ABSTRACT
West Nile virus (WNV) is an emerging zoonotic mosquito-borne flavivirus responsible for outbreaks of febrile illness and meningoencephalitis. The replication of WNV takes place on virus-modified membranes from the endoplasmic reticulum of the host cell, and virions acquire their envelope by budding into this organelle. Consistent with this view, the cellular biology of this pathogen is intimately linked to modifications of the intracellular membranes, and the requirement for specific lipids, such as cholesterol and fatty acids, has been documented. In this study, we evaluated the impact of WNV infection on two important components of cellular membranes, glycerophospholipids and sphingolipids, by mass spectrometry of infected cells. A significant increase in the content of several glycerophospholipids (phosphatidylcholine, plasmalogens, and lysophospholipids) and sphingolipids (ceramide, dihydroceramide, and sphingomyelin) was noticed in WNV-infected cells, suggesting that these lipids have functional roles during WNV infection. Furthermore, the analysis of the lipid envelope of WNV virions and recombinant virus-like particles revealed that their envelopes had a unique composition. The envelopes were enriched in sphingolipids (sphingomyelin) and showed reduced levels of phosphatidylcholine, similar to sphingolipid-enriched lipid microdomains. Inhibition of neutral sphingomyelinase (which catalyzes the hydrolysis of sphingomyelin into ceramide) by either pharmacological approaches or small interfering RNA-mediated silencing reduced the release of flavivirus virions as well as virus-like particles, suggesting a role of sphingomyelin-to-ceramide conversion in flavivirus budding and confirming the importance of sphingolipids in the biogenesis of WNV.
IMPORTANCE West Nile virus (WNV) is a neurotropic flavivirus spread by mosquitoes that can infect multiple vertebrate hosts, including humans. There is no specific vaccine or therapy against this pathogen licensed for human use. Since the multiplication of this virus is associated with rearrangements of host cell membranes, we analyzed the effect of WNV infection on different cellular lipids that constitute important membrane components. The levels of multiple lipid species were increased in infected cells, pointing to the induction of major alterations of cellular lipid metabolism by WNV infection. Interestingly, certain sphingolipids, which were increased in infected cells, were also enriched in the lipid envelope of the virus, thus suggesting a potential role during virus assembly. We further verified the role of sphingolipids in the production of WNV by means of functional analyses. This study provides new insight into the formation of flavivirus infectious particles and the involvement of sphingolipids in the WNV life cycle.
INTRODUCTION
West Nile virus (WNV) is a mosquito-borne flavivirus that is distributed worldwide and that is responsible for recurrent outbreaks of febrile illness and encephalitis. The virus is maintained in nature in an enzootic infectious cycle between birds and mosquitoes, which act as its vectors, although it can also infect multiple vertebrate hosts, including horses and humans (1, 2). The continuing spread of WNV due to a variety of ecological factors, combined with the lack of specific therapeutics or vaccines for human use, makes the identification of the viral and host processes that control the biology of this pathogen important to improve the design of specific antiviral strategies (3).
As a flavivirus, WNV is an enveloped plus-strand RNA virus (1, 2). A feature that it shares with other plus-strand RNA viruses is replication in the cytoplasm of infected cells in tight association with intracellular membrane rearrangements (4). In the case of flaviviruses and, hence, WNV, the membranes associated with virus replication are from the endoplasmic reticulum (ER) (5). Even more, flavivirus particles are assembled by invagination and budding of the ER membrane into the lumen of this organelle, so the ER also provides the membrane source for WNV envelopment (6). Although the lipid composition of the viral envelopes of a few distinct viruses has been characterized (7–10), to our knowledge, the molecular composition of the flavivirus envelope has not been analyzed. Only certain lipids in the flavivirus envelope (cholesterol and phosphatidylserine) have been identified, and these have been identified by functional studies rather than by analytical approaches (11, 12).
Lipids are the main components of cellular membranes, playing key roles in viral infections by acting as signaling molecules, as well as by determining the physical properties of the membranes, such as fluidity, thickness, or shape (13, 14). In fact, the strong manipulation of cellular lipid metabolism by different viruses has recently been documented (15–17). Accordingly, enrichment in specific lipids contributes to the generation of a membrane curvature adequate for the correct assembly of the replication complex or virus budding, and the cone-shaped or inverted cone-shaped lipids, which can govern membrane bending, budding processes, or fusion events, are of particular interest (18–20). Although differences in the lipid requirements between viruses of the same family have been noted (21), the dependence on certain lipids, such as fatty acids (15, 22, 23) or phosphatidylinositol 4-phosphate (PI4P) (21, 24), can be shared by unrelated enveloped and nonenveloped viruses. These observations suggest that each virus creates its own characteristic cellular microenvironment for replication by developing specialized virus-induced organelle-like structures within infected cells (4, 24, 25). Regarding flaviviruses, major rearrangements of cellular lipid metabolism have been observed in dengue virus (DENV)-infected cells (26, 27), and several studies have highlighted the importance of both fatty acids and cholesterol for WNV and DENV infection (22, 28–31), as well as a lack of a requirement for PI4P (21, 29).
In this study, we have analyzed the effect of WNV infection on the cellular content of 11 lipid classes. Our results revealed an increase in the cellular content of multiple lipid species upon WNV infection, which is consistent with major lipid metabolic changes in WNV-infected cells. The proportions of these metabolites in the envelope of WNV virions and virus-like particles were also determined, unveiling a unique composition of the viral envelope and pointing to specific roles of some of these lipids for virus assembly. Supporting this view, a direct link between specific lipids (sphingolipids) and WNV biogenesis was found by means of functional analyses.
MATERIALS AND METHODS
Cells, viruses, infections, and virus titrations.
All infectious virus manipulations were performed in our biosafety level 3 (BSL-3) facilities. The origins of WNV strain NY99, Usutu virus (USUV) strain SAAR-1776, and Sindbis virus (SINV) have been previously described (32). Vero and HeLa cells were grown at 37°C in a 5% CO2 atmosphere in Dulbecco's modified minimum essential medium (DMEM) supplemented with 2 mM glutamine, penicillin-streptomycin, and 5 or 10% fetal bovine serum, respectively. Cells of a HeLa cell line stably transfected with plasmid pcDNA 3.1 (+) (Invitrogen, Carlsbad, CA) encoding the 25 last amino acids of the WNV NY99 C protein followed by the sequence of premembrane/membrane (prM) and envelope (E) proteins (HeLa3-WNV cells) were obtained by limiting dilution and grown in complete culture medium supplemented with 500 μg/ml G-418 (T. Merino-Ramos, A. B. Blázquez, E. Escribano-Romero, R. Cañas-Arranz, F. Sobrino, J. C. Saiz, and M. A. Martín-Acebes, submitted for publication). This line constitutively secreted WNV recombinant subviral particles (RSPs). C6/36 mosquito cells were cultured in M3 medium supplemented with 4 mM glutamine, gentamicin, penicillin-streptomycin, nonessential amino acids, amphotericin B, and 10% fetal bovine serum at 28°C without CO2. For infections in liquid medium, the viral inoculum was incubated with cell monolayers for 1 h at the temperature appropriate for cell growth, and then the inoculum was removed and fresh medium containing or not containing 1% fetal bovine serum was added. The viral titer was determined at 24 h postinfection (p.i.) for WNV and USUV and 8 h p.i. for SINV by plaque assay on Vero cells (29, 32, 33). Cell-associated virus was extracted from infected cells by three cycles of freeze and thaw and titrated as described above. The multiplicity of infection (MOI) used in each experiment was expressed as the number of PFU/cell and is indicated in the corresponding figure legend.
Antibodies.
Mouse monoclonal antibodies against the WNV envelope (E) protein (Millipore, Temecula, CA), β-actin (Sigma, St. Louis, MO), and GM130 (ECM Biosciences, Versailles, KY) and rabbit sera against WNV M protein (Imgenex, San Diego, CA), calnexin (StressMarq Biosciences Inc., Victoria, Canada), mannosidase II (Millipore), and sphingomyelin phosphodiesterase 3 (SMPD3), also named neutral sphingomyelinase 2 (nSMase2; ECM Biosciences), were used as primary antibodies. Anti-mouse or anti-rabbit IgG coupled to Alexa Fluor 594 or 488 (Invitrogen, Carlsbad, CA) and goat anti-mouse or anti-rabbit IgG coupled to horseradish peroxidase (Pierce Biotechnology, Rockford, IL) were used as secondary antibodies.
Drug treatments.
Brefeldin A (BFA), golgicide A (GCA), GW4869, and glutathione were from Sigma. Spiroepoxide was from Santa Cruz Biotechnology (Santa Cruz, CA). BFA, GCA, GW4869, and spiroepoxide stock solutions were prepared in dimethyl sulfoxide, while glutathione was directly dissolved in culture medium. Working concentrations of GW4869 were prepared as described previously (34). Control cells were treated in a parallel manner with the same amount of drug vehicle. For estimation of RSP release, the medium from HeLa3-WNV cells was replaced by serum-free medium containing the drugs. Cells were incubated for 4 h, and the amount of RSPs released to the culture medium was determined by an enzyme-linked immunodot assay using a monoclonal antibody directed against WNV E glycoprotein, as described below. In the case of virus infections, drugs were added after the first hour of infection, when the viral inoculum was replaced by medium containing 1% fetal bovine serum. The lack of toxicity of drug concentrations during the assays was evaluated by determination of the cellular ATP content with a CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI).
siRNA experiments.
