ABSTRACT
Many plant viruses without 5′caps or 3′ poly(A) tails contain 3′ proximal, cap-independent translation enhancers (3′CITEs) that bind to ribosomal subunits or translation factors thought to assist in ribosome recruitment. Most 3′CITEs participate in a long-distance kissing-loop interaction with a 5′ proximal hairpin to deliver ribosomal subunits to the 5′ end for translation initiation. Pea Enation Mosaic Virus (PEMV) contains two adjacent 3′CITEs in the center of its 703-nucleotide 3′ untranslated region (3′UTR), the ribosome-binding, kissing-loop T-shaped structure (kl-TSS) and eukaryotic translation initiation factor 4E-binding Panicum mosaic virus-like translation enhance (PTE). We now report that PEMV contains a third, independent 3′CITE located near the 3′ terminus. This 3′CITE is composed of three hairpins and two pseudoknots, similar to the TSS 3′CITE of the carmovirus Turnip crinkle virus (TCV). As with the TCV TSS, the PEMV 3′TSS is predicted to fold into a T-shaped structure that binds to 80S ribosomes and 60S ribosomal subunits. A small hairpin (kl-H) upstream of the 3′TSS contains an apical loop capable of forming a kissing-loop interaction with a 5′ proximal hairpin and is critical for the accumulation of full-length PEMV in protoplasts. Although the kl-H and 3′TSS are dispensable for the translation of a reporter construct containing the complete PEMV 3′UTR in vitro, deleting the normally required kl-TSS and PTE 3′CITEs and placing the kl-H and 3′TSS proximal to the reporter termination codon restores translation to near wild-type levels. This suggests that PEMV requires three 3′CITEs for proper translation and that additional translation enhancers may have been missed if reporter constructs were used in 3′CITE identification.
IMPORTANCE The rapid life cycle of viruses requires efficient translation of viral-encoded proteins. Many plant RNA viruses contain 3′ cap-independent translation enhancers (3′CITEs) to effectively compete with ongoing host translation. Since only single 3′CITEs have been identified for the vast majority of individual viruses, it is widely accepted that this is sufficient for a virus's translational needs. Pea Enation Mosaic Virus possesses a ribosome-binding 3′CITE that can connect to the 5′ end through an RNA-RNA interaction and an adjacent eukaryotic translation initiation factor 4E-binding 3′CITE. We report the identification of a third 3′CITE that binds weakly to ribosomes and requires an upstream hairpin to form a bridge between the 3′ and 5′ ends. Although both ribosome-binding 3′CITEs are critical for virus accumulation in vivo, only the CITE closest to the termination codon of a reporter open reading frame is active, suggesting that artificial constructs used for 3′CITE identification may underestimate the number of CITEs that participate in translation.
INTRODUCTION
Protein biosynthesis is principally regulated at the stage of translation initiation, allowing for rapid and efficient posttranscriptional control of gene expression (1–3). The 5′ m7GpppN cap and 3′ poly(A) tail at the termini of most eukaryotic mRNAs are required to recruit the ribosome with the assistance of numerous eukaryotic initiation factors (eIFs). The 5′ cap structure is recognized by eIF4E, a subunit of the eukaryotic translation initiation factor complex eIF4F (4). eIF4G, another subunit of eIF4F, is the core scaffolding protein that simultaneously interacts with eIF4E and 3′terminal-bound poly(A)-binding protein to bridge the cap and poly(A) tail (5). Bridging the ends of an mRNA is generally thought to facilitate reinitiation by post-termination ribosomes (5, 6), although circularized polyribosomes on a mRNA template are also visible in the absence of a 5′ cap and 3′ poly(A) tail (7). The 43S ribosome preinitiation complex, which is recruited to the 5′ end of mRNA via eIF4G and additional eIFs, transits the template in the 3′ direction until recognizing an initiation codon in the proper context. After the 60S ribosomal subunit joins the preinitiation complex, the 80S ribosome is formed, and translation initiation commences.
Many positive-strand RNA viruses do not possess a cap structure and/or a 3′ poly(A) tail but instead have evolved a variety of effective noncanonical mechanisms to directly recruit eIFs or ribosomes to the viral RNA for translation initiation (8). Most animal RNA viruses have replaced the 5′ cap with a highly structured, cis-acting element known as an internal ribosome entry site (IRES), which is located in the 5′ untranslated region (5′UTR) and may overlap the initiation codon and extend into the translated open reading frame (ORF). Viral IRESs are capable of recruiting ribosomes directly or via the assistance of a subset of eIFs (9, 10). In contrast, most uncapped and nonpolyadenylated plant viruses contain cap-independent translation enhancers (CITEs) in their 3′UTRs that can extend into nearby coding regions (11, 12). Currently, 3′CITEs have been predominantly characterized in genomes of viruses throughout the family Tombusviridae, in the Luteovirus genus of the family Luteoviridae and in the genus Umbravirus (no family assignment). 3′CITEs adopt diverse secondary structures, such as I-shaped, Y-shaped, and T-shaped, or radiate multiple helices from a central hub (12). Most 3′CITEs stimulate cap-independent translation by recruiting either eIFs or ribosomes directly and then relocating the bound eIFs/ribosomes to the 5′ end via long-distance, kissing-loop interactions with a hairpin loop located in the 5′UTR or nearby coding region (13–15). Since nearly all of these 3′CITEs are functional only in cis, it is generally assumed that they assist in recycling ribosomes to the 5′ end after translation termination.
Several classes of 3′ CITEs have been shown to bind preferably to different components of the eIF4F complex to efficiently initiate translation in host cells. The Barley yellow dwarf virus-like translation element (BTE) binds to the eIF4G subunit of eIF4F and does not require eIF4E for translation enhancement in vitro (16). The Panicum mosaic virus-like translational enhancer (PTE) in Pea Enation Mosaic Virus (PEMV) RNA2 binds to eIF4E for efficient translation enhancement in vitro (17). The Satellite tobacco necrosis virus translation enhancer domain, the Y-shaped 3′CITE of Carnation Italian ringspot virus, and the I-shaped 3′CITE of Maize necrotic streak virus have been shown to bind to eIF4F (18–20).
3′CITEs that bind directly to ribosomes/ribosomal subunits were first discovered in the carmovirus Turnip crinkle virus (TCV). The TCV 3′ CITE, consisting of three hairpins and two pseudoknots, forms a T-shaped structure (TSS) that binds to the P-site of 80S ribosomes and 60S subunits, and binding is important for efficient translation enhancement (21). In contrast to most 3′CITEs, no long-distance RNA:RNA interaction connects the TCV TSS with the 5′ end, leading to a ribosome-bridging model whereby TSS-bound 60S ribosomal subunits join with 40S subunits bound to a pyrimidine-rich sequence in the 5′UTR to recycle 60S subunits and enhance translation (22). A similar set of hairpins and pseudoknots was also predicted for the carmoviruses Cardamine chorotic fleck virus (CCFV) and Japanese iris necrosis ringspot virus (JINRV) (23), suggesting that this type of TSS 3′CITE may also exist in other viruses.
A second type of ribosome-binding 3′CITE was recently discovered in the central 3′UTR of PEMV RNA2. PEMV is composed of two taxonomically distinct, single-stranded, plus-sense viral RNAs: luteovirus PEMV RNA1 and umbravirus PEMV RNA2 (24). The 4,252-nucleotide (nt) genome of PEMV RNA2 lacks a 5′ cap and a 3′ poly(A) tail and relies on PEMV RNA1-encoded capsid protein for encapsidation and transmission but not infection of single cells (25). The carmovirus-like RNA-dependent RNA polymerase (RdRp; 94 kDa) encoded by PEMV RNA2 (referred to as “PEMV” here) is synthesized following a −1 ribosomal frameshift that extends the length of p33, the product of ORF1 (25). Two overlapping proteins (p26 and p27) are expressed from ORF3 and ORF4 via at least one subgenomic RNA (26, 27). PEMV contains two adjacent 3′CITEs in the center of its 703-nt 3′UTR: eIF4E-binding PTE and the ribosome-binding, kissing-loop TSS (kl-TSS) (13, 28). Unlike PTEs in carmoviruses (12), the PEMV PTE is not associated with any long-distance RNA-RNA interaction and instead likely functions to assist in ribosomes binding to the adjacent, upstream kl-TSS (28). The kl-TSS is a two hairpin, three-way branched element that forms a three-dimensional (3-D) T-shaped structure similar to the three hairpin/two pseudoknot TCV TSS. Unlike the TCV TSS, the kl-TSS binds to 40S ribosomal subunits in addition to 60S and 80S ribosomes and can engage in a simultaneous long-distance kissing-loop interaction with a 5′ end coding sequence hairpin (13, 28). It has been proposed that the kl-TSS binds to ribosomal subunits following translation termination to facilitate reinitiation at the 5′ end.
With the possible exception of the paired PTE/kl-TSS elements of PEMV and an isolate of Melon necrotic spot virus that acquired an additional 3′CITE by recombination (29), only single 3′CITEs have been discovered in 3′ regions of plant viruses, leading to the general assumption that a single element is normally sufficient for all necessary translation enhancement. We now report that PEMV contains a second T-shaped structure downstream from the kl-TSS/PTE, located in the same position in the genome as the TCV TSS. The 3′ proximal PEMV TSS (3′TSS) is composed of a similar combination of three hairpins and two pseudoknots as the TCV TSS and also binds to 80S and 60S ribosomes but with lower affinity. A hairpin upstream of the PEMV 3′TSS (kl-H) can participate in a long-distance interaction with the same 5′ proximal coding region hairpin that is utilized by the kl-TSS in reporter constructs in vivo and in vitro, and together they can effectively substitute for the kl-TSS/PTE if placed proximal to the translation termination site. The kl-H and 3′TSS are critical for accumulation of PEMV in vivo, but the kl-H does not connect with the 5′ proximal coding region hairpin as it does in the reporter construct. This suggests that the kl-H/3′TSS must enhance the translation of a critical factor other than the 5′ proximal ORF.
MATERIALS AND METHODS
In-line structure probing.
In-line probing was performed as previously described (23). Briefly, PEMV TSS RNA transcripts were 5′ end labeled with [γ-32P]ATP and purified by electrophoresis through 5% denaturing polyacrylamide gels. End-labeled RNA was heated at 75°C and allowed to slow cool to room temperature. For in-line probing, 5 pmol of end-labeled RNA was allowed to self-cleave at 25°C for 14 h in 1× in-line probing buffer (50 mM Tris-HCl [pH 8.5], 20 mM MgCl2). Reaction products were resolved on an 8% denaturing polyacrylamide gel. At least three independent in-line probing assays were performed, and only reproducible differences are described.
Accumulation of PEMV in protoplasts.
