ABSTRACT
Little is known about virus adaptation in immunocompromised patients with chronic genotype 3 hepatitis E virus (HEV3) infections. Virus-host recombinant strains have been isolated recently from chronically infected patients. The nature and incidence of such recombinant events occurring during infections of solid-organ transplant (SOT) recipients are essentially unknown. The polyproline region (PPR) of strains isolated from SOT patients was sequenced during the acute-infection phase (n = 59) and during follow-up of patients whose infections became chronic (n = 27). These 27 HEV strains included 3 (11%) that showed recombinant events 12, 34, 48, or 88 months after infection. In one strain, parts of the PPR and the RNA-dependent RNA polymerase were concomitantly inserted. In the second, a fragment of a human tyrosine aminotransferase (TAT) gene was inserted first, followed by a fragment of PPR. A fragment of the human inter-α-trypsin inhibitor (ITI) gene was inserted in the third. All the inserted sequences were rich in aliphatic and basic amino acids. In vitro growth experiments suggest that the ITI insertion promoted more vigorous virus growth. In silico studies showed that the inserted sequences could provide potential acetylation, ubiquitination, and phosphorylation sites. We found that recombinant events had occurred in the HEV PPR in approximately 11% of the strains isolated from chronically infected transplant patients followed up in Toulouse University Hospital. These inserted fragments came from the HEV genome or a human gene and could enhance virus replication.
IMPORTANCE Hepatitis E virus (HEV) can cause chronic infections in immunocompromised patients, including solid-organ transplant (SOT) recipients. Two strains that had undergone recombination with human ribosomal genes were described recently. The strains with inserted sequences replicated better in vitro. Little is known about the frequency of such recombinant events or how such an insertion enhances replication. We therefore investigated 59 SOT patients infected with HEV and found 3 strains with 4 recombinant events in 27 of these patients whose infection became chronic. The 4 inserted sequences were of different origins (human gene or HEV genome), but all were enriched in aliphatic and basic amino acids and provided potential regulation sites. Our data indicate that recombinant events occur in approximately 11% of strains isolated from chronically infected patients. The structures of the inserted sequences provide new clues as to how the inserted sequences could foster virus replication.
INTRODUCTION
The hepatitis E virus (HEV), which is responsible for hepatitis E, is the cause of epidemic jaundice in developing countries and sporadic cases in developed countries (1, 2). HEV is a member of the genus Hepevirus, family Hepeviridae (3), and, like all RNA viruses, exists as a mixture of closely related variants defining a quasispecies (4). The HEV genome is a single-strand, approximately 7.2-kb-long, positive-sense RNA. It contains 3 open reading frames (ORFs), ORF1, ORF2, and ORF3, flanked by noncoding regions. ORF1 encodes a nonstructural protein with at least four putative functional domains: methyltransferase, papain-like cysteine protease (PCP), helicase, and RNA-dependent RNA polymerase (RdRp). It also has domains that are homologous to those of other plant and animal positive-strand RNA viruses: a Y domain, the polyproline region (PPR) previously called the hypervariable region (HVR), and a macro domain (or X domain). ORF2 encodes the 660-amino-acid (aa) capsid protein. Its 3 domains are S (shell), M (middle), and P (protruding). Lastly, ORF3 encodes a small (113- or 114-aa) phosphoprotein that is involved in virus egress (5).
HEV causes chronic hepatitis in immunocompromised patients, especially in solid-organ transplant (SOT) patients (6). The mechanisms leading to HEV persistence are largely unknown but seem to involve a complex interplay between virus diversity and the host immune response. Recent reports indicate that great quasispecies heterogeneity at the acute phase of the infection is associated with HEV persistence (7, 8). The development of a chronic HEV infection also seems to be associated with a weak inflammatory response, poor T-cell activation, and high serum concentrations of the chemokines involved in leukocyte recruitment to the liver (7). Another study reported that the evolution to chronic infection is associated with an impaired specific T-cell response (9).
Little is known about how the HEV genome evolves during a chronic infection. Segments of human genes were recently identified in the PPR of HEV RNA obtained from cell culture systems (portions of the S17 ribosomal gene) (10, 11) and from a chronically infected patient (part of the S19 ribosomal gene) (12). These fragments of human genes could reflect some as-yet-unknown aspect of HEV replication.
We therefore characterized the PPR of HEV strains infecting SOT patients at the acute phase and during follow-up to obtain information about the incidence of insertion and the nature of the inserted sequences.
MATERIALS AND METHODS
Patients and samples.
We studied 59 consecutive SOT patients who became acutely infected with hepatitis E virus between January 2004 and May 2012 and who were followed up at the Toulouse University Hospital, France. Infections were diagnosed by detecting HEV RNA using real-time PCR and IgM/IgG anti-HEV antibodies by the use of a commercial enzyme-linked immunosorbent assay (Wantai Biological Pharmacy Enterprise Co., China). While 32 patients had self-limiting infections, 27 patients developed a chronic infection, defined by persistently elevated liver enzyme activity and serum that contained HEV RNA for more than 6 months after diagnosis. Plasma samples were collected from all patients at the acute phase of infection and during the follow-up when available for patients whose infection became chronic. All samples were stored at −80°C.
Data were analyzed using an anonymized database. Such a protocol does not require written informed consent according to French Public Health law (CSP Art. L 1121-1.1).
Plasma HEV RNA concentrations and genotype determination.
The concentrations of HEV RNA in the plasma samples were measured by real-time PCR based on ORF3 data as previously described (13). HEV genotypes were determined by sequencing a 305-nucleotide (nt) fragment within the ORF2 gene (14). The sequences were compared to reference HEV strain sequences (GenBank).
Sequencing the PPR. (i) HEV-RNA extraction, HEV-cDNA synthesis, and PCR.
RNA was isolated from serum samples and purified using a QIAamp Viral RNA kit (Qiagen, Courtaboeuf, France). A reverse transcriptase PCR (RT-PCR) was used to amplify a 5,400-nt fragment of the HEV genome. RT-PCR was performed with a SuperScript III One-Step RT-PCR system (Invitrogen, Cergy-Pontoise, France), under the following conditions: 30 min at 50°C, 2 min at 94°C, and 50 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min 30 s at 68°C. The primer pair consisted of 10-sense (10-S) primer ATGTGGTCGATGCCATGGAGGCCCA and 5500 antisense (5500-AS) primer CTTGGCGYGACCAGTCCCA.
