Abstract
Iteradensoviruses are 5-kb parvoviruses with typical J-shaped inverted terminal repeats of about 250 nucleotides and terminal hairpins of about 165 nucleotides. The single-stranded DNA genome contains several open reading frames, but their expression strategy is still unknown. Here the transcription maps and expression of the viruses in this genus were explored. As for brevidensoviruses, the two nonstructural (NS) genes were expressed by overlapping promoters with alternate transcription starts at both sides of the NS1 start codon.
TEXT
Invertebrate densoviruses (DVs) form a separate subfamily (Densovirinae) within the Parvoviridae family (1–4) and have recently been reclassified (1). They share many physicochemical and genome properties with Parvovirinae of vertebrates (5–7), and most viruses of these subfamilies have a phospholipase A2 (PLA2) activity for cell entry (8–10). The left-hand side of the genome contains the nonstructural (NS) genes with the rolling-circle replication endonuclease and the superfamily 3 helicase in NS1 (11, 12). Open-reading frames (ORFs) for structural proteins (VPs) are on the right-hand side of the genomes (5, 6).
Densovirinae include the Ambidensovirus, Brevidensovirus, Hepandensovirus, Penstyldensovirus, and Iteradensovirus genera, which have distinct genome sizes and organizations and the presence or absence of inverted terminal repeats (ITRs) and PLA2 (1, 2). Iteradensoviruses have a 5-kb genome with ITRs, a monosense genome organization, and PLA2 activity. Four previously known iteradensoviruses are those from Bombyx mori (BmDV) (13), Casphalia extranea (CeDV) (14), Dendrolimus punctatus (DpDV) (15), and Helicoverpa armigera (HaDV2) (16). The reported sequence of HaDV2 is incomplete, differs significantly from the sequences of the others, and remains to be confirmed. Additionally, we isolated iteradensoviruses from Papilio polyxenes (black swallowtail butterfly) (PpDV) (17), Sibine fusca (oil palm pest) (SfDV) (18), and the monarch butterfly (DpIDV) (19) (Fig. 1A). Based on NS1 identities, the taxons of these viruses have been defined as lepidopteran iteradensovirus (LI) 1 with BmDV, the LI 2 species with CeDV, SfDV, and DpIDV, the LI 3 species with DpDV, the LI 4 species with PpDV, and the LI 5 species with HaDV2 (1). NS1 and NS2 proteins of the Chinese isolates (LI 3 and LI 5) had <40% identity scores with other iteradensoviruses. In contrast, except for HaDV2, the VP proteins of iteradensoviruses all have similar identities of 70 to 80%, independent of the species.
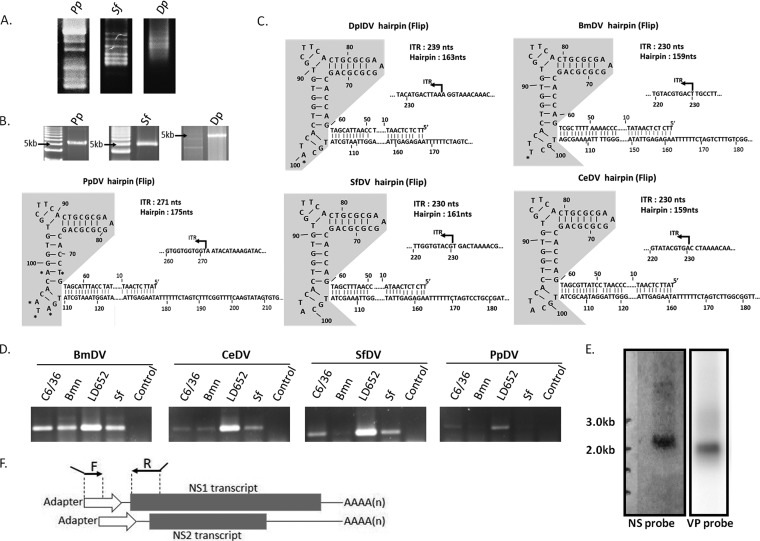
FIG 1.
