Abstract
Escherichia coli is a highly adaptive microorganism, and its ability to form biofilms under certain conditions can be critical for antimicrobial resistance. The adhesion of four E. coli isolates from bovine mastitis to bovine mammary alveolar (MAC-T) cells, biofilm production on a polystyrene surface, and the expression profiles of the genes fliC, csgA, fimA, and luxS in the presence of enrofloxacin, gentamicin, co-trimoxazole, and ampicillin at half of the MIC were investigated. Increased adhesion of E. coli isolates in the presence of antimicrobials was not observed; however, increased internalization of some isolates was observed by confocal microscopy. All of the antimicrobials induced the formation of biofilms by at least one isolate, whereas enrofloxacin and co-trimoxazole decreased biofilm formation by at least one isolate. Quantitative PCR analysis revealed that all four genes were differentially expressed when bacteria were exposed to subinhibitory concentrations of antimicrobials, with expression altered on the order of 1.5- to 22-fold. However, it was not possible to associate gene expression with induction or reduction of biofilm formation in the presence of the antimicrobials. Taken together, the results demonstrate that antimicrobials could induce biofilm formation by some isolates, in addition to inducing MAC-T cell invasion, a situation that might occur in vivo, potentially resulting in a bacterial reservoir in the udder, which might explain some cases of persistent mastitis in herds.
INTRODUCTION
Mastitis is the most important disease in the dairy industry and causes a decrease in milk production, increased health care costs, and even animal death (1). The major mastitis pathogens include Staphylococcus aureus, Escherichia coli, Streptococcus uberis, S. dysgalactiae, and S. agalactiae (2). These bacteria are capable of biofilm formation in vitro, which is an important mechanism of protection and resistance to antimicrobials (1, 3) and plays an important role in the virulence of the microorganisms (4). Adhesion of microorganisms to a surface, followed by cell division under suitable growth conditions (nutrients and temperature), results in colonization of the surface and is considered biofilm establishment (5).
Biofilms probably represent the most important mechanism of microorganism attachment and colonization in nature (6) and are structured communities of microorganisms organized in a complex structure that adheres to an inert or living surface (6). Field strains of E. coli from cases of bovine mastitis demonstrate a large variation in biofilm formation in vitro, and their significance in the pathogenesis of mastitis is still unknown (7).
Treatment with antimicrobials is able to kill only planktonic cells, which abandon the sessile form to spread when treatment ceases (8). Furthermore, some antimicrobial agents can stimulate the production of biofilms by microorganisms (9–11). Subinhibitory concentrations of aminoglycosides, such as gentamicin, tobramycin, and amikacin, induce biofilm production in Pseudomonas aeruginosa and E. coli isolates from patients with cystic fibrosis (12). Antimicrobials used in the treatment of clinical mastitis, such as gentamicin (aminoglycoside) and enrofloxacin (quinolone), at subinhibitory concentrations induce biofilm formation by E. coli isolates from cases of mastitis (9). This might have clinical relevance, as bacteria are exposed to sub-MICs of antimicrobials at the beginning and end of a dosing regimen (between doses) or continuously during low-dose therapy (13).
In response to external stimuli, biofilm production is related to several genes that encode various structures, such as proteins and extracellular polymeric substances (EPS), involved in the development and establishment of biofilms (14, 15). Previous studies have examined the genes that are differentially expressed during biofilm formation by E. coli by using DNA microarrays (15, 16). However, little is known about the induction of biofilm formation by antimicrobial agents used for the treatment of diseases.
In E. coli, a molecule similar to autoinducer 2 (AI-2) produced by Vibrio harveyi (the luxS gene encodes AI-2 synthase) is involved in biofilm formation. When present in the extracellular medium at a limiting concentration, it can initiate a cascade of signal transduction that culminates in a change in the behavior of the cell population (17, 18). Other structures play important roles during biofilm formation in different phases on the bacterial surface, such as flagella (the fliC gene encodes flagellin, the main component), which are responsible for motility, and drive the cells toward a favorable colonization surface (19). In addition, the flagella might be directly associated with fixation, allowing bacteria to reach the surface to be colonized by facilitating proliferation and dissemination (20). The type I fimbriae (the fimA gene encodes the larger subunit) is the most common adhesin produced by E. coli and mediates adhesion to mannose-containing glycoproteins found on the surface of many eukaryotic cells (21). This has proven to be important for the initial interaction with abiotic surfaces, since it is known that its absence results in virtually no attachment to the surface to be colonized (20). The curli fimbriae (csgA encodes the major subunit) play a role in the adherence of E. coli K-12 C600 when in contact with the host and interact specifically with matrix proteins such as fibronectin, laminin, and plasminogen to start accession and colonization (22–24).
Generally, mastitis caused by E. coli has a short duration, resulting in either bacterial clearance or death of the host (25). However, persistent intramammary infections (IMI) caused by the same clone (26) are observed, with prevalence estimates between 5 and 24% of all mastitis cases caused by E. coli (26, 27). One of our possible hypotheses to explain resistance to therapy is the ability of E. coli to grow in biofilms in infected tissues, thus developing a physiological resistance to almost all therapeutic agents. The aims of this study were to evaluate the ability of E. coli obtained from the milk of mastitic cows to adhere to bovine mammary alveolar cells (MAC-T) and to form biofilms on a polystyrene surface, as well as to determine the expression profile of the fliC, csgA, luxS, and fimA genes in the presence of antimicrobials used for the treatment of mastitis.
MATERIALS AND METHODS
Bacterial isolates.
Four E. coli strains were isolated from milk samples obtained from cases of clinical bovine mastitis at dairy farms located in the regions of Viçosa and Juiz de Fora, in Minas Gerais, Brazil. These isolates were previously characterized phenotypically and genotypically, including biofilm production, virulence markers, and serology (Table 1) (28). Four isolates were selected for further study, two weak biofilm producers (WBP), E. coli 5 and 30, and two strong biofilm producers (SBP), E. coli 51 and 53. Bacterial isolates were stored at −80°C and subcultured on MacConkey agar (Oxoid, Powai, Mumbai, India) for 24 h before use in experiments. The isolates were routinely grown in brain heart infusion (BHI; Oxoid, Powai, Mumbai, India) broth at 37°C for 6 h (to mid-log phase) without shaking.
TABLE 1.
Phenotypic and genotypic characteristics of the four strains used in this studya
| E. coli isolate | Phenotypic characteristics |
Genotypic characteristics |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Biofilm production | Serogroup | K1 capsular antigen | Type 1 fimbriae | K99 | F41 | F17 | F165 | CS31A | eae | ipaH | |
| 5 | WBP | O12:HNT | − | + | − | − | − | − | − | − | − |
| 30 | WBP | O93:H4 | − | + | − | − | − | − | + | − | − |
| 51 | SBP | ONT:H7 | − | + | − | − | − | − | + | − | − |
| 53 | SBP | OR:H21 | − | + | − | − | − | − | − | − | − |
Table adapted from reference 28. +, gene present; −, gene absent. K99, F41, F17, and F165 are fimbrial adhesins; CS31A is a surface antigen; eae is the E. coli attaching and effacing gene; ipaH is the invasion antigen gene.