The nested PCR was performed with an Expand High Fidelity PCR system (Roche, Mannheim, Germany) under the following conditions: 30 s at 98°C and 30 cycles of 30 s at 98°C, 10 s at 55°C, and 35 s at 72°C. The sense primer was 1710-S (GAGTGCCGYACKGTGCTYGGGAATAA), and the antisense primer was 3050-AS (ACATCRACATCCCCCTGYTGTATRGA). PCR fragments were analyzed by capillary electrophoresis with Caliper (Life Sciences, Villepinte, France) and purified using Qiaquick PCR purification (Qiagen, Villepinte, France) as specified by the manufacturer.
(ii) Nucleotide sequencing.
Nested PCR products were sequenced on both strands by the dideoxy chain termination method (PRISM Ready Reaction AmpliTaq Fs and BigDye Terminator; Applied Biosystems, Paris, France) on an ABI 3130XL analyzer (Applied Biosystems, Foster City, CA) using primers 1710-S, 2080-S (GCAYATCTGGGAGTCTGCTAACCC), 3050-AS, and 2150-AS (GGGGAGAAGTCGCTAGAGAAACCTGATGT). Electropherogram data were analyzed using Sequencher 4.8 (Gene Codes Corporation).
Sequencing the full-length genome. (i) HEV-RNA extraction, HEV-cDNA synthesis, and PCR.
The extraction procedure and RT-PCR were performed with primers 10-S and 5500-AS as described above. The last 1,900-nt fragment of the HEV genome was amplified with the same protocol using primer pair 5254-S (TGCCTATGCTGCCCGCGCCACCGGC) and 7250-AS (TTTTTTTTTTTTCCGGGGRGCGMGRAACC).
Nested PCR was performed on the 5,400- and 1,900-nt fragments with an Expand High Fidelity PCR system (Roche, Mannheim, Germany) under the following conditions: 30 s at 98°C and 35 cycles of 30 s at 98°C, 30 s at 55°C, and 3 min at 72°C. PCR fragments were analyzed by capillary electrophoresis with Caliper (Life Sciences, Villepinte, France) and purified using Qiaquick PCR purification (Qiagen, Courtaboeuf, France) as specified by the manufacturer.
(ii) Nucleotide sequencing.
Nested PCR products were sequenced on both strands by the dideoxy chain termination method (PRISM Ready Reaction AmpliTaq Fs and BigDye Terminator; Applied Biosystems, Paris, France) on an ABI 3130XL analyzer (Applied Biosystems, Foster City, CA). The sequences of the primer pairs used to sequence the complete genome are available on request.
Electropherogram data were analyzed using Sequencher 4.8 (Gene Codes Corporation). Sequencher 4.8 was used to determine the final assembly of the complete genomic sequences.
Sequence analysis.
Sequences were aligned using ClustalX (version 1.83). The similarities of the aligned nucleotide or deduced amino acid sequences were analyzed using Bioedit (version 7.0.5.3).
Potential ubiquitination sites were identified using the BDM-PUB server (http://bdmpub.biocuckoo.org/prediction.php) with a threshold value of a >0.3 average potential score.
Potential phosphorylation sites were identified using the NetPhos 2.0 server (www.cbs.dtu.dk/services/NetPhos/) with a threshold value of a >0.5 average potential score (15).
Potential acetylation sites were identified using the Prediction of Acetylation on Internal Lysines (PAIL) server (http://bdmpail.biocuckoo.org/prediction.php) with a threshold value of a >0.2 average potential score.
Potential N-linked glycosylation sites were identified using the NetNGlyc 1.0 server (www.cbs.dtu.dk/services/NetNGlyc) with a threshold value of a >0.5 average potential score.
Potential methylation sites were identified using the BPB-PPMS server (www.bioinfo.bio.cuhk.edu.hk/bpbppms/index.jsp) with a threshold value of a >0.5 average potential score (16).
Disordered regions in proteins were predicted using the Protein Disorder Predictors DISOPRED2 server (http://bioinf.cs.ucl.ac.uk/psipred/?disopred=1) with a prediction threshold set at 5% false positives (17).
Cell culture and virus inoculation.
HepG2/C3A cells were obtained from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS; Invitrogen), 100 U/ml penicillin G, 100 U/ml streptomycin, and 0.25 μg/ml amphotericin B (culture medium) at 37°C in a humidified 5% CO2 atmosphere.
Inoculum titers were measured by the limiting dilution method. Briefly, serial 10-fold dilutions of HEV strains were diluted in 100 μl phosphate-buffered saline (PBS) containing 0.2% bovine serum albumin (BSA) and inoculated onto monolayers of HepG2/C3A to identify the doses of HEV inocula infecting cell cultures prior to a growth assay.
Cells to be infected were placed in 24-well plates (5 · 105 cells per well) and cultured for 2 days. They were then washed 3 times with 1.5 ml PBS, inoculated, and incubated for 1 h at room temperature. The inoculum was removed and replaced with 1.5 ml cell culture medium, and the cells were cultured at 35.5°C in a humidified 5% CO2 atmosphere. The maintenance medium used for virus culture was a 1:1 mixture of DMEM and medium 199 (Invitrogen) containing 2% (vol/vol) heat-inactivated FBS and 30 mM MgCl2 (final concentration); the other supplements were the same as those in the growth medium. Inoculations were made in triplicate for every strain.
On day 1 postinfection, the cells were washed 5 times with 1.5 ml PBS and 1.5 ml cell culture medium was added. Subsequently, half the medium was replaced with fresh cell culture medium every 2 days, and the harvested medium samples were stored at −80°C. The virus in the culture supernatants was measured by real-time PCR (13).
Statistical analysis.
All statistical calculations were performed using Stata software (version 9.2; Stata Corp.). The characteristics of strains were compared using Fisher's exact test for categorical variables. The Mann-Whitney test was used to compare quantitative variables. P values of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
PPR sequences have been deposited in GenBank under accession numbers KJ917667 to KJ917774. Full-length genomes have been deposited in GenBank under accession numbers KC166967 to KC166971.