HeLa or HeLa3-WNV cells were transfected in serum-free culture medium with 100 nM endoribonuclease-prepared small interfering RNAs (esiRNAs) against human SMPD3 (MISSION; Sigma) or an equivalent amount of MISSION small interfering RNA (siRNA) universal negative control 1 (MISSION; Sigma) using an siRNA transfection reagent (MISSION; Sigma), as indicated by the manufacturer. To estimate the release of WNV RSPs, the culture medium of HeLa3-WNV cells was replaced by fresh serum-free medium, and the amount of RSPs in the culture medium was determined after 4 h of incubation by an enzyme-linked immunodot assay. For viral infections, transfected cells were infected (MOI, 1 PFU/cell) as described above for infections in liquid medium, and subsequent experiments were performed at 48 h posttransfection. The extent of siRNA silencing was analyzed by Western blotting (29, 35) using specific antibodies against nSMase2.
Purification of RSPs and virions.
HeLa3-WNV cells were incubated for 48 h in serum-free medium (to avoid the possible interference of serum with subsequent lipid determinations), and RSPs were purified by sucrose gradient centrifugation essentially as described previously (36). Briefly, culture medium from HeLa3-WNV cells was clarified by centrifugation at 15,000 × g for 30 min at 4°C and then centrifuged through a 20% sucrose cushion for 3.5 h at 112,000 × g at 4°C. The pellet containing RSPs was resuspended in phosphate-buffered saline (PBS), loaded onto a 12-ml 20 to 60% linear sucrose gradient, and centrifuged at 256,000 × g for 18 h at 4°C. For purification of WNV particles, HeLa cells were infected with WNV (MOI, 5 PFU/cell) in serum-free medium and incubated for 48 h. The culture medium was cleared of cell debris by centrifugation at 850 × g for 15 min at 4°C and centrifuged through a 20% sucrose cushion at 112,000 × g for 3.5 h at 4°C. The pellet containing the virions was loaded onto a six-step discontinuous 20 to 60% sucrose gradient and centrifuged at 55,500 × g for 18 h at 4°C. Gradients were fractioned from the top, and the E-protein content of each fraction was determined by enzyme-linked immunodot assay as described below. Only the fraction with the highest E-protein content was selected for lipid analyses.
Immunofluorescence and confocal microscopy.
Cells grown on glass coverslips were fixed with 4% paraformaldehyde in PBS for 15 min and processed for immunofluorescence as described previously (29, 35). Cells were observed using a Leica TCS SPE confocal laser scanning microscope and an HCX PL Apo ×63/1.4 oil immersion objective. Images were acquired using Leica advanced fluorescence software and processed with Adobe Photoshop CS2 software (Adobe Inc., San Jose, CA). The optical slice thickness for all confocal images displayed was 1 airy unit.
Transmission electron microscopy.
Negative staining of RSPs was performed in dialyzed samples containing the peak amount of E protein within the gradient, as described previously (36). For immunolabeling and negative staining, samples were adsorbed to ionized collodion-carbon-coated grids, washed with PBS, and blocked with 10% fetal bovine serum in PBS for 5 min. The grids were incubated with primary antibodies diluted in 5% fetal bovine serum in PBS for 30 min, washed five times with PBS, and incubated with protein A coupled to 5-nm colloidal gold diluted in 5% fetal bovine serum in PBS for 30 min. Samples were then fixed with 1% glutaraldehyde in PBS for 5 min, washed three times with bidistilled water, and negatively stained with 1% uranyl acetate for 2 min. HeLa or Vero cells infected with WNV were fixed in 4% paraformaldehyde–2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) at 24 h p.i. and processed for electron microscopy as described previously (29, 35). Samples were examined using a JEOL JEM-1010 electron microscope (JEOL, Japan) operated at 80 kV, and images were acquired using a digital camera (4K64K TemCam-F416; Tietz Video and Image Processing Systems GmbH, Gauting, Germany).
Enzyme-linked immunodot assay.
Cell culture medium (usually 5 to 10 μl) was adsorbed to a nitrocellulose membrane by vacuum using a Bio-Dot apparatus (Bio-Rad, Hercules, CA). For quantification of the amount of RSPs in the culture medium and in order to verify that samples were within the linear range, a standard curve was prepared using different dilutions. The membrane was blocked with 3% skimmed milk in PBS and incubated with monoclonal anti-E antibody diluted in 1% skimmed milk in PBS. After three washes with PBS, the membranes were incubated with secondary antibodies coupled to horseradish peroxidase and washed, and proteins were detected by chemiluminescence using ImageQuant LAS 4000 mini equipment (GE Healthcare, Buckinghamshire, United Kingdom).
Quantitative RT-PCR.
Viral RNA was extracted with a NucleoSpin viral RNA isolation kit (Macherey-Nagel, Düren, Germany). The number of viral RNA copies was determined by quantitative reverse transcription-PCR (RT-PCR) (37) and is given as the number of genomic equivalents corresponding to the number of PFU/ml by comparison with the amount of RNA extracted from previously titrated samples (38).
Lipid analysis.
HeLa cells were infected or not infected (mock infected) with WNV (MOI, 50 PFU/cell) in serum-free medium as described above for infections in liquid medium, detached from the flasks, and resuspended in PBS at 24 h p.i. The number of cells in each sample was determined, and aliquots containing 2.7 × 106 cells were subjected to lipid extractions. The amount of protein in each sample was also determined by the Bradford assay. Lipids were extracted using a modified Bligh and Dyer protocol. Cell pellets were resuspended in 100 μl of PBS and 750 μl of a methanol-chloroform (1:2, vol/vol) solution containing 0.01% butylated hydroxytoluene (BHT) and internal standards (1,2-diheptadecanoyl-sn-glycero-3-phosphocholine, 1,2-diheptadecanoyl-sn-glycero-3-phosphoethanolamine, 1,2-diheptadecanoyl-sn-glycero-3-phosphoserine, 1-heptadecanoyl-2-hydroxy-sn-glycero-3-phosphocholine, 1-heptadecanoyl-2-hydroxy-sn-glycero-3-phosphoethanoamine, and 1-heptadecanoyl-2-hydroxy-sn-glycero-3-phosphoserine, 200 pmol each, from Avanti Polar Lipids). Samples were extracted at 48°C overnight and centrifuged at 13,000 × g for 5 min. Then, the supernatant was transferred to a new vial, evaporated to dryness, and stored at −20°C in an argon atmosphere until the analysis of glycerophospholipids (39). On the other hand, 100 μl of PBS and 750 μl of a methanol-chloroform (2:1, vol/vol) solution containing 0.01% BHT and internal standards (N-lauroyl-d-erythro-sphingosine, N-lauroyl-d-erythro-sphingosylphosphorylcholine, and d-glucosyl-β-1,1′-N-lauroyl-d-erythro-sphingosine, 200 pmol each, from Avanti Polar Lipids) were added to the cell pellets. Samples were extracted at 48°C overnight and cooled, 75 μl of 1 M KOH in methanol was added, and the mixture was incubated for 2 h at 37°C. Following addition of 75 μl of 1 M acetic acid, samples were centrifuged at 13,000 × g for 5 min and the supernatant was transferred to a new vial, evaporated to dryness, and stored at −20°C in an argon atmosphere until the analysis of sphingolipids (40).
Lipids were measured with an Acquity ultraperformance liquid chromatography (UPLC) system (Waters, USA) connected to a time-of-flight (TOF; LCT Premier XE) detector controlled with Waters/Micromass MassLynx (v.4.1) software. An Acquity UPLC BEH C8 column (particle size, 1.7 μm; 100 mm by 2.1 mm; Waters, Ireland), a flow rate of 0.3 ml/min, and a column temperature of 30°C were used. The mobile phase was methanol with 1 mM ammonium formate and 0.2% formic acid (solution A)–water with 2 mM ammonium formate and 0.2% formic acid (solution B). Gradient elution started at 80% solution A, was increased to 90% solution A over 3 min, was held for 3 min, was increased to 99% solution A over 9 min, and was held for 3 min. Initial conditions were attained in 2 min, and the system was stabilized for 3 min. The acquisition range of the TOF detector was m/z 50 to 1,500, the capillary voltage was set to 3.0 kV, the desolvation temperature was 350°C, and the desolvation gas flow rate was 600 liters/h (41).
Lipid identification was confirmed by the analysis of one sample from each group with an ultra-high-performance liquid chromatography system (Accela) coupled to a Thermo Fisher Scientific LTQ Orbitrap Velos mass spectrometer (MS) controlled with Thermo Fisher Scientific/Xcalibur software using the same column and eluted with the following conditions: gradient elution started at 85% solution A, was increased to 90% solution A over 9 min, was held for 2 min, was increased to 99% solution A over 6 min, and was held for 2 min. Initial conditions were attained in 2 min, and the system was stabilized for 3 min. The acquisition range of the Orbitrap detector was m/z 200 to 100, the source voltage was set to 3.5 kV, the capillary temperature was 350°C, the sheath gas flow rate was 50 liters/h, the auxiliary gas flow rate of 20 liters/h, and the sweep gas flow rate was 2 liters/h (42).