Mammalian cell culture.
A bovine cell line, MAC-T (29), was used for adhesion assays. Monolayers of MAC-T cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL, Grand Island, NY) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Cultilab, Campinas, SP, Brazil), penicillin (100 μg/ml), and streptomycin (100 μg/ml) (Sigma-Aldrich, St. Louis, MO) at 37°C in 5% (vol/vol) CO2. For adhesion assays and biotic surface inoculations, 106 MAC-T cells were plated into each well of 24-well polystyrene plates (Sarstedt AG & Co., Nümbrecht, Germany) without the addition of penicillin and streptomycin. Epithelial cell viability was unchanged (as determined by visual inspection) during the experiments.
Antimicrobial agents.
Ampicillin, gentamicin, enrofloxacin, and co-trimoxazole (trimethoprim-sulfamethoxazole), which are commonly used in the treatment of E. coli bovine mastitis, were purchased from a pharmacy (Formullarium Farmácia de Manipulação, Viçosa, Minas Gerais, Brazil). Stock solutions (1 mg/ml) of all antimicrobial agents were prepared in sterile water, sterile filtered (0.22-μm pore diameter; Schleicher & Schuell BioScience GmbH, Dassel, Germany), aliquoted, and stored at −20°C for 1 month. The concentrations tested are shown in Table 2.
TABLE 2.
Antimicrobial concentrations used for the strains in this study
| E. coli isolate | 0.5× MIC (μg/ml) of: |
|||
|---|---|---|---|---|
| Ampicillin | Enrofloxacin | Gentamicin | Co-trimoxazole | |
| 5 | 2 | 0.10 | 4 | 0.25 |
| 30 | 3 | 0.07 | 4 | 0.50 |
| 51 | 3 | 0.03 | 0.50 | 0.50 |
| 53 | 3 | 0.02 | 0.50 | 0.25 |
Antimicrobials were tested at half the MIC (0.5× MIC), which was previously selected by using the antimicrobials at five concentrations, one-fourth, one-third, one-half, three-fifths, and three-fourths of the MIC, to evaluate biofilm production after 24 h of incubation (data not shown). It was found that 0.5× MIC was the highest concentration at which biofilm formation occurred. This concentration was also used in several previous studies (30–32).
Adhesion assay.
To quantify the total cell-associated (intracellular and surface-adherent) bacteria, an adhesion assay was performed as previously described (33), with modifications. Briefly, bacteria were grown on MacConkey agar overnight and one colony was incubated for 6 h (to mid-log phase) at 37°C in 5 ml BHI broth without shaking. Bacteria were harvested by centrifugation (5 min at 5,000 × g), washed twice with 2 ml of 1 M phosphate-buffered saline (PBS, pH 7.4; Laborclin, Produtos para Laboratórios Ltd., Paraná, Brazil), and resuspended in cell culture medium to ∼108 CFU/ml, and antimicrobials were added to obtain a final concentration equal to 0.5× MIC. Confluent monolayers of MAC-T cells grown in 24-well polystyrene plates (Sarstedt AG & Co., Nümbrecht, Germany) were washed three times with 2 ml of PBS and infected with a multiplicity of infection (MOI) of 100 (the ratio of infectious agents to infection targets was 100:1). After 1 h of incubation at 37°C in 5% (vol/vol) CO2, culture supernatants were removed and monolayers were washed three times with PBS and lysed with 1 ml of 0.25% (wt/vol) trypsin (Sigma-Aldrich, St. Louis, MO) and 0.25% (vol/vol) Triton X-100 (Amersham, Arlington Heights, IL) in PBS for 10 min. Lysates were serially diluted and plated on BHI agar, and bacterial concentrations were determined from the colony counts after incubation at 37°C for 24 h. Control wells (without antimicrobials) were incubated as described above to determine bacterial adhesion to MAC-T cells. Adhesion was expressed as the total number of CFU/ml recovered per well. Each assay was performed with triplicate samples.
Cytotoxicity.
Antimicrobials (each at a concentration of 10 μg/ml) were added to MAC-T cells to determine cytotoxicity. Briefly, confluent monolayers of MAC-T cells grown in 24-well polystyrene plates (Sarstedt AG & Co., Nümbrecht, Germany) were washed three times with 2 ml of PBS and then added to 1 ml of DMEM supplemented with 10% (vol/vol) FBS, and an antimicrobial at 10 μg/ml. After 24 h of incubation at 37°C in 5% (vol/vol) CO2, MAC-T cells were observed through an Olympus IX70 inverted microscope (Olympus Corporation, Shinjuku, Tokyo, Japan). Cytotoxicity was assessed qualitatively by microscopic examination for changes in the general morphology, vacuolization, detachment, or cell lysis.
Differential staining of extra- and intracellular E. coli.
MAC-T cells were seeded onto coverslips, grown to confluence overnight, and then infected with E. coli 5 (WBP) or 51 (SBP) under the same conditions described for the adhesion assay. After 1 h of incubation, cells were washed three times with PBS and fixed with 3.7% (wt/vol) formaldehyde (Sigma-Aldrich, St. Louis, MO) for 15 min. Immunofluorescence staining of extracellular and intracellular bacteria was performed as previously described (25), with the following modifications. Polyclonal rabbit anti-E. coli antibody (B65001R; Biodesign, Saco, ME) was diluted 1:50 in PBS containing 10% (vol/vol) FBS (primary antibody) and incubated for 30 min at room temperature. Extracellular bacteria were stained with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG antibodies (F0382; Sigma-Aldrich, St. Louis, MO) diluted 1:50, and intracellular bacteria were stained with tetramethylrhodamine isothiocyanate-conjugated goat anti-rabbit IgG antibodies (T677; Sigma-Aldrich, St. Louis, MO) diluted 1:50 (secondary antibodies) and incubated for 30 min at room temperature. After staining, the coverslip was sealed with nail varnish onto glass slides and viewed with a confocal laser scanning microscope (LSM510 META; Carl Zeiss Inc., Thornwood, NY).
Biofilm production. (i) Quantitative evaluation by spectrophotometry.
Biofilm formation in the presence of subinhibitory antimicrobial concentrations (0.5× MIC, based on pre-established MICs) was evaluated with a static system as in other studies (34, 35). The procedure was performed according to the methodology described by Hoffman et al. (12) and Moreira et al. (36), both with modifications. Each antimicrobial agent (60 μl of a predetermined concentration) was mixed with 190 μl of E. coli culture (approximately 5.0 × 105 CFU/ml) and added to 96-well microtiter plates (Nunc-Immuno MaxiSorp Plates; Nalge Nunc International, Rochester, NY). Distilled water (60 μl) was used instead of antimicrobials as the positive control for bacterial growth, and BHI broth (190 μl), without bacteria, supplemented with 60 μl of antimicrobial solution was used as the negative control. The microplates were incubated at 37°C for 24 h, and biofilm production was analyzed by taking readings of optical density at 550 nm (ELx800; BioTek, Winooski, VT) and comparing the values of the samples with those of the positive control. All procedures were performed with triplicate samples.
