ABSTRACT
Respiratory syncytial virus (RSV) is the leading infectious cause of severe respiratory disease in infants and a major cause of respiratory illness in the elderly. There remains an unmet vaccine need despite decades of research. Insufficient potency, homogeneity, and stability of previous RSV fusion protein (F) subunit vaccine candidates have hampered vaccine development. RSV F and related parainfluenza virus (PIV) F proteins are cleaved by furin during intracellular maturation, producing disulfide-linked F1 and F2 fragments. During cell entry, the cleaved Fs rearrange from prefusion trimers to postfusion trimers. Using RSV F constructs with mutated furin cleavage sites, we isolated an uncleaved RSV F ectodomain that is predominantly monomeric and requires specific cleavage between F1 and F2 for self-association and rearrangement into stable postfusion trimers. The uncleaved RSV F monomer is folded and homogenous and displays at least two key RSV-neutralizing epitopes shared between the prefusion and postfusion conformations. Unlike the cleaved trimer, the uncleaved monomer binds the prefusion-specific monoclonal antibody D25 and human neutralizing immunoglobulins that do not bind to postfusion F. These observations suggest that the uncleaved RSV F monomer has a prefusion-like conformation and is a potential prefusion subunit vaccine candidate.
IMPORTANCE RSV is the leading infectious cause of severe respiratory disease in infants and a major cause of respiratory illness in the elderly. Development of an RSV vaccine was stymied when a clinical trial using a formalin-inactivated RSV virus made disease, following RSV infection, more severe. Recent studies have defined the structures that the RSV F envelope glycoprotein adopts before and after virus entry (prefusion and postfusion conformations, respectively). Key neutralization epitopes of prefusion and postfusion RSV F have been identified, and a number of current vaccine development efforts are focused on generating easily produced subunit antigens that retain these epitopes. Here we show that a simple modification in the F ectodomain results in a homogeneous protein that retains critical prefusion neutralizing epitopes. These results improve our understanding of RSV F protein folding and structure and can guide further vaccine design efforts.
INTRODUCTION
Respiratory syncytial virus (RSV) is a member of the Paramyxoviridae family of RNA viruses, which also includes human metapneumovirus, measles virus, mumps virus, Newcastle disease virus (NDV), human parainfluenzavirus 1 (PIV1) to PIV4, and PIV5. RSV is the major cause of bronchiolitis and pneumonia in infants. It is the leading cause of infant hospitalization in developed countries and is responsible for an estimated 200,000 infant deaths in developing countries each year (1, 2). RSV also causes substantial morbidity and mortality among the elderly (3, 4). There is no specific antiviral treatment recommended for RSV infection, and the only currently available prophylactic is a monoclonal antibody, palivizumab (Synagis), used to prevent disease in the highest-risk infants (5). The cost of palivizumab prevents general use, and the need for a vaccine is clear. However, despite decades of research there remains no licensed vaccine for RSV. Development of a vaccine was stymied in the 1960s when a formalin-inactivated RSV vaccine candidate made subsequent RSV disease more severe (6). Increased structural understanding of key RSV neutralization epitopes has supported a resurgence of interest in developing an RSV subunit-based vaccine.
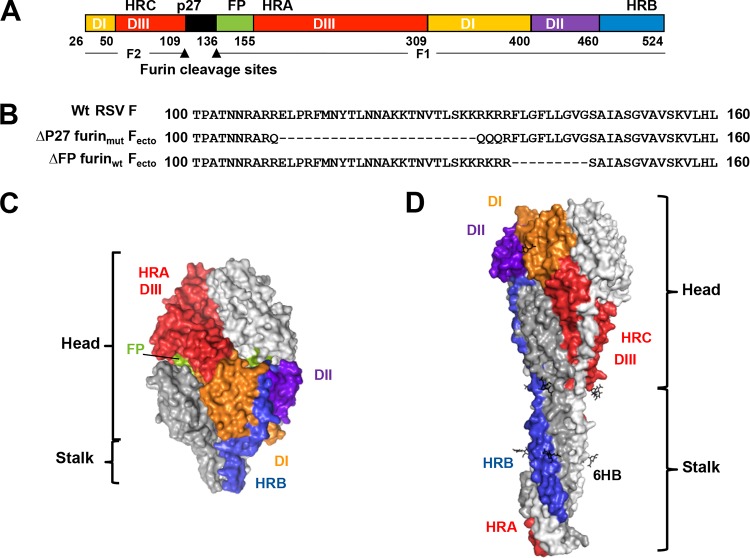
RSV-neutralizing antibodies target the two major RSV surface antigens, the attachment protein (G) and the fusion protein (F) (7). G is variable in sequence, whereas F is highly conserved among strains, making F the more attractive vaccine antigen. RSV F is a type I viral fusion protein responsible for driving fusion of the viral envelope with host cell membranes during viral entry. Crystal structures of RSV F ectodomain trimers have documented two conformational states—prefusion and postfusion (Fig. 1C and D) (8–11). In the prefusion conformation (Fig. 1C), the heptad repeat A (HRA) region is associated with the globular head and the tip of the fusion peptide is mostly buried in the center of the protein. In the postfusion conformation (Fig. 1D), HRA and the fusion peptide (not present in published crystal structures) have extended from the globular head to attach to the target membrane and the heptad repeat B (HRB) region has rearranged to associate with the HRA region, forming a stable 6-helix bundle. This rearrangement places the host membrane bound by the fusion peptide and the viral membrane bound by the transmembrane region in close proximity to drive membrane fusion. The commercial product RespiGam (RSV immune globulin; Medimmune), made by purifying antibodies from human sera with high RSV-neutralizing titers, was shown to include antibodies specific for the prefusion F conformation (12). Indeed, depleting RSV immune globulin of antibodies that bind G and postfusion F demonstrated that the prefusion-specific F antibodies were predominantly responsible for virus neutralization by the product. The crystal structure of an RSV F ectodomain (Fecto) stabilized by the C-terminal addition of a trimerization tag (foldon) bound to the FAb of the prefusion-specific antibody D25 identified a new antigenic site designated site Ø (9). Site Ø is formed in prefusion RSV Fecto by the packing of the HRA region against the rest of the F globular head (a structural feature not shared by postfusion F) (Fig. 1C and D). In addition, a prefusion RSV Fecto antigen with a trimerization tag and mutations that stabilized the HRA-globular head interactions elicited a higher neutralizing titer than did postfusion Fecto in mice and nonhuman primates (11).