RESULTS
Strain characteristics.
The strains isolated from patients who developed a chronic HEV infection (n = 27) did not differ from those whose acute hepatitis E was resolved (n = 32) in terms of their HEV RNA concentrations at the acute phase or their genotype profiles (P > 0.05). The median (interquartile range) HEV RNA concentration was 6.3 (5.3 to 6.7) base 10 logarithm (log) copies/ml for the chronic infections and 6.1 (5.5 to 6.6) log copies/ml for the acute infections. There were 18 HEV3f, 7 HEV3c, 1 HEV3e, and 1 rabbit HEV strains isolated from the patients whose infection became chronic and 20 HEV3f, 10 HEV3c, and 2 HEV3e strains isolated from those who cleared the virus. The rabbit strain was characterized by sequencing the X domain, in which a 93-nt insertion sequence was found (18).
PPR analysis.
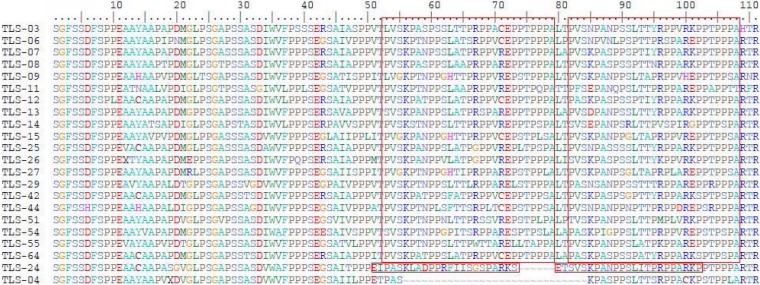
The PPRs in the HEV3c strains at the acute phase of hepatitis E were all 243 nt long, and those in the HEV3e strains were all 246 nt long. In contrast, the lengths of the PPRs of the HEV3f strains differed. The PPRs of almost half of the HEV3f strains (n = 17/38) were 246 nt long, while those of the others (n = 20/38) were 333 nt long. The length of the PPR of one strain was intermediate (315 nt). Analysis of the HEV3f PPR sequences showed that the longer PPR sequences were due to a 27-aa head-to-tail duplication and insertion (Fig. 1). Finally, the PPR of the rabbit strain was 207 nt long.
FIG 1.
Sequence duplications in HEV3f. Twenty strains of HEV3f had a head-to-tail duplication of 27 aa. TLS-24 had a 23-aa duplication. TLS-04 had no duplications in the PPR.
The PPRs of strains from patients who developed a chronic HEV infection were sequenced every year after the acute phase when samples were available. The length of the PPRs of most strains (n = 24/27) did not change during follow-up. But the viruses (TLS-20, TLS-26, and TLS-09) in 3 patients who were chronically infected with HEV3f had insertions in their PPRs during follow-up. The lengths of the PPRs before the recombinant events were 246, 333, and 333 nt, respectively. The presence of these inserted sequences was confirmed with specific primers and independent PCRs.
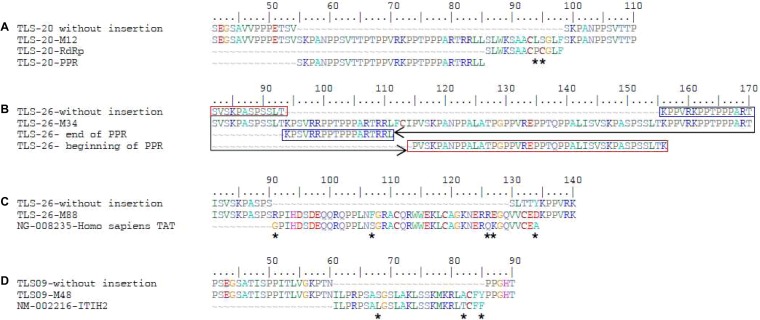
The 132-nt insertion in the PPR of the TLS-20 strain was composed of 2 distinct fragments of the HEV genome: the first 93 nt were derived from the PPR and the last 39 nt from the HEV RdRp. Of these, 36 nt were identical to the RdRp sequence of the TLS-20 strain, as were 11 of the corresponding 13 aa (Fig. 2A). The inserted sequence was detected at month 12 (M12), M36, and M48.
FIG 2.
Positions of inserted sequences in the polyproline region and comparison with reference sequences. Stars indicate differences between the reference sequences and the insertions.
The 186-nt insertion in the PPR of the TLS-26 strain was derived from the PPR of the HEV genome. The 186 nucleotides included 177 that were identical to the PPR, as were 59 of the corresponding 62 aa (Fig. 2B). This inserted sequence was detected at M34. Direct sequencing of the PPR at M47, M64, M88, and M96 did not detect this insertion. However, another inserted sequence of 132 nt was found at M88. A BLAST search identified the 132-nt insertion, out of frame, as similar to a fragment of the human tyrosine aminotransferase (TAT) gene. Its 132 nt included 126 that were identical to those of the Homo sapiens TAT (GenBank NG_008235.1), as were 39 of the corresponding 44 aa (Fig. 2C).
A 75-nt insertion in the PPR of the TLS-09 strain was detected at M48. A BLAST search identified the 75-nt insertion, in frame, as similar to a fragment of the human inter-alpha-trypsin inhibitor (ITI) 2 gene. Its 75 nt included 71 that were identical to those of Homo sapiens inter-alpha-trypsin inhibitor heavy chain 2 (ITI-H2) (GenBank NM_002216.2), as were 22 of the corresponding 25 aa (Fig. 2D).
Analysis of inserted sequences.