(A) Detection of pathogens by sequence-independent single-primer amplification in larvae from Papilio polyxenes (Pp) and Sibine fusca (Sf) and in pupae from the monarch butterfly (Dp) on a 2% agarose gel. (B) DNA purification of new iteradensoviruses on 1% electrophoresis agarose gels. The DNA ladder is a 1-kb plus DNA ladder. (C) Inverted terminal repeats (ITRs) of five iteradensoviruses with J-shaped terminal hairpins. The shaded areas in the ITRs occur in two alternative sequences, named “flip” and its reverse complement “flop.” Iteradensoviruses' ITR sequences are highly similar to each other. Only a few nucleotides differ and are indicated with an asterisk. (D) Infectious clones of CeDV, BmDV, PpDV, and SfDV were transfected into four different insect cell lines (C6/36, Bmn, Ld652, and Sf), and total RNA was extracted 48 h posttransfection. Gene-specific primers were used for RT-PCR identification. PCR products were checked by electrophoresis in a 1% agarose gel. Total RNAs without reverse transcription were used as a control template for PCR. (E) Northern blotting of PpDV NS and VP transcripts. Both NS and VP yielded single transcript bands. However, further results showed that the 2.3-kb transcripts for NS consisted of two transcripts with almost identical sizes. (F) Scheme of a PCR method for distinguishing NS1 and NS2 transcription starts. A forward primer with ∼20 nt of complementary sequence with the RACE adapter at the 3′ end and an unspecific extension with restriction sites at the 5′ end. The 3′ ends of the reverse primers were extended with the sequence complementary to the putative NS1 ATG.
Infectious clones were created and sequenced for BmDV (pIN919), CeDV (pSMART-CeDV), PpDV (pCR2.1-PpDV), SfDV (pBlue-SfDV), and DpIDV (pBlue-DpIDV) (14, 17–20). The completely sequenced genomes all have typical J-shaped ITRs with lengths ranging from 230 to 271 nucleotides (nt) and terminal hairpins of 159 to 175 nt (Fig. 1B and C). The 5′- and 3′-terminal hairpins occurred in two orientations, “flip” and its reverse-complement orientation, “flop” (Fig. 1C). These were identical among the three LI 2 viruses and had few differences in PpDV and BmDV. The positions and sizes of the ORFs were virtually identical in all iteradensoviruses (Fig. 2), except for HaDV2 (which had a truncated NS). The typical parvoviral phospholipase A2 motif (8) was present in all VP1s.
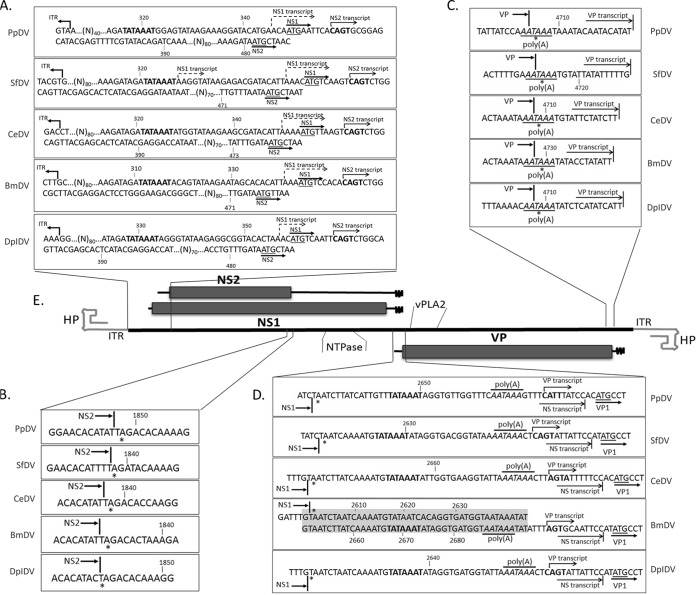
FIG 2.
Genome organization and transcription profiles of five iteradensoviruses. (A) The 5′ ends of NS transcripts and the ATG sequences of NS1 and NS2 ORFs are underlined. CAGT motifs of NS2 transcription starts are indicated in bold. NS1 transcription starts are marked with dashed arrows. (B) Stop codons of NS2 are indicated with an asterisk. (C) Ends of VP translation and 3′ ends of VP transcripts. (D) The 3′ ends of NS transcripts and 5′-end starts of VP transcripts. Poly(A) motifs of NS transcripts are in italics. Translation starts of VP are underlined. (E) The genome organization of iteradensoviruses is deduced from the ORFs of all five iteradensoviruses described in this report.