FIG 1.
RSV F ectodomain structure. (A) Linear diagram listing residue numbers corresponding to the N terminus of each segment. The furin cleavage sites (arrowheads) divide the protein into F1 and F2. DI to DIII, domains I to III; p27, excised peptide; FP, fusion peptide; HRA, -B, and -C, heptad repeats A, B, and C. (B) Sequences showing the mutations introduced into p27 and the FP region of Fecto constructs. (Top) Wild-type RSV F sequence with p27 and fusion peptide unaltered. (Middle) ΔP27 furinmut Fecto sequence with mutations to the furin sites and deletion of p27 residues. (Bottom) ΔFP furinwt Fecto sequence with fusion peptide residues deleted. Deleted residues are shown as hyphens. (C) Surface representation of the prefusion RSV Fecto (9). One subunit shows domains and heptad repeat regions colored as in panel A; the other two subunits are white and gray. (D) Surface representation of the postfusion RSV Fecto (8), colored as in panel C to highlight the rearrangement of domains and heptad repeat regions after conformational change. Note the HRA (red) largely buried by the HRB (blue) upon formation of the 6-helix bundle (6HB).
Fusion proteins are expressed as uncleaved F0 precursors (Fig. 1A and B). Crystal structures of uncleaved PIV3, PIV5, and NDV Fecto revealed that furin cleavage is not required for these proteins to trimerize (13–15). PIV3 Fecto was crystalized in the postfusion conformation, whereas PIV5 Fecto was trapped in the prefusion conformation by the addition of a C-terminal GCN trimerization tag (14). Unlike PIV Fs, each of which contains a single furin cleavage site, RSV F has two sites separated by a 27-amino-acid fragment, p27 (Fig. 1B). Activation of RSV F for membrane fusion requires cleavage by furin at the two sites (16). Our initial attempts to generate a prefusion RSV Fecto trimer relied on mutating the furin cleavage sites and adding a GCN trimerization tag, similar to the strategy used to generate the prefusion PIV5 Fecto (14). However, addition of the GCN trimerization domain to the uncleaved RSV Fecto greatly reduced the amount of secreted protein. In the course of these manipulations, we observed that uncleaved RSV Fecto was predominantly monomeric in solution. We demonstrate that this monomeric species is a folded, soluble protein which harbors key neutralizing epitopes, including the site Ø prefusion epitope. This uncleaved species might represent a prefusion Fecto vaccine candidate.
MATERIALS AND METHODS
Fusion protein construct design, expression, and purification.
RSV Fecto constructs with mutations to the furin cleavage sites and fusion peptide region (residues 109 to 148) were designed (Fig. 1A and B). Δp27 furinmut Fecto contains a deletion of 23 residues from the p27 fragment and point mutations to the furin cleavage sites. These point mutations prevent cleavage by intercellular furin but permit in vitro cleavage by trypsin. ΔFP furinwt Fecto retains the wild-type furin cleavage sites but has a fusion peptide deletion. Unlike Δp27 furinmut Fecto, which is purified as an uncleaved F0 species, ΔFP furinwt Fecto is cleaved by the cell into the F1/F2 species. DNA constructs encoding Fecto with these mutations and a C-terminal histidine tag were synthesized (GeneArt) and cloned into the pFastBac baculovirus system (Invitrogen). RSV Fecto constructs were expressed in 1-liter cultures for 3 days and purified by nickel affinity followed by size exclusion chromatography (Superdex P200; GE Healthcare). For limited proteolysis studies, Δp27 furinmut RSV Fecto was digested with trypsin (Sigma) at a weight ratio of 1:1,000 trypsin to Δp27 furinmut RSV Fecto, and the resulting cleaved protein was purified from a size exclusion column void volume.
Size exclusion chromatography and multiangle light scattering (SEC-MALS).
Size exclusion high pressure liquid chromatography (HPLC) was performed using a Waters 2685 separation module HPLC instrument with an isocratic flow rate of 0.75 ml/min coupled to a multiangle light scattering (MALS) system. The mobile phase consisted of 2× phosphate-buffered saline (PBS). The size exclusion column (WTC-030S5; Wyatt Technology) was used at ambient temperature with a run time of 30 min. UV was detected at 280 nm (PDA 2996; Waters), and a light-scattering detector (miniDawn TREOS; Wyatt Technology) using a laser wavelength of 690.0 nm and a refractive-index detector (Optilab rEX; Wyatt Technology) were added in series. A gel filtration standard solution (Bio-Rad) was used to generate a Stokes radius standard curve. Immediately prior to the measurement of the samples, the system was calibrated using a known bovine serum albumin (BSA) solution.
A 100-μl injection for a target column load of 100 μg protein (RSV Fecto alone or a 1:1 molar ratio of RSV Fecto:FAb) was used. Peak retention times were used to estimate the molecular mass of RSV Fecto or RSV Fecto:FAb complexes based on the Stokes radius standard curve. Protein concentrations for identified peaks were calculated using calculated UV extinction coefficients, and the molecular masses for RSV Fecto or RSV Fecto:FAb complexes were determined from multiangle light scattering data using Astra 5.3.4 software (Wyatt Technologies).
Binding studies by surface plasmon resonance.