(ii) Qualitative assessment by SEM.
The scanning electron microscopy (SEM) methodology used was an adaptation of that of Abdi-Ali et al. (37), and polystyrene coverslips (0.5 by 0.5 cm) were used to simulate the surface of the microtiter plates used. The coverslips were cleaned by immersion in 2% (vol/vol) sodium hypochlorite for 30 min, rinsed in autoclaved distilled water, immersed in 70% (vol/vol) ethanol, and washed with autoclaved distilled water. After cleaning, they were dried for 5 min and then exposed to UV radiation for 20 min in a laminar-flow cabinet.
E. coli 5 (WBP) and 51 (SBP) biofilm formation was assessed by SEM. Cleaned coverslips were placed on 24-well plates containing 1 ml of culture at 1.0 × 105 CFU/ml in BHI broth with antimicrobials (0.5× MIC) and incubated at 37°C for 24 h. Bacterial cultures without antimicrobials served as the controls. Slides were fixed in 2.5% (vol/vol) glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 1 h at room temperature, postfixed in 1% (wt/vol) osmium tetroxide (Sigma-Aldrich, St. Louis, MO) for 1 h at room temperature, dehydrated, critical point dried (Balzers CPD 020; BAL-TEC AG, Balzers, Liechtenstein), mounted onto a support, and sputter coated with aluminum-gold (Balzers ACS 010; BAL-TEC AG, Balzers, Liechtenstein). Coverslips were analyzed by SEM (Leo 1430VP; LEO Electron Microscopy Ltd., Singapore) with magnification at ×3,000 or ×5,000.
Expression of genes luxS, fliC, csgA, and fimA. (i) Abiotic surface inoculation.
Bacterial cultures (5.0 × 105 CFU/ml) with or without antibiotics (0.5× MIC) were added to wells of 24-well polystyrene plates (abiotic surface) and incubated at 37°C for 0, 6, 12, and 24 h. Planktonic cells were harvested at 0, 6, 12, and 24 h, while biofilm samples were harvested at 24 h and all were processed for quantitative PCR (qPCR). All procedures were performed with triplicate samples, and BHI plus an antimicrobial was used as a negative control.
(ii) Biotic surface inoculation.
MAC-T cells (biotic surface) were washed three times each with 2 ml of PBS (to remove nonadherent cells from the surface of the culture plate) and infected at an MOI of 100. The cell culture medium contained DMEM supplemented with 10% (vol/vol) FBS and an antimicrobial at 0.5× MIC. After 0 h, 30 min, 1 h, and 2 h of incubation at 37°C in 5% (vol/vol) CO2, well supernatants were removed, the monolayers were washed three times with PBS, and cells were processed for qPCR.
(iii) Total RNA extraction, cDNA synthesis, and qPCR analysis.
Total bacterial RNA was isolated from planktonic cells, biofilm, and adherent cells. Briefly, biofilm or cells were washed three times with PBS to remove nonadherent cells (33) and RNA samples were prepared with TRIzol reagent (Invitrogen, Grand Island, NY) according to the enclosed protocol. RNA samples were quantitated with the NanoDrop Lite (Thermo Fisher Scientific, Inc., Waltham, MA), and the concentration was confirmed by electrophoresis in a 1% (wt/vol) agarose gel and staining with GelRed (Biotium, Hayward, CA). A minimum of three samples were harvested and extracted.
cDNA was synthesized from 300 ng of total RNA as follows. RNA was mixed with 0.5 μl Random Hexamer Primer (Fermentas Inc., Glen Burnie, MD), 1 μl of a deoxynucleoside triphosphate mixture (10 mM dATP, dCTP, dGTP, and dTTP), and 10.5 μl of nuclease-free water (Fermentas Inc., Glen Burnie, MD) and then incubated at 65°C for 5 min and on ice for 1 min. Reagents (4 μl of 5× First-Strand Buffer [Invitrogen, Washington, DC], 2 μl of 0.1 M dithiothreitol [Invitrogen, Washington, DC], and 1 μl of RNaseOUT [Invitrogen, Washington, DC]) were then added, and the mixture was incubated at 37°C for 2 min. The RNase was inactivated according to the manufacturer's instructions, 1 μl of the Moloney murine leukemia virus reverse transcriptase (M-MLV RT) (Invitrogen, Grand Island, NY) was added, and the mixture was incubated at 25°C for 10 min, 37°C for 50 min, and 70°C for 15 min. qPCR for the quantification of cDNA was executed with the Maxima SYBR Green/ROX qPCR master mix (Fermentas Inc., Glen Burnie, MD) and the Eco real-time PCR system (Illumina Inc., San Diego, CA) according to the procedure recommended by the manufacturer. The primers designed for qPCR are shown in Table 3. The following PCR conditions were used: 50°C for 2 min, 95°C for 10 min; 40 cycles of 95°C for 30 s and 60°C for 1 min; followed by 95°C for 15 s, 55°C for 15 s, and 95°C for 15 s. The glyceraldehyde-3-phosphate dehydrogenase gene was used as an internal control for normalization (38). The fold changes in the AI-2 synthase (luxS), flagellin (fliC), major curli subunit (csgA), and major type 1 subunit fimbrin (fimA) expression levels were calculated by the comparative cycle threshold (CT) method (38).
TABLE 3.
Genes, primers, and amplicons used for qPCR analysis
| Genea | Primerb | Amplicon size (bp) |
|---|---|---|
| luxS | F, 5′-ATGAGCAGCGTGTTGCTGATGC-3′ | 122 |
| R, 5′-CAACGAGTGCATCTGGTAAGTGC-3′ | ||
| fliC | F, 5′-ATTAACAGCGCGAAGGATGACG-3′ | 180 |
| R, 5′-TACCGTCAGTTCACGCACACG-3′ | ||
| csgA | F, 5′-GATCTGACCCAACGTGGCTTCG-3′ | 178 |
| R, 5′-GATGAGCGGTCGCGTTGTTACC-3′ | ||
| fimA | F, 5′-CTCTGGCAATCGTTGTTCTGTCG-3′ | 119 |
| R, 5′-GCAAGCGGCGTTAACAACTTCC-3′ | ||
| gapA* | F, 5′-GATTACATGGCATACCTG-3′ | 244 |
| R, 5′-CAGACGAACGTTCAGGTCAA-3′ |
luxS, AI-2 synthase gene; fliC, flagellin gene; csgA, major curli subunit gene; fimA, major type 1 subunit fimbrin gene; gapA*, glyceraldehyde-3-phosphate dehydrogenase gene (internal control for qPCR).
F, forward; R, reverse.
P values were calculated by two-way analysis of variance (ANOVA), followed by Bonferroni's multiple-comparison test (P < 0.05) (GraphPad Prism version 5.00 for Windows; GraphPad Software, San Diego, CA).
Statistical analysis.
