ABSTRACT
Protein-protein and protein-nucleic acid interactions within subcellular compartments are required for viral genome replication. To understand the localization of the human cytomegalovirus viral replication factor UL84 relative to other proteins involved in viral DNA synthesis and to replicating viral DNA in infected cells, we created a recombinant virus expressing a FLAG-tagged version of UL84 (UL84FLAG) and used this virus in immunofluorescence assays. UL84FLAG localization differed at early and late times of infection, transitioning from diffuse distribution throughout the nucleus to exclusion from the interior of replication compartments, with some concentration at the periphery of replication compartments with newly labeled DNA and the viral DNA polymerase subunit UL44. Early in infection, UL84FLAG colocalized with the viral single-stranded DNA binding protein UL57, but colocalization became less prominent as infection progressed. A portion of UL84FLAG also colocalized with the host nucleolar protein nucleolin at the peripheries of both replication compartments and nucleoli. Small interfering RNA (siRNA)-mediated knockdown of nucleolin resulted in a dramatic elimination of UL84FLAG from replication compartments and other parts of the nucleus and its accumulation in the cytoplasm. Reciprocal coimmunoprecipitation of viral proteins from infected cell lysates revealed association of UL84, UL44, and nucleolin. These results indicate that UL84 localization during infection is dynamic, which is likely relevant to its functions, and suggest that its nuclear and subnuclear localization is highly dependent on direct or indirect interactions with nucleolin.
IMPORTANCE The protein-protein interactions among viral and cellular proteins required for replication of the human cytomegalovirus (HCMV) DNA genome are poorly understood. We sought to understand how an enigmatic HCMV protein critical for virus replication, UL84, localizes relative to other viral and cellular proteins required for HCMV genome replication and replicating viral DNA. We found that UL84 localizes with viral proteins, viral DNA, and the cellular nucleolar protein nucleolin in the subnuclear replication compartments in which viral DNA replication occurs. Unexpectedly, we also found localization of UL84 with nucleolin in nucleoli and showed that the presence of nucleolin is involved in localization of UL84 to the nucleus. These results add to previous work showing the importance of nucleolin in replication compartment architecture and viral DNA synthesis and are relevant to understanding UL84 function.
INTRODUCTION
Virus replication requires subcellular compartmentalization and protein-protein and protein-nucleic acid interactions. In the nuclei of cells infected with herpesviruses, nonmembranous replication compartments form. Within these compartments, viral DNA synthesis occurs concomitant with the accumulation of DNA replication proteins. In cells infected with the betaherpesvirus human cytomegalovirus (HCMV), these replication proteins include the accessory subunit of the viral DNA polymerase UL44 and the viral single-stranded DNA (ssDNA) binding protein UL57 (1–3). The HCMV replication compartment architecture has several distinguishing features (2, 4). In particular, UL44 concentrates at the periphery of replication compartments. Viral DNA synthesis occurs within this peripheral layer, and newly replicated DNA is subsequently localized within the interior of replication compartments, which is where most UL57 is found (2). In contrast, studies of the alphaherpesvirus herpes simplex virus 1 (HSV-1) have revealed little evidence of comparable replication compartment organization (5, 6).
Several factors are involved in maintaining the architecture of HCMV replication compartments. One is viral DNA synthesis, as treatment of infected cells with an inhibitor of viral DNA synthesis leads to loss of compartment organization (1, 2), and compartment formation is delayed in cells infected with a virus mutant in which viral genome replication is delayed (1). Notably, a host nucleolar component, nucleolin (Ncl), colocalizes with UL44 at the periphery of replication compartments (3). Based on studies using small interfering RNA (siRNA), nucleolin is required to maintain the concentration of UL44 at this location, which facilitates viral DNA synthesis (3, 7).
The HCMV UL84 protein is involved in viral DNA synthesis (8–11), although its role in the process is unclear. It has been reported that UL84 is a member of the DExD/H family of helicase proteins that includes DNA replication origin binding proteins (12), which would be consistent with the results of studies utilizing chromatin immunoprecipitation and proteomics that place UL84 at the viral origin of replication. It has therefore been proposed that UL84 serves as an origin binding protein (12–15). Furthermore, UL44 and UL84 associate with each other in infected cell lysate (16, 17), and in experiments in which a plasmid encoding an enhanced green fluorescent protein (EGFP)-UL84 fusion has been cotransfected with plasmids encoding other viral replication proteins, the EGFP signal colocalizes with UL44 (18). Analysis of UL84 localization with a monoclonal antibody (MAb) found the protein to be diffusely distributed throughout the nucleus at day 1 postinfection (p.i.), before DNA synthesis has commenced, and at day 5 p.i., at the end of the replication cycle (14). Neither intermediate time points nor localization relative to other replication proteins was assessed. Thus, whether UL84 colocalizes with other proteins within replication compartments in virus-infected cells is unknown.
The presence of both nuclear localization and nuclear export signals in UL84 suggests that UL84 shuttles between the nucleus and the cytoplasm in the infected cell (19, 20). Electroporation of a bacterial artificial chromosome (BAC) containing the HCMV genome in which a UL84 nuclear export signal had been deleted failed to produce detectable virus replication, suggesting that nucleocytoplasmic shuttling of UL84 is necessary for virus replication (21). UL84 can interact with the viral transcriptional transactivator IE2, potentially modulating IE2 function (22, 23) and stabilizing UL84 levels (24, 25) in infected cells. UL84 also associates with a number of other viral and cellular proteins (7, 16), although it is unclear what functions the majority of these protein-protein interactions have in the infected cell. Finally, UL84 may play a role in capsid trafficking within the nucleus, as a deletion that removes sequences encoding a portion of the UL84 amino terminus causes a modest nuclear egress defect in which capsids accumulate around nucleoli in the infected cell (26).
To gain a better understanding of UL84 localization, particularly in terms of replication compartment architecture and UL84 function, we generated a recombinant HCMV expressing a tagged version of UL84 and analyzed the association of UL84 with replicating viral DNA and proteins involved in viral genome replication in infected cells.
MATERIALS AND METHODS
Cells and viruses.
Human foreskin fibroblasts (HFFs) (American Type Culture Collection CRL-1684) were used in all experiments. The HCMV laboratory strain AD169 was used where indicated. The virus derived from AD169-BAC, AD169 recombinant virus (AD169rv) (27), and the virus AD169-FLAGUL44, which expresses FLAG-tagged UL44, were described previously (7). To generate the bacterial artificial chromosome AD169-BACUL84FLAG and the virus AD169-UL84FLAG, briefly, a single FLAG epitope (DYKDDDDK) was inserted before the termination codon of the UL84 coding sequence in the bacterial artificial chromosome AD169-BAC (27) using the two-step Red recombination method described by Tischer and coworkers (28) in Escherichia coli strain DY380 (29) and the PCR primers Fw (5′-GAGAAAACGAGTCCTGTCACTCTAGCCATGGTCTGCGGCGATCTCGACTACAAGGATGACGACGATAAGTAAACAGAGGACTAGGGATAACAGGGTAATCGATTT-3′) and Rv (5′-GCGGACGCCTAGTGTCCGTTTTCCATCACCAGGGTCCTCTGTTTACTTATCGTCGTCATCCTTGTAGTCGAGATCGCCGCCAGTGTTACAACCAATTAACC-3′). Insertion of the sequence encoding FLAG was confirmed by sequencing. The resulting BAC, AD169-BACUL84FLAG, was electroporated into HFFs as described previously (28) with plasmids pCGN71 (30), expressing the viral transcriptional transactivator pp71, and pBRep-Cre (27) to generate the virus AD169-UL84FLAG. To assess virus replication, titration by plaque assay of virus produced by infection of HFFs was carried out as previously described (7).
Western blotting.
Western blotting of proteins separated on 4 to 15% or 4 to 20% polyacrylamide gels was carried out as described previously (31), using monoclonal antibodies recognizing UL44 (antibody [Ab] CA006; Virusys), UL84 (Virusys), nucleolin (antibody 4E2; Abcam), FLAG (Sigma), and β-actin (Sigma) as primary antibodies (all used at 1:1,000 dilution). Goat anti-mouse–horseradish peroxidase (HRP)-conjugated antibody or goat anti-rabbit–HRP-conjugated antibody (both Southern Biotech; 1:1,000 dilution) and chemiluminescence solution (Pierce) were used to detect primary antibodies, and the images were captured using film.
Immunofluorescence and “click chemistry.”
HFFs (5 × 104) were plated on glass coverslips in 24-well plates. The cells were mock infected or infected with the indicated viruses (multiplicity of infection [MOI] = 3). Where indicated, 520 μM phosphoformic acid (PFA) (Sigma) was added to infected cell cultures following removal of inocula. To label replicating DNA, cells were incubated for the times indicated with 10 μM 5-ethynyl-2′-deoxyuridine (EdU) (Invitrogen). The cells were fixed, permeabilized, and stained as previously described (2). Murine MAbs recognizing UL44 (antibody CA006; Virusys), UL57 (Virusys), and FLAG (Sigma) were used at a dilution of 1:1,000. A rabbit antibody recognizing nucleolin (Abcam; ab50279) was used at a dilution of 1:100. A rabbit antibody recognizing FLAG and directly conjugated to Alexa 488 (Sigma) was used at a dilution of 1:500. Fluorescently labeled secondary goat anti-mouse and anti-rabbit antibodies labeled with Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 647 were obtained from Molecular Probes and used at a dilution of 1:1,000.
Samples were imaged at the Nikon Imaging Facility of Harvard Medical School on a Nikon Ti motorized inverted microscope with a Perfect Focus System and Yokagawa CSU-X1 spinning-disk confocal microscope with Spectral Applied Research Aurora Borealis modification. Images were acquired using MetaMorph image acquisition software. Analyses of control samples showed no detection of signal from one fluorescent reagent while assaying for signal from another (no “bleedthrough”).
IP of FLAG-tagged proteins.
Immunoprecipitation (IP) assays were carried out as previously described (7); however, in each reaction mixture, 400 U Benzonase (Sigma) was added to the mixture containing clarified cell lysate and beads.
Transfection of siRNA into HFFs.
Transfection of HFFs (5 × 104 per well on glass coverslips in 24-well plates) with siRNA (siControl nontargeting siRNA pool or On-Targetplus Smartpool Human Ncl [both Dharmacon]) was carried out essentially as described previiously (7).
RESULTS
Generation of a recombinant HCMV expressing FLAG-tagged UL84.
Our initial efforts to visualize UL84 by immunofluorescence in infected cells using commercially available antibodies were not successful, nor were our efforts to produce quantities of glutathione S-transferase-tagged UL84 in E. coli (17) sufficient for the generation of antibodies recognizing UL84 (data not shown). We therefore generated a recombinant HCMV expressing a FLAG-tagged version of UL84. Toward this end, we inserted a single FLAG epitope (DYKDDDDK) immediately before the termination codon of the UL84 coding sequence in the BAC AD169-BAC (27) using Red two-step recombination (28). This new BAC was termed AD169-BACUL84FLAG. AD169-BACUL84FLAG was electroporated into HFFs to generate the virus AD169-UL84FLAG. The replication kinetics of AD169-UL84FLAG and AD169rv (the virus derived from AD169-BAC [27]) were comparable following infection at an MOI of 1 (Fig. 1). AD169-UL84FLAG was not defective for expression of the tagged UL84 or for expression of UL44 or UL57 (data not shown) (see below). Thus, we deemed this virus suitable for studies of UL84 localization.
FIG 1.
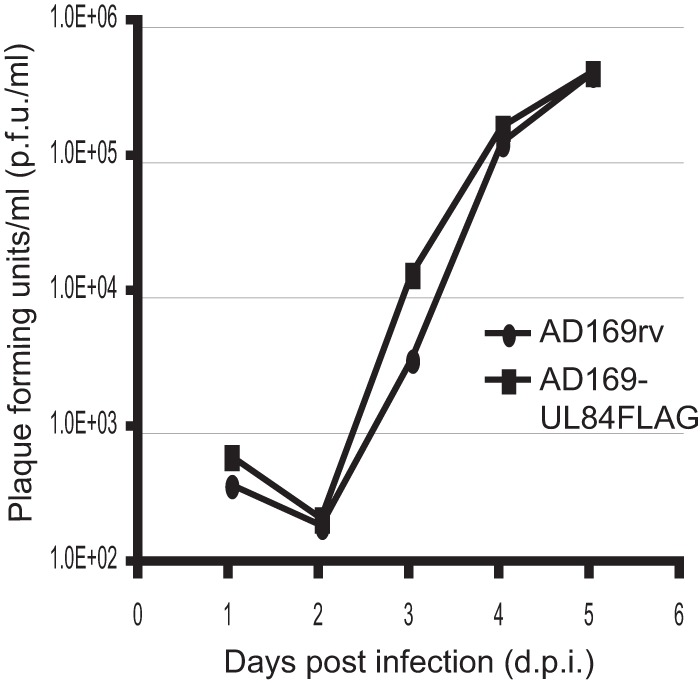
Replication kinetics of AD169-UL84FLAG virus. HFFs were infected at an MOI of 1 with either AD169rv or AD169-UL84FLAG, and the virus supernatant was harvested at the indicated time points. The virus titer in the supernatant was assayed and is represented as PFU/ml on HFFs.
Localization of UL84FLAG in infected cells.
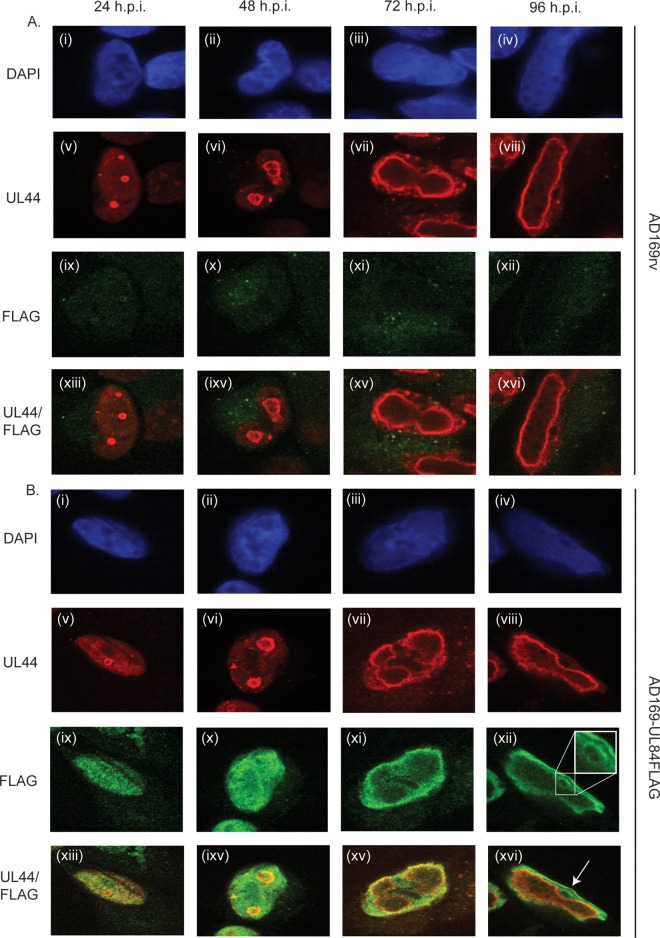
Using immunofluorescence confocal microscopy, we assayed the localization of UL84FLAG in relation to the nucleus and UL44 (Fig. 2). Cells were mock infected or infected with either AD169rv or AD169-UL84FLAG and fixed at several time points (as indicated in Fig. 2) postinfection. The cells were stained with DAPI (4′,6-diamidino-2-phenylindole) to visualize nuclei and with MAbs recognizing UL44 and FLAG. Only background levels of staining were observed in mock-infected cells using these antibodies (data not shown) (see below). Consistent with previous results (2, 4, 7), UL44 accumulated at the periphery of replication compartments throughout infection with both viruses (Fig. 2A and B, v to viii). Also consistent with previous observations (2), the replication compartments changed over time, from multiple small compartments at early times to a single large compartment at late time points (Fig. 2A and B, v to viii). FLAG staining was at background levels in AD169rv-infected cells (Fig. 2A, ix to xii). In AD169-UL84FLAG-infected cells, UL84FLAG was observed predominantly within the nucleus at all time points, with only background levels in the cytoplasm. However, intranuclear localization of UL84FLAG changed over time (Fig. 2B, ix to vii). At early times postinfection (24 to 48 h p.i.) (Fig. 2B, ix and x), UL84FLAG was distributed throughout the nucleus, consistent with observations made using a UL84 MAb (14), and some colocalized with UL44. Later in infection (72 to 96 h p.i.) (Fig. 2B, xi to xiii), UL84FLAG appeared to be largely excluded from the interior of replication compartments, with some colocalizing with UL44 at the periphery of these compartments at 72 h p.i. and less at 96 h p.i., when most UL84 was found at the margins of the nucleus and in ring-like structures (indicated in Fig. 2B, xii and xvi). Thus, the intranuclear localization of UL84 varied over time, but at all time points, there was at least some colocalization with UL44.
FIG 2.
Detection of UL84FLAG in infected cells. HFFs were infected at an MOI of 3 with either AD169rv (A) or AD169-UL84FLAG (B), fixed at the time points indicated at the top, and stained for IF analysis with DAPI reagent and MAbs recognizing either UL44 or FLAG. MAbs recognizing UL44 or FLAG were detected by using secondary antibody conjugated to a red or green fluorophore, respectively, and the images were acquired using spinning-disk confocal microscopy. (A and B) (i to iv) DNA detected by DAPI (shown in blue). (v to viii and ix to xii) Proteins detected by UL44 and FLAG MAbs. (xiii to xvi) Images v to viii and ix to xii combined. (B) (xii) The inset shows an enlarged version of the boxed portion of the image. (B) (xvi) The arrow indicates an example of a nucleolus in an infected cell. The antibody or stain detected in each row is shown on the left.
We also assayed UL84FLAG and UL44 colocalization at 48 h p.i. in infected cells treated with the viral DNA synthesis inhibitor PFA (Fig. 3). As previously observed (3), continuous PFA treatment of AD169-UL84FLAG-infected cells caused UL44 staining in the nucleus to become more diffuse, with a few areas of concentration (Fig. 3F), consistent with a failure to form replication compartments. PFA treatment caused UL84FLAG staining to become more dispersed (Fig. 3C and G). Some puncta of colocalized UL44-UL84FLAG could be observed in PFA-treated infected cells (Fig. 3H and D, respectively). Thus, inhibition of viral DNA synthesis can prevent accumulation of both UL44 and UL84FLAG in replication compartments but did not completely abrogate colocalization of the proteins.
FIG 3.
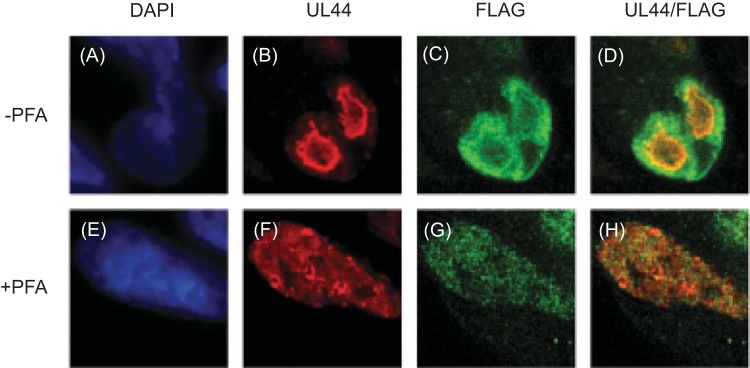
Localization of UL44 and UL84FLAG in PFA-treated cells. HFFs were infected with AD169-UL84FLAG at an MOI of 3 in the presence (+) or absence (−) of PFA (520 μM). Forty-eight hours p.i., the infected cells were stained for IF analysis with DAPI reagent and either a MAb recognizing FLAG conjugated to a green fluorophore or a MAb recognizing UL44 followed by a secondary antibody conjugated to a red fluorophore and analyzed using spinning-disk confocal microscopy. (A and E) DNA detected by DAPI (shown in blue). (B and F) Protein detected by UL44 MAb. (C and G) Protein detected by FLAG MAb. (D and H) Images of UL44 (B and F) and FLAG (C and G) proteins combined. The antibody or stain detected in each column is shown at the top.
Localization of UL84FLAG relative to UL57.
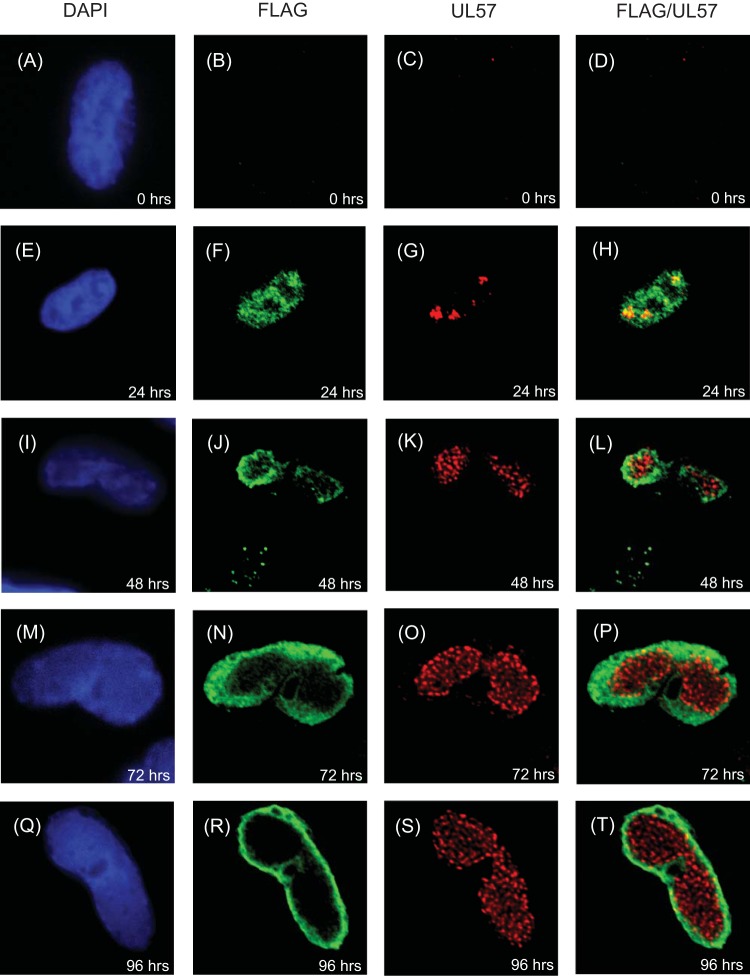
To investigate the localization of UL84 further, we assayed the localization of UL84FLAG relative to UL57, the viral ssDNA binding protein, in AD169-UL84FLAG-infected cells over time (Fig. 4). We have previously observed punctate staining of UL57 throughout replication compartments, with a subpopulation at the periphery colocalizing with UL44 (2). Consistent with these observations (2), we observed punctate UL57 staining in the infected cell that, like UL44 staining, grew from several clusters at early times to occupy nearly the entire nucleus at late times (Fig. 4C, G, K, O, and S). At 24 h p.i., small puncta of UL57 staining colocalized with diffuse FLAG staining (Fig. 4H). Only a very small number of UL57 puncta were observed colocalizing with FLAG staining at the periphery of replication compartments by 72 and 96 h postinfection (Fig. 4P and T, respectively). Thus, colocalization of UL84FLAG with UL57 was more prominent early in infection than later.
FIG 4.
Analysis of UL84FLAG and UL57 localization in infected cells. HFFs were infected at an MOI of 3 with HCMV-FLAG, fixed at the indicated time points, stained for IF analysis using DAPI reagent and MAbs recognizing FLAG or UL57, and analyzed by spinning-disk confocal microscopy. The images in the first column shows cells stained with DAPI (shown in blue). The second column shows cells stained with Ab recognizing FLAG directly conjugated to a green fluorophore. The images in the third column show cells stained with Ab recognizing UL57 and a secondary antibody conjugated to a red fluorophore. The fourth column shows images in the second and third columns merged. The antibody or stain detected in each column is shown at the top.
Localization of UL84FLAG relative to replicating HCMV DNA.
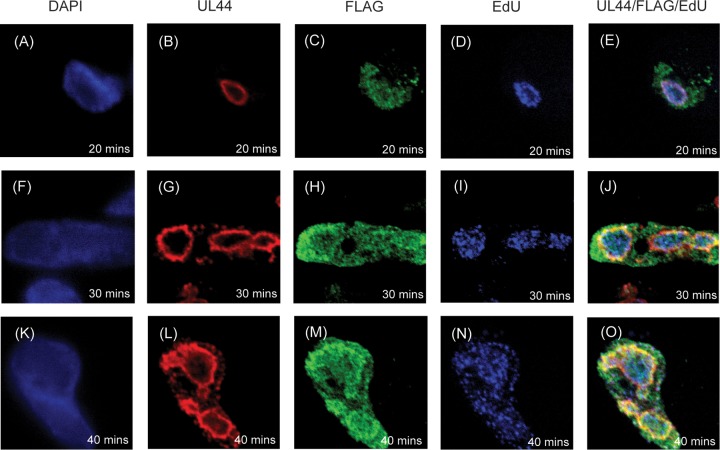
We next assayed the localization of UL84FLAG relative to sites of viral DNA synthesis by visualizing incorporation of EdU into newly synthesized DNA using click chemistry in cells infected with AD169-UL84FLAG (Fig. 5). At 48 h p.i., cells were incubated with EdU for 20, 30, or 40 min and then fixed for analysis. The cells were then chemically treated to detect EdU and stained with MAbs to detect UL84FLAG and UL44. The localization of UL84FLAG relative to UL44 was similar to that shown in Fig. 2 (Fig. 5C, H, and M, and B, G, and L, respectively). Consistent with our previous observations (2), at the earliest time point assayed after addition of EdU to infected cells (20 min) (Fig. 5A to E), puncta of EdU-labeled DNA colocalized with a layer of UL44 at the periphery of replication compartments. Over time (30 and 40 min after addition of EdU) (Fig. 5F to J and 4K to O, respectively), Edu-labeled viral DNA accumulated within the interior of replication compartments. We found points of colocalization of UL84FLAG and UL44 with EdU at all time points assayed (Fig. 5E, J, and O). Similar results were observed by assaying AD169-UL84FLAG-infected cells prepared for analysis at 24 h p.i. (data not shown). Thus, we observed colocalization of UL84FLAG with viral DNA labeled with EdU in viral replication compartments.
FIG 5.
Localization of EdU-labeled viral DNA in infected cells. HFFs were infected at an MOI of 3 with AD169 and fixed at 48 h p.i. after incubation with EdU for the indicated times. The cells were stained using DAPI reagent and MAbs recognizing either UL44 or FLAG, treated to detect EdU incorporated into DNA with a green fluorophore, and analyzed using spinning-disk confocal microscopy. The images in the first column show cells stained with DAPI (shown in blue). The second column shows cells stained with MAb recognizing UL44 and detected with a secondary antibody conjugated to a far-red fluorophore. The images in the third column show cells stained with MAb detecting FLAG conjugated to a green fluorophore. The images in the fourth column show cells in which EdU is detected using a red fluorophore (here colored blue). The fifth column shows images in the second, third, and fourth columns merged. The antibody or stain detected in each column is shown at the top. The treatment time with EdU (20, 30, or 40 min) is shown in each panel.
Localization of UL84FLAG and nucleolin within virus-infected cells.
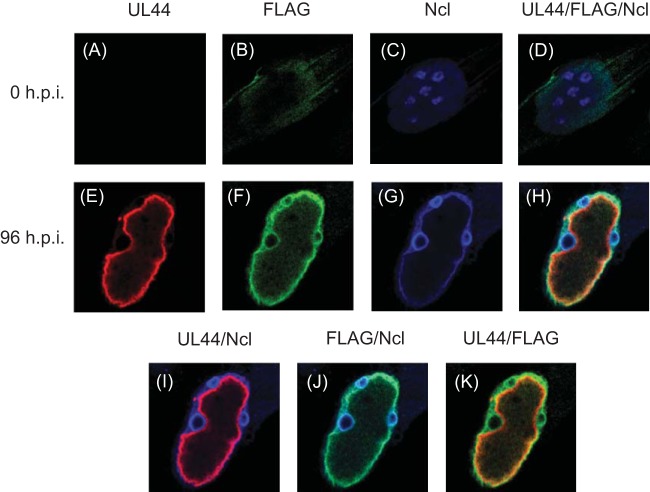
As noted above, UL84FLAG accumulates not only at the periphery of replication compartments, but also at the periphery of ring-like structures within the nucleus outside replication compartments (Fig. 2). Furthermore, during HCMV infection, nucleolin accumulates both at the periphery of replication compartments and at the periphery of nucleoli, giving nucleoli a ring-like appearance (3). We, therefore, hypothesized that UL84FLAG might associate with nucleolin during infection. Cells infected with AD169-UL84FLAG were stained with DAPI and analyzed by immunofluorescence at 0 and 96 h p.i. using antibodies recognizing UL44, FLAG, or nucleolin (Fig. 6). Only background staining was observed at 0 h p.i. (Fig. 6A to D). Notable colocalization of the UL44, UL84FLAG, and nucleolin was found at the periphery of replication compartments (Fig. 6H), and we found colocalization of nucleolin and UL84FLAG, but not UL44, at the periphery of the nucleoli of infected cells (Fig. 6H and J). Consistent with previous results (3), we also observed changes in nucleoli during infection, as more nucleolin accumulated at the periphery of nucleoli than in the interior in AD169-UL84FLAG-infected cells (contrast panels 6C and G). Colocalization of diffuse UL84FLAG staining with nucleolin in nucleoli was observed as early as 48 h p.i. (data not shown). Thus, in HCMV-infected cells, nucleolin colocalizes with UL84FLAG both at the periphery of replication compartments and the periphery of nucleoli.
FIG 6.
Localization of UL84FLAG and nucleolin in HCMV-infected cells. (A to H) HFFs were fixed at 0 h p.i. (A to D) or infected with AD169-UL84FLAG (MOI = 3) and fixed at 96 h p.i. (E to H). The cells were stained with primary antibodies recognizing UL44 or nucleolin or a MAb recognizing FLAG conjugated to a green fluorophore (FLAG). Secondary antibodies conjugated to red (UL44) or far-red (here colored blue) (Ncl) fluorophores are shown. The cells were analyzed using spinning-disk confocal microscopy. (D and H) Images from panels A to C and E to G, respectively, combined. (I to K) Images in panels E to G merged. The antibody or stain detected in each panel is shown at the top.
Immunoprecipitation of viral and cellular proteins with UL84FLAG.
To further investigate the association of UL44, UL84FLAG, and nucleolin in infected cells, we performed coimmunoprecipitation assays (Fig. 7). Protein was immunoprecipitated from the lysate of cells infected with AD169rv, AD169-FLAGUL44 (7) (which expresses a FLAG-tagged version of UL44 [FLAGUL44]), or AD169-UL84FLAG using a MAb recognizing FLAG in the presence of Benzonase to remove nucleic acids. Using Western blotting, we assayed the presence of viral and cellular proteins in the immunoprecipitate. FLAGUL44, UL84, and nucleolin were found in FLAG immunoprecipitates from AD169-FLAGUL44-infected cell lysate, while UL84FLAG, UL44, and nucleolin were found in FLAG immunoprecipitates from AD169-UL84FLAG-infected cell lysate (Fig. 7, lanes 4 and 6, respectively). β-Actin was not observed in immunoprecipitates from any cell lysate (Fig. 7, lanes 2, 4, and 6), and none of the proteins were found in immunoprecipitates from lysate of cells infected with AD169rv (Fig. 7, lane 2), indicating that the proteins observed in the immunoprecipitations from AD169-FLAGUL44- or AD169-UL84FLAG-infected cell lysates are unlikely to be the result of nonspecific interactions. In sum, these results indicate that UL44, UL84, and nucleolin associate in the infected cell, consistent with their colocalization in immunofluorescence studies.
FIG 7.
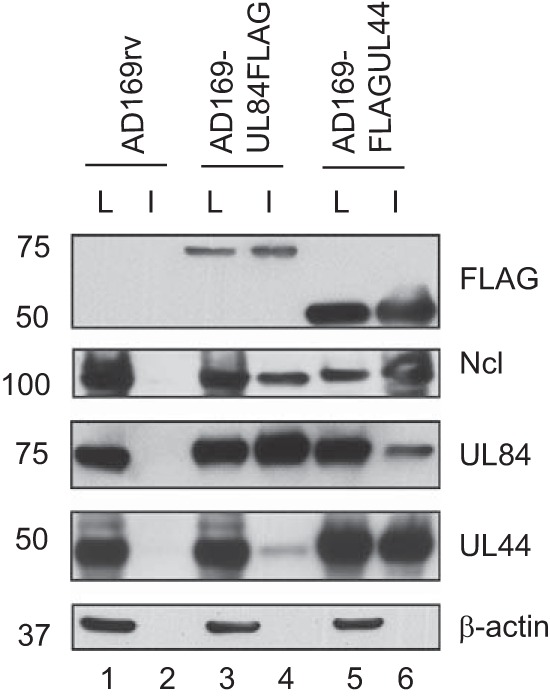
Coimmunoprecipitation of viral and cellular proteins. HFFs were mock infected or infected with AD169rv, AD169-UL84FLAG, or AD169-FLAGUL44 (MOI = 3), and cell lysates were prepared 72 h postinfection. IP of the lysates was carried out in the presence of Benzonase using a MAb recognizing FLAG immobilized on agarose beads. Proteins were analyzed by Western blotting using the antibodies recognizing the proteins indicated on the right. The positions of molecular mass markers (kDa) are indicated on the left. L, lysate; I, immunoprecipitated protein.
Localization of UL44 and UL84FLAG in cells treated with nucleolin siRNA.
As nucleolin is required for the concentrated pattern of localization of UL44 at the periphery of replication compartments (3), we hypothesized that nucleolin would also influence the localization of UL84FLAG. We, therefore, treated cells with siRNA targeting nucleolin mRNA (Ncl siRNA) or nontargeting control siRNA (Ctrl siRNA). Cells infected with AD169-UL84FLAG were stained at 96 h p.i. with Abs recognizing either UL44, FLAG, or nucleolin (Fig. 8). Consistent with previous results (3) (Fig. 6), in infected cells treated with Ctrl siRNA, we found concentration of UL44 together with UL84FLAG and Ncl at the periphery of replication compartments, and also colocalization of UL84FLAG and Ncl in nucleoli (Fig. 8F). In infected cells treated with Ncl siRNA, again consistent with previous results (3), we observed very small amounts of Ncl staining (Fig. 8J) and only diffuse and disrupted staining of UL44 at the periphery of replication compartments (Fig. 8H). Remarkably, compared to cells treated with Ctrl siRNA, we found more UL84FLAG in the cytoplasm than in the nuclei of infected cells treated with Ncl siRNA (compare Fig. 8E and K). We also assayed infected cells treated with Ncl siRNA at earlier time points after infection and found accumulation of UL84FLAG in the cytoplasm of infected cells from 48 h p.i. onward (data not shown). Using antibodies recognizing Ncl, UL44, and UL84, we also assayed protein levels in infected cells treated with either Ctrl or Ncl siRNA by Western blotting. Compared to cells treated with Ctrl siRNA, treatment of cells with Ncl siRNA resulted in a drastic decrease in Ncl protein levels but no obvious decrease in either UL44 or UL84 protein levels (data not shown). Thus, treatment of cells with Ncl siRNA dramatically affects UL84FLAG localization.
FIG 8.
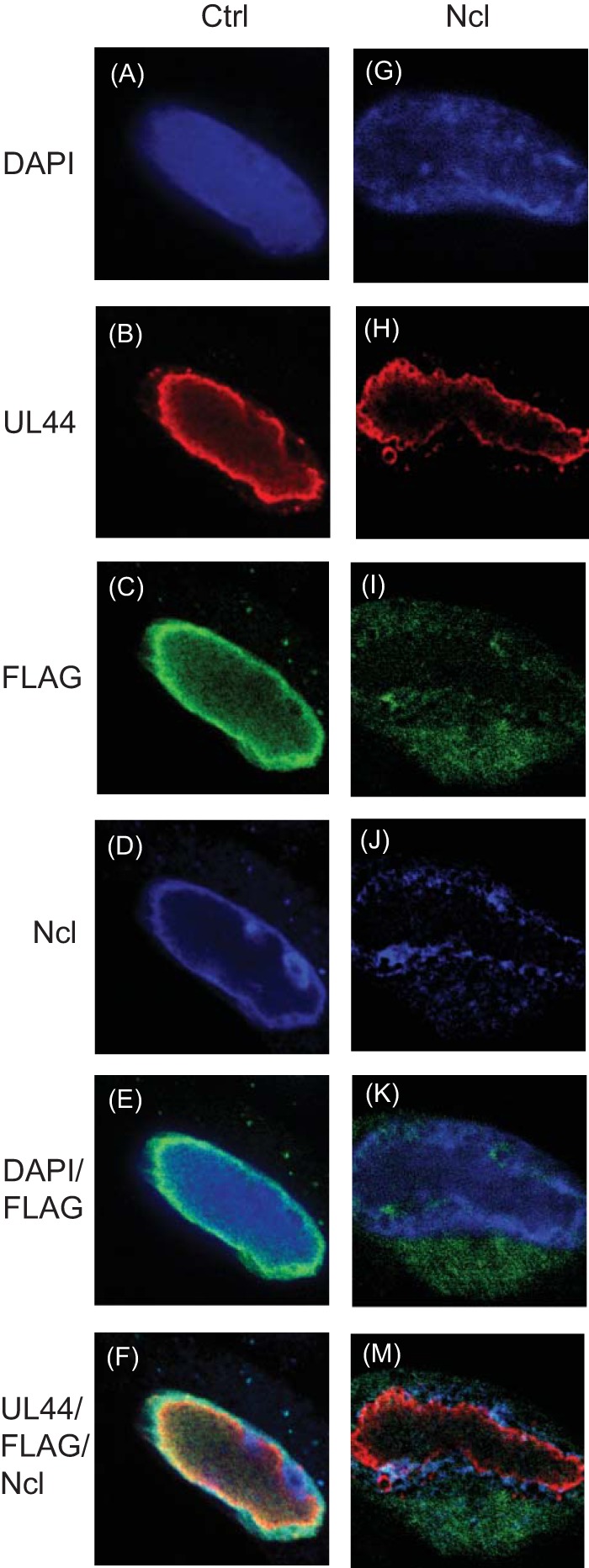
Localization of proteins in siRNA-treated cells. HFFs transfected with Ctrl (A to F) or Ncl (G to M) siRNA were infected at an MOI of 3 with virus AD169-UL84FLAG. Ninety-six hours p.i., the infected cells were stained for immunofluorescence analysis using DAPI reagent (A and G) and primary antibodies recognizing UL44 or Ncl or a MAb recognizing FLAG conjugated to a green fluorophore (C and I). Primary antibodies to UL44 (B and H) and Ncl (D and J) were detected with secondary antibodies conjugated to red and far-red (here colored blue) fluorophores, respectively. The images were acquired using spinning-disk confocal microscopy. (F and M) Images from panels B to D and H to K, respectively, merged. The siRNAs used are shown at the top. The antibody or stain detected in each row of panels is shown on the left.
DISCUSSION
The localization of UL84 early and late in HCMV infection has been previously reported (14), but it has been unclear how UL84 localizes with other viral and cellular proteins throughout the course of HCMV replication. Here, we show that the distribution of a FLAG-tagged version of the viral replication factor UL84 changes during infection from a diffuse pattern within the nucleus at early time points to a more defined pattern at later times. In particular, subpopulations of UL84 colocalize with UL44 and with UL57 within the infected cell starting by 24 h p.i. As replication compartments form, portions of UL84 colocalize with UL44, UL57, and nucleolin at the periphery of these compartments, where UL44 and nucleolin concentrate, but less so in the interior of the compartments, where most UL57 is found. A portion of UL84 and nucleolin also colocalizes at the periphery of nucleoli. Consistent with these findings, immunoprecipitation of either UL84 or UL44 also precipitates the other and nucleolin. Finally, and most unexpectedly, we found that depletion of nucleolin not only eliminates the localization of UL84 at these subnuclear compartments, but also eliminates it from in the nucleus. To our knowledge, the requirement for nucleolin in UL84 localization, or the localization of UL84 with nucleolin in nucleoli, has not been previously reported.
The colocalization of UL84 with a portion of the viral DNA polymerase subunit UL44, puncta of the viral ssDNA binding protein UL57, and newly synthesized viral DNA at the periphery of replication compartments is consistent with a role for UL84 in viral genome replication. It has been suggested that UL84 acts during the initiation of viral DNA replication as an origin binding protein (12). Consistent with this suggestion, we note the colocalization of UL84 and UL44 throughout infection, the more prominent colocalization of UL84 with the single-stranded DNA binding protein UL57 at early versus late time points of infection, and the eventual exclusion of UL84 from the interior of replication compartments, where previously synthesized DNA accumulates. Additionally, there appears to be more extensive colocalization of UL84 with the layer of UL44 at the periphery of replication compartments at early versus later times of infection. These results can all be viewed as consistent with a role for UL84 in initiation of DNA replication. On the other hand, the colocalization of UL84 with nucleolin at the periphery of replication compartments might also be viewed as consistent with a role in maintaining replication compartment architecture, similar to nucleolin. Thus, UL84 may have an indirect role in viral DNA synthesis. It is unclear why so little UL84 staining is localized to replication compartments. It is possible that the UL84 staining localized outside replication compartments is due to roles in modulating the function of the HCMV transcriptional transactivator IE2 (22, 23) and/or virus capsid movement within the nucleus (26).
To our knowledge, this study provides the first evidence that UL84 can localize to the nucleolus. How this occurs is unclear, but a direct interaction with nucleolin would provide a simple explanation. Alternatively, it has been reported that several viral proteins, including pp65, can localize to the nucleolus during infection (32–35). UL84 and pp65 have been reported to interact within the infected cell (16), and thus, pp65 may recruit UL84 to the nucleolus or vice versa, or other viral or cellular factors may be involved.
Although it has been suggested that UL84 shuttles between the nucleus and the cytoplasm (21), to our knowledge, appreciable accumulation of UL84 in the cytoplasm of infected cells has not been previously observed. Here, we observe that UL84FLAG can be readily visualized in the cytoplasm of infected cells treated with Ncl siRNA. This suggests that nucleolin is involved in efficient UL84FLAG nuclear localization. Association of nucleolin and UL84 may retain UL84 in the nucleus. Alternatively, nucleolin may be required, directly or indirectly, for efficient entry of UL84 into the nucleus.
The association of UL44, UL84, and nucleolin seen here and elsewhere (3, 7) in immunofluorescence and immunoprecipitation assays can be explained in models in which one of the proteins bridges the association of the other two or each protein interacts with the other two directly. Given the structural similarities between UL44 and the cellular protein PCNA (36–39), which can bind multiple proteins simultaneously (40, 41), it is tempting to speculate that UL44 acts to organize protein binding in either of the aforementioned models. It has yet to be determined what protein binding surfaces on UL44, UL84, and nucleolin are required for their association. Furthermore, although each of the proteins is capable of binding nucleic acid, it is unlikely that either RNA or DNA is required for protein-protein binding, as association of all three proteins can be observed during immunoprecipitation in the presence of nuclease here and elsewhere (7).
In summary, this report further highlights the complexity of the replication compartment organization and the hitherto unappreciated relationship between the critical viral replication factor UL84 and replication compartments, nucleoli, and nucleolin. Indeed, the importance of nucleolin for viral DNA synthesis (7) may be at least as much due to its obvious importance in retaining UL84 in the nucleus as to its more subtle effect of maintaining UL44 concentration at the periphery of the replication compartment. To maintain the replication compartment architecture, nucleolin may play a direct structural role. As discussed above, the data described here suggest there may be direct protein-protein interactions between nucleolin and viral proteins, which maintain the integrity of viral replication compartments. Further study of UL44, UL84, and nucleolin will uncover further details of not only their roles in genome replication, but also poorly understood processes such as localization of proteins required for viral replication and the modification of nucleoli in infected cells.
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance and facilities provided by the Nikon Imaging Center at Harvard Medical School for acquisition and analysis of immunofluorescence data. Thanks are also due to all members of the Coen laboratory, in particular Mayuri Sharma, for their insights and support.
This work was supported by NIH grants RO1 AI0I19838 and RO1 AI0I26077 to D.M.C. and start-up funds provided by St George's, University of London, to B.L.S.
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1. Penfold ME, Mocarski ES. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46–61. 10.1006/viro.1997.8848 [DOI] [PubMed] [Google Scholar]
- 2. Strang BL, Boulant S, Chang L, Knipe DM, Kirchhausen T, Coen DM. 2012. Human cytomegalovirus UL44 concentrates at the periphery of replication compartments, the site of viral DNA synthesis. J. Virol. 86:2089–2095. 10.1128/JVI.06720-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Strang BL, Boulant S, Kirchhausen T, Coen DM. 2012. Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments. mBio 3:e00301-11. 10.1128/mBio.00301-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, Golan DE, Marto JA, Coen DM. 2009. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5:e1000275. 10.1371/journal.ppat.1000275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liptak LM, Uprichard SL, Knipe DM. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quinlan MP, Chen LB, Knipe DM. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857–868. 10.1016/0092-8674(84)90035-7 [DOI] [PubMed] [Google Scholar]
- 7. Strang BL, Boulant S, Coen DM. 2010. Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication. J. Virol. 84:1771–1784. 10.1128/JVI.01510-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pari GS, Anders DG. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979–6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pari GS, Kacica MA, Anders DG. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Y, Cei SA, Huete AR, Pari GS. 2004. Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth. J. Virol. 78:10360–10369. 10.1128/JVI.78.19.10360-10369.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarisky RT, Hayward GS. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colletti KS, Xu Y, Yamboliev I, Pari GS. 2005. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J. Biol. Chem. 280:11955–11960. 10.1074/jbc.C400603200 [DOI] [PubMed] [Google Scholar]
- 13. Colletti KS, Smallenburg KE, Xu Y, Pari GS. 2007. Human cytomegalovirus UL84 interacts with an RNA stem-loop sequence found within the RNA/DNA hybrid region of oriLyt. J. Virol. 81:7077–7085. 10.1128/JVI.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kagele D, Rossetto CC, Tarrant MT, Pari GS. 2012. Analysis of the interactions of viral and cellular factors with human cytomegalovirus lytic origin of replication, oriLyt. Virology 424:106–114. 10.1016/j.virol.2011.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kagele D, Gao Y, Smallenburg K, Pari GS. 2009. Interaction of HCMV UL84 with C/EBPalpha transcription factor binding sites within oriLyt is essential for lytic DNA replication. Virology 392:16–23. 10.1016/j.virol.2009.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao Y, Colletti K, Pari GS. 2008. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J. Virol. 82:96–104. 10.1128/JVI.01559-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strang BL, Sinigalia E, Silva LA, Coen DM, Loregian A. 2009. Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84. J. Virol. 83:7581–7589. 10.1128/JVI.00663-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu Y, Colletti KS, Pari GS. 2002. Human cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments. J. Virol. 76:8931–8938. 10.1128/JVI.76.17.8931-8938.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lischka P, Rauh C, Mueller R, Stamminger T. 2006. Human cytomegalovirus UL84 protein contains two nuclear export signals and shuttles between the nucleus and the cytoplasm. J. Virol. 80:10274–10280. 10.1128/JVI.00995-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lischka P, Sorg G, Kann M, Winkler M, Stamminger T. 2003. A nonconventional nuclear localization signal within the UL84 protein of human cytomegalovirus mediates nuclear import via the importin alpha/beta pathway. J. Virol. 77:3734–3748. 10.1128/JVI.77.6.3734-3748.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao Y, Kagele D, Smallenberg K, Pari GS. 2010. Nucleocytoplasmic shuttling of human cytomegalovirus UL84 is essential for virus growth. J. Virol. 84:8484–8494. 10.1128/JVI.00738-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spector DJ, Tevethia MJ. 1994. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J. Virol. 68:7549–7553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gebert S, Schmolke S, Sorg G, Floss S, Plachter B, Stamminger T. 1997. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol. 71:7048–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanders RL, Del Rosario CJ, White EA, Spector DH. 2008. Internal deletions of IE2 86 and loss of the late IE2 60 and IE2 40 proteins encoded by human cytomegalovirus affect the levels of UL84 protein but not the amount of UL84 mRNA or the loading and distribution of the mRNA on polysomes. J. Virol. 82:11383–11397. 10.1128/JVI.01293-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanders RL, Spector DH. 2010. Human cytomegalovirus IE2 86 and IE2 40 proteins differentially regulate UL84 protein expression posttranscriptionally in the absence of other viral gene products. J. Virol. 84:5158–5170. 10.1128/JVI.00090-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strang BL, Bender BJ, Sharma M, Pesola JM, Sanders RL, Spector DH, Coen DM. 2012. A mutation deleting sequences encoding the amino terminus of human cytomegalovirus UL84 impairs interaction with UL44 and capsid localization. J. Virol. 86:11066–11077. 10.1128/JVI.01379-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720–7729. 10.1128/JVI.74.17.7720-7729.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- 29. Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56–65. 10.1006/geno.2000.6451 [DOI] [PubMed] [Google Scholar]
- 30. Baldick CJ, Jr, Marchini A, Patterson CE, Shenk T. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strang BL, Stow ND. 2005. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 79:12487–12494. 10.1128/JVI.79.19.12487-12494.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arcangeletti MC, De Conto F, Ferraglia F, Pinardi F, Gatti R, Orlandini G, Calderaro A, Motta F, Medici MC, Martinelli M, Valcavi P, Razin SV, Chezzi C, Dettori G. 2003. Human cytomegalovirus proteins PP65 and IEP72 are targeted to distinct compartments in nuclei and nuclear matrices of infected human embryo fibroblasts. J. Cell. Biochem. 90:1056–1067. 10.1002/jcb.10655 [DOI] [PubMed] [Google Scholar]
- 33. Arcangeletti MC, Rodighiero I, De Conto F, Gatti R, Orlandini G, Ferraglia F, Motta F, Covan S, Razin SV, Dettori G, Chezzi C. 2009. Modulatory effect of rRNA synthesis and ppUL83 nucleolar compartmentalization on human cytomegalovirus gene expression in vitro. J. Cell. Biochem. 108:415–423. 10.1002/jcb.22268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arcangeletti MC, Rodighiero I, Mirandola P, De Conto F, Covan S, Germini D, Razin S, Dettori G, Chezzi C. 2011. Cell-cycle-dependent localization of human cytomegalovirus UL83 phosphoprotein in the nucleolus and modulation of viral gene expression in human embryo fibroblasts in vitro. J. Cell. Biochem. 112:307–317. 10.1002/jcb.22928 [DOI] [PubMed] [Google Scholar]
- 35. Salsman J, Zimmerman N, Chen T, Domagala M, Frappier L. 2008. Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog. 4:e1000100. 10.1371/journal.ppat.1000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Appleton BA, Loregian A, Filman DJ, Coen DM, Hogle JM. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell 15:233–244. 10.1016/j.molcel.2004.06.018 [DOI] [PubMed] [Google Scholar]
- 37. Loregian A, Appleton BA, Hogle JM, Coen DM. 2004. Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit. J. Virol. 78:9084–9092. 10.1128/JVI.78.17.9084-9092.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loregian A, Appleton BA, Hogle JM, Coen DM. 2004. Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44. J. Virol. 78:158–167. 10.1128/JVI.78.1.158-167.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Appleton BA, Brooks J, Loregian A, Filman DJ, Coen DM, Hogle JM. 2006. Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44. J. Biol. Chem. 281:5224–5232. 10.1074/jbc.M506900200 [DOI] [PubMed] [Google Scholar]
- 40. Maga G, Hubscher U. 2003. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116:3051–3060. 10.1242/jcs.00653 [DOI] [PubMed] [Google Scholar]
- 41. Moldovan GL, Pfander B, Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell 129:665–679. 10.1016/j.cell.2007.05.003 [DOI] [PubMed] [Google Scholar]