1126/science.1187936 [DOI] [PubMed] [Google Scholar]
- 8.Mendez-Perez D, Begemann MB, Pfleger BF. 2011. Modular synthase-encoding gene involved in α-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 77:4264–4267. 10.1128/AEM.00467-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ladygina N, Dedyukhina EG, Vainshtein MB. 2006. A review on microbial synthesis of hydrocarbons. Process Biochem. 41:1001–1014. 10.1016/j.procbio.2005.12.007 [DOI] [Google Scholar]
- 10.Chapman D. 1975. Phase transitions and fluidity characteristics of lipids and cell membranes. Q. Rev. Biophys. 8:185–235. 10.1017/S0033583500001797 [DOI] [PubMed] [Google Scholar]
- 11.Stevens SEJ, Patterson CP, Myers J. 1973. The production of hydrogen peroxide by blue-green algae: a survey. J. Phycol. 9:427–430 [Google Scholar]
- 12.Frigaard N-U, Sakuragi Y, Bryant DA. 2004. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol. Biol. 274:325–340. 10.1385/1-59259-799-8:325 [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Alvey RM, Byrne PO, Graham JE, Shen G, Bryant DA. 2011. Expression of genes in cyanobacteria: adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002. Methods Mol. Biol. 684:273–293. 10.1007/978-1-60761-925-3_21 [DOI] [PubMed] [Google Scholar]
- 14.Gibson DG, Young L, Chuang R, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 15.Begemann MB, Zess EK, Walters EM, Schmitt EF, Markley AL, Pfleger BF. 2013. An organic acid based counter selection system for cyanobacteria. PLoS One 8:e76594. 10.1371/journal.pone.0076594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lennen RM, Braden DJ, West RA, Dumesic JA, Pfleger BF. 2010. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol. Bioeng. 106:193–202. 10.1002/bit.22660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murata N, Wada H, Gombos Z. 1992. Modes of fatty-acid desaturation in cyanobacteria. Plant Cell Physiol. 33:933–941 [Google Scholar]
- 18.Sakamoto T, Wada H, Nishida I, Ohmori M, Murata N. 1994. Identification of conserved domains in the Δ12 desaturases of cyanobacteria. Plant Mol. Biol. 24:643–650. 10.1007/BF00023560 [DOI] [PubMed] [Google Scholar]
- 19.Murata N, Wada H. 1995. Acyl-lipid desaturases and their importance in the tolerance and acclimatization to cold of cyanobacteria. Biochem. J. 308:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakamoto T, Shen G, Higashi S, Murata N, Bryant DA. 1998. Alteration of low-temperature susceptibility of the cyanobacterium Synechococcus sp. PCC 7002 by genetic manipulation of membrane lipid unsaturation. Arch. Microbiol. 169:20–28 [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto T, Wada H, Nishida I, Ohmori M, Murata N. 1994. Identification of conserved domains in the Δ12 desaturase of cyanobacteria. Plant Mol. Biol. 24:643–650. 10.1007/BF00023560 [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto T, Bryant D. 1997. Temperature-regulated mRNA accumulation and stabilization for fatty acid desaturase genes in the cyanobacterium Synechococcus sp. strain PCC 7002. Mol. Microbiol. 23:1281–1292. 10.1046/j.1365-2958.1997.3071676.x [DOI] [PubMed] [Google Scholar]
- 23.Tasaka Y, Gombos Z, Nishiyama Y, Mohanty P, Ohba T, Ohki K, Murata N. 1996. Targeted mutagenesis of acyl-lipid desaturases in Synechocystis: evidence for the important roles of polyunsaturated membrane lipids in growth, respiration and photosynthesis. EMBO J. 15:6416–6425 [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig M, Pandelia M-E, Chew CY, Zhang B, Golbeck JH, Krebs C, Bryant DA. 2014. ChlR protein of Synechococcus sp. PCC 7002 is a transcription activator that uses an oxygen-sensitive [4Fe-4S] cluster to control genes involved in pigment biosynthesis. J. Biol. Chem. 289:16624–16639. 10.1074/jbc.M114.561233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakamoto T, Stirewalt VL, Bryant DA. 1997. Two acyl-lipid Δ9 desaturase genes of the cyanobacterium, Synechococcus sp. strain PCC 7002, p 380–382 In Williams JP, Khan MU, Lem NW. (ed), Physiology, biochemistry, and molecular biology of plant lipids. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 26.Kaczmarzyk D, Fulda M. 2010. Fatty acid activation in cyanobacteria mediated by acyl-acyl carrier protein synthetase enables fatty acid recycling. Plant Physiol. 152:1598–1610. 10.1104/pp.109.148007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. 2005. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science 307:1317–1321. 10.1126/science.1106392 [DOI] [PubMed] [Google Scholar]
- 28.Ludwig M, Bryant DA. 2012. Synechococcus sp. strain PCC 7002 transcriptome: acclimation to temperature, salinity, oxidative stress, and mixotrophic growth conditions. Front. Microbiol. 3:354. 10.3389/fmicb.2012.00354 [DOI] [PMC free article] [PubMed] [Google Scholar]