A significance level of 5% (P < 0.05) was used for statistical analyses. The Kolmogorov-Smirnov normality test was used to evaluate the normal distribution of data. Differences in CFU/ml in adhesion assays were analyzed by the Kruskal-Wallis test, and Dunn's test was used to determine differences between groups. Separately, the Mann-Whitney test was used to identify possible differences between the numbers of CFU/ml obtained in the adhesion assay with WBPs and SBPs.
A one-way ANOVA for one factor was used to identify possible differences in the production of biofilms in the presence of antimicrobials, and Tukey's test was used to find the differences between the groups. All graphic evaluations were made with GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego CA).
RESULTS AND DISCUSSION
The capacity of E. coli isolates to adhere to MAC-T cells was not significantly increased by the presence of antimicrobials (Table 4). However, in the absence of antimicrobial agents, SBP isolates showed greater adhesion (P < 0.05) than WBP isolates. There were no cytotoxic effects of antimicrobials at a concentration of 10 μg/ml on MAC-T cells.
TABLE 4.
Adhesion assay results obtained in the presence of different antimicrobials
| E. coli isolate | Biofilm production | No. of E. coli CFU/ml adhered to bovine MAC-Ts |
||||
|---|---|---|---|---|---|---|
| Control | Ampicillin | Enrofloxacin | Co-trimoxazole | Gentamicin | ||
| 5 | WBP | 4.50 × 105 ± 3.5 × 104 | 2.20 × 105(48.89%) ± 5.3 × 104 | 1.90 × 105(42.22%) ± 1.4 × 104 | 4.10 × 105(91.11%) ± 4.2 × 104 | 5.00 × 105(111.11%) ± 5.7 × 104 |
| 30 | WBP | 3.13 × 105 ± 3.0 × 104 | 2.15 × 105(68.69%) ± 6.4 × 104 | 6.00 × 104(19.17%) ± 2.8 × 103 | 4.37 × 105(139.62%) ± 8.5 × 104 | 1.20 × 105(38.34%) ± 2.8 × 104 |
| 51 | SBP | 3.06 × 106 ± 4.5 × 105 | 5.95 × 105(19.44%) ± 4.4 × 104 | 7.34 × 105(23.99%) ± 3.3 × 104 | 8.18 × 105(26.73%) ± 2.9 × 104 | 7.41 × 105(24.22%) ± 2.9 × 104 |
| 53 | SBP | 1.83 × 106 ± 4.6 × 105 | 6.78 × 105(37.05%) ± 3.7 × 104 | 6.92 × 105(37.81%) ± 4.5 × 104 | 5.78 × 105(31.58%) ± 3.5 × 104 | 6.55 × 105(35.79%) ± 3.8 × 104 |
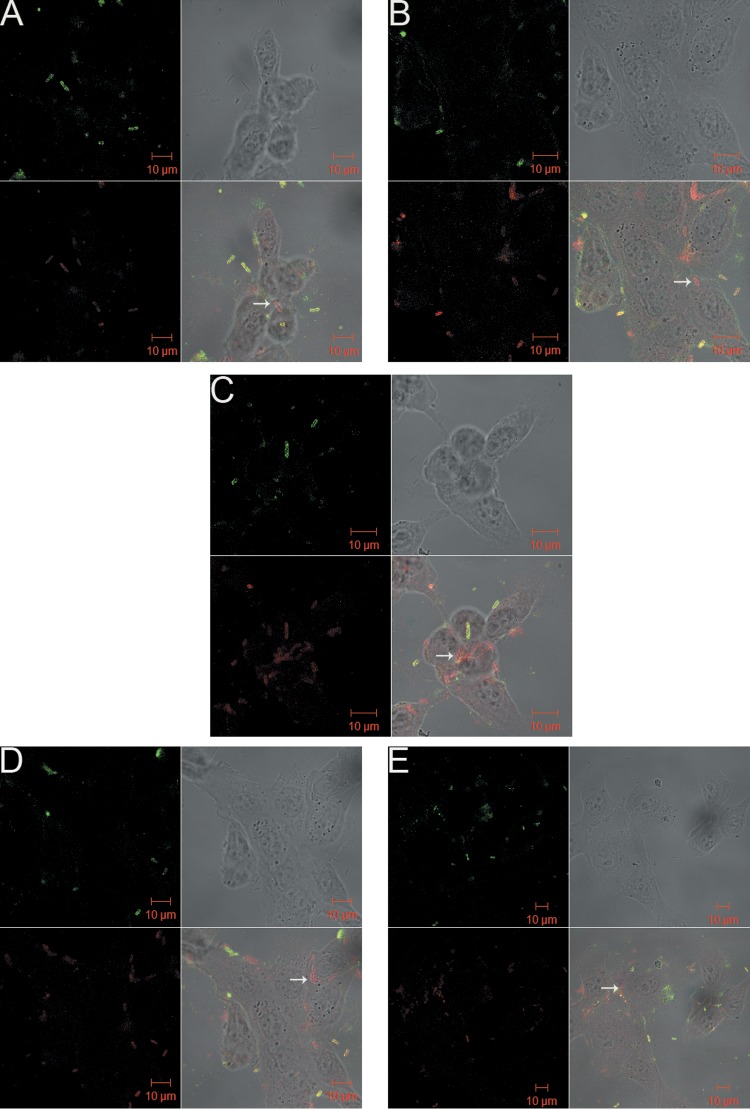
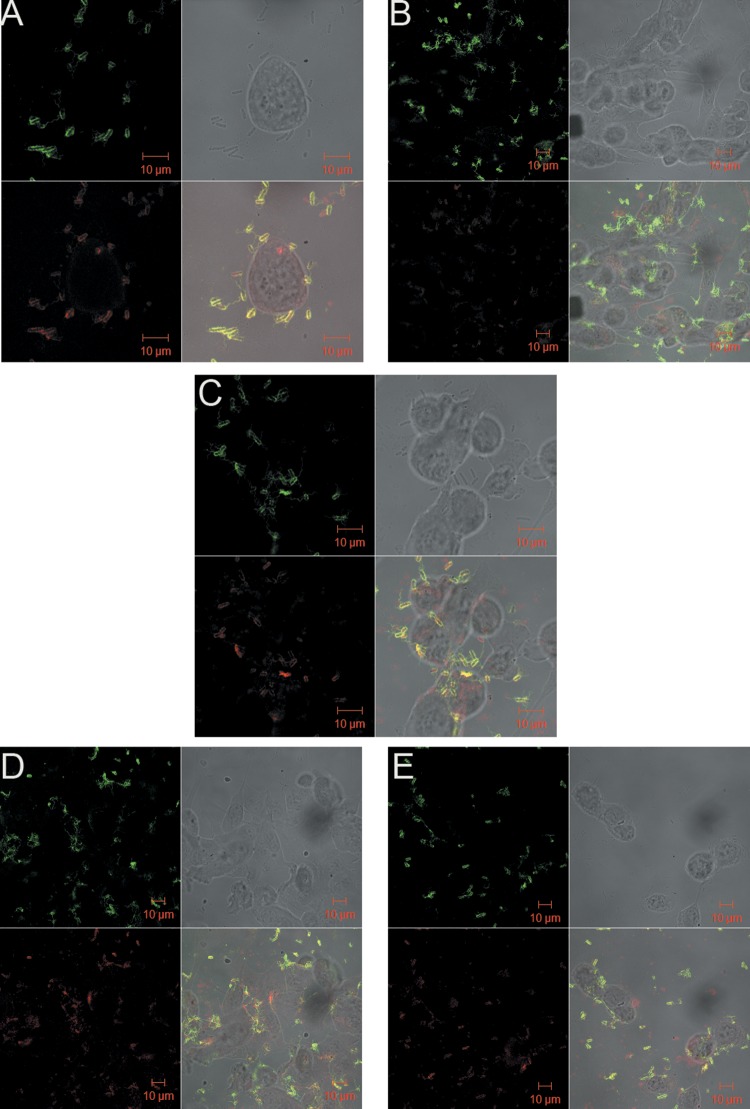
Qualitative analysis of confocal microscopy results (Fig. 1 and 2) confirmed the results obtained for adhesion assays and showed that E. coli 51 (SBP) had more adherent bacterial cells than E. coli 5 (WBP) in the absence of antimicrobials. Flagella were noted on planktonic E. coli 51 cells but not on E. coli 5 cells. Figure 1B to E shows increased internalization of E. coli 5 (WBP), which suggests that internalization was induced by the antimicrobials present in the medium. Internalization of E. coli 51 (SBP) was not observed in either the control (Fig. 2A) or cultures treated with any of the (four) antimicrobials (Fig. 2B to E). These results agree with data obtained by Pratt and Kolter (20) suggesting that flagella might be related to the adhesion capacity of E. coli obtained from mastitic milk. Furthermore, flagella may not be required for invasion, because the absence of flagella from intracellular bacteria was demonstrated (Fig. 1). A similar finding has been reported for uropathogenic E. coli (39).
