ABSTRACT
Antigen-specific CD4+ T cells are essential for effective virus-specific host responses, with recent human challenge studies (in volunteers) establishing their importance for influenza A virus (IAV)-specific immunity. However, while many IAV CD4+ T cell epitopes have been identified, few are known to stimulate immunodominant CD4+ T cell responses. Moreover, much remains unclear concerning the major antigen(s) responded to by the human CD4+ T cells and the extents and magnitudes of these responses. We initiated a systematic screen of immunodominant CD4+ T cell responses to IAV in healthy individuals. Using in vitro expanded-multispecificity IAV-specific T cell lines and individual IAV protein antigens produced by recombinant vaccinia viruses, we found that the internal matrix protein 1 (M1) and nucleoprotein (NP) were the immunodominant targets of CD4+ T cell responses. Ten epitopes derived from M1 and NP were definitively characterized. Furthermore, epitope sequence conservation analysis established that immunodominance correlated with an increased frequency of mutations, reflecting the fact that these prominent epitopes are under greater selective pressure. Such evidence that particular CD4+ T cells are important for protection/recovery is of value for the development of novel IAV vaccines and for our understanding of different profiles of susceptibility to these major pathogens.
IMPORTANCE Influenza virus causes half a million deaths annually. CD4+ T cell responses have been shown to be important for protection against influenza and for recovery. CD4+ T cell responses are also critical for efficient CD8+ T cell response and antibody response. As immunodominant T cells generally play a more important role, characterizing these immunodominant responses is critical for influenza vaccine development. We show here that the internal matrix protein 1 (M1) and nucleoprotein (NP), rather than the surface proteins reported previously, are the immunodominant targets of CD4+ T cell responses. Interestingly, these immunodominant epitope regions accumulated many mutations over time, which likely indicates increased immune pressure. These findings have significant implications for the design of T cell-based influenza vaccines.
INTRODUCTION
Influenza virus infection causes half a million deaths annually worldwide and remains one of the biggest global threats to human health. Neutralizing antibodies that bind to the virion surface proteins hemagglutinin (HA) and neuraminidase (NA) and prevent the virus from entering host cells are considered to be the key point of the protective immunity against influenza A virus (IAV) infection (1). However, frequent mutation in HA and NA of the circulating viruses renders such antibody-mediated protective immunity ineffective. Increasing evidence shows that T cell immunity plays a pivotal role in anti-IAV protective immunity. CD8+ T cells directly clear virus-infected cells via perforin-, Fas ligand-, and TRAIL-mediated cytotoxicity and indirectly help recruit other immune cells to the infection site by secreting multiple cytokines and chemokines (2, 3). CD4+ T cells provide “help” for B cell responses by facilitating B cell activation, differentiation, and subsequent antibody production and isotype switching. CD4+ T cells also play an important role in the initiation and persistence of CD8+ T cell responses by enhancing CD8+ T cell proliferation and memory generation (3). Interestingly, increasing evidence suggests that CD4+ T cells do more than simply help B cells and CD8+ T cells (4). Like CD8+ T cells, they can also kill virus-infected cells directly and recruit other immune cells to the infection site by producing cytokines (4, 5). Studies in healthy volunteers with no detectable anti-IAV antibodies to the challenging IAV strain even demonstrated that the presence of IAV-specific memory CD4+, but not CD8+, T cells correlated with less virus shedding and less severe illness upon reinfection (6).