Individual chromatographic peaks of distinct lipid species were isolated from full-scan MS spectra when the theoretical exact masses of the lipid species were selected. The spectra were extracted from a database that was previously generated using the spectrum simulation tool of Micromass MassLynx software on the basis of our previous results (39). Then, a list of possible candidates fitting the specific exact mass was generated using formula determination tools (elemental composition search) of Micromass MassLynx software. The elemental number was restricted to include C, H, O, N, and P. The formula constraints were as follows: C, H, and O were ≥1, P was ≥0, and N was ≥1, following the nitrogen rule. The number of double-bond equivalents (DBEs) was set to be between −0.5 and 15.0. The search was based on single mass analysis and considered only the m/z value of the monoisotopic peak. Positive identification of the lipids was based on accurate mass measurement with an error <5 ppm, a low i-Fit parameter in the spectrum window, and its relative retention time by liquid chromatography compared to that of the standard (±2%) (39). The following lipids from Avanti Polar Lipids were used: 1,2-dihexadecanoyl-sn-glycero-3-phosphocholine, 1,2-dioctadecanoyl-sn-glycero-3-phosphocholine, 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine, 1-hexadecanoyl-2-(4Z,7Z,10Z,13Z,16Z,19Z-docosahexaenoyl)-sn-glycero-3-phosphocholine, 1-(9Z-octadecenoyl)-2-hydroxy-sn-glycero-3-phosphocholine, 1-(1Z-octadecenyl)-2-(9Z-octadecenoyl9-sn-glycero-3-phosphocholine, 1-octadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoethanolamine, 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-l-serine, N-hexadecanoyl-d-erythro-sphingosine, N-hexadecanoyl-d-erythro-sphingosylphosphorylcholine, and d-glucosyl-β-1,1′-N-hexadecanoyl-d-erythro-sphingosine In addition, identifications of lipids were made by searching their molecular weights against entries in the Lipid MAPS database. As mentioned above, this identification procedure was confirmed by the analysis of selected samples using Thermo Fisher Scientific Orbitrap analyzers (resolution, 30,000 full width at half maximum) (42).
The data presented are the products of 4 to 5 independent lipid extractions and determinations.
Annotation of lipid species.
Glycerophospholipids were annotated as lipid subclass and total fatty acyl chain length:total number of unsaturated bonds. Plasmalogens (P) were annotated as described above, except that P was added. Sphingolipids were annotated as lipid subclass and total fatty acyl chain length:total number of unsaturated bonds. If the sphingoid base residue was dihydrosphingosine (dihydro sphingomyelin or dihydro ceramide), the lipid class contains the prefix dh (dihydro).
Data analysis.
Analysis of variance (ANOVA) was performed with the SPSS (v.15) statistical package (SPSS Inc., Chicago IL), with Bonferroni's correction applied for multiple comparisons. Nonparametric comparisons were performed using the statistical package GraphPad Prism (v.2.01; GraphPad Software Inc., La Jolla, CA). Data are presented as the mean ± standard deviation (SD). Differences with P values of <0.05 were considered statistically significant.
RESULTS
Alteration of lipid content in WNV-infected cells.
To analyze the effect of WNV infection on the lipid content of the host cell, HeLa cells were infected or not infected at a high multiplicity of infection (MOI) and processed for lipid analysis at 24 h p.i. At this time point, infectious virus release into the culture medium was already detected (Fig. 1A). When the cells were observed by transmission electron microscopy, they exhibited intracellular membrane rearrangements associated with flavivirus replication and assembly, such as membrane sacs containing small virus-induced vesicles, referred to as vesicle packets (VPs) (5, 29, 43), as well as electron-dense virions (Fig. 1B). As expected, these rearrangements were not observed in control uninfected cells. Eleven lipid classes (seven glycerophospholipids [GPLs] and four sphingolipids [SLs]) were included in the analysis. The GPLs analyzed were phosphatidylcholine (PC), 1-alkenyl,2-acylglycerolphosphocholine (referred to as plasmalogen-PC [p-PC; plasmenylcholine]), 2-acylglycero-3-phosphocholine (lyso-PC), phosphatidylethanolamine (PE), plasmalogen-PE (p-PE), 2-acylglycero-3-phosphoethanolamine (lyso-PE), and phosphatidylserine (PS). The four SLs analyzed were ceramide (CER), dihydro-CER (dhCER), sphingomyelin (SM), and dihydro-SM (dhSM). A tendency for an increase in the content of most of the lipid classes analyzed per cell (except for lyso-PE, a minor lipid class present in the extracts) was noticed in WNV-infected cells (Fig. 1C). In fact, statistically significant increases in the amounts of PC (1.7-fold), p-PC (1.9-fold), lyso-PC (1.9-fold), p-PE (1.6-fold), CER (1.8-fold), dhCER (1.3-fold), and SM (1.4-fold) were noted when infected cells were compared to uninfected cells. Among the 11 lipid classes analyzed, a total of 162 different molecular species were identified in HeLa cells (the complete list of molecular species identified in this and subsequent analyses is included in Data Set S1 in the supplemental material), with 54 (33.3%) molecular species being significantly increased upon WNV infection (Fig. 1D). Interestingly, although a tendency for a reduction of some lipid species was found, at the level of resolution of this analysis, no statistically significant reduction in the content of individual lipid species could be determined, so we assumed that no major modifications in the level of the rest of the lipid species occurred in infected cells. The molecular species significantly increased in infected cells included 44 GPLs (13 PCs, 9 p-PCs, 3 lyso-PCs, 5 PEs, and 14 p-PEs) and 10 SLs (5 CERs, 1 dhCERs, and 4 SMs). Therefore, WNV infection markedly alters the lipid metabolism of host cells.
FIG 1.
WNV infection alters lipid metabolism. (A) Time course analysis of WNV infection. HeLa cells were infected with WNV (MOI, 50 PFU/cell), and the amount of infectious virus released into the culture medium was determined by plaque assay at different times p.i. (B) Intracellular membrane rearrangements in WNV-infected HeLa cells observed by transmission electron microscopy. A micrograph showing the ER from uninfected cells is included for comparison. Characteristic features of flavivirus-infected cells, vesicle packets (VPs) and virions (Vi), were observed at 24 h p.i. (MOI, 50 PFU/cell). Bars, 200 nm. (C) Relative amount of different classes of GPLs and SLs in HeLa cells infected or not infected with WNV (MOI, 50 PFU/cell) determined by mass spectrometry at 24 h p.i. Statistically significant differences are indicated: *, P < 0.05; **, P < 0.005. (D) Fold differences in individual lipid species significantly increased in infected cells analyzed as described in the legend to panel C. Dashed line, mean value for each lipid in uninfected cells. Data are presented as the mean ± SD.
GPL and SL content of the envelope of flavivirus virions and RSPs.
The lipid composition of viral envelopes can provide novel clues to understand the biology of these pathogens (7, 9, 10). Therefore, we studied the GPL and SL content of the viral envelope of WNV. Since the coexpression of flavivirus structural glycoproteins (premembrane/membrane [prM/M] and envelope [E]) results in the assembly and secretion of a sort of virus-like particle termed a recombinant subviral particle (RSP) that provides a noninfectious model system to study the assembly and secretion of flavivirus particles (44, 45), the analysis of the lipid composition of RSPs was included in this study. A stable HeLa clone (HeLa3-WNV cells) expressing WNV structural glycoproteins (Fig. 2A) that constitutively secrete WNV RSPs of about 30 nm in diameter into the culture medium (Fig. 2B) was used as a source of RSPs. The identity of these structures was confirmed by immunogold electron microscopy using an antibody against the E glycoprotein (Fig. 2B inset). In this cell line, immunofluorescence and confocal analysis revealed that the WNV envelope glycoprotein colocalized with markers of the ER (calnexin) and the Golgi complex (mannosidase II) (Fig. 2C and D). The molecular weights of the proteins were analyzed by Western blotting using anti-E and anti-M antibodies (Fig. 2E). As expected, using anti-E antibody, a single band (about 53 kDa [46]) was observed in samples containing purified RSPs. In the case of anti-M antibody, two bands, one band compatible with prM protein and the other band compatible with mature M protein (about 20 and 6 kDa, respectively [46]), were observed. These results confirmed the correct expression of WNV glycoproteins by cells of the HeLa3-WNV cell line.
FIG 2.