FIG 1.
E. coli 5 (WBP) adherence after 1 h of incubation in cell culture with antimicrobials at 0.5× MIC, visualized by confocal microscopy. Panels: A, control (without antimicrobial); B, ampicillin; C, enrofloxacin; D, gentamicin; E, co-trimoxazole. White arrows indicate internalized E. coli cells.
FIG 2.
E. coli 51 (SBP) adherence after 1 h of incubation in cell culture with antimicrobials at 0.5× MIC, visualized by confocal microscopy. Panels: A, control (without antimicrobial); B, co-trimoxazole; C, gentamicin; D, enrofloxacin; E, ampicillin.
It was observed that internalization ability may not be associated with bacterial adhesion. This event might provide a distinct survival advantage that allows the pathogens to avoid the action of antimicrobials present in the medium and to resist detection and elimination by the defense mechanisms of the innate and adaptive immune systems, as well as give them access to a more nutrient-rich environment. Döpfer et al. (33) reported the invasion of cultured MAC-T cells by E. coli strains associated with mastitis and suggested that this phenomenon might be responsible for the development of an E. coli reservoir during the pathogenesis of chronic IMIs.
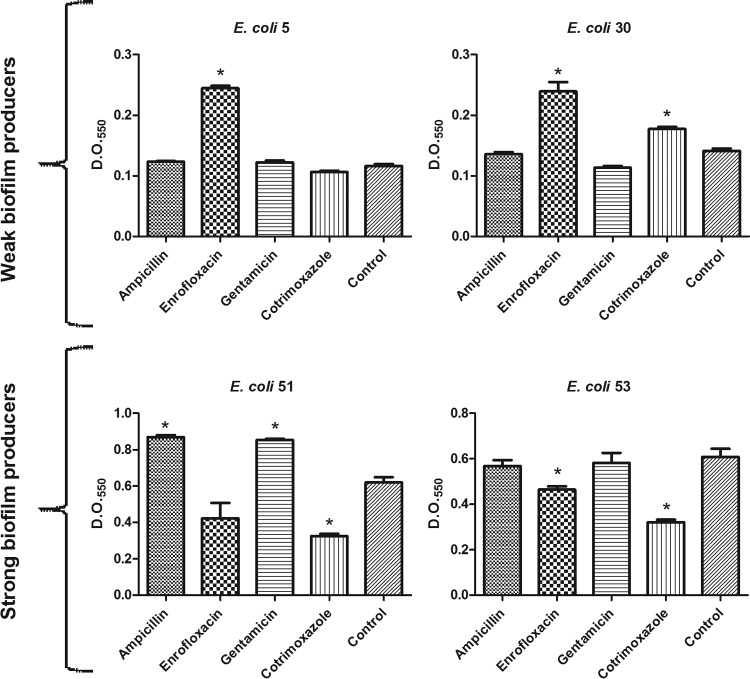
Biofilm formation by some isolates was induced by the presence of antimicrobials (Fig. 3). This induction was different among the isolates, even within groups of WBP and SBP. Induction was observed in the presence of ampicillin and gentamicin in E. coli 51 (SBP) (P < 0.05) (Fig. 3C) and in the presence of enrofloxacin in the two WBP isolates (E. coli 5 and 30) (P < 0.05) (Fig. 3A and B). However, enrofloxacin also caused a significant reduction in the biofilm-forming capacity of the SBP E. coli 53 (P < 0.05) (Fig. 3D). Increased biofilm production by E. coli 30 in the presence of co-trimoxazole was observed (Fig. 3A) (P < 0.05), whereas a reduction in that by E. coli 51 and 53 was observed (Fig. 3C and D) (P < 0.05).
FIG 3.
Biofilm production of four E. coli isolates treated with ampicillin, enrofloxacin, gentamicin, and co-trimoxazole at 0.5× MIC. Control, without antimicrobial. *, significant at P <0.05.
Boehm et al. (40) reported an induction of biofilm formation by strains derived from E. coli K-12 MG1655 exposed to β-lactams (ampicillin, amoxicillin, and penicillin G), fluoroquinolones (enoxacin and ciprofloxacin), and trimethoprim (sulfonamide present in the antimicrobial co-trimoxazole). Previous studies showed that subinhibitory concentrations of gentamicin (aminoglycoside) and enrofloxacin (quinolone), both used in the treatment of clinical mastitis, induced the formation of E. coli biofilms (9, 12). Therefore, these results, together with the data in this study, suggest that antimicrobials present at sub-MICs can significantly induce in vitro biofilm formation by E. coli obtained from mastitic milk. This might have clinical relevance, as bacteria are exposed to sub-MICs of antimicrobials at the beginning and end of a dosing regimen (between doses) or continuously during low-dose therapy (13).
SEM analysis confirmed the biofilm production results obtained. Differences were observed in biofilm development with time, with variations in the number of adherent cells, as well as in extracellular matrix production (Fig. 4 and 5). The micrographs of biofilms formed by E. coli 5 (WBP) revealed an increase in surface-adherent cells in the presence of antimicrobials without visible extracellular matrix formation compared to the control (Fig. 4). For E. coli 51 (SBP), treatment with gentamicin resulted in a higher biofilm biomass, while a greater number of adherent bacterial cells and significant amounts of deformed fibrous structures, possibly exopolysaccharides (EPS), surrounding the cells were observed (Fig. 5B).