T cells exert their effect mainly in an antigen-specific manner. Epitope identification has been the first step in investigating the antigen specificity of IAV-specific T cell responses. The Immune Epitope Database (IEDB) has recorded 251 human CD8+ T cell epitopes for IAV so far; 42% are derived from nucleoprotein (NP), 17% from matrix protein 1 (M1), 13% from polymerase basic protein 1 (PB1), and the remainder from the other IAV gene products. These data from the IEDB indicate that IAV-specific CD8+ T cell responses focus on the internal proteins NP, M1, and PB1, especially NP. Using in vitro expanded-multispecificity IAV-specific T cell lines and synthetic overlapping peptides, we further demonstrated systematically that NP was the major target of immunodominant CD8+ T cell responses, regardless of the host HLA background (HLA-A2+ [7] or HLA-A2− [8]). However, the immunodominant epitopes were quite different between individuals with different HLA alleles (7, 8). On the other hand, 774 human CD4+ T cell epitopes for IAV are currently indexed in the IEDB, three times the number of indexed CD8+ T cell epitopes; 51% of these are shown as derived from HA, 15% from NP, 13% from M1, and 11% from NA. It seems that HA is the primary target of IAV-specific CD4+ T cell responses. A recent genome-wide T cell epitope screen using synthetic peptides found that HA and M1 contained more CD4+ T cell epitopes, which seemed to support the above statistics (9). However, an earlier study found that M1 and NP, not HA, were the major targets of IAV-specific CD4+ T cells (10), and another found that PB1 was the major target for both CD4+ and CD8+ T cell responses (11). Thus, the antigen specificity of IAV-specific CD4+ T cell responses is still controversial. Moreover, due to the limited availability of peripheral blood mononuclear cells (PBMCs), the above-mentioned studies (9–11) used peptide pools to screen T cell responses and, as a result, did not determine the immunodominance hierarchy at the single-epitope and HLA levels in each individual.
Immunodominance refers to the phenomenon in which the cellular immunity tends to focus on a very limited number of antigenic epitopes even during immune responses to complex antigens or pathogens in immunized or infected individuals. Generally, immunodominant T cells are not only more prevalent, but also provide better protection than subdominant ones (12, 13). Although many human CD4+ T cell epitopes from IAV have been identified in the past, few have been shown to stimulate immunodominant CD4+ T cell responses. Moreover, the antigen(s) responded to by the human immunodominant CD4+ T cells and the extents and magnitudes of such responses remain unclear. The present study, therefore, conducted a systematic screen of immunodominant CD4+ T cell responses to IAV strain PR8 [A/Puerto Rico/8/1934(H1N1)] in healthy individuals. Using in vitro expanded-multispecificity IAV-specific T cell lines and individual IAV protein antigens produced by recombinant vaccinia viruses (rVVs), we demonstrate that M1 and NP are the immunodominant targets of CD4+ T cell responses. The immunodominance hierarchies at the single-epitope and HLA levels in four individuals were definitively characterized. Interestingly, variants of these epitopes existed widely among the influenza strains circulating previously, indicating these immunodominant epitopes might have been subjected to greater selective pressure. The potential implications for T cell-based vaccine development are discussed further below.
MATERIALS AND METHODS
Ethics statement.
Buffy coats were obtained with informed written consent from Australian Red Cross donors under 12-07VIC-17 Material Supply Agreement V15.1. The proposed work was approved by the La Trobe University Faculty Human Ethics Committee (FHEC) under project number FHEC12/NR81. Influenza virus propagation using 10-day-old embryonated chicken eggs was approved by the La Trobe University Animal Ethics Committee under project number AEC12-64.
PBMC samples.
Buffy coat PBMCs were isolated by Ficoll-Hypaque gradient and stored in liquid nitrogen until use. HLA typing was performed by the Victorian Transplantation and Immunogenetics Service (VTIS) (Melbourne, Australia).
Viruses.
The Mount Sinai strain of IAV PR8 was propagated in 10-day-old embryonated chicken eggs provided by Research Poultry Farm and Hatchery (Melbourne, Australia). Allantoic fluids were harvested 2 days after infection, and aliquots were stored at −80°C until use. rVVs containing individual HA, NA, NP, M1, M2, PB1, PB1F2, PB2, acidic polymerase (PA), nonstructural protein 1 (NS1), and NS2 IAV genes were gifts from Jonathan Yewdell and Jack Bennink (National Institutes of Health, Bethesda, MD). The viruses were propagated using a thymidine kinase-negative (TK−) cell line and were stored at −80°C until use. These proteins are all derived from IAV PR8.