Densovirus transcription has not been studied extensively. So far, VP transcripts of different densoviruses with one VP ORF are not spliced but sets of N-terminally extended proteins are generated by leaky scanning (6, 21–23). Among others, densoviruses from Blattella germanica (BgDV) and Acheta domestica (AdDV) have two VP ORFs that are joined by splicing and generate proteins with alternate N termini in addition to nested N-terminally extended sets by leaky scanning (4, 24). Leaky scanning is also important for the NS proteins of densoviruses from different genera (4, 6, 21–24). However, splicing determines whether NS3 or NS1/NS2 is expressed for the ambisense densoviruses, whereas the two NS genes of the brevidensoviruses have overlapping promoters so that transcripts start at either side of the NS1 start codon, allowing either NS1 or the downstream NS2 to be expressed (21–24). Transcription of viruses in the Iteradensovirus genus has not yet been studied.
Production of iteradensovirus mRNAs and Northern blotting.
A major impediment to iteradensovirus studies has been the lack of suitable insect cells that support their replication. Four different insect cell lines were compared for their efficacy to produce iteradensovirus mRNA (Fig. 1D). LD652, Sf, and Bmn cells (in Sf-900 II medium) and mosquito C6/36 cells (in RPMI 1640 medium) were propagated with 5% and 10% fetal bovine serum, respectively. Transfections with the infectious clones of CeDV, BmDV, PpDV, and SfDV were performed using DOTAP liposomal transfection reagent (Roche). PpDV was purified as previously described (25). Third-instar Papilio larvae, the only host insect available, were fed pesticide-free parsley leaves that had been submerged in a PpDV suspension and served also as a source of PpDV mRNA. Reverse transcription (RT)-PCR results (Fig. 1D) showed that LD652 cells were most suitable for mRNA production of these iteradensoviruses. Total RNA was isolated from Papilio larvae 72 h postinfection and 48 h posttransfection from LD652 cells, which had been transfected with SfDV, BmDV, CeDV, or DpIDV infectious clones, by use of the NucleoSpin RNA II kit (Clontech). An extra DNase I treatment was added after RNA extraction with the Turbo DNA-free kit (Ambion). An RT-negative PCR test was included to verify the absence of DNA. Total RNA was subjected to an mRNA purification step using a Poly(A)Purist kit (Ambion). Northern blots were obtained as described previously (21). RNA probes for NS and VP genes were transcribed from NS and VP amplicons obtained by PCR using primers PpNSRpF700 and PpNSNpR850 for NS and PpVPRpF3200 and PpVPNpR3400 for VP (Table 1). Northern blotting of PpDV mRNA revealed one 2.3-kb band for the NS gene and one 2.0-kb band for the VP gene (Fig. 1E). Since the NS2 ORF overlapped the N-terminal half of NS1, it is expected to share the same poly(A) motif in the 3′ ends of their transcripts.
TABLE 1.
Primers
| Primera | Sequence (5′→3′)b | Purpose |
|---|---|---|
| Nbam24 | AGGCAACTGTGCTATCCGAGGGAG | SISPA adapter and primer |
| NCsp11 | TACTCCCTCGG | SISPA adapter and primer |
