ABSTRACT
The identification of viroid-derived small RNAs (vd-sRNAs) of 21 to 24 nucleotides (nt) in plants infected by viroids (infectious non-protein-coding RNAs of just 250 to 400 nt) supports their targeting by Dicer-like enzymes, the first host RNA-silencing barrier. However, whether viroids, like RNA viruses, are also targeted by the RNA-induced silencing complex (RISC) remains controversial. At the RISC core is one Argonaute (AGO) protein that, guided by endogenous or viral sRNAs, targets complementary RNAs. To examine whether AGO proteins also load vd-sRNAs, leaves of Nicotiana benthamiana infected by potato spindle tuber viroid (PSTVd) were agroinfiltrated with plasmids expressing epitope-tagged versions of AGO1, AGO2, AGO3, AGO4, AGO5, AGO6, AGO7, AGO9, and AGO10 from Arabidopsis thaliana. Immunoprecipitation analyses of the agroinfiltrated halos revealed that all AGOs except AGO6, AGO7, and AGO10 associated with vd-sRNAs: AGO1, AGO2, and AGO3 preferentially with those of 21 and 22 nt, while AGO4, AGO5, and AGO9 additionally bound those of 24 nt. Deep-sequencing analyses showed that sorting of vd-sRNAs into AGO1, AGO2, AGO4, and AGO5 depended essentially on their 5′-terminal nucleotides, with the profiles of the corresponding AGO-loaded vd-sRNAs adopting specific hot spot distributions along the viroid genome. Furthermore, agroexpression of AGO1, AGO2, AGO4, and AGO5 on PSTVd-infected tissue attenuated the level of the genomic RNAs, suggesting that they, or their precursors, are RISC targeted. In contrast to RNA viruses, PSTVd infection of N. benthamiana did not affect miR168-mediated regulation of the endogenous AGO1, which loaded vd-sRNAs with specificity similar to that of its A. thaliana counterpart.
IMPORTANCE To contain invaders, particularly RNA viruses, plants have evolved an RNA-silencing mechanism relying on the generation by Dicer-like (DCL) enzymes of virus-derived small RNAs of 21 to 24 nucleotides (nt) that load and guide Argonaute (AGO) proteins to target and repress viral RNA. Viroids, despite their minimal genomes (non-protein-coding RNAs of only 250 to 400 nt), infect and incite disease in plants. The accumulation in these plants of 21- to 24-nt viroid-derived small RNAs (vd-sRNAs) supports the notion that DCLs also target viroids but does not clarify whether vd-sRNAs activate one or more AGOs. Here, we show that in leaves of Nicotiana benthamiana infected by potato spindle tuber viroid, the endogenous AGO1 and distinct AGOs from Arabidopsis thaliana that were overexpressed were associated with vd-sRNAs displaying the same properties (5′-terminal nucleotide and size) previously established for endogenous and viral small RNAs. Overexpression of AGO1, AGO2, AGO4, and AGO5 attenuated viroid accumulation, supporting their role in antiviroid defense.
INTRODUCTION
Viroids, despite having minimal genomes restricted to a non-protein-coding single-stranded RNA (ssRNA) of only ∼250 to 400 nucleotides (nt), can parasitize the higher plants they infect and replicate, spread systemically, and frequently incite disease (1–3). Most of the approximately 30 viroids characterized so far, including the type species potato spindle tuber viroid (PSTVd) (4, 5), have been assigned to the family Pospiviroidae on the basis of a rod-like (or quasi-rod-like) secondary structure with a central conserved region (CCR) and replication in the nucleus through an asymmetric rolling-circle mechanism with double-stranded RNA (dsRNA) intermediates (6–10). This mechanism is catalyzed by the RNA polymerase II forced to transcribe RNA templates (11–13), an RNase most likely of class III (14), and the DNA ligase 1 redirected to act as an RNA ligase (15), with the CCR playing a critical role in the cleavage of the oligomeric plus strands generated by the rolling-circle mechanism (14) and in the ligation (circularization) of the resulting monomeric plus strands (16). The remaining four viroids, members of the family Avsunviroidae, display a quasi-rod-like or clearly branched secondary structure without a CCR but with hammerhead ribozymes in both polarity strands; these catalytic RNA motifs mediate self-cleavage of the oligomeric strands produced by a symmetric rolling-circle mechanism occurring in plastids, mostly chloroplasts (17). To complete their infectious cycle, viroids must move at short (cell-to-cell) and long distances through the plasmodesmata and phloem, respectively. Some of the RNA motifs that mediate these movements have been finely dissected in PSTVd and include trafficking from the bundle sheath to the mesophyll (18), entry of PSTVd from nonvascular into phloem tissue (19), and trafficking from palisade mesophyll to spongy mesophyll (20). A genome-wide mutational analysis has mapped loops/bulges in the secondary structure of PSTVd crucial or relevant for replication in single cells (protoplasts) or for systemic movement (21).
In addition to replicating and moving, viroids need to overcome their host surveillance responses, conspicuous among which is RNA silencing (22, 23). This regulatory mechanism, which functions at the transcriptional and posttranscriptional levels and is particularly sophisticated in plants, tunes endogenous gene expression and plays a defensive role, restricting pathogen invasion and transposon proliferation. The key elicitors of RNA silencing, dsRNAs and snap-folded ssRNA, are processed by specific Dicer-like (DCL) isozymes into small RNAs (sRNAs), mainly small interfering RNAs (siRNAs) of 21, 22, and 24 nt and microRNAs (miRNAs) of 21 and 22 nt (24–26). One strand of these sRNAs, and of some secondary siRNAs resulting from an amplification pathway catalyzed by endogenous RNA-directed RNA polymerases (RDRs) and DCLs (27, 28), preferentially loads and guides the RNA-inducing silencing complex (RISC), particularly its core, consisting of an Argonaute (AGO) protein, to target and inactivate their cognate RNAs or DNAs (29).
Arabidopsis thaliana contains 10 AGO proteins classified into three clades: (i) AGO1, AGO5, and AGO10; (ii) AGO2, AGO3, and AGO7; and (iii) AGO4, AGO6, AGO8, and AGO9. This phylogenetic grouping, however, does not necessarily imply functional similarity (29, 30). AGO1, AGO2, and AGO7 associate with 21- or 22-nt sRNAs (and AGO5 additionally with 24-nt sRNAs), either endogenous or exogenous (such as those derived from viruses), and at least AGO1, AGO2, and AGO5 operate in posttranscriptional silencing through cleavage or translation arrest of their RNA targets (31–37). AGO4, AGO6, and AGO9 associate with 24-nt siRNAs to mediate transcriptional silencing through RNA-directed DNA methylation (38, 39), and at least AGO4 binds to diverse classes of sRNAs, including siRNAs originating from transposable and repetitive elements, and cleaves target RNA transcripts (40). AGO10 acts in the translational control of several miRNA targets, like the mRNAs coding for AGO1 (41, 42). Finally, the roles of AGO3 and AGO8 in sRNA-directed regulation remain unclear (29, 30). A recent transcriptome assembly has identified in Nicotiana benthamiana homologues for each AGO from A. thaliana, except for AGO3 and AGO9 (returned as AGO2 and as a variant of AGO4, respectively) (43).
Several independent lines suggest that, in addition to playing a key role in antiviral defense, RNA silencing is also involved in antiviroid defense. First, viroid infection results in the genomic RNA progeny folding upon itself into collapsed secondary structures and in the production of dsRNAs, either replicative intermediates or RDR products. Northern hybridizations have detected viroid-derived sRNAs (vd-sRNAs), with properties akin to those of the host and virus sRNAs generated by DCLs, accompanying infections by representative members of the two viroid families; these vd-sRNAs have been subsequently sequenced by conventional and high-throughput approaches (44, 45). Second, the expression of a reporter gene and the accumulation in infected plants of the genomic viroid RNA appear to be repressed by vd-sRNAs in a sequence-specific mode (46–49). Third, increased levels of the genomic PSTVd RNA have been detected in early infection stages of N. benthamiana deficient in RDR6, in which, additionally, PSTVd entry into floral and vegetative meristems is facilitated (50). Fourth, two vd-sRNAs containing the albinism determinant of a chloroplast-replicating viroid guide cleavage of the host mRNA coding for the chloroplastic heat shock protein 90 (cHSP90) as predicted by RNA silencing, thus providing a feasible mechanism of pathogenesis (51). Fifth, the titer of a viroid is enhanced in coinfections with a virus, with this effect occurring via the expression of virus-encoded silencing suppressors (52). Last but not least, early pioneering research discovered that PSTVd-cDNA introduced into the tobacco genome via Agrobacterium tumefaciens became methylated only following viroid RNA-RNA replication, thus revealing an RNA-directed and sequence-specific mechanism for de novo methylation of genomic sequences in plants (53); this mechanism of transcriptional silencing is now known to be mediated by specific DCL-dependent siRNAs and AGOs. More recently, viroid infection has been associated with changes in DNA methylation of host rRNA genes (54), but whether those changes result from direct or indirect effects is not known. However, despite all these data, no direct evidence exists supporting the notion that AGOs recruit vd-sRNAs. We report here that certain members of the AGO family, specifically AGO1, AGO2, AGO4, and AGO5, load vd-sRNAs from PSTVd and eventually attenuate the viroid titer.
MATERIALS AND METHODS
PSTVd inoculation.
N. benthamiana seedlings were grown in a controlled chamber (28°C with fluorescent light for 16 h and 25°C in darkness for 8 h), and at the cotyledon/first-true-leaf stage, they were infiltrated with cultures of A. tumefaciens C58 carrying a binary plasmid, empty or with a head-to-tail dimeric insert of PSTVd (NB variant; GenBank accession number AJ634596.1) under the control of the 35S promoter of cauliflower mosaic virus (CaMV) (55).
Agroinfiltration assays.
Binary plasmids for expressing, under the control of the 35S promoter of CaMV, AGO versions from A. thaliana tagged at their N-terminal regions with three tandem repeats of the hemagglutinin (HA) epitope (AGO1, AGO2, AGO7, and AGO10) or with a single HA epitope (AGO3, AGO4, AGO5, AGO6, and AGO9) were described previously (34, 37). Binary plasmids for expressing miR390 from A. thaliana and the beta-glucuronidase (GUS) from Escherichia coli, both under the control of the 35S promoter of CaMV, were also described previously (34). Transient-expression assays in N. benthamiana leaves with cultures of A. tumefaciens were performed as described previously (56, 57).