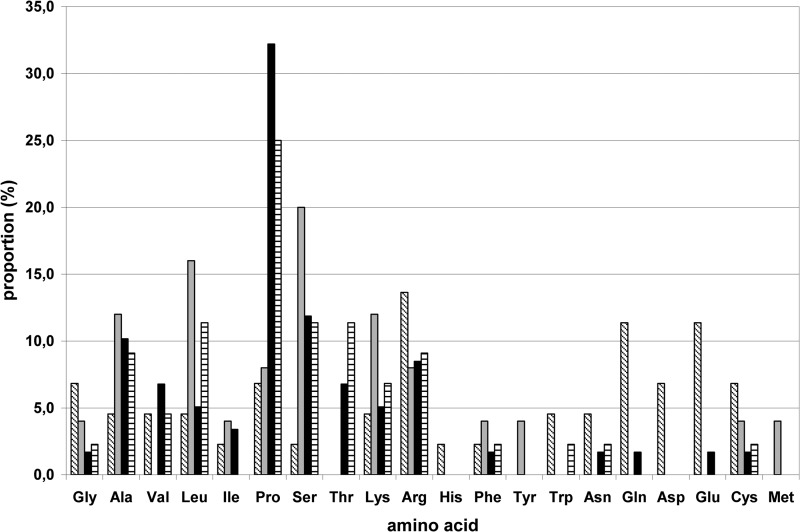
The inserted sequences encoded peptides rich in aliphatic hydrophobic amino acids (particularly alanine, leucine, and proline) and polar, charged amino acids (serine, lysine, and arginine) (Fig. 3). They also had at least 1 site suitable for acetylation, ubiquitination, or phosphorylation (especially serine) but no new glycosylation or methylation sites (Table 1). Even though the whole sequence of the PPR was tested, the potential regulation sites were detected only in the inserted sequences.
FIG 3.
Amino acid composition of inserted sequences. Boxes with diagonal lines, TAT; gray filled boxes, ITI-H2; black filled boxes, PPR; boxes with horizontal lines, RdRp-PPR.
TABLE 1.
Potential new regulation sites provided by inserted sequencesa
| Strain | Insertion length (nt) | Origin(s) | Acetylation site | Ubiquitination site | Glycosylation site | Methylation site | Phosphorylation site |
||
|---|---|---|---|---|---|---|---|---|---|
| Ser | Thr | Tyr | |||||||
| TLS-09 | 75 | ITI | +3 | +3 | 0 | 0 | +3 | 0 | 0 |
| TLS20 | 132 | PPR + RdRp | +3 | +3 | 0 | +1 | +3 | 0 | 0 |
| TLS-26 | 184 | PPR | +4 | +4 | 0 | 0 | +5 | 0 | 0 |
| TLS-26 | 132 | TAT | +1 | +1 | 0 | 0 | 0 | −2 | 0 |
ITI, fragment of human inter-α-trypsin inhibitor gene; TAT, tyrosine amino transferase gene; PPR, polyproline region; RdRp, RNA-dependent RNA polymerase. Numbers in the site columns indicate numbers of potential regulation sites provided by the inserted sequences.
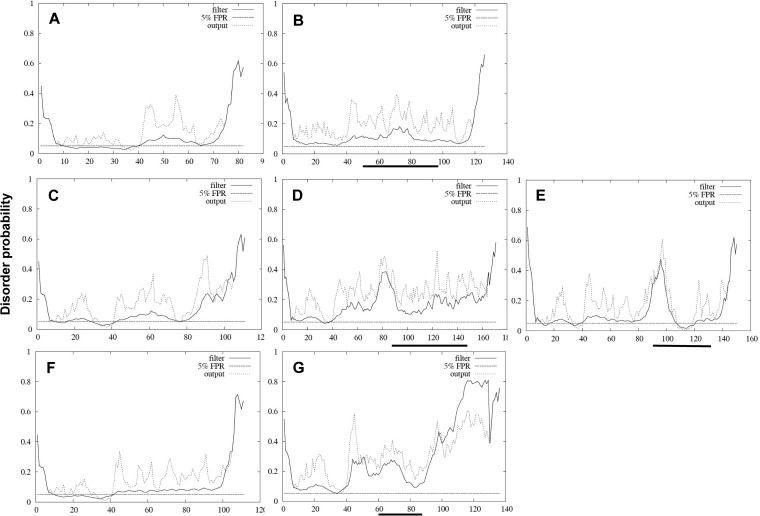
As the PPR is believed to be an intrinsically disordered region (IDR) (19), we determined the impact of the inserted sequences on the disorder of the PPR. The 4 inserted sequences did not systematically increase disorder (Fig. 4). PPR/RdRp insertion did not increase disorder probability (Fig. 4B). TAT and PPR insertion seemed to result in increased disorder probability (Fig. 4D and E). ITI insertion moderately increased disorder probability (Fig. 4G).
FIG 4.
Disordered-profile plots of the PPR with and without inserted sequences. (A) PPR sequence of TLS-20 without the insertion. (B) PPR of TLS-20 with the insertion. (C to E) PPR sequence of TLS-26 without inserted TAT sequence (C), with TAT (D), and with PPR insertion (E). (F and G) PPR sequence of TLS-09 without the inserted ITI sequence (F) and with the ITI insertion (G). Solid lines indicate the filtered disorder probabilities (FDP). Dashed lines indicate the raw disorder probabilities. The horizontal dashed line indicates the prediction threshold at 5% FPR. The horizontal solid bold line indicates the location of the inserted sequence. FDP > 30% was considered to represent a potential disordered region.
Complete genome sequence analysis of TLS-09.
In order to determine the extent of genomic evolution during chronic infection, we sequenced the full genome of the TLS-09 strain at the time of the acute phase (TLS-09/M0) and every year during the chronic phase (TLS-09/M12, TLS-09/M24, TLS-09/M36, and TLS-09/M48). The full genomes of the TLS-20 and TLS-24 strains were not sequenced because of a lack of material.
The complete genome sequence of TLS-09/M0 was 7,290 nt long, as were those of TLS-09/M12, TLS-09/M24, and TLS-09/M36. ORF1 extended from nucleotide 1 to 5205 (5,205 nt) and encoded a 1,735-residue protein. ORF2 extended from nucleotide 5240 to 7219 (1,980 nt) and encoded a 660-aa-long protein. ORF3, which partially overlaps ORF2, extended from nucleotide 5229 to 5567 (339 nt) and encoded a 113-residue protein. The complete sequence of the TLS-09/48 genome was 75 nucleotides longer due to the insertion in the PPR.