WNV RSP as a noninfectious system to study flavivirus biogenesis. (A) Immunofluorescence analysis of the expression of the WNV E glycoprotein in HeLa3-WNV cells. HeLa or HeLa3-WNV cells were subjected to immunofluorescence analysis using a monoclonal antibody directed against the E glycoprotein (green) and observed by confocal microscopy. Nuclei were stained with ToPro3 (blue). Bars, 10 μm. (B) RSPs produced by HeLa3-WNV cells. The RSPs released into the culture medium were purified by sucrose gradient centrifugation and observed by negative staining and transmission electron microscopy. (Inset) Immunogold staining of RSPs using a monoclonal antibody against the E protein. RSPs were detected using protein A coupled to 5-nm colloidal gold. Bars, 100 nm. (C and D) Localization of the WNV E glycoprotein at the ER (C) and the Golgi complex (D) of HeLa3-WNV cells. Immunofluorescence was performed using a monoclonal antibody directed against the E glycoprotein (green) in combination with rabbit polyclonal antibodies against calnexin (red) to stain the ER or mannosidase II (red) to stain the Golgi complex. Nuclei were stained with ToPro3 (blue). HeLa cells not expressing viral proteins were included as negative control. Bars, 10 μm. (E) Western blot analysis of RSPs purified through a sucrose cushion from the culture medium of HeLa3-WNV cells. Cell culture medium from control HeLa cells was processed and analyzed in parallel as a negative control. Western blotting was performed using anti-E or anti-M antibodies. (F) Enzyme-linked immunodot assay using a monoclonal antibody against the E glycoprotein of culture supernatants from HeLa3-WNV cells. Culture medium from HeLa cells not expressing viral glycoproteins was included as a negative control.
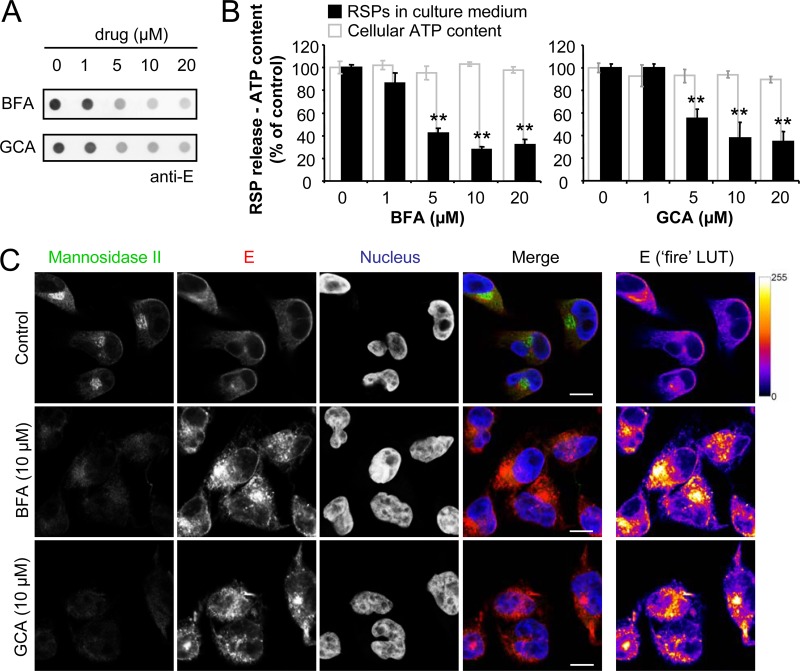
Enzyme-linked immunodot assay is faster than Western blotting and allows the analysis of up to 96 samples at once. This kind of assay has successfully been applied to quantify the amount of RSPs released into the culture medium by other flaviviruses (47). Thus, a dot blot assay was used to detect the WNV RSPs released into the culture medium from HeLa3-WNV cells (Fig. 2F). To further validate this methodology, the amount of RSPs released into the culture medium upon treatment with the Golgi complex-disturbing agents brefeldin A (BFA) or golgicide A (GCA) was studied. Treatment of HeLa3-WNV cells with BFA or GCA impaired the release of RSPs into the culture medium (Fig. 3A). The quantification of the blots revealed that BFA or GCA at concentrations of ≥5 μM significantly inhibited the release of RSPs into the culture medium without exerting noticeable toxic effects on the cells (Fig. 3B). Immunofluorescence analysis of BFA- or GCA-treated cells confirmed the disruption of the Golgi complex architecture that these drugs induced (Fig. 3C). Golgi complex disassembly was concomitant with the accumulation of E glycoprotein inside the cells (Fig. 3C), an observation consistent with a reduction in the release of RSPs into the culture medium observed in Fig. 3A and B. Overall, these observations are consistent with previous data showing that flavivirus RSPs, as well as infectious virions, are assembled by budding into the ER and then traffic through the secretory pathway (44, 45). Therefore, in the HeLa3-WNV cell model, the lipid envelope of RSPs is acquired from the ER in a manner parallel to that observed for infectious virus (6).
FIG 3.
Treatment with BFA or GCA impairs the secretion of RSPs into the culture medium. (A) RSP release into the culture medium by HeLa3-WNV cells treated (4 h) with the drugs was analyzed by enzyme-linked immunodot assay using a monoclonal antibody against the E glycoprotein. (B) Quantification of the amount of RSPs released into the culture medium by treated HeLa3-WNV cells from panel A. For each drug concentration, the cellular ATP content is indicated as an indicator of cell viability upon drug treatment. (C) Localization of the WNV E glycoprotein in infected HeLa cells treated with Golgi complex-disrupting agents. HeLa3-WNV cells were treated with 10 μM BFA or GCA for 4 h. Immunofluorescence was performed using a monoclonal antibody directed against the WNV E glycoprotein (red) in combination with a rabbit polyclonal antibody against mannosidase II (green) to stain the Golgi complex. Nuclei were stained with ToPro3 (blue). The panels in the rightmost column display E glycoprotein staining with false coloring from dark purple to bright yellow by use of the fire lookup table (LUT) scheme to highlight differences in the intensities of the signals. Bars, 10 μm.
Hence, the lipid composition of the envelope from sucrose gradient-purified WNV and RSPs was analyzed by mass spectrometry and compared to that of total cellular membranes of HeLa cells (Fig. 4A) following an approach similar to that described for other viruses (7–10). The most abundant GPL in virions, RSPs, and total cellular membranes was PC, although both RSPs and virions displayed significantly reduced contents of PC relative to those of the total cellular membranes (about 0.57-fold for both RSPs and virions). Among the SLs, a significant enrichment in SM was noticed in the envelope of RSPs and virions compared to the amount in total cellular membranes (about 3.9- and 2.5-fold, respectively). These analyses also revealed that the lipid composition of both virions and RSPs was very similar for all lipid classes analyzed, except for p-PC, which appeared to be enriched in virions compared with the amounts in both RSPs and total cellular membranes. This could be related to the differences in the sizes between virions (50 nm) and RSPs (30 nm), since vesicles formed with p-PC are larger than those formed with PC (48).
FIG 4.
Lipid composition of WNV RSPs and virions. (A) GPL and SL compositions of WNV RSPs and virions. The content of individual lipid classes of total HeLa cells and WNV RSPs or virions was determined by summing the absolute abundances of all species identified. Values are standardized to the mole percentage of all membrane lipids detected in the sample. Values multiplied by 10 for visibility are indicated. (B) Individual lipid species whose levels were significantly altered in virions or RSPs in comparison to their levels in total cellular membranes. Data are presented as the mean ± SD. Statistically significant differences are indicated: *, P < 0.05.
Of the 162 lipid species identified in total cellular membranes, about 95% and 93% were also detected in RSPs (n = 155) and virions (n = 152), respectively (see Data Set S1 in the supplemental material). Significant differences in the proportion of lipid species between RSPs and virions were restricted to 3 lipid species (lyso-PC, 18:2; CER, 18:0; dhSM, 24:0; these represented only about 2% of the total species identified) (see Data Set S1 in the supplemental material), supporting the suggestion that the lipid envelopes of RSPs and virions share multiple common features. Compared to the lipid species in total cellular membranes, virions and RSPs displayed significantly altered proportions of 30 (19%) and 36 (24%) lipid species, respectively, with 15 of them being significantly altered in both virions and RSPs (Fig. 4B). Among these common species altered, 5 corresponded to PE or its derivative, p-PE (4 were enriched [PE, 32:1; p-PE, 32:2; p-PE, 34:3; p-PE, 36:3] and 1 was reduced [PE, 36:4]), and 6 corresponded to PC (34:1, 34:3, 36:2, 38:5, 38:6, and 44:3, all of which were reduced). Regarding SLs, the enrichment of dhCER (24:0) and three SM species (16:0, 24:0, and 24:2) was noticed. When lipids enriched in both RSPs and virions (Fig. 4B) were compared with those increased in infected cells (Fig. 1D), only 5 matches were found (p-PE, 34:3; p-PE, 36:3; dhCER, 24:0; SM, 24:0; SM, 24:1). Interestingly, three of these species were SLs, suggesting that the enrichment of these molecular species in infected cells could be related to an increasing demand for these SLs for the assembly of the viral envelope.
Involvement of SM and CER in flavivirus biogenesis.