Protoplasts were prepared from callus cultures of Arabidopsis thaliana (ecotype Col-0) and were transformed with PEMV gRNA transcripts using polyethylene glycol as previously described (30). First-strand cDNA was synthesized from total RNA prepared from infected protoplasts using MMLV reverse transcriptase (Invitrogen) and subjected to TaqMan real-time PCR assays using a LightCycler 480 real-time PCR system (Roche Applied Science), as previously described (13). Raw data were analyzed using LightCycler 480 real-time PCR system software. Each sample was assayed in triplicate in every experiment, and at least three independent experiments were performed.
Purification of 80S ribosomes and 40S/60S ribosomal subunits from Arabidopsis thaliana protoplasts and filter binding assays.
Plant 80S ribosomes and 40S/60S ribosomal subunits were purified as previously described (31). Briefly, Arabidopsis protoplasts prepared from callus cultures were lysed and centrifuged, and the resulting supernatants were subjected to centrifugation through a 25% glycerol cushion. Ribosome pellets were resuspended in storage buffer [50 mM HEPES-KOH (pH 7.6), 5 mM Mg(CH3COO)2, 50 mM NH4Cl, 25% glycerol, 1 mM dithiothreitol] and subjected to salt washing to remove any associated tRNAs and translation factors, as described previously for yeast ribosomes (22). For purification of 40S/60S ribosomal subunits, purified 80S ribosomes were subjected to sucrose gradient centrifugation and 40S- or 60S-containing fractions applied to Amicon ultra (100k) columns (Millipore) for buffer exchange. Purified 40S/60S ribosomal subunits were stored in storage buffer (2 to 10 pmol/μl) at −80°C. Filter binding assays were performed as described previously (21).
SHAPE structure probing.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) structure probing was performed essentially as previously described (32). Briefly, full-length PEMV gRNA was synthesized in vitro by using T7 RNA polymerase and subjected to phenol-chloroform extraction and ethanol precipitation. RNA was denatured at 95°C for 3 min and snap-cooled on ice for 2 min. The RNA was then folded in SHAPE folding buffer [80 mM Tris-HCl (pH 8.0), 11 mM Mg(CH3COO)2, 160 mM NH4Cl] at 37°C for 20 min. The folded RNA was divided into two aliquots, and each aliquot was incubated with either 15 mM N-methylisatoic anhydride (NMIA) or dimethyl sulfoxide (DMSO) at 37°C for 40 min. Oligonucleotides labeled at their 5′ ends with [γ-32P]ATP were used for primer extension reactions with SuperScript III reverse transcriptase (Invitrogen) as previously described (32). Six oligonucleotides (positions 3595 to 3614, 3830 to 3849, 4018 to 4037, 4121 to 4140, 4235 to 4252, and 4302 to 4318) were used to probe the structure of the 3′UTR of PEMV. To provide structural data for the 3′ terminus of the 3′UTR, 214 additional nucleotides from the pUC19 vector located downstream of the 3′ end were included in the in vitro synthesis of full-length PEMV gRNA to generate a 4,466-nt RNA (used only to structurally probe the 3′ terminus). Reaction products were then subjected to electrophoresis through 8% denaturing polyacrylamide gels, and radioactive bands were visualized with a phosphorimager. The intensities of individual bands were inspected visually and assigned values of weak/moderate and moderate strong/strong. The nucleotide positions were identified by reference to the sequencing ladder generated from unmodified RNA by Sanger methods. RNA secondary structural maps were generated from structure probing results and the best-fitting Mfold predictions (33).
In vitro translation and trans inhibition assays.
In vitro translation and trans inhibition assays were performed as previously described (28). Briefly, 3 pmol of in vitro-synthesized RNA transcripts from designated translation reporter constructs were used for a 15-μl translation reaction using wheat germ extracts (WGE; Promega) according to the manufacturer's instructions. The luciferase activity was measured by using a luciferase assay reporter system (Promega) and a Modulus microplate multimode reader (Turner BioSystems). For trans inhibition assays, 30 pmol of competitor RNAs was added to the translation reactions prior to incubation.
In vivo translation assays.
In vivo translation assays were performed as previously described (13). Briefly, 30 μg of uncapped parental and mutant Firefly luciferase transcripts and 10 μg of Renilla luciferase control transcripts were introduced into 5 × 106 A. thaliana (ecotype Col-0) protoplasts using polyethylene glycol-mediated transformation. Luciferase activity was measured 18 h later using a luciferase assay reporter system (Promega) and a Modulus microplate multimode reader (Turner BioSystems).
Molecular modeling.
Three-dimensional models are based on the 3′TSS SHAPE-probed secondary structure information for the 82-nt fragment spanning positions 4128 through 4209. Two models of the 3-D structure were built: one with a 3-bp pseudoknot ψ3 (residues 4128, 4129, and 4130 pairing with residues 4140, 4139, and 4138, respectively) and one with 4 bp (adding pair 4131-4137). RNAComposer (34, 35) web server (http://euterpe.man.poznan.pl/Home) was first used to build the preliminary models, and RNA2D3D (36) was used to edit certain structural collisions in those models. The corrected models (two variants for 3-bp ψ3 and one for 4-bp ψ3) were energy minimized and then placed in explicit solvent boxes, equilibrated, and subjected to molecular dynamics (MD) simulations. Since the initial models and a standard MD protocol showed potential problems with the stability of ψ3, several MD protocols with some restraints meant to temporarily stabilize the base pairs in ψ3 were run (see “Molecular dynamics simulation protocol,” below, for details). Multiple 100-ns-long MD simulations were conducted.
Molecular dynamics simulation protocol.
Amber 12 with the ff10 Cornell force field for RNA was used to perform MD simulations, using the Particle Mesh Ewald (PME) summation method to calculate the electrostatic interactions (37–39). The energy-minimized 3′TSS models (82 nt, 2,625 atoms) were solvated in a TIP3P water box with a buffer zone of 15.0 Å. The RNA was neutralized with 81 Na+ ions. Additional Na+/Cl− ion pairs were added to reach a relative salt concentration of 0.1 M (varying from 64 to 69, depending on the 3′TSS model used). The total sizes of the systems ranged from 90,560 to 97,272 atoms. A multistep equilibration protocol was used that started with solvent equilibration (minimization, heating, and short dynamics stages), while the RNA was being subjected to motion restraints (holding) decreased at every stage. The last phase of the equilibration was performed for 2.0 ns without or with (separate equilibrations) distance restraints placed on the hydrogen bonds of the 3- or 4-bp ψ3 (depending on the initial model). The entire system was equilibrated at 300K using a Berendsen thermostat (40). Other parameters included use of periodic boundary condition, a cutoff of 9 Å for the nonbonded interactions, and SHAKE applied to all hydrogen bonds in the system. Pressure was maintained at 1.0 Pa utilizing the Berendsen algorithm (40). Production runs of 100-ns duration (excluding the equilibration phase) were conducted without or with the ψ3 base pair restraints lasting 10 and 25 ns of the total production run.
RESULTS
The PEMV 3′ proximal region contains a series of hairpins and pseudoknots similar to those present in several carmoviruses.
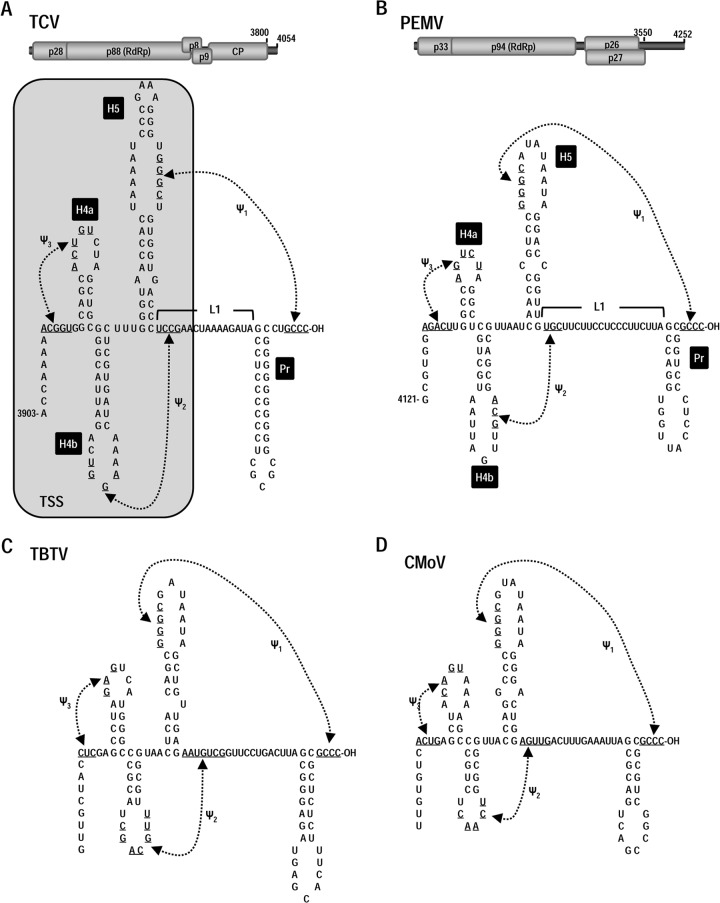
Examination of the 3′ terminus of PEMV revealed sequences that could potentially fold into a 3′ terminal hairpin (Pr), an upstream long linker region (L1), and an adjacent hairpin (H5). This configuration of elements is very similar to those found in identical locations in all members of the Carmovirus genus (23). Complementary sequences in the PEMV H5 apical loop (5′ GGGC) and at the 3′ terminus (3′ OH-CCCG) also suggested that a pseudoknot (ψ1) connects these two motifs, similar to pseudoknots found in all members of the Tombusviridae (41, 42). Three carmoviruses—TCV, Cardamine chlorotic fleck virus (CCFV), and Japanese iris necrosis ringspot virus (JINRV)—also contain juxtaposed hairpins H4a and H4b just upstream of H5, with two additional pseudoknots (ψ2 and ψ3) connecting their apical loops with nearby single-stranded sequences (23) (Fig. 1A). Disrupting any of these hairpins and pseudoknots in TCV results in significantly reduced levels of virus accumulation in vivo (23). Visual inspection of sequences upstream of H5 in PEMV revealed that a similar configuration of H4a, H4b, ψ2, and ψ3 was also a possibility (Fig. 1B). Comparable TCV-like structural organization was also possible for the umbraviruses Carrot mottle virus (CMoV) and Tobacco bushy top virus (TBTV) (Fig. 1C and D). In contrast, Carrot mottle mimic virus may only contain H5, Pr and ψ1, and all elements were absent from the 3′ end of Groundnut rosette virus (data not shown).
FIG 1.