RNA extraction and analysis by PAGE and Northern blot hybridization.
Total RNA preparations (RNA-Input) or immunoprecipitates (RNA-IP [see below]) from PSTVd-infected upper, noninoculated leaves of N. benthamiana (and from mock-inoculated controls) were obtained by double extraction with buffer-saturated phenol, precipitated with ethanol, and resuspended. The RNAs were then separated by denaturing PAGE (in 1× Tris-borate-EDTA [TBE] buffer and 8 M urea) on either 5% gels (for the monomeric circular [MC] and linear [ML] PSTVd forms) or 17% gels (for vd-sRNAs and miRNAs) that were stained with ethidium bromide for assessing equal loading by the fluorescence emitted by the 5S or 4S RNAs. Following electrotransference of RNAs to Hybond-N+ membranes (Roche Diagnostics GmbH), they were hybridized with internally radiolabeled full-length riboprobes (synthesized by in vitro transcription) for detecting PSTVd plus strands or with 5′-radiolabeled deoxyribonucleotide probes (prepared according to standard procedures [58]) for detecting specific miRNAs. Hybridization was at 70°C in the presence of 50% formamide (for detecting MC and ML forms) or at 42°C with PerfectHyb Plus hybridization buffer (Sigma) (for detecting vd-sRNAs) (50). After washing at 60°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% SDS (MC and ML forms) or at 55°C in 1× SSC with 0.1% SDS (vd-sRNAs and miRNAs), the membranes were analyzed with a phosphorimager (Fujifilm FLA-5100) using the programs Image Reader FLA-5100 and Image Gauge 4.0.
RNA immunoprecipitation assays.
RNA-IP were obtained with a rabbit polyclonal antibody (GenScript) against the N-terminal region of AGO1 from N. benthamiana (MVRKKRTDVPGGAESFESHEC), which is characteristic of this AGO isoform, and with a mouse monoclonal antibody (12CA5; Roche Diagnostics GmbH) against an epitope of the HA from human influenza virus. Immunoprecipitation assays were performed as reported previously (37) with minor modifications. In brief, N. benthamiana leaves (0.5 to 1 g) were homogenized with liquid nitrogen and mixed with 12 ml/g (HA antibody) or 3 ml/g (AGO1 antibody) lysis buffer (50 mM Tris-HCl, pH 7.4, 2.5 mM MgCl2, 100 mM KCl, 0.1% Nonidet P-40, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 0.5 mM phenylmethylsulfonyl fluoride, and one tablet of complete proteinase inhibitor cocktail [Roche Diagnostics GmbH]). The insoluble material was removed by centrifugation at 8,000 × g for 5 min at 4°C, and after taking two aliquots (20 μl and 1 ml) for analysis of the protein- and RNA-Input fractions, respectively, the remaining clarified lysate was incubated at 4°C for 15 min with 4 μg/ml of one (HA) or the other (AGO1) antibody and then at 4°C for 30 min with 100 μl/ml of protein A agarose beads (Roche Diagnostics GmbH). The beads were washed six times for 10 min each time with lysis buffer at 4°C, and after taking one aliquot (20 μl) of the final bead suspension for protein-IP analysis, the rest was used for RNA-IP analysis. RNAs were released by incubating the beads at 65°C for 15 min in 0.5 volume of proteinase K buffer (0.1 M Tris-HCl, pH 7.4, 10 mM EDTA, 300 mM NaCl, 2% SDS, and 1 μg/μl proteinase K [Roche Diagnostics GmbH]). RNA-Input and RNA-IP aliquots were extracted with saturated phenol, pH 4.5 (Amresco), phenol-chloroform-isoamyl alcohol, and chloroform and recovered by ethanol precipitation. For sRNA gel blot assays, 2 to 5 μg of RNA of the RNA-Input fraction, and one-half of the RNA-IP fraction, were used as indicated above. For protein blot assays, 20 μl of the protein-Input and protein-IP fractions was mixed with the same volume of 2× PDB buffer (1.25 M Tris, pH 6.8, 10% SDS, 80% glycerol, 10% β-mercaptoethanol, and 0.02% bromophenol blue) and heated at 100°C for 3 min; aliquots (10 and 2.5 μl, respectively) were applied onto NuPAGE Bis-Tris minigels (4 to 12%; Novex, Life Technologies), and equal loading was assessed by the intensity generated by the large subunit of RubisCO after staining with Ponceau S (Sigma). HA-AGOs were electrotransferred to polyvinylidene difluoride (PVDF) membranes and detected by chemiluminescence with anti-HA-peroxidase antibody (Roche Diagnostics GmbH) at 1:1,000 dilution and Western Lighting plus-ECL substrate (Perkin-Elmer). AGO1 from N. benthamiana was similarly detected, but using the antibody against its N-terminal region at 1:1,000 dilution and a goat anti-rabbit IgG (H+L) secondary antibody conjugated to horseradish peroxidase (Agrisera) at 1:20,000 dilution and the same substrate.
Deep-sequencing analysis of sRNAs.
Experimental details of sRNA purification, adapter ligation, RT-PCR amplification, library purification, and high-throughput DNA sequencing on the Illumina Genome Analyzer Hi-Seq 2000 (Fasteris SA, Plan-les-Ouates, Switzerland) have been described previously (50). In the first deep sequencing, four bar-coded samples, corresponding total sRNAs (inputs) and immunoprecipitates (IPs) (obtained with an anti-HA monoclonal antibody) from PSTVd-infected N. benthamiana overexpressing HA-tagged AGO1 and AGO2 were run in the same channel. In the second deep sequencing, we proceeded as in the first, but the four bar-coded samples corresponded to sRNA inputs and IPs from PSTVd-infected N. benthamiana overexpressing HA-tagged AGO4 and AGO5. In the third deep sequencing, we proceeded again as in the first, but the four bar-coded samples corresponded to sRNA inputs and IPs (obtained with an anti-AGO1 polyclonal antibody) from mock-inoculated and PSTVd-infected N. benthamiana. The resulting reads, after bar code identification, were processed by removing the adaptor and grouping them into different files according to their sequence lengths. Only 18- to 26-nt reads were mapped against the PSTVd sequence (variant NB) (59). To filter, analyze, and visualize the mapping data, a set of Perl scripts was developed. IP enrichment or depletion was calculated for each unique 21-, 22-, or 24-nt vd-sRNA as log2 [(IP reads + 1)/(input reads + 1)] and plotted for each size class as the fraction (percent) of unique vd-sRNA sequences enriched >2-fold (log2 > 1) or depleted >2-fold (log2 < 1) in the IP compared to the input.
RESULTS
PSTVd infection of N. benthamiana does not affect miR168-mediated AGO1 accumulation.
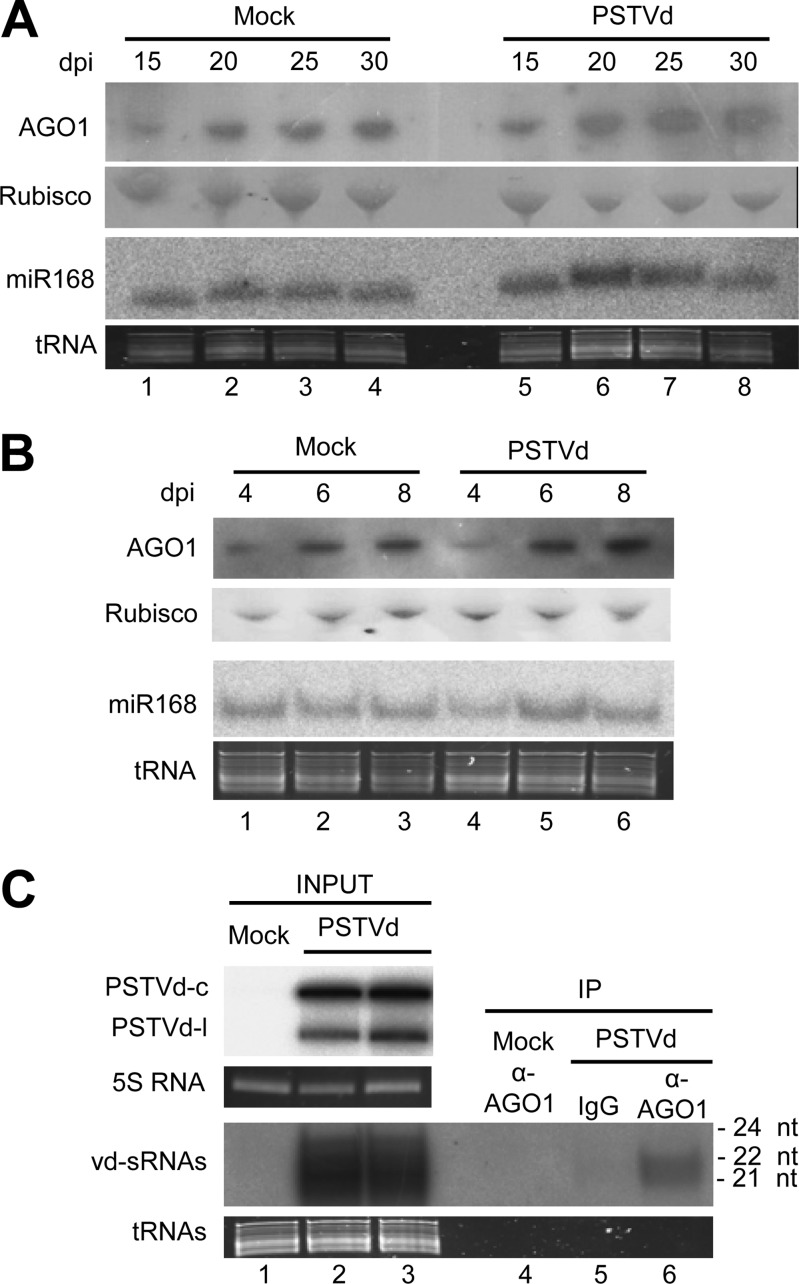
As indicated above, AGO1 is a key player in the antiviral defense mediated by RNA silencing. Infection by plant RNA viruses induces and reduces the accumulation of AGO1 mRNA and AGO1 protein, respectively (42, 60). The explanation for this apparent paradox is that expression of AGO1 mRNA is translationally repressed by miR168 (61), the accumulation of which is elicited by virus infection (42, 60). In view of these results, we first checked whether a similar situation occurs during a viroid infection. For this purpose, plants of N. benthamiana were inoculated by leaf infiltration with cultures of A. tumefaciens carrying binary plasmids for expressing a head-to-tail dimeric (plus) insert of PSTVd [35S:dPSTVd(+)] or the beta-glucuronidase (GUS) (35S:GUS), both under the control of the 35S promoter of CaMV. Western blot analysis of PSTVd-infected upper, noninoculated leaves—using a polyclonal antibody raised specifically against the N-terminal region of AGO1 from N. benthamiana—revealed that infection by PSTVd induced a slight increase in the accumulation of AGO1 compared with mock-inoculated controls (expressing GUS) at the same developmental stage. This effect was observed in leaves collected at early (15 days postinoculation [p.i.], when symptoms start appearing) and later (20, 25, and 30 days p.i.) infection stages (Fig. 1A). However, no significant effect on AGO1 was detected when the leaves of N. benthamiana agroinfiltrated with the plasmid for expressing the head-to-tail dimeric insert of PSTVd were directly examined at 4, 6, and 8 days p.i. (again using as a control leaves agroinfiltrated with 35S:GUS) (Fig. 1B).