The affinity of RSV Fecto constructs for motavizumab or 101F FAbs was measured by surface plasmon resonance (SPR) with a Biacore T100 instrument. FAbs were directly immobilized on CM5 sensor chips using amine coupling at very low levels (75 response units), and RSV Fecto construct preparations at various concentrations were injected at a high flow rate (50 μl/min) to avoid avidity effects and higher than 1:1 binding interactions. The data were processed using Biacore T100 evaluation software and double referenced by subtraction of the blank surface and buffer-only injection values before global fitting of the data to a 1:1 binding model.
Depletion of RSV immune globulin antibodies.
Antibody depletion was carried out by a method similar to that previously described (17). Briefly, the Δp27 furinmut Fecto and ΔFP furinwt Fecto preparations were bound covalently to CNBr-activated Sepharose 4B (GE Healthcare) according to the manufacturer's instructions (500 μg of protein/160 mg Sepharose). The resins were packed into columns that were equilibrated in PBS. Three milligrams of RSV immune globulin was loaded on each of these columns. The unbound material (referred to as Fecto-depleted RSV immune globulin) was collected. After the column was washed with PBS, bound antibodies were eluted with glycine-HCl (pH 2.5), immediately neutralized with saturated Tris, and concentrated, and the buffer was exchanged for PBS. The eluted antibodies are referred to as Fecto-captured RSV immune globulin.
Enzyme-linked immunosorbent assay with depleted RSV immune globulin antibodies.
Enzyme-linked immunosorbent assay (ELISA) of depleted antibodies was performed as previously described (12). Briefly, purified 101F antibody was used to coat 96-well microtiter plates overnight at 4°C. Nonspecific antibody binding was blocked with 2% porcine serum in PBS with 0.05% Tween 20. An excess of purified proteins (either Δp27 furinmut Fecto or ΔFP furinwt Fecto, 2 μg/well) was added to each well, and incubation continued for 1 h at 37°C followed by the addition of serially diluted antibodies and incubation for 1 h at 37°C. The plates were extensively washed with water after each step. Finally, bound antibodies were detected with peroxidase-labeled rabbit anti-human immunoglobulins and o-phenylenediamine (OPD) as the substrate (GE Healthcare). The reaction was stopped with 2 N sulfuric acid, and absorbance was read at 490 nm.
Neutralization assay using depleted RSV immune globulin antibodies.
RSV neutralization by depleted RSV immune globulin antibodies was performed using a microneutralization assay described originally by Anderson et al. in 1988 and modified by Martínez and Melero in 1998 (18, 19). Briefly, immunoglobulin dilutions were incubated with 1 × 105 PFU of the RSV Long strain for 30 min at 37°C in a total volume of 50 μl. These mixtures were used to infect 1 × 105 HEp-2 cells growing in 96-well plates with Dulbecco's modified Eagle's medium (DMEM) supplemented with 2.5% fetal calf serum (FCS) that had been inactivated 30 min at 60°C. After 1 h of adsorption, DMEM with 2.5% FCS was added, and the cultures were incubated 72 h at 37°C with 5% CO2. The plates were washed three times with 0.05% Tween 20 in PBS and fixed with 80% cold acetone in PBS. After air drying, viral antigen production in the fixed monolayers was measured by ELISA with a pool of anti-G and anti-F murine antibodies and a horseradish peroxidase (HRP)-labeled anti-mouse immunoglobulin preparation (Sigma).
Electron microscopy (EM).
RSV Fecto preparations (30 μg/ml in 25 mM Tris, 300 mM NaCl) were absorbed onto a 400-mesh-sized carbon-coated grid (Electron Microscopy Sciences) and stained with 0.75% uranyl formate. A JEOL 1200EX microscope operated at 80 kV was used to analyze the samples. Micrographs were taken at a magnification of ×65,000.
RESULTS
ΔP27 furinmut and ΔFP furinwt Fecto oligomerization states by SEC-MALS.
PIV F ectodomains with furin cleavage site mutations form trimers in the postfusion F trimer conformation (13, 15, 20). To determine if ΔP27 furinmut Fecto similarly formed trimers, we analyzed its oligomerization state with analytical SEC coupled with MALS (Fig. 2). ΔP27 furinmut Fecto principally eluted from the analytical column at a retention volume indicating a monomer, as estimated by Stokes radius. In addition, MALS analysis of the principal ΔP27 furinmut Fecto peak gave a mass consistent with a monomer. A minor peak in the ΔP27 furinmut Fecto analysis ran consistent with the retention time of a trimer, suggesting some protein in this preparation was competent for trimerization in solution (Fig. 2, red arrow). For comparison, we performed SEC-MALS analysis of ΔFP furinwt Fecto, which is known by its crystal structure and EM analysis to be a postfusion trimer (8). ΔFP furinwt Fecto eluted at a retention time consistent with a trimer, and analysis of the protein peak by MALS gave a mass consistent with a trimer (Fig. 2).
FIG 2.