The nucleotide sequences of the various regions of the five HEV genomes obtained during 4 years of chronic infection were more than 98% identical (Table 2). The greatest differences (at least 0.9% of the nucleotides between two consecutive sequences) were in ORF1, especially in the PPR, Y domain, and PCP gene. We found no mutations in the ORF3 gene of any of the strains analyzed.
TABLE 2.
Nucleotide identities of strains isolated in contiguous years
| Sequencea | % nucleotide identity |
|||
|---|---|---|---|---|
| M12 vs M0 | M24 vs M12 | M36 vs M24 | M48 vs M36 | |
| Complete genome | 99.7 | 99.7 | 99.8 | 99.3 |
| Complete ORF1 | 99.9 | 99.7 | 99.9 | 99.2 |
| Methyltransferase | 99.6 | 99.3 | 98.5 | 98.5 |
| Y domain | 99.7 | 98.8 | 99.7 | 99.1 |
| Papain-like cysteine protease | 99.8 | 100 | 99.8 | 98.8 |
| PPR | 100 | 99.4 | 99.4 | 98.5 |
| X domain | 100 | 99.6 | 99.0 | 99.2 |
| Helicase | 100 | 100 | 100 | 99.6 |
| RdRp | 99.9 | 100 | 100 | 99.5 |
| Complete ORF2 | 99.9 | 99.9 | 100 | 99.9 |
| ORF2-S | 100 | 100 | 100 | 99.9 |
| ORF2-M | 99.8 | 99.7 | 100 | 99.8 |
| ORF2-P | 99.7 | 99.8 | 99.8 | 99.8 |
| ORF3 | 100 | 100 | 100 | 100 |
PPR, polyproline region; RdRp, RNA-dependent RNA polymerase.
Analysis of the mutations over the entire genome and the predicted amino acid differences within the 3 ORFs between the strains isolated at the acute phase and thereafter revealed that only 19 of the 59 mutations observed were nonsynonymous (Table 3). There were few nonsynonymous mutations in regions encoding methyl transferase, helicase, RdRp, or the X domain, while there were 5 mutations in the PPR. Three of the 5 nonsynonymous mutations detected in the PPR were detected at the same time as the insertion. Only 1 nonsynonymous mutation was detected in the P domain of the capsid protein. Over half (27/40) of the synonymous mutations were detected 4 years after the acute phase.
TABLE 3.
Evolution of the entire genome sequences of the 5 virus strainsa
| Region | Nucleotide position(s) | Nucleotide/corresponding amino acid(s) from strain: |
||||
|---|---|---|---|---|---|---|
| TLS-09/M0 | TLS-09/M12 | TLS-09/M24 | TLS-09/M36 | TLS-09/M48 | ||
| Me-transferase | 589 | G/Val | R/Val-Ile | G/Val | G/Val | G/Val |
| Me-transferase | 647 | A/Asn | A/Asn | A/Asn | A/Asn | R/Asn-Ser |
| Me-transferase | 695 | M/His-Pro | M/His-Pro | A/His | A/His | M/His-Pro |
| Y domain | 730 | M/Pro-Thr | M/Pro-Thr | A/Thr | A/Thr | M/Pro-Thr |
| Y domain | 778 | M/Ile-Leu | M/Ile-Leu | A/Ile | A/Ile | M/Ile-Leu |
| Y domain | 788 | M/His-Pro | M/His-Pro | A/His | A/His | M/His-Pro |
| Y domain | 896 | C/Ser | Y/Ser-Leu | C/Ser | C/Ser | C/Ser |
| Y domain | 1006 | T/Phe | T/Phe | T/Phe | Y/Leu-Phe | T/Phe |
| PPR | 2168 | C/Ala | C/Ala | S/Ala-Gly | C/Ala | C/Ala |
| PPR | 2474 | T/Leu | T/Leu | T/Leu | T/Leu | C/Pro |
| PPR | 2476 | A/Thr | A/Thr | A/Thr | A/Thr | G/Ala |
| PPR | 2495 | C/Val | C/Val | C/Val | C/Val | T/Ala |
| PPR | 2500 | G/Glu | G/Glu | R/Glu-Lys | G/Glu | G/Glu |
| PPR | 2301–2376 | Insertion | ||||
| X domain | 2573 | Y/Val-Ala | Y/Val-Ala | Y/Val-Ala | Y/Val-Ala | T/Val |
| X domain | 2686 | C/His | C/His | Y/His-Tyr | C/His | C/His |
| Helicase | 3040 | A/Ile | R/Ile-Val | R/Ile-Val | R/Ile-Val | G/Val |
| RdRp | 4147 | A/Thr | A/Thr | A/Thr | A/Thr | R/Thr-Ala |
| RdRp | 4943 | G/Arg | G/Arg | G/Arg | G/Arg | R/Gln-Arg |
| P domain | 6730 | G/Trp | T/Cys | G/Trp | G/Trp | G/Trp |
Sequences from the complete genome were aligned and analyzed. PPR, polyproline region; Me-transferase, methyl transferase; PCP, papain-like cysteine protease; RdRp, RNA-dependent RNA polymerase. Bold entries represent nonsynonymous mutations.
Culture of strain TLS-09 with and without the inserted ITI sequence.
