Abstract
We describe endogenous viral elements (EVEs) derived from parvoviruses (family Parvoviridae) in the genomes of the long-tailed chinchilla (Chinchilla lanigera) and the degu (Octodon degus). The novel EVEs include dependovirus-related elements and representatives of a clearly distinct parvovirus lineage that also has endogenous representatives in marsupial genomes. In the degu, one dependovirus-derived EVE was found to carry an intact reading frame and was differentially expressed in vivo, with increased expression in the liver.
TEXT
Parvoviruses are small, nonenveloped viruses containing a single-stranded DNA (ssDNA) genome ∼5 kb in length. In recent years, several reports have described endogenous viral elements (EVEs) derived from parvoviruses in animal genomes (1–4). We performed an in silico screen of recently published low-coverage genome sequence assemblies using the Database-Integrated Genome Screening (DIGS) tool, version 1.0 (http://paleovirology.org.uk/). Screening identified novel parvovirus-related EVEs in two caviomorph rodents, the long-tailed chinchilla (Chinchilla lanigera) and the degu (Octodon degus). A total of 12 novel EVEs were identified in these two species (Table 1).
TABLE 1.
Parvovirus EVEs in C. lanigera and O. degus genomes
| Elementa | GenBank accession no. | Orientationb | Structure | Groupc | Match coordinate |
|||
|---|---|---|---|---|---|---|---|---|
| Scaffold |
Viral genomed |
|||||||
| Start | End | Start | End | |||||
| C. lanigera-1 | JH721894.1 | −ve | NS-VP | Parvo | 14636996 | 14641202 | 1137 | 4398 |
| C. lanigera-2 | JH721993.1 | −ve | NS-VP | Parvo | 459340 | 462818 | 1065 | 4428 |
| C. lanigera-3 | JH721905.1 | −ve | NS | Dependo | 7599480 | 7600235 | 717 | 896 |
| C. lanigera-4* | JH721911.1 | −ve | NS | Dependo | 18094486 | 18095973 | 330 | 1684 |
| C. lanigera-5a | JH721873.1 | +ve | NS | Dependo | 22669255 | 22669578 | 1428 | 1724 |
| C. lanigera-5b | JH721873.1 | +ve | NS | Dependo | 22702412 | 22702712 | 1428 | 1724 |
| C. lanigera-6 | JH721896.1 | −ve | VP | Dependo | 13652894 | 13654833 | 4054 | 4407 |
| O. degus-1 | JH651603.1 | +ve | NS-VP | Parvo | 365906 | 369616 | 507 | 4443 |
| O. degus-2 | JH651624.1 | −ve | NS-VP | Parvo | 3857091 | 3858404 | 1089 | 2652 |
| O. degus-3 | JH651827.1 | −ve | NS-VP | Parvo | 1343057 | 1345620 | 295 | 3142 |
| O. degus-4* | JH651579.1 | +ve | NS | Dependo | 8427679 | 8429061 | 321 | 1784 |
| O. degus-5 | JH651577.1 | +ve | VP | Parvo | 12296699 | 12296983 | 4135 | 4419 |
| O. degus-6 | JH651549.1 | −ve | VP | Dependo | 16377593 | 16377937 | 4063 | 4401 |
Asterisks denote elements carrying intact open reading frames.
Orientation of elements in scaffold (−ve, negative orientation; +ve, positive orientation).
Dependo, dependovirus-related EVEs; parvo, parvovirus-related EVEs.
Viral genome coordinates are based on pairwise alignment to genus-specific reference sequences of adeno-associated virus 2 (GenBank accession no. AF043303.1) for dependoviruses and mouse minute virus (GenBank accession no. J02275.1) for parvovirus-like elements.
Chinchillas are medium-sized, crepuscular rodents that live at high altitude in the Andes mountains. Degus are small rodents endemic to the Chilean Matorral ecoregion. Although both species are indigenous to South America, they are relatively distantly related, having diverged ∼37 million years ago (MYA) (5).