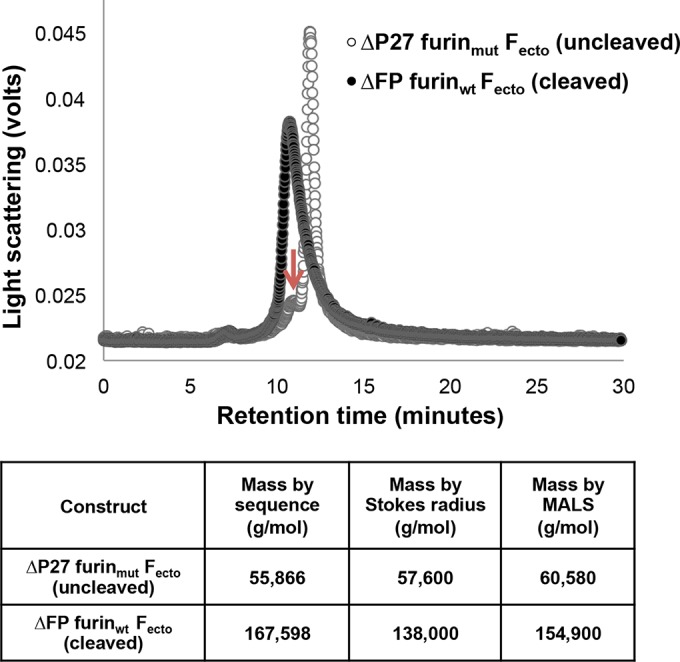
Analytical size exclusion chromatography analysis of RSV Fecto constructs. (Top) Open circles represent the chromatogram of ΔP27 furinmut Fecto, and closed circles represent the chromatogram of ΔFP furinwt Fecto. The principal peak of the cleaved ΔFP furinwt Fecto is at approximately 10.5 min. The principal peak of the uncleaved ΔP27 furinmut Fecto is at approximately 11.2 min, with a minor shoulder peak at approximately 10.5 min (arrow). (Bottom) Masses of Fecto constructs measured by SEC-MALS. The molecular masses estimated according to the protein sequence for a monomer ΔP27 furinmut Fecto and trimer ΔFP furinwt Fecto are shown in the second column. The masses for the RSV Fecto constructs as measured by Stokes radius and MALS analysis are shown in the third and fourth columns, respectively.
Motavizumab and 101F FAb binding to ΔP27 furinmut Fecto by SPR and SEC.
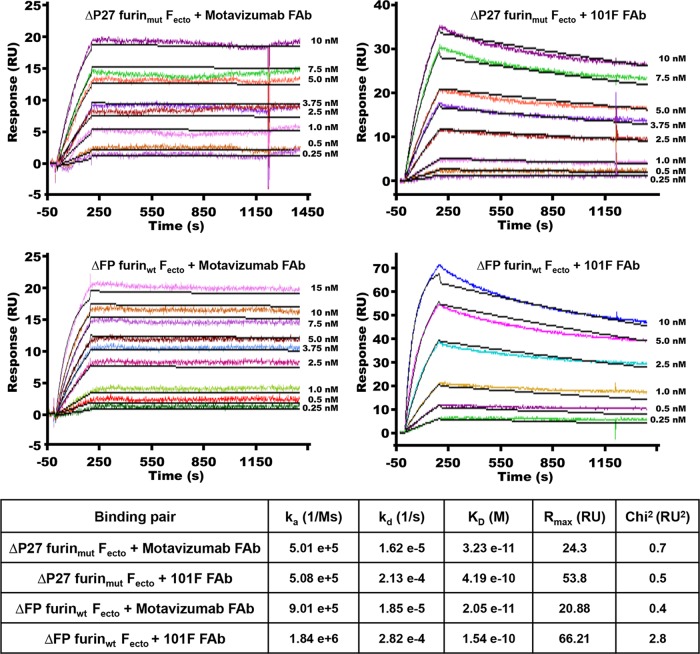
Using SPR, we tested whether ΔP27 furinmut Fecto binds FAbs from two structurally characterized neutralizing antibodies, motavizumab, which binds site II (defined as site A in reference 5), and 101F, which binds site IV (defined as site C in reference 5) (Fig. 3). Both ΔP27 furinmut and ΔFP furinwt Fecto bound tightly to motavizumab and 101F FAbs. The KD values (equilibrium dissociation constants) for the ΔP27 furinmut Fecto-motavizumab FAb interaction (3.23 × 10−11 M) and the ΔFP furinwt Fecto-motavizumab FAb interaction (2.05 × 10−11 M) were similar, as were the KD values for the ΔP27 furinmut Fecto-101F FAb (4.19 × 10−10 M) and ΔFP furinwt Fecto-101F FAb (1.54 × 10−10 M) interactions.
FIG 3.
Binding studies by SPR of Fecto and FAbs. 101F and motavizumab FAbs were immobilized on a sensor chip, various concentrations of Fecto were injected, and mass absorption response units (RU) were monitored as a function of time (sensorgrams are labeled according to Fecto construct, FAb chip, and concentration, as indicated). Fitting the resulting curves to a 1:1 binding stoichiometry resulted in ka (association rate constant), kd (dissociation rate constant), and fit values as indicated in the table. Rmax, maximum analyte binding capacity of the surface in RU. Ms, mol × seconds.
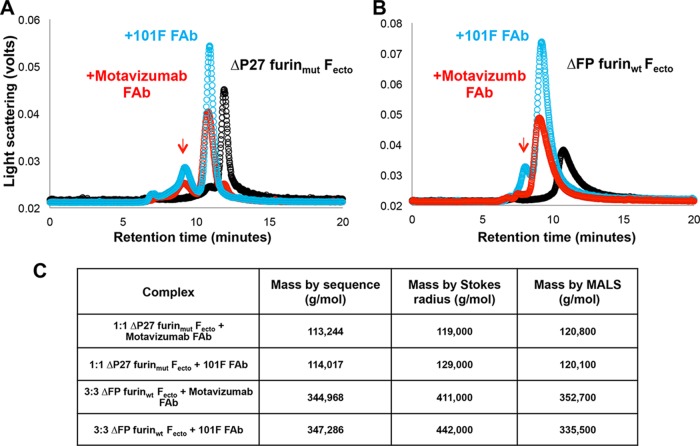
To determine if FAb binding could stabilize trimerization of ΔP27 furinmut Fecto, we incubated the ΔP27 furinmut Fecto with a molar equivalent of either motavizumab or 101F FAb and analyzed the resulting complex by SEC-MALS (Fig. 4). The retention times of the principal peaks for the ΔP27 furinmut Fecto-motavizumab FAb complex and for the ΔP27 furinmut Fecto-101F FAb complex were consistent with 1:1 F monomer:FAb species as determined by Stokes radius (Fig. 4A). MALS analysis also gave masses consistent with 1:1 F monomer:FAb complexes. In comparison, Stokes radius and MALS analysis results for the peaks from ΔFP furinwt Fecto-FAb complexes were consistent with those for 3:3 F-FAb complexes (or an F trimer bound to three FAbs) (Fig. 4B). As previously discussed, a shoulder peak with a retention time consistent with an F trimer was observed in ΔP27 furinmut Fecto preparations and corresponding shoulder peaks (Fig. 4B, red arrow) were observed in the ΔP27 furinmut Fecto-FAb complex preparations with a retention time consistent with F trimers bound by three FAbs.