Recombinant IAV proteins, synthetic peptides, and antibodies.
Four recombinant IAV proteins, HA, NA, M1, and NP, were purchased from Sino Biological Inc. (Beijing, China). All the peptides were synthesized by Mimotopes (Melbourne, Australia); IAV M1 and NP overlapping 18-mers with 6-amino-acid (aa) shifts and 13-mers with 2-aa shifts were synthesized as cleaved pin peptides. All peptides were dissolved in dimethyl sulfoxide (DMSO). Anti-CD3 (fluorescein isothiocyanate [FITC]), anti-CD4 (phycoerythrin [PE]), anti-CD4 (allophycocyanin [APC]), anti-gamma interferon (IFN-γ) (PE-Cy7), anti-tumor necrosis factor alpha (TNF-α) (PE), and anti-CD107a (PE) monoclonal antibodies (MAbs) were purchased from eBioscience. Pan-anti-DR (L243), anti-DP (B7/21), and anti-DQ (SPV-L3) antibodies were used as culture supernatants (14). Anti-rVV RNA-binding protein antibody TW2.3 (15) and anti-HA antibody H28-E23 (16) were kind gifts from Jonathan Yewdell and Jack Bennink (National Institutes of Health, Bethesda, MD).
Cell culture.
All cells were cultured in RF-10 consisting of RPMI 1640 supplemented with 10% fetal calf serum (FCS), 2-mercaptoethanol (2-ME) (5 × 10−5 M), and antibiotics. Donor Epstein-Barr virus-transformed B lymphoblast lines (BLCLs) were established using standard EBV transformation. The other human BLCLs were made available from the International HLA Workshop and the VTIS. P815 cells were kind gifts from Jonathan Yewdell and Jack Bennink (National Institutes of Health, Bethesda, MD).
Preparation of IAV- and rVV-infected P815 cell lysates.
For IAV infection, P815 cells were infected with IAV at a multiplicity of infection (MOI) of 10 for 1 h at 37°C in acidified FCS-free RPMI 1640 (pH 6.8), followed by addition of 10 volumes of RF-10 for overnight incubation. For rVV infection, P815 cells were infected with rVV at an MOI of 10 for 1 h at 37°C in phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA), followed by addition of 10 volumes of RF-10 for overnight incubation. Infected cells were pelleted and lysed with 8 M urea. The lysates were aliquoted and preserved at −20°C until use.
Antigen-specific T cell bulk cultures.
PBMCs (5 × 106) were pulsed with 5 μl IAV-infected P815 cell lysates (equivalent to 105 infected cells) in 200 μl RF-10 for 1 h in 24-well tissue culture plates. Two milliliters of RF-10 with 20 U/ml recombinant human interleukin 2 (rIL-2) (Peprotech, Brisbane, Australia) were then added, and the cell lines were cultured in the rIL-2 containing RF-10 until use. Peptide-specific CD4+ T cell lines were generated as previously described (14). In brief, PBMCs (1 × 106 to 2 × 106) were pulsed with 5 μM peptide and cultured in 1 ml RP-5, consisting of RPMI 1640 (Gibco) supplemented with 5% human AB serum, l-glutamine (2 mM), 2-ME (5 × 105 mM), and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) in 48-well tissue culture plates. The medium was 50% replaced by RP-5 containing 10 U/ml rIL-2 on day 5 and then 50% replaced by RP-5 containing 20 U/ml rIL-2 when required.
Intracellular cytokine staining (ICS).