| Pa140 | GTT GTG GTG GTA TAT ATT GAG AAT AT | Highly conserved primer in iteradensoviruses' ITR |
| Pp5NS-1 | GTCAAGCACTTTCCTGAGGTTAA | 5′-RACE primer for PpDV NS |
| Pp3NS2–1 | ATAGAACAGAGCCAATACCACATGG | 3′-RACE primer for PpDV NS2 |
| Pp3NS1–1 | ATGCCCATATGGGGTGCTACAGGT | 3′-RACE primer for PpDV NS1 |
| Pp5VP-1 | CTCCTGTACGTGGATCAACTTGTGC | 5′-RACE primer for PpDV VP |
| Pp3VP-1 | GTAATGTCATGACTCCTATACCAGGT | 3′-RACE primer for PpDV VP |
| Sf5NS-1 | GTTTCTTCGAAGCTCAATGGAGTC | 5′-RACE primer for SfDV NS |
| Sf3NS2–1 | AGCCAGCAATCGAGCCAGTCGGAAG | 3′-RACE primer for SfDV NS2 |
| Sf3NS1–1 | AACATACAATGCCAATATGGGGTGC | 3′-RACE primer for SfDV NS1 |
| Sf5VP-1 | TGCTGCTGTATCCATAGGTACTTC | 5′-RACE primer for SfDV VP |
| Sf3VP-1 | ATGACTCCAATTCCAGGTTTAGAA | 3′-RACE primer for SfDV VP |
| Bm5NS-1 | GTTCCATTTGTTGATAACTGACAGGG | 5′-RACE primer for BmDV NS |
| Bm3NS1–1 | CAGTACTTGCATTTTATTGGAACACTG | 3′-RACE primer for BmDV NS2 |
| Bm3NS2–1 | GGCCAACATGGCCCTTGGAACAC | 3′-RACE primer for BmDV NS1 |
| Bm5VP-1 | CGTCTTGCGGCTCTAGCTCTATTAATC | 5′-RACE primer for BmDV VP |
| Bm3VP-1 | GCCTAAGTTTATGATTGGATTTGTAAAC | 3′-RACE primer for BmDV VP |
| Ce5NS-1 | GCTCAATGGAGTCGTGTAGAATTC | 5′-RACE primer for CeDV NS |
| Ce3NS1–1 | CAATGAATATGACCTACAGCGAAC | 3′-RACE primer for CeDV NS2 |
| Ce3NS2–1 | CCACATACTTCACGCCTGCCAA | 3′-RACE primer for CeDV NS1 |
| Ce5VP-1 | CTCCTCCAACATCAGTGGCTCC | 5′-RACE primer for CeDV VP |
| Ce3VP-1 | GCTTATACTGCAACTAAATAT | 3′-RACE primer for CeDV VP |
| Dpp5NS-1 | GCTGAGAATTCCTCGGTTAAGTATTC | 5′-RACE primer for DpIDV NS |
| Dpp3NS-1 | GGAAGCCAACGTTCAGAAGACACCTG | 3′-RACE primer for DpIDV NS |
| Dpp5VP-1 | CTCGTCTAGCTGTTCTCGCTCTATTG | 5′-RACE primer for DpIDV VP |
| Dpp3VP-1 | CGAAGACAATTCCCTGTTAACTGC | 3′-RACE primer for DpIDV VP |
| PpNSRpF700 | GCTTATCAACAAATGGAACTATGC | NS probe primer for PpDV |
| PpNSNpR850 (+T7 sequence) | CAAATTAATACGACTCACTATAGGCTCGGTATTCGATGGAGTCG | NS probe primer for PpDV |
| PpVPRpF3200 | GCTCCTCAGCCTAATCAACATCATC | VP probe primer for PpDV |
| PpVPNpR3400 (+T7 sequence) | CAAATTAATACGACTCACTATAGGGAGAGCTGTGTAATCTCTG | VP probe primer for PpDV |
| pIZ5-SfNS1FK | CGGGGTACCGTATAAGAGACGATACATTAAACATG | pIZT/V5-SfNS cloning primer |
| pIZ5-SfNS2RX | GCTCTAGACCAAATGTGTTAAAGGGGCCATG | pIZT/V5-SfNS cloning primer |
| pIZ5-SfNS2FA | CCACCGGTATACAAAAGATTGGCTAGAAAGTG | pIZT/V5-SfNS cloning primer |
| pIZ5-SfNS1RAha (+ HA tag) | CCACCGGTAGCGTAATCTGGAACATCGTATGGGTAGGAGATATCGATAACATACTTATC | pIZT/V5-SfNS cloning primer |
| SfNS1mATGFn | GACGATACATTAAACACCTCAAGTCAGTCTGGCAG | pIZT/V5-SfNS mutagenesis primer |
| SfNS1mATGRn | CTGCCAGACTGACTTGAGGTGTTTAATGTATCGTC | pIZT/V5-SfNS mutagenesis primer |
| SfNS2mATGFn | CTCATTTGTTTAATAACCCTAATACTACTGGTG | pIZT/V5-SfNS mutagenesis primer |
| SfNS2mATGRn | CACCAGTAGTATTAGGGTTATTAAACAAATGAG | pIZT/V5-SfNS mutagenesis primer |
| pIZ5-V5muFn | CCCGCGGTTCGAAGGCAAGCCTATCCCAAACCCTCTCCTCGG | pIZT/V5-SfNS mutagenesis primer |
| pIZ5-V5muRn | CCGAGGAGAGGGTTTGGGATAGGCTTGCCTTCGAACCGCGGG | pIZT/V5-SfNS mutagenesis primer |
In the primer names, “R” and “F” represent reverse and forward primers, respectively. The numbers in the primer names indicate the 5′ end of the primer sequence in virus.