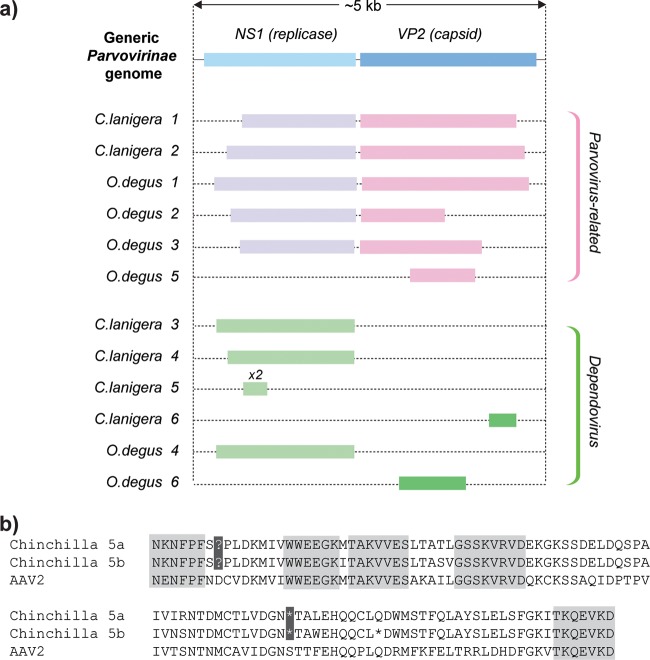
Parvovirus genomes comprise two major gene cassettes that separately encode nonstructural (NS) and structural (VP) proteins. The genetic structures of previously described parvovirus-derived EVEs have included complete viral genomes, intact individual genes, isolated genome fragments, and rearranged complete genomes (2, 3). A similar range of genetic structures was observed here, with five of the novel EVEs representing complete or nearly complete viral genomes spanning both major gene cassettes, while the remainder represented single gene cassettes or fragments of genes (Fig. 1a).
FIG 1.
(a) Genetic structures of parvovirus-derived EVEs in C. lanigera and O. degus genomes. Open reading frames (ORFs) were inferred by manual comparison of putative peptide sequences to those of closely related exogenous parvoviruses. (b) Alignment of a pair of duplicated EVEs in the Chinchilla lanigera genome against an adeno-associated virus type 2 (AAV2) reference sequence (GenBank accession no. AF043303.1). Blocks of six or more amino acids that are conserved between all three sequences are highlighted in gray. Nonsense mutations shared between the two duplicated EVEs are highlighted in black. The two EVEs differed at 8 of 323 nucleotide positions (i.e., an average of 4 nucleotide substitutions have occurred in each insertion), indicating that these EVEs arose in a duplication event that occurred between 2.8 and 5.6 MYA (assuming a range of mammalian neutral substitution rates from 2.2 × 109 to 4.5 × 109 per site per year [17]).
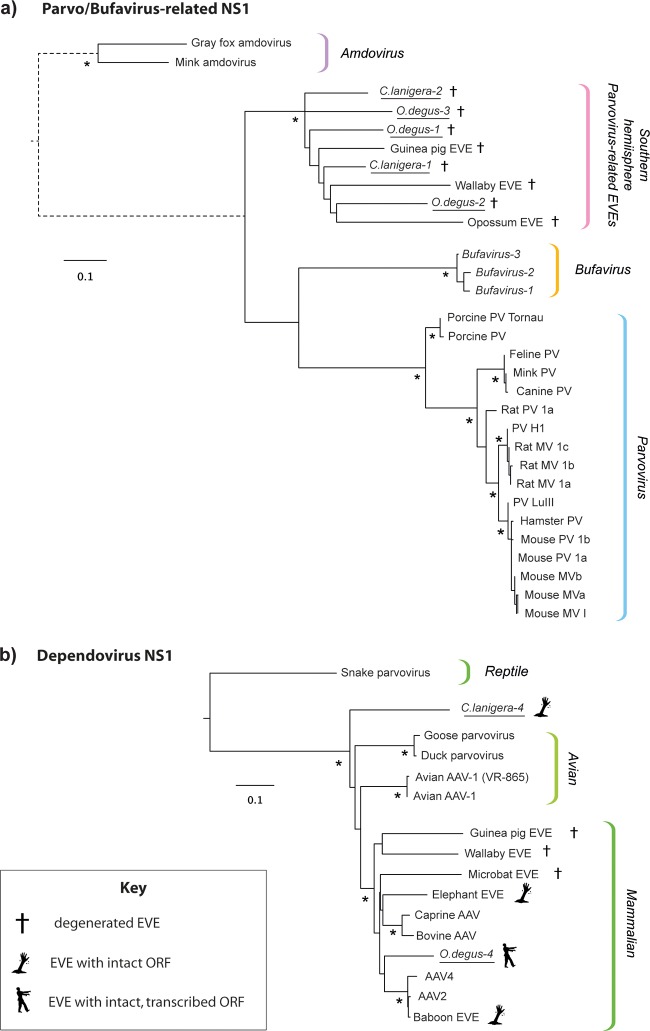
Phylogenetic analysis of the newly identified EVEs revealed that half of them grouped robustly within the diversity of avian and mammalian dependoviruses, while the others clustered in a single, well-defined clade composed exclusively of EVEs obtained from the genomes of South American and Australian mammals (Fig. 2). In phylogenies, this lineage of EVEs formed a sister clade with exogenous viruses in the Parvovirus and “Bufavirus” (6) genera.