FIG 4.
Analytical size exclusion chromatography analysis of RSV Fecto constructs bound to FAbs. (A) Black circles represent the chromatogram of ΔP27 furinmut Fecto, and blue and red circles represent the chromatogram of 101F FAb-bound and motavizumab FAb-bound ΔP27 furinmut Fecto, respectively. Upon FAb binding, the principal peak of ΔP27 furinmut Fecto shifts from approximately 11.2 min to 10.7 min. An arrow shows a shoulder peak shifting to a retention time of approximately 9.5 min. (B) Chromatogram of ΔFP furinwt Fecto with or without FAb binding, colored as in panel A. Upon FAb binding, the principal peak of ΔFP furinwt Fecto shifts from approximately 10.5 min to approximately 9.5 min. (C) Masses of Fecto constructs predicted by the protein sequence for a monomer ΔFP furinwt Fecto and trimer ΔP27 furinmut Fecto bound to a single Fab or to three FAbs, respectively, are shown in the second column. Masses for the RSV Fecto constructs bound to FAbs as measured by Stokes radius and MALS analysis are shown in columns three and four, respectively.
ΔP27 furinmut Fecto binding to D25 FAb by SPR and SEC.
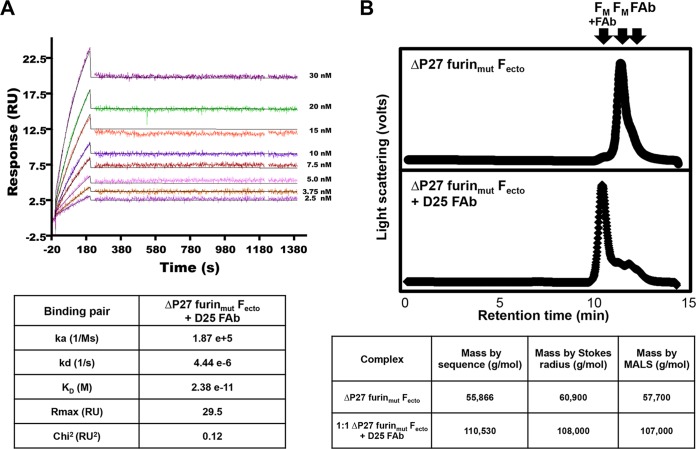
D25 FAb binds the prefusion Fecto but does not bind postfusion Fecto (9). To determine if ΔP27 furinmut Fecto is competent to bind the prefusion site Ø-specific D25 FAb, we tested binding by SPR and SEC (Fig. 5). The KD for the ΔP27 furinmut Fecto-D25 FAb interaction was 2.38 × 10−11 M. In the RSV Fecto -D25 crystal structure, the D25 FAb has principal contacts with a single protomer and modest contacts with an adjacent protomer, suggesting that binding of D25 might occur with F trimers rather than monomers (9). To determine if binding to D25 FAb induced trimerization of ΔP27 furinmut Fecto, we incubated the F protein with a molar equivalent of D25 FAb and analyzed the complex by SEC-MALS (Fig. 5B). The retention time and Stokes radius of the D25-bound ΔP27 furinmut Fecto was consistent with 1:1 F monomer:FAb species. Similarly, MALS analysis of the D25-bound ΔP27 furinmut Fecto gave a mass consistent with 1:1 F monomer:FAb species. We conclude that binding to D25 FAb does not induce trimerization of ΔP27 furinmut Fecto.
FIG 5.
Prefusion-specific D25 FAb binding to ΔP27 furinmut Fecto by SPR and SEC-MALS. (A) D25 FAb was immobilized on a sensor chip, various concentrations of ΔP27 furinmut Fecto were injected, and mass absorption response units (RU) were monitored as a function of time (Fecto concentrations are indicated). Fitting the resulting curves to 1:1 binding stoichiometry resulted in ka, kd, and fit values as indicated in the table below. The resulting KD for D25 FAb binding to ΔP27 furinmut Fecto is 2.38 × 10−11 M. (B) (Top) SEC chromatogram of ΔP27 furinmut Fecto. (Bottom) Chromatogram of D25 FAb-bound ΔP27 furinmut Fecto. Arrows indicate the expected retention times of RSV Fecto monomer (FM), unbound FAb (FAb), and RSV Fecto monomer bound to FAb (FM+FAb). Below, the masses of unbound ΔP27 furinmut Fecto or FAb-bound ΔP27 furinmut Fecto predicted by sequence are shown in the second column. The measured masses for ΔP27 furinmut Fecto and D25 FAb-bound ΔP27 furinmut Fecto by Stokes radius and MALS analysis are shown in the third and fourth columns, respectively.
RSV immune globulin antibodies binding to ΔP27 furinmut Fecto.