Conserved structural features at the 3′ ends of TCV and three umbraviruses. (A) Top, genome organization of the carmovirus TCV. p28 is a replication-required protein, and its readthrough product p88 is the viral RdRp. p8 and p9 movement proteins are expressed from one sgRNA, and the capsid protein (CP) is expressed from a second sgRNA. Bottom, secondary and tertiary structure elements at the 3′ terminus of TCV. All 15 carmoviruses contain Pr, H5, and ψ1. Most carmoviruses contain H4b and fewer contain both H4b and ψ2 (63). Only TCV, CCFV, and JINRV contain all hairpins and pseudoknots shown (23). The boxed, shaded region denotes that hairpins H4a, H4b, H5, and pseudoknots ψ2 and ψ3 fold into a 3-D T-shaped structure (TSS). (B) Top, Genome organization of PEMV. p33 is a putative replication-required protein, and its −1 ribosomal frameshift protein is the RdRp. p26 and p27 are movement required proteins expressed from the single sgRNA. Bottom, possible structural organization at the 3′ terminus of PEMV based on the structures found in TCV. (C and D) Phylogenetically conserved elements at the 3′ end of TBTV (C) and CMoV (D).
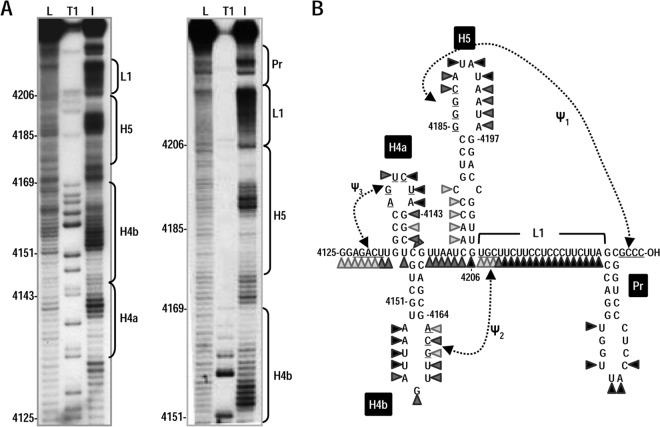
A 170-nt 3′ terminal PEMV fragment was subjected to in-line probing to determine whether an experimentally derived structure was consistent with H4a, H4b, H5, ψ2, and ψ3. In-line probing interrogates the flexibility of each nucleotide within an RNA fragment, since only flexible (i.e., non-base-paired) nucleotides can position their 2′ oxygen, backbone phosphorus, and adjacent 5′ oxygen in the in-line configuration that is needed for nucleophilic attack of the 2′ oxygen on the phosphorus and subsequent backbone cleavage. The level of cleavage at each nucleotide in a radiolabeled fragment is detected following denaturing gel electrophoresis and autoradiography and is directly proportional to the flexibility of the nucleotide (32, 43) within an averaged population of structures.
The cleavage pattern for the 3′ region of PEMV is presented in Fig. 2B, and typical in-line cleavage gels are shown in Fig. 2A. The flexibility profiles for Pr and H4b residues were consistent with hairpins in these locations, with inflexible stem residues and highly flexible loop residues. In the apical loop of H4b, residues 4161GCA showed reduced cleavages compared to the remaining apical loop residues, a finding consistent with their participation in ψ2. L1 was also highly flexible with the exception of putative ψ2 residues 4207UGC. The flexibility of H5 residues was mainly consistent with the predicted hairpin. All apical loop residues were highly flexible, with the exception of 4185GGG, which are proposed to participate in ψ1. The residues on the 5′ side of the lower H5 stem were weakly flexible, similar to the pattern found for the same H5 residues in a fragment containing the TCV 3′ region that was subjected to in-line probing (23). Unlike H5 and H4b, the flexibility pattern for PEMV H4a within this fragment was not consistent with a hairpin, with 3′ side “stem” residues exhibiting considerable flexibility. These results suggest that this PEMV fragment contains H4b, H5, Pr, ψ1, and ψ2.
FIG 2.
In-line probing of the 3′ end of PEMV. (A) Susceptibility of residues at the 3′ end of PEMV to in-line cleavage. The 170-nt 3′ terminal fragment was radiolabeled at the 5′ end and incubated at 25°C for 14 h, followed by denaturing gel electrophoresis. The locations of different putative 3′ elements are indicated to the right of each autoradiogram. L, OH− treated ladder; T1, partial RNase T1 digest to denote location of guanylates; I, in-line cleavage of the fragment. The intensity of each band is proportional to the flexibility of the residue at that location. (B) Susceptibility of residues in the putative structure of PEMV 3′ region to in-line cleavage. Darker triangles denote stronger cleavage.
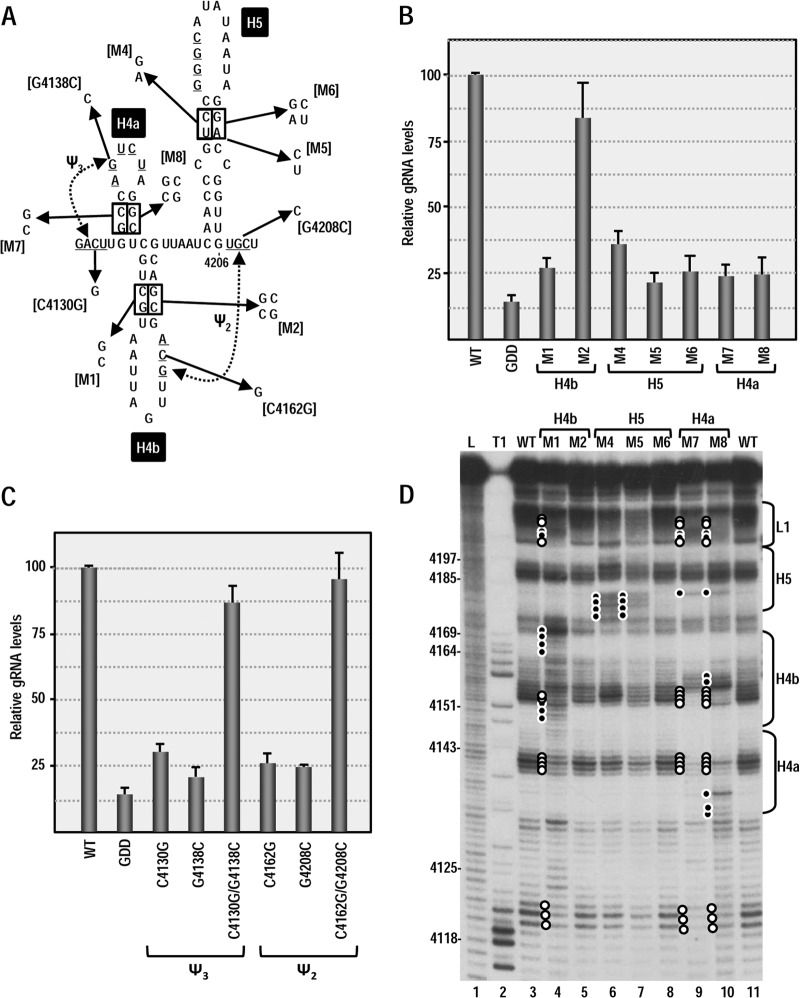
Since it is possible that RNA structures exist in the full-length virus in vivo that are not present or discernible in the averaged population of in vitro folded molecules (and vice versa), single and compensatory mutations were generated in each putative hairpin and pseudoknot in full-length PEMV, and virus levels were assayed in Arabidopsis thaliana protoplasts using quantitative PCR (Fig. 3). Altering 4150CG in the H4b stem to GC (construct M1) reduced viral RNA levels to 27% of the wild type (WT) and combining these mutations with compensatory alterations across the stem (construct M2) restored accumulation to 82% of the WT (Fig. 3B). The C-to-G transversion at position 4162 (C4162G) in the H4b apical loop that was designed to disrupt ψ2 also reduced accumulation to 27% of the WT. Disruption of the putative partner residue in the L1 linker region (G4308C) had a similar negative effect on virus accumulation (27% of the WT), whereas the two compensatory mutations together restored PEMV accumulation to 94% of the WT. These results support the presence of H4b and ψ2 and their importance for PEMV accumulation in vivo.
FIG 3.
Effect of alterations in 3′ elements on PEMV accumulation in protoplasts. (A) Single and compensatory mutations generated in the 3′ hairpins and pseudoknots that may comprise a TSS. The names of the alterations are indicated in brackets. (B) Relative levels of full-length WT PEMV and PEMV containing alterations in 3′ hairpins accumulating in Arabidopsis protoplasts at 24 h after inoculation. Standard deviations from three independent experiments are shown. GDD, PEMV nonreplicating control with an altered GDD RdRp active-site motif. (C) Relative levels of full-length PEMV accumulating in Arabidopsis protoplasts containing alterations in ψ2 or ψ3. (D) In-line probing of PEMV 3′ terminal fragment containing alterations in hairpins. Residues that are more susceptible to cleavage in fragments containing mutations are denoted by a solid circle. Open circles denote residues showing reduced cleavage levels. The locations of hairpins and linker regions are indicated on the right.
Disruption of either the 5′ or the 3′ sides of the H5 stem (M4 and M5) reduced accumulation to 34 and 21% of WT, respectively. The compensatory alteration that combined these two mutations (M6), however, did not restore PEMV accumulation. Likewise, altering the stem of H4a (construct M7) reduced accumulation to 24% of the WT, which was not restored by inclusion of the compensatory mutations (construct M8) (Fig. 3B). In contrast, disrupting ψ3 by altering either the loop of H4a (G4138C) or the partner residue upstream of H4a (C4130G) reduced accumulation to 30 and 19% of the WT, respectively, whereas the two compensatory mutations together restored accumulation to 83% of the WT.
The inability of H4a and H5 compensatory mutations to restore accumulation could indicate (i) the absence of these structures, (ii) a requirement for specific sequences within the stems, or (iii) that the stem-disrupting alterations caused stable, inadvertent structural changes that were not corrected by the compensatory mutations. To distinguish among these possibilities, in-line probing was conducted for the 3′ fragment of PEMV in the presence of the single and compensatory stem alterations in H4b, H5, and H4a (Fig. 3D). Disrupting the H4b stem (M1) caused significant changes in the flexibility of residues throughout the H4a, H4b, and L1 regions (Fig. 3D, compare lane 3 with lane 4), suggesting that this hairpin is part of an integrated 3-D structure. The compensatory mutations in the stem of H4b (M2) restored the WT flexibility pattern, indicating that the H4b structure refolded properly. Altering either side of the upper stem of H5 (M4 and M5) disrupted the structure in that region (Fig. 3D, lanes 6 and 7). The compensatory alterations (M6) restored the structure of H5 to its WT configuration (Fig. 3D, lane 8), indicating that a hairpin is present at this location. Therefore, the compensatory M6 alterations that did not restore virus accumulation may either disrupt a necessary sequence requirement or affect the structure of the hairpin within the full-length virus.