FIG 2.
Maximum likelihood phylogenies showing the relationships of parvovirus-related (a) and dependovirus-related (b) EVEs in the C. lanigera and O. degus genomes to exogenous parvoviruses and previously described EVEs. Phylogenies are based on alignments of NS1 proteins and putative NS1 proteins encoded by EVE pseudogenes and were constructed using PHYML (18) and the JTT+ protein substitution model as selected by ProtTest (19). EVEs identified in this study are underlined. Phylogenies are midpoint rooted for clarity of presentation. The scale bar indicates evolutionary distance in numbers of substitutions per amino acid site. Branches shown as dashed lines have been shortened and are not shown to scale. Asterisks indicate nodes with maximum likelihood bootstrap support levels above 75% for 100 bootstrap replicates. EVE, endogenous viral element; AAV, adeno-associated virus; MV, minute virus; PV, parvovirus; NS, nonstructural protein gene cassette; VP, viral capsid protein gene cassette.
None of the novel EVEs were orthologous in the species examined; thus, we could not infer minimum dates of integration based on orthology. However, we did identify a pair of endogenous dependovirus elements (C. lanigera-5a and -5b [Table 1]) that had apparently been duplicated after integrating into the chinchilla genome. This pair of nearly identical elements shared at least two nonsense mutations (Fig. 1b), indicating that the coding sequence had degenerated prior to its duplication, and can thus be assumed to have been evolving neutrally in the subsequent period. These elements are at least as old as the duplication event that generated them, which we estimated to have occurred between 2.8 and 5.6 MYA (data not shown).
Although most of the novel EVEs contained frameshifts and/or stop codons, insertions encoding apparently intact NS1 proteins were identified in both the chinchilla and the degu genomes. To exclude the possibility that it was somehow derived from contaminating viral DNA (7, 8), one intact element (O. degus-4) was independently amplified from genomic DNA by PCR. Tissue was obtained from a fresh male headless O. degus cadaver (kindly donated by Adrian Palacios from Universidad de Valparaiso, Chile). Genomic DNA was extracted from liver tissue, and PCR using primers targeting the 5′ and 3′ flanking regions of O. degus-4 (Fig. 3a) confirmed the presence of this EVE locus in a second, outbred degu individual, demonstrating that O. degus-4 is a genuinely endogenous sequence.
FIG 3.
