ABSTRACT
The phenomenon of prion strains with distinct biological characteristics has been hypothesized to be involved in the structural diversity of abnormal prion protein (PrPSc). However, the molecular basis of the transmission of strain properties remains poorly understood. Real-time quaking-induced conversion (RT-QUIC) is a cell-free system that uses Escherichia coli-derived recombinant PrP (rPrP) for the sensitive detection of PrPSc. To investigate whether the properties of various prion strains can be transmitted to amyloid fibrils consisting of rPrP (rPrP fibrils) using RT-QUIC, we examined the secondary structure, conformational stability, and infectivity of rPrP fibrils seeded with PrPSc derived from either the Chandler or the 22L strain. In the first round of the reaction, there were differences in the secondary structures, especially in bands attributed to β-sheets, as determined by infrared spectroscopy, and conformational stability between Chandler-seeded (1st-rPrP-fibCh) and 22L-seeded (1st-rPrP-fib22L) rPrP fibrils. Of note, specific identifying characteristics of the two rPrP fibril types seen in the β-sheets resembled those of the original PrPSc. Furthermore, the conformational stability of 1st-rPrP-fibCh was significantly higher than that of 1st-rPrP-fib22L, as with Chandler and 22L PrPSc. The survival periods of mice inoculated with 1st-rPrP-fibCh or 1st-rPrP-fib22L were significantly shorter than those of mice inoculated with mixtures from the mock 1st-round RT-QUIC procedure. In contrast, these biochemical characteristics were no longer evident in subsequent rounds, suggesting that nonspecific uninfected rPrP fibrils became predominant probably because of their high growth rate. Together, these findings show that at least some strain-specific conformational properties can be transmitted to rPrP fibrils and unknown cofactors or environmental conditions may be required for further conservation.
IMPORTANCE The phenomenon of prion strains with distinct biological characteristics is assumed to result from the conformational variations in the abnormal prion protein (PrPSc). However, important questions remain about the mechanistic relationship between the conformational differences and the strain diversity, including how strain-specific conformations are transmitted. In this study, we investigated whether the properties of diverse prion strains can be transmitted to amyloid fibrils consisting of E. coli-derived recombinant PrP (rPrP) generated by real-time quaking-induced conversion (RT-QUIC), a recently developed in vitro PrPSc formation method. We demonstrate that at least some of the strain-specific conformational properties can be transmitted to rPrP fibrils in the first round of RT-QUIC by examining the secondary structure, conformational stability, and infectivity of rPrP fibrils seeded with PrPSc derived from either the Chandler or the 22L prion strain. We believe that these findings will advance our understanding of the conformational basis underlying prion strain diversity.
INTRODUCTION
Prion diseases, or transmissible spongiform encephalopathies (TSEs), are infectious and fatal neurodegenerative disorders characterized by progressive spongiform changes and the accumulation of abnormal prion protein (PrPSc) in the central nervous system. Although the pathogenic mechanisms have not been fully elucidated, prion disease is thought to occur through autocatalytic conversion of normal prion protein (PrPC) to PrPSc (1, 2), known as the protein-only hypothesis. Some biophysical properties are known to differ between PrPC and PrPSc. PrPC is monomeric, detergent soluble, and protease sensitive, while PrPSc is polymeric, detergent insoluble, and partially protease resistant (3). These differences are most likely due to the different conformations of the two isoforms. PrPC is largely α-helical, whereas PrPSc is substantially enriched in β-sheets (4, 5), frequently resulting in amyloid fibril formation.
The existence of diverse prion strains in mammalian species manifesting as phenotypic differences is well-known. The strain-specific characteristics are usually maintained upon serial passage in the same species and may be explained by conformational variations in PrPSc. Indeed, strain-dependent differences in β-sheet-rich structures of PrPSc have been demonstrated by infrared spectroscopy (6–9). In addition, the conformational stability of PrPSc differs among prion strains, as demonstrated by a guanidine hydrochloride (GdnHCl) denaturation assay followed by protease digestion (10, 11). However, the mechanistic relationship between PrPSc conformational differences and the molecular basis of prion strains remains poorly understood.
Various in vitro PrPSc formation methods have been developed to elucidate the pathogenesis of prion diseases. One of these methods, protein misfolding cyclic amplification (PMCA), enabled the exponential amplification of PrPSc in vitro by sonication-induced fragmentation of large PrPSc polymers into smaller units (12). An increase in the infectivity of PrPSc amplified by PMCA was obtained by using brain homogenate (BH) from healthy mice (normal brain homogenate [NBH]) as a source of PrPC substrates (BH-PMCA) (13). Furthermore, PrPSc generated by BH-PMCA from five different mouse prion strains retained the strain-specific properties (14). In addition, prion infectivity could be propagated when purified brain-derived PrPC or baculovirus-derived PrPC was used as the substrate in the presence of certain cofactors, such as nucleic acids and BH from PrP-deficient mice (15–17). These results provide strong evidence to support the protein-only hypothesis, but the structural basis of prion pathogenesis, including the tertiary structure of PrPSc, has not been fully clarified.
On the other hand, the use of Escherichia coli-derived purified recombinant PrP (rPrP) offers an advantage over the use of conformational analyses, which generally require target protein of high purity and a large quantity of the target protein. Spontaneously polymerized amyloid fibrils of rPrP have been reported to induce the accumulation of PrPSc in the brains of PrP-overexpressing transgenic (Tg) mice (18–20) and some wild-type hamsters (21); however, the incubation periods spanned no less than several hundred days, and none of the wild-type hamsters developed any neurological signs at first passage, indicating that the level of infectivity generated in these studies is very low. More recently, wild-type mice developed clinical disease typical of TSE at about 130 days after injection of proteinase K (PK)-resistant rPrP fibrils generated by unseeded PMCA in the presence of 1-palmitoyl-2-oleoylphosphatidylglycerol (POPG), a synthetic lipid molecule, and total liver RNA (22). Although these results were reproduced by the same group (23), others have reported that rPrP fibrils generated by the same method were unable to induce either neuropathological changes or the accumulation of PrPSc (24). Thus, the role of POPG and RNA in the de novo generation of infectious rPrP fibrils remains controversial.
Meanwhile, two different seeded PMCA reaction studies using rPrP as a substrate (rPrP-PMCA) have demonstrated the propagation of moderate levels of prion infectivity. One study showed that hamster rPrP can be converted to rPrP fibrils capable of inducing TSE in the presence of SDS, a synthetic anionic detergent, but there were great variations in the attack rate and the incubation period, which ranged from 119 to 401 days (25). Another study revealed that phosphatidylethanolamine (PE), a phospholipid found in biological membranes, enhances the conversion of mouse rPrP into rPrP fibrils capable of inducing TSE after about 400 days of incubation with a 100% attack rate (26, 27). Of note, three different strains used as a seed were converted into a single strain with unique properties during the serial rPrP-PMCA experiments (27). These studies suggest that a certain amphipathic molecule, such as PE, is a cofactor required for the propagation of prion infectivity in vitro but not for the transmission of strain-specific properties.
The recently developed real-time (RT) quaking-induced conversion (QUIC) is a sensitive prion detection method (28, 29) in which intermittent shaking enhances the conversion of soluble rPrP into amyloid fibrils in the presence of PrPSc. The aim of the present research was to investigate whether properties of diverse prion strains can be transmitted to rPrP fibrils generated in the RT-QUIC system. We produced proteinase K-resistant rPrP fibrils seeded with minute quantities of mouse-adapted scrapie (Chandler or 22L strain) PrPSc and investigated the secondary structure, conformational stability, and infectivity.
MATERIALS AND METHODS
Mouse rPrP expression and purification.
Recombinant PrP (rPrP) equivalent to residues 23 to 231 of the mouse PrP sequence was expressed, refolded into a soluble form, and purified essentially as previously described (30). The concentration of rPrP was determined by measuring the absorbance at 280 nm. The purity of the final protein preparations was ≥99%, as estimated by SDS-PAGE, immunoblotting, and liquid chromatography-mass spectrometry (data not shown). After purification, aliquots of the proteins were stored at −80°C in 10 mM phosphate buffer, pH 6.8, or distilled water.
Preparation of brain homogenates.
Brain tissues were homogenized at 10% (wt/vol) in ice-cold phosphate-buffered saline (PBS) supplemented with a protease inhibitor mixture (Roche) using a multibead shocker (Yasui Kikai, Osaka, Japan). After centrifugation at 2,000 × g for 2 min, supernatants were collected and frozen at −80°C until use. Total protein concentrations were determined by the bicinchoninic acid protein assay (Pierce). The PrPSc concentrations in the brain homogenates were estimated by dot blot analysis using a reference standard of rPrP, as previously described (31).
RT-QUIC experiments.
We prepared reaction mixtures in a 96-well, optical, black-bottom plate (catalog number 265301; Nunc) to a final total volume of 100 μl. To avoid contamination, we prepared noninfectious materials inside a biological safety cabinet in a prion-free laboratory and used aerosol-resistant tips. The final concentrations of the reaction buffer components were 300 mM NaCl, 50 mM HEPES, pH 7.5, and 10 μM thioflavin T (ThT). The concentration of rPrP was 50 or 100 μg/ml, and only freshly thawed rPrP was used. Brain homogenate was diluted with reaction buffer prior to the reactions. The 96-well plate was covered with sealing tape (catalog number 236366; Nunc) and incubated at 40°C in a plate reader (Infinite M200 fluorescence plate reader; Tecan) with intermittent shaking consisting of 30 s of circular shaking at the highest speed and no shaking for 30 s and then with a 2-min pause to measure the fluorescence. The kinetics of amyloid formation was monitored by reading of the fluorescence intensity on the bottom of the plate every 10 min using monochromators with 440-nm excitation and 485-nm emission wavelengths.
RT-QUIC product analysis.
For detection of protease-resistant rPrP, 10 μl of the QUIC samples (1 μg of rPrP) was diluted with 40 μl of buffer (300 mM NaCl, 50 mM HEPES, pH 7.5) and digested with 10 μg/ml of PK at 37°C for 1 h. After adding 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (Pefabloc; Roche) at a final concentration of 4 mM and 20 μg of thyroglobulin, the proteins were precipitated with 4 volumes of methanol. The samples were heated in sample buffer (2% SDS, 5% β-mercaptoethanol, 5% sucrose, 0.005% bromophenol blue, 62.5 mM Tris-HCl, pH 6.8) at 95°C for 5 min and then loaded onto 10% bis-Tris NuPAGE gels (Invitrogen). Proteins were transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). The membranes were probed with polyclonal anti-PrP antibody R20 (the epitope located at mouse PrP amino acids 218 to 231) or ICSM35 (D-Gen, London, United Kingdom).
TEM.
Negative staining was done on carbon supporting film grids, which were glow discharged before staining. The 10-μl samples were adsorbed to the grids for 3 min, and then the residual solution was absorbed by filter paper. The grids were stained with 20 μl of fleshly filtered stain (2% uranyl acetate). Once they were dry, the samples were viewed in a transmission electron microscope (TEM; JEM-1200EX; JEOL, Japan).
FTIR.
Fourier transform infrared spectroscopy (FTIR) spectra were measured with a Bruker Tensor 27 FTIR instrument (Bruker Optics) equipped with a mercuric cadmium telluride (MCT) detector cooled with liquid nitrogen. Three hundred microliters of each of the QUIC samples (30 μg of rPrP) was pelleted by centrifugation for 1 h at 77,000 × g and resuspended in 20 μl buffer (300 mM NaCl, 50 mM HEPES, pH 7.5). The slurry was loaded into a BioATRcell II attenuated total reflectance-type reflectance unit. PrPSc was purified from the brains of mice infected with the mouse-adapted Chandler and 22L prions using a combination of detergent solubilization, centrifugation at ultrahigh speeds, and PK digestion (4, 32), and 15 μl of purified PrPSc was directly loaded. One hundred twenty-eight scans at a 4-cm−1 resolution were collected for each sample under constant purging with nitrogen, corrected for water vapor, and the background spectra of the buffer were subtracted.
Conformational stability assay.
Ten microliters of the QUIC products (equivalent to 1 μg of rPrP) and brain homogenates (80 μg of total proteins) was mixed with 22 μl of various concentrations of guanidine hydrochloride (GdnHCl) at final concentrations of 0 to 5 M and 0 to 3.5 M, respectively, and the mixed samples were incubated at 37°C for 1 h. After adjustment of the final GdnHCl concentration of the QUIC products to 1 M and the brain homogenates to 0.6 M, the samples were digested with PK (10 μg/ml) at 37°C for 1 h and analyzed by Western blotting following methanol precipitation. The bands were visualized using an Attophos AP fluorescent substrate system (Promega) and quantified using a Molecular Imager FX imager (Bio-Rad). The sigmoidal patterns of the denaturation curves were plotted using a Boltzmann curve fit. The concentration of GdnHCl required to denature 50% of PK-resistant fragments ([GdnHCl]1/2) was estimated from the denaturation curves.
Bioassay.
Four-week-old male ddY mice were intracerebrally inoculated with 40 μl of QUIC products (equivalent to 4 μg rPrP). As controls for rPrP fibrils, we performed a mock QUIC procedure using seed-only solutions that contained the same concentration of PrPSc as that from the 1st round of QUIC with the rPrP fibril (1st-rPrP-fibril; 1 pg/μl) or the 5th round of QUIC with the rPrP fibril (5th-rPrP-fibril; 1 × 10−8 pg/μl) and then added the same amount of rPrP and inoculated the mixtures into mice. Brain homogenates were serially diluted from 100 to 10−7 with PBS, and 20 μl of each dilution was intracerebrally inoculated. Mice were monitored weekly until the terminal stage of disease or sacrifice. Clinical onset was determined as the presence of 3 or more of the following signs: greasy and/or yellowish hair, hunchback, weight loss, yellow pubes, ataxic gait, and nonparallel hind limbs. The 50% lethal dose (LD50) was determined according to the Behrens-Karber formula. Animals were cared for in accordance with the guidelines for animal experimentation of Nagasaki University.
Histopathology and lesion profiles.
The brain tissue was fixed in 4% paraformaldehyde, and 5-μm paraffin sections were prepared on poly-l-lysine (PLL) coat slides using a microtome. After deparaffinization and rehydration, the tissue sections were stained with hematoxylin-eosin. The pattern of vacuolation was examined in 8 fields per slice from the hippocampus (HI), cerebral cortex, hypothalamus, pons, and cerebellum (CE). Spongiform degeneration was scored using the following scale: 0, no vacuoles; 1, a few vacuoles widely and unevenly distributed; 2, a few vacuoles evenly scattered; 3, moderate numbers of vacuoles evenly scattered; 4, many vacuoles with some confluences; and 5, dense vacuolation.
Statistical analysis.
The fibril length or width determined by electron microscopy analysis was subjected to one-way analysis of variance (ANOVA) followed by the Tukey-Kramer test. Data from the conformational stability test were analyzed by one-way ANOVA followed by Student's t test. Analysis of the data for the survival times was evaluated by the log-rank test. The vacuolation scores were analyzed by Mann-Whitney's U test.
RESULTS
Conversion of the soluble form of mouse rPrP into amyloid fibrils by RT-QUIC.
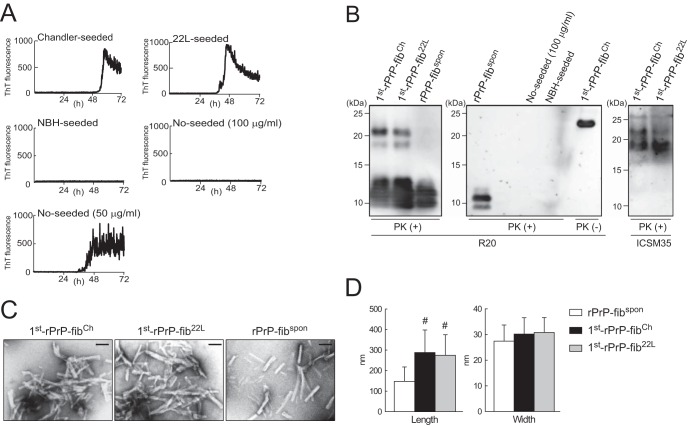
We first tested whether formation of mouse rPrP amyloid fibrils could be induced in the RT-QUIC by monitoring the levels of ThT fluorescence. We observed positive ThT fluorescence in the presence of diluted strain Chandler-seeded brain homogenate (BH) or strain 22L-seeded BH containing 100 pg of PrPSc (Fig. 1A), whereas negative-control reactions seeded with comparable dilutions of BH from healthy mice (normal brain homogenate [NBH]) or not seeded resulted in no increase in ThT fluorescence over 72 h (Fig. 1A). However, because an inverse correlation existed between the rate of fibril formation and the concentration of rPrP (28, 33), the spontaneous formation of rPrP fibrils (rPrP-fibspon) was induced by decreasing the concentration of rPrP from 100 to 50 μg/ml (Fig. 1A).
FIG 1.
Formation of rPrP fibrils in RT-QUIC reactions. (A) The formation of rPrP fibrils in the presence of diluted Chandler or 22L BH containing 100 pg of PrPSc or a comparable amount of NBH or in the absence of seed (No-seeded) was monitored by measurement of ThT fluorescence. The graphs depict representative results of the RT-QUIC reactions. No-seed reactions were performed at two different concentrations (100 or 50 μg/ml) of rPrP. (B) The QUIC reaction mixtures were digested with PK and immunoblotted using polyclonal anti-PrP antibody R20 (specific for the epitope located at mouse PrP amino acids 218 to 231) or ICSM35 (specific for the epitope located at mouse PrP amino acids 93 to 102). For comparison, 1st-rPrP-fibCh (50 ng of total rPrP) without PK digestion [PK (−)] is shown. Molecular mass markers are indicated in kilodaltons (kDa) on the left side of each panel. (C) Samples (1st-rPrP-fibCh, 1st-rPrP-fib22L, and rPrP-fibspon) were examined by TEM. Bars, 100 nm. (D) The bar graph shows the length and width of rPrP-fibspon, 1st-rPrP-fibCh, and 1st-rPrP-fib22L. The results are the mean ± SD for 30 rPrP fibrils each. Statistical significance was determined using one-way ANOVA, followed by the Tukey-Kramer test. *, P < 0.01.
We next examined the PK resistance of rPrP fibrils by immunoblotting using anti-PrP antibody R20 directed toward C-terminal residues 218 to 231. Although the ThT-negative reactions seeded with NBH or not seeded produced no PK-resistant bands (Fig. 1B, middle), the Chandler-seeded rPrP fibrils (rPrP-fibCh) and 22L-seeded rPrP fibrils (rPrP-fib22L) produced several (21-, 18-, 12-, 11-, and 10-kDa) PK-resistant fragments (Fig. 1B, left). In contrast, the PK digestion of rPrP-fibspon generated only 10- to 12-kDa fragments. It should be noted that anti-PrP monoclonal antibody ICSM35 (directed toward an epitope consisting of residues 93 to 102) specifically recognized the 21- and 18-kDa fragments derived from PrPSc-seeded rPrP fibrils in the first round (1st-rPrP-fibSc), indicating that they contained mouse PrP from about residues 93 to 231 (Fig. 1B, right).
To further characterize the structure of 1st-rPrP-fibSc and rPrP-fibspon, the samples were examined using a TEM with negative staining. The electron micrographs of samples of 1st-rPrP-fibCh and 1st-rPrP-fib22L revealed bundles of irregularly rod-shaped and branched fibrils, while most samples of rPrP-fibspon displayed smooth and nonbranched rod-shaped fibrils (Fig. 1C). Moreover, the lengths of 1st-rPrP-fibCh and 1st-rPrP-fib22L were significantly longer than those of rPrP-fibspon (Fig. 1D). Thus, the results of TEM analysis suggest that 1st-rPrP-fibSc are structurally distinct from spontaneous rPrP-fibspon.
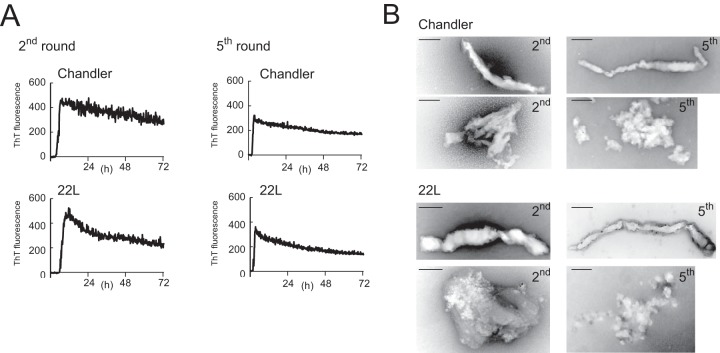
We next examined the morphology of PrPSc-seeded rPrP fibrils in the 2nd- and 5th-round reactions (2nd- and 5th-rPrP-fibSc, respectively) by TEM. In contrast to samples of 1st-rPrP-fibSc, samples of 2nd- and 5th-rPrP-fibSc displayed spindly and nonbranched fibrils or amorphous aggregates (Fig. 2). These data support the view that 1st-rPrP-fibSc are structurally distinct from 2nd- and 5th-rPrP fibSc.
FIG 2.
Formation of PrPSc-seeded rPrP fibrils in the 2nd round (2nd-rPrP-fibSc) and 5th round (5th-rPrP-fibSc) of RT-QUIC. (A) Between each round, the reaction mixtures from the previous reaction were diluted 100-fold into fresh rPrP. The reaction buffer contained 300 mM NaCl, 50 mM HEPES (pH 7.5), and 10 μM ThT. The rPrP concentration was 100 μg/ml. (B) TEM analysis of PrPSc-seeded rPrP fibrils generated in the second and fifth rounds of RT-QUIC. Bars, 100 nm.
Structural characterization of rPrP fibrils by FTIR.
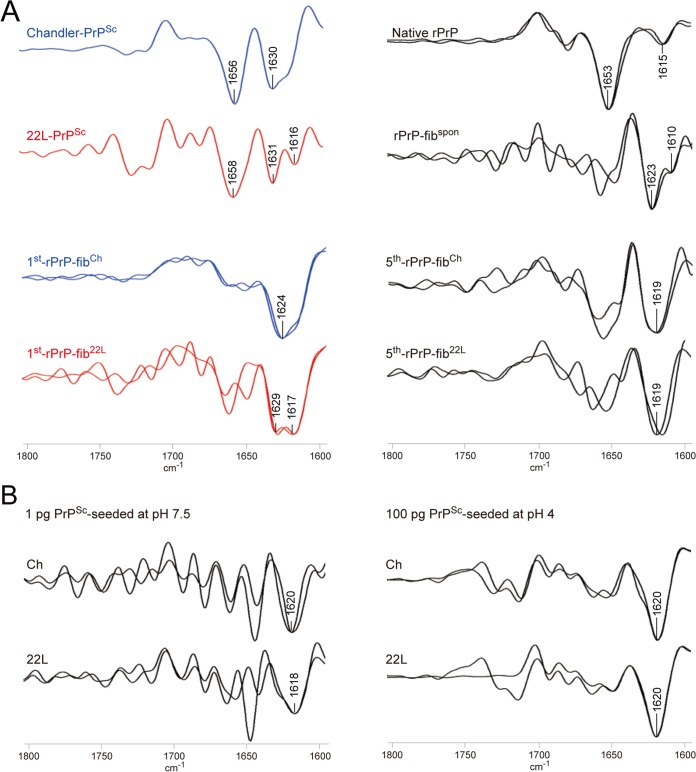
We next examined the secondary structure of rPrP fibrils and purified PrPSc from the brains of mice infected with Chandler or 22L scrapie by FTIR. A silver-stained SDS-polyacrylamide gel analysis revealed that Chandler and 22L PrPSc preparations were highly purified (Fig. 3A). Furthermore, TEM analysis demonstrated that the PrPSc preparations consisted exclusively of amyloid-like fibrils (Fig. 3B). FTIR analysis showed that Chandler PrPSc was characterized by a major band at 1,630 cm−1 in the β-sheet region of second-derivative spectra, while 22L PrPSc was characterized by two absorbance bands at 1,631 and 1,616 cm−1 (Fig. 4A), indicating that there were conformational differences in the β-sheet structures between Chandler and 22L PrPSc, as previously reported (7). Consistent with previous reports (6–9), bands of about 1,656 to 1,658 cm−1 were observed in both Chandler and 22L PrPSc. Although these bands were formerly attributed to an α-helix, recent studies using direct mass spectrometric analysis of hydrogen-deuterium exchange and FTIR analysis have suggested that purified PrPSc has little α-helix content and the bands probably result from turns (9, 34). Native rPrP had a maximum absorbance at 1,653 cm−1, which was congruent with that of prominent α-helical structures. In contrast, all rPrP fibrils displayed prominent bands at lower wave numbers (1,630 to 1,610 cm−1), indicating a predominantly β-sheet content (Fig. 4A). The β-sheet spectra revealed conformational differences among rPrP-fibspon, 1st-rPrP-fibCh, and 1st-rPrP-fib22L. rPrP-fibspon had a prominent band at 1,623 cm−1 and a modest band at 1,610 cm−1. While the 1st-rPrP-fibCh were characterized by a single major band at 1,624 cm−1, the 1st-rPrP-fib22L had two major maxima at 1,629 and 1,617 cm−1 (Fig. 4A). Although 1st-rPrP-fibSc lacked the bands at about 1,656 to 1,658 cm−1, the strain-specific shapes (one peak in Chandler versus two peaks in 22L) in the β-sheet spectrum of the purified PrPSc resembled those of 1st-rPrP-fibSc.
FIG 3.
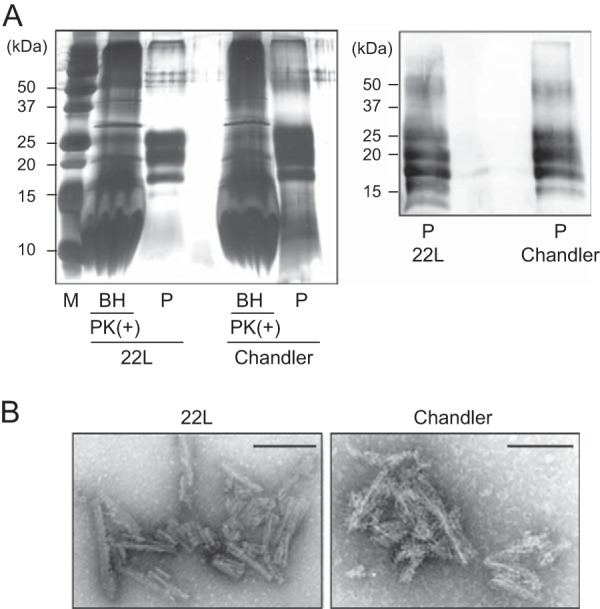
Silver staining and Western blot analysis of purified PrPSc. (A) The purified PrPSc samples (P) were examined by silver-stained SDS-polyacrylamide gel analysis (left). For comparison, the electrophoretic pattern of prion-infected BHs containing 100 μg total protein digested with PK (20 μg/ml, 37°C for 1 h) is shown (left). The purified PrPSc samples were immunoblotted with polyclonal anti-PrP antibody M20 (right). Molecular mass markers (lane M) are indicated in kilodaltons (kDa) on the left side of each panel. (B) Electron microscopy analysis of purified 22L PrPSc (left) and Chandler PrPSc (right). Bars, 100 nm.
FIG 4.
FTIR spectroscopic characterization of rPrP fibrils and purified PrPSc. (A) Second-derivative FTIR spectra are shown for purified PrPSc, 1st-rPrP-fibSc, 5th-rPrP-fibSc, spontaneously formed rPrP fibrils (rPrP-fibspon), and native rPrP. Overlaid spectra are from independent preparations. (B) FTIR spectra of rPrP fibrils generated at pH 7.5 in the presence of a small amount (1 pg) of PrPSc and rPrP fibrils generated at pH 4 in the presence of 100 pg of PrPSc.
To test whether the strain-specific infrared spectra observed in 1st-rPrP-fibCh and 1st-rPrP-fib22L are transmitted to sequential QUIC reactions, we performed 5 serial rounds of QUIC (Fig. 2A). There was little difference in the β-sheet spectra between 5th-rPrP-fibCh and 5th-rPrP-fib22L (Fig. 3), suggesting that strain-specific conformations were lost in the 5th-rPrP-fibSc. Furthermore, additional experiments revealed that the infrared spectra of rPrP fibrils produced in the presence of a small amount of PrPSc (1 pg) or under acidic conditions (pH 4) displayed few differences between strains (Fig. 4B).
Conformational stability analysis of rPrP fibrils and PrPSc.
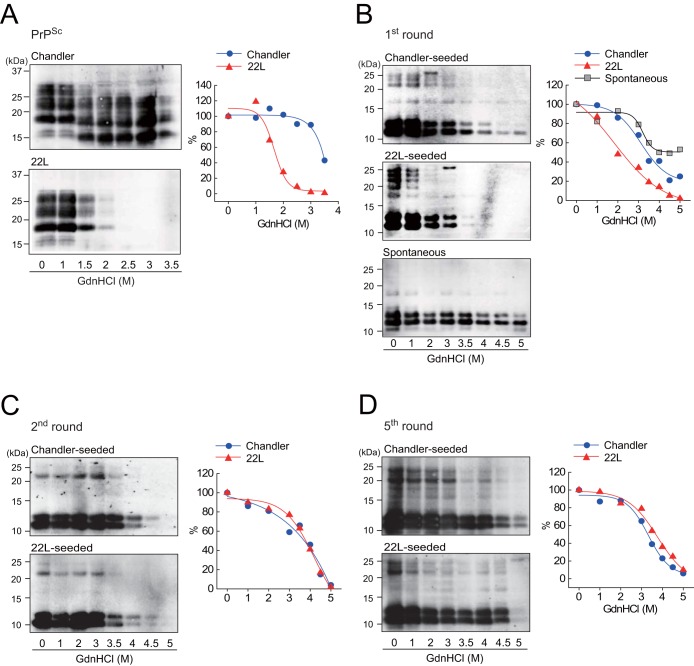
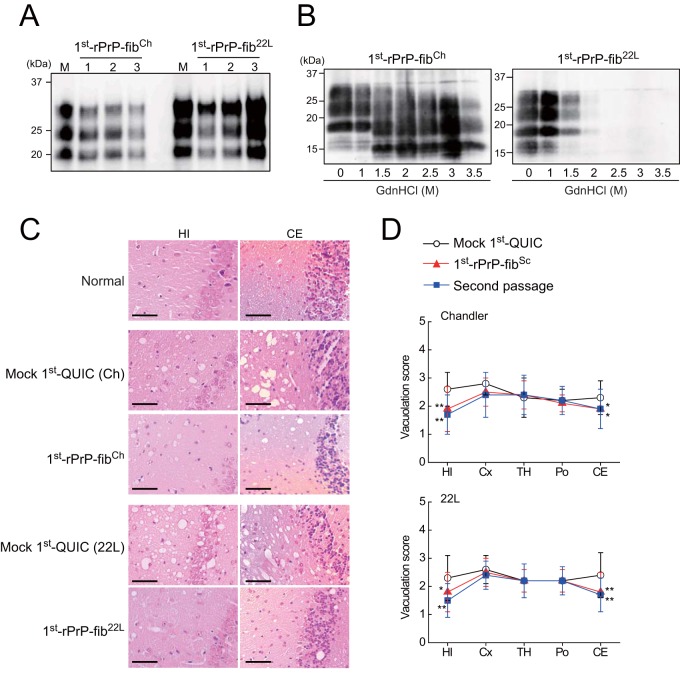
To examine the biochemical differences of rPrP fibrils and PrPSc in BH between strains, we performed a conformational stability assay, which combines GdnHCl denaturation with PK digestion. The [GdnHCl]1/2 values for Chandler and 22L PrPSc were 3.3 ± 0.4 and 1.7 ± 0.3 M, respectively (Fig. 5A and Table 1), indicating that the conformational stability of Chandler-PrPSc was significantly higher than that of 22L-PrPSc. Consistent with previous work (11), Chandler PrPSc bands treated with more than 1.5 M GdnHCl were approximately 5 kDa smaller than those treated with lower concentrations (Fig. 5A, top). The [GdnHCl]1/2 values of 1st-rPrP-fibCh and 1st-rPrP-fib22L were 3.3 ± 0.1 and 2.3 ± 0.6 M, respectively (Fig. 5B and Table 1), showing that the stability of 1st-rPrP-fibCh was significantly higher than that of 1st-rPrP-fib22L, as with Chandler and 22L PrPSc. Thus, the relationship between Chandler and 22L in terms of conformational stability was common to both the original PrPSc and 1st-rPrP-fibSc. In contrast, the [GdnHCl]1/2 of rPrP-fibspon was more than 5 M, which was markedly higher than that of 1st-rPrP-fibSc (Fig. 5B and Table 1). Additionally, we tested the conformational stability of 2nd- and 5th-rPrP-fibSc but found no significant differences between strains (Fig. 5C and D and Table 1).
FIG 5.
Conformational stability assay for PrPSc in BH and rPrP fibrils. (A) Chandler-infected (top left) or 22L-infected (bottom left) BHs were treated with 0 to 3.5 M GdnHCl and subjected to PK digestion. PrPSc was detected by R20 anti-PrP polyclonal antibody. The denaturation curves were plotted using Boltzmann curve fitting (right). (B to D) PK-digested 1st-rPrP-fibSc (generated as described in the legend to Fig. 1) and rPrP-fibspon (B), 2nd-rPrP-fibSc (C), or 5th-rPrP-fibSc (D) were analyzed by Western blotting following GdnHCl treatment (0 to 5 M). The PK-resistant fragments of the rPrP fibrils were detected by antibody R20.
TABLE 1.
Conformational stabilities of purified PrPSc and rPrP fibrilsa
| Strain | [GdnHCl]1/2 (mol/liter) |
|||
|---|---|---|---|---|
| Purified PrPSc | rPrP fibrils |
|||
| 1st | 2nd | 5th | ||
| Chandler | 3.3 ± 0.4** | 3.3 ± 0.1* | 3.7 ± 0.1 | 3.3 ± 0.3 |
| 22L | 1.7 ± 0.3 | 2.3 ± 0.6 | 3.8 ± 0.2 | 3.5 ± 1.0 |
| Spontaneous | >5 | |||
The [GdnHCl]1/2 values (mol/liter) are means ± standard deviations from three independent experiments. Statistical significance was determined using one-way ANOVA, followed by Student's t test. **, P < 0.01 (compared with 22L); *, P < 0.05 (compared with 22L).
Bioassay for rPrP fibrils generated in QUIC reactions.
To determine whether the infectivity was transmitted to the rPrP fibrils, we performed a bioassay using wild-type mice. To prepare the control materials, seed-only solutions containing the same concentration of PrPSc as that in 1st- or 5th-rPrP-fibSc were subjected to a mock RT-QUIC procedure and then mixed with the same amount of soluble rPrP (Table 2). The survival periods of mice inoculated with 40-μl aliquots containing rPrP fibrils were 185.5 ± 4.0 days postinoculation (dpi) for 1st-rPrP-fibCh and 213.0 ± 8.9 dpi for 1st-rPrP-fib22L (Table 2). In contrast, the attack rate of these control mice was only 50% (2/4) for Chandler and 20% (1/5) for 22L. Moreover, the survival period of the affected mice was much longer than that of the mice inoculated with 1st-rPrP-fibSc (Table 2). For comparison with the 50% lethal dose (LD50) of the original PrPSc, the LD50 of 1st-rPrP-fibSc was determined from the linear regression relationship between infectious titers and survival periods. The infectious titers (per 40 μl) of 1st-rPrP-fibCh and 1st-rPrP-fib22L were estimated to be 407.2 ± 226.6 and 1,067.0 ± 678.7 LD50s, respectively, whereas the titers of the Chandler and 22L prions were 20.2 and 28.9 LD50 units/40 pg of PrPSc, respectively. Because the QUIC reaction in the first round resulted in a 20- to 37-fold increase in the infectious titer, the seed contribution to infectivity is estimated to be about 3 to 5%. In contrast, none of the mice inoculated with 5th-rPrP-fibSc developed symptoms related to TSE (Table 2), suggesting that the 5th-rPrP-fibSc has no substantial infectivity.
TABLE 2.
Bioassay for rPrP fibrils generated in QUIC reactions in wild-type micea
| Inoculum | Concn of seed PrPSc (pg/μl) | Survival period (dpi)b | Mortality (no. of dead mice/total no. tested) |
|---|---|---|---|
| 1st-rPrP-fibCh | 1 | 185.5 ± 4.0*d | 4/4 |
| Mock 1st QUIC (Ch)c | 1 | 201, 220e | 2/4 |
| 1st-rPrP-fib22L | 1 | 213.0 ± 8.9**d | 6/6 |
| Mock 1st QUIC (22L)c | 1 | 333e | 1/5 |
| 5th-rPrP-fibCh | 1 × 10−8 | >660f | 0/4 |
| Mock 5th QUIC (Ch)c | 1 × 10−8 | >660f | 0/4 |
| 5th-rPrP-fib22L | 1 × 10−8 | >660f | 0/6 |
| Mock 5th QUIC (22L)c | 1 × 10−8 | >660f | 0/6 |
| rPrP-fibspon | 0 | >660f | 0/6 |
| Second passage of 1st-rPrP-fibCh | 152.0 ± 8.5d | 5/5 | |
| Second passage of mock 1st QUIC (Ch)g | 148.4 ± 5.9d | 5/5 | |
| Second passage of 1st-rPrP-fib22L | 153.5 ± 0.6d | 5/5 | |
| Second passage of mock 1st QUIC (22L)h | 149.6 ± 10.4d | 4/4 |
Mice were intracerebrally inoculated with 40 μl of each inoculum. For the second passage, 10% BH was used.
Statistical significance was determined using the log rank test. **, P < 0.01 (compared with the controls); *, P < 0.05 (compared with the controls). dpi, days postinoculation.
After subjecting seed-only mixtures containing the same concentration of PrPSc as 1st- or 5th-rPrP-fibSc to a mock QUIC procedure, the same amount of rPrP was added. The solutions were inoculated into mice as controls for rPrP fibrils.
Data represent means ± standard deviations.
Data represent the survival periods of the TSE-positive mice. All nonsymptomatic mice were negative for PrPSc at 660 dpi.
Data represent the day postinoculation when the experiment was ended.
A sample from a mouse obtained at 201 dpi was used.
A sample from a mouse obtained at 333 dpi was used.
We analyzed by Western blotting the levels of PrPSc in the brain tissues of mice in the terminal stage that had been inoculated with 1st-rPrP-fibSc or control materials (mock 1st QUIC) and found no apparent differences in the accumulation of PrPSc between them and mice inoculated with mock 1st QUIC (Fig. 6A). In addition, a conformational stability assay with GdnHCl revealed that the strain-specific digestion pattern was preserved in mice inoculated with 1st-rPrP-fibSc (Fig. 6B).
FIG 6.
Bioassay of rPrP fibrils in mice. (A) PrPSc in the brains of prion-affected mice inoculated with 1st-rPrP-fibCh or 1st-rPrP-fib22L was analyzed by Western blotting using anti-PrP antibody M20. Lanes M, mock 1st QUIC for strains Chandler and 22L. (B) The strain-specific properties of PrPSc in the brains of mice inoculated with 1st-rPrP-fibSc were examined by a conformational stability assay with GdnHCl (0 to 3.5 M). (C) Sections of the hippocampus (HI) and cerebellum (CE), stained with hematoxylin-eosin, from healthy mice, mice inoculated with 1st-rPrP-fibSc, and mock 1st QUIC-inoculated mice at terminal stages are shown. Bars, 50 μm. (D) Lesion profiles of spongiform changes in the hippocampus, cerebral cortex (Cx), thalamus (TH), pons (Po), and cerebellum were compared. Data are expressed as means ± SDs (n = 3). Statistical significance was determined using Mann-Whitney's U test. **, P < 0.01; *, P < 0.05.
Next, the degree of vacuolation in brain sections, including the hippocampus (HI), cerebral cortex, thalamus, pons, and cerebellum (CE), from affected mice inoculated with 1st-rPrP-fibSc or mock 1st QUIC and those inoculated with the second passage of 1st-rPrP-fibSc was examined histologically (Fig. 6C and D). Of note, we found that the spongiform change in mice inoculated with 1st-rPrP-fibSc was less severe in the HI and CE than that in the HI and CE of mice inoculated with mock 1st QUIC strains (Fig. 6C and D). Furthermore, these different lesion profiles observed in mice inoculated with 1st-rPrP-fibSc were preserved upon second passage (Fig. 6D), suggesting that the characters of 1st-rPrP-fibSc are partially distinct from those of the original strains. These findings support the notion that 1st-rPrP-fibSc provoke the emergence of a mutant strain beyond seed-derived infectivity.
DISCUSSION
Recent studies show that RT-QUIC assays are useful for the sensitive detection of PrPSc in most species and strains, including Creutzfeldt-Jakob disease (CJD) in humans (28, 35–37), scrapie in rodents (29, 38), and chronic wasting disease (CWD) in cervids (39). In the RT-QUIC reaction, soluble rPrP is converted to amyloid fibrils in a seed-dependent fashion in the presence of PrPSc. Previous studies using FTIR and hydrogen-deuterium exchange have shown that there are structural differences between PrPSc-seeded fibrils and spontaneous rPrP fibrils generated in rPrP amplified by PMCA (7, 40). We also found that the structural morphology (Fig. 1C), secondary structure (Fig. 3), and conformational stability (Fig. 4B and Table 1) distinguish 1st-rPrP-fibSc from rPrP-fibspon. However, it has been unknown whether rPrP retains the conformational properties of the original PrPSc in the RT-QUIC. Consistent with previous reports (7, 11), we observed strain differences in the β-sheet structure and conformational stability of PrPSc between the Chandler and 22L strains. Likewise, the differences in the shape of the β-sheet spectrum between strains were common to both PrPSc and 1st-rPrP-fibSc. Furthermore, the conformational stability of 1st-rPrP-fib22L was significantly lower than that of 1st-rPrP-fibCh, as was the case with Chandler and 22L PrPSc. Since the original PrPSc remaining in 1st-rPrP-fibSc was equivalent to only about 0.01 to 0.02% of the PK-resistant 1st-rPrP-fibSc (1 to 2 μg/10 μg of total PrP) in our estimation, the contribution to the FTIR spectra and the conformational stability of 1st-rPrP-fibSc are considered to be negligible. Taken together, these studies demonstrate that at least some strain-specific conformational features, especially in the β-sheet region, are conserved between PrPSc and 1st-rPrP-fibSc. However, these unique structural features disappeared in subsequent rounds.
One of the reasons for the loss of strain specificity may be due to differences between E. coli-derived rPrP and brain-derived PrPC. Studies using circular dichroism and 1H nuclear magnetic resonance spectroscopy showed that the tertiary structure and the thermal stability of bovine rPrP from positions 23 to 230 are essentially identical to those of healthy calf brain-derived PrPC (41). However, it should be noted that E. coli-derived rPrP lacks posttranslational modifications of PrPC, such as glycosylation and a glycosylphosphatidylinositol (GPI) anchor. PrP has two N-linked glycosylation sites at amino acids 180 and 196, resulting in di-, mono-, and unglycosylated forms. Mature PrPC is rich in the diglycosylated form, whereas the glycoform ratio of PrPSc is known to vary among strains (42–44). Studies using PrP glycan-lacking Tg mice revealed that the strain-specific characteristics of strain 79A were affected by the glycosylation status of PrPC, but those of strains ME7 and 301C were not (45). Meanwhile, enzymatic deglycosylation of PrPC failed to affect strain-specific pathological changes in serial PMCA experiments seeded with two murine strains, RML and 301C (46). However, the same two strains were converted into a new single strain during serial rPrP-PMCA in the presence of synthetic PE (27). Similarly, the emergence of mutant strains whose lesion profiles differed from the lesion profile of the seed strain was also observed in a bioassay using hamster rPrP fibrils generated in seeded rPrP-PMCA (25) or 1st-rPrP-fibSc (Fig. 6C and D). These results raise the possibility that the lack of a GPI anchor in rPrP leads to alterations in strain-specific characteristics. Furthermore, the cell tropisms determined by the cell panel assay were altered in strains RML, 139A, 79A, and ME7 but not in strain 22L when the strains were propagated in Tg mice expressing PrP devoid of a GPI anchor (47). These studies demonstrate that glycosylation and a GPI anchor are not necessarily required for the propagation of prion infectivity but can influence the strain properties. Although the molecular basis of the emergence of mutant strains remains elusive, we can speculate that the posttranslational changes to PrP might affect the conformation of PrPSc or the interaction with some cofactor(s) in a strain-specific manner.
Another possible explanation is that nonspecific rPrP fibrils are generated during the serial RT-QUIC and replicate more rapidly than the fibrils with strain-specific conformations. The term “nonspecific rPrP fibrils” arises from our findings that there was little difference in the infrared spectra and conformational stability of 5th-rPrP-fibSc between strains. It has been reported that the propagation of prion strains in cells cultured under different environmental conditions often leads to the formation of quasispecies that are assumed to be composed of a variety of conformational variants (48, 49). Once generated, the competition among the variants is thought to occur during propagation. Indeed, two conformational variants of rPrP fibrils have been shown to be mutually exclusive and compete for monomeric rPrP as a substrate in fibril formation (30). Furthermore, competitive amplification of two prion strains was demonstrated by BH-PMCA (50). Similarly, nonspecific rPrP fibrils would be expected to become the majority if they had a selective growth advantage in the RT-QUIC. We found that the β-sheet spectra of rPrP fibrils generated in the presence of a small amount (1 pg) of PrPSc or rPrP fibrils generated at pH 4 in the first round were similar to those seen for 5th-rPrP-fibSc (Fig. 4B). These observations also support this hypothesis and suggest that the amplification of nonspecific rPrP fibrils is accelerated by certain conditions, such as an acidic environment. Further studies are needed to investigate whether unknown cofactors or environmental conditions are required to maintain the strain-specific conformations in subsequent rounds. On the other hand, this hypothesis also explains why prion infectivity was lost in the fifth round of RT-QUIC, as nonspecific rPrP fibrils generated during the serial RT-QUIC would be noninfectious. Although there remains the question as to what exactly the conformational differences between the noninfectious and infectious forms of rPrP fibrils are, the lack of cofactor molecules, such as SDS and synthetic PE, in the RT-QUIC might enhance the amplification of nonspecific rPrP fibrils lacking prion infectivity. Moreover, the fact that prion infectivity is sometimes too low to be detected and, more frequently, the fact that prion infectivity declines in the serial rPrP-PMCA (24, 25) or BH-PMCA (51–53) are consistent with the hypothesis.
ACKNOWLEDGMENTS
We thank Takashi Suematsu for help with the electron microscopy study and Matsuo Atsuko and Ayumi Yamakawa for technical assistance.
This work was supported by a grant-in-aid for young scientists (B; grant no. 21790846) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, a grant for BSE research, and a grant-in-aid from the Research Committee of Prion Disease and Slow Virus Infection from the Ministry of Health, Labor and Welfare of Japan.
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1. Prusiner SB. 1991. Molecular biology of prion diseases. Science 252:1515–1522. 10.1126/science.1675487 [DOI] [PubMed] [Google Scholar]
- 2. Weissmann C, Enari M, Klohn PC, Rossi D, Flechsig E. 2002. Molecular biology of prions. Acta Neurobiol. Exp. (Wars.) 62:153–166 [DOI] [PubMed] [Google Scholar]
- 3. Meyer RK, McKinley MP, Bowman KA, Braunfeld MB, Barry RA, Prusiner SB. 1986. Separation and properties of cellular and scrapie prion proteins. Proc. Natl. Acad. Sci. U. S. A. 83:2310–2314. 10.1073/pnas.83.8.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. 1991. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry 30:7672–7680. 10.1021/bi00245a003 [DOI] [PubMed] [Google Scholar]
- 5. Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, Prusiner SB. 1993. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U. S. A. 90:10962–10966. 10.1073/pnas.90.23.10962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caughey B, Raymond GJ, Bessen RA. 1998. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J. Biol. Chem. 273:32230–32235. 10.1074/jbc.273.48.32230 [DOI] [PubMed] [Google Scholar]
- 7. Atarashi R, Sim VL, Nishida N, Caughey B, Katamine S. 2006. Prion strain-dependent differences in conversion of mutant prion proteins in cell culture. J. Virol. 80:7854–7862. 10.1128/JVI.00424-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thomzig A, Spassov S, Friedrich M, Naumann D, Beekes M. 2004. Discriminating scrapie and bovine spongiform encephalopathy isolates by infrared spectroscopy of pathological prion protein. J. Biol. Chem. 279:33847–33854. 10.1074/jbc.M403730200 [DOI] [PubMed] [Google Scholar]
- 9. Baron GS, Hughson AG, Raymond GJ, Offerdahl DK, Barton KA, Raymond LD, Dorward DW, Caughey B. 2011. Effect of glycans and the glycophosphatidylinositol anchor on strain dependent conformations of scrapie prion protein: improved purifications and infrared spectra. Biochemistry 50:4479–4490. 10.1021/bi2003907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peretz D, Scott MR, Groth D, Williamson RA, Burton DR, Cohen FE, Prusiner SB. 2001. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 10:854–863. 10.1110/ps.39201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shindoh R, Kim CL, Song CH, Hasebe R, Horiuchi M. 2009. The region approximately between amino acids 81 and 137 of proteinase K-resistant PrPSc is critical for the infectivity of the Chandler prion strain. J. Virol. 83:3852–3860. 10.1128/JVI.01740-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saborio GP, Permanne B, Soto C. 2001. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411:810–813. 10.1038/35081095 [DOI] [PubMed] [Google Scholar]
- 13. Castilla J, Saa P, Hetz C, Soto C. 2005. In vitro generation of infectious scrapie prions. Cell 121:195–206. 10.1016/j.cell.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 14. Castilla J, Morales R, Saa P, Barria M, Gambetti P, Soto C. 2008. Cell-free propagation of prion strains. EMBO J. 27:2557–2566. 10.1038/emboj.2008.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deleault NR, Harris BT, Rees JR, Supattapone S. 2007. Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. U. S. A. 104:9741–9746. 10.1073/pnas.0702662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imamura M, Kato N, Yoshioka M, Okada H, Iwamaru Y, Shimizu Y, Mohri S, Yokoyama T, Murayama Y. 2011. Glycosylphosphatidylinositol anchor-dependent stimulation pathway required for generation of baculovirus-derived recombinant scrapie prion protein. J. Virol. 85:2582–2588. 10.1128/JVI.02098-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imamura M, Kato N, Okada H, Yoshioka M, Iwamaru Y, Shimizu Y, Mohri S, Yokoyama T, Murayama Y. 2013. Insect cell-derived cofactors become fully functional after proteinase K and heat treatment for high-fidelity amplification of glycosylphosphatidylinositol-anchored recombinant scrapie and BSE prion proteins. PLoS One 8:e82538. 10.1371/journal.pone.0082538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. 2004. Synthetic mammalian prions. Science 305:673–676. 10.1126/science.1100195 [DOI] [PubMed] [Google Scholar]
- 19. Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, Prusiner SB. 2009. Design and construction of diverse mammalian prion strains. Proc. Natl. Acad. Sci. U. S. A. 106:20417–20422. 10.1073/pnas.0910350106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raymond GJ, Race B, Hollister JR, Offerdahl DK, Moore RA, Kodali R, Raymond LD, Hughson AG, Rosenke R, Long D, Dorward DW, Baron GS. 2012. Isolation of novel synthetic prion strains by amplification in transgenic mice coexpressing wild-type and anchorless prion proteins. J. Virol. 86:11763–11778. 10.1128/JVI.01353-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. 2010. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol. 119:177–187. 10.1007/s00401-009-0633-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang F, Wang X, Yuan CG, Ma J. 2010. Generating a prion with bacterially expressed recombinant prion protein. Science 327:1132–1135. 10.1126/science.1183748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Z, Zhang Y, Wang F, Wang X, Xu Y, Yang H, Yu G, Yuan C, Ma J. 2013. De novo generation of infectious prions with bacterially expressed recombinant prion protein. FASEB J. 27:4768–4775. 10.1096/fj.13-233965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Timmes AG, Moore RA, Fischer ER, Priola SA. 2013. Recombinant prion protein refolded with lipid and RNA has the biochemical hallmarks of a prion but lacks in vivo infectivity. PLoS One 8:e71081. 10.1371/journal.pone.0071081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim JI, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, Race B, Qing L, Gambetti P, Caughey B, Surewicz WK. 2010. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J. Biol. Chem. 285:14083–14087. 10.1074/jbc.C110.113464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deleault NR, Piro JR, Walsh DJ, Wang F, Ma J, Geoghegan JC, Supattapone S. 2012. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc. Natl. Acad. Sci. U. S. A. 109:8546–8551. 10.1073/pnas.1204498109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR, Supattapone S. 2012. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc. Natl. Acad. Sci. U. S. A. 109:E1938–E1946. 10.1073/pnas.1206999109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Atarashi R, Satoh K, Sano K, Fuse T, Yamaguchi N, Ishibashi D, Matsubara T, Nakagaki T, Yamanaka H, Shirabe S, Yamada M, Mizusawa H, Kitamoto T, Klug G, McGlade A, Collins SJ, Nishida N. 2011. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat. Med. 17:175–178. 10.1038/nm.2294 [DOI] [PubMed] [Google Scholar]
- 29. Wilham JM, Orru CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. 2010. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 6:e1001217. 10.1371/journal.ppat.1001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, Priola SA, Caughey B. 2007. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat. Methods 4:645–650. 10.1038/nmeth1066 [DOI] [PubMed] [Google Scholar]
- 31. Fujihara A, Atarashi R, Fuse T, Ubagai K, Nakagaki T, Yamaguchi N, Ishibashi D, Katamine S, Nishida N. 2009. Hyperefficient PrP Sc amplification of mouse-adapted BSE and scrapie strain by protein misfolding cyclic amplification technique. FEBS J. 276:2841–2848. 10.1111/j.1742-4658.2009.07007.x [DOI] [PubMed] [Google Scholar]
- 32. Kocisko DA, Lansbury PT, Jr, Caughey B. 1996. Partial unfolding and refolding of scrapie-associated prion protein: evidence for a critical 16-kDa C-terminal domain. Biochemistry 35:13434–13442. 10.1021/bi9610562 [DOI] [PubMed] [Google Scholar]
- 33. Atarashi R, Sano K, Satoh K, Nishida N. 2011. Real-time quaking-induced conversion: a highly sensitive assay for prion detection. Prion 5:150–153. 10.4161/pri.5.3.16893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. 2011. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 18:504–506. 10.1038/nsmb.2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McGuire LI, Peden AH, Orru CD, Wilham JM, Appleford NE, Mallinson G, Andrews M, Head MW, Caughey B, Will RG, Knight RS, Green AJ. 2012. Real time quaking-induced conversion analysis of cerebrospinal fluid in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 72:278–285. 10.1002/ana.23589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Orru CD, Wilham JM, Raymond LD, Kuhn F, Schroeder B, Raeber AJ, Caughey B. 2011. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. mBio 2(3):e00078-00011. 10.1128/mBio.00078-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sano K, Satoh K, Atarashi R, Takashima H, Iwasaki Y, Yoshida M, Sanjo N, Murai H, Mizusawa H, Schmitz M, Zerr I, Kim YS, Nishida N. 2013. Early detection of abnormal prion protein in genetic human prion diseases now possible using real-time QUIC assay. PLoS One 8:e54915. 10.1371/journal.pone.0054915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vascellari S, Orru CD, Hughson AG, King D, Barron R, Wilham JM, Baron GS, Race B, Pani A, Caughey B. 2012. Prion seeding activities of mouse scrapie strains with divergent PrPSc protease sensitivities and amyloid plaque content using RT-QuIC and eQuIC. PLoS One 7:e48969. 10.1371/journal.pone.0048969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Henderson DM, Manca M, Haley NJ, Denkers ND, Nalls AV, Mathiason CK, Caughey B, Hoover EA. 2013. Rapid antemortem detection of CWD prions in deer saliva. PLoS One 8:e74377. 10.1371/journal.pone.0074377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smirnovas V, Kim JI, Lu X, Atarashi R, Caughey B, Surewicz WK. 2009. Distinct structures of scrapie prion protein (PrPSc)-seeded versus spontaneous recombinant prion protein fibrils revealed by hydrogen/deuterium exchange. J. Biol. Chem. 284:24233–24241. 10.1074/jbc.M109.036558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hornemann S, Schorn C, Wuthrich K. 2004. NMR structure of the bovine prion protein isolated from healthy calf brains. EMBO Rep. 5:1159–1164. 10.1038/sj.embor.7400297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Clarke AR, Jackson GS, Collinge J. 2001. The molecular biology of prion propagation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:185–195. 10.1098/rstb.2000.0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lawson VA, Collins SJ, Masters CL, Hill AF. 2005. Prion protein glycosylation. J. Neurochem. 93:793–801. 10.1111/j.1471-4159.2005.03104.x [DOI] [PubMed] [Google Scholar]
- 44. Aguzzi A, Heikenwalder M, Polymenidou M. 2007. Insights into prion strains and neurotoxicity. Nat. Rev. Mol. Cell Biol. 8:552–561. 10.1038/nrm2204 [DOI] [PubMed] [Google Scholar]
- 45. Cancellotti E, Mahal SP, Somerville R, Diack A, Brown D, Piccardo P, Weissmann C, Manson JC. 2013. Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties. EMBO J. 32:756–769. 10.1038/emboj.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Piro JR, Harris BT, Nishina K, Soto C, Morales R, Rees JR, Supattapone S. 2009. Prion protein glycosylation is not required for strain-specific neurotropism. J. Virol. 83:5321–5328. 10.1128/JVI.02502-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mahal SP, Jablonski J, Suponitsky-Kroyter I, Oelschlegel AM, Herva ME, Oldstone M, Weissmann C. 2012. Propagation of RML prions in mice expressing PrP devoid of GPI anchor leads to formation of a novel, stable prion strain. PLoS Pathog. 8:e1002746. 10.1371/journal.ppat.1002746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. 2010. Darwinian evolution of prions in cell culture. Science 327:869–872. 10.1126/science.1183218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weissmann C, Li J, Mahal SP, Browning S. 2011. Prions on the move. EMBO Rep. 12:1109–1117. 10.1038/embor.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shikiya RA, Ayers JI, Schutt CR, Kincaid AE, Bartz JC. 2010. Coinfecting prion strains compete for a limiting cellular resource. J. Virol. 84:5706–5714. 10.1128/JVI.00243-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bieschke J, Weber P, Sarafoff N, Beekes M, Giese A, Kretzschmar H. 2004. Autocatalytic self-propagation of misfolded prion protein. Proc. Natl. Acad. Sci. U. S. A. 101:12207–12211. 10.1073/pnas.0404650101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klingeborn M, Race B, Meade-White KD, Chesebro B. 2011. Lower specific infectivity of protease-resistant prion protein generated in cell-free reactions. Proc. Natl. Acad. Sci. U. S. A. 108:E1244–E1253. 10.1073/pnas.1111255108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gonzalez-Montalban N, Lee YJ, Makarava N, Savtchenko R, Baskakov IV. 2013. Changes in prion replication environment cause prion strain mutation. FASEB J. 27:3702–3710. 10.1096/fj.13-230466 [DOI] [PMC free article] [PubMed] [Google Scholar]