For dominant-protein identification, autologous BLCLs were pulsed with IAV- or rVV-infected P815 cell lysates overnight and then cocultured with IAV-specific T cell lines for 5 h in the presence of 10 μg/ml brefeldin A (BFA). For dominant 18-mer and 13-mer peptide identification, antigen-specific T cell cultures were incubated with peptide at 10 μg/ml at 37°C for 5 h in the presence of BFA. For identifying restriction HLA, BLCLs were pulsed with the peptide of interest at 10 μg/ml for 1 h, washed extensively, and then cocultured with peptide-specific T cells for 5 h in the presence of BFA. For ex vivo assessment of the epitope-specific memory CD4+ T cell precursor frequencies, PBMCs were thawed, and dominant and subdominant peptides at 10 μg/ml were used to stimulate the PBMCs for 18 h in the presence of BFA. For antibody-blocking assays, T cells were incubated with 10 μl of anti-HLA class II (HLA-II) antibody supernatant for 30 min before addition of peptide and BFA. The coincubated cells were harvested and stained first with anti-CD3 (FITC) and anti-CD4 (PE), and then washed, fixed, and stained with anti-IFN-γ (PE-Cy7), as described previously (7). For IFN-γ and TNF-α costaining, anti-TNF-α (PE) was also included. For CD107a staining, the immunodominant 18-mer-specific T cell lines were restimulated with the corresponding dominant 13-mer peptides at 10 μg/ml at 37°C for 5 h in the presence of monensin and anti-CD107a (PE) (17). Cells were harvested and stained first with anti-CD3 (FITC) and anti-CD4 (APC) and then washed, fixed, and stained with anti-IFN-γ (PE-Cy7). Samples were acquired on a FACSCanto II flow cytometer (Becton Dickinson), and fluorescence-activated cell sorter (FACS) data were analyzed with FlowJo software (Tree Star, Ashland, OR).
Bioinformatics analysis.
Protein sequences were aligned, and amino acid differences were scored to determine the sequence conservation between local circulating and vaccine strains for the newly identified M1- and NP-derived peptides. The National Center for Biotechnology Information (NCBI) influenza virus database (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=1) was used (accessed on 25 April 2014) with the search criteria set as Australia, M1/NP, and H1N1/H3N2; identical sequences were represented by the oldest sequence in the group, full length only, which identified H1N1 (n = 20 for M1; n = 45 for NP) and H3N2 (n = 25 for M1; n = 72 for NP) sequences. Protein sequences were aligned using the NCBI database, peptide regions were mapped, and the frequency of mutation was determined across the various sequence groups.
Statistical analysis.
The Student t test was used to analyze the differences between two groups. Differences were considered significant when the P value was less than 0.05.
RESULTS
Identification of immunodominant CD4+ T cell responses with a systematic approach.
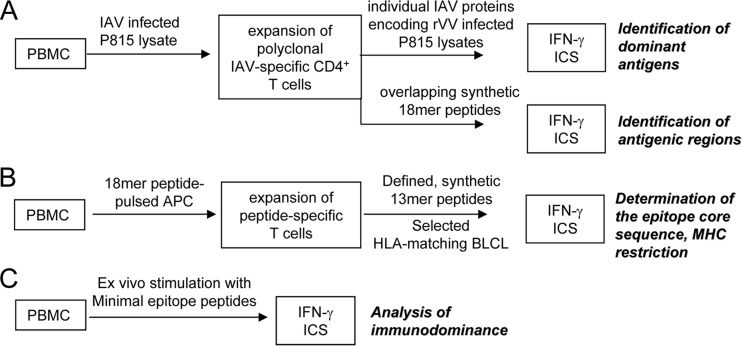
We previously developed a systematic protocol for identifying immunodominant CD8+ T cell responses by using in vitro expanded-multispecificity IAV-specific T cell lines, rVVs encoding 11 individual IAV proteins, and synthetic overlapping peptides (7, 8). Based on that protocol, we developed a similar approach for identifying immunodominant CD4+ T cell responses with minor modification (Fig. 1). As CD8+ T cells mainly recognize endogenous antigens produced in the infected antigen-presenting cells (APCs), in the CD8+ T cell expansion protocol, IAV-specific CD8+ T cell lines were generated by infecting 1/10 donor PBMCs with IAV and then cocultured with the remaining uninfected PBMCs for 12 to 15 days, as IAV-infected APCs would stimulate all IAV-specific memory CD8+ T cells. However, CD4+ T cells primarily recognize exogenous antigens captured from the extracellular environment by APCs. Therefore, in the CD4+ T cell expansion protocol, a soluble IAV antigen source was generated by lysing IAV-infected P815 cells in 8 M urea. This IAV lysate was then used to stimulate PBMCs in the presence of IL-2 to establish IAV-specific CD4+ T cell lines. Twelve rVV (11 rVVs encoding 11 individual IAV proteins and 1 wild-type rVV)-infected P815 lysates were prepared similarly to reveal the dominant viral protein. After confirmation of the dominant protein, the immunodominant epitope regions within the protein were determined by screening 18-mer overlapping peptides covering the full protein length (Fig. 1A). Dominant-18-mer-specific T cell lines were then generated to define the core sequence of the epitope using 13-mer overlapping peptides and the presenting HLA molecule by HLA class II-blocking antibodies and partially HLA-matched APC lines (Fig. 1B). Finally, ex vivo memory precursor frequencies of the dominant and subdominant epitope-specific CD4+ T cells from the same PBMC sample were compared using ICS (Fig. 1C).