The T7 and HA tag sequences are underlined. The first two primers are intended for sequence-independent single-primer amplification (SISPA) (see Fig. 1A).
Transcript mapping.
Transcription maps for RNA isolated from PpDV-infected Papilio larvae or for RNAs isolated from transfected LD652 cells were established by a FirstChoice RLM rapid amplification of cDNA ends (RACE) kit (Ambion) according to the supplier's instructions (see primers in Table 1). All RACE PCR products, cloned into pGEM-T or pGEM-T Easy vectors (Promega), were sequenced by Sanger's method and demonstrated that 5′ ends of NS2 transcripts started at the conserved CAGT sites downstream of the expected NS1 ATG (Fig. 2A).
Due to the >40-fold NS2 transcript excess, both in cells and larvae, it was challenging to obtain NS1 transcripts for conventional RACE methods and RNase protection assays. Therefore, a reverse primer (Fig. 1F, labeled R) that extended upstream of the NS2 transcript to the ATG of the putative NS1 start was designed to obtain NS1-specific amplicons. Sequencing of these amplicons revealed the 5′-end start of the NS1 transcripts (Fig. 2A). No differences were observed between mRNA from transfected cells and that from PpDV-infected larvae. The 5′ ends of NS1 transcripts usually started 2 to 4 nt upstream of the NS1 ATG (Fig. 2A). Unexpectedly, 5′-untranslated regions of either 4 or 26 nt were obtained by RACE experiments for SfDV NS1 transcripts. NS1 and NS2 3′ ends shared the same poly(A) motif downstream of the TATA box of VP transcripts. The BmDV NS transcript poly(A) was also located downstream of the TATA box of VP transcript, even though this virus contained an intergenic direct repeat of 45 nt between NS1 and VP (Fig. 2B).
All VP transcripts had short 5′-untranslated regions, and the transcripts started only 10 to 15 nt upstream of the first ATG (Fig. 2D) that, moreover, had unfavorable Kozak sequences promoting a leaky scanning translation mechanism for VPs as for ambidensoviruses (6, 22, 23, 26, 27). Previously, we expressed the VP gene of BmDV to generate viruslike particles in order to obtain the three-dimensional structure by X-ray crystallography (28). Poly(A) motifs of the 2.0-kb VP transcripts, determined by 3′-end RACE, overlapped with stop codons of VP proteins for all iteradensoviruses. It would be interesting to obtain transcript maps for LI 3 and 5 viruses as well.
Expression of nonstructural proteins NS1 and NS2.
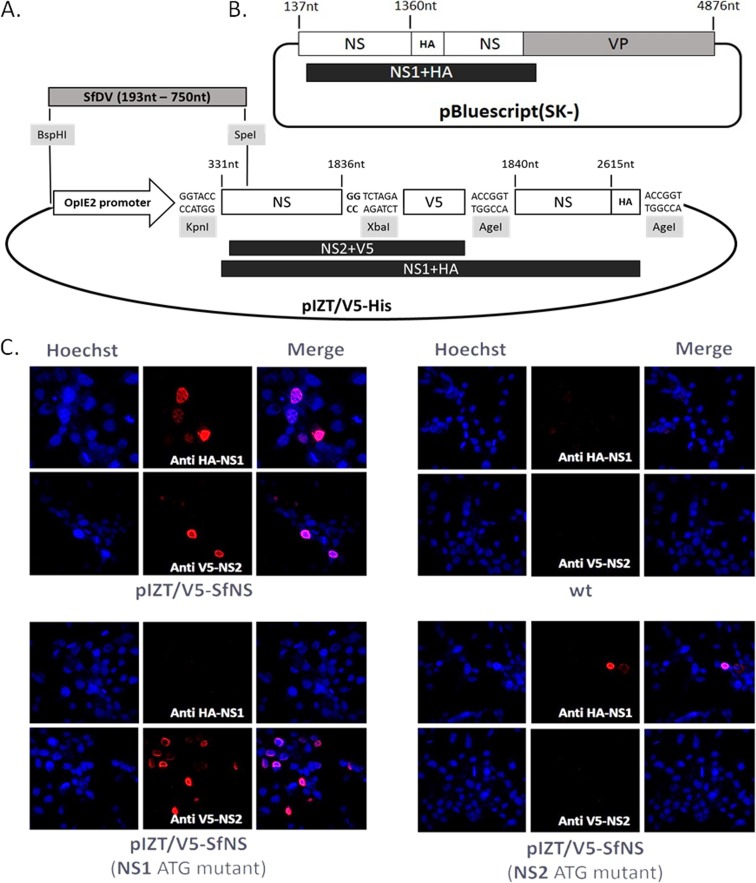