In-line probing of the fragment containing H4a alteration M7 revealed significant loss of flexibility in the putative apical loop of H4a and the apical loop of H4b, as well as alterations in H5, and L1. Inclusion of H4a alterations designed to be compensatory (M8) did not restore the WT structure but rather caused additional flexibility changes in H4a and H4b. One possible explanation is that the M7 alterations created conditions allowing for inadvertent pairing between the apical loops of H4a (5′ CUAG) and H4b (3′GAUU), and this pairing was maintained in M8. Although these results do not demonstrate the existence of H4a, the presence of all other elements that together with H4a form a TSS in TCV (H4b, H5, ψ2, and ψ3), along with conservation of a hairpin at the H4a location in CMoV and TBTV, suggest that H4a may be present in PEMV.
The SHAPE-derived flexibility profile of H4a residues in full-length PEMV is consistent with a hairpin in this location.
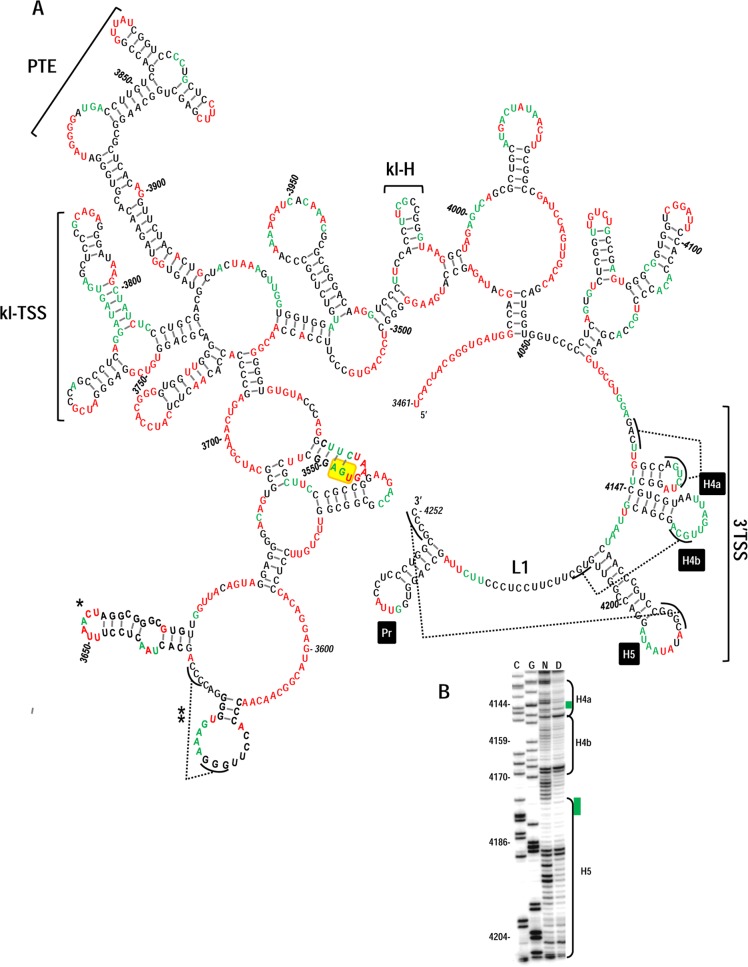
In a further attempt to provide evidence for the presence of H4a in PEMV, selective 2′OH acylation analyzed by primer extension (SHAPE) was used to probe the structure of the 3′UTR of PEMV within full-length gRNA. To probe the structure of the 3′ terminal region only, 214 additional plasmid-derived nucleotides were present at the 3′ end of full-length PEMV gRNA. SHAPE interrogates the flexibility of residues in full-length RNAs by their level of conjugation to NMIA, which restricts primer extension by reverse transcriptase. By prohibiting Mfold (33) from pairing moderate and strong NMIA-reactive nucleotides, a secondary structure emerged that was a good fit with most of the SHAPE structural data (Fig. 4A). The structure contains several known elements within the 3′UTR, including the kl-TSS and adjacent PTE 3′CITEs, and a hairpin at position 3635 of unknown function that is conserved in all umbraviruses (A. E. Simon, unpublished results). The region of the structure least supported by the SHAPE data lies just upstream of this conserved hairpin (positions 3612 to 3636). Originally, Mfold predictions suggested that the conserved hairpin lower stem should contain three additional base pairs (A:U, C:G, and C:G). The poor support for these pairings by the SHAPE data could be explained by the presence of an H-type pseudoknot connecting the loop of the adjacent hairpin with at least two of these residues, a structure predicted by pknotsRG (44).
FIG 4.
Proposed secondary structure for the 3′UTR of PEMV. SHAPE was conducted on full-length PEMV gRNA as described in Materials and Methods. (A) Residues with moderately high and high reactivity to NMIA are colored red, and residues with low or moderate reactivity are colored green. The locations of the 3′TSS, kl-TSS, and PTE are shown. kl-H is a hairpin identified as also engaging in a long-distance interaction (see the text). Single asterisk denotes a hairpin that is sequence, structure, and positionally conserved in all sequenced umbraviruses (A. E. Simon, unpublished results). A double asterisk denotes a potential pseudoknot predicted by pknotsRG (44). The termination codon for p27 is boxed in yellow. (B) Portion of one of the SHAPE phosphorimages used for the structural prediction. Lanes C and G, ladder lanes of cytidylates and guanylate positions; lane N, NMIA-treated sample; lane D, control DMSO-treated sample denoting reverse transcriptase stops in the absence of NMIA. Green bars denote regions in H4a or H5 that are susceptible to in-line cleavages but not reactive with NMIA.
At the 3′ end of PEMV, the structures of Pr, H5, and H4b are compatible with the SHAPE-derived data, with flexible bases restricted to apical loops. Residues in the 5′ side lower stem of H5 were no longer weakly flexible according to SHAPE (Fig. 4B), suggesting that the weak cleavages at these residues detected by in-line probing were a function of either the different procedure or that in-line probing used a shorter fragment that affected the structure of the hairpin. A major difference was found in the flexibility of residues within L1, most of which were no longer flexible in the full-length gRNA, suggesting a possible interaction between these residues and upstream sequences not present in the fragment used for in-line probing. Searches of upstream sequences for a likely pairing partner, however, did not reveal any obvious candidates.
Importantly, the flexibility pattern of residues within H4a as assayed by SHAPE in full-length PEMV was now consistent with a hairpin at this location. Unlike the in-line probing data, the upper three residues in the 3′ side of the H4a stem were not flexible in the SHAPE assay, suggesting that these residues are paired across the stem. The SHAPE data also suggest that the putative base pair 4133G:U4146 at the base of the stem may not be stable and that ψ3 residue 4127A is not pairing with 4141U in the averaged population.
In the SHAPE-derived structure of the 3′ region of PEMV, the 3′UTR, together with 73 upstream residues, was predicted to form a fold-back RNA domain. Since the parental reporter construct (5′89+3U) used to identify the kl-TSS as a 3′CITE contained only the precise 3′UTR downstream of the firefly luciferase reporter ORF, the possibility existed that adding 73 nt of upstream coding sequence would enhance translation by improving the formation of important structures in the 3′UTR. However, the addition of these 73 residues to the reporter construct had no significant effect on luciferase activity (data not shown), suggesting that the entire domain is not necessary for proper translation enhancement in the reporter construct.
The PEMV 3′ region is predicted to form a TSS.
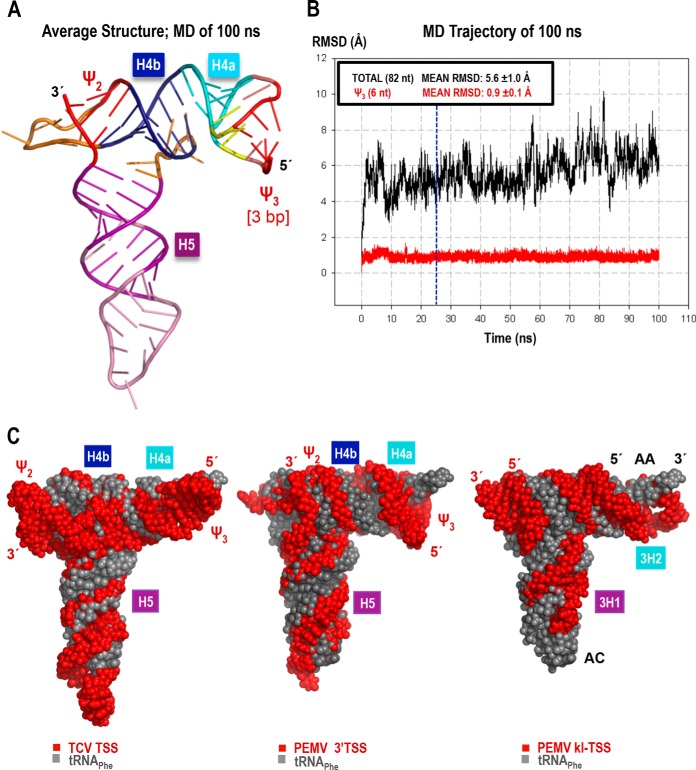
In TCV, H4a, H4b, H5, ψ3, and ψ2 fold into a tRNA-shaped structure (TSS) that binds to 80S ribosomes and 60S ribosomal subunits and functions as a 3′CITE (22, 23). To determine whether the region flanked by ψ3 and ψ2 in PEMV also adopts a 3-D T-shaped structure, molecular models were built for this region of PEMV, and their stability was evaluated with the aid of MD simulations (see Materials and Methods for details). The smaller size of the PEMV H4a hairpin loop (relative to that of TCVs) made it difficult to accommodate a 4-bp ψ3. Therefore, a 3-bp ψ3 model was also constructed. In MD simulations, the two variants of the latter model converged to nearly identical T-shaped structures reminiscent of the TCV TSS and similar in size to a tRNA, as illustrated for one model in Fig. 5A and C. All helices (H4a, H4b, and H5) and ψ2 were stable in all MD protocols used. MD simulations also revealed a lower stability for ψ3 base pairs than was previously observed for the TCV TSS (23). An MD protocol using ψ3 base pair restraints in the equilibration phase and during the first 25 ns of the production run was necessary to maintain a stable 3-bp ψ3 in the remainder of the 100-ns simulation (Fig. 5B). Restraints applied for only the first 10 ns of the production MD yielded an average structure maintaining the second and third base pairs of ψ3; however, the first two base pairs opened by the end of the run. The PEMV TSS model with 4 bp in ψ3 also had problems maintaining all base pairs despite a 25-ns-long restrained phase of the production MD run (the first and second base pairs opened intermittently after ∼72 ns of the run; data not shown). Although it is possible that the quality of the initial models (built from a database of relatively large fragments fit together) affected the MD results, the long equilibration protocol and the extended stabilization with base pair restraints applied to ψ3 should have overcome any initial structural strain. Considering that no special measures were necessary to maintain the stability of the TCV ψ3, these results suggest that PEMV ψ3 is less stable. This structural weakness may be contributing to the experimentally observed weaker ribosome binding relative to the TCV TSS (see below).
FIG 5.