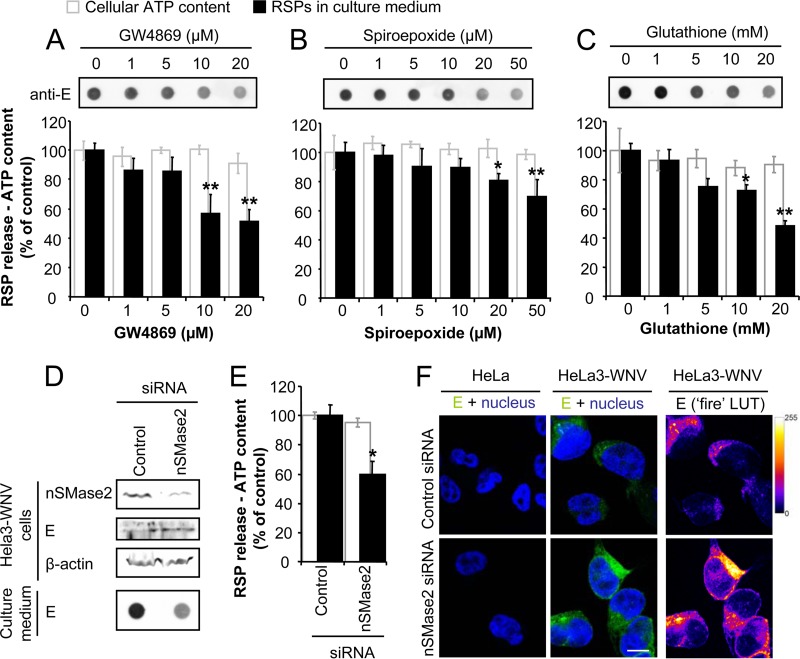
The SLs are derived from sphingosine, a long-chain amino alcohol, which is acylated with a long-chain fatty acid to give CER, which in turn is the central core of SM. These lipids are important components for membrane organization and shape, playing key roles in multiple cellular processes (49). Our analysis showed that different SLs (CER, dhCER, and SM) were increased in infected cells (Fig. 1C); this was particularly the case for SM, which was found to be enriched in the lipid envelope of both RSPs and virions (Fig. 4A). The hydrolytic removal of the phosphocholine moiety of SM by sphingomyelinases (SMases) to render CER has been associated with the induction of membrane curvature in SL-enriched membrane domains (50, 51). In fact, compounds that inhibit the activity of the neutral SMase 2 (nSMase2; also named sphingomyelin phosphodiesterase 3 [SMPD3]) can inhibit budding processes (18). Since these observations are compatible with the potential roles of SLs during WNV infection, i.e., in virion budding and/or assembly, we first addressed the effect of nSMase inhibition in HeLa3-WNV cells, used as a model system to study flavivirus biogenesis in the absence of virus replication. To this end, we selected three structurally unrelated nSMase inhibitors (18): GW4869, a noncompetitive inhibitor of nSMase (34); spiroepoxide, a selective irreversible inhibitor of nSMase (52); and glutathione, a cellular regulator of nSMase activity (53). Treatment of these cells with either of these inhibitors significantly reduced the amount of RSPs released into the culture medium without exerting noticeable toxic effects on the cells (Fig. 5A to C). GW4869 was the drug that produced significant inhibition at a lower concentration, so this compound was selected for further experiments involving virus infections (see below). As a complementary approach, the effect of nSMase2 depletion using RNA interference was also analyzed. Silencing of the expression of nSMase2 by transfection with siRNA, which was verified by Western blotting (Fig. 5D), significantly inhibited the release of RSPs into the culture medium (Fig. 5D and E). When the E-protein content in siRNA-transfected cells was analyzed by Western blotting (Fig. 5D) or immunofluorescence (Fig. 5F), no reduction was observed in cells transfected with siRNA against nSMase2. These results point to an accumulation of viral proteins inside cells depleted of nSMase2, which is consistent with the reduction in the release of RSPs from cells depleted of nSMase2 that was observed. The extent of the inhibition of RSP release in cells depleted of nSMase2 by siRNA (Fig. 5E) was comparable to that exerted by treatment with nSMase inhibitors (Fig. 5A to C), further supporting the requirement for nSMase function during the biogenesis of flaviviruses.
FIG 5.
Impairment of nSMase function inhibits the release of RSPs. (A to C) Treatment with the nSMase inhibitor GW4869 (A), spiroepoxide (B), or glutathione (C) impairs the release of RSPs by HeLa3-WNV cells. The content of RSPs in the culture medium of cells treated (for 4 h) with the drugs was determined by enzyme-linked immunodot assay using a monoclonal antibody against the E glycoprotein. The amount of RSPs determined by enzyme-linked immunodot assay was quantified, and the cellular ATP content for each drug concentration is shown in the graphs. (D) Depletion of nSMase2 by RNA interference. HeLa3-WNV cells were transfected with universal negative-control siRNA or with siRNA against nSMase2 for 48 h, and the amount of nSMase2, E glycoprotein, and β-actin in cell lysates was analyzed by Western blotting using specific antibodies. RSP release into the culture medium after 4 h of incubation in cells transfected for 48 h with siRNA was analyzed by enzyme-linked immunodot assay using an anti-E antibody. (E) Quantification of the amount of RSPs released into the culture medium by HeLa3-WNV cells transfected with siRNA control or against nSMase2 from panel D. The cellular ATP content for each drug concentration is shown as an indicator of cell viability upon drug treatment. (F) Immunofluorescence analysis of the expression of the WNV E glycoprotein in HeLa3-WNV cells transfected with siRNAs. HeLa or HeLa3-WNV cells transfected as described in the legend to panel D were subjected to immunofluorescence analysis using an antibody directed against E glycoprotein (green) and observed by confocal microscopy. Nuclei were stained with ToPro3 (blue). The panels on the right display E glycoprotein staining with false coloring from dark purple to bright yellow by use of the fire lookup table (LUT) to highlight differences in the intensity of the signal. Bars, 10 μm. Data are presented as the mean ± SD. Statistically significant differences are indicated: *, P < 0.05; **, P < 0.005.
Interference with nSMase function reduces flavivirus release from infected cells.
The effect of the nSMase inhibitor GW4869 on WNV infection was assayed in two mammalian cell lines (HeLa and Vero) and one mosquito cell line (C6/36) (Fig. 6A to C). Treatment with GW4869 at concentrations of ≥5 μM significantly inhibited the release of infectious WNV in these three cell lines, confirming the requirement for nSMase for the production of WNV in both mammalian and insect cells. The effect of this inhibitor on infection with Usutu virus (USUV), a related emerging flavivirus, was also tested in HeLa cells (Fig. 6D). In these experiments, inhibition of USUV similar to that of WNV at drug concentrations of ≥5 μM was observed. To analyze whether the requirement for nSMase function for virus production was shared by other unrelated arboviruses, the effect of GW4869 on infection with the alphavirus Sindbis virus (SINV) was analyzed (Fig. 6E). Interestingly, the release of SINV infectious particles from HeLa cells was not inhibited but was significantly increased by treatment with GW4869. This enhancing effect on virus titer was similar to that previously reported when SINV infection was performed in cells exhibiting an altered SL content induced by depletion of acid SMase, which was associated with an increase in virion infectivity (54). We next addressed the effect of depletion of nSMase2 on infection with these three viruses by transfection with specific siRNA (Fig. 6F). Silencing of nSMase2 expression significantly inhibited the production of both WNV and USUV to an extent comparable to that produced by treatment with GW4869. Conversely, a significant increase in SINV titer was found in cells treated with siRNA against nSMase2. Therefore, these observations confirm the results obtained with GW4869 and highlight the involvement of nSMase function during flavivirus infection.
FIG 6.
Impairment of nSMase function inhibits the release of WNV. (A to C) Treatment with the nSMase inhibitor GW4869 reduces WNV production in mammalian and insect cells. HeLa (A), Vero (B), or C6/36 (C) cells were infected with WNV (MOI, 1 PFU/cell) and treated with the indicated drug, and the amount of infectious virus released into the culture medium was determined by plaque assay at 24 h p.i. The cellular ATP content of uninfected cells treated with the different concentrations of the drug is indicated in all graphs as an estimate of cell viability upon drug treatment. (D) Treatment with GW4869 reduces USUV production. HeLa cells were infected with USUV (MOI, 1 PFU/cell), and the amount of infectious virus released was determined as described in the legends to panels A to C. (E) Treatment with GW4869 increases SINV production. HeLa cells were infected with SINV (MOI, 1 PFU/cell), and the amount of infectious virus released into the culture medium was determined by plaque assay (8 h p.i.). (F) Effect of nSMase2 depletion by siRNA on WNV, USUV, and SINV infection. HeLa cells were transfected with a universal negative siRNA control or with siRNA against nSMase2 for 48 h and infected with WNV, USUV, or SINV, and the amount of virus released into the culture medium was analyzed as described in the legend to panels A to C. Data are presented as the mean ± SD. Statistically significant differences are indicated: *, P < 0.05; **, P < 0.005.
Inhibition of nSMase activity reduces WNV-induced membrane budding.