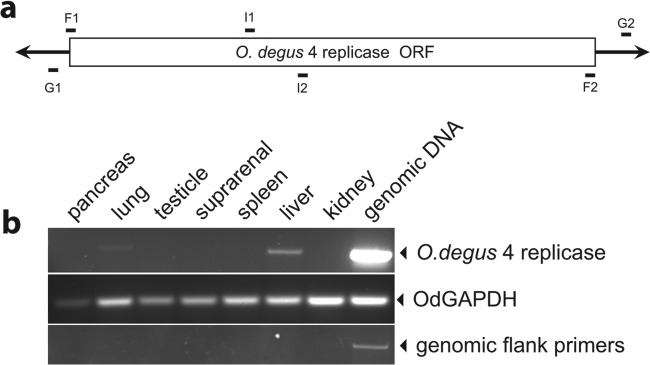
(a) Schematic representation of EVE O. degus-4. The replicase ORF is shown as a white box, and the 5′ and 3′ genomic sequences are shown as black lines. The relative positions of primers used in this study are shown. Primers F1 and F2, positioned at the extreme ends of the EVE, were used to amplify genomic DNA and sequence PCR products. Primers G1 and G2, positioned in the genomic flanking sequence, were used to control for genomic contamination of the RNA preparations. The internal primers I1 and I2 were used to detect O. degus-4 replicase mRNA. (b) Tissue-specific expression of a dependovirus-related EVE mRNA in the degu. Total RNA was extracted from O. degus pancreas, lung, testicle, suprarenal gland, spleen, liver, and kidney. After cDNA generation, expression of the O. degus-4 EVE was detected using primers designed to amplify the region comprising nucleotides (nt) 519 to 689 of the predicted mRNA (top panel). O. degus glyceraldehyde-3-phosphate dehydrogenase (OdGAPDH) mRNA (GenBank accession no. XM_004643553.1) was used as a positive control for mRNA presence (middle panel). To discard genomic DNA contamination in the cDNA preparations, primers aligning 35 nt upstream and 85 nt downstream of the O. degus-4 element were used (bottom panel). The different tissues analyzed are indicated above the top panel. Genomic DNA was used as a positive amplification control.
O. degus rodents were obtained from a breeding colony at the animal facility of the University of Valparaiso. All experiments were approved by the bioethics committee of the Universidad de Valparaiso and complied with the international NIH Approved Animal Welfare Assurance A5823-01.
Reverse transcription-PCR (RT-PCR) was used to investigate expression of O. degus-4 in distinct degu tissues. RNA was extracted from pancreas, liver, testicle, kidney, suprarenal, spleen, and lung tissues (Fig. 3b). This analysis revealed that the O. degus-4 replicase is differentially expressed in vivo, with markedly elevated expression of mRNA in the liver and little or no expression in other tissues.
Scientifically, EVEs can be approached from two overlapping but distinct perspectives. First, they can be viewed a kind of genomic “fossil record” from which the long-term, coevolutionary relationships of viruses and hosts can be inferred. In this respect, the presence of a specific, monophyletic lineage of EVEs in both South American and Australian marsupial genomes, and the apparent absence of this lineage from the genomes of Old World rodents, suggests the existence of an ancient parvovirus lineage that evolved in the indigenous mammal populations of biogeographically isolated Southern Hemisphere continents (marsupials and xenarthrans) and was acquired by caviomorph rodents subsequent to their colonization of the South American continent (estimated to have occurred ∼40 MYA [9]).
The second way in which EVEs can be viewed is as host genes. While most EVE sequences are highly degenerated, it is clear that at least a proportion of these elements have been coopted or “exapted” (i.e., adapted for a function distinct from that for which they originally evolved [10]) to perform physiological functions in their host species (11–14). Dependovirus-derived EVEs encoding intact replicase proteins have previously been identified in mammalian genomes, including those of the African elephant (Loxodonta africana) and the Hamadryas baboon (Papio hamadryas). However, this is the first study to demonstrate expression of such elements in vivo. While no physiological function has yet been demonstrated for the numerous parvovirus-related EVEs in mammalian genomes, the identification of an intact element with differential expression across tissues provides further indication that such functions may exist. Since degus are experimental organisms that are used currently to research mammalian pathologies and behaviors (15, 16), the identification of an intact, expressed parvovirus-derived EVE in this species suggests a possible path forward for research in this area.
ACKNOWLEDGMENTS
This work was supported by FONDECYT 1130852 (to G.A.) and United Kingdom Medical Research Council grant MC_UU_12014 (to R.G.).