3-D model and molecular dynamics simulation of the PEMV 3′TSS. The model with a 3-bp ψ3 is shown (see the text for details). (A) Energy-minimized average structure based on the 100-ns-long MD simulation. (B) The all-atom root mean square deviation (RMSD; 82 nt; 2,625 atoms), measured relative to the first structure of the MD simulation, is 5.6 Å (black). RMSD values plotted in red were calculated for all atoms of the 6 nt involved in ψ3 (3 bp long in this model, 191 atoms). The blue vertical line at the 25-ns point in the MD indicates when restraints on the first three base pairs were released (see the text for details). The low mean RMSD and standard deviation (0.9 ± 0.1 Å) of ψ3 illustrates the stability of the base pairs after the restraints were lifted. (C) Comparison of the 3-D structure models of the TCV TSS (left), PEMV 3′TSS (center), and PEMV kl-TSS (13) (right), all shown in red, aligned with the tRNAPhe structure (PDB 1EHZ) (gray). The TSS and kl-TSS 5′ and 3′ positions are labeled in red, while the tRNA's 5′ and 3′ positions, the anticodon loop (AC), and the acceptor stem (AA) are labeled in black over the most exposed tRNA (right).
The PEMV 3′TSS binds to 80S ribosomes and 60S ribosomal subunits.
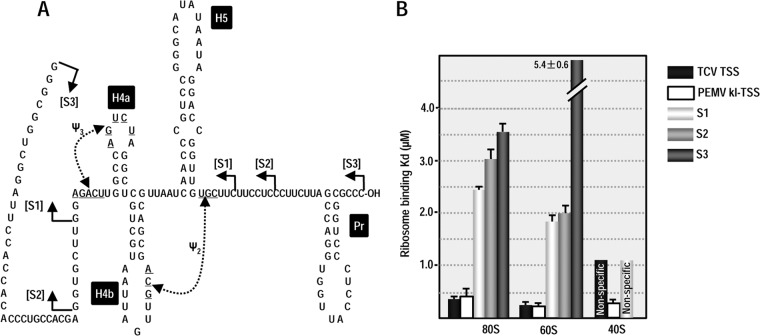
TCV TSS binding to 80S ribosomes and 60S ribosomal subunits is important for its function as a 3′CITE (22). The kl-TSS, which also binds to 80S ribosomes and 60S subunits, additionally binds to 40S subunits, although binding is strongest to 60S subunits (13). To determine whether the 3′TSS of PEMV is a ribosome-binding element, filter binding assays were conducted using Arabidopsis ribosomes purified from callus-derived protoplasts (31). The initial fragment used in the assay (S1) extended from just before ψ3 to just after ψ2 (Fig. 6A). Fragment S1 bound 80S ribosomes with a Kd of 2.4 μM, which is 5-fold weaker than the TCV TSS and PEMV kl-TSS (Fig. 6B). To determine whether a larger fragment might improve binding (i.e., might better support the TSS structure), the fragment was extended by 7 nt on the 5′ side and 6 nt on the 3′ side. This fragment (S2) was a weaker template for 80S ribosome binding (Kd = 3.1 μM). Using a larger fragment that was previously used for in-line probing (S3) further weakened binding (Kd = 3.6 μM).
FIG 6.
Ribosome binding to the 3′TSS. Filter-binding assays were conducted using three different-sized fragments that contain the 3′TSS (S1, S2, and S3). (A) Locations of the endpoints of the fragments used for filter binding. (B) One to 60 pmol of [32P] 5′-end-labeled fragments were combined with 15 pmol of salt-washed ribosomes or separated subunits purified from Arabidopsis protoplasts. The standard deviations are shown for three experiments. The values for the TCV TSS (31) and PEMV kl-TSS (13) were previously published and are presented here for comparison.
We also subjected all three PEMV fragments to 60S subunit binding (Fig. 6B). Binding to 60S by fragments S1 and S2 was slightly stronger compared to 80S (Kd = 1.8 and 2.0 μM, respectively), as was previously found for the TCV TSS and PEMV kl-TSS. Fragment S3 binding to 60S was very weak (Kd = 5.4 μM), suggesting that the TSS structure (i.e., H4a or ψ3) may not be properly forming.
Since PEMV fragment S1 achieved the best binding to 80S and 60S, this fragment was also subjected to 40S filter binding. Similar to the TCV TSS, fragment S1 was unable to bind specifically to 40S subunits. Altogether, these results suggest that the PEMV 3′TSS can bind to 80S and 60S ribosomes, similar to the TCV TSS. However, reduced binding compared to the TCV TSS could reflect either weaker binding by the element, a need for additional factors to bind more efficiently, or that the 3′TSS structure is not forming properly within the truncated fragments, as suggested by the in-line probing data for H4a and the MD simulation data for ψ3.
The 3′TSS and upstream hairpin kl-H comprise a functional 3′CITE.
In PEMV, the adjacent kl-TSS and PTE are 3′CITEs thought to function together to efficiently attract ribosomes to the kl-TSS (28). As described above, the kl-TSS can bind to ribosomes and simultaneously engage in a long-distance kissing-loop interaction between its 5′ side hairpin (3H1) and a stable coding region hairpin (5H2) near the gRNA 5′ end (28). Major questions therefore are whether the PEMV 3′TSS functions as a third 3′CITE and, if so, whether an additional kissing-loop interaction connects the 3′TSS with the 5′ end. It should be noted that lack of a specific connection between the 3′TSS and the 5′ end would not abrogate the hypothesis that the 3′TSS is an independent 3′CITE since no discernible RNA-RNA interaction connects the TCV TSS with its 5′ end (22).
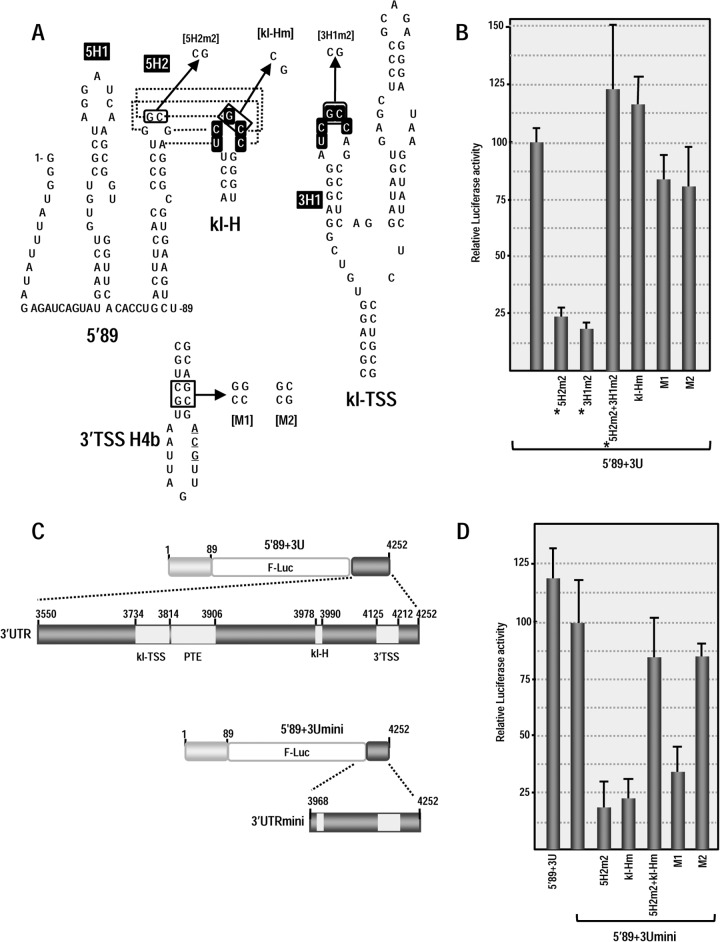
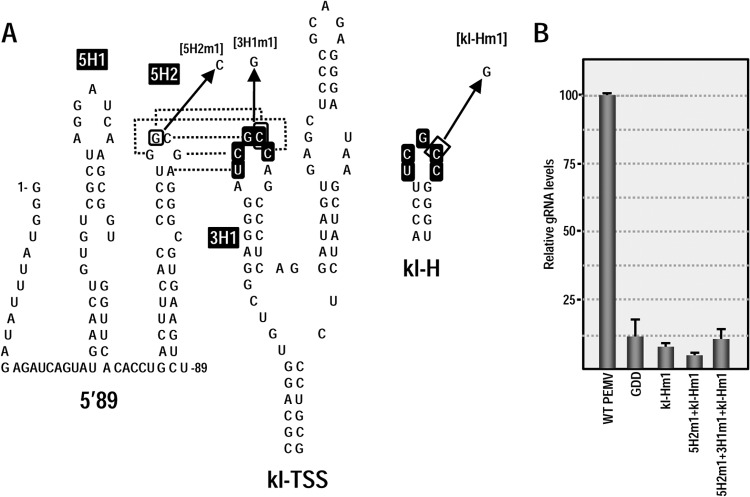
To begin answering these questions, we searched the PEMV 3′UTR for a second sequence that might putatively engage in a kissing-loop interaction with either of the hairpins in the 5′ 89 nt of PEMV (5H2 or upstream hairpin 5H1; Fig. 7A) or other nearby downstream sequences. One possibility, the apical loop of hairpin kl-H, contains the same five kissing-loop residues (UCGCC) found in the apical loop of kl-TSS hairpin 3H1, the hairpin that interacts with 5H2 (Fig. 7A). Discounting the structures branching off the central “backbone” of the 3′UTR, the kl-H is located ∼30 nt upstream of the 3′TSS (Fig. 4A), suggesting that the kl-H might also be capable of engaging in a long-distance interaction with 5H2 to position the 3′TSS (and bound ribosome/ribosomal subunit) proximal to the gRNA 5′ end (Fig. 7A).
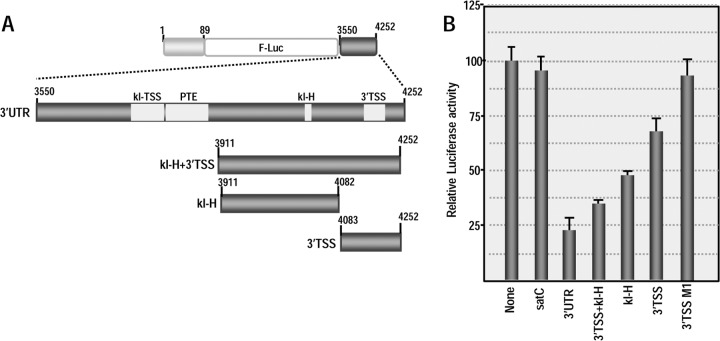
FIG 7.