FIG 1.
Schematic representation of the systematic CD4+ T cell epitope screening method.
M1 and NP were the most dominant targets of IAV-specific CD4+ T cell responses.
To investigate the immunodominance hierarchy of IAV-specific CD4+ T cell responses, we needed to first determine the immunodominant IAV proteins recognized by IAV-specific CD4+ T cell responses. Multispecificity IAV-specific CD4+ T cell lines were generated and cocultured with autologous BLCLs pulsed with lysates of P815 cells infected with rVVs encoding a single IAV antigen in an IFN-γ ICS assay. As shown in Fig. 2A, M1-specific CD4+ T cell responses were observed in all eight of the investigated donors. These responses were dominant in two donors (donors 3 and 4) and were comparable to other antigen-specific CD4+ T cell responses in four other donors (donors 5, 6, 7, and 8). The other two of the eight donors (donors 1 and 2) showed immunodominant CD4+ T cell responses to NP. NP-specific CD4+ T cell responses were also observed in 5 of the other 6 donors (donors 3, 5, 6, 7, and 8) and were comparable to M1-specific CD4+ T cell responses in three donors (donors 6, 7, and 8). Except for M1- and NP-specific CD4+ T cell responses, PB1-specific CD4+ T cell responses were found in 5 donors (donors 2, 3, 5, 6, and 7), mostly subdominant (donors 2, 3, 5, and 7). Other CD4+ T cell responses specific for IAV internal proteins, including NS1 (donors 2, 7, and 8), PA (donor 5), and PB2 (donor 7), were also observed. Surprisingly, we did not observe obvious HA-specific CD4+ T cell responses in any of the investigated donors, although half of the reported epitopes were HA derived (IEDB). NA-specific CD4+ T cell responses were found to be subdominant in donors 3, 6, and 7 and comparable to the M1-specific CD4+ T cell response in donor 5. Taken together, IAV-specific CD4+ T cell responses mainly targeted internal proteins rather than surface proteins, and, M1 and NP were the most dominant targets of IAV-specific CD4+ T cell responses. This was further confirmed by the statistical analysis of compiled single responses to the 11 IAV proteins (Fig. 2B).
FIG 2.
Most immunodominant T cells focused on M1 and NP. (A) Multispecificity T cell lines were raised using IAV-infected P815 cell lysate; 12 to 15 days later, these lines were tested for their reactivity to autologous BLCLs pulsed with individual lysates of P815 cells infected with 11 rVVs encoding single IAV antigens in an IFN-γ ICS assay. BLCLs not pulsed with any lysate (Nil) or pulsed with lysate from uninfected P815 cells (Uninfected) or wild-type (WT) (empty-vector) rVV-infected P815 cells were used as background and specificity controls. Total IAV-specific CD4+ T cell responses stimulated by autologous BLCLs pulsed with IAV-infected P815 cell lysate are shown as black bars for easier comparison. The donors' HLA class II alleles are shown in the inset text boxes. (B) The responses of all 8 donors shown in panel A were compiled and statistically analyzed. Statistically significant differences between groups were determined by the Student t test: *, P < 0.05. The error bars indicate the standard error of the mean.