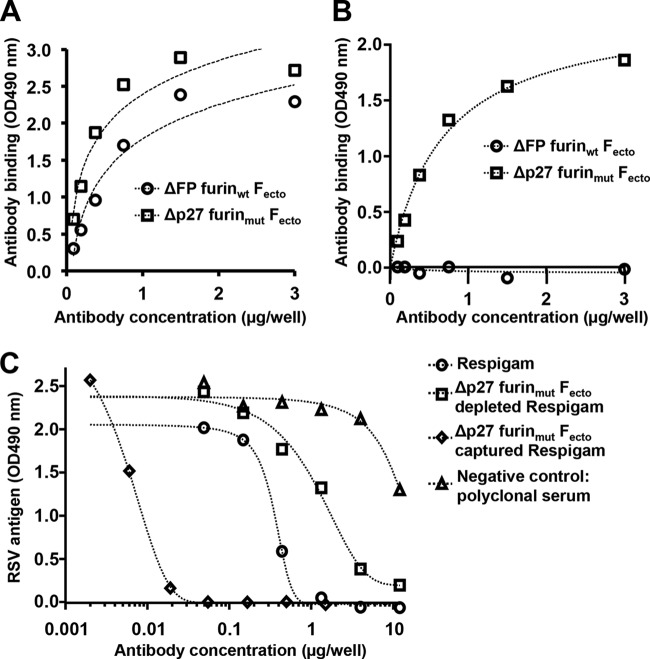
To determine if ΔP27 furinmut Fecto is competent to bind prefusion-specific human antibodies, we performed binding experiments using RSV immune globulin-depletion assays similar to those published in Magro et al. (12). ΔP27 furinmut Fecto or ΔFP furinwt Fecto (postfusion F trimer) was immobilized on resin, and the protein-bound resin was incubated with RSV immune globulin. Antibodies that did not bind the F protein were collected in the column flow-through (referred to as ΔP27 furinmut Fecto-depleted RSV immune globulin or ΔFP furinwt Fecto-depleted RSV immune globulin, respectively). Antibodies that bound ΔP27 furinmut Fecto and were eluted are referred to as ΔP27 furinmut Fecto-captured RSV immune globulin. Untreated RSV immune globulin is able to bind both ΔP27 furinmut Fecto and ΔFP furinwt Fecto (Fig. 6A). To demonstrate that antibodies that specifically bind ΔP27 furinmut exist in RSV immune globulin, we tested ΔFP furinwt Fecto-depleted RSV immune globulin in ELISA for its ability to bind ΔP27 furinmut Fecto (Fig. 6B). Although ΔFP furinwt Fecto-depleted RSV immune globulin had no significant binding to ΔFP furinwt Fecto (demonstrating sufficient depletion of the sera), ΔFP furinwt Fecto-depleted RSV immune globulin retained binding for ΔP27 furinmut Fecto (i.e., monomeric F). In comparison, ΔP27 furinmut Fecto-depleted RSV immune globulin showed no significant binding to either ΔFP furinwt Fecto or ΔP27 furinmut Fecto (data not shown).
FIG 6.
ΔP27 furinmut Fecto is recognized by unique neutralizing antibodies of RSV immune globulin (Respigam). (A) ΔFP furinwt Fecto or ΔP27 furinmut Fecto was adsorbed on an ELISA plate, and different concentrations of RSV immune globulin were analyzed for binding. RSV immune globulin bound both ΔFP furinwt Fecto (circles) and ΔP27 furinmut Fecto (squares) similarly. (B) ΔFP furinwt Fecto or ΔP27 furinmut Fecto was adsorbed on an ELISA plate, and ΔFP furinwt Fecto-depleted RSV immune globulin was analyzed at different concentrations for binding. ΔFP furinwt Fecto-depleted RSV immune globulin did not significantly bind ΔFP furinwt Fecto (circles) but retained significant binding to ΔP27 furinmut Fecto (squares). (C) ΔP27 furinmut Fecto-depleted and -captured RSV immune globulins were tested for neutralizing titers. Relative to those from the strongly neutralizing, untreated RSV immune globulin (circles), antibodies from ΔP27 furinmut Fecto-depleted RSV immune globulin (squares) retain a lower neutralization titer, whereas antibodies from ΔP27 furinmut Fecto-captured RSV immune globulin (diamonds) retain a higher neutralizing titer per mass of IgG (plotted on the x axis).
It has been previously shown that postfusion F-depleted RSV immune globulin retained the majority of its neutralizing titer and that prefusion F-depleted RSV immune globulin had a greatly diminished neutralizing titer (12). Furthermore, the antibodies that bind prefusion F represent a greater fraction of RSV immune globulin neutralizing titer (12). To test whether the antibodies that bind ΔP27 furinmut Fecto represent the majority of the neutralizing titer of RSV immune globulin, we tested the neutralizing titer of ΔP27 furinmut Fecto-depleted versus that of ΔP27 furinmut Fecto-captured RSV immune globulin (Fig. 6C). Untreated RSV immune globulin neutralized RSV infectivity well relative to the poorly neutralizing polyclonal antibodies purified from a nonimmunized rabbit (negative control). ΔP27 furinmut Fecto-depleted RSV immune globulin has a dramatically lower neutralizing titer than RSV immune globulin, similar to observations for prefusion F-depleted RSV immune globulin (12). ΔP27 furinmut Fecto-captured RSV immune globulin had a higher neutralizing titer per mass of IgG than untreated RSV immune globulin. Taken together, the data indicate that most of the neutralizing activity of RSV immune globulin is from antibodies that bind to ΔP27 furinmut Fecto (i.e., monomeric F) but not to the postfusion ΔFP furinwt Fecto and that this monomeric RSV F ectodomain retains prefusion F-specific epitopes.
Limited proteolysis of ΔP27 furinmut Fecto.
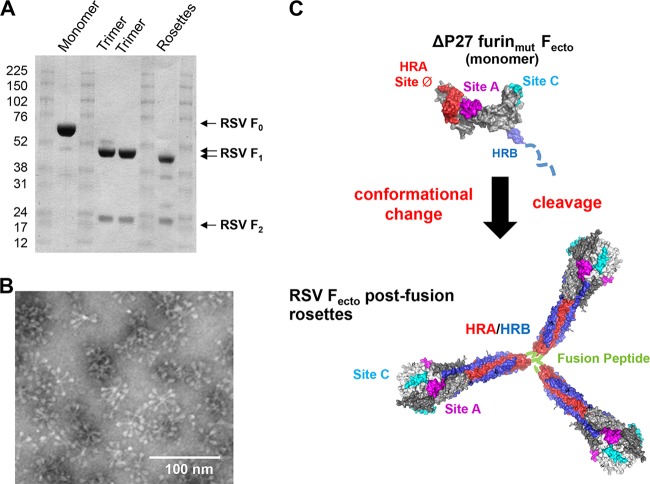
Uncleaved postfusion PIV trimers (F0) form rosettes, self-associated by aggregation of their fusion peptides, after in vitro cleavage into the F1/F2 species (20). To determine if uncleaved ΔP27 furinmut Fecto was competent to form rosettes of postfusion trimers upon in vitro cleavage, we digested ΔP27 furinmut Fecto with trypsin (Fig. 7A). Upon treatment with trypsin, uncleaved ΔP27 furinmut Fecto was cleaved into the native F1/F2 species of wild-type Fecto, further suggesting that the protein retains a mostly globular fold. Cleaved ΔP27 furinmut Fecto was further purified from the SEC void volume and analyzed by electron microscopy (Fig. 7B). ΔFP furinwt Fecto was previously described as mono-dispersed elongated crutches (8); however, cleaved ΔP27 furinmut Fecto forms rosettes of elongated crutches similar to the postfusion rosettes described for RSV and paramyxoviruses (20, 21). The observations that ΔP27 furinmut Fecto is resistant to degradation by limited proteolysis and is competent to form rosettes of postfusion F trimers suggest that the uncleaved ΔP27 furinmut Fecto (i.e., monomeric F) is a well-folded prefusion-like molecule structurally competent to rearrange into the postfusion conformation (Fig. 7C). We also attempted to observe the monomeric RSV F by electron microscopy, but we were unsuccessful, which is likely due to the protein's relatively small size (i.e., 65 kDa).
FIG 7.
Limited proteolysis of ΔP27 furinmut Fecto induces protein trimerization and association by the fusion peptide. (A) Sodium dodecyl sulfate-polyacrylamide gel showing limited proteolysis of RSV F constructs. RSV ΔP27 furinmut Fecto prior to digestion with trypsin is labeled “Monomer” and produces the F0 band. Two lots of RSV ΔFP furinwt Fecto are labeled “Trimer” and produce F1 and F2 bands in the absence of trypsin. RSV ΔP27 furinmut Fecto is labeled “Rosettes” and produces F1 and F2 bands after digestion with trypsin. (B) Electron microscopy image of the RSV Fecto rosettes formed spontaneously after trypsin treatment of ΔP27 furinmut Fecto. The rosettes have the elongated crutch shape of postfusion RSV Fecto, and proteins are associated at the end of the stalk where the hydrophobic fusion peptide is located. (C) Hypothetical model of ΔP27 furinmut Fecto as it undergoes conformational rearrangement after cleavage into the F1/F2 species. (Top) Model of uncleaved ΔP27 furinmut Fecto based on prefusion structure with Site A (sites II), Site C (site IV) and site Ø formed on the protein surface. HRB in blue is likely unfolded and is represented as a dashed line. A black arrow represents trypsin digestion of the monomer into F1/F2, leading to a conformational change of the prefusion-like monomer into the postfusion trimer. (Bottom) A model of RSV Fecto rosettes formed by postfusion trimers associated by interactions between their fusion peptides (green). Two neutralizing epitopes, Sites A and C, remain on the protein surface, while the prefusion site Ø epitope is lost as the HRA (red) is largely buried by the HRB (blue).
DISCUSSION
RSV remains one of the most important pathogens for which a vaccine is needed. Renewed interest in developing an RSV F subunit-based vaccine was triggered first by information from the structurally characterized PIV F proteins and now from structural studies of RSV F itself. Recent data have demonstrated that RSV Fecto stabilized in a prefusion conformation elicits higher neutralizing titers than postfusion RSV Fecto (11, 12), and there is high interest in developing a prefusion RSV F subunit vaccine. The novel prefusion-specific site Ø epitope, recognized by the D25 FAb and formed by the HRA region folding against the globular head, appears to be largely responsible for the improved immunogenicity (9). For this reason, antigens presenting the site Ø epitope will likely become components of next-generation RSV vaccine candidates.
Our initial attempts to generate a stable prefusion RSV Fecto antigen focused on the strategy used to stabilize PIV5 F in its metastable, prefusion conformation (data not shown) (14). We attempted to trimerize an uncleaved (with mutated furin sites) or cleaved (with wild-type furin sites) RSV Fecto using a C-terminal GCN trimerization domain but found the protein secretion from the cell was greatly reduced. We similarly tried expressing a cleaved RSV Fecto using a C-terminal foldon tag, but this protein formed rosettes similar to those of a postfusion Fecto. For this reason, we examined the oligomerization states of RSV F proteins without trimerization tags. SEC-MALS demonstrated that uncleaved ΔP27 furinmut Fecto is predominantly a monomer in solution (Fig. 2). This observed difference in the cleavage dependence of PIV F and RSV F trimerization may be linked to the presence of a single furin cleavage site in PIV F but two cleavage sites in RSV F flanking a p27 insertion (Fig. 1). The uncleaved, monomeric Fecto (ΔP27 furinmut Fecto) has some features consistent with the postfusion trimer (ΔFP furinwt Fecto). Specifically, two neutralizing epitopes present on both prefusion and postfusion Fecto, sites II and IV, are available for binding by their respective FAbs from motavizumab and 101F, as demonstrated by SPR (Fig. 3) and SEC-MALS (Fig. 4). A linear peptide harboring the motavizumab epitope is capable of transiently adopting its native conformation and being stabilized by FAb binding; however, the affinity of the FAb-peptide interaction is ∼6,000-fold less than folded postfusion protein-FAb affinity (10). Thus, the tight binding of uncleaved, monomeric Fecto to the motavizumab FAb suggests the epitope is preformed on the protein surface, as it is on the trimer (Fig. 3). SEC-MALS indicates that, upon binding either the motavizumab FAb or the 101F FAb, the monomer does not self-assemble into a trimer (Fig. 4). This finding suggests that the constraints preventing trimerization of the uncleaved monomer are not relieved by FAb binding.
Further evidence suggests that the RSV Fecto monomer is a folded species. When the RSV Fecto monomer is treated with trypsin, it is cleaved into its biologically relevant F1/F2 species. This limited and specific proteolysis suggests that the RSV Fecto monomer has a tight fold that protects it from nonspecific trypsin digestion. Upon cleavage into F1/F2, RSV Fecto spontaneously self-associates into rosettes of postfusion trimers (Fig. 7B). This behavior is analogous to the rosette formation observed for the PIV Fs (20).
If the RSV Fecto monomer is in a prefusion-like conformation, the HRA region would be packed against the globular head, presenting novel HRA epitopes lost in the postfusion conformation (Fig. 7C). Unlike the postfusion Fecto trimer, the Fecto monomer is capable of binding the site Ø, prefusion-specific FAb D25 (Fig. 5). Furthermore, the Fecto monomer binds antibodies in the human serum-derived RSV immune globulin commercial product that do not bind the postfusion Fecto antigen (Fig. 6B). Finally, previous studies have shown that postfusion Fecto antigens are unable to bind and deplete the majority of neutralizing activity from RSV immune globulin (12). However, the Fecto monomer is able to bind and deplete the majority of neutralizing antibodies from RSV immune globulin, suggesting that the Fecto monomer retains the important site Ø epitopes present in other prefusion antigens and presumably on the RSV F that is present on the surface of the virion.
Retention of site Ø is a desired characteristic of next-generation RSV F antigens, and therefore the uncleaved RSV F monomer here described represents a potential prefusion subunit vaccine candidate. However, the RSV F monomer described here readily refolds into the postfusion conformation in the presence of trace amounts of trypsin or contaminant trypsin-like proteases and therefore is not stable enough for evaluation in immunization studies. Future work will be aimed at stabilizing the RSV F monomer by introducing additional mutations that lock it more stably in a prefusion conformation amenable to in vivo studies.
ACKNOWLEDGMENT
Work in Madrid was supported in part by grant SAF2012–31217 from Plan Nacional I+D+I (Ministerio de Economía y Competitividad).
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1. Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. 10.1016/S0140-6736(10)60206-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hall CB. 2010. Respiratory syncytial virus, p 2207–2221 In Mandell GL, Bennett JE, Dolin R. (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 7th ed. Churchill Livingstone, New York, NY [Google Scholar]
- 3. Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179–186. 10.1001/jama.289.2.179 [DOI] [PubMed] [Google Scholar]
- 4. Han LL, Alexander JP, Anderson LJ. 1999. Respiratory syncytial virus pneumonia among the elderly: an assessment of disease burden. J. Infect. Dis. 179:25–30. 10.1086/314567 [DOI] [PubMed] [Google Scholar]
- 5. Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, Dormitzer M, O'Grady J, Koenig S, Tamura JK, Woods R, Bansal G, Couchenour D, Tsao E, Hall WC, Young JF. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 176:1215–1224. 10.1086/514115 [DOI] [PubMed] [Google Scholar]
- 6. Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89:422–434 [DOI] [PubMed] [Google Scholar]
- 7. Anderson R, Huang Y, Langley JM. 2010. Prospects for defined epitope vaccines for respiratory syncytial virus. Future Microbiol. 5:585–602. 10.2217/fmb.10.22 [DOI] [PubMed] [Google Scholar]
- 8. Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. 2011. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc. Natl. Acad. Sci. U. S. A. 108:9619–9624. 10.1073/pnas.1106536108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. 10.1126/science.1234914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McLellan JS, Yang Y, Graham BS, Kwong PD. 2011. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 85:7788–7796. 10.1128/JVI.00555-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342:592–598. 10.1126/science.1243283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, Terron MC, Melero JA, Palomo C. 2012. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc. Natl. Acad. Sci. U. S. A. 109:3089–3094. 10.1073/pnas.1115941109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 102:9288–9293. 10.1073/pnas.0503989102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44. 10.1038/nature04322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Swanson K, Wen X, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2010. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology 402:372–379. 10.1016/j.virol.2010.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. González-Reyes L, Ruiz-Arguello MB, Garcia-Barreno B, Calder L, Lopez JA, Albar JP, Skehel JJ, Wiley DC, Melero JA. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 98:9859–9864. 10.1073/pnas.151098198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sastre P, Melero JA, Garcia-Barreno B, Palomo C. 2005. Comparison of affinity chromatography and adsorption to vaccinia virus recombinant infected cells for depletion of antibodies directed against respiratory syncytial virus glycoproteins present in a human immunoglobulin preparation. J. Med. Virol. 76:248–255. 10.1002/jmv.20349 [DOI] [PubMed] [Google Scholar]
- 18. Anderson LJ, Bingham P, Hierholzer JC. 1988. Neutralization of respiratory syncytial virus by individual and mixtures of F and G protein monoclonal antibodies. J. Virol. 62:4232–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martinez I, Melero JA. 1998. Enhanced neutralization of human respiratory syncytial virus by mixtures of monoclonal antibodies to the attachment (G) glycoprotein. J. Gen. Virol. 79:2215–2220 [DOI] [PubMed] [Google Scholar]
- 20. Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. 2006. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 103:17903–17908. 10.1073/pnas.0608678103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Calder LJ, Gonzalez-Reyes L, Garcia-Barreno B, Wharton SA, Skehel JJ, Wiley DC, Melero JA. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271:122–131. 10.1006/viro.2000.0279 [DOI] [PubMed] [Google Scholar]