The mechanism behind the reduction of WNV growth upon nSMase inhibition using GW4869 was further studied. The amount of viral particles released into the culture medium showed a significant reduction, as determined by an enzyme-linked immunodot assay analyzing the levels of E glycoprotein secreted in cells treated with GW4869 (Fig. 7A). This result was consistent with the decrease in RSP and infectious virus release observed (Fig. 5A and 6A). Analysis of infected cells treated with GW4869 by immunofluorescence and confocal microscopy showed an accumulation of E glycoprotein colocalized with the ER marker calnexin (Fig. 7B). To rule out the possibility that the effect of GW4869 on the biogenesis of WNV occurred in a step after virus assembly or maturation, the amount of cell-associated infectious virus or infectious virus released into the medium was determined for cells treated with GW4869 (Fig. 7C). Whereas treatment with GW4869 significantly reduced the amount of infectious WNV released, it did not induce an accumulation of cell-associated infectious virus. When the amounts of cell-associated viral RNA and viral RNA released into the medium were analyzed (Fig. 7D), a significant reduction of viral RNA levels in the culture medium of cells treated with GW4869 was observed. Conversely, a significant accumulation of cell-associated viral RNA was noticed. Overall, these results are compatible with a reduction in the amount of infectious virus released with a concomitant accumulation of viral RNA but not the amount of infectious particles inside GW4869-treated cells, a result which suggests that inhibition of nSMase function alters a process of flavivirus biogenesis before the assembly and/or maturation of infectious particles. Interestingly, the extent of the inhibition of PFU release (about 60%; Fig. 7C) was higher than the extent of the inhibition of total particle or viral RNA release (about 40%; Fig. 7A and D). This could suggest that GW4869 not only decreases the amount of released viral particles but also elevates the proportion of secreted noninfectious particles.
FIG 7.
Inhibition of nSMase function reduces WNV-induced budding. (A) Treatment with the nSMase inhibitor GW4869 reduces WNV release. HeLa cells were infected with WNV (MOI, 1 PFU/cell) and treated with the drug, and the amount of virus particles released into the culture medium was estimated by an enzyme-linked immunodot assay using a monoclonal antibody against the E glycoprotein at 24 h p.i. (B) Localization of the WNV E glycoprotein in HeLa cells infected with WNV (MOI, 1 PFU/cell) and treated with 10 μM GW4869. Immunofluorescence was performed using a monoclonal antibody directed against the E glycoprotein (green) in combination with a rabbit polyclonal antibody against calnexin (red) to stain the ER at 24 h p.i. Nuclei were stained with ToPro3 (blue). The panels in the rightmost column display E glycoprotein staining with false coloring from dark purple to bright yellow by use of the fire lookup table (LUT) scheme to highlight differences in the intensities of the signals. Bars, 10 μm. (C) Impairment of nSMase function with GW4869 does not increase the amount of cell-associated WNV. HeLa cells were infected with WNV (MOI, 1 PFU/cell), and the amount of virus either released or cell associated was determined by plaque assay at 24 h p.i. (D) The amount of viral RNA released into the culture medium or cell associated was determined by quantitative RT-PCR in cells infected and treated as described in the legend to panel C. (E) Representative electron micrographs at 24 h p.i. of cells infected with WNV (MOI, 10 PFU/cell) and treated or not treated (control) with 10 μM GW4869 showing the formation of vesicle packets (VPs) containing virus-induced vesicles (Ve) and electron-dense virions (Vi). Bar, 200 nm. (F) Diameter of virus-induced vesicles detected in the VPs of cells infected and treated or not treated with 10 μM GW4869. Each point represents an individual virus-induced vesicle. Solid lines denote the mean vesicle diameter. (G) Number of virus-induced vesicles per VP detected in the VPs of cells treated or not treated with 10 μM GW4869. Each point represents the number of virus-induced vesicles detected in an individual VP. Solid lines denote the mean number of virus-induced vesicles per VP. Unless specified otherwise, data are presented as the mean ± SD. Statistically significant differences are indicated: **, P < 0.005.
As the inhibition of nSMase function can interfere with intracellular budding processes, the effect of GW4869 on virus-mediated budding was analyzed. To this end, infected cells were treated with GW4869 and analyzed by transmission electron microscopy (Fig. 7E). Cells treated with the nSMase inhibitor displayed ultrastructural features associated with flavivirus infection, i.e., the formation of VPs and the presence of electron-dense virions. VPs are produced by budding into the ER, and the abundance of these structures can be quantified by transmission electron microscopy in flavivirus-infected cells (55). According to this view, these structures were analyzed to directly evaluate the effect of nSMase inhibition on viral budding. The mean diameter of the virus-induced vesicles detected inside the VPs (about 80 nm) in control and GW4869-treated cells was indistinguishable (Fig. 7F). However, when the number of vesicles inside VPs was scored, a significant reduction in the number of vesicles inside each VP in GW4869-treated cells relative to the number in control infected cells was noted (mean numbers of vesicles per VP, 1.9 ± 0.2 and 5.8 ± 0.6, respectively; Fig. 7G), supporting the suggestion that viral budding into the ER was reduced upon inhibition of nSMase function.
DISCUSSION
Our results illustrate the alteration of the content of a wide variety of lipid metabolites in WNV-infected cells. The increase in the cellular content of PC, lysophospholipids (lyso-PC), SM, and CER in HeLa cells infected with WNV is consistent with previous results with mosquito cells infected with DENV (26). Our analysis also revealed the enrichment of p-PC, p-PE, and dhCER in WNV-infected cells. Apart from PC, which is a cylindrical lipid, the rest of the lipids whose levels were found to be increased in WNV-infected cells were conical. The increase in the levels of these conical lipids could be related to their roles in the establishment and maintenance of the cellular membrane curvature necessary for proper viral replication complex assembly, as well as for virion envelopment. The enrichment of unsaturated PC species in DENV-infected cells has been suggested to be associated with the development of more fluidic membranes (26). Supporting this idea, all PC species whose levels were increased in WNV-infected cells were unsaturated. Likewise, the observed enrichment of SLs (CER, dhCER, and SM) in WNV-infected cells is also compatible with the requirement for these molecules in the membranes where virus replication and assembly take place. According to this hypothesis, it has been documented that DENV and hepatitis C virus replication complexes are enriched in SLs (17, 26).
To identify the potential roles of the lipid classes analyzed in the biogenesis of WNV particles, the proportion of each lipid in both purified virions and RSPs was analyzed. All lipid classes analyzed, including PS, which has been shown to play a functional role during flavivirus entry, were detected in the envelopes of WNV and RSPs (11). The high degree of similarity between the lipid composition of RSPs and virions, along with their parallel mechanisms of formation, reinforces the view that RSPs are a valuable tool for the study of lipid involvement during the biogenesis of flaviviruses.
We next addressed the question of whether the lipid composition of the WNV envelope was similar to that of total host cell membranes or if it reflected that of a particular membrane domain, following an approach similar to the approaches used for the characterization of other viral envelopes (7, 10). This analysis indicated that the envelopes of both RSPs and virions were enriched in SM and exhibited a reduction in the content of PC compared to that of total cellular membranes. Similar features, such as an increase in the SM (or other SL) content and a depletion of PC, have also been observed in the lipid envelopes of human immunodeficiency virus type 1 (HIV-1) and influenza virus (7, 9). On the contrary, the rhabdovirus vesicular stomatitis virus and the alphavirus Semliki Forest virus exert little lipid selection when they acquire their envelopes (8). The characteristics of the WNV envelope also differed from those of the hepatitis C virus envelope, which more closely resembles the composition of very low- and low-density lipoproteins (10). Enrichment in SLs and a reduction of PC levels are characteristics of liquid ordered membrane microdomains or membrane rafts (7, 9, 18, 56). Since membrane raft-like microdomains, including those located in the ER (57), are usually cholesterol enriched (7, 9, 18, 56), our findings could be connected to the role(s) of membrane microdomains enriched in cholesterol in flavivirus infections (28, 31, 58–60). SL-enriched membrane microdomains have functional properties directly linked to their lipid composition, such as coalescence and the promotion of budding processes during virus assembly (7, 9) or exosome biogenesis (18). Indeed, flavivirus assembly by invagination of ER membrane shares topological features with the formation of exosomes, since both processes mainly consist of budding outward from the cell cytoplasm. Supporting this hypothesis, the parallelism between viral budding processes and exosome biogenesis has already been documented (61, 62). An important point that must also be considered in these analyses is that in the RSP system, HeLa3-WNV cells express viral prM and E proteins in the absence of any other viral protein. Since viral nonstructural proteins (such as NS4A) induce membrane proliferation (63, 64), their expression is expected to modify cellular lipids in infected cells. In fact, modification of cellular lipids upon infection with WNV was confirmed by analysis of the lipids in WNV-infected cells. Therefore, it is foreseeable that the lipid components from WNV-infected cells can differ from those of RSP-producing cells, which may explain some of the minor differences between RSPs and virions found in the analysis, such as the amount of p-PC or variations in the specific lipid species found between RSPs and virions. However, the major features observed (a reduction of PC and enrichment in SM) were common between the two systems, supporting the use of RSPs as a model to study flavivirus biogenesis.