The pIZT/V5-His insect cell expression vector was used since it contained an OpIE2 promoter for constitutive expression of the NS gene, the Zeocin resistance gene for colony selection, and the cycle 3 green fluorescent protein (GFP) gene for cell line transfection efficiency detection. A V5 epitope was added to the C terminus of NS2, and an HA epitope was added to the C terminus of NS1 (Fig. 3A). An amplicon from NS1 ATG to the NS2 stop codon with CC added to the reverse primer was cloned into the KpnI and XbaI cloning sites of the pIZT/V5-His vector to obtain an in-frame NS2 gene/V5 epitope construct. A second amplicon from the NS2 stop codon to the NS1 stop codon was obtained by PCR with an HA tag added to the reverse primer and a mutation in the NS2 stop codon in the forward primer (Table 1). ATG of the NS1 and NS2 genes was substituted for ACC (Table 1, primers). To test the expression of SfDV NS1 and NS2 genes by their own promoter, the OpIE2 promoter of the vector was replaced by a fragment between the BspHI and SpeI sites of the viral genome (nt 193 to 750) (Fig. 3A). Two additional mutants were created with the TATA box changed to GAGA and the CAGT motif changed to TTGT. LD652 cells in 24-well plates were transfected with 0.5 μg DNA and 5 μl DOTAP reagent. Cell fixation, indirect immunofluorescence (IF) with 1:200 of V5 antibody (Invitrogen) for staining of NS2 or 1:50 of HA antibody (Santa Cruz) for staining of NS1, and DNA staining with Hoechst 33258 were done as described elsewhere (29).
FIG 3.
(A) SfDV NS gene expression in the insect expression vector pIZT/V5-His was studied after the addition of HA and V5 tags in frame to NS1 and NS2, respectively. A fragment from nt 331 to 1836, containing the NS2 and N-terminal half of NS1, of the SfDV genome was first cloned between the KpnI and XbaI sites of the pIZT/V5 multiple-cloning site (MCS), CC in bold was added to have the NS2 in frame with V5 epitope. After the V5 sequence, the fragment from nt 1840 to 2615 of the SfDV genome, containing the remainder of NS1, was cloned into the AgeI site and an HA tag sequence was added in frame with NS1 to the 3′ end of the insert. To investigate the NS promoter element, the OpIE2 promoter of vector was replaced in alternate constructs by a fragment from nt 193 to 750 of the SfDV genome, between the BspHI and SpeI restriction sites. (B) Scheme for introducing an HA tag into the SfDV genome clone to be in frame with the NS1 ORF. SfDV genome sequence from nt 137 to 4876 (lacking the hairpins) was cloned into pBluescript(SK−) vector. An HA tag was added at nt 1360 of the virus genome. Introducing a stop codon in frame immediately downstream of the NS1 start codon blocked NS1 expression, confirming the position of the NS1 translation start. (C) Detection of NS1 and NS2 gene expression in LD652 cells after 48 h posttransfection by immunofluorescence using the HA tag antibody (for NS1) and V5 antibody (for NS2) and an Alexa Fluor 568 goat anti-mouse secondary antibody. pIZT/V5-SfNS constructs allowed detection of both NS1 and NS2 (not possible with the wild type). Mutating the ATG of either the NS1 or NS2 gene blocked their expression.