FIG 4.
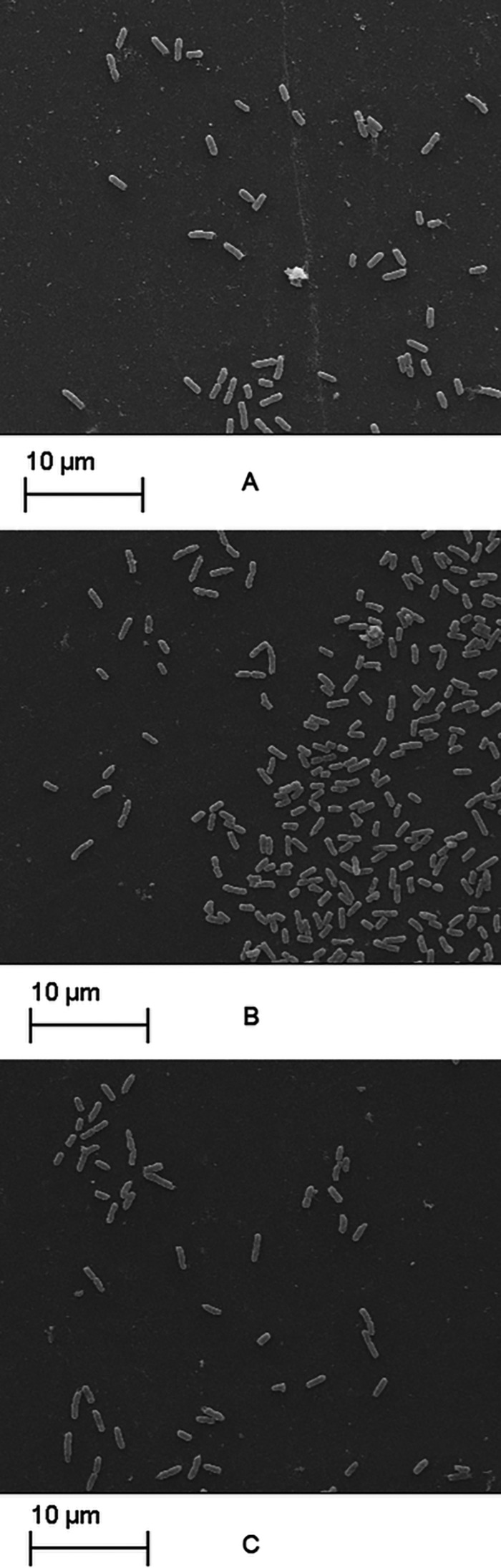
Adherent E. coli 5 cells (WBP) after 24 h of incubation in BHI broth with antimicrobials at 0.5× MIC. Panels: A, control (without antimicrobials); B, enrofloxacin; C, co-trimoxazole. Magnification, ×3,000.
FIG 5.
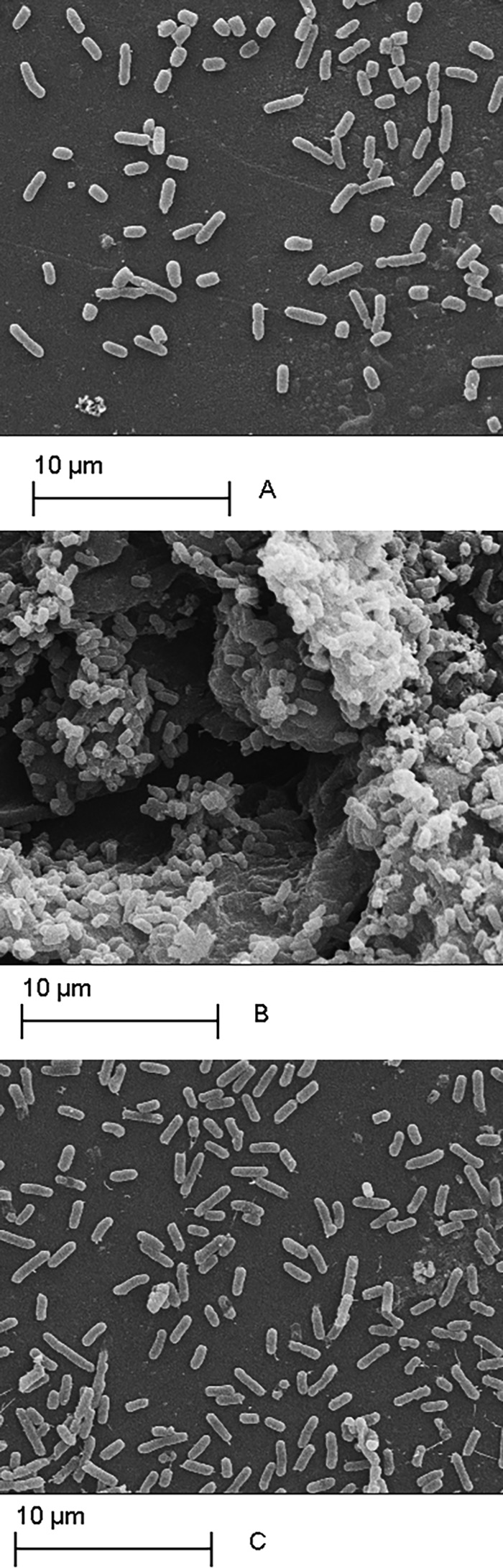
Adherent E. coli 51 cells (SBP) after 24 h of incubation in BHI broth with antimicrobials at 0.5× MIC. Panels: A, control (without antimicrobials); B, gentamicin; C, ampicillin. Magnification, ×5,000.
The luxS, fliC, csgA, and fimA genes showed differences in expression during biofilm formation. Alterations in gene expression on the order of 1.5- to 22-fold occurred, which varied according to the ability of bacteria to produce biofilm, the target gene, and the collection time (see Fig. S1 to S8 in the supplemental material).
Most of the genes were upregulated in expression when cells were treated with antimicrobials at 0.5× MIC. Increased luxS (AI-2 synthase) and fliC (flagellin) gene expression (P < 0.05) was observed in planktonic WBP and SBP cells in the presence of antimicrobials (see Fig. S1 and S3 in the supplemental material). Expression varied with respect to the collection time, with larger increases detected after 12 and 24 h of incubation with antimicrobials. Sessile cells also showed an increased in luxS gene expression, with ∼22-fold (P < 0.05) higher expression observed in cultures of E. coli 51 treated with enrofloxacin than in the untreated control (see Fig. S1C). Unlike the results of luxS gene expression in cells on an abiotic surface, expression in bacterial cells that adhered to cells in culture increased (P < 0.05) in the presence of antimicrobials at all of the time points tested (0.5, 1, and 2 h) (see Fig. S2). In the presence of the antimicrobial agents, luxS expression in bacterial cells that adhered to MAC-T cells in culture differed from that seen when bacteria were attached to an abiotic surface.
