ABSTRACT
Passage of hepatitis C virus (HCV) in human hepatoma cells resulted in populations that displayed partial resistance to alpha interferon (IFN-α), telaprevir, daclatasvir, cyclosporine, and ribavirin, despite no prior exposure to these drugs. Mutant spectrum analyses and kinetics of virus production in the absence and presence of drugs indicate that resistance is not due to the presence of drug resistance mutations in the mutant spectrum of the initial or passaged populations but to increased replicative fitness acquired during passage. Fitness increases did not alter host factors that lead to shutoff of general host cell protein synthesis and preferential translation of HCV RNA. The results imply that viral replicative fitness is a mechanism of multidrug resistance in HCV.
IMPORTANCE Viral drug resistance is usually attributed to the presence of amino acid substitutions in the protein targeted by the drug. In the present study with HCV, we show that high viral replicative fitness can confer a general drug resistance phenotype to the virus. The results exclude the possibility that genomes with drug resistance mutations are responsible for the observed phenotype. The fact that replicative fitness can be a determinant of multidrug resistance may explain why the virus is less sensitive to drug treatments in prolonged chronic HCV infections that favor increases in replicative fitness.
INTRODUCTION
Selection of viral mutants resistant to antiviral agents is a major problem for the successful treatment of viral diseases. In the case of RNA viruses, high mutation rates during genome replication provide viral populations with an ample reservoir of phenotypic variants, including mutants that can escape selective constraints. Resistance to a single drug that targets a viral protein develops at a rate that depends on the genetic barrier (number and types of mutations needed to acquire resistance) and the phenotypic barrier (fitness cost) imposed by the resistance mutations (1–16). When drug resistance mutations do not entail a significant fitness cost—either because the mutations per se do not critically affect viral functions or because compensatory mutations are acquired—they may reach detectable levels despite no prior exposure of the viral population to the drug (1, 16–27).
Control of hepatitis C virus (HCV) infections is hampered by the complexity of HCV quasispecies replicating in the liver (16, 28, 29). Directly acting antiviral agents (DAAs)—some currently in use and others under development—offer great promise for control of HCV either as a substitute for or complement of the standard-of-care (SOC) therapy based on treatment using a combination of pegylated alpha interferon (IFN-α) and ribavirin (30–36). Combinations that include the polymerase inhibitor sofosbuvir have produced sustained viral responses that in some cases have been higher than 90% in clinical trials (37–40), but the possible impact of resistance mutations is not known; sofosbuvir resistance substitution S282T in NS5B is present in the sequence of HCV reference isolate ED43 of genotype 4a and L159F is present in the mutant spectrum of HCV quasispecies following treatment of HCV p100 with ribavirin (I. Gallego, E. Domingo, and C. Perales, unpublished results).
The advent of cell culture systems designed to achieve replication of full-length, infectious HCV (41–43) has opened the way to studies on antiviral agents for HCV in cell culture. Using this system (44), we performed up to 100 serial passages in the human hepatoma Huh-7.5 cell line, either in the absence or the presence of different concentrations of IFN-α (45). In the course of these studies, we made the unexpected observations that populations of HCV passaged in the absence of IFN-α acquired partial resistance to IFN-α and that their capacity to shut off host cell protein synthesis was increased relative to that of the parental virus HCV p0 (where “HCV p0” represents the HCV population before the first passage in Huh-7.5 cells) (45). It was unlikely that selection for partial IFN-α resistance was due to endogenous IFN produced by the host cell since the Huh-7.5 cells used for the infections are defective in IFN production (46, 47). This observation raised three issues: (i) what the difference is in replicative parameters between HCV p0 and the passaged populations, (ii) whether the partial resistance is unique to IFN-α or whether it extends to other anti-HCV drugs, and (iii) what the molecular basis is of either a specific or general partial resistance to drugs. We addressed these issues in the present study and show that passaged HCV displays increased replicative capacity and diminished sensitivity not only to IFN-α but also to several other anti-HCV drugs. Furthermore, independent HCV evolutionary lineages and biological clones display the same behavior. Mutant spectrum analysis and viral replication in the absence and presence of drugs render unlikely the possibility that the presence of drug resistance mutations in the passaged populations is responsible for the expanded drug resistance. The results provide evidence that increased replicative HCV fitness results in a multidrug resistance phenotype. Implications for treatment of acute versus chronic HCV infections are discussed.
MATERIALS AND METHODS
Cells, viruses, and drugs.
The origin of Huh-7.5, Huh-7 Lunet, and Huh-7.5 reporter cell lines and procedures for cell growth in Dulbecco's modification of Eagle's medium (DMEM) have been previously described (45, 48, 49); cells were cultured at 37°C and 5% CO2. Huh-7.5 cells were used for titration of virus infectivity, and Huh-7.5 reporter cells were used for standard infections and serial passages of HCV.
The viruses used in the experiments are those rescued from plasmids Jc1FLAG2(p7-nsGluc2A) (a chimera of J6 and JFH-1 from genotype 2a), termed HCV p0, and from plasmid GNNFLAG2(p7-nsGluc2A), termed GNN (which carries a mutation in NS5B that renders the virus replication defective); GNN has been used as negative infection control, and both viruses have been previously described (44). Preparation of the initial HCV p0 population from the progeny of the initial electroporation with the RNA transcripts and passage conditions in Huh-7.5 cells were previously described (45). Each virus passage involved infection of a monolayer of 4 × 105 Huh-7.5 reporter cells with the number of infectious units (measured as a 50% tissue culture infective dose [TCID50] value) indicated in each experiment. To control for the absence of contamination, the supernatants of mock-infected cells maintained in parallel with the infected cultures were titrated; no infectivity in the mock-infected cultures was detected in any of the experiments.
Biological clones were isolated from HCV populations by limiting dilution followed by growth in Huh 7.5 cells. Briefly, HCV p0 and HCV p100 populations were treated with sodium deoxycholate (0.01%) for 10 min at room temperature. Dilutions (10−3 to 10−7) were used to infect Huh-7.5 cells in 96-well plates, and at 72 h postinfection (hpi), supernatants were collected and infected cells visualized by immunostaining with anti-NS5A antibody (9E10). Viruses collected from wells containing a single infected-cell focus were passaged twice on fresh Huh-7.5 cells, yielding clonal populations with infectious titers of 104 to 105 TCID50/ml. Each virus was then subjected to another round of limiting dilution as described above, yielding final infectious virus stocks with titers of 105 to 106 TCID50/ml.
Stock solutions of 0.1 M ribavirin (Sigma), 10 mM telaprevir (Selleck Chemicals), 10 mM cyclosporine (Sigma), and 10 mM daclatasvir (Selleck Chemicals) were prepared in phosphate-buffered saline (PBS) or H2O and stored at −70°C. Prior to use, the solutions were diluted in DMEM to reach the desired concentration.
Virus titration.
For titration of infectious HCV, samples were serially diluted and applied to Huh-7.5 cells in 96-well plates (6,400 cells/well seeded 16 h earlier). Three days later, cells were washed with PBS, fixed with ice-cold methanol, and stained for the presence of NS5A using anti-NS5A monoclonal antibody 9E10, as described previously (41, 45). The virus titer was expressed as TCID50/ml (50).
Drug toxicity and inhibitory activity.
Drug toxicity was measured in Huh-7.5 reporter cells by seeding 96-well plates at 70% confluence and exposing the cells to different drug concentrations for 72 h. Then, MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] was added to each well at a final concentration of 500 μg/ml; following 4 h of incubation, 100 μl of dimethyl sulfoxide (DMSO) was added, and the optical density at 550 nm was measured. The drug concentration required for 50% cell killing (CC50) was calculated as described previously (51). The drug concentration required for 50% inhibition of infectious HCV yield (IC50) was determined by seeding Huh-7.5 reporter cells in 96-well plates to 70% confluence and infecting with HCV at a multiplicity of infection (MOI) of 0.03 TCID50/cell. After incubation at 37°C for 5 h, the inoculum was removed and medium with or without dilutions of antiviral drugs was added to each well. Infected cells were immunostained at 48 hpi using anti-NS5A antibody (9E10), and the number of infected foci present in each well was counted. IC50s were calculated by comparison of the number of infected foci in each dilution of antiviral drug with the number in the drug-untreated controls, using the program Sigma Plot. CC50 and IC50 values were calculated as the averages of the results of at least three determinations. The therapeutic index (TI = CC50/IC50) was calculated for each drug.
Cell culture system for long-term serial passage of HCV.
To initiate serial passages of HCV, 4 × 105 Huh-7.5 reporter cells were infected with HCV p0 at an MOI of 0.5 TCID50/cell, and virus adsorption was allowed to proceed for 5 h at 37°C. Then, the inoculum was removed and 2 ml of medium added to the cell monolayer. The infected cells were further incubated at 37°C for 72 to 96 h. For each subsequent passage, 4 × 105 Huh-7.5 reporter cells were infected as described above using 0.5 ml of cell culture supernatant from the previous passage. The MOI for each infection was calculated from the infectivity values given for each experiment, and it ranged between 0.1 and 0.5 TCID50/cell. Viral populations are identified with a “p” followed by the passage number (e.g., HCV p10 is HCV p0 passaged 10 times in Huh-7.5 cells). Mock-infected cells and cells infected with HCV p0 were maintained for 100 passages in parallel to control for the absence of contamination of cells with virus.
Infection in the presence of antiviral compounds.
A fixed TCID50 of 12,800 was used to infect 4 × 105 Huh-7.5 cells (MOI = 0.03 TCID50/cell). After 5 h of virus adsorption, supernatants were replaced with fresh media with or without the antiviral compound. For subsequent passages, 0.5 ml of the previously infected cell culture supernatant was used to infect 4 × 105 Huh-7.5 reporter cells; infections were allowed to proceed for 72 to 96 h. For infections in the presence of ribavirin, cells were pretreated with ribavirin for 16 h prior to infection.
RNA extraction, cDNA synthesis, PCR amplification, and nucleotide sequencing.
Intracellular viral RNA was extracted from infected cells after each passage using a Qiagen RNeasy kit (Qiagen, Valencia, CA), according to the manufacturer's instructions. Reverse transcription (RT) was performed using avian myeloblastosis virus (AMV) reverse transcriptase (Promega), and PCR amplification of specific HCV genomic regions was carried out using AccuScript (Agilent Technologies), with specific oligonucleotide primers (45) (see Table S1 [http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf]). Amplification products were analyzed by agarose gel electrophoresis, with the DNAs produced by digestion of Φ-29 DNA with HindIII employed as molecular mass standards. Negative controls without template RNA were included in parallel to ascertain the absence of cross-contamination by template nucleic acids. Nucleotide sequences of genomic HCV RNA were determined on the two strands of an amplified cDNA copy (45, 52). To evaluate the complexity of mutant spectra, HCV RNA was extracted as described above and subjected to RT-PCR to amplify the NS5A-coding regions as previously described (53). To ensure an excess of template in the RT-PCR amplifications for quasispecies analysis, and to avoid complexity biases due to redundant amplifications of the same initial RNA templates, amplifications were carried out with template preparations diluted 1:10, 1:100, and 1:1,000; only when the template diluted 1:100 produced a visible DNA band was molecular cloning pursued using the DNA amplified from the undiluted template. Controls to ascertain that the mutation frequencies were not affected by the basal error rate during amplification have been previously described (54).
For the ultradeep pyrosequencing (UDPS) analysis (GS-FLX platform; 454 Life Sciences-Roche), RT-PCR was performed using Accuscript (Agilent). To cover the complete NS5A region, and considering that the GS-FLX Titanium chemistry allows sequencing fragments of 400 to 500 nucleotides, this genomic region was divided into six overlapping amplicons, and amplification products were obtained using specific primers (see Table S1 [http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf]). To minimize the errors due to PCRs, RT-PCR amplifications were performed in triplicate, and the amplification products were mixed equimolarly prior to the analysis. Then, PCR products were purified (Ampure beads), quantified (Pico green assay), and analyzed for quality (Bioanalyzer) prior to the UDPS procedure. Negative controls (without template RNA) were run in parallel to ascertain the absence of contamination with undesired templates. Procedures for the data analysis have been previously described (53). The genomic nucleotide sequences of the HCV described in the present study have been deposited in GenBank (see below).
Quantification of HCV RNA using real-time RT-PCR.
Real-time quantitative RT-PCR (qRT-PCR) of HCV RNA was carried out using a LightCycler RNA Master SYBR green I kit (Roche) (41, 45). The 5′ untranscribed region (5′-UTR) of the HCV genome was amplified using as primers oligonucleotides HCV-5UTR-F2 and HCV-5UTR-R2 (see Table S1 [http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf]). Quantification was performed relative to a standard curve obtained with known amounts of HCV RNA, synthesized by in vitro transcription of plasmid GNNFLAG2(p7-nsGluc2A). The specificity of the reaction was monitored by the denaturation curve of the amplified DNAs. Negative controls (without template RNA and RNA from mock-infected cells) were run in parallel with each amplification reaction to ascertain the absence of contamination with undesired templates. Quantifications were carried out in triplicate.
Relative-fitness assays.
Relative fitness was measured by growth-competition experiments between two viruses in Huh-7.5 cells. Competitions were established between HCV p0 and either HCV p45 or HCV p100 and also between HCV p45 and HCV p100. To this end, 4 × 105 Huh-7.5 cells were infected with a 1:1 mixture of the two competing viruses based on infectivity values (total, 1.2 × 104 TCID50; MOI of 0.03 TCID50/cell). RNA extracted from the initial mixtures and from infected cells at 24, 48, and 72 h postinfection was sequenced in regions that contained mutations diagnostic of each population (45). The ratio of the two competing viruses at each time point was estimated by measuring the areas of the relevant chromatogram peaks, as described previously (45, 52). The logarithm of this ratio was plotted against the time postinfection, and the fitness vector was adjusted to the following exponential equation: y = a × ebx. The antilogarithm of the vector slope gives the relative replicative fitness of the competing viruses.
Pulse-labeling of infected cells and Western blot assays.
Protein synthesis was analyzed by metabolic labeling of uninfected or infected cells with [35S]methionine (Met)-cysteine (Cys), followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of protein extracts and fluorography, using previously described procedures (45). Proteins were labeled by the addition of 60 μCi of [35S]Met-Cys (Amersham) per ml contained in methionine-free DMEM at 72 h postinfection. After 1 h of incubation of the cell monolayers with the radioactive medium, the latter was removed and the cells were harvested in 0.2 ml of sample buffer (160 mM Tris-HCl [pH 6.8], 2% SDS, 11% glycerol, 0.1 M dithiothreitol [DTT], 0.033% bromophenol blue). The samples were boiled for 5 min, and aliquots were analyzed by SDS-PAGE at 200 V. Fluorography and autoradiography of the gels were done as previously described (55). The amount of cell extract used for electrophoretic analysis was normalized to the amount of cellular actin, measured by reactivity with a specific monoclonal antibody (anti-β-actin clone AC-15; Sigma).
For Western blot assays, proteins were transferred to a 0.45-μm-pore-size nitrocellulose membrane (Bio-Rad); blots were developed with the following antibodies: mouse monoclonal anti-NS5A (9E10) at a dilution of 1:1,000; mouse monoclonal anti-core (Santa Cruz Biotechnology) at a dilution of 1:200; mouse monoclonal anti-β-actin (clone AC-15; Sigma) at a 1:1,000 dilution; rabbit polyclonal anti-eukaryotic initiation factor 3 subunit a (anti-eIF3a), anti-eIF3b, and anti-eIF4B antibodies (Novus) at a 1:2,000 dilution; rabbit polyclonal anti-eIF4G and anti-poly(A)-binding protein (anti-PABP) antibodies at a 1:2,000 dilution; anti-polypyrimidine tract-binding protein (anti-PTB) at a 1:3,000 dilution (56); mouse monoclonal anti-tubulin (Sigma) at a 1:5,000 dilution; rabbit polyclonal anti-eIF2α phosphospecific antibody (Invitrogen) at a dilution of 1:300; and rabbit polyclonal anti-eIF2α (Santa Cruz Biotechnology) at a dilution of 1:200. Goat anti-mouse IgG (Pierce) and donkey anti-rabbit IgG (Amersham) coupled to peroxidase were used at a 1:10,000 dilution. The amount of protein extract used to determine the relative intensities of viral and cellular proteins corresponded to the linear region in the relationship between concentrations of protein extract and band intensity determined by Western blot analysis.
Measurements of infectivity decay.
Equal TCID50s of viral samples of HCV p0, HCV p45, and HCV p100 were incubated at 45°C, and aliquots were collected at different time points and titrated. The values corresponding to the decay of virus titer over time were fitted to an exponential curve, and the inactivation rate constant was calculated as previously described (57).
Statistical analyses.
The statistical significance of differences between mutation frequencies was evaluated by the chi-square test (χ2 test). To determine the statistical significance of differences in infectivity decay and IC50 values, one-way analysis of variance (ANOVA) was performed with the SPSS 13.0 statistical package (SPSS, Inc.). For the differences determined in serial-passage experiments, two-way ANOVA was used. For multiple comparisons, Bonferroni's correction was applied.
Nucleotide sequence accession numbers.
The genomic nucleotide sequences of the HCV described in the present study have been deposited in GenBank with accession numbers KC595606 to KC595624.
RESULTS
Intracellular replication and particle stability of HCV p0 and HCV p100.
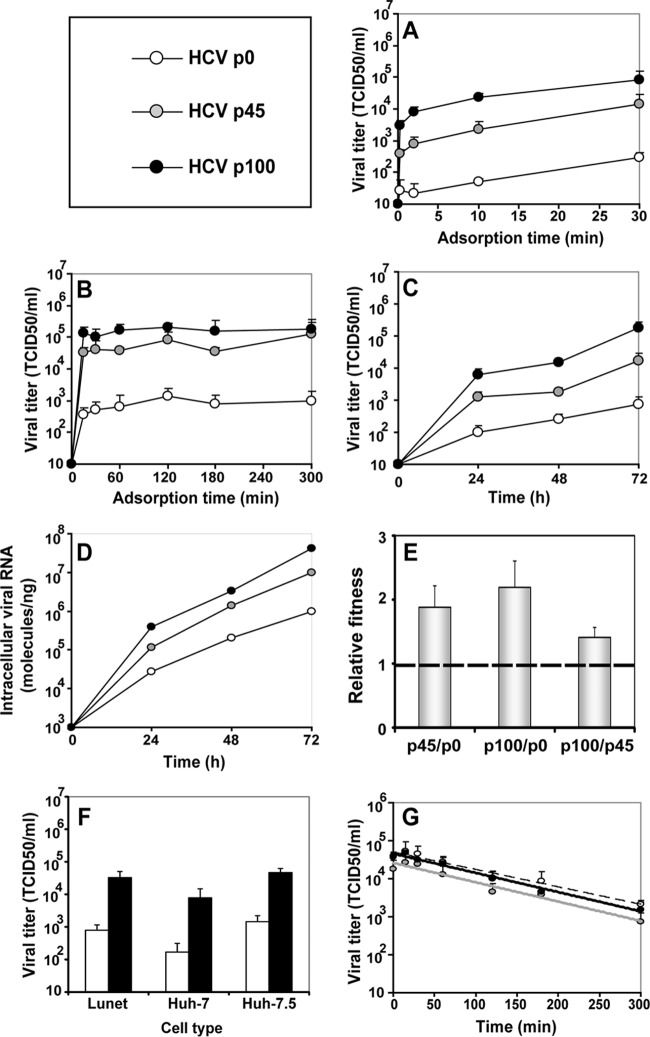
HCV p0 was subjected to 100 serial passages in Huh-7.5 cells, and infectivity in the cell culture supernatant, as well as intracellular and extracellular viral RNA levels, was measured at each passage (Fig. 1). By passage 60, the viral population displayed increased cytopathology that could alter the intracellular-to-extracellular-RNA ratios and diminish the number of cells and duration of replication cycles. To avoid confounding effects derived from cell killing, beginning at passage 60, the MOI was reduced about 10-fold (Fig. 1). HCV passaged in Huh-7.5 cells in the absence of IFN-α acquired partial resistance to IFN-α (45). To explore the basis of such resistance, the replication dynamics, fitness, and thermal stability characteristics of HCV p0, HCV p45, and HCV p100 were compared (Fig. 2). Viral progeny production increased with passage number (HCV p100 > HCV p45 > HCV p0) independently of the time allowed for virus adsorption to cells (Fig. 2A and B). The selective advantage of passaged HCV was associated with increased infectious particle production (P < 0.03 in all comparisons; ANOVA test) and intracellular replication (Fig. 2C and D), an advantage further confirmed by a relative HCV p100/HCV p0 replicative-fitness ratio of 2.2 ± 0.4 (Fig. 2E; see also Table S2 [http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf]). Since Lunet, Huh-7, and Huh-7.5 cells may allow viral multiplication at different rates (58), we examined whether extensive passage of HCV resulted in a differences in the capacity to infect the three cell lines. The ratios of the yield of HCV p100 to that of HCV p0 were very similar in all cell lines, as evaluated in a single infectious cycle (Fig. 2F). The three HCV populations displayed indistinguishable thermal-stability results, with inactivation rate constants at 45°C of k = 0.00756 ± 0.00658 (corresponding to a half-life of 91 min) for HCV p0, k = 0.00813 ± 0.00691 for HCV p45 (corresponding to a half-life of 85 min), and k = 0.00733 ± 0.00631 for HCV p100 (corresponding to a half-life of 94 min); the differences were not statistically significant (P = 0.13 to 1; ANOVA test) (Fig. 2G). This result excluded the possibility that an increase in thermal stability (selected during passaging) influenced the replicative-fitness values. Thus, the parameter associated with partial resistance of HCV p100 to IFN-α was a more efficient intracellular replication process that resulted in increased progeny production.
FIG 1.
Evolution of HCV infectivity and RNA during 100 passages in cell culture. (Top gray panel) Scheme of the origin of HCV populations. The initial clonal population (HCVcc) obtained by electroporation of Lunet cells by an HCV transcript is depicted as a filled square, and subsequent population passages are represented by empty circles; GNN is a replication defective mutant of HCVcc used as negative control. (White panels) Infectious viral titers of the supernatants, extracellular and intracellular viral RNA levels (quantified by real-time RT-PCR), and multiplicity of infection (M.O.I.) as a function of passage number. The crosses in the abscissae of the first panel indicate measurements upon serial infection with mutant GNN. Arrows indicate the passage number (passage 60) after which the MOI was decreased to prevent cell lysis. The origin of HCV, conditions for infections, determination of HCV infectivity, and quantification of HCV RNA, as well as positive and negative controls included in the assays, are described in Materials and Methods.
FIG 2.
Comparison of phenotypic traits of HCV p0, HCV p45, and HCV p100 (symbols in upper left box). (A and B) The effect of adsorption time on the yield of infectious progeny. Huh-7.5 cells were either mock infected or infected with HCV p0, HCV p45, or HCV p100 at an MOI of 0.03 TCID50/cell (4 × 105 Huh-7.5 cells infected with 1.2 × 104 TCID50). The virus was adsorbed to cells for the indicated times, the cells were washed twice with PBS, and DMEM was added to the monolayers. Infectivity levels in the cell culture supernatant were determined at 72 h postinfection. (C and D) HCV p0, HCV p45, and HCV p100 progeny production in Huh-7.5 cells. Infections were performed as described for panels A and B, using a 5-h adsorption period. Infectivity levels in the cell culture supernatant and intracellular viral RNA levels were determined at 72 h postinfection. (E) Relative fitness of HCV p0, HCV p45, and HCV p100. A single growth-competition experiment was performed by infecting Huh-7.5 cells with the viral mixtures indicated in the abscissa (MOI = 0.03 TCID50/cell). The relative amounts of the competing viruses were determined at 24, 48, and 72 h postinfection. Viral RNA present in the initial mixtures and at each passage was sequenced, and the proportions of the two competing populations were estimated and fitness data were calculated as detailed in Materials and Methods. Numerical values are given in Table S2 (http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf). (F) Infection of Lunet, Huh-7, and Huh-7.5 cells by HCV p0 and p100. Infections were performed as described for panels A and B, with a 5-h adsorption period. Infectivity levels in the cell culture supernatant were determined at 72 h postinfection. (G) Thermal stability of HCV p0, HCV p45, and HCV p100. Virus samples in DMEM were incubated at 45°C for the indicated amounts of time and titrated. Values are averages of the results of triplicate determinations. Procedures are further detailed in Materials and Methods.
Response of passaged HCV to antiviral inhibitors.
To test whether the partial resistance of HCV p45 and p100 to IFN-α (45) was a consequence of specific features of the antiviral activity evoked by IFN-α (59), resistance to ribavirin, telaprevir, daclatasvir, and cyclosporine, drugs that display unrelated mechanisms of activity, was investigated. The drug concentrations required to achieve a 50% inhibition of infectious HCV progeny production (IC50) were higher for the passaged HCV populations than for the parental HCV p0 populations (Table 1). The differences between the HCV p45 or HCV p100 values and the HCV p0 IC50 values were statistically significant (P < 0.01; ANOVA test), but the difference between the HCV p45 and HCV p100 values was not (P > 0.05; ANOVA test). The therapeutic index (TI) for each drug and each virus was calculated from the CC50 and IC50 values (see Table S3 [http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf]). As expected, passaged HCV displayed lower TI values than the parental HCV p0.
TABLE 1.
Quantification of inhibition of HCV p0, HCV p45, and HCV p100 infectious progeny production by antiviral agentsa
| Antiviral agent | CC50 ± SD | IC50 ± SD |
||
|---|---|---|---|---|
| HCV p0 | HCV p45 | HCV p100 | ||
| Ribavirin | 108 ± 4.2 μM | 6.9 ± 0.9 μM | 23 ± 2 μM | 20 ± 4 μM |
| Telaprevir | 22,000 ± 3,200 nM | 87 ± 18 nM | 273 ± 67 nM | 302 ± 75 nM |
| Daclatasvir | 14,900 ± 600 nM | 10 ± 0.3 pM | 63 ± 3 pM | 78 ± 18 pM |
| Cyclosporine | >50,000 nM | 83 ± 22 nM | 343 ± 130 nM | 358 ± 95 nM |
Determinations of 50% cytotoxic concentrations (CC50) and 50% inhibitory concentrations (IC50) were carried out in triplicate. Average values and standard deviations were calculated using the program Sigma Plot. Experimental conditions for cell growth, HCV infection, and determinations of cell viability and HCV infectivity are described in Materials and Methods.
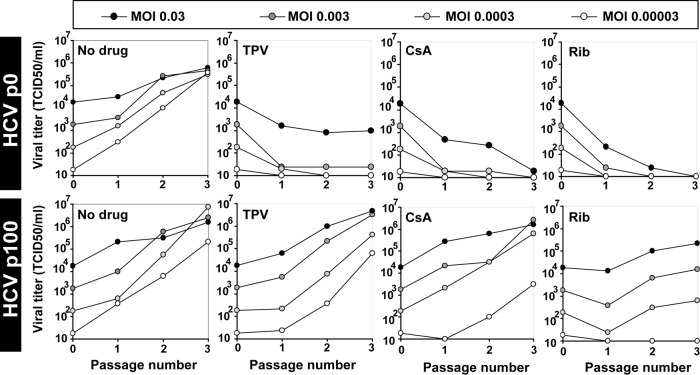
The increased resistance of HCV p45 and HCV p100 to the drugs relative to that of HCV p0 was also evident upon serial viral passages (Fig. 3), with loss of detectable infectivity of HCV p0 by passage 5 in the presence of ribavirin, telaprevir, or cyclosporine and yields of HCV p100 above 105 TCID50/ml; differences among the three viruses were statistically significant in the absence of drug and in the presence of IFN-α or ribavirin (P < 0.01; ANOVA test). In the presence of telaprevir, daclatasvir, and cyclosporine, the differences seen with comparisons of HCV p45 or HCV p100 to HCV p0 were statistically significant (P < 0.001; ANOVA test), but the differences seen with comparisons of HCV p45 to HCV p100 were not statistically significant (P > 0.05; ANOVA test). The sensitivity of HCV p0 to daclatasvir decreased with passage number (Fig. 3). HCV p100 displayed resistance to higher telaprevir and ribavirin concentrations also (see Fig. S1).
FIG 3.
Response of HCV p0, HCV p45, and HCV p100 to several antiviral agents. Huh-7.5 cells were either mock infected or infected with HCV p0, HCV p45, or HCV p100 at an MOI of 0.03 TCID50/cell (4 × 105 Huh-7.5 cells infected with 1.2 × 104 TCID50); the virus was adsorbed to cells for 5 h and the infection allowed to proceed for 72 h. Each of the populations was subjected to 10 passages in the absence or presence of IFN-α (HCV p45 passaged in the presence of 2 IU/ml and HCV p100 with 12 IU/ml, as previously described [45]), ribavirin (Rib) (50 μM), telaprevir (TPV) (600 nM), daclatasvir (DCV) (500 pM), or cyclosporine (CsA) (800 nM). The passages in the presence of IFN-α that had been previously described are included here for comparison. For the successive passages, the same numbers of cells were infected with the virus contained in 0.5 ml of the supernatant from the previous infection, maintaining the same drug concentration, and viral titers in the cell culture supernatants were determined. Parallel infections were carried out with HCV GNN as a negative control. Procedures are described in Materials and Methods.
Sequence analysis of the consensus sequence of the NS5A-coding region at passage 10 revealed the presence of substitution F28C, a position involved in daclatasvir resistance. To determine whether resistance mutations were selected upon passages of HCV p45 and HCV p100 in the presence of the drugs, the consensus sequences corresponding to NS3-coding regions (for telaprevir), NS5A-coding regions (for daclatasvir and cyclosporine), and NS5B-coding regions (for ribavirin) at passage 10 were analyzed. Mutations conferring resistance to telaprevir and daclatasvir were frequently selected (see Tables S3 to S6 [http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf]), as expected from the low barrier to resistance of these drugs (60, 61). HCV lineages, other than the ones depicted in Fig. 1, that were described in our previous study (45) also displayed increased resistance to the drugs relative to HCV p0 (see Fig. S2 at the URL mentioned above). Thus, serial passage of HCV p0 in Huh-7.5 cells led to populations that displayed resistance to several anti-HCV agents that target different viral or cellular factors, despite no prior exposure of the viral populations to the corresponding drugs.
Molecular basis of expanded HCV drug resistance.
Replication rate, population complexity, and replicative fitness are interconnected parameters that have been linked to the adaptability and pathogenic potential of viral populations (16, 62–66). To determine whether extensive passage of HCV in Huh-7.5 cells resulted in an expanded mutant spectrum, mutation frequency, Shannon entropy, mutation types, and numbers of polymorphic sites and haplotypes at the NS5A-coding region were determined for populations of HCV p0, HCV p45, and HCV p100, using molecular cloning and Sanger sequencing and ultradeep pyrosequencing (UDPS). The mutation frequency among components of the mutant spectrum increased 2.6-fold from HCV p0 to HCV p100 (P < 0.0001; χ2 test), and the Shannon entropy reached its maximum possible value (Table 2). UDPS indicated an increase of the numbers of different mutations, polymorphic sites, and haplotypes in HCV p45 and HCV p100 relative to HCV p0 (Table 3). Transitions amounted to 75% to 77% of the total number of mutations, and the ratio [(G→A)+(C→U)]/[(A→G)+(U→C)] was maintained in the range of 0.21 to 0.69 (see Fig. S3), as expected for HCV not subjected to a mutagenic agent (53). The amino acid substitutions deduced from the NS5A mutant spectra of HCV p45 and HCV p100 did not reveal any substitutions (F28S, L31M, C92R, and Y93H) currently catalogued as conferring resistance to daclatasvir (67–73) (see Tables S7 and S8). However, substitution F28I was present at a 0.75% level in the mutant spectrum of HCV p100 (see Table S9 [http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf]) but did not become dominant upon passage of HCV p100 in the presence of daclatasvir (see Table S6 at the URL mentioned above). In the case of cyclosporine, the mutations that lead to resistance substitution V464L were present in the quasispecies of HCV p0 and HCV p45 but not in HCV p100 (see Table S9 at the URL mentioned above). Furthermore, this substitution did not become dominant upon passage in the presence of cyclosporine (see Table S7 at the URL mentioned above). Therefore, an F28I substitution or a V464L substitution cannot explain the resistance of HCV p45 and HCV p100 to these drugs.
TABLE 2.
Quasispecies analysis of HCV p0, HCV p45, and HCV p100 populationsa
| Virus | No. of nucleotides analyzed (no. of clones/no. of haplotypes)b | Total no. of mutationsc | No. of different mutationsd | Mutation frequency |
Shannon entropyg | |
|---|---|---|---|---|---|---|
| Maximume | Minimumf | |||||
| HCV p0 | 37,746 (27/19) | 32 | 30 | 8.5 × 10−4 | 7.9 × 10−4 | 0.78 |
| HCV p45 | 41,940 (30/30) | 133 | 57 | 3.2 × 10−3 | 1.4 × 10−3 | 1 |
| HCV p100 | 40,542 (29/29) | 298 | 83 | 7.4 × 10−3 | 2.1 × 10−3 | 1 |
The populations analyzed correspond to passages 0, 45, and 100 of the infections described in the Fig. 1 legend. The genomic region analyzed was that of NS5A (nt 6269 to 7666). The HCV genome residue numbering corresponds to the JFH-1 genome (GenBank accession number AB047639).
The values in parentheses indicate the number of clones analyzed, followed by the number of haplotypes (numbers of different RNA sequences).
Data represent the total number of mutations found by comparing the sequence of each individual clone with the reference (parental) sequence.
Data represent the number of different mutations found by comparing the sequence of each individual clone with the reference (parental) sequence.
Data represent the average number of total mutations per nucleotide relative to the reference (parental) sequence.
Data represent the average number of different mutations per nucleotide relative to the reference (parental) sequence.
Shannon entropy (S) is a measure of the number of different molecules in the mutant spectrum of the quasispecies. It is calculated by the formula S = −[∑i (pi × ln pi)]/ln N, in which pi is the frequency of each sequence in the quasispecies and N is the total number of sequences compared.
TABLE 3.
Ultradeep pyrosequencing analysis of hepatitis C virus populations HCV p0, HCV p45, and HCV p100a
| Parameter | Virus | Value(s) for indicated NS5A amplicon (no. of haplotypes with 1, 2, 3, 4, 5, and 6 mutations)b |
|||||
|---|---|---|---|---|---|---|---|
| A1 (residues 6152–6454) | A2 (residues 6446–6767) | A3 (residues 6737–6954) | A4 (residues 6910–7252) | A5 (residues 7224–7550) | A6 (residues 7432–7725) | ||
| No. of different mutations | HCV p0 | 1 | 6 | 4 | 9 | 6 | 9 |
| HCV p45 | 6 | 12 | 10 | 14 | 13 | 19 | |
| HCV p100 | 21 | 14 | 16 | 29 | 21 | 17 | |
| No. of polymorphic sites | HCV p0 | 1 | 6 | 4 | 9 | 6 | 7 |
| HCV p45 | 6 | 11 | 10 | 13 | 13 | 18 | |
| HCV p100 | 20 | 14 | 15 | 29 | 20 | 16 | |
| No. of haplotypesc | HCV p0 | 2 (1/0/0/0/0/0) | 7 (6/0/0/0/0/0) | 5 (4/0/0/0/0/0) | 9 (7/1/0/0/0/0) | 6 (4/1/0/0/0/0) | 10 (9/0/0/0/0/0) |
| HCV p45 | 7 (6/0/0/0/0/0) | 13 (11/1/0/0/0/0) | 11 (10/0/0/0/0/0) | 14 (10/3/0/0/0/0) | 14 (7/2/3/1/0/0) | 26 (9/6/4/6/0/0) | |
| HCV p100 | 29 (9/6/9/3/1/0) | 18 (7/6/3/0/1/0) | 17 (15/1/0/0/0/0) | 29 (8/13/5/2/0/0) | 19 (10/3/5/0/0/0) | 26 (6/6/5/5/2/1) | |
The populations analyzed correspond to passages 0, 45, and 100 of the infections described in the Fig. 1 legend.
The HCV genome residue numbering corresponds to the JFH-1 genome (GenBank accession number AB047639). Amplicon (A) length ranged between 216 and 339 nucleotides. The number of nucleotides sequenced was 2.1 × 106 to 3.3 × 106, and the number of reads on which the parameters were calculated was 10,000 for each amplicon. Procedures are described in Materials and Methods.
The numbers of haplotypes with one, two, three, four, five, and six mutations are given in parentheses; no haplotypes with a higher number of mutations were found. Mutation types are summarized elsewhere (see Fig. S3), and their positions in the HCV genome and deduced amino acid substitutions are given in Table S9 (the relevant tables can be found at http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf).
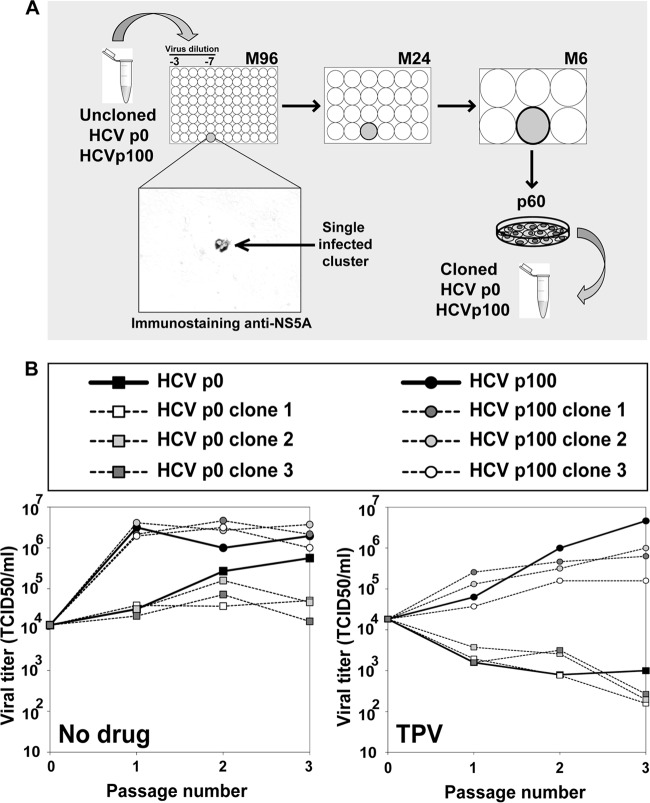
It is probable that not all drug resistance mutations in HCV have been identified. To further exclude the possibility that the presence of inhibitor resistance mutations may underlie the observed decrease of viral sensitivity to inhibitors, the MOI in the serial infections was decreased up to 1,000-fold. The kinetics of HCV progeny production followed an MOI-independent pattern (Fig. 4). The differences between the increases in viral titers in the passages in the absence of drug and either telaprevir or cyclosporine were not significant at any MOI tested (P = 0.724 to 1 and P = 0.187 to 1, respectively; ANOVA). The difference in the case of ribavirin reached significance at all MOI (P = 0.01 to 0.032) except at an MOI of 0.03 TCID50/cell (P = 0.065; ANOVA). This result is not compatible with an increase of the frequency of mutations conferring resistance to telaprevir and daclatasvir in HCV p100. The variation of statistical significance with MOI in the case of ribavirin is not easy to interpret since under our infection conditions ribavirin acts as both an inhibitor and a mutagen (53). Resistance to telaprevir was examined with more detail. Infectious progeny production by individual biological clones derived from HCV p0 and HCV p100 in the absence or presence of telaprevir was indistinguishable from that seen with the corresponding parental populations (P = 0.24 to 1 for HCV p0 clones; P = 1 for HCV p100 clones, in the absence of telaprevir; P = 1 for HCV p0 clones; P = 0.14 to 1 for HCV p100 clones, in the presence of telaprevir [ANOVA]); the difference between the uncloned populations of HCV p0 and HCV p100 was significant (P = 0.045; ANOVA) (Fig. 5). Thus, the multidrug resistance phenotype of passaged HCV is associated with its replicative fitness and is not due to the presence of inhibitor resistance mutations.
FIG 4.
Effect of MOI on the response of HCV p0 and HCV p100 to several antiviral agents in cell culture. Huh-7.5 cells were either mock infected or infected with HCV p0 or HCV p100 at an MOI of 0.03, 0.003, 0.0003, and 0.00003 TCID50/cell (top boxes) (4 × 105 Huh-7.5 cells infected with 1.2 × 104, 1.2 × 103, 1.2 × 102, and 1.2 × 101 TCID50, respectively); the virus was adsorbed to cells for 5 h and the infection allowed to proceed for 72 h. Each of the populations was subjected to 3 passages in the absence or presence of telaprevir (TPV) (600 nM), cyclosporine (CsA) (800 nM), or ribavirin (Rib) (50 μM). For the successive passages, the same numbers of cells were infected with the virus contained in 0.5 ml of the supernatant from the previous infection, maintaining the same drug concentration. Viral titers in the cell culture supernatants were determined. Parallel infections were carried out with HCV GNN as a negative control. Procedures are described in Materials and Methods.
FIG 5.
Replication of biological clones derived from HCV p0 and HCV p100 in the absence and presence of telaprevir. (A) Scheme of the procedure to obtain clonal preparations from populations of HCV p0 and HCV p100. Mild-detergent-treated virus was diluted and applied to M96 wells with a Huh-7.5 cell monolayer. Cell culture supernatant from wells with a single cluster of infected cells was used to infect Huh-7.5 cells in M24 wells, and the supernatant was again transferred to a M6 dish with Huh-7.5 cells. The final round of amplification consisted of infecting 5.2 × 105 Huh-7.5 cells in a p60 dish under the standard conditions detailed in Materials and Methods. (B) Huh-7.5 cells were either mock infected or infected with uncloned populations HCV p0 and HCV p100, and with three biological clones from each population, at an MOI of 0.03 TCID50/cell (4 × 105 Huh-7.5 cells infected with 1.2 × 104 TCID50); the virus was adsorbed to cells for 5 h and the infection allowed to proceed for 72 h in the absence and presence of telaprevir (TPV; 600 nM). For the successive passages, the same numbers of cells were infected with the virus contained in 0.5 ml of the supernatant from the previous infection, maintaining the same drug concentration. Viral titers in the cell culture supernatants were determined. Parallel infections were carried out with HCV GNN as a negative control. Procedures are described in Materials and Methods.
Alterations of host cell protein synthesis upon HCV infection.
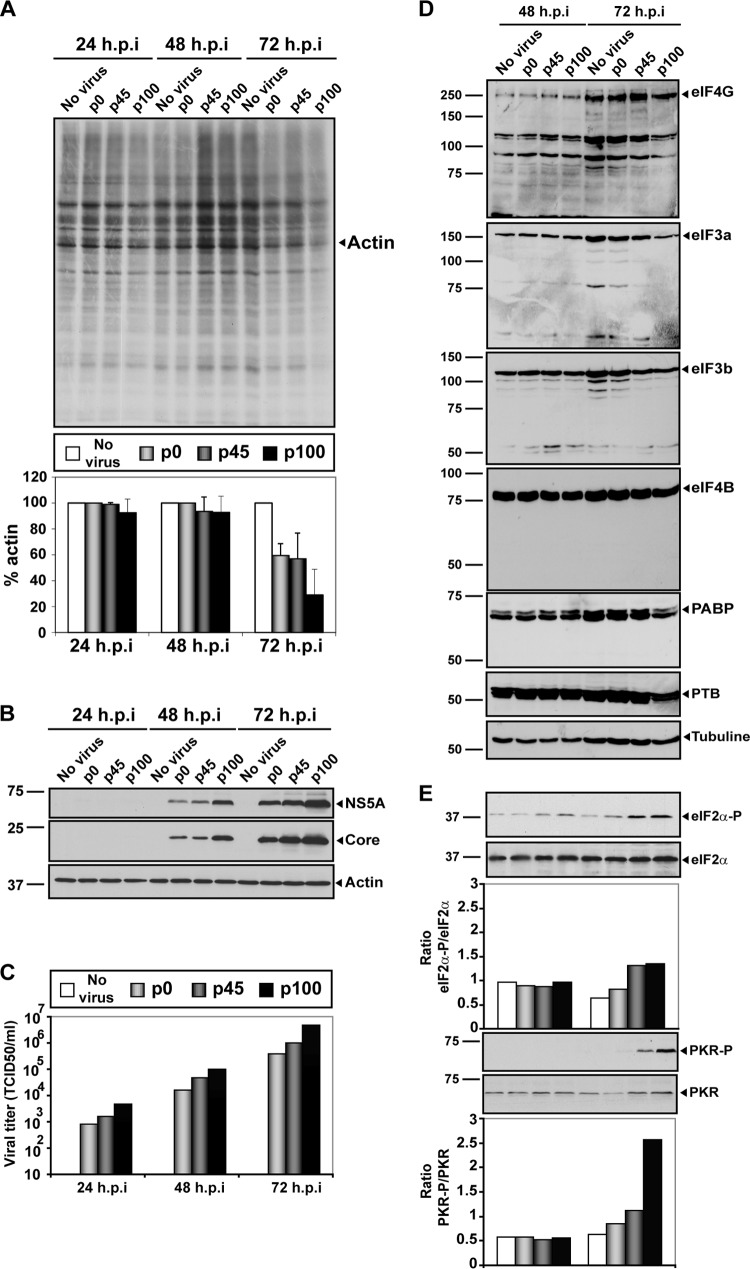
The replicative capacity of a virus is often determined by the presence of multiple viral expression products in interaction with host factors. Virus infection, as well as apoptosis, may result in cleavage of several translation initiation factors (eIFs) and thus in a decrease of 5′-cap-dependent protein synthesis without affecting ribosome entry site (IRES)-dependent viral protein synthesis, as is the case with HCV and picornavirus RNA (74, 75). This is partly mediated by disruption of critical components of the protein synthesis preinitiation complex consisting of eIF4G, eIF3, and poly(A)-binding protein (PABP) (56, 76–82). Previous work indicated that HCV p100 produced a larger shutoff of host cell protein synthesis than the parental HCV p0, and the change was associated with increased phosphorylation of protein kinase R (PKR) and eIF2α (45). To study whether cleavage of other host factors required for translation control was involved in the replicative advantage of passaged HCV, we compared results revealing the shutoff of host cell protein synthesis and the integrity of initiation factors eIF4G, eIF3a, eIF3b, eIF4B, PABP (80), and IRES-transacting factor polypyrimidine tract-binding protein (PTB) (83, 84) in Huh-7.5 cells, following infection with HCV p0, HCV p45, or HCV p100. Enhanced shutoff of host cell protein synthesis at late times postinfection by passaged HCV (Fig. 6A) was accompanied by increased virus-specific protein synthesis (Fig. 6B) and production of infectious progeny (Fig. 6C). No cleavage of any of the factors tested was observed (Fig. 6D). In the same infections, phosphorylation of eIF2α and PKR was increased at 72 h postinfection (Fig. 6E), in agreement with previous results (45, 46, 85). Thus, the enhanced replication of HCV and the diminished host cell protein synthesis achieved upon extensive passage in Huh-7.5 hepatoma cells was associated with increased phosphorylation of eIF2α and PKR and did not require cleavage of translation initiation factors controlling 5′-cap-dependent translation initiation. The enhanced phosphorylation of eIF2α and PKR was established independently of the degree of adaptation of HCV to the host cell (45, 46, 85).
FIG 6.
Effect of HCV p0, HCV p45, and HCV p100 infection on host cell protein synthesis and translation initiation factors. (A) Huh-7.5 cell were either mock infected or infected with HCV p0, HCV p45, or HCV p100 at an MOI of 0.03 TCID50/cell (4 × 105 Huh-7.5 cells infected with 1.2 × 104 TCID50), and the virus was allowed to adsorb to cells for 5 h. At 24, 48, and 72 h postinfection (h.p.i), cells were labeled with [35S]Met-Cys for 1 h, and cell extracts were subjected to SDS-PAGE and visualized by autoradiography. The total amount of labeled protein was calculated by densitometry measurements of the corresponding autoradiograms and is expressed as a percentage of the amount of actin protein taken as 100%. (B) HCV protein expression at 24, 48, and 72 h postinfection. Infected Huh-7.5 protein extracts were those of the experiment described for panel A. HCV NS5A and core were stained by Western blotting using monoclonal antibodies specific for the proteins indicated on the right side of the panel. The amount of cellular proteins was normalized to the amount of actin, visualized by Western blotting (Actin panel). (C) Viral titers in the cell culture supernatants determined at 24, 48, and 72 h postinfection. (D) Effect of HCV infection (top of each lane) on some host factors involved in translation control. Huh-7.5 cells were infected as indicated for panel A, and at 48 and 72 h postinfection, cell extracts were analyzed by Western blotting using specific antibodies against eIF4G, eIF3a, eIF3b, eIF4B, PABP, and PTB proteins (as indicated to the right of each panel). (E) Phosphorylation levels of PKR and eIF2-α determined in the same protein extracts used as described for panels A, B, C, and D. Procedures are detailed in Materials and Methods.
DISCUSSION
Serial passage of HCV in Huh-7.5 cells resulted in populations that display increased replicative fitness and partial resistance to inhibitors that target viral or cellular proteins. The partial resistance extends also to ribavirin, which acts as an HCV mutagen in our experimental system (53). Despite an expansion of the mutant spectrum complexity during HCV passage in Huh-7.5 cells (Tables 2 and 3), the drug resistance phenotype was not due to the presence of any inhibitor resistance mutations that might have accumulated in the expanded mutant spectrum of HCV. This conclusion is supported by different observations. (i) Several well-characterized inhibitor resistance mutations of HCV were not detected in the mutant spectra of HCV p45 and HCV p100 (see Tables S8 and S9 [http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf]). Substitution F28I in NS5A affected the same amino acid of the F28S daclatasvir resistance mutation (73). UDPS identified F28I as present at a 0.75% frequency in the mutant spectrum of HCV p100. Upon passage of HCV p100 in the presence of daclatasvir, F28I did not become dominant, but F28S did. It cannot be argued that F28I was an intermediate substitution required to reach F28S since substitution I28S requires two mutations while F28S requires only one (see Tables S6 and S9 at the URL mentioned above). Concerning cyclosporine, resistance substitution V464L (86) in NS5A was present in the mutant spectrum of HCV p0 and HCV p45 but not in that of HCV p100 (see Table S9 at the URL mentioned above). Furthermore, V464L did not become dominant upon passage of HCV p45 and HCV p100 in the presence of cyclosporine (see Table S7 at the URL mentioned above). (ii) Reductions in the infecting MOI did not affect the kinetics of viral production in the presence of inhibitors (Fig. 4). If progeny production were dependent on the presence of resistance mutations, a 1,000-fold decrease in MOI should have delayed viral production in the presence of the inhibitors (5, 87–89). (iii) Biological clones isolated from HCV p0 and HCV p100 populations in the absence of inhibitors responded to telaprevir in a manner similar to that seen with their parental, uncloned populations (Fig. 5). The biological clones were obtained by limiting dilution and were grown to form a minimal working stock (see Materials and Methods). The total number of virus production rounds undergone by the biological clones was at least 3-fold lower than the number of rounds involved in the preparation of HCV p0, a population that did not display drug resistance. Thus, the cloning procedure used should have excluded genomes with drug resistance mutations.
The drug resistance phenotype was associated with increased replicative fitness (Fig. 2). Fitness is expected to increase upon virus replication in a constant environment (90–92). High foot-and-mouth disease virus (FMDV) and human immunodeficiency virus type 1 (HIV-1) fitness rendered the viruses partially resistant to lethal mutagenesis (93–95), but a fitness effect in general drug resistance has not been reported. Reduced replication fitness of genotype 2a NS2-5B replicons increased sensitivity to cyclosporine and some other inhibitors but not sensitivity to telaprevir (96). High HCV replicative fitness increased the levels of eIF2α and PKR phosphorylation produced by the parental HCV but, in contrast to other viruses using IRES-dependent translation initiation of their genomic RNA (56, 79–82), did not induce cleavage of factors needed for host cell protein synthesis (Fig. 6). Thus, at a high level of replicative fitness, HCV maintained the mechanisms that result in shutoff of host cell protein synthesis and preferential IRES-dependent translation of the RNA when eIF2α is phosphorylated (46, 85, 97, 98). The increase in the intracellular replicative load that enhances eIF2α and PKR phosphorylation may also underlie the drug resistance phenotype. High levels of replicating RNA and RNA expression products may confer a selective advantage in competition with the number of inhibitor molecules that reach the target viral or cellular proteins at the replication complex.
The results of the present study suggest that cross-resistance among antiviral inhibitors may come about not only through specific amino acid substitutions that confer resistance to more than one drug but also through a general increase in viral fitness, without involving specific drug resistance mutations. Studies are now in progress to attempt to map those mutations that, among the many accumulated in the consensus sequence of HCV p100 and its mutant spectrum (see Tables S8 and S9 [http://www2.cbm.uam.es:8080/cv-303/SupplMatSheldon.pdf]) (45), might be involved in replicative-fitness enhancement. One of the mutants under study includes substitutions N34D in E2, N17D in p7, and Y618F in NS3 and displays increased resistance to IFN-α, telaprevir, and daclatasvir but does not reach the same resistance level as that seen with HCV p100 population.
The results presented here may help interpretation of HCV treatment responses in vivo. High replicative fitness may allow persistence of functional HCV RNAs in the face of antiviral treatment, hence requiring higher drug concentrations or longer treatment duration to achieve antiviral effects comparable to those attained with a virus with low replicative fitness. A study performed with patients subjected to daclatasvir monotherapy suggested that viral fitness, rather than the presence of daclatasvir resistance mutations, determined the dominance and persistence of HCV variants following therapy (99). Prolonged replication of HCV in the liver environment during chronic infection is expected to lead to an increase in viral fitness, thus suggesting that chronicity and advanced liver disease may render the virus less sensitive to antiviral treatment not only through accumulation of inhibitor resistance mutations in an expanded mutant spectrum but also through an increase in replicative fitness.
ACKNOWLEDGMENTS
We thank R. Bartenschlager for the supply of Huh-7 Lunet cells. We also thank M. E. Soria, A. I. de Ávila, and D. García-Cehic for expert technical assistance.
This work was supported by grants BFU2011-23604, SAF2009-10403, PI10/01505, BFU2011-25437, PI13/00456, and reference no. IDI-20110115 CDTI (Centro para el Desarrollo Tecnológico Industrial) from the Ministerio de Ciencia e Innovación, Fundación Ramon Areces, and Marie Curie International outgoing fellowship 219570. CIBERehd (Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas) is funded by Instituto de Salud Carlos III. N.M.B. is supported by a JAE-DOC contract from Consejo Superior de Investigaciones Científicas (CSIC) and J.S. by a Juan de la Cierva contract from CSIC. Support for C.M.R. was from NIH grants AI072613, CA57973, and AI099284, the Greenberg Medical Research Institute, and the Starr Foundation.
Footnotes
Published ahead of print 13 August 2014
Dedicated to John J. Holland (1929-2013).
REFERENCES
- 1. Suzuki F, Akuta N, Suzuki Y, Yatsuji H, Sezaki H, Arase Y, Kawamura Y, Hosaka T, Kobayashi M, Ikeda K, Kobayashi M, Watahiki S, Kumada H. 2007. Selection of a virus strain resistant to entecavir in a nucleoside-naive patient with hepatitis B of genotype H. J. Clin. Virol. 39:149–152. 10.1016/j.jcv.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 2. Domingo E. 1989. RNA virus evolution and the control of viral disease. Prog. Drug Res. 33:93–133 [DOI] [PubMed] [Google Scholar]
- 3. Domingo E, Holland JJ. 1992. Complications of RNA heterogeneity for the engineering of virus vaccines and antiviral agents. Genet. Eng. (N Y) 14:13–31 [DOI] [PubMed] [Google Scholar]
- 4. Le Moing V, Chene G, Carrieri MP, Alioum A, Brun-Vezinet F, Piroth L, Cassuto JP, Moatti JP, Raffi F, Leport C. 2002. Predictors of virological rebound in HIV-1-infected patients initiating a protease inhibitor-containing regimen. AIDS 16:21–29. 10.1097/00002030-200201040-00004 [DOI] [PubMed] [Google Scholar]
- 5. Ribeiro RM, Bonhoeffer S. 2000. Production of resistant HIV mutants during antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 97:7681–7686. 10.1073/pnas.97.14.7681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eggink D, Bontjer I, Langedijk JP, Berkhout B, Sanders RW. 2011. Resistance of human immunodeficiency virus type 1 to a third-generation fusion inhibitor requires multiple mutations in gp41 and is accompanied by a dramatic loss of gp41 function. J. Virol. 85:10785–10797. 10.1128/JVI.05331-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu Z, Kuritzkes DR. 17 August 2011. Interaction of reverse transcriptase (RT) mutations conferring resistance to lamivudine and etravirine: effects on fitness and RT activity of human immunodeficiency virus type 1. J. Virol. 10.1128/JVI.05578-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez-Picado J, Martinez MA. 2008. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res. 134:104–123. 10.1016/j.virusres.2007.12.021 [DOI] [PubMed] [Google Scholar]
- 9. Menéndez-Arias L, Martínez MA, Quiñones-Mateu ME, Martinez-Picado J. 2003. Fitness variations and their impact on the evolution of antiretroviral drug resistance. Curr. Drug Targets Infect. Disord. 3:355–371. 10.2174/1568005033481033 [DOI] [PubMed] [Google Scholar]
- 10. Nijhuis M, Schuurman R, de Jong D, Erickson J, Gustchina E, Albert J, Schipper P, Gulnik S, Boucher CA. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349–2359. 10.1097/00002030-199912030-00006 [DOI] [PubMed] [Google Scholar]
- 11. Deng L, Tang H. 2011. Hepatitis B virus drug resistance to current nucleos(t) ide analogs: mechanisms and mutation sites. Hepatol. Res. 41:1017–1024. 10.1111/j.1872-034X.2011.00873.x [DOI] [PubMed] [Google Scholar]
- 12. Durantel D. 2010. Fitness and infectivity of drug-resistant and cross-resistant hepatitis B virus mutants: why and how is it studied? Antivir. Ther. 15:521–527. 10.3851/IMP1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hiraga N, Imamura M, Abe H, Hayes CN, Kono T, Onishi M, Tsuge M, Takahashi S, Ochi H, Iwao E, Kamiya N, Yamada I, Tateno C, Yoshizato K, Matsui H, Kanai A, Inaba T, Tanaka S, Chayama K. 2011. Rapid emergence of telaprevir resistant hepatitis C virus strain from wild type clone in vivo. Hepatology 54:781–788. 10.1002/hep.24460 [DOI] [PubMed] [Google Scholar]
- 14. Chayama K, Suzuki Y, Kobayashi M, Kobayashi M, Tsubota A, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Murashima N, Ikeda K, Kumada H. 1998. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology 27:1711–1716. 10.1002/hep.510270634 [DOI] [PubMed] [Google Scholar]
- 15. Welsch C, Zeuzem S. 2012. Clinical relevance of HCV antiviral drug resistance. Curr. Opin. Virol. 2:651–655. 10.1016/j.coviro.2012.08.008 [DOI] [PubMed] [Google Scholar]
- 16. Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76:159–216. 10.1128/MMBR.05023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nájera I, Holguín A, Quiñones-Mateu ME, Muñoz-Fernández MA, Nájera R, López-Galíndez C, Domingo E. 1995. Pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J. Virol. 69:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Havlir DV, Eastman S, Gamst A, Richman DD. 1996. Nevirapine-resistant human immunodeficiency virus: kinetics of replication and estimated prevalence in untreated patients. J. Virol. 70:7894–7899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson JA, Li JF, Wei X, Lipscomb J, Irlbeck D, Craig C, Smith A, Bennett DE, Monsour M, Sandstrom P, Lanier ER, Heneine W. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 5:e158. 10.1371/journal.pmed.0050158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lech WJ, Wang G, Yang YL, Chee Y, Dorman K, McCrae D, Lazzeroni LC, Erickson JW, Sinsheimer JS, Kaplan AH. 1996. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J. Virol. 70:2038–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nájera I, Richman DD, Olivares I, Rojas JM, Peinado MA, Perucho M, Najera R, Lopez-Galindez C. 1994. Natural occurrence of drug resistance mutations in the reverse transcriptase of human immunodeficiency virus type 1 isolates. AIDS Res. Hum. Retroviruses 10:1479–1488. 10.1089/aid.1994.10.1479 [DOI] [PubMed] [Google Scholar]
- 22. Toni TA, Asahchop EL, Moisi D, Ntemgwa M, Oliveira M, Masquelier B, Brenner BG, Wainberg MA. 2009. Detection of human immunodeficiency virus (HIV) type 1 M184V and K103N minority variants in patients with primary HIV infection. Antimicrob. Agents Chemother. 53:1670–1672. 10.1128/AAC.01494-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsibris AM, Korber B, Arnaout R, Russ C, Lo CC, Leitner T, Gaschen B, Theiler J, Paredes R, Su Z, Hughes MD, Gulick RM, Greaves W, Coakley E, Flexner C, Nusbaum C, Kuritzkes DR. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683. 10.1371/journal.pone.0005683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cubero M, Esteban JI, Otero T, Sauleda S, Bes M, Esteban R, Guardia J, Quer J. 2008. Naturally occurring NS3-protease-inhibitor resistant mutant A156T in the liver of an untreated chronic hepatitis C patient. Virology 370:237–245. 10.1016/j.virol.2007.10.006 [DOI] [PubMed] [Google Scholar]
- 25. Sarrazin C, Zeuzem S. 2010. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447–462. 10.1053/j.gastro.2009.11.055 [DOI] [PubMed] [Google Scholar]
- 26. Verbinnen T, Van Marck H, Vandenbroucke I, Vijgen L, Claes M, Lin TI, Simmen K, Neyts J, Fanning G, Lenz O. 2010. Tracking the evolution of multiple in vitro hepatitis C virus replicon variants under protease inhibitor selection pressure by 454 deep sequencing. J. Virol. 84:11124–11133. 10.1128/JVI.01217-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simen BB, Simons JF, Hullsiek KH, Novak RM, Macarthur RD, Baxter JD, Huang C, Lubeski C, Turenchalk GS, Braverman MS, Desany B, Rothberg JM, Egholm M, Kozal MJ. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693–701. 10.1086/596736 [DOI] [PubMed] [Google Scholar]
- 28. Martell M, Esteban JI, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. 1992. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J. Virol. 66:3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farci P. 2011. New insights into the HCV quasispecies and compartmentalization. Semin. Liver Dis. 31:356–374. 10.1055/s-0031-1297925 [DOI] [PubMed] [Google Scholar]
- 30. Aghemo A, De Francesco R. 2013. New horizons in hepatitis C antiviral therapy with direct-acting antivirals. Hepatology 58:428–438. 10.1002/hep.26371 [DOI] [PubMed] [Google Scholar]
- 31. Heim MH. 2013. 25 Years of interferon-based treatment of chronic hepatitis C: an epoch coming to an end. Nat. Rev. Immunol. 13:535–542. 10.1038/nri3463 [DOI] [PubMed] [Google Scholar]
- 32. Chatel-Chaix L, Germain MA, Gotte M, Lamarre D. 2012. Direct-acting and host-targeting HCV inhibitors: current and future directions. Curr. Opin. Virol. 2:588–598. 10.1016/j.coviro.2012.08.002 [DOI] [PubMed] [Google Scholar]
- 33. Cummings KJ, Lee SM, West ES, Cid-Ruzafa J, Fein SG, Aoki Y, Sulkowski MS, Goodman SN. 2001. Interferon and ribavirin vs interferon alone in the re-treatment of chronic hepatitis C previously nonresponsive to interferon: a meta-analysis of randomized trials. JAMA 285:193–199. 10.1001/jama.285.2.193 [DOI] [PubMed] [Google Scholar]
- 34. Di Bisceglie AM, Thompson J, Smith-Wilkaitis N, Brunt EM, Bacon BR. 2001. Combination of interferon and ribavirin in chronic hepatitis C: re-treatment of nonresponders to interferon. Hepatology 33:704–707. 10.1053/jhep.2001.22346 [DOI] [PubMed] [Google Scholar]
- 35. McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339:1485–1492 [DOI] [PubMed] [Google Scholar]
- 36. Lange CM, Jacobson IM, Rice CM, Zeuzem S. 9 January 2014. Emerging therapies for the treatment of hepatitis C. EMBO Mol. Med. 10.1002/emmm.201303131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zeuzem S, Dusheiko GM, Salupere R, Mangia A, Flisiak R, Hyland RH, Illeperuma A, Svarovskaia E, Brainard DM, Symonds WT, Subramanian GM, McHutchison JG, Weiland O, Reesink HW, Ferenci P, Hezode C, Esteban R. 2014. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N. Engl. J. Med. 370:1993–2001. 10.1056/NEJMoa1316145 [DOI] [PubMed] [Google Scholar]
- 38. Gane EJ, Stedman CA, Hyland RH, Ding X, Svarovskaia E, Symonds WT, Hindes RG, Berrey MM. 2013. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N. Engl. J. Med. 368:34–44. 10.1056/NEJMoa1208953 [DOI] [PubMed] [Google Scholar]
- 39. Jacobson IM, Gordon SC, Kowdley KV, Yoshida EM, Rodriguez-Torres M, Sulkowski MS, Shiffman ML, Lawitz E, Everson G, Bennett M, Schiff E, Al-Assi MT, Subramanian GM, An D, Lin M, McNally J, Brainard D, Symonds WT, McHutchison JG, Patel K, Feld J, Pianko S, Nelson DR. 2013. Sofosbuvir for hepatitis C genotype 2 or 3 in patients without treatment options. N. Engl. J. Med. 368:1867–1877. 10.1056/NEJMoa1214854 [DOI] [PubMed] [Google Scholar]
- 40. Afdhal N, Reddy KR, Nelson DR, Lawitz E, Gordon SC, Schiff E, Nahass R, Ghalib R, Gitlin N, Herring R, Lalezari J, Younes ZH, Pockros PJ, Di Bisceglie AM, Arora S, Subramanian GM, Zhu Y, Dvory-Sobol H, Yang JC, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Sulkowski M, Kwo P. 2014. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N. Engl. J. Med. 370:1483–1493. 10.1056/NEJMoa1316366 [DOI] [PubMed] [Google Scholar]
- 41. Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. 10.1126/science.1114016 [DOI] [PubMed] [Google Scholar]
- 42. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796. 10.1038/nm1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299. 10.1073/pnas.0503596102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB. 2008. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 48:1843–1850. 10.1002/hep.22550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perales C, Beach NM, Gallego I, Soria ME, Quer J, Esteban JI, Rice C, Domingo E, Sheldon J. 2013. Response of hepatitis C virus to long-term passage in the presence of alpha interferon: multiple mutations and a common phenotype. J. Virol. 87:7593–7607. 10.1128/JVI.02824-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, Wakita T, Meurs EF. 2010. Hepatitis C virus controls interferon production through PKR activation. PLoS One 5:e10575. 10.1371/journal.pone.0010575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689–2699. 10.1128/JVI.79.5.2689-2699.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014. 10.1128/JVI.76.24.13001-13014.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 28:167–171. 10.1038/nbt.1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reed LJ, Muench H. 1938. A simple method for estimating fifty per cent endpoint. Am. J. Hyg. (Lond) 27:493–497 [Google Scholar]
- 51. Vandamme AM, Witvrouw M, Pannecouque C, Balzarini J, Van Laethem K, Schmit JC, Desmyter J, De Clercq E. 2000. Evaluating clinical isolates for their phenotypic and genotypic resistance against anti-HIV drugs. Methods Mol. Med. 24:223–258. 10.1385/1-59259-245-7:223 [DOI] [PubMed] [Google Scholar]
- 52. Agudo R, Ferrer-Orta C, Arias A, de la Higuera I, Perales C, Perez-Luque R, Verdaguer N, Domingo E. 2010. A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape. PLoS Pathog. 6:e1001072. 10.1371/journal.ppat.1001072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ortega-Prieto AM, Sheldon J, Grande-Perez A, Tejero H, Gregori J, Quer J, Esteban JI, Domingo E, Perales C. 2013. Extinction of hepatitis C virus by ribavirin in hepatoma cells involves lethal mutagenesis. PLoS One 8:e71039. 10.1371/journal.pone.0071039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sánchez G, Bosch A, Gómez-Mariano G, Domingo E, Pintó RM. 2003. Evidence for quasispecies distributions in the human hepatitis A virus genome. Virology 315:34–42. 10.1016/S0042-6822(03)00483-5 [DOI] [PubMed] [Google Scholar]
- 55. Perales C, Mateo R, Mateu MG, Domingo E. 2007. Insights into RNA virus mutant spectrum and lethal mutagenesis events: replicative interference and complementation by multiple point mutants. J. Mol. Biol. 369:985–1000. 10.1016/j.jmb.2007.03.074 [DOI] [PubMed] [Google Scholar]
- 56. Rodríguez Pulido M, Serrano P, Sáiz M, Martínez-Salas E. 2007. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology 364:466–474. 10.1016/j.virol.2007.03.013 [DOI] [PubMed] [Google Scholar]
- 57. Ojosnegros S, Garcia-Arriaza J, Escarmis C, Manrubia SC, Perales C, Arias A, Mateu MG, Domingo E. 2011. Viral genome segmentation can result from a trade-off between genetic content and particle stability. PLoS Genet. 7:e1001344. 10.1371/journal.pgen.1001344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588–598. 10.1128/JVI.01534-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Perales C, Beach NM, Sheldon J, Domingo E. 2014. Molecular basis of interferon resistance in hepatitis C virus. Curr. Opin. Virol. 8:38–44. 10.1016/j.coviro.2014.05.003 [DOI] [PubMed] [Google Scholar]
- 60. Pawlotsky JM. 2014. What are the pros and cons of the use of host-targeted agents against hepatitis C? Antiviral Res. 105:22–25. 10.1016/j.antiviral.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lange CM, Zeuzem S. 2013. Perspectives and challenges of interferon-free therapy for chronic hepatitis C. J. Hepatol. 58:583–592. 10.1016/j.jhep.2012.10.019 [DOI] [PubMed] [Google Scholar]
- 62. Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348. 10.1038/nature04388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pfeiffer JK, Kirkegaard K. 2005. Increased fidelity reduces poliovirus fitness under selective pressure in mice. PLoS Pathog. 1:e11. 10.1371/journal.ppat.0010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rozen-Gagnon K, Stapleford KA, Mongelli V, Blanc H, Failloux AB, Saleh MC, Vignuzzi M. 2014. Alphavirus mutator variants present host-specific defects and attenuation in mammalian and insect models. PLoS Pathog. 10:e1003877. 10.1371/journal.ppat.1003877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gnädig NF, Beaucourt S, Campagnola G, Bordería AV, Sanz-Ramos M, Gong P, Blanc H, Peersen OB, Vignuzzi M. 2012. Coxsackievirus B3 mutator strains are attenuated in vivo. Proc. Natl. Acad. Sci. U. S. A. 109:E2294–E2303. 10.1073/pnas.1204022109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Coffey LL, Beeharry Y, Borderia AV, Blanc H, Vignuzzi M. 2011. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc. Natl. Acad. Sci. U. S. A. 108:16038–16043. 10.1073/pnas.1111650108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Robida JM, Nelson HB, Liu Z, Tang H. 2007. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J. Virol. 81:5829–5840. 10.1128/JVI.02524-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fernandes F, Ansari IU, Striker R. 2010. Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis C NS5A. PLoS One 5:e9815. 10.1371/journal.pone.0009815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu Z, Robida JM, Chinnaswamy S, Yi G, Robotham JM, Nelson HB, Irsigler A, Kao CC, Tang H. 2009. Mutations in the hepatitis C virus polymerase that increase RNA binding can confer resistance to cyclosporine A. Hepatology 50:25–33. 10.1002/hep.22987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fernandes F, Poole DS, Hoover S, Middleton R, Andrei AC, Gerstner J, Striker R. 2007. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology 46:1026–1033. 10.1002/hep.21809 [DOI] [PubMed] [Google Scholar]
- 71. Puyang X, Poulin DL, Mathy JE, Anderson LJ, Ma S, Fang Z, Zhu S, Lin K, Fujimoto R, Compton T, Wiedmann B. 2010. Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors. Antimicrob. Agents Chemother. 54:1981–1987. 10.1128/AAC.01236-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Arai M, Tsukiyama-Kohara K, Takagi A, Tobita Y, Inoue K, Kohara M. 2014. Resistance to cyclosporin A derives from mutations in hepatitis C virus nonstructural proteins. Biochem. Biophys. Res. Commun. 448:56–62. 10.1016/j.bbrc.2014.04.053 [DOI] [PubMed] [Google Scholar]
- 73. Nakamoto S, Kanda T, Wu S, Shirasawa H, Yokosuka O. 2014. Hepatitis C virus NS5A inhibitors and drug resistance mutations. World J. Gastroenterol. 20:2902–2912. 10.3748/wjg.v20.i11.2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Martínez-Salas E, Pacheco A, Serrano P, Fernandez N. 2008. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 89:611–626. 10.1099/vir.0.83426-0 [DOI] [PubMed] [Google Scholar]
- 75. Niepmann M. 2009. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim. Biophys. Acta 1789:529–541. 10.1016/j.bbagrm.2009.05.002 [DOI] [PubMed] [Google Scholar]
- 76. Marissen WE, Triyoso D, Younan P, Lloyd RE. 2004. Degradation of poly(A)-binding protein in apoptotic cells and linkage to translation regulation. Apoptosis 9:67–75. 10.1023/B:APPT.0000012123.62856.20 [DOI] [PubMed] [Google Scholar]
- 77. Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 277:50286–50292. 10.1074/jbc.M203029200 [DOI] [PubMed] [Google Scholar]
- 78. Karbstein K. 2011. Inside the 40S ribosome assembly machinery. Curr. Opin. Chem. Biol. 15:657–663. 10.1016/j.cbpa.2011.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dougherty JD, Park N, Gustin KE, Lloyd RE. 2010. Interference with cellular gene expression, p 165–180 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 80. Walsh D, Mohr I. 2011. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 9:860–875. 10.1038/nrmicro2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gradi A, Foeger N, Strong R, Svitkin YV, Sonenberg N, Skern T, Belsham GJ. 2004. Cleavage of eukaryotic translation initiation factor 4GII within foot-and-mouth disease virus-infected cells: identification of the L-protease cleavage site in vitro. J. Virol. 78:3271–3278. 10.1128/JVI.78.7.3271-3278.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 24:1779–1790. 10.1128/MCB.24.4.1779-1790.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gosert R, Chang KH, Rijnbrand R, Yi M, Sangar DV, Lemon SM. 2000. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 20:1583–1595. 10.1128/MCB.20.5.1583-1595.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. U. S. A. 90:7642–7646. 10.1073/pnas.90.16.7642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Garaigorta U, Chisari FV. 2009. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 6:513–522. 10.1016/j.chom.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 5:e1000546. 10.1371/journal.ppat.1000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Perales C, Agudo R, Manrubia SC, Domingo E. 2011. Influence of mutagenesis and viral load on the sustained low-level replication of an RNA virus. J. Mol. Biol. 407:60–78. 10.1016/j.jmb.2011.01.026 [DOI] [PubMed] [Google Scholar]
- 88. Bonhoeffer S, May RM, Shaw GM, Nowak MA. 1997. Virus dynamics and drug therapy. Proc. Natl. Acad. Sci. U. S. A. 94:6971–6976. 10.1073/pnas.94.13.6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hatano H, Lampiris H, Fransen S, Gupta S, Huang W, Hoh R, Martin JN, Lalezari J, Bangsberg D, Petropoulos C, Deeks SG. 2010. Evolution of integrase resistance during failure of integrase inhibitor-based antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 54:389–393. 10.1097/QAI.0b013e3181c42ea4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Novella IS, Duarte EA, Elena SF, Moya A, Domingo E, Holland JJ. 1995. Exponential increases of RNA virus fitness during large population transmissions. Proc. Natl. Acad. Sci. U. S. A. 92:5841–5844. 10.1073/pnas.92.13.5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Domingo E, Holland JJ. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151–178. 10.1146/annurev.micro.51.1.151 [DOI] [PubMed] [Google Scholar]
- 92. Escarmís C, Dávila M, Domingo E. 1999. Multiple molecular pathways for fitness recovery of an RNA virus debilitated by operation of Muller's ratchet. J. Mol. Biol. 285:495–505. 10.1006/jmbi.1998.2366 [DOI] [PubMed] [Google Scholar]
- 93. Sierra S, Dávila M, Lowenstein PR, Domingo E. 2000. Response of foot-and-mouth disease virus to increased mutagenesis. Influence of viral load and fitness in loss of infectivity. J. Virol. 74:8316–8323. 10.1128/JVI.74.18.8316-8323.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pariente N, Sierra S, Lowenstein PR, Domingo E. 2001. Efficient virus extinction by combinations of a mutagen and antiviral inhibitors. J. Virol. 75:9723–9730. 10.1128/JVI.75.20.9723-9730.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tapia N, Fernandez G, Parera M, Gomez-Mariano G, Clotet B, Quinones-Mateu M, Domingo E, Martinez MA. 2005. Combination of a mutagenic agent with a reverse transcriptase inhibitor results in systematic inhibition of HIV-1 infection. Virology 338:1–8. 10.1016/j.virol.2005.05.008 [DOI] [PubMed] [Google Scholar]
- 96. Madan V, Paul D, Lohmann V, Bartenschlager R. 2014. Inhibition of HCV replication by cyclophilin antagonists is linked to replication fitness and occurs by inhibition of membranous web formation. Gastroenterology 146:1361–1372.e9. 10.1053/j.gastro.2014.01.055 [DOI] [PubMed] [Google Scholar]
- 97. Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. 2008. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 15:836–841. 10.1038/nsmb.1445 [DOI] [PubMed] [Google Scholar]
- 98. Kim JH, Park SM, Park JH, Keum SJ, Jang SK. 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 30:2454–2464. 10.1038/emboj.2011.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wang C, Sun JH, O'Boyle DR, II, Nower P, Valera L, Roberts S, Fridell RA, Gao M. 2013. Persistence of resistant variants in hepatitis C virus-infected patients treated with the NS5A replication complex inhibitor daclatasvir. Antimicrob. Agents Chemother. 57:2054–2065. 10.1128/AAC.02494-12 [DOI] [PMC free article] [PubMed] [Google Scholar]