3′TSS and kl-H function as 3′CITEs in WGE when proximal to the luciferase reporter termination codon. (A) Location of mutations in 3′TSS, kl-H, and 5′89. The putative kissing-loop interaction between hairpin 5H2 in the 5′89 nt of PEMV and the kl-H is shown. Sequences shared between the kl-H and hairpin 3H1 of the kl-TSS (which is known to interact with 5H2) are boxed. (B) Relative luciferase activity in WGE programmed with 5′89+3U containing mutations in the kl-H and 3′TSS. Asterisks denote data previously published (13) and presented here for comparison. The data are from three independent experiments performed in triplicate, and the standard deviations are shown. (C) Diagram of 5′89+3Umini. This construct contains a deletion that removes the kl-TSS and PTE and places the kl-H just downstream of the luciferase ORF. (D) Relative luciferase activity of 5′89+3Umini and 5′89+3Umini that contains mutations in the kl-H and 3′TSS shown in panel A.
Using a reporter construct containing firefly luciferase flanked by the 5′ 89 nt and 3′UTR of PEMV (5′89+3U), we previously reported that disrupting the kissing-loop interaction between 3H1 and 5H2 reduced translation in protoplasts and wheat germ extracts (WGE) by at least 4-fold, whereas compensatory mutations restored translation to greater than WT levels (13, 28) (Fig. 7A and B). To determine whether the kl-H contributes to translation, mutations were generated in the kl-H loop in 5′89+3U that would disrupt any kissing-loop interaction with 5H2 (Fig. 7A). Translation of 5′89+3U containing this alteration (kl-Hm) in WGE was slightly enhanced compared to the parental construct (116% of the WT) (Fig. 7B), suggesting that any interaction between kl-H and 5H2 is not contributing positively to translation. Mutations in H4b of the 3′TSS that disrupt and reform the hairpin structure (M1 and M2 alterations in the H4b stem) produced only 15 and 20% reductions in luciferase activity (Fig. 7B). These results suggest that neither the kl-H nor the 3′TSS are important for translation of the parental reporter construct in WGE.
In 5′89+3U, the kl-TSS/PTE are located spatially proximal to the 3′ end of the luciferase ORF (see Fig. 4A). If the purpose of a TSS is to recycle ribosomes back to the 5′ end following translation termination, then proximity to where ribosomes terminate translation may be an important factor for TSS function. To determine whether the kl-H and 3′TSS might function as translational enhancers if they are located proximal to the 3′ end of the luciferase ORF, the sequence between the 5′ end of the 3′UTR and the kl-H was deleted, generating construct 5′89+3Umini. In addition to placing the kl-H and 3′TSS proximal to the luciferase ORF termination site, this deletion eliminated both the kl-TSS and PTE, which are critical elements for translation of 5′89+3U. The luciferase activity of 5′89+3Umini was 84% of the 5′89+3U levels in WGE (Fig. 7D), indicating that the kl-TSS/PTE were no longer needed for translation. Mutations in the kl-H (kl-Hm) and 3′TSS (M1) that previously had little effect on the translation of 5′89+3U now reduced the translation of 5′89+3Umini in WGE by 78 and 65%, respectively. The compensatory mutations in H4b (M2) enhanced translation to 84% of the 5′89+3Umini levels, strongly suggesting that the 3′TSS is being utilized as a translation enhancer. Mutation in the 5H2 loop in 5′89-U3mini reduced translation to 18% of 5′89+3Umini levels, suggesting that a long-distance interaction with this hairpin is still important in 5′89-U3mini. Combining the mutations in 5H2 and kl-H that were designed to be compensatory restored translation to 84% of the 5′89-U3mini levels, strongly suggesting that a kissing-loop interaction was forming between 5H2 and kl-H in WGE.
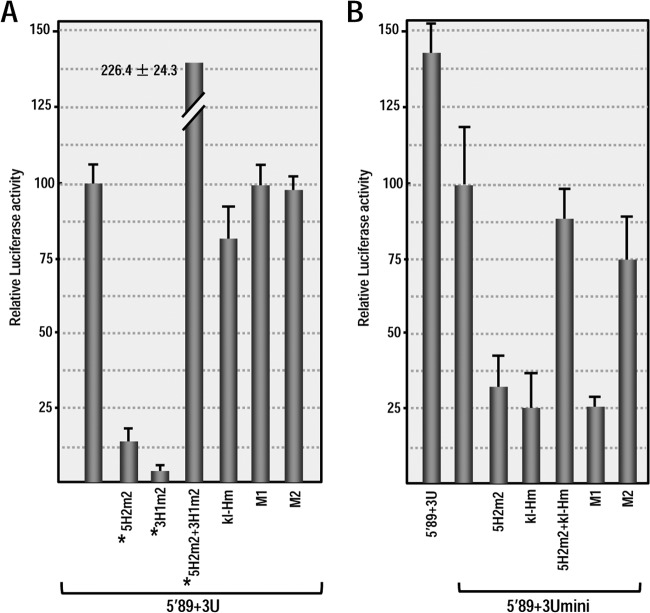
To confirm these results using a more natural in vivo system, Arabidopsis protoplasts were transformed with RNA transcripts of parental (5′89+3U and 5′89+3Umini) and mutant constructs, and the luciferase activity was measured 18 h later. As in WGE, mutations disrupting the kl-H or 3′TSS had no significant effect on the translation of 5′89+3U (Fig. 8A). In contrast, mutating the kl-H in 5′89+3Umini reduced translation to 28% of the parental construct (Fig. 8B). When the kl-H mutations were combined with 5H2 mutations that restored the putative long-distance RNA-RNA interaction, the luciferase activity increased to 90% of the parental construct. Disrupting the 3′TSS by mutating H4b (construct M1) reduced luciferase activity to similar levels as the kl-H mutation (to 27% of the parental construct) and the compensatory H4b M2 mutations increased the luciferase activity to 76% of the parental construct (Fig. 8B). Altogether, these results strongly suggest that when placed proximal to where ribosomes are terminating translation, the 3′TSS and kl-H function to enhance translation in WGE and in protoplasts in the absence of the kl-TSS and PTE. In addition, a long-distance RNA-RNA interaction with the 5′ end is likely important for kl-H function in enhancing translation.
FIG 8.
3′TSS and kl-H function as 3′CITEs in protoplasts when proximal to the luciferase reporter termination codon. (A) Relative luciferase activity in protoplasts at 18 h after transformation with 5′89+3U transcripts containing mutations in the kl-H and 3′TSS that were described in Fig. 7A. Asterisks denote data previously published (13) and presented here for comparison. The data are from three independent experiments performed in triplicate, and the standard deviations are shown. (B) Relative luciferase activity in protoplasts of 5′89+3Umini and 5′89+3Umini containing mutations in the kl-H and 3′TSS.
The kl-H is important for PEMV accumulation in vivo but not for interaction with the 5′ end of the genome.
As shown in Fig. 2, mutations in 3′TSS hairpins and pseudoknots strongly reduced accumulation of PEMV in vivo. To determine whether the kl-H also impacts virus accumulation in vivo, a single base alteration was generated in the kl-H loop (kl-Hm1; Fig. 9A) in full-length PEMV gRNA, and virus levels were assayed in protoplasts. kl-Hm1 reduced PEMV viral RNA to near background (PEMV-GDD) levels (Fig. 9B), indicating that this hairpin is essential for PEMV viability in protoplasts.
FIG 9.
The kl-H is critical for gRNA accumulation but does not connect with 5′ proximal hairpin 5H2. (A) Location of mutations in 5H2, kl-TSS (13), and kl-H. The known kissing-loop interaction between 5H2 and the kl-TSS is shown. Sequences shared between the kl-H and hairpin 3H1 of the kl-TSS are boxed. (B) PEMV accumulation in protoplasts at 24 h postinfection as measured by quantitative PCR. GDD, PEMV containing mutations in the RdRp active site that abrogate PEMV replication. The data are from three independent experiments performed in triplicate, and the standard deviations are shown.
We previously demonstrated that single base alterations in hairpin loops that disrupted the kl-TSS/5H2 interaction in PEMV gRNA reduced accumulation to background levels, and the mutations together that were designed to be compensatory restored accumulation to 88% of wt (13). Given this previous result, it was unlikely that the critical function of the kl-H was to also interact with 5H2 in the viral gRNA, since this interaction would be disrupted in the virus containing the kl-TSS/5H2 compensatory alterations. In support of this hypothesis, PEMV containing kl-Hm1 and 5H2m1, which would disrupt the kl-TSS/5H2 interaction but allow the kl-H/5H2 interaction (Fig. 9A), accumulated at background levels (Fig. 9B). Furthermore, virus with all three mutations that would restore the kl-TSS/5H2 and kl-H/5H2 interactions also accumulated at near background levels (Fig. 9B). These results strongly suggest that the 3′CITE activity of the kl-H/3′TSS supports the translation of a factor other than ORF1.
The kl-H and 3′TSS inhibit translation in trans.
The addition of a 10-fold molar excess of the entire 3′UTR or a fragment containing only the kl-TSS and PTE to WGE programmed with 5′89+3U reduced luciferase activity by 75% compared to levels obtained in the absence of any added fragments (28). The kl-TSS alone reduced translation by 57% and the PTE by 15%. To determine whether the kl-H and 3′TSS could also inhibit 5′89+3U translation when added in trans, fragments containing either the kl-H or 3′TSS or both were added in a 10-fold molar excess to WGE programmed with 5′89+3U (Fig. 10A). The addition of transcripts containing both elements reduced translation by 66%, whereas the individual kl-H and 3′TSS reduced translation by 52 and 33%, respectively (Fig. 10). This reduction in translation mediated by the 3′TSS was also negated when the fragment contained the H4b M1 mutations. These results suggest that the kl-H is nearly as inhibitory as the kl-TSS alone, possibly due to interference with the cis long-distance kissing-loop interaction. The 3′TSS inhibited translation ∼2-fold better than the PTE alone but only 50% as effectively as was previously found for the TCV TSS (28). The ability to trans-inhibit the translation of 5′89+3U supports the hypothesis that the kl-H and 3′TSS also function as elements important for the translation of PEMV.
FIG 10.
kl-H and 3′TSS repress translation in trans. (A) Diagram of the 3′UTR fragments added to WGE containing 5′89+3U. (B) Relative luciferase activity of 5′89+3U in the presence of the fragments indicated. None, no fragments added; satC, 356-nt untranslated satellite RNA of TCV used as a control; 3′TSS M1, 3′TSS fragment containing mutations that disrupt hairpin H4b (see Fig. 3A). Values represent three independent experiments, and the standard deviations are shown.