The hydrolysis of SM by nSMases converts SM into CER, a compound with a smaller head size (49). CER induces an asymmetric membrane tension and segregates into highly ordered domains triggering modifications of membrane shape (65). These properties have been associated with the promotion of membrane bending and budding processes both on model membranes (50) and in living cells (18, 51, 66, 67). In this scenario, pharmacological inhibition of nSMase activity, which catalyzes the hydrolysis of SM to CER, impaired the release of RSPs into the culture medium and reduced the level of production of infectious particles of WNV and the related flavivirus USUV, which parallel the effects on the production of certain types of exosomes that have been documented (18, 66, 67). Supporting this observation, depletion of nSMase2 by RNA interference inhibited the release of RSPs and the production of WNV and USUV. On the contrary, an increase in the titer of another arbovirus (SINV) was observed upon inhibition of nSMase function, therefore indicating that the requirement for the conversion of SM to CER during the biogenesis of flavivirus is not shared by this alphavirus. Consistent with these results, lipid recruitment during alphavirus envelopment has been proposed to be a process with a low level of selection (8), in contrast to the enrichment of SM and the reduction in the level of PC described here for the flavivirus envelope. Alphavirus biogenesis is dependent on the functionality of the secretory pathway, since viral envelope glycoproteins must traffic from the ER through the Golgi complex to the plasma membrane for viral budding (68). Thus, the lack of inhibition of SINV release upon pharmacological inhibition of nSMase2 or siRNA-mediated depletion of this enzyme shows the functionality of the secretory pathway in cells treated with GW4869 or siRNA against nSMAse2. This observation further supports the specific involvement of nSMase2 in the biogenesis of flavivirus. Although other nSMases are expected to be functional in cells depleted of nSMase2 by siRNA, the degree of inhibition (of RSPs and virion release) achieved with specific siRNA against nSMase2 was similar to that observed using the nSMase inhibitor GW4869, thus reinforcing the idea that nSMase2 is the main nSMase involved in flavivirus biogenesis.
When the effect of nSMase inhibition on virus biogenesis was analyzed by several approaches, a reduction of the amount of virus-induced vesicles inside VPs in cells treated with an inhibitor of nSMase function was noticed. Since these vesicles are associated with flavivirus replication and virion envelopment (5, 43, 55), this observation supports the role of SM conversion to CER in virus-induced budding. The diameter of the vesicles found in cells treated with the nSMase inhibitor was similar to that of the vesicles observed in untreated cells. A possible explanation for this phenomenon could be that vesicle components others than lipids (presumably, viral proteins) also contribute to vesicle formation, although the inhibition of nSMase activity makes this assembly less efficient. Accordingly, an important role for viral glycoproteins in the induction and maintenance of membrane curvature in flavivirus virions has been proposed (69). Since budding mechanisms are usually driven by a regulated combination of proteins and lipids (70), our results indicate that, indeed, the lipid composition of the flavivirus envelope also contributes to this process.
Interestingly, the dhCER content was found to be increased in WNV-infected cells, as it was one of the molecular species enriched in the viral envelope (dhCER, 24:0). This metabolite is an intermediate in the SL biosynthetic pathway (49), and the presence of increased levels of dhCER points to the upregulation of the de novo synthesis of SLs within WNV-infected cells, as reported in other virus models (17, 26). On the other hand, the involvement of nSMase in WNV and USUV biogenesis indicates that the production of CER by the hydrolysis of SM is also required during infection with these pathogens, providing evidence of the functionality of these two pathways for CER production in flavivirus-infected cells. However, one must be cautious with the interpretation of the reason for the increase in CER levels in WNV-infected cells, since this can be due to different factors, such as the induction of ER stress (71) mediated by the activation of the unfolded protein response in flavivirus-infected cells (72).
In summary, our results provide additional evidence of the complex degree of manipulation of host cell lipid metabolism by WNV. These results also unveil a connection between SL metabolism and flavivirus biogenesis. The findings presented here could contribute to the further development of lipid-based antiviral strategies to combat these pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Fabrias and E. Dalmau for help with lipid analyses, M. Guerra and M. T. Rejas for help with electron microscopy, and M. Calvo for technical assistance.
This work was supported by grants RTA 00036-2011 (to J.-C.S.), BIO2011-24351 (to F.S.), and E-RTA2013-0013 (to J.-C. S. and F.S.) and by the Fundacio la Marato de TV3 (grant 112130 to J.C.). M.A.M.-A. is the recipient of a Junta de Ampliación de Estudios (JAE) postdoctoral fellowship from the Spanish Research Council (CSIC). T.M.-R. is the recipient of a Formación de Personal Investigador (FPI) predoctoral fellowship from INIA.
Footnotes
Published ahead of print 13 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02061-14.
REFERENCES
- 1. Martin-Acebes MA, Saiz JC. 2012. West Nile virus: a re-emerging pathogen revisited. World J. Virol. 1:51–70. 10.5501/wjv.v1.i2.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brinton MA. 2013. Replication cycle and molecular biology of the West Nile virus. Viruses 6:13–53. 10.3390/v6010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suthar MS, Diamond MS, Gale M., Jr 2013. West Nile virus infection and immunity. Nat. Rev. Microbiol. 11:115–128. 10.1038/nrmicro2950 [DOI] [PubMed] [Google Scholar]
- 4. den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64:241–256. 10.1146/annurev.micro.112408.134012 [DOI] [PubMed] [Google Scholar]
- 5. Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 84:10438–10447. 10.1128/JVI.00986-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13–22. 10.1038/nrmicro1067 [DOI] [PubMed] [Google Scholar]
- 7. Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. 2006. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. U. S. A. 103:2641–2646. 10.1073/pnas.0511136103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kalvodova L, Sampaio JL, Cordo S, Ejsing CS, Shevchenko A, Simons K. 2009. The lipidomes of vesicular stomatitis virus, Semliki Forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J. Virol. 83:7996–8003. 10.1128/JVI.00635-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerl MJ, Sampaio JL, Urban S, Kalvodova L, Verbavatz JM, Binnington B, Lindemann D, Lingwood CA, Shevchenko A, Schroeder C, Simons K. 2012. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 196:213–221. 10.1083/jcb.201108175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Merz A, Long G, Hiet MS, Brugger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 286:3018–3032. 10.1074/jbc.M110.175018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. 2012. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12:544–557. 10.1016/j.chom.2012.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carro AC, Damonte EB. 2013. Requirement of cholesterol in the viral envelope for dengue virus infection. Virus Res. 174:78–87. 10.1016/j.virusres.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 13. Heaton NS, Randall G. 2011. Multifaceted roles for lipids in viral infection. Trends Microbiol. 19:368–375. 10.1016/j.tim.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin-Acebes MA, Vazquez-Calvo A, Caridi F, Saiz JC, Sobrino F. 2013. Lipid involvement in viral infections: present and future perspectives for the design of antiviral strategies, p 291–321 In Valenzuela R. (ed), Lipid metabolism. InTech, Rijeka, Croatia [Google Scholar]
- 15. Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 26:1179–1186. 10.1038/nbt.1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Q, Pene V, Krishnamurthy S, Cha H, Liang TJ. 2013. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat. Med. 19:722–729. 10.1038/nm.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, Ohta M, Tobita Y, Okano K, Ozeki K, Kawasaki K, Tsukuda T, Katsume A, Aoki Y, Umehara T, Sekiguchi S, Toyoda T, Shimotohno K, Soga T, Nishijima M, Taguchi R, Kohara M. 2012. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog. 8:e1002860. 10.1371/journal.ppat.1002860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- 19. Lorizate M, Krausslich HG. 2011. Role of lipids in virus replication. Cold Spring Harb. Perspect. Biol. 3:a004820. 10.1101/cshperspect.a004820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sprong H, van der Sluijs P, van Meer G. 2001. How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2:504–513. 10.1038/35080071 [DOI] [PubMed] [Google Scholar]
- 21. Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Buhler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. 10.1016/j.chom.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. U. S. A. 107:17345–17350. 10.1073/pnas.1010811107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nchoutmboube JA, Viktorova EG, Scott AJ, Ford LA, Pei Z, Watkins PA, Ernst RK, Belov GA. 2013. Increased long chain acyl-CoA synthetase activity and fatty acid import is linked to membrane synthesis for development of picornavirus replication organelles. PLoS Pathog. 9:e1003401. 10.1371/journal.ppat.1003401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. 10.1016/j.cell.2010.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. 2013. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 87:10612–10627. 10.1128/JVI.01370-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn RJ. 2012. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 8:e1002584. 10.1371/journal.ppat.1002584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heaton NS, Randall G. 2010. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8:422–432. 10.1016/j.chom.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mackenzie JM, Khromykh AA, Parton RG. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229–239. 10.1016/j.chom.2007.09.003 [DOI] [PubMed] [Google Scholar]