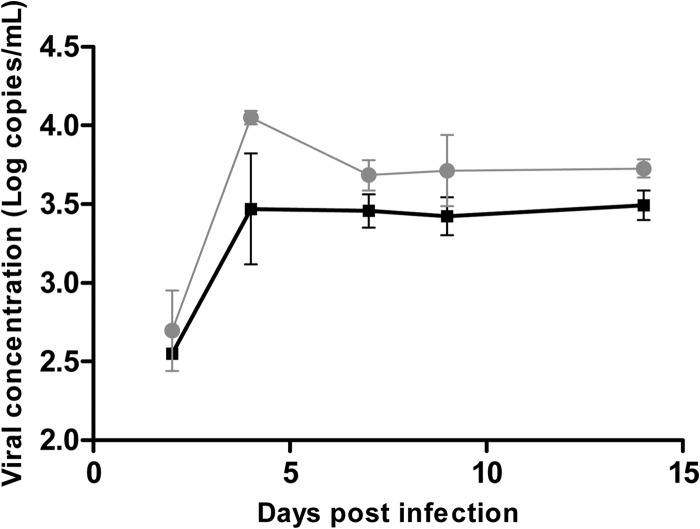
We inoculated triplicate wells containing HepG2/C3A cells with the TLS-09/M48 and TLS-09/M36 strains, with and without the inserted sequence, to see whether the inserted sequence conferred a growth advantage in culture. Strains isolated from plasma and from the feces were used, but only the strains isolated from the feces grew in our cell culture system. The presence of the inserted sequence in the inoculum was verified using primer 1710-S and specific primer INS-AS (GCTGGACAGTTTTGCCAAGCTCCCCGAA). Monolayers of HepG2/C3A cells were inoculated with serial 10-fold dilutions of the HEV inoculum to determine the 50% tissue culture infective dose (TCID50). The titer obtained was 103 TICD50/ml for both strains. Using 100 TCID50 per well for the growth assay, the TLS-09/M48 strain with the human sequence insertion seemed to grow better in culture than did the TLS-09/M36 (Fig. 5). The virus concentration in the supernatant after 4 days in culture was higher in the wells inoculated with the strain containing the inserted sequence than in the wells containing virus without the insertion. The virus RNA concentration also seemed to be higher on the other days. Cell culture was stopped at day 15. At the end of the culture experiment, the presence of the ITI-inserted sequence in TLS-09/M48 but not in TLS-09/M36 was confirmed with specific primers.
FIG 5.
Growth of TLS-09/M48 and TLS-09/M36 in cell culture. Data represent growth of TLS09/M48 (with human-sequence insertion) and TLS09/M36 (without insertion) strains on HepG2/C3A cells. Inoculations were performed in triplicate for both strains. Gray circles, TLS-09/M48 strain; black squares, TLS-09/M36 strain. Error bars show standard errors of the means (SEM).
DISCUSSION
We have analyzed the sequences of the PPRs of HEV strains isolated from 59 SOT patients at the acute phase and during follow-up. The length of the PPR varies according to the HEV genotype 3 strain. Recombinant events were detected in 3 strains (11%) over a lengthy period of infection. One strain underwent 2 distinct recombinant events in which fragments of a human gene and the virus genome were inserted.
The PPR of HEV3c was 243 nt long, that of HEV3e was 246 nt long, and that of HEV3f was 246 or 333 nt long. Although subgenotypes are not recognized by the 2012 ICTV 9th Report, complete genomes of HEV3f, HEV3c, and HEV3e are available and belong to the most prevalent strains circulating in Europe (20). While the function of the PPR is not well understood, its deletion does not abolish HEV infectivity in vivo or in vitro but leads to virus attenuation (21, 22). Koonin et al. suggested that this region is a putative proline hinge (23). This suggestion is supported by a recent study suggesting that the PPR is an intrinsically disordered region (IDR) that probably contributes to virus adaptation (19). IDRs evolve rapidly by recombinatorial repeat expansion as observed in HEV3f. IDRs have no stable tertiary structure, and their functions include regulating translation, cellular signal transduction, and protein phosphorylation (24, 25).
We have identified 3 strains with 4 different fragments inserted in their PPRs. Two fragments were derived from the HEV genome (PPR and/or RdRp) and two others from human genes TAT and ITI-H2. Insertion was not due to an artifact of PCR amplification as it was detected with specific primers targeting the inserted sequence. The HEV strain of another chronically infected patient also had a PPR containing a segment of the S17 human ribosomal protein gene (10). This variant was isolated by inoculating a cell culture system with fecal samples (10). Serum samples collected 10 months after the acute phase from another chronically infected patient were found to have virus whose PPR contained a segment of the human S19 ribosomal protein gene (12). Unlike those two cases, we found insertions derived from the human TAT and ITI-H2 genes. All these recombinant events occurred at different positions in the PPR. TAT is a nuclear gene encoding a liver enzyme which catalyzes the conversion of l-tyrosine to 4-hydroxyphenylbutyrate (26). The ITI is composed of 3 chains, two heavy (HC1 and HC2) and one light (bikunin), which confers the protease inhibitor function (27). ITI stabilizes the extracellular matrix in humans. However, the role of the sequences inserted in the HEV genome is unknown.
We were able to compare the growth of TLS-09/M48 with the ITI-derived insertion to that of TLS-09/M36 without the insertion from stool samples. The population of viruses in the feces might have differed from that in the plasma, but the ITI-derived insertion was present in both samples. The ITI-derived insertion seems to promote the growth of TLS-09/M48 virus in HepG2/C3A cells on the basis of analysis of the HEV RNA concentration in the supernatant. Focus-forming units (FFU) were not studied, but previous work showed that the numbers of viruses released into the medium increased in parallel with FFU (10). Although the inserted sequence could promote replication, as previously described for other recombinant strains (10, 12), nonsynonymous mutations that occur in the HEV genome could also influence virus replication. Just how the inserted sequence improved HEV replication has yet to be elucidated. The effect of the insertion in the Kernow-C1 strain was due to the encoded peptide rather than to the RNA, since introducing synonymous mutations into the insertion did not affect virus replication (11). The inserted sequence encodes additional hydrophobic amino acids which are also found in IDRs (28). The insertion also provides a few phenylalanine and tyrosine residues. These new hydrophobic residues could enhance peptide disorder and finally improve virus replication. In silico studies showed that disorder was not always improved by inserted sequences, but as disorder prediction programs may not reliably predict the impact of the insertion on the disorder, study of the crystal structure of the protein encoded by the PPR is needed. The insertions could also provide new regulation sites. We used bioinformatic prediction tools to show that the amino acid sequence of the inserted fragment provides new putative regulation sites. The peptide encoded by the inserted sequence of the Kernow-C1 strain (GenBank accession no. HQ70917) and the LBPR-0379 strain (GenBank accession no. JN564006) also has such sites. The inserted sequences provided new ubiquitination, acetylation, or phosphorylation sites but no glycosylation or methylation sites. The peptides derived from the Kernow strain with reversed or reversed complementary insertions had fewer regulation sites, especially acetylation or ubiquitination regulation sites, and they had lost their in vitro replicative advantage (10). The fact that no new glycosylation sites were detected suggests that acquisition of regulation sites does not seem to be done randomly. Indeed, glycosylation sites are useless for cytoplasmic protein.