FIG 1.
(A and B) PSTVd infection of N. benthamiana has no significant effect on AGO1 or miR168 accumulation in the upper, noninoculated leaves (A) or in the agroinfiltrated leaves (B). Western blot analyses were performed with a rabbit polyclonal antibody against the N-terminal region of AGO1 from N. benthamiana and a goat anti-rabbit secondary antibody conjugated to horseradish peroxidase. Total proteins were separated by PAGE in 4 to 12% gels, and equal loading was assessed by the intensity of the large subunit of RubisCO after staining with Ponceau S. Northern blot hybridizations were carried out with a 5′-radiolabeled oligodeoxyribonucleotide complementary to miR168. Total RNAs were separated by denaturing PAGE in 17% gels, and equal loading was assessed by the intensity of tRNA after staining with ethidium bromide. (A) Samples were collected at 15 (lanes 1 and 5), 20 (lanes 2 and 6), 25 (lanes 3 and 7), and 30 (lanes 4 and 8) days p.i. (dpi). (B) Samples were collected at 4 (lanes 1 and 4), 6 (lanes 2 and 5), and 8 (lanes 3 to 6) days p.i. (C) Endogenous AGO1 loads vd-sRNAs during PSTVd infection of N. benthamiana. Aliquots of total sRNA (INPUT) and of the sRNA fraction immunoprecipitated with a rabbit polyclonal antibody against the N-terminal region of AGO1 from N. benthamiana (α-AGO1) (IP) were separated by denaturing PAGE in 17% gels and revealed by Northern blot hybridization with a radiolabeled riboprobe for detecting PSTVd plus strands. Lanes 1 and 4, mock-inoculated control; lanes 2, 3, 5, and 6, PSTVd-infected upper, noninoculated leaves collected at 25 days postinoculation. IPs were obtained with the antibody against AGO1 (lanes 4 and 6) or with a preimmune rabbit immunoglobulin fraction (IgG) (lane 5). Mock inoculations were performed with cultures of A. tumefaciens with a binary plasmid expressing GUS instead of the head-to-tail dimeric plus transcript of PSTVd. Accumulation of the PSTVd MC and ML forms was also examined in the RNA inputs after denaturing PAGE in 5% gels (upper gel). Equal loading was assessed by the intensity of the bands generated by the 5S RNA and tRNAs after staining with ethidium bromide.
Although these results anticipated that miR168 levels were most likely not affected by PSTVd infection, we confirmed that this was indeed the case by examining total RNA preparations from the same samples by Northern blot hybridization to detect miR168 (Fig. 1A and B). Altogether, our data showed that, in contrast with the situation observed for representative RNA viruses (42, 60), the effect of viroid infection on AGO1, if any, is not mediated by miR168 targeting of AGO1 mRNA.
Endogenous AGO1 loads vd-sRNAs during PSTVd infection of N. benthamiana.
Using the polyclonal antibody against AGO1 from N. benthamiana, we next tested whether the protein interacts in vivo with vd-sRNA. Approximately 3 weeks after inoculation, plants displayed a stunted phenotype with curling and reduction of the foliar area compared with the mock-inoculated controls, as reported previously for this and other PSTVd strains (50, 62). Analysis of total RNA preparations by denaturing PAGE (in 5% gels) and Northern blot hybridization confirmed the accumulation of the MC and ML PSTVd plus RNAs in upper, noninoculated leaves collected at 20, 25, and 30 days p.i. (Fig. 1C). Parallel analysis of the same samples, fractionated by denaturing PAGE (in 17% gels), revealed intense hybridization signals in the gel region corresponding to vd-sRNAs of 21 to 24 nt (Fig. 1C). Moreover, a minor fraction of the vd-sRNAs of 21 or 22 nt were also detected in the immunoprecipitates generated by the AGO1-specific antibody (Fig. 1C), showing that the host AGO1 indeed interacts with PSTVd-sRNAs with the expected size specificity (29). A more detailed description of the PSTVd-sRNAs loaded by AGO1 from N. benthamiana is provided below.
Setting up a system for the study of AGO–vd-sRNA interactions: agroexpressed AGO1 and AGO2, but neither AGO7 nor AGO10, bind PSTVd-sRNAs.
To circumvent the problem posed by the inability of PSTVd to infect A. thaliana (10) and by the lack of specific antibodies against most other AGOs from N. benthamiana, we tested whether overexpressing AGO1 and other AGO members from A. thaliana in PSTVd-infected N. benthamiana resulted in their loading with vd-sRNAs. To increase recovery in immunoprecipitates, we used AGO versions tagged in their N-terminal regions with three tandem repeats of the HA epitope (AGO1, AGO2, AGO7, and AGO10) (34, 37) or with a single HA epitope (AGO3, AGO4, AGO5, AGO6, and AGO9 [kindly provided by A. Takeda and Y. Watanabe]). First, we reproduced results obtained with a protocol reported previously showing that AGO7 and miR390 from A. thaliana interact specifically when overexpressed in N. benthamiana (34). On one hand, leaves of noninfected N. benthamiana were agroinfiltrated with a culture of A. tumefaciens with the construct 35S:miR390 for expressing miR390 under the control of the 35S promoter. On the other hand, the same culture was coagroinfiltrated with others carrying plasmids for expressing either AGO1 or AGO7, both tagged with the HA epitope and under the control of the same promoter (35S:HA-AGO1 and 35S:HA-AGO7, respectively). Northern blot hybridizations with a 5′-radiolabeled oligodeoxyribonucleotide complementary to miR390 revealed that while the RNA accumulated to high levels in the total RNA fraction from infiltrated halos, it was detected only in AGO7 immunoprecipitate (data not shown). Given that AGO7, but not AGO1, specifically binds miR390 (34) and that Western blot analysis with an anti-HA-peroxidase monoclonal antibody showed similar accumulation of AGO1 and AGO7 in the total protein and in the immunoprecipitate fractions (data not shown), these results provided the support needed for extending the same approach to investigate whether one or more AGOs interact specifically with vd-sRNAs.
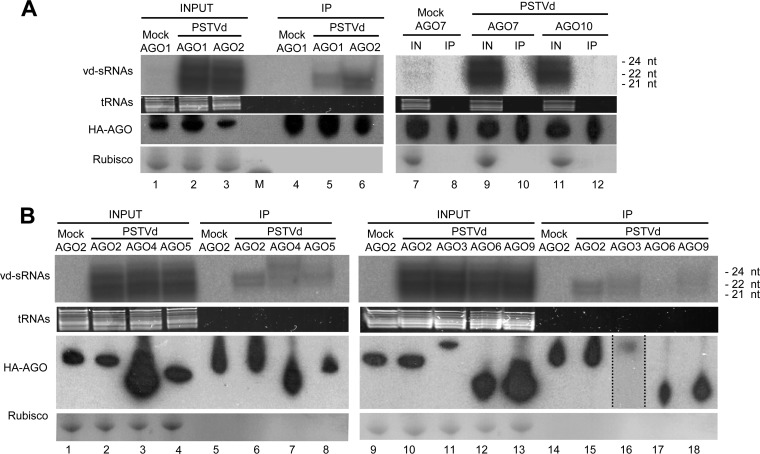
We initially examined AGO1, AGO2, AGO7, and AGO10 from A. thaliana because some of them have been involved in defense against RNA viruses—namely, AGO1, AGO2, and AGO7 against turnip crinkle virus (TCV) (31, 36); AGO2 against turnip mosaic virus (TuMV) (37); and AGO1 and AGO2 against cucumber mosaic virus (CMV) (31, 33, 36)—and also because they display distinct size and sequence specificity for some sRNAs (29, 30). Moreover, because when the present work was started association of AGO10 with sRNAs had not been shown (29), the protein was taken as a non-sRNA-binding AGO control. To begin with, each plasmid for expressing the HA-tagged AGOs was coagroinfiltrated with the 35S:dPSTVd(+) construct to trigger infection. Analysis by denaturing PAGE and Northern blot hybridization revealed the presence of vd-sRNAs in the total RNA fractions from the coagroinfiltrated halos—with higher intensity at 3 than at 2 days p.i.—but not in the corresponding immunoprecipitates (data not shown). In view of these results, we took an alternative approach: plants were first agroinfiltrated with the 35S:dPSTVd(+) construct, and 19 days later, when PSTVd had spread systemically, upper, noninoculated leaves were agroinfiltrated with the HA-tagged AGO constructs (35S:3×HA-AGO1, 35S:3×HA-AGO2, 35S:3×HA-AGO7, or 35S:3×HA-AGO10). Analysis, as described above, of the corresponding halos at 2 days after the second agroinfiltration showed high accumulation of vd-sRNAs of 21 to 24 nt in the total RNA fractions of the four samples, in contrast with the immunoprecipitates, in which only vd-sRNAs of 21 or 22 nt were detected, exclusively associated with AGO1 and AGO2 (Fig. 2A). Western blot analyses showed that the four agroinfiltrated AGOs (AGO1, AGO2, AGO7, and AGO10) were expressed (Fig. 2A). Altogether, these results indicated that vd-RNAs behave like other sRNAs of viral and endogenous origin (29) and are specifically loaded in some, but not in all, AGOs, mainly according to their sizes (and possibly to other structural properties [see below]).
FIG 2.
(A) AGO1 and AGO2, but not AGO7 or AGO10, specifically bind certain vd-sRNAs. Shown are Northern blot hybridizations with a full-length radiolabeled riboprobe for detecting PSTVd plus total RNA strands (INPUT or IN) from mock- and PSTVd-inoculated N. benthamiana agroinfiltrated with cultures of A. tumefaciens with binary plasmids for expressing HA-tagged AGO1 (lanes 1 and 2), AGO2 (lane 3), AGO7 (lanes 7 and 9), and AGO10 (lane 11). A size marker was included in lane M. RNA immunoprecipitates (IP) generated with an anti-HA monoclonal antibody from the halos agroexpressing HA-tagged AGO1 (lanes 4 and 5), AGO 2 (lane 6), AGO7 (lanes 8 and 10), and AGO10 (lane 12) were similarly analyzed. (B) Other agroinfiltrated AGOs, apart from AGO1 and AGO2, also bind vd-sRNAs with different affinities. Shown are Northern blot hybridizations with a full-length radiolabeled riboprobe for detecting PSTVd plus strands of total RNAs (INPUT) from mock- and PSTVd-inoculated N. benthamiana agroinfiltrated with cultures of A. tumefaciens with binary plasmids for expressing HA-tagged AGO2 (lanes 1, 2, 9, and 10), AGO4 (lane 3), AGO5 (lane 4), AGO3 (lane 11), AGO6 (lane 12), and AGO9 (lane 13). RNA immunoprecipitates (IP) generated with an anti-HA monoclonal antibody from the halos agroexpressing HA-tagged AGO2 (lanes 5, 6, 14, and 15), AGO4 (lane 7), AGO5 (lane 8), AGO3 (lane 16, overexposed to make the band visible), AGO6 (lane 17), and AGO9 (lane 18) were similarly analyzed. RNAs were separated by denaturing PAGE in 17% gels, and equal loading was assessed by the intensity of tRNA after staining with ethidium bromide. Western blot analyses of total proteins from halos were carried out with the anti-HA monoclonal antibody following protein separation by PAGE in 4 to 12% gels; equal loading was assessed by the intensity of the large subunit of RubisCO after staining with Ponceau S. Mock inoculations were performed with cultures of A. tumefaciens with a binary plasmid expressing GUS instead of the head-to-tail dimeric transcript of PSTVd. In all cases, samples were processed 2 days after agroinfiltration of plants that were PSTVd infected, or mock inoculated, 19 days before.
Agroexpressed AGO3, AGO4, AGO5, and AGO9 also bind vd-sRNAs with different affinities.
We then extended these analyses to other available constructs, also under the control of the 35S promoter, for expressing the single HA-tagged AGOs from A. thaliana: 35S:HA-AGO3, 35S:HA-AGO4, 35S:HA-AGO5, 35S:HA-AGO6, and 35S:HA-AGO9; 35S:3×HA-AGO2 was used as an internal control for linking results from these experiments with those of the previous one. Analysis by denaturing PAGE and Northern blot hybridization of the corresponding halos at 2 days after the second agroinfiltration revealed high levels of vd-sRNAs of 21, 22, and 24 nt in the total RNA fractions of all samples, except in a negative control in which the 35S:dPSTVd(+) construct used in the first agroinfiltration for triggering PSTVd infection was replaced by the 35S:GUS construct (Fig. 2B). However, the immunoprecipitate fractions behaved in a different manner: the vd-sRNAs of 21 or 22 nt were detected in samples expressing AGO2 and AGO3, while in the samples expressing AGO4 and AGO5 (and to a lesser extent in that expressing AGO9), vd-sRNAs of 24 nt were additionally detected; in contrast, essentially no vd-sRNAs were observed in the sample expressing AGO6 (Fig. 2B). Recovery of AGO-bound vd-sRNAs from immunoprecipitates could be influenced by the HA epitope being single or triple and by the distinct stability of AGOs against endogenous proteases, although Western blot analyses with the anti-HA-peroxidase monoclonal antibody showed that, albeit with different extensions, all agroinfiltrated AGOs were expressed (Fig. 2B). The inability of AGO6 to bind vd-sRNAs was nevertheless confirmed in further experiments (data not shown). Collectively, these results are consistent with vd-RNAs being differentially sorted into specific AGOs according to the sizes reported previously for other viral and endogenous sRNAs (29, 30).
Deep sequencing reveals that sorting of vd-sRNAs into AGO1, AGO2, AGO4, and AGO5 mainly depends on their sizes and 5′-terminal nucleotides.
To better understand the differential AGO affinity for vd-sRNAs, AGO1 and AGO2 (as representatives of those members of the family that associate preferentially with the sRNAs of 21 or 22 nt) and AGO4 and AGO5 (as representatives of those members of the family that associate additionally with the sRNAs of 24 nt) (29, 30) were selected for further examination. Moreover, while AGO1, AGO2, and AGO5 are able to bind virus-derived sRNAs and have been involved in antiviral defense, presumably via posttranscriptional gene silencing (31, 33, 36, 37), AGO4 mostly mediates transcriptional silencing (38). Previous immunoprecipitation assays have also revealed that the identity of the 5′-terminal nucleotide and the lengths of the sRNAs contribute to their sorting into these four AGO proteins (34, 35, 63).
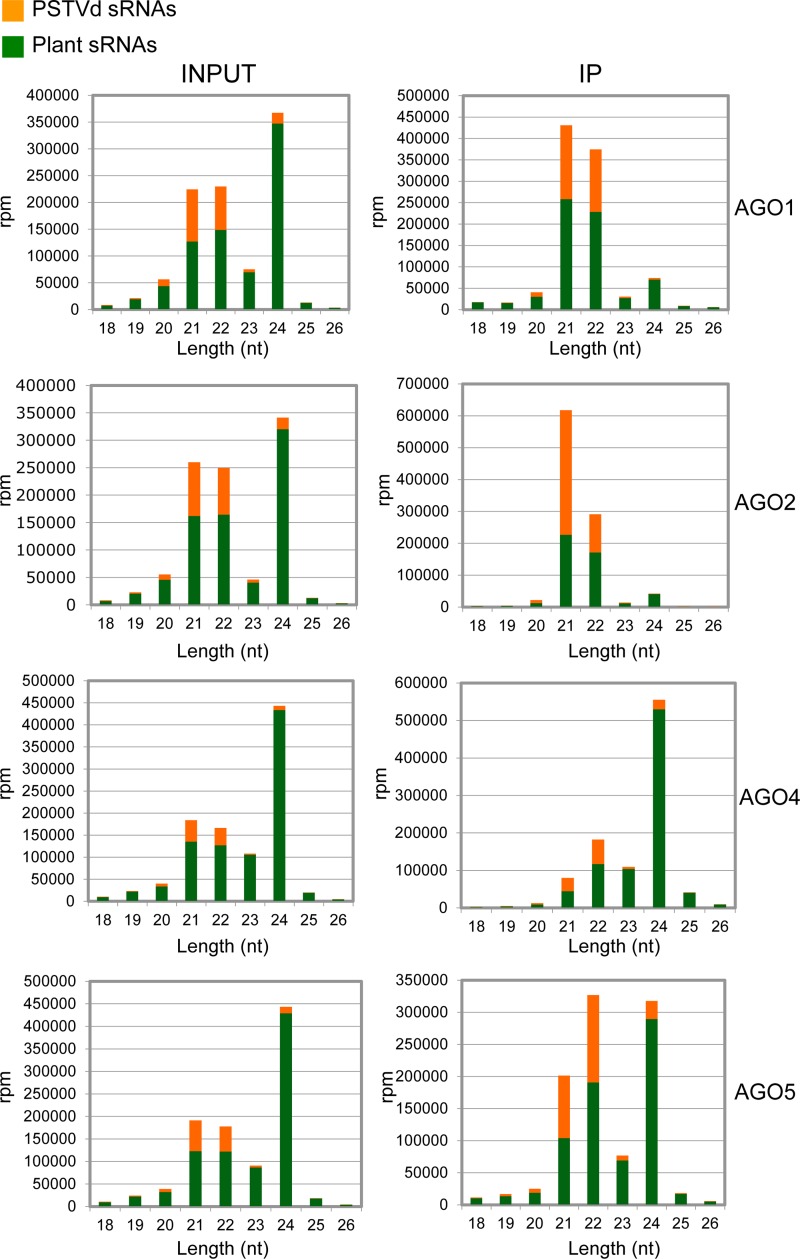
The first deep sequencing of sRNAs resulted in approximately 137,500,000 reads, 95.6% of which corresponded to the four bar-coded samples run in the same channel: the total sRNAs (inputs) and IPs from PSTVd-infected N. benthamiana overexpressing AGO1 and AGO2, with the fraction of each sample representing 23 to 29% of the total number of reads. Within the range of 18 to 26 nt, vd-sRNAs amounted to about 22% in the inputs and 34% and 53% in AGO1-IP and AGO2-IP, respectively. The second deep sequencing of sRNAs generated roughly 209,300,000 reads, 96.6% of which corresponded to the four bar-coded samples run in the same channel: the inputs and IPs from PSTVd-infected N. benthamiana overexpressing AGO4 and AGO5, with the fraction of each sample representing 19 to 29% of the total number of reads. Within the range of 18 to 26 nt, vd-sRNAs amounted to 11 to 15% in the inputs and 14% and 28% in AGO4-IP and AGO5-IP, respectively. Therefore, with respect to the inputs, the proportion of vd-sRNA was enriched in the IPs, making up a significant fraction of the total. This bias was also observed (particularly in AGO4 and AGO5) when the vd-sRNAs were disaggregated into size classes (Fig. 3).
FIG 3.
Size distribution of PSTVd and plant sRNAs in total RNAs (INPUT) and immunoprecipitates (IP) from halos of PSTVd-infected N. benthamiana agroinfiltrated with cultures of A. tumefaciens with binary plasmids for expressing HA-tagged versions of AGO1, AGO2, AGO4, and AGO5 from A. thaliana. The histograms compare the distributions of 18- to 26-nt total sRNA reads. The IP fractions were generated with an anti-HA monoclonal antibody. Note that the scales are not identical in the different histograms and that the fraction of PSTVd-sRNAs could be higher considering that the viroid may not invade all cells.
Analysis of the vd-sRNA reads from the inputs revealed similar size distributions in the four samples: 44 to 45% were 21 nt, 36 to 38% were 22 nt, and 9 to 9.5% were 24 nt, in agreement with previous results (50). However, the situation was somewhat different in the IPs, dominated in AGO1 and AGO2 by vd-sRNAs of 21 and, to a lesser extent, 22 nt (with those of 24 nt essentially absent), while the preponderant vd-sRNA species in AGO4 and AGO5 were 22 nt (and to a lesser extent 21 nt), with those of 24 nt amounting to 18 and 10%, respectively. Hence, the AGO proteins exert some size-based selection on the vd-sRNAs they capture. On the other hand, the bias between plus and minus vd-sRNAs (derived from the most abundant and less abundant viroid strands accumulating in vivo, respectively) was minor in inputs and IPs, except in the AGO2-IP, in which the fraction of vd-sRNAs of plus polarity was significantly higher (data not shown).
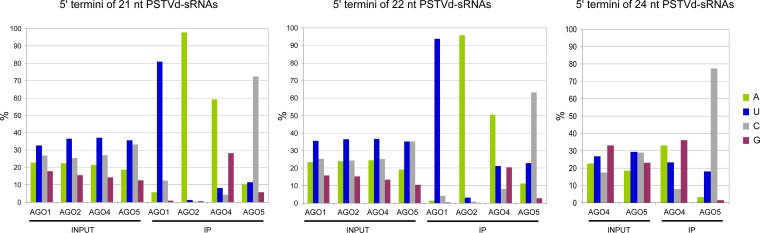
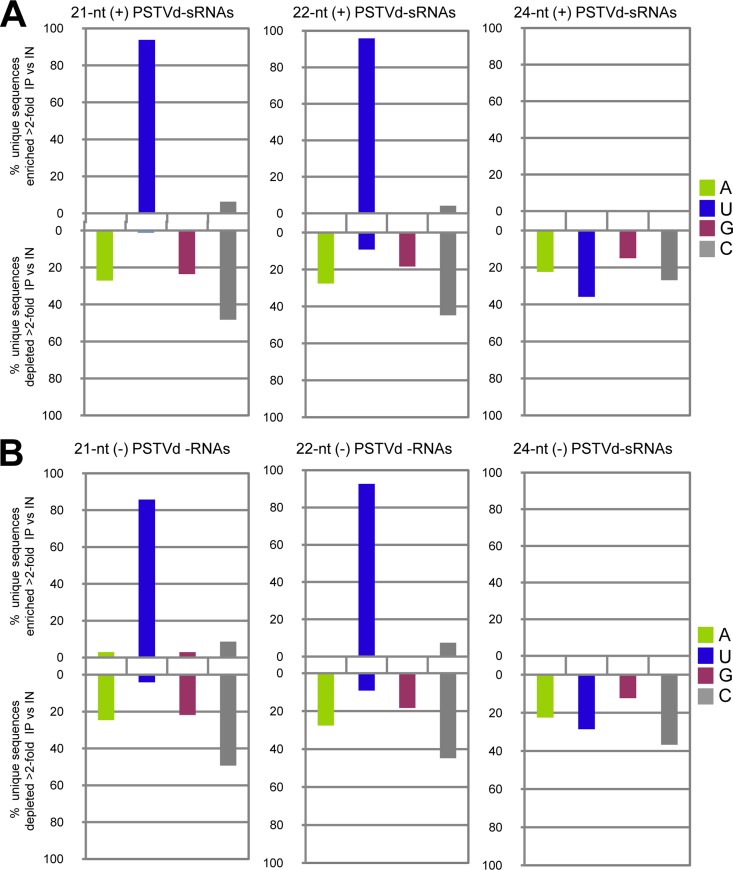
We next compared the distributions of the vd-sRNAs with respect to their 5′-terminal nucleotides, considering that in Arabidopsis this feature has a crucial role in sorting the sRNAs into the different AGOs (34, 35, 63). While 21- and 22-nt vd-sRNAs with a 5′-terminal U and C were moderately predominant in the inputs (up to 37% and 33%, respectively), the corresponding IP patterns were highly biased in their 5′-terminal nucleotides: 81% U, 98% A, 59% A, and 72% C in AGO1, AGO2, AGO4, and AGO5, respectively (Fig. 4). As for the 24-nt vd-sRNAs, those with a 5′-terminal G were moderately prevalent (33%) in the AGO4 input, as also were those with a 5′-terminal U or C in the AGO5 input (29%); however, the distribution in their IP counterparts was clearly biased for vd-sRNAs with a 5′-terminal C, which were underrepresented in AGO4-IP (8%) but prevalent in AGO5-IP (77%). This result indicates that the strong preference of AGO5 for binding vd-sRNAs with a 5′-terminal C is size independent. Similar distributions were obtained when the plus and minus vd-sRNAs from the IPs were examined separately (data not shown). Altogether, these results support the notion that the 5′-terminal nucleotide of vd-sRNAs is a major determinant for AGO sorting, following rules similar to those governing AGO sorting of endogenous and viral sRNAs (34, 35, 63). However, as proposed for certain endogenous sRNAs (39), characteristics other than the 5′-terminal nucleotide may also contribute to vd-sRNA loading.
FIG 4.
Sorting of PSTVd-sRNAs into AGO1, AGO2, AGO4, and AGO5 mainly depends on their 5′-terminal nucleotides. The histograms display, in total RNA (INPUT) and in RNA immunoprecipates (IP), the fraction (%) of total reads corresponding to the 21-, 22-, and 24-nt PSTV-sRNAs with distinct 5′ termini.
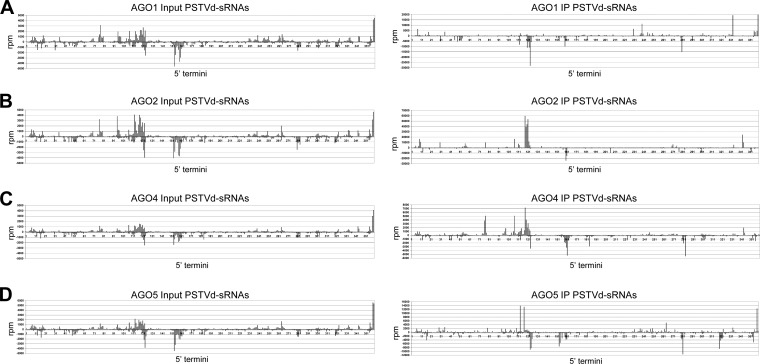
The profiles of AGO-loaded vd-sRNAs adopt specific hot spot distributions along the viroid genome.
Analysis of the vd-sRNA reads from AGO IPs revealed that they mapped at numerous positions of the genomic plus and minus viroid strands, with a significant fraction of the reads accumulating in specific regions (hot spots) and displaying some peculiarities (Fig. 5). First, the hot spot profiles of the vd-sRNAs immunoprecipitated by each of the four AGOs tested were different, as a consequence of their affinities for specific 5′-terminal nucleotides and of the uneven distribution of the four nucleotides in the PSTVd plus and minus genomic strands. However, the IP profiles were not a direct reflection of their corresponding input counterparts (e.g., the vd-sRNAs with their 5′-terminal U mapping at positions ∼240 and ∼333 in the plus strand and ∼123 and ∼281 in the minus strand, which are overrepresented in the AGO1 IP with respect to the AGO1 input), thus indicating the existence of some bias (Fig. 5). Second, hot spots in both input and IP profiles mapped in regions with a high G+C content, a likely consequence of the preference of DCLs for such regions (64, 65). Third, focusing on the vd-sRNAs mapping around positions 45 to 50 and 308 to 318 of the pathogenic (P) domain, 119 to 122 in the boundary between the central (C) and variable (V) domains, and 257 and 259 of the C domain—all associated with pathogenesis (66, 67)—striking accumulation of those of minus and plus polarity around positions 119 to 122 were observed in the AGO1 and AGO2 IPs, respectively (Fig. 5). This result may have implications for the mechanism of symptom induction, despite the observation that low-abundance vd-sRNAs can be functionally relevant (see Discussion).
FIG 5.
AGO-loaded vd-sRNAs adopt hot spot distributions along the viroid genome that are specific for each of the four HA-tagged AGOs from A. thaliana agroexpressed in PSTVd-infected N. benthamiana. Shown are the locations and frequencies in the genomic PSTVd RNA of the 5′ termini of the plus-strand (positive values) and minus-strand (negative values) vd-sRNA reads per million (rpm) from total RNAs (Input) and from immunoprecìpitates (IP) generated with an anti-HA monoclonal antibody. Note that the same numbers are used in the plus polarity (the 5′-to-3′ orientation is from left to right) and in the minus polarity (the 5′-to-3′ orientation is from right to left).
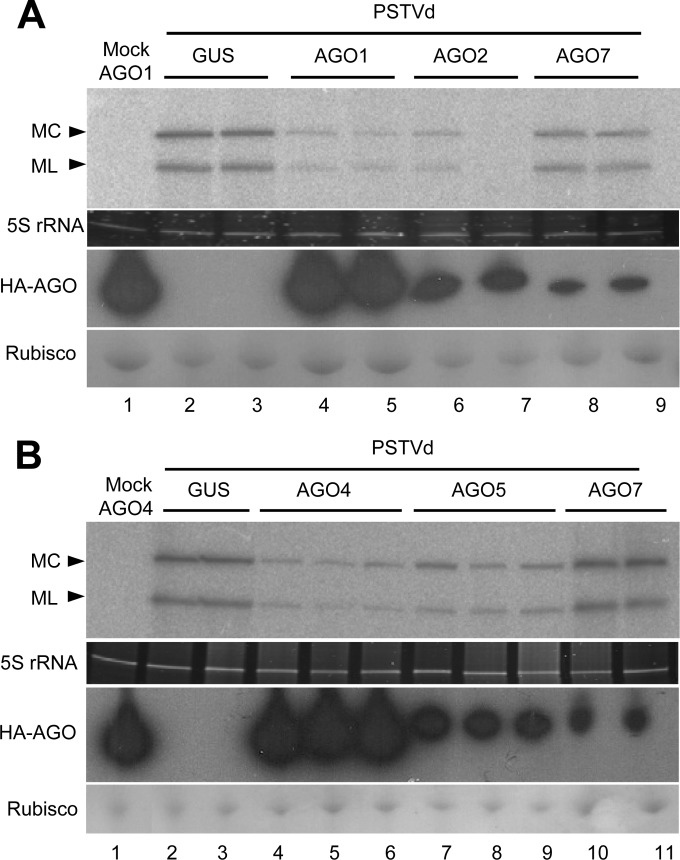
Agroexpression of AGO1, AGO2, AGO4, and AGO5 on PSTVd-infected tissue attenuates the level of the genomic RNAs.
To test whether AGO-loaded vd-sRNAs could have a functional effect, we next examined the titers of the MC and ML PSTVd plus forms. For this purpose, plants of N. benthamiana were agroinfected with PSTVd as indicated previously, and 8 days later, upper, noninoculated leaves were agroinfiltrated with A. tumefaciens cultures for expressing HA-AGO1 or HA-AGO2; total RNA preparations from the corresponding halos were extracted 2 days afterward. After some preliminary experiments, we chose this early sampling time to avoid the possibility that the potential effects of AGOs could be masked at later infectious stages, when the accumulation levels of the genomic viroid RNA increase very rapidly. Plants of N. benthamiana agroinfiltrated with cultures for expressing HA-AGO7 and GUS, as well as mock-inoculated plants, were included as controls based on previous results (Fig. 2). Analysis by denaturing PAGE and Northern blot hybridization showed that the titers of MC and ML PSTVd plus RNAs in plants expressing HA-AGO1 and HA-AGO2 were lower than those in the controls expressing HA-AGO7 and GUS (Fig. 6A). Similar effects (attenuation of the titer of MC and ML PSTVd plus RNAs with respect to the same controls) were observed in plants expressing HA-AGO4 and HA-AGO5 (Fig. 6B). These results are consistent with the view that AGO1, AGO2, AGO4, and AGO5, loaded with vd-sRNAs and forming part of RISC, target PSTVd RNAs. Thus, not only DCLs—as revealed by the generation of vd-sRNAs in viroid-infected tissues (see above)—but also RISCs, seem to operate in maintaining viroid titers below certain levels.
FIG 6.
Agroexpression of AGO1, AGO2, AGO4, and AGO5, but not of AGO7 or of GUS, attenuates viroid accumulation. (A and B) Northern blot hybridizations with a full-length radiolabeled riboprobe for detecting PSTVd plus strands of total RNAs from halos of mock- and PSTVd-inoculated N. benthamiana agroinfiltrated with cultures of A. tumefaciens with binary plasmids for expressing HA-tagged AGO1 (A, lanes 1, 4, and 5), AGO2 (A, lanes 6 and 7), AGO4 (B, lanes 1, 4, 5, and 6), AGO5 (B, lanes 7, 8, and 9), AGO7 (A, lanes 8 and 9, and B, lanes 10 and 11), and GUS (A and B, lanes 2 and 3). Mock inoculations were performed as indicated in the legend to Fig. 3. Total RNAs, extracted 2 days after agroinfiltration, were separated by denaturing PAGE in 5% gels, and equal loading was assessed by the intensity of tRNA after staining with ethidium bromide. Western blot analyses of total proteins from halos were carried out with the anti-HA monoclonal antibody following protein separation by PAGE in 4 to 12% gels; equal loading was assessed by the intensity of the large subunit of RubisCO after staining with Ponceau S. In all cases, samples were processed 2 days after agroinfiltration of plants that were PSTVd infected, or mock inoculated, 8 days before.
Deep sequencing confirms loading of vd-sRNA by the endogenous AGO1 of N. benthamiana infected by PSTVd.
As a final control in an experimental context excluding agroexpression of AGO proteins from A. thaliana, we also performed a deep-sequencing analysis of the vd-sRNAs in the input and IP generated by a polyclonal antibody against the endogenous AGO1 from PSTVd-infected N. benthamiana (Fig. 1C). The analysis of the two bar-coded samples run in the same channel resulted in 3,839,392 reads of 18- to 26-nt vd-sRNAs (8.5% of the total reads of the input) and 4,270,376 reads of 18- to 26-nt vd-sRNAs (9.3% of the total reads of the IP). While the proportions of vd-sRNAs of 21, 22, and 24 nt in the input were similar (27 to 32%), the 21- and 22-nt vd-sRNAs in the IP amounted to 63 and 28%, respectively, with those of 24 nt representing only 2%. Therefore, AGO1 from N. benthamiana exerted size selection on the vd-sRNAs similar to that of its homologue from A. thaliana, displaying a clear preference for binding those of 21 and (to a lesser extent) 22 nt. Regarding polarity, the plus and minus vd-sRNAs in the input represented 66 and 34%, respectively, while the ratio in the IP was reversed (39 and 61%, respectively). The size selection exerted by AGO1 from N. benthamiana on vd-sRNAs (see above) was not significantly influenced by their polarity.
An enrichment/depletion analysis of vd-sRNA reads in the IP versus the input showed significant enrichment in the IPs of plus and minus vd-sRNAs of 21 and 22 nt (but not of 24 nt) with a 5′-terminal U (but not with the other three 5′-terminal nucleotides) (Fig. 7). Therefore, regarding binding specificity for vd-sRNAs, AGO1 from N. benthamiana behaved similarly to its agroexpressed homologue from A. thaliana.
FIG 7.
Analysis of vd-sRNAs in the IP versus the input generated by a polyclonal antibody against AGO1 from N. benthamiana reveals a clear enrichment in the IP of plus (A) and minus (B) vd-sRNAs of 21 and 22 nt (but not 24 nt) with a 5′-terminal U. IP enrichment or depletion was determined for each unique 21-, 22-, or 24-nt vd-sRNA as log2 [(IP reads + 1)/(input reads + 1)] and plotted for each size class as the fraction (%) of unique vd-sRNA sequences enriched >2-fold (log2 >1) or depleted >2-fold (log2 < 1) in the IP compared.
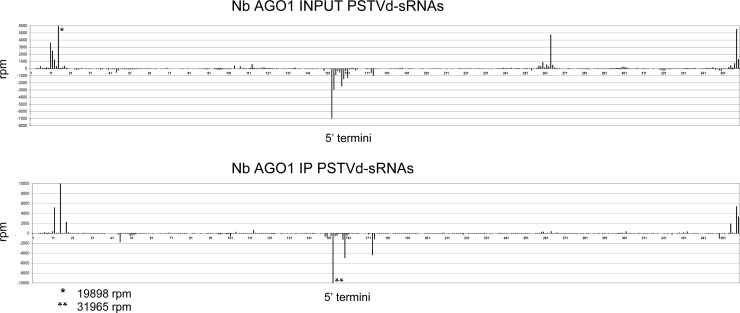
Finally, the profile of vd-sRNAs loaded by AGO1 from N. benthamiana presented a specific hot spot distribution along the viroid genome (Fig. 7 and 8), thus recapitulating the situation previously observed with the agroexpressed AGOs from A. thaliana. However, the vd-sRNA profiles corresponding to the agroinfiltrated AGO1 from A. thaliana and the endogenous AGO1 from N. benthamiana differed, most likely because of the different experimental conditions: in the first instance, the AGO1 from A. thaliana was overexpressed, while in the second instance, the accumulation of the endogenous AGO1 was considerably lower and possibly subjected to developmental regulation.
FIG 8.
AGO1-loaded vd-sRNAs adopt a hot spot distribution along the viroid genome in PSTVd-infected N. benthamiana. Shown are the locations and frequencies in the genomic PSTVd RNA of the 5′ termini of the plus-strand (positive values) and minus-strand (negative values) vd-sRNA reads per million (rpm) from total RNAs (INPUT) and from the immunoprecipitate (IP) generated with an anti-N. benthamiana AGO1 (Nb AGO1) polyclonal antibody. See the legend to Fig. 5 for details.
DISCUSSION
Our first immunoprecipitation assays, using PSTVd-infected leaves of N. benthamiana and a polyclonal antibody specific for its endogenous AGO1, showed that the protein indeed preferentially binds vd-sRNAs with the expected size (21 or 22 nt). However, PSTVd infection of N. benthamiana did not significantly affect the accumulation of either endogenous AGO1 or miR168 (which regulates AGO1 mRNA expression), as opposed to the situation observed in the same host following infection by different RNA viruses (42, 60). Considering that in the latter case the specific induction of miR168 is promoted by virus-encoded protein suppressors of RNA silencing and that PSTVd is a non-protein-coding RNA, this result is not surprising. Moreover, it does not favor the idea that vd-sRNAs, as proposed previously for transgene siRNAs and endogenous siRNAs and miRNAs (68), could compete to bind to AGO1 and lead to a reduction in AGO1-miR168 complexes and an increase in AGO1 mRNA translation. However, recent data indicate that infection by citrus exocortis viroid, a close relative of PSTVd, induces the accumulation of other enzymes mediating RNA-silencing steps in tomato (69). Whether this accumulation is a direct or indirect effect, and what is the nature of the underlying mechanism, remains unknown.
Previous reports indicated that viroids are significantly resistant to RISC-mediated degradation (47, 70, 71), suggesting that they may have evolved their secondary structures as a response against this selection pressure. In such a scenario, the compact secondary structure of PSTVd plus strands may hinder their targeting (and inactivation) by AGO proteins loaded with vd-sRNAs, while targeting PSTVd minus strands is even more difficult, because they mostly form part of double-stranded replicative complexes (8). From an alternative perspective, the secondary structure of viroids could have emerged as a compromise between resistance to DCL and to RISC, which act preferentially against RNAs with compact and relaxed conformations, respectively (48). Indeed, data obtained in other experimental contexts indicate that viroids are RISC sensitive (46, 48, 49, 52), and recent results show that RISC promotes cleavage of viral RNAs with a packed secondary structure—resembling that of viroids—by targeting bulged regions within the structure (72). However, the evidence that one or more AGOs are loaded with vd-sRNAs and function in antiviroid RISC is circumstantial, with no data providing direct support for this view.
The finding in tissues infected by typical members of both viroid families of vd-sRNAs with the characteristic features of DCL products (see above) does not necessarily entail their loading in one or more AGO proteins. Previous data from a study with an RNA virus have shown that the bulk of virus-derived sRNAs in latently infected Drosophila cells are not loaded into any AGO member, suggesting that dicing of viral dsRNAs by itself plays a key function in maintaining the latent state (73). Although dicing of the snap-folded genomic viroid ssRNA (or, more likely, of its dsRNA replication intermediates) could play a role in containing infection below a threshold value, extension of the “dicing-only” model to PSTVd does not seem justified. In support of this view, when 9 of the 10 AGOs from A. thaliana were agroexpressed in PSTVd-infected leaves of N. benthamiana, all except AGO6, AGO7, and AGO10 bound vd-sRNAs: AGO1, AGO2, and AGO3 bound those of 21 and 22 nt, while AGO4, AGO5, and AGO9 additionally bound those of 24 nt. Deep sequencing showed that, when agroexpressed in PSTVd-infected N. benthamiana leaves, AGO1, AGO2, AGO4, and AGO5 bound the vd-sRNAs, particularly those of 21 and 22 nt, primarily according to their 5′-terminal nucleotides, as reported previously for endogenous and viral sRNAs (34, 35, 63). Moreover, the ratio of vd-sRNA to total sRNAs in the AGO-IPs was higher than that in the inputs, indicating that vd-sRNAs were loaded into these AGO proteins with some preference. Therefore, DCLs could function as the first defensive barrier against viroid infection and, additionally, provide vd-sRNAs for priming the second RISC-based defensive barrier.
Viroids, lacking protein-coding ability, also might have evolved a sort of RNA-mediated decoy mechanism protecting them against RNA silencing, similar to that developed by alphaviruses, like Semliki Forest virus (SFV), which do not encode RNA-silencing suppressors. More specifically, alphaviruses have been proposed to produce decoy virus-derived sRNAs to hamper the RNA-silencing machinery and to provide the virus time for replication before being eventually silenced (74). In consonance with this view, the predominant virus sRNAs derived from hot spots are less effective at silencing SFV accumulation than those derived from cold spots (74). Regarding viroids, infections by PSTVd and the chloroplast-replicating peach latent mosaic viroid (PLMVd) (75) are accompanied by large amounts of vd-sRNAs (references 44 and 45 and this work). Moreover, the experimental evidence available supports the idea that vd-sRNAs mapping at cold spots are biologically active. Specifically, variants of PLMVd inducing severe albinism have a particular hairpin insertion of 12 to 14 nt (76, 77), and two low-abundance minus vd-sRNAs containing this insertion target for cleavage—as predicted by RNA silencing—the peach mRNA coding for cHSP90 involved in plastid-to-nucleus signal transduction (51). The two vd-sRNAs are 21 nt, fulfill the criteria for being functional sRNAs (78, 79), and have a 5′-terminal U, indicating that they are most likely loaded in AGO1; these criteria are also met by a 22-nt RNA that contains the region responsible for the yellow phenotype incited by the Y satellite RNA of CMV and directs cleavage, via RNA silencing, of the mRNA of a gene involved in chlorophyll biosynthesis (80, 81). A similar mechanism has been proposed for the phenotypes induced by artificial miRNAs (amiRNAs) derived from the virulence-modulating region of PSTVd (82), although the evidence is indirect and the amiRNAs do not fulfill all the above-mentioned criteria. The finding that host mRNAs are targeted by AGOs loaded with sRNAs derived from viroids and satellite RNAs supports the notion that these subviral replicons, like RNA viruses, are also targets of RISC.
Previously, hypomorphic ago1 mutants have been tested against virus infection, with their hypersensitive reaction and overaccumulation of viral RNA being interpreted as a confirmation of the involvement of RNA silencing, and particularly of AGO1, in antiviral defense (31). Here, we have taken the opposite approach: to overexpress certain AGO proteins and examine whether they result in viroid underaccumulation. Specifically, the synchronized overexpression of AGO1, AGO2, AGO4, and AGO5 in leaves of N. benthamiana at early stages of PSTVd infection has facilitated the observation of the attenuating effects of these proteins on the viroid titer. These results, together with the specific loading of vd-sRNAs with the expected size and 5′-terminal nucleotide by agroinfiltrated AGO1, AGO2, AGO4, and AGO5 from A. thaliana, as well as by the endogenous AGO1 of N. benthamiana, are consistent with the view that those members of the AGO family may play a role in anti-PSTVd defense.
ACKNOWLEDGMENTS
We are grateful to A. Takeda and Y. Watanabe for kindly providing 35S:HA-AGO3, 35S:HA-AGO4, 35S:HA-AGO5, 35S:HA-AGO6, 35S:HA-AGO8, and 35S:HA-AGO9 constructs; to N. Fahlgren for helping in developing scripts for the deep-sequencing analyses; to M. Cambra for a rabbit preimmune serum; to J. Burgyan for a rabbit polyclonal antibody against the N-terminal region of AGO1 from N. benthamiana that we used in preliminary experiments; and to A. Ahuir for excellent technical assistance.
Research in the laboratory of R.F. is currently funded by grant BFU2011-28443 from the Ministerio de Economía y Competitividad (MINECO, Spain). S.M. has been supported by a fellowship and a predoctoral contract from MINECO. Research in the laboratory of B.N. and F.D.S. has been funded by a dedicated grant from the Ministero dell'Economia e Finanze Italiano to the CNR (CISIA; Legge no. 191/2009). Research in the laboratory of J.C.C. was supported by grants from the National Science Foundation (MCB-0956526 and MCB-1231726) and the National Institutes of Health (AI043288).
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1. Flores R, Hernández C, Martínez de Alba AE, Daròs JA, Di Serio F. 2005. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117–139. 10.1146/annurev.phyto.43.040204.140243 [DOI] [PubMed] [Google Scholar]
- 2. Tsagris EM, Martínez de Alba AE, Gozmanova M, Kalantidis K. 2008. Viroids. Cell Microbiol. 10:2168–2179. 10.1111/j.1462-5822.2008.01231.x [DOI] [PubMed] [Google Scholar]
- 3. Ding B. 2009. The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47:105–131. 10.1146/annurev-phyto-080508-081927 [DOI] [PubMed] [Google Scholar]
- 4. Diener TO. 1967. Potato spindle tuber virus: a plant virus with properties of a free nucleic acid. Science 158:378–381. 10.1126/science.158.3799.378 [DOI] [PubMed] [Google Scholar]
- 5. Gross HJ, Domdey H, Lossow C, Jank P, Raba M, Alberty H, Sänger HL. 1978. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273:203–208. 10.1038/273203a0 [DOI] [PubMed] [Google Scholar]
- 6. Grill LK, Semancik JS. 1978. RNA sequences complementary to citrus exocortis viroid in nucleic acid preparations from infected Gynura aurantiaca. Proc. Natl. Acad. Sci. U. S. A. 75:896–900. 10.1073/pnas.75.2.896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ishikawa M, Meshi T, Ohno T, Okada Y, Sano T, Ueda I, Shikata E. 1984. A revised replication cycle for viroids: the role of longer than unit RNA in viroid replication. Mol. Gen. Genet. 196:421–428. 10.1007/BF00436189 [DOI] [PubMed] [Google Scholar]
- 8. Branch AD, Benenfeld BJ, Robertson HD. 1988. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. U. S. A. 85:9128–9132. 10.1073/pnas.85.23.9128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feldstein PA, Hu Y, Owens RA. 1998. Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc. Natl. Acad. Sci. U. S. A. 95:6560–6565. 10.1073/pnas.95.11.6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daròs JA, Flores R. 2004. Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc. Natl. Acad. Sci. U. S. A. 101:6792–6797. 10.1073/pnas.0401090101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mühlbach HP, Sänger HL. 1979. Viroid replication is inhibited by α-amanitin. Nature 278:185–188. 10.1038/278185a0 [DOI] [PubMed] [Google Scholar]
- 12. Flores R, Semancik JS. 1982. Properties of a cell-free system for synthesis of citrus exocortis viroid. Proc. Natl. Acad. Sci. U. S. A. 79:6285–6288. 10.1073/pnas.79.20.6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schindler IM, Mühlbach HP. 1992. Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci. 84:221–229. 10.1016/0168-9452(92)90138-C [DOI] [Google Scholar]
- 14. Gas ME, Hernández C, Flores R, Daròs JA. 2007. Processing of nuclear viroids in vivo: an interplay between RNA conformations. PLoS Pathog. 3:e182. 10.1371/journal.ppat.0030182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nohales MA, Flores R, Daròs JA. 2012. Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. U. S. A. 109:13805–13810. 10.1073/pnas.1206187109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gas ME, Molina-Serrano D, Hernández C, Flores R, Daròs JA. 2008. Monomeric linear RNA of citrus exocortis viroid resulting from processing in vivo has 5′-phosphomonoester and 3′-hydroxyl termini: implications for the ribonuclease and RNA ligase involved in replication. J. Virol. 82:10321–10325. 10.1128/JVI.01229-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flores R, Daròs JA, Hernández C. 2000. The Avsunviroidae family: viroids with hammerhead ribozymes. Adv. Virus Res. 55:271–323. 10.1016/S0065-3527(00)55006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qi Y, Pélissier T, Itaya A, Hunt E, Wassenegger M, Ding B. 2004. Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 16:1741–1752. 10.1105/tpc.021980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhong X, Tao X, Stombaugh J, Leontis N, Ding B. 2007. Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J. 26:3836–3846. 10.1038/sj.emboj.7601812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takeda R, Petrov AI, Leontis NB, Ding B. 2011. A three-dimensional RNA motif in potato spindle tuber viroid mediates trafficking from palisade mesophyll to spongy mesophyll in Nicotiana benthamiana. Plant Cell 23:258–272. 10.1105/tpc.110.081414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhong X, Archual AJ, Amin AA, Ding B. 2008. A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 20:35–47. 10.1105/tpc.107.056606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ding SW. 2010. RNA-based antiviral immunity. Nat. Rev. Immunol. 10:632–644. 10.1038/nri2824 [DOI] [PubMed] [Google Scholar]
- 24. Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64:137–159. 10.1146/annurev-arplant-050312-120043 [DOI] [PubMed] [Google Scholar]
- 25. Incarbone M, Dunoyer P. 2013. RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 18:382–392. 10.1016/j.tplants.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 26. Pumplin N, Voinnet O. 2013. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 11:745–760. 10.1038/nrmicro3120 [DOI] [PubMed] [Google Scholar]
- 27. Schiebel W, Pélissier T, Riedel L, Thalmeir S, Schiebel R, Kempe D, Lottspeich F, Sänger HL, Wassenegger M. 1998. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10:2087–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voinnet O. 2008. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 13:317–328. 10.1016/j.tplants.2008.05.004 [DOI] [PubMed] [Google Scholar]
- 29. Mallory A, Vaucheret H. 2010. Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22:3879–3889. 10.1105/tpc.110.080671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bologna NG, Voinnet O. 2014. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65:473–503. 10.1146/annurev-arplant-050213-035728 [DOI] [PubMed] [Google Scholar]
- 31. Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14:629–639. 10.1105/tpc.010358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baumberger N, Baulcombe DC. 2005. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 102:11928–11933. 10.1073/pnas.0505461102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qu F, Ye X, Morris TJ. 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U. S. A. 105:14732–14737. 10.1073/pnas.0805760105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. 2008. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133:128–141. 10.1016/j.cell.2008.02.033 [DOI] [PubMed] [Google Scholar]
- 35. Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. 2008. The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol. 49:493–500. 10.1093/pcp/pcn043 [DOI] [PubMed] [Google Scholar]
- 36. Alvarado VY, Scholthof HB. 2011. AGO2: a new Argonaute compromising plant virus accumulation. Front. Plant Sci. 2:112. 10.3389/fpls.2011.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carbonell A, Fahlgren N, García-Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, Cuperus JT, Carrington JC. 2012. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24:3613–3629. 10.1105/tpc.112.099945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zilberman D, Cao X, Jacobsen SE. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716–719. 10.1126/science.1079695 [DOI] [PubMed] [Google Scholar]
- 39. Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. 2010. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22:321–334. 10.1105/tpc.109.072199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. 2006. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443:1008–1012. 10.1038/nature05198 [DOI] [PubMed] [Google Scholar]
- 41. Mallory AC, Hinze A, Tucker MR, Bouché N, Gasciolli V, Elmayan T, Lauressergues D, Jauvion V, Vaucheret H, Laux T. 2009. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 5:e1000646. 10.1371/journal.pgen.1000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Várallyay E, Valoczi A, Agyi A, Burgyán J, Havelda Z. 2010. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 29:3507–3519. 10.1038/emboj.2010.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM. 2013. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One 8:e59534. 10.1371/journal.pone.0059534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Navarro B, Gisel A, Rodio ME, Delgado S, Flores R, Di Serio F. 2012. Viroids: how to infect a host and cause disease without encoding proteins. Biochimie 94:1474–1480. 10.1016/j.biochi.2012.02.020 [DOI] [PubMed] [Google Scholar]
- 45. Hammann C, Steger G. 2012. Viroid-specific small RNA in plant disease. RNA Biol. 9:809–819. 10.4161/rna.19810 [DOI] [PubMed] [Google Scholar]
- 46. Vogt U, Pelissier T, Putz A, Razvi F, Fischer R, Wassenegger M. 2004. Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. 38:107–118. 10.1111/j.1365-313X.2004.02029.x [DOI] [PubMed] [Google Scholar]
- 47. Itaya A, Zhong X, Bundschuh R, Qi Y, Wang Y, Takeda R, Harris AR, Molina C, Nelson RS, Ding B. 2007. A structured viroid RNA is substrate for Dicer-like cleavage to produce biologically active small RNAs but is resistant to RISC-mediated degradation. J. Virol. 81:2980–2994. 10.1128/JVI.02339-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carbonell A, Martínez de Alba AE, Flores R, Gago S. 2008. Double-stranded RNA interferes in a sequence-specific manner with infection of representative members of the two viroid families. Virology 371:44–53. 10.1016/j.virol.2007.09.031 [DOI] [PubMed] [Google Scholar]
- 49. Schwind N, Zwiebel M, Itaya A, Ding B, Wang MB, Krczal G, Wassenegger M. 2009. RNAi-mediated resistance to potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. Plant Pathol. 10:459–469. 10.1111/j.1364-3703.2009.00546.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Di Serio F, Martínez de Alba AE, Navarro B, Gisel A, Flores R. 2010. RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a nuclear-replicating viroid. J. Virol. 84:2477–2489. 10.1128/JVI.02336-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Navarro B, Gisel A, Rodio ME, Delgado S, Flores R, Di Serio F. 2012. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 70:991–1003. 10.1111/j.1365-313X.2012.04940.x [DOI] [PubMed] [Google Scholar]
- 52. Serra P, Bani-Hashemian SM, Fagoaga C, Romero J, Ruiz-Ruiz S, Gorris MT, Bertolini E, Duran-Vila N. 2014. Virus-viroid interactions: citrus tristeza virus enhances the accumulation of citrus dwarfing viroid in Mexican lime via the viral-encoded silencing suppressors. J. Virol. 88:1394–1397. 10.1128/JVI.02619-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wassenegger M, Heimes S, Riedel L, Sänger HL. 1994. RNA-directed de novo methylation of genomic sequences in plants, Cell 76:567–576 [DOI] [PubMed] [Google Scholar]
- 54. Martínez G, Castellano M, Tortosa M, Pallás V, Gómez G. 2014. A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic Acids Res. 42:1553–1562. 10.1093/nar/gkt968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grimsley N, Hohn B, Hohn T, Walden R. 1986. Agroinfection, an alternative route for viral-infection of plants by using the Ti plasmid. Proc. Natl. Acad. Sci. U. S. A. 83:3282–3286. 10.1073/pnas.83.10.3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Llave C, Xie Z, Kasschau KD, Carrington JC. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056. 10.1126/science.1076311 [DOI] [PubMed] [Google Scholar]
- 57. Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. 2010. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17:997–1003. 10.1038/nsmb.1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 59. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Várallyay E, Havelda Z. 2013. Unrelated suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol. Plant Pathol. 14:567–575. 10.1111/mpp.12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mallory AC, Vaucheret H. 2009. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 10:521–526. 10.1038/embor.2009.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Matousek J, Kozlová P, Orctová L, Schmitz A, Pesina K, Bannach O, Diermann N, Steger G, Riesner D. 2007. Accumulation of viroid-specific small RNAs and increase of nucleolytic activities linked to viroid-caused pathogenesis. Biol. Chem. 388:1–13. 10.1515/BC.2007.001 [DOI] [PubMed] [Google Scholar]
- 63. Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, Chen S, Hannon GJ, Qi Y. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116–127. 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ho T, Wang H, Palletta D, Dalmay T. 2007. Evidence for targeting common siRNA hotspots and GC preference by plant Dicer-like proteins. FEBS Lett. 581:3267–3272. 10.1016/j.febslet.2007.06.022 [DOI] [PubMed] [Google Scholar]
- 65. Zhang Y, Chenghuan Y, Hanhui K. 2014. GC content fluctuation around plant small RNA-generating sites. FEBS Lett. 588:764–769. 10.1016/j.febslet.2014.01.023 [DOI] [PubMed] [Google Scholar]
- 66. Qi Y, Ding B. 2003. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15:1360–1374. 10.1105/tpc.011585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schnölzer M, Haas B, Ramm K, Hofmann H, Sänger HL. 1985. Correlation between structure and pathogenicity of potato spindle tuber viroid (PSTV). EMBO J. 4:2181–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Martínez de Alba AE, Jauvion V, Mallory AC, Bouteiller N, Vaucheret H. 2011. The miRNA pathway limits AGO1 availability during siRNA-mediated PTGS defense against exogenous RNA. Nucleic Acids Res. 39:9339–9344. 10.1093/nar/gkr590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Campos L, Granell P, Tárraga S, López-Gresa P, Conejero V, Bellés JM, Rodrigo I, Lisón P. 2014. Salicylic acid and gentisic acid induce RNA silencing-related genes and plant resistance to RNA pathogens. Plant Physiol. Biochem. 77:35–43. 10.1016/j.plaphy.2014.01.016 [DOI] [PubMed] [Google Scholar]
- 70. Wang MB, Bian XY, Wu LM, Liu LX, Smith NA, Isenegger D, Wu RM, Masuta C, Vance VB, Watson JM, Rezaian A, Dennis ES, Waterhouse PM. 2004. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. U. S. A. 101:3275–3280. 10.1073/pnas.0400104101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gómez G, Pallás V. 2007. Mature monomeric forms of hop stunt viroid resist RNA silencing in transgenic plants. Plant J. 51:1041–1049. 10.1111/j.1365-313X.2007.03203.x [DOI] [PubMed] [Google Scholar]
- 72. Schuck J, Gursinsky T, Pantaleo V, Burgyan J, Behrens SE. 2013. AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res. 41:5090–5103. 10.1093/nar/gkt193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Flynt A, Liu N, Martin R, Lai EC. 2009. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc. Natl. Acad. Sci. U. S. A. 106:5270–5275. 10.1073/pnas.0813412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Siu RW, Fragkoudis R, Simmonds P, Donald CL, Chase-Topping ME, Barry G, Attarzadeh-Yazdi G, Rodriguez-Andres J, Nash AA, Merits A, Fazakerley JK, Kohl A. 2011. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J. Virol. 85:2907–2917. 10.1128/JVI.02052-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hernández C, Flores R. 1992. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. U. S. A. 89:3711–3715. 10.1073/pnas.89.9.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Malfitano M, Di Serio F, Covelli L, Ragozzino A, Hernández C, Flores R. 2003. Peach latent mosaic viroid variants inducing peach calico contain a characteristic insertion that is responsible for this symptomatology. Virology 313:492–501. 10.1016/S0042-6822(03)00315-5 [DOI] [PubMed] [Google Scholar]
- 77. Rodio ME, Delgado S, De Stradis AE, Gómez MD, Flores R, Di Serio F. 2007. A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell 19:3610–3626. 10.1105/tpc.106.049775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Iwakawa HO, Tomari Y. 2013. Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell 52:591–601. 10.1016/j.molcel.2013.10.033 [DOI] [PubMed] [Google Scholar]
- 79. Liu Q, Wang F, Axtell MJ. 2014. Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell 26:741–753. 10.1105/tpc.113.120972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba JI, Sueda K, Burgyán J, Masuta C. 2011. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7:e1002021. 10.1371/journal.ppat.1002021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Smith NA, Eamens AL, Wang MB. 2011. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 7:e1002022. 10.1371/journal.ppat.1002022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Eamens AL, Smith NA, Dennis ES, Wassenegger M, Wang MB. 2014. In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology 450–451:266–277. 10.1016/j.virol.2013.12.019 [DOI] [PubMed] [Google Scholar]