- 29. Martin-Acebes MA, Blazquez AB, Jimenez de Oya N, Escribano-Romero E, Saiz JC. 2011. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS One 6:e24970. 10.1371/journal.pone.0024970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rothwell C, Lebreton A, Young Ng C, Lim JY, Liu W, Vasudevan S, Labow M, Gu F, Gaither LA. 2009. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 389:8–19. 10.1016/j.virol.2009.03.025 [DOI] [PubMed] [Google Scholar]
- 31. Poh MK, Shui G, Xie X, Shi PY, Wenk MR, Gu F. 2012. U18666A, an intra-cellular cholesterol transport inhibitor, inhibits dengue virus entry and replication. Antiviral Res. 93:191–198. 10.1016/j.antiviral.2011.11.014 [DOI] [PubMed] [Google Scholar]
- 32. Vazquez-Calvo A, Saiz JC, Sobrino F, Martin-Acebes MA. 2011. Inhibition of enveloped virus infection of cultured cells by valproic acid. J. Virol. 85:1267–1274. 10.1128/JVI.01717-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin-Acebes MA, Saiz JC. 2011. A West Nile virus mutant with increased resistance to acid-induced inactivation. J. Gen. Virol. 92:831–840. 10.1099/vir.0.027185-0 [DOI] [PubMed] [Google Scholar]
- 34. Luberto C, Hassler DF, Signorelli P, Okamoto Y, Sawai H, Boros E, Hazen-Martin DJ, Obeid LM, Hannun YA, Smith GK. 2002. Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J. Biol. Chem. 277:41128–41139. 10.1074/jbc.M206747200 [DOI] [PubMed] [Google Scholar]
- 35. Blazquez AB, Escribano-Romero E, Merino-Ramos T, Saiz JC, Martin-Acebes MA. 2013. Infection with Usutu virus induces an autophagic response in mammalian cells. PLoS Negl. Trop. Dis. 7:e2509. 10.1371/journal.pntd.0002509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schalich J, Allison SL, Stiasny K, Mandl CW, Kunz C, Heinz FX. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 38:4066–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blazquez AB, Saiz JC. 2010. West Nile virus (WNV) transmission routes in the murine model: intrauterine, by breastfeeding and after cannibal ingestion. Virus Res. 151:240–243. 10.1016/j.virusres.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 39. Garanto A, Mandal NA, Egido-Gabas M, Marfany G, Fabrias G, Anderson RE, Casas J, Gonzalez-Duarte R. 2013. Specific sphingolipid content decrease in Cerkl knockdown mouse retinas. Exp. Eye Res. 110:96–106. 10.1016/j.exer.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. 2005. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 36:207–224. 10.1016/j.ymeth.2005.01.009 [DOI] [PubMed] [Google Scholar]
- 41. Canals D, Mormeneo D, Fabrias G, Llebaria A, Casas J, Delgado A. 2009. Synthesis and biological properties of pachastrissamine (jaspine B) and diastereoisomeric jaspines. Bioorg. Med. Chem. 17:235–241. 10.1016/j.bmc.2008.11.026 [DOI] [PubMed] [Google Scholar]
- 42. Gorrochategui E, Casas J, Perez-Albaladejo E, Jauregui O, Porte C, Lacorte S. 28 June 2014. Characterization of complex lipid mixtures in contaminant exposed JEG-3 cells using liquid chromatography and high-resolution mass spectrometry. Environ. Sci. Pollut Res. Int. 10.1007/s11356-014-3172-5 [DOI] [PubMed] [Google Scholar]
- 43. Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. 10.1016/j.chom.2009.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lorenz IC, Kartenbeck J, Mezzacasa A, Allison SL, Heinz FX, Helenius A. 2003. Intracellular assembly and secretion of recombinant subviral particles from tick-borne encephalitis virus. J. Virol. 77:4370–4382. 10.1128/JVI.77.7.4370-4382.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Calvert AE, Huang CY, Blair CD, Roehrig JT. 2012. Mutations in the West Nile prM protein affect VLP and virion secretion in vitro. Virology 433:35–44. 10.1016/j.virol.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mukherjee S, Lin TY, Dowd KA, Manhart CJ, Pierson TC. 2011. The infectivity of prM-containing partially mature West Nile virus does not require the activity of cellular furin-like proteases. J. Virol. 85:12067–12072. 10.1128/JVI.05559-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang PG, Kudelko M, Lo J, Siu LY, Kwok KT, Sachse M, Nicholls JM, Bruzzone R, Altmeyer RM, Nal B. 2009. Efficient assembly and secretion of recombinant subviral particles of the four dengue serotypes using native prM and E proteins. PLoS One 4:e8325. 10.1371/journal.pone.0008325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hermetter A, Lohner K, Degovics G, Laggner P, Paltauf F. 1985. Effect of cholesterol on vesicle bilayer geometry of choline plasmalogen and comparison with dialkyl-, alkylacyl- and diacyl-glycerophosphocholines. Chem. Phys. Lipids 38:353–364 [DOI] [PubMed] [Google Scholar]
- 49. Merrill AH., Jr 2011. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 111:6387–6422. 10.1021/cr2002917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Holopainen JM, Angelova MI, Kinnunen PK. 2000. Vectorial budding of vesicles by asymmetrical enzymatic formation of ceramide in giant liposomes. Biophys. J. 78:830–838. 10.1016/S0006-3495(00)76640-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zha X, Pierini LM, Leopold PL, Skiba PJ, Tabas I, Maxfield FR. 1998. Sphingomyelinase treatment induces ATP-independent endocytosis. J. Cell Biol. 140:39–47. 10.1083/jcb.140.1.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arenz C, Giannis A. 2000. Synthesis of the first selective irreversible inhibitor of neutral sphingomyelinase. Angew. Chem. Int. Ed. Engl. 39:1440–1442. [DOI] [PubMed] [Google Scholar]
- 53. Liu B, Hannun YA. 1997. Inhibition of the neutral magnesium-dependent sphingomyelinase by glutathione. J. Biol. Chem. 272:16281–16287. 10.1074/jbc.272.26.16281 [DOI] [PubMed] [Google Scholar]
- 54. Ng CG, Coppens I, Govindarajan D, Pisciotta J, Shulaev V, Griffin DE. 2008. Effect of host cell lipid metabolism on alphavirus replication, virion morphogenesis, and infectivity. Proc. Natl. Acad. Sci. U. S. A. 105:16326–16331. 10.1073/pnas.0808720105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Junjhon J, Pennington JG, Edwards TJ, Perera R, Lanman J, Kuhn RJ. 2014. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 88:4687–4697. 10.1128/JVI.00118-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. 2003. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 278:10963–10972. 10.1074/jbc.M207550200 [DOI] [PubMed] [Google Scholar]
- 57. Browman DT, Resek ME, Zajchowski LD, Robbins SM. 2006. Erlin-1 and Erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 119:3149–3160. 10.1242/jcs.03060 [DOI] [PubMed] [Google Scholar]
- 58. Noisakran S, Dechtawewat T, Avirutnan P, Kinoshita T, Siripanyaphinyo U, Puttikhunt C, Kasinrerk W, Malasit P, Sittisombut N. 2008. Association of dengue virus NS1 protein with lipid rafts. J. Gen. Virol. 89:2492–2500. 10.1099/vir.0.83620-0 [DOI] [PubMed] [Google Scholar]
- 59. Garcia Cordero J, Leon Juarez M, Gonzalez-y-Merchand JA, Cedillo Barron L, Gutierrez Castaneda B. 2014. Caveolin-1 in lipid rafts interacts with dengue virus NS3 during polyprotein processing and replication in HMEC-1 cells. PLoS One 9:e90704. 10.1371/journal.pone.0090704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pena J, Harris E. 2012. Early dengue virus protein synthesis induces extensive rearrangement of the endoplasmic reticulum independent of the UPR and SREBP-2 pathway. PLoS One 7:e38202. 10.1371/journal.pone.0038202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morita E, Sundquist WI. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425. 10.1146/annurev.cellbio.20.010403.102350 [DOI] [PubMed] [Google Scholar]
- 62. Hanson PI, Cashikar A. 2012. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 28:337–362. 10.1146/annurev-cellbio-092910-154152 [DOI] [PubMed] [Google Scholar]
- 63. Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 80:4623–4632. 10.1128/JVI.80.9.4623-4632.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873–8882. 10.1074/jbc.M609919200 [DOI] [PubMed] [Google Scholar]
- 65. Lopez-Montero I, Monroy F, Velez M, Devaux PF. 2010. Ceramide: from lateral segregation to mechanical stress. Biochim. Biophys. Acta 1798:1348–1356. 10.1016/j.bbamem.2009.12.007 [DOI] [PubMed] [Google Scholar]
- 66. Yuyama K, Sun H, Mitsutake S, Igarashi Y. 2012. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-beta by microglia. J. Biol. Chem. 287:10977–10989. 10.1074/jbc.M111.324616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. 2013. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 288:10849–10859. 10.1074/jbc.M112.446831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jose J, Snyder JE, Kuhn RJ. 2009. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 4:837–856. 10.2217/fmb.09.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang W, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. 2013. Membrane curvature in flaviviruses. J. Struct. Biol. 183:86–94. 10.1016/j.jsb.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hurley JH, Boura E, Carlson LA, Rozycki B. 2010. Membrane budding. Cell 143:875–887. 10.1016/j.cell.2010.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Epstein S, Kirkpatrick CL, Castillon GA, Muniz M, Riezman I, David FP, Wollheim CB, Riezman H. 2012. Activation of the unfolded protein response pathway causes ceramide accumulation in yeast and INS-1E insulinoma cells. J. Lipid Res. 53:412–420. 10.1194/jlr.M022186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Blazquez AB, Escribano-Romero E, Merino-Ramos T, Saiz JC, Martin-Acebes MA. 2014. Stress responses in flavivirus-infected cells: activation of unfolded protein response and autophagy. Front. Microbiol. 5:266. 10.3389/fmicb.2014.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.