The conjugation of ubiquitin with a substrate usually leads to the degradation of a peptide by the proteasome, and viruses seem to be able to hijack the ubiquitin/proteasome system (UPS). It was shown recently that replication of the rotavirus genome requires an active ubiquitin proteasome system to ensure the efficient recruitment of virus protein to the viroplasm (29). Ubiquitination of the polymerase of coxsackievirus B3 is also necessary for completion of the virus life cycle (30). HEV may also manipulate the UPS to regulate its life cycle, especially since inhibiting an active UPS affects HEV replication, possibly by inhibiting transcription or translation (31). Overexpression of ubiquitin in inhibitor-treated cells partially reverses the inhibitor effect on HEV replication (31). Although the influence of PPR ubiquitination on the regulation of virus replication must still be demonstrated, these findings suggest that the UPS regulates HEV replication. The function of cellular enzymes can be modified by phosphorylation (32, 33). Virus proteins can also be phosphorylated. Phosphorylation of NS5B of the hepatitis C virus improves RNA replication (34). Similarly, acetylation of histone and nonhistone proteins modulates protein function or the intracellular distribution of the protein (35, 36). Acetylation of virus proteins can improve their function. The affinity of the integrase of HIV for DNA is increased by acetylation, and its enzymatic activity is enhanced (37). While the life cycle of HEV is not yet clear, the PPR could regulate transcription and translation through ubiquitination, acetylation, or phosphorylation. Knowledge of the crystal structure of the protein encoded by the PPR region could also provide useful information about its molecular surface accessibility to enzymes.
Finally, complete genome sequence analysis of the TLS-09 strain showed that only 19 of the 59 mutations found were nonsynonymous, indicating great conservation of the major variant. Artifactual changes were limited using high-fidelity polymerase, but the possibility of amplification-derived misincorporation cannot be excluded. On the other side, observed mutations over a 5-year period may reflect shifting dominance in a mixed population. The PPR was the region most prone to nonsynonymous mutations. Mutations may appear to adapt to the host environment or as a result of polymerase error. Lorenzo et al. examined mutations of the HEV genome in primary cell culture (38) and found few (0.3%) genetic changes. Similarly, no nucleotide mutations were found in the full-length consensus sequence amplified after the transmission of the strain from human to swine, which shows adaptation of genotype 3 HEV to both hosts (39). These results suggest that virus adaptation is associated with a limited number of mutations. We found few mutations in the TLS-09 genome during the first year of the chronic infection, suggesting that the strain adapted to a new host with only a limited number of mutations.
In conclusion, we have found 3 recombinant strains among 27 strains isolated from chronically infected SOT patients. We have described inserted fragments derived from human genes that are different from ribosomal genes. Finally, we hypothesize that an insertion could confer a replicative advantage by providing new regulation sites rather than by increasing disorder in the PPR.
ACKNOWLEDGMENTS
We thank Owen Parkes for editing the English text.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1. Hoofnagle JH, Nelson KE, Purcell RH. 2012. Hepatitis E. N. Engl. J. Med. 367:1237–1244. 10.1056/NEJMra1204512 [DOI] [PubMed] [Google Scholar]
- 2. Kamar N, Bendall R, Legrand-Abravanel F, Xia NS, Ijaz S, Izopet J, Dalton HR. 2012. Hepatitis E. Lancet 379:2477–2488. 10.1016/S0140-6736(11)61849-7 [DOI] [PubMed] [Google Scholar]
- 3. Meng XJ, Anderson D, Arankalle A, Emerson S, Harrison TJ, Jameel S, Okamoto H. 2012. Hepeviridae, p 1021–1028 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee of Taxonomy of Viruses. Elsevier, San Diego, CA [Google Scholar]
- 4. Grandadam M, Tebbal S, Caron M, Siriwardana M, Larouze B, Koeck JL, Buisson Y, Enouf V, Nicand E. 2004. Evidence for hepatitis E virus quasispecies. J. Gen. Virol. 85:3189–3194. 10.1099/vir.0.80248-0 [DOI] [PubMed] [Google Scholar]
- 5. Holla RP, Ahmad I, Ahmad Z, Jameel S. 2013. Molecular virology of hepatitis E virus. Semin. Liver Dis. 33:3–14. 10.1055/s-0033-1338110 [DOI] [PubMed] [Google Scholar]
- 6. Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel JP, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N. Engl. J. Med. 358:811–817. 10.1056/NEJMoa0706992 [DOI] [PubMed] [Google Scholar]
- 7. Lhomme S, Abravanel F, Dubois M, Sandres Saune K, Rostaing L, Kamar N, Izopet J. 2012. HEV quasispecies and the outcome of acute hepatitis E in solid-organ transplant patients. J. Virol. 86:10006–10014. 10.1128/JVI.01003-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lhomme S, Garrouste C, Kamar N, Sandres Saune K, Abravanel F, Mansuy JM, Dubois M, Rostaing L, Izopet J. 2014. Influence of polyproline region and macro domain genetic heterogeneity on HEV persistence in immunocompromised patients. J. Infect. Dis. 209:300–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suneetha PV, Pischke S, Schlaphoff V, Grabowski J, Fytili P, Gronert A, Bremer B, Markova A, Jaroszewicz J, Bara C, Manns MP, Cornberg M, Wedemeyer H. 2012. Hepatitis E virus (HEV)-specific T-cell responses are associated with control of HEV infection. Hepatology 55:695–708 [DOI] [PubMed] [Google Scholar]