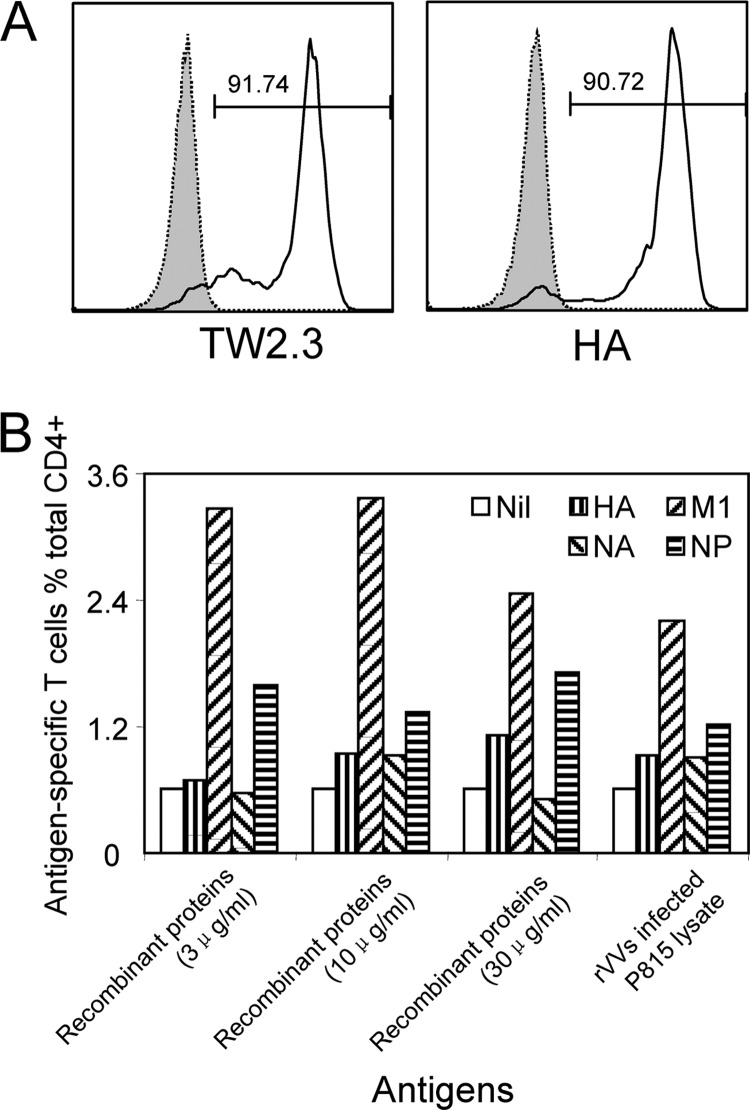
As the individual IAV antigens were produced as lysates of rVV-IAV-infected P815 cells, it was possible that the lack of CD4+ T cell response to individual antigens, such as HA, was due to a poorer rVV infection and therefore a lower antigen concentration in the lysate. To assess such a possibility, we determined HA expression in the rVV HA-infected P815 cells. As shown in Fig. 3A, we found that HA was effectively produced by rVV HA-infected cells, as the percentage of HA-positive cells (shown by anti-HA [16]) was similar to that of rVV HA-infected cells (recognized by antibody TW2.3 [15]). Although we did not have individual antibodies for each IAV protein in the P815 cell lysates, we demonstrated that these rVVs have similar infectious capacities in our previous report (7). As every rVV-infected cell produced the recombinant IAV protein effectively, the lysates should contain similar levels of IAV proteins. This was further confirmed, as commercial pure recombinant IAV proteins and rVV-infected P815 cell lysates stimulated similar immunodominance hierarchies (Fig. 3B).
FIG 3.
Lysates of rVVs encoding individual IAV proteins stimulated the same immunodominance hierarchy that was stimulated by purified IAV antigens. (A) P815 cells were infected with rVVs encoding each of the 11 IAV proteins at an MOI of 10 for 1 h at 37°C in PBS containing 0.1% BSA (Sigma). Following overnight incubation, rVV HA-infected P815 cells were stained with mouse MAb TW2.3, specific for rVV RNA-binding protein (15), and H28-E23, specific for HA (16) (kind gifts from Jonathan Yewdell and Jack Bennink, National Institutes of Health, Bethesda, MD), and representative FACS plots are shown. Uninfected P815 cells were used as a control (dotted lines). (B) Autologous BLCLs pulsed with individual recombinant IAV proteins, HA, NA, M1, and NP, at different concentration were used to compare their activities to activate IAV-specific CD4+ T cell responses in the multispecificity T cell line with that of the lysates from P815 cells infected by rVVs encoding corresponding individual IAV proteins.
Identification of the core sequences of immunodominant epitopes presented by various HLA molecules.
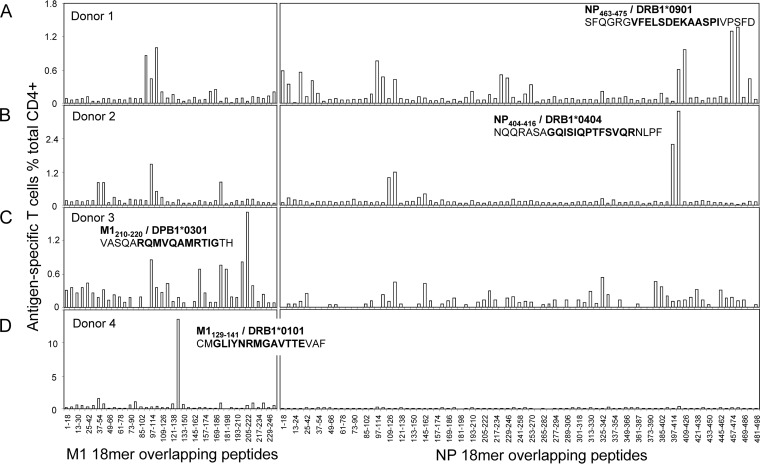
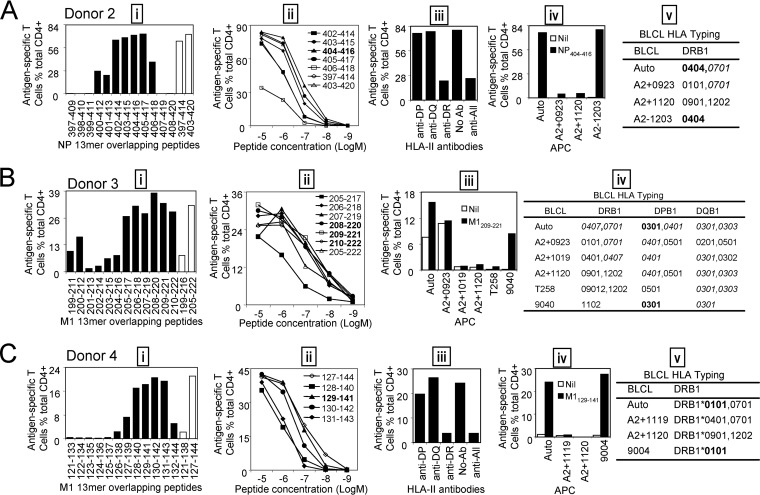
M1 and NP were demonstrated to be the most dominant targets of IAV-specific CD4+ T cell responses. Thus, further screening with the M1 and NP 18-mer overlapping peptides were conducted in donors 1, 2, 3, and 4, whose IAV-specific CD4+ T cell responses were dominated by those specific to either M1 or NP. As shown in Fig. 4, like the IAV-specific CD8+ T cell responses, which generally focused on one or two antigenic regions (7, 8), the IAV-specific CD4+ T cell responses seemed to also target two regions in both NP and M1. As expected, these regions differed between donors (Fig. 4A to D), especially the most dominant regions, including NP457-480 (donor 1) (Fig. 4A), NP397-420 (donor 2) (Fig. 4B), M1199-222 (donor 3) (Fig. 4C), and M1121-144 (donor 4) (Fig. 4D). (All published and defined minimal epitopes are shown as subscript amino acid positions, such as NP463-475 or M1209-221; other peptide sequences are shown as normal text, such as NP457-474 or M1(205-222).)
FIG 4.
Identification of the immunodominant T cell epitope-containing 18-mer regions. The same IAV-specific T cell lines used in Fig. 2, derived from four donors whose IAV-specific CD4+ T cell responses were dominated by M1 or NP, were further screened for their specific responses to the 121 overlapping 18-mer peptides from M1 (40 peptides) and NP (81 peptides) at a final concentration of approximately 1 μg/ml in an ICS assay. The identified 18-mer sequences are shown, and the subsequently identified minimal epitopes and their HLA restrictions are in boldface. Panels A to D correspond to donors 1 to 4 in Fig. 2.
The IAV-specific CD4+ T cell lines mentioned above were polyclonal, and the response specific to one individual peptide was relatively weak (Fig. 4). To define the core sequence of immunodominant epitopes, peptide-specific CD4+ T cell lines with much stronger responses were generated with the identified 18-mer peptides and subsequently used to screen overlapping 13-mer peptides within the 18-mer sequences (Fig. 5, 6, and 7). Peptide titrations were used to further confirm the epitope core sequences quantitatively. To identify HLA restriction of the immunodominant epitopes, partially HLA-matched BLCLs were pulsed with the corresponding 13-mer peptides, and then the excess peptides were washed away before assessment with the 18-mer-peptide-specific T cell lines in an ICS assay.
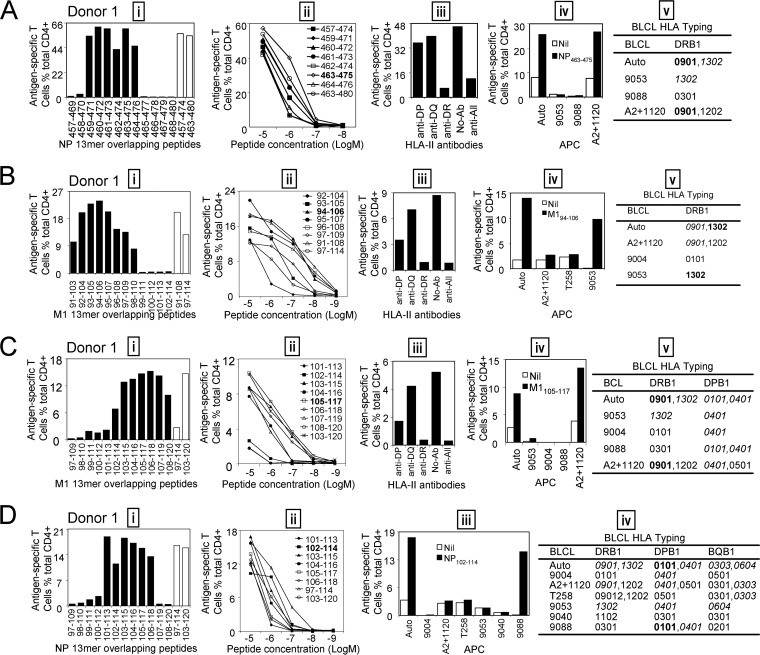
FIG 5.
Identification of the core sequences and HLA restrictions of epitopes in donor 1. (A) (i) The 13-mer peptides within 18-mer NP457-474 and NP463-480 observed in donor 1 were screened by ICS; the control 18-mer results are shown by open bars. (ii) Several potential dominant 13-mers and corresponding 18-mers were titrated to compare their T cell-stimulating capacities, which led to the identification of the core peptide NP463-475. (iii to v) HLA restriction of the