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1. Belyi VA, Levine AJ, Skalka AM. 2010. Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the parvoviridae and circoviridae are more than 40 to 50 million years old. J. Virol. 84:12458–12462. 10.1128/JVI.01789-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kapoor A, Simmonds P, Lipkin WI. 2010. Discovery and characterization of mammalian endogenous parvoviruses. J. Virol. 84:12628–12635. 10.1128/JVI.01732-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6:e1001191. 10.1371/journal.pgen.1001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu H, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Peng Y, Yi X, Jiang D. 2011. Widespread endogenization of densoviruses and parvoviruses in animal and human genomes. J. Virol. 85:9863–9876. 10.1128/JVI.00828-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Opazo JC. 2005. A molecular timescale for caviomorph rodents (Mammalia, Hystricognathi). Mol. Phylogenet. Evol. 37:932–937. 10.1016/j.ympev.2005.05.002 [DOI] [PubMed] [Google Scholar]
- 6. Phan TG, Vo NP, Bonkoungou IJ, Kapoor A, Barro N, O'Ryan M, Kapusinszky B, Wang C, Delwart E. 2012. Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J. Virol. 86:11024–11030. 10.1128/JVI.01427-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naccache SN, Greninger AL, Lee D, Coffey LL, Phan T, Rein-Weston A, Aronsohn A, Hackett J, Jr., Delwart EL, Chiu CY. 2013. The perils of pathogen discovery: origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J. Virol. 87:11966–11977. 10.1128/JVI.02323-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smuts H, Kew M, Khan A, Korsman S. 2014. Novel hybrid parvovirus-like virus, NIH-CQV/PHV, contaminants in silica column-based nucleic acid extraction kits. J. Virol. 88:1398. 10.1128/JVI.03206-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Antoine PO, Marivaux L, Croft DA, Billet G, Ganerod M, Jaramillo C, Martin T, Orliac MJ, Tejada J, Altamirano AJ, Duranthon F, Fanjat G, Rousse S, Gismondi RS. 2012. Middle Eocene rodents from Peruvian Amazonia reveal the pattern and timing of caviomorph origins and biogeography. Proc. Biol. Sci. 279:1319–1326. 10.1098/rspb.2011.1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gould SJ, Vrba ES. 1982. Exaptation: a missing term in the science of form. Paleobiology 8:4–15 [Google Scholar]
- 11. Arnaud F, Caporale M, Varela M, Biek R, Chessa B, Alberti A, Golder M, Mura M, Zhang YP, Yu L, Pereira F, Demartini JC, Leymaster K, Spencer TE, Palmarini M. 2007. A paradigm for virus-host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog. 3:e170. 10.1371/journal.ppat.0030170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bezier A, Annaheim M, Herbiniere J, Wetterwald C, Gyapay G, Bernard-Samain S, Wincker P, Roditi I, Heller M, Belghazi M, Pfister-Wilhem R, Periquet G, Dupuy C, Huguet E, Volkoff AN, Lanzrein B, Drezen JM. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323:926–930. 10.1126/science.1166788 [DOI] [PubMed] [Google Scholar]
- 13. Rowe HM, Trono D. 2011. Dynamic control of endogenous retroviruses during development. Virology 411:273–287. 10.1016/j.virol.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 14. Dewannieux M, Heidmann T. 2013. Endogenous retroviruses: acquisition, amplification and taming of genome invaders. Curr. Opin. Virol. 3:646–656. 10.1016/j.coviro.2013.08.005 [DOI] [PubMed] [Google Scholar]
- 15. Okanoya K, Tokimoto N, Kumazawa N, Hihara S, Iriki A. 2008. Tool-use training in a species of rodent: the emergence of an optimal motor strategy and functional understanding. PLoS One 3:e1860. 10.1371/journal.pone.0001860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ardiles AO, Tapia-Rojas CC, Mandal M, Alexandre F, Kirkwood A, Inestrosa NC, Palacios AG. 2012. Postsynaptic dysfunction is associated with spatial and object recognition memory loss in a natural model of Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 109:13835–13840. 10.1073/pnas.1201209109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar S, Subramanian S. 2002. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. U. S. A. 99:803–808. 10.1073/pnas.022629899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 19. Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics (Oxford, England) 21:2104–2105. 10.1093/bioinformatics/bti263 [DOI] [PubMed] [Google Scholar]