In comparison to positive controls, NS2 protein expression could be detected only when NS1 ATG was mutated, whereas after NS2 ATG mutant transfection, only NS1 protein expression could be detected (Fig. 3C). IF results showed that after transfection of LD652 cells with plasmids containing the original presumed viral promoter element, only NS2 protein expression was detected. After introduction of a stop codon after NS2 ATG or mutation of the CAGT of NS2 transcript, NS2 expression became undetectable. Mutation of the TATA box did not change NS2 expression, which may mean that this TATA box may not be an essential element of the NS2 gene promoter. The viral genome fragment that replaced the OpIE2 promoter may not contain the complete NS1 promoter, since NS1 could not be expressed in LD652 cells after transfection. After introduction of an HA tag in frame with NS1 into the SfDV whole-genome clone (except the hairpins) (Fig. 3B), the plasmid expressed NS1 in LD652 cells after transfection. However, NS1 became undetectable after introduction of a stop codon after NS1 ATG, confirming its authenticity as the NS1 start codon and its location upstream of the NS2 transcription start. Combined, these results indicate that iteradensoviruses have, like brevidensoviruses (21), overlapping NS gene promoters that are responsible for different transcript starts and likely dictate the relative amounts of these transcripts.
ACKNOWLEDGMENTS
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERCC) grant to P.T. Q.Y. acknowledges support from a scholarship from the People's Republic of China and tuition waivers from INRS-IAF.
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1. Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch. Virol. 159:1239–1247. 10.1007/s00705-013-1914-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tijssen P, Agbandje-McKenna M, Almendral JM, Bergoin M, Flegel TW, Hedman K, Kleinschmidt JA, Li Y, Pintel DJ, Tattersall P. 2011. Parvoviridae, p 375–395 In King AMQ, Adams MJ, Carstens E, Lefkowitz EJ. (ed), Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, CA [Google Scholar]
- 3. Gudenkauf BM, Eaglesham JB, Aragundi WM, Hewson I. 2014. Discovery of urchin-associated densoviruses (family Parvoviridae) in coastal waters of the Big Island, Hawaii. J. Gen. Virol. 95:652–658. 10.1099/vir.0.060780-0 [DOI] [PubMed] [Google Scholar]
- 4. Kapelinskaya TV, Martynova EU, Schal C, Mukha DV. 2011. Expression strategy of densonucleosis virus from the German cockroach, Blattella germanica. J. Virol. 85:11855–11870. 10.1128/JVI.05523-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cotmore SF, Tattersall P. 2006. Structure and organization of the viral genome, p 73–94 In Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Oxford University Press, London, United Kingdom [Google Scholar]
- 6. Tijssen P, Bando H, Li Y, Jousset FX, Zadori Z, Fediere G, El-Far M, Szelei J, Bergoin M. 2006. Evolution of densoviruses, p 55–68 In Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Hodder Arnold, London, England [Google Scholar]
- 7. Chapman MS, Agbandje-McKenna M. 2006. Atomic structure of viral particles, p 107–123 In Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Hodder Arnold, London, England [Google Scholar]
- 8. Zadori Z, Szelei J, Lacoste MC, Li Y, Gariepy S, Raymond P, Allaire M, Nabi IR, Tijssen P. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291–302. 10.1016/S1534-5807(01)00031-4 [DOI] [PubMed] [Google Scholar]
- 9. Canaan S, Zadori Z, Ghomashchi F, Bollinger J, Sadilek M, Moreau ME, Tijssen P, Gelb MH. 2004. Interfacial enzymology of parvovirus phospholipases A2. J. Biol. Chem. 279:14502–14508. 10.1074/jbc.M312630200 [DOI] [PubMed] [Google Scholar]
- 10. Farr GA, Zhang LG, Tattersall P. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. U. S. A. 102:17148–17153. 10.1073/pnas.0508477102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nüesch JPF. 2006. Regulation of non-structural protein functions by differential synthesis, modification and trafficking, p 275–289 In Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Hodder Arnold, London, England [Google Scholar]