González Barrios et al. (41) showed that upregulation of AI-2 increases biofilm formation. However, it was not possible in this study to associate increased expression/induction of AI-2 synthase with biofilm formation or adhesion to MAC-T cells, as the gene was also expressed under conditions that induced or reduced the formation of biofilms. When E. coli 30 and 51 were treated with enrofloxacin and gentamicin, respectively, luxS gene expression increased (P < 0.05) (see Fig. S1B and C in the supplemental material), concomitant with an increase in biofilm formation (P < 0.05) (Fig. 3B and C). However, co-trimoxazole treatment also increased luxS (P < 0.05) expression in E. coli 51 and 53 (see Fig. S1C and D), even though biofilm formation decreased (P < 0.05) (Fig. 3C and D).
The highest relative level of fliC gene expression was observed in E. coli 5 treated with co-trimoxazole, which was 13-fold (P < 0.05) higher than that in the untreated control (see Fig. S3A in the supplemental material). Unlike planktonic cells, sessile cells showed a low level of fliC expression (P < 0.05), whereas bacteria that adhered to cells in culture showed increased expression (P < 0.05) in the presence of antimicrobials at all collection times (0.5, 1, and 2 h) (see Fig. S4).
The expression of fliC was similar to that reported by Domka et al. (15), verifying flagellar gene induction, and a 10-fold increase in the expression of the flagellar biosynthesis operon flgBCEF was observed. Flagella facilitate the dispersion process, allowing the cells to spread over the surface and the absence or suspension of flagellum synthesis might lead to decreased biofilm formation (20). Furthermore, expression of the flagellar gene fliC was virtually unchanged in cells present in biofilms, as the cells were fixed to a surface and did not need to move.
Planktonic cells of WBP and SBP isolates showed greater csgA (major curli subunit) expression (P < 0.05) in the presence of the antimicrobials tested than the controls did. Expression varied according to collection time, with a major increase (P < 0.05) observed at 24 h (see Fig. S5 in the supplemental material). A 13-fold (P < 0.05) increase in csgA expression was observed in E. coli 5 when it was treated with gentamicin at 0.5× MIC (see Fig. S5A). A high level of expression of this gene was noted in sessile cells, which increased up to 5-fold in E. coli 30 (P < 0.05) treated with enrofloxacin (see Fig. S5B). Expression of csgA in bacterial cells that adhered to cell cultures was similar to that observed for bacteria on abiotic surfaces (see Fig. S6), with increased expression (P < 0.05) at all collection times (0.5, 1, and 2 h).
Expression of the csgA gene increased (P < 0.05) even before cells attached to the inert surface, suggesting that this structure might play an important role in increasing bacterial attachment. Research has shown that curli fimbriae are required for biofilm formation and bacterial autoaggregation (35, 42). The presence of antimicrobials in the growth medium might have provided conditions that favored the expression of this gene, which might assist in the formation of biofilms. These structures are essential for initial adhesion to the surface to be colonized and mediate cell-cell interactions in strains of E. coli (35, 42, 43).
The expression of fimA (which encodes the large subunit of type I fimbriae) increased (P < 0.05) in WBP and SBP planktonic cells that were treated with antimicrobials and was dependent upon the collection time. Larger increases were observed at 12 and 24 h (P < 0.05) (see Fig. S7 in the supplemental material), whereas sessile cells had low levels of expression of this gene (P < 0.05). Ampicillin and gentamicin induced the expression of fimA by 8-fold in E. coli 51 and 22-fold (P < 0.05) in E. coli 5 (see Fig. S7A and C). The expression of fimA in bacterial cells that adhered to cells in culture was higher (P < 0.05) than in those on abiotic surfaces (see Fig. S8) and increased (P < 0.05) at all collection times (0.5, 1, and 2 h). Expression of the fimA gene was greater (P < 0.05) in bacteria that adhered to cells in culture than in those that adhered to an abiotic surface. This difference might be due to the presence of glycoprotein on the MAC-T cell surface, which stimulated the expression of this gene, since type I fimbriae mediate adhesion to mannose-containing glycoproteins found on the surfaces of many eukaryotic cells (21).
The fimA gene showed increased expression (P < 0.05) at subinhibitory doses of antimicrobials. This resulted in biofilm formation by gentamicin-treated E. coli 51 (Fig. 3C), whereas co-trimoxazole inhibited biofilm formation by E. coli 53 (Fig. 3D). fimA expression (P < 0.05) was much higher when biofilm formation was induced than during antimicrobial inhibition. An increase in fimA of up to 16-fold at the first collection times (6 and 12 h) indicates the possible involvement of this structure in initial attachment and bacterial autoaggregation. Previous studies have also reported fimA induction in planktonic cells during the process of E. coli biofilm formation (15, 16).
Data from this study showed that genes involved in adhesion (csgA and fimA), motility (fliC), and quorum sensing (luxS) were expressed significantly more highly (P < 0.05) during biofilm development in E. coli obtained from mastitic bovine milk in the presence of antimicrobial agents at 0.5× MIC. However, it was not possible to correlate the gene expression profile with biofilm formation or adherence/invasion of MAC-T cells, which indicated that other factors might influence these phenotypes. One major factor might be a lack of correlation between mRNA levels and functional protein for the same gene (44). This phenomenon was observed in a WBP isolate in which fliC gene expression increased (P < 0.05) even though flagella were not observed (Fig. 1A to C).
The results of this study suggest that greater care should be taken in choosing the correct antimicrobial treatment for mammary infections in cattle, because subinhibitory concentrations of antimicrobials can induce biofilm formation by some isolates, in addition to stimulating invasion of host cells. This is of concern, considering that these protective mechanisms might promote the creation of a bacterial reservoir in the udder of the cow, which might explain some cases of persistent mastitis in herds.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and the Agência Brasileira da Inovação (FINEP). Maria Aparecida S. Moreira, Abelardo Silva Júnior, and Hilário C. Mantovani are supported by CNPq.
Footnotes
Published ahead of print 25 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01953-14.
REFERENCES
- 1.Melchior MB, Vaarkamp H, Fink-Gremmels J. 2006. Biofilms: a role in recurrent mastitis infections? Vet. J. 171:398–407. 10.1016/j.tvjl.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 2.Bradley A. 2002. Bovine mastitis: an evolving disease. Vet. J. 164:116–128. 10.1053/tvjl.2002.0724 [DOI] [PubMed] [Google Scholar]
- 3.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35:322–332. 10.1016/j.ijantimicag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 4.Van Houdt R, Michiels CW. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156:626–633. 10.1016/j.resmic.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 5.Flint S, Palmer J, Bremer P, Seale B, Brooks J, Lindsay D, Burgess S. 2011. Biofilm formation, p 445–450 In Fuquay JW. (ed), Encyclopedia of dairy sciences, 2nd ed. Elsevier, Boston, MA [Google Scholar]
- 6.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 7.Shpigel NY, Elazar S, Rosenshine I. 2008. Mammary pathogenic Escherichia coli. Curr. Opin. Microbiol. 11:60–65. 10.1016/j.mib.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 8.Schwarz S, Kehrenberg C, Walsh TR. 2001. Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 17:431–437. 10.1016/S0924-8579(01)00297-7 [DOI] [PubMed] [Google Scholar]
- 9.Costa JC, Espeschit Ide F, Pieri FA, Benjamin Ldos A, Moreira MA. 2012. Increased production of biofilms by Escherichia coli in the presence of enrofloxacin. Vet. Microbiol. 160:488–490. 10.1016/j.vetmic.2012.05.036 [DOI] [PubMed] [Google Scholar]
- 10.Bradley AJ, Green MJ. 2009. Factors affecting cure when treating bovine clinical mastitis with cephalosporin-based intramammary preparations. J. Dairy Sci. 92:1941–1953. 10.3168/jds.2008-1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357–3363. 10.1128/AAC.44.12.3357-3363.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. 10.1038/nature03912 [DOI] [PubMed] [Google Scholar]
- 13.Odenholt I. 2001. Pharmacodynamic effects of subinhibitory antibiotic concentrations. Int. J. Antimicrob. Agents 17:1–8. 10.1016/S0924-8579(00)00243-0 [DOI] [PubMed] [Google Scholar]
- 14.Landini P. 2009. Cross-talk mechanisms in biofilm formation and responses to environmental and physiological stress in Escherichia coli. Res. Microbiol. 160:259–266. 10.1016/j.resmic.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 15.Domka J, Lee J, Bansal T, Wood TK. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9:332–346. 10.1111/j.1462-2920.2006.01143.x [DOI] [PubMed] [Google Scholar]
- 16.Schembri MA, Kjaergaard K, Klemm P. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253–267. 10.1046/j.1365-2958.2003.03432.x [DOI] [PubMed] [Google Scholar]
- 17.Bassler BL. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582–587. 10.1016/S1369-5274(99)00025-9 [DOI] [PubMed] [Google Scholar]
- 18.de Kievit TR, Iglewski BH. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839–4849. 10.1128/IAI.68.9.4839-4849.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prüss BM, Besemann C, Denton A, Wolfe AJ. 2006. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J. Bacteriol. 188:3731–3739. 10.1128/JB.01780-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285–293. 10.1046/j.1365-2958.1998.01061.x [DOI] [PubMed] [Google Scholar]
- 21.Connell I, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 93:9827–9832. 10.1073/pnas.93.18.9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gophna U, Oelschlaeger TA, Hacker J, Ron EZ. 2002. Role of fibronectin in curli-mediated internalization. FEMS Microbiol. Lett. 212:55–58. 10.1111/j.1574-6968.2002.tb11244.x [DOI] [PubMed] [Google Scholar]
- 23.Olsén A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655. 10.1038/338652a0 [DOI] [PubMed] [Google Scholar]
- 24.Sjöbring U, Pohl G, Olsén A. 1994. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA). Mol. Microbiol. 14:443–452. 10.1111/j.1365-2958.1994.tb02179.x [DOI] [PubMed] [Google Scholar]
- 25.Dogan B, Klaessig S, Rishniw M, Almeida RA, Oliver SP, Simpson K, Schukken YH. 2006. Adherent and invasive Escherichia coli are associated with persistent bovine mastitis. Vet. Microbiol. 116:270–282. 10.1016/j.vetmic.2006.04.023 [DOI] [PubMed] [Google Scholar]
- 26.Döpfer D, Barkema HW, Lam TJ, Schukken YH, Gaastra W. 1999. Recurrent clinical mastitis caused by Escherichia coli in dairy cows. J. Dairy Sci. 82:80–85. 10.3168/jds.S0022-0302(99)75211-2 [DOI] [PubMed] [Google Scholar]
- 27.Bradley AJ, Green MJ. 2001. Adaptation of Escherichia coli to the bovine mammary gland. J. Clin. Microbiol. 39:1845–1849. 10.1128/JCM.39.5.1845-1849.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandes JB, Zanardo LG, Galvao NN, Carvalho IA, Nero LA, Moreira MA. 2011. Escherichia coli from clinical mastitis: serotypes and virulence factors. J. Vet. Diagn. Invest. 23:1146–1152. 10.1177/1040638711425581 [DOI] [PubMed] [Google Scholar]
- 29.Huynh HT, Robitaille G, Turner JD. 1991. Establishment of bovine mammary epithelial cells (MAC-T): an in vitro model for bovine lactation. Exp. Cell Res. 197:191–199. 10.1016/0014-4827(91)90422-Q [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Sun FJ, Liu Y, Xiong LR, Xie LL, Xia PY. 2010. Enhancement of biofilm formation by subinhibitory concentrations of macrolides in icaADBC-positive and -negative clinical isolates of Staphylococcus epidermidis. Antimicrob. Agents Chemother. 54:2707–2711. 10.1128/AAC.01565-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majtán J, Majtánová L, Xu M, Majtán V. 2008. In vitro effect of subinhibitory concentrations of antibiotics on biofilm formation by clinical strains of Salmonella enterica serovar Typhimurium isolated in Slovakia. J. Appl. Microbiol. 104:1294–1301. 10.1111/j.1365-2672.2007.03653.x [DOI] [PubMed] [Google Scholar]
- 32.Dong L, Tong Z, Linghu D, Lin Y, Tao R, Liu J, Tian Y, Ni L. 2012. Effects of sub-minimum inhibitory concentrations of antimicrobial agents on Streptococcus mutans biofilm formation. Int. J. Antimicrob. Agents 39:390–395. 10.1016/j.ijantimicag.2012.01.009 [DOI] [PubMed] [Google Scholar]
- 33.Döpfer D, Almeida RA, Lam TJ, Nederbragt H, Oliver SP, Gaastra W. 2000. Adhesion and invasion of Escherichia coli from single and recurrent clinical cases of bovine mastitis in vitro. Vet. Microbiol. 74:331–343. 10.1016/S0378-1135(00)00191-7 [DOI] [PubMed] [Google Scholar]
- 34.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304. 10.1046/j.1365-2958.1998.01062.x [DOI] [PubMed] [Google Scholar]
- 35.Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450–464. 10.1046/j.1462-2920.2000.00128.x [DOI] [PubMed] [Google Scholar]
- 36.Moreira MA, Oliveira JA, Teixeira LM, Moraes CA. 2005. Detection of a chloramphenicol efflux system in Escherichia coli isolated from poultry carcass. Vet. Microbiol. 109:75–81. 10.1016/j.vetmic.2005.04.012 [DOI] [PubMed] [Google Scholar]
- 37.Abdi-Ali A, Mohammadi-Mehr M, Agha Alaei Y. 2006. Bactericidal activity of various antibiotics against biofilm-producing Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 27:196–200. 10.1016/j.ijantimicag.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 39.Wright KJ, Seed PC, Hultgren SJ. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 73:7657–7668. 10.1128/IAI.73.11.7657-7668.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. 2009. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 72:1500–1516. 10.1111/j.1365-2958.2009.06739.x [DOI] [PubMed] [Google Scholar]
- 41.González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305–316. 10.1128/JB.188.1.305-316.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. 2005. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 49:875–884. 10.1111/j.1348-0421.2005.tb03678.x [DOI] [PubMed] [Google Scholar]
- 44.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329:533–538. 10.1126/science.1188308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.