DISCUSSION
Termination of translation in eukaryotes occurs when a translating ribosome encounters a stop codon in the A-site. A ternary complex containing release factors and GTP binds to the vacant A-site through tRNA mimicry, prompting GTP hydrolysis and binding of the ATPase ABCE1/Rli1p. After peptide release, ATP hydrolysis triggers ribosome subunit dissociation (45, 46), leading to two presumptive fates: (i) separated subunits diffuse into the cytosol and combine with initiation factors for reinitiation on the same or a different template (45, 47), and (ii) inefficient subunit dissociation by ABCE1/Rli1p allows ribosomes to continue scanning through the 3′UTR until release by rescue factors DoM24 and Hbs1 (48). In the latter scenario, continued association with the 3′UTR might favor reinitiation on the same template due to canonical bridges between the poly(A) tail and the 5′ cap. Our discovery of T-shaped, ribosome-binding elements in the 3′UTR of TCV (TSS) and PEMV (kl-TSS) suggest an additional fate for terminating ribosomes: (iii) ribosomes or separated subunits remain with the template by interacting with internal ribosome-binding structures located proximal to the termination site (13, 21, 22). In the case of the kl-TSS, ribosomes/subunits are then transferred to the 5′ end through the kl-TSS's long-distance kissing-loop interaction that mimics the closed loop topology of canonical mRNAs (28). Maintaining template association with ribosomal subunits following translation termination should enhance protein synthesis by increasing the rate of reinitiation, as was recently shown for a 40S binding element in the 3′UTR of hepatitis C virus (49), which would be positioned proximal to the 5′ IRES through nearby long-distance RNA-RNA interactions (50).
Although efficient 3′CITEs had been identified for PEMV (the kl-TSS and adjacent PTE), visual inspection of the virus's 3′ terminal region suggested the presence of hairpins and pseudoknots (H4a, H4b, H5, ψ2, and ψ3) that in TCV form a well-studied TSS (23). These structures in PEMV were located upstream of a 3′ terminal hairpin (the Pr) and extended linker sequence, elements also found in similar locations in TCV and other carmoviruses. A PEMV Pr hairpin was consistent with both in-line probing and SHAPE structure mapping data (Fig. 2 and 4). As with the Pr of TCV and most other carmoviruses, the PEMV Pr apical loop contains sequence that is complementary with a bulged loop in a hairpin just downstream from the ribosome-recoding site at the end of the p33 ORF, an interaction that is required for ribosome readthrough in TCV or −1 frameshifting in PEMV (51). ψ1 connecting the 3′ terminal 4 residues and the loop of hairpin H5 is conserved in all carmoviruses and tombusviruses (52), and both SHAPE of the full-length gRNA and in-line probing of a 3′ fragment suggests that this pseudoknot also forms in PEMV (Fig. 2 and 4). The presence of H5 was confirmed structurally by in-line probing and SHAPE, and mutations disrupting the H5 upper stem in full-length gRNA reduced virus accumulation in protoplasts. H5 is also conserved in all carmoviruses (53) and is critical for the accumulation of a nontranslated TCV satellite RNA, satC (54). TCV H5 has been proposed to function as an RdRp chaperone (55) independent of its role in the TCV TSS (23), and the analogous hairpin in the tombusvirus Tomato bushy stunt virus binds to viral replicase proteins and has been proposed to also function as a replication repressor (41, 53). The existence and importance of H4b in PEMV was supported both genetically and structurally, and single and compensatory mutations also indicate the presence and importance of ψ2 and ψ3 (Fig. 3). H4a was consistent with the SHAPE-derived structural data (Fig. 4A) but not in-line probing of a 3′ fragment (Fig. 2), and mutations designed to be compensatory did not restore the WT structure in the fragment, as assayed by in-line probing (Fig. 3). However, the conservation of all of these TCV-like 3′ elements in two other umbraviruses (Fig. 1) suggests that all are probably present in the PEMV gRNA and altering any of these elements reduced PEMV accumulation, indicating that they are important for PEMV viability.
PEMV H4a, H4b, H5, ψ2, and ψ3 combine to fold into a T-shaped structure (3′TSS) with structural similarities to the TCV TSS and PEMV kl-TSS (Fig. 5C). There was no identifiable structural similarity with a recently determined high-resolution structure for the cricket paralysis virus tRNA-like element that functions as an IRES (56). Although the model and SHAPE data are mainly consistent, we did note that the lower base pair in H4a (4133G:U4146) was stable in the MD simulations, but the residues showed flexibility by SHAPE (Fig. 4). Similar to the TCV TSS, the PEMV 3′TSS binds to 80S ribosomes and 60S subunits but not to 40S subunits, although binding was ∼4-fold weaker than that of the TCV TSS. There are many possible reasons why ribosome binding to the PEMV 3′TSS was less robust compared to the TCV TSS: (i) the limited stability of ψ3 formation revealed by structure modeling and MD simulations suggests that fewer RNA transcripts synthesized in vitro might adopt the proper folding necessary for ribosome binding, (ii) efficient ribosome binding to the PEMV 3′TSS may require additional factors not present in the purified components, or (iii) the function of the PEMV 3′TSS may necessitate weaker ribosome binding.
Although the TCV TSS is not associated with any obvious RNA bridge connecting it to the 5′ end (22), we propose that the kl-H hairpin performs this function for the PEMV 3′TSS. The kl-H apical loop was critical for PEMV accumulation in vivo, and a fragment containing the kl-H was able to partially repress translation of a reporter construct, most likely by interfering with the kl-TSS kissing-loop interaction with hairpin 5H2 (Fig. 10). In the absence of the kl-TSS/PTE and when the kl-H (and 3′TSS) are positioned proximal to the 3′ end of the luciferase ORF, a kissing-loop interaction between the kl-H and 5H2 was important for efficient translation in WGE (Fig. 7D) and protoplasts (Fig. 8B). Altogether, these results indicate that the kl-H/3′TSS can function as a 3′CITE for translation of the reporter mRNA and thus likely comprise an additional 3′CITE for PEMV. As was previously reported for the TCV TSS and the PEMV kl-TSS/PTE (13), inhibition of 5′89+3U translation by fragments containing the kl-H and 3′TSS added in trans to WGE implies that the elements enhance translation only in cis. Thus, translation enhancement mediated by the kl-H/3′TSS and the kl-TSS/PTE likely impact translation reinitiation and not the pioneer round of translation initiation.
An important question is why a virus would contain two (presumably) independently functioning, ribosome-binding 3′CITEs and why would both be associated with a motif that could potentially connect with the same 5′ hairpin (5H2)? Some possibilities for why a virus might have multiple 3′CITEs include the following: (i) redundancy to support a critical aspect of translation; (ii) ribosomes could be recycled from different translation termination sites; (iii) ribosomes could be recycled to different initiation sites, such as the sgRNA initiation site; and (iv) ability to overcome translation-based resistance in different hosts (29). Since mutations in the kl-TSS that disrupt the kissing-loop interaction are highly detrimental despite maintaining the potential kl-H/5H2 interaction, the answer is unlikely to be redundancy of a single function. In addition, compensatory alterations between the kl-TSS and 5H2 apical loops in full-length PEMV gRNA resulted in near WT levels of PEMV accumulation in protoplasts (13), despite the mutations in 5H2 disrupting a putative interaction with the kl-H. Furthermore, since alteration of the kl-H apical loop in full-length PEMV alone or in combination with mutations that maintain the kl-TSS/5H2 interaction and possible kl-H/5H2 interaction still produced background levels of virus in protoplasts (Fig. 9), it is unlikely that the function of the kl-H in PEMV is to form a kissing-loop interaction with 5H2. Rather, the kl-H may connect the 3′TSS to an alternative interacting sequence. We previously found that nearly all carmovirus kissing loops between their 3′CITEs and 5′ ends involve the same sequence motifs (5′YGCCA and 5′UGGCR) (12), and thus the presence of these motifs in both the kl-TSS and kl-H may reflect an unknown requirement for these particular sequences and not necessarily interaction with the same 5′ hairpin (5H2).
A hairpin just downstream from the translation start site of PEMV p26/p27 can also potentially pair with either the kl-H or kl-TSS (2829GGAGCUACUGGCAGCUCU; the putative stem is underlined, and the kl-H/kl-TSS complementary sequence is in boldface). An additional possible interacting sequence exclusive to the kl-H is located just downstream at 2866CGGCGA. In addition to possibly enhancing translation of the subgenomic RNA template for p26/p27, connecting the kl-H to either of these sequences might allow for putative internal ribosome entry site in the gRNA that could be used for translation of p26/p27 prior to sgRNA synthesis, as was found for the coat protein of the carmovirus Pelargonium flower break virus (57). Since mutations in kl-H reduced PEMV accumulation in protoplasts to background levels, both of these latter possibilities require that p26 and/or p27 be critical for PEMV accumulation in single cells (or that the kl-H is important for PEMV replication). The p27 orthologue of the umbravirus Groundnut rosette virus functions as a cell-to-cell movement protein (27, 58), while the p26 orthologue is a multifunctional protein that transits to the nucleolus through its association with fibrillarin and is then exported to cytoplasmic granules for binding to the gRNA and long-distance movement (59, 60). p26 is also important for stabilization of the gRNA via its cooperative binding (61), which may be critical in the apparent absence of an encoded umbravirus silencing suppressor (27). The importance of these products for PEMV accumulation in protoplasts and possible translational regulation mediated by the kl-H/3′TSS is currently being investigated.
An important question is whether PEMV is unique in having three 3′ CITEs or whether additional 3′CITEs remain to be discovered in other cap-independently translated plant viruses. Umbravirus TBTV has a BTE-like element in the same location as the kl-TSS (62) and also contains a similar collection of hairpins and pseudoknots at the 3′ end that form the PEMV 3′TSS (Fig. 1C), suggesting that it too has multiple 3′CITEs. Since 3′CITEs have mainly been elucidated using reporter constructs, the proximity of a 3′CITE to the reporter termination codon may have guided identification, suggesting that additional translation enhancers may remain to be elucidated.