- 12. Cotmore SF, Tattersall P. 2006. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses, p 171–188 In Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Hodder Arnold, London, England [Google Scholar]
- 13. Nakagaki M, Kawase S. 1980. Structural proteins of densonucleosis virus isolated from the silk worm, Bombyx mori, infected with flacherie virus. J. Invertebr. Pathol. 36:166–171. 10.1016/0022-2011(80)90020-8 [DOI] [Google Scholar]
- 14. Fediere G, Li Y, Zadori Z, Szelei J, Tijssen P. 2002. Genome organization of Casphalia extranea densovirus, a new iteravirus. Virology 292:299–308. 10.1006/viro.2001.1257 [DOI] [PubMed] [Google Scholar]
- 15. Wang J, Zhang J, Jiang H, Liu C, Yi F, Hu Y. 2005. Nucleotide sequence and genomic organization of a newly isolated densovirus infecting Dendrolimus punctatus. J. Gen. Virol. 86:2169–2173. 10.1099/vir.0.80898-0 [DOI] [PubMed] [Google Scholar]
- 16. Xu P, Cheng P, Liu Z, Li Y, Murphy RW, Wu K. 2012. Complete genome sequence of a monosense densovirus infecting the cotton bollworm, Helicoverpa armigera. J. Virol. 86:10909. 10.1128/JVI.01912-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu Q, Hajek AE, Bergoin M, Tijssen P. 2012. Papilio polyxenes densovirus has an iteravirus-like genome organization. J. Virol. 86:9534–9535. 10.1128/JVI.01368-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu Q, Fediere G, Abd-Alla A, Bergoin M, Tijssen P. 2012. Iteravirus-like genome organization of a densovirus from Sibine fusca Stoll. J. Virol. 86:8897–8898. 10.1128/JVI.01267-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu Q, Tijssen P. 2014. Iteradensovirus from the monarch butterfly, Danaus plexippus plexippus. Genome Announc. 2(2):e00321–14. 10.1128/genomeA.00321-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Zadori Z, Bando H, Dubuc R, Fediere G, Szelei J, Tijssen P. 2001. Genome organization of the densovirus from Bombyx mori (BmDNV-1) and enzyme activity of its capsid. J. Gen. Virol. 82:2821–2825 [DOI] [PubMed] [Google Scholar]
- 21. Pham HT, Jousset FX, Perreault J, Shike H, Szelei J, Bergoin M, Tijssen P. 2013. Expression strategy of Aedes albopictus densovirus. J. Virol. 87:9928–9932. 10.1128/JVI.01259-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fediere G, El-Far M, Li Y, Bergoin M, Tijssen P. 2004. Expression strategy of densonucleosis virus from Mythimna loreyi. Virology 320:181–189. 10.1016/j.virol.2003.11.033 [DOI] [PubMed] [Google Scholar]
- 23. Tijssen P, Li Y, El-Far M, Szelei J, Letarte M, Zadori Z. 2003. Organization and expression strategy of the ambisense genome of densonucleosis virus of Galleria mellonella. J. Virol. 77:10357–10365. 10.1128/JVI.77.19.10357-10365.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu K, Li Y, Jousset FX, Zadori Z, Szelei J, Yu Q, Pham HT, Lepine F, Bergoin M, Tijssen P. 2011. The Acheta domesticus densovirus, isolated from the European house cricket, has evolved an expression strategy unique among parvoviruses. J. Virol. 85:10069–10078. 10.1128/JVI.00625-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tijssen P, Tijssen-van der Slikke T, Kurstak E. 1977. Biochemical, biophysical, and biological properties of densonucleosis virus (paravovirus). II. Two types of infectious virions. J. Virol. 21:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huynh OT, Pham HT, Yu Q, Tijssen P. 2012. Pseudoplusia includens densovirus genome organization and expression strategy. J. Virol. 86:13127–13128. 10.1128/JVI.02462-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El-Far M, Szelei J, Yu Q, Fediere G, Bergoin M, Tijssen P. 2012. Organization of the ambisense genome of the Helicoverpa armigera densovirus. J. Virol. 86:7024. 10.1128/JVI.00865-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaufmann B, El-Far M, Plevka P, Bowman VD, Li Y, Tijssen P, Rossmann MG. 2011. Structure of Bombyx mori densovirus 1, a silkworm pathogen. J. Virol. 85:4691–4697. 10.1128/JVI.02688-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boisvert M, Fernandes S, Tijssen P. 2010. Multiple pathways involved in porcine parvovirus cellular entry and trafficking toward the nucleus. J. Virol. 84:7782–7792. 10.1128/JVI.00479-10 [DOI] [PMC free article] [PubMed] [Google Scholar]