- 10. Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU. 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc. Natl. Acad. Sci. U. S. A. 108:2438–2443. 10.1073/pnas.1018878108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shukla P, Nguyen HT, Faulk K, Mather K, Torian U, Engle RE, Emerson SU. 2012. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J. Virol. 86:5697–5707. 10.1128/JVI.00146-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nguyen HT, Torian U, Faulk K, Mather K, Engle RE, Thompson E, Bonkovsky HL, Emerson SU. 2012. A naturally occurring human/hepatitis E recombinant virus predominates in serum but not in faeces of a chronic hepatitis E patient and has a growth advantage in cell culture. J. Gen. Virol. 93:526–530. 10.1099/vir.0.037259-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abravanel F, Sandres-Saune K, Lhomme S, Dubois M, Mansuy JM, Izopet J. 2012. Genotype 3 diversity and quantification of hepatitis E virus RNA. J. Clin. Microbiol. 50:897–902. 10.1128/JCM.05942-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Legrand-Abravanel F, Mansuy JM, Dubois M, Kamar N, Peron JM, Rostaing L, Izopet J. 2009. Hepatitis E virus genotype 3 diversity, France. Emerg. Infect. Dis. 15:110–114. 10.3201/eid1501.080296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blom N, Gammeltoft S, Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351–1362. 10.1006/jmbi.1999.3310 [DOI] [PubMed] [Google Scholar]
- 16. Shao J, Xu D, Tsai SN, Wang Y, Ngai SM. 2009. Computational identification of protein methylation sites through bi-profile Bayes feature extraction. PLoS One 4:e4920. 10.1371/journal.pone.0004920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. 2004. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337:635–645. 10.1016/j.jmb.2004.02.002 [DOI] [PubMed] [Google Scholar]
- 18. Izopet J, Dubois M, Bertagnoli S, Lhomme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guérin JL. 2012. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg. Infect. Dis. 18:1274–1281. 10.3201/eid1808.120057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Purdy MA, Lara J, Khudyakov YE. 2012. The hepatitis E virus polyproline region is involved in viral adaptation. PLoS One 7:e35974. 10.1371/journal.pone.0035974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kamar N, Dalton HR, Abravanel F, Izopet J. 2014. Hepatitis E virus infection. Clin. Microbiol. Rev. 27:116–138. 10.1128/CMR.00057-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pudupakam RS, Kenney SP, Cordoba L, Huang YW, Dryman BA, Leroith T, Pierson FW, Meng XJ. 2011. Mutational analysis of the hypervariable region of hepatitis E virus reveals its involvement in the efficiency of viral RNA replication. J. Virol. 85:10031–10040. 10.1128/JVI.00763-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pudupakam RS, Huang YW, Opriessnig T, Halbur PG, Pierson FW, Meng XJ. 2009. Deletions of the hypervariable region (HVR) in open reading frame 1 of hepatitis E virus do not abolish virus infectivity: evidence for attenuation of HVR deletion mutants in vivo. J. Virol. 83:384–395. 10.1128/JVI.01854-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U. S. A. 89:8259–8263. 10.1073/pnas.89.17.8259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dyson HJ, Wright PE. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6:197–208. 10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- 25. Tompa P. 2003. Intrinsically unstructured proteins evolve by repeat expansion. Bioessays 25:847–855. 10.1002/bies.10324 [DOI] [PubMed] [Google Scholar]
- 26. Dietrich JB. 1992. Tyrosine aminotransferase: a transaminase among others? Cell. Mol. Biol. 38:95–114 [PubMed] [Google Scholar]
- 27. Bost F, Diarra-Mehrpour M, Martin JP. 1998. Inter-alpha-trypsin inhibitor proteoglycan family–a group of proteins binding and stabilizing the extracellular matrix. Eur. J. Biochem. 252:339–346. 10.1046/j.1432-1327.1998.2520339.x [DOI] [PubMed] [Google Scholar]
- 28. Purdy MA. 18 July 2012. Evolution of the hepatitis E virus polyproline region: order from disorder. J. Virol. 10.1128/JVI.01374-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. López T, Silva-Ayala D, López S, Arias CF. 2011. Replication of the rotavirus genome requires an active ubiquitin-proteasome system. J. Virol. 85:11964–11971. 10.1128/JVI.05286-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Si X, Gao G, Wong J, Wang Y, Zhang J, Luo H. 2008. Ubiquitination is required for effective replication of coxsackievirus B3. PLoS One 3:e2585. 10.1371/journal.pone.0002585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karpe YA, Meng XJ. 2012. Hepatitis E virus replication requires an active ubiquitin-proteasome system. J. Virol. 86:5948–5952. 10.1128/JVI.07039-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X, Chuang HC, Li JP, Tan TH. 2012. Regulation of PKC-theta function by phosphorylation in T cell receptor signaling. Front. Immunol. 3:197. 10.3389/fimmu.2012.00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Derouiche A, Cousin C, Mijakovic I. 2012. Protein phosphorylation from the perspective of systems biology. Curr. Opin. Biotechnol. 23:585–590. 10.1016/j.copbio.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 34. Kim SJ, Kim JH, Kim YG, Lim HS, Oh JW. 2004. Protein kinase C-related kinase 2 regulates hepatitis C virus RNA polymerase function by phosphorylation. J. Biol. Chem. 279:50031–50041. 10.1074/jbc.M408617200 [DOI] [PubMed] [Google Scholar]
- 35. Glozak MA, Sengupta N, Zhang X, Seto E. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363:15–23. 10.1016/j.gene.2005.09.010 [DOI] [PubMed] [Google Scholar]
- 36. Sterner DE, Berger SL. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435–459. 10.1128/MMBR.64.2.435-459.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. 2005. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 24:3070–3081. 10.1038/sj.emboj.7600770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lorenzo FR, Tanaka T, Takahashi H, Ichiyama K, Hoshino Y, Yamada K, Inoue J, Takahashi M, Okamoto H. 2008. Mutational events during the primary propagation and consecutive passages of hepatitis E virus strain JE03–1760F in cell culture. Virus Res. 137:86–96. 10.1016/j.virusres.2008.06.005 [DOI] [PubMed] [Google Scholar]
- 39. Bouquet J, Cheval J, Rogee S, Pavio N, Eloit M. 2012. Identical consensus sequence and conserved genomic polymorphism of hepatitis E virus during controlled interspecies transmission. J. Virol. 86:6238–6245. 10.1128/JVI.06843-11 [DOI] [PMC free article] [PubMed] [Google Scholar]