ACKNOWLEDGMENTS
This study was supported by the NSF (MCB 1157906) to A.E.S. This publication has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN 261200800001E to W.K.K. This research was supported in part by the Intramural Research Program of the National Institutes of Health, Center for Cancer Research, to B.A.S.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1. Aitken CE, Lorsch JR. 2012. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19:568–576. 10.1038/nsmb.2303 [DOI] [PubMed] [Google Scholar]
- 2. Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sonenberg N. 2008. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem. Cell Biol. 86:178–183. 10.1139/O08-034 [DOI] [PubMed] [Google Scholar]
- 5. Amrani N, Ghosh S, Mangus DA, Jacobson A. 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453:1276–U1285. 10.1038/nature06974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135–140. 10.1016/S1097-2765(00)80122-7 [DOI] [PubMed] [Google Scholar]
- 7. Afonia ZA, Myasnikov AG, Shirokov VA, Klaholz BP, Spirin AS. 2014. Formation of circular polyribosomes on eukaryotic mRNA without cap-structure and poly(A)-tail: a cryo electron tomography study. Nucleic Acids Res. 42:9461–9469. 10.1093/nar/gku599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J. Gen. Virol. 93:1385–1409. 10.1099/vir.0.042499-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doudna JA, Sarnow P. 2007. Translation initiation by viral internal ribosome entry sites, p 129–154 In Mathews NS, Hershey JWB. (ed), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 10. de Quinto SL, Saiz M, de la Morena D, Sobrino F, Martinez-Salas E. 2002. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acid Res. 30:4398–4405. 10.1093/nar/gkf569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chattopadhyay M, Shi K, Yuan X, Simon AE. 2011. Long-distance kissing loop interactions between a 3′ proximal Y-shaped structure and apical loops of 5′ hairpins enhance translation of Saguaro cactus virus. Virology 417:113–125. 10.1016/j.virol.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simon AE, Miller WA. 2013. 3′ Cap-independent translation enhancers of plant viruses. Annu. Rev. Microbiol. 67:21–42. 10.1146/annurev-micro-092412-155609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao F, Kasprzak W, Stupina VA, Shapiro BA, Simon AE. 2012. A ribosome-binding, 3′ translational enhancer has a T-shaped structure and engages in a long-distance RNA-RNA interaction. J. Virol. 86:9828–9842. 10.1128/JVI.00677-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fabian MR, White KA. 2004. 5′-3′ RNA-RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: a potential common mechanism for Tombusviridae. J. Biol. Chem. 279:28862–28872. 10.1074/jbc.M401272200 [DOI] [PubMed] [Google Scholar]
- 15. Nicholson BL, White KA. 2011. 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr. Opin. Virol. 1:373–380. 10.1016/j.coviro.2011.10.002 [DOI] [PubMed] [Google Scholar]
- 16. Truniger V, Nieto C, Gonzalez-Ibeas D, Aranda M. 2008. Mechanism of plant eIF4E-mediated resistance against a carmovirus (Tombusviridae): cap-independent translation of a viral RNA controlled in cis by a virulence determinant. Plant J. 56:716–727. 10.1111/j.1365-313X.2008.03630.x [DOI] [PubMed] [Google Scholar]
- 17. Wang Z, Treder K, Miller WA. 2009. Structure of a viral cap-independent translation element that functions via high-affinity binding to the eIF4E subunit of eIF4F. J. Biol. Chem. 284:14189–14202. 10.1074/jbc.M808841200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicholson BL, Wu B, Chevtchenko I, White KA. 2010. Tombusvirus recruitment of host translational machinery via the 3′UTR. RNA 16:1402–1419. 10.1261/rna.2135210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gazo BM, Murphy P, Gatchel JR, Browning KS. 2004. A novel interaction of cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 279:13584–13592. 10.1074/jbc.M311361200 [DOI] [PubMed] [Google Scholar]
- 20. Nicholson BL, Zaslaver O, Mayberry LK, Browning KS, White KA. 2013. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J. Virol. 87:1872–1883. 10.1128/JVI.02711-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stupina VA, Meskauskas A, McCormack JC, Yingling YG, Shapiro BA, Dinman JD, Simon AE. 2008. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA 14:2379–2393. 10.1261/rna.1227808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stupina VA, Yuan X, Meskauskas A, Dinman JD, Simon AE. 2011. Ribosome binding to a 5′ translational enhancer is altered in the presence of the 3′UTR in cap-independent translation of Turnip crinkle virus. J. Virol. 85:4638–4653. 10.1128/JVI.00005-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCormack JC, Yuan X, Yingling YG, Kasprzak W, Zamora RE, Shapiro BA, Simon AE. 2008. Structural domains within the 3′ untranslated region of Turnip crinkle virus. J. Virol. 82:8706–8720. 10.1128/JVI.00416-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Zoeten GA, Skaf JS. 2001. Pea enation mosaic and the vagaries of a plant virus. Adv. Virus Res. 57:323–350. 10.1016/S0065-3527(01)57007-4 [DOI] [PubMed] [Google Scholar]
- 25. Demler SA, Rucker DG, de Zoeten GA. 1993. The chimeric nature of the genome of pea enation mosaic virus: the independent replication of RNA 2. J. Gen. Virol. 74:1–14. 10.1099/0022-1317-74-1-1 [DOI] [PubMed] [Google Scholar]
- 26. Ryabov EV, Robinson DJ, Taliansky M. 2001. Umbravirus-encoded proteins both stabilize heterologous viral RNA and mediate its systemic movement in some plant species. Virology 288:391–400. 10.1006/viro.2001.1078 [DOI] [PubMed] [Google Scholar]
- 27. Taliansky ME, Robinson DJ. 2003. Molecular biology of umbraviruses: phantom warriors. J. Gen. Virol. 84:1951–1960. 10.1099/vir.0.19219-0 [DOI] [PubMed] [Google Scholar]
- 28. Gao F, Gulay SP, Kasprzak W, Dinman JD, Shapiro BA, Simon AE. 2013. The kissing-loop T-shaped structure translational enhancer of Pea enation mosaic virus can bind simultaneously to ribosomes and a 5′ proximal hairpin. J. Virol. 87:11987–12002. 10.1128/JVI.02005-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miras M, Sempere RN, Kraft JJ, Miller WA, Aranda MA, Truniger V. 2014. Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 202:233–246. 10.1111/nph.12650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang JC, Zhang GH, Guo R, Shapiro BA, Simon AE. 2006. A pseudoknot in a preactive form of a viral RNA is part of a structural switch activating minus-strand synthesis. J. Virol. 80:9181–9191. 10.1128/JVI.00295-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stupina VA, Simon AE. 2013. Preparation of biologically active Arabidopsis ribosomes and comparison with yeast ribosomes for binding to a tRNA-mimic that enhances translation of plant plus-strand RNA viruses. Front. Plant Sci. 4:271. 10.3389/fpls.2013.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610–1616. 10.1038/nprot.2006.249 [DOI] [PubMed] [Google Scholar]
- 33. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Popenda M, Szachniuk M, Blazewicz M, Wasik S, Burke EK, Blazewicz J, Adamiak RW. 2010. RNA FRABASE 2.0: an advanced web-accessible database with the capacity to search the three-dimensional fragments within RNA structures. BMC Bioinform 11:231. 10.1186/1471-2105-11-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, Blazewicz J, Adamiak RW. 2012. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 40:e112. 10.1093/nar/gks339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martinez HM, Maizel JV, Shapiro BA. 2008. RNA2D3D: A program for generating, viewing, and comparing 3-dimensional models of RNA. J. Biomol. Struct. Dyn. 25:669–683. 10.1080/07391102.2008.10531240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Case DA, Darden TA, Cheatham TEI, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A. 2012. AMBER 12. University of California, San Francisco, CA [Google Scholar]
- 38. Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. 1995. A smooth particle mesh EWALD method. J. Chem. Phys. 103:8577–8593 [Google Scholar]
- 39. Wang J, Cieplak P, Kollman PA. 2000. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21:1049–1074. [DOI] [Google Scholar]
- 40. Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690 [Google Scholar]
- 41. Panaviene Z, Panavas T, Nagy PD. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 79:10608–10618. 10.1128/JVI.79.16.10608-10618.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang JC, Simon AE. 2005. Importance of sequence and structural elements within a viral replication repressor. Virology 333:301–315. 10.1016/j.virol.2004.12.015 [DOI] [PubMed] [Google Scholar]
- 43. Low JT, Weeks KM. 2010. SHAPE-directed RNA secondary structure prediction. Methods 52:150–158. 10.1016/j.ymeth.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reeder J, Giegerich R. 2004. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics 5:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dever TE, Green R. 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harbor Perspect Biol. 4:a013706. 10.1101/cshperspect.a013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache J-P, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, Karcher A, Thomm M, Hopfner K-P, Green R, Beckmann R. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482:501–506. 10.1038/nature10829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pisarev AV, Hellen CUT, Pestova TV. 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131:286–299. 10.1016/j.cell.2007.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guydosh NR, Green R. 2014. Dom34 rescues ribosomes in 3′ untranslated regions. Cell 156:950–962. 10.1016/j.cell.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bai Y, Zhou K, Doudna JA. 2013. Hepatitis C virus 3′UTR regulates viral translation through direct interactions with the host translation machinery. Nucleic Acids Res. 41:7861–7874. 10.1093/nar/gkt543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Romero-Lopez C, Berzal-Herranz A. 2009. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA. 15:1740–1752. 10.1261/rna.1680809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cimino PA, Nicholson BL, Wu B, Xu W, White KA. 2011. Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS Pathog. 7:e1002423. 10.1371/journal.ppat.1002423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Na H, White KA. 2006. Structure and prevalence of replication silencer-3′ terminus RNA interactions in Tombusviridae. Virology 345:305–316. 10.1016/j.virol.2005.09.008 [DOI] [PubMed] [Google Scholar]
- 53. Zhang GH, Zhang JC, Simon AE. 2004. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J. Virol. 78:7619–7633. 10.1128/JVI.78.14.7619-7633.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang GH, Simon AE. 2003. A multifunctional turnip crinkle virus replication enhancer revealed by in vivo functional SELEX. J. Mol. Biol. 326:35–48. 10.1016/S0022-2836(02)01366-9 [DOI] [PubMed] [Google Scholar]
- 55. McCormack JC, Simon AE. 2004. Biased hypermutagenesis associated with mutations in an untranslated hairpin of an RNA virus. J. Virol. 78:7813–7817. 10.1128/JVI.78.14.7813-7817.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fernandez IS, Bai X-C, Murshudov G, Scheres SHW, Ramakrishnan V. 2014. Initiation of translation by Cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157:823–831. 10.1016/j.cell.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fernandez-Miragall O, Hernandez C. 2011. An internal ribosome entry site directs translation of the 3′-gene from Pelargonium flower break virus genomic RNA: implications for infectivity. PLoS One 6:e22617. 10.1371/journal.pone.0022617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ryabov EV, Roberts IM, Palukaitis P, Taliansky M. 1999. Host-specific cell-to-cell and long-distance movements of cucumber mosaic virus are facilitated by the movement protein of groundnut rosette virus. Virology 260:98–108. 10.1006/viro.1999.9806 [DOI] [PubMed] [Google Scholar]
- 59. Kim SH, MacFarlane S, Kalinina NO, Rakitina DV, Ryabov EV, Gillespie T, Haupt S, Brown JWS, Taliansky M. 2007. Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. U. S. A. 104:11115–11120. 10.1073/pnas.0704632104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JWS, Taliansky M. 2007. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 26:2169–2179. 10.1038/sj.emboj.7601674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Taliansky M, Roberts IM, Kalinina N, Ryabov EV, Raj SK, Robinson DJ, Oparka KJ. 2003. An umbraviral protein, involved in long-distance RNA movement, binds viral RNA and forms unique, protective ribonucleoprotein complexes. J. Virol. 77:3031–3040. 10.1128/JVI.77.5.3031-3040.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Z, Kraft JJ, Hui AY, Miller WA. 2010. Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology 402:177–186. 10.1016/j.virol.2010.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yuan X, Shi K, Young MYL, Simon AE. 2010. The terminal loop of a 3′ proximal hairpin plays a critical role in replication and the structure of the 3′ region of Turnip crinkle virus. Virology 402:271–280. 10.1016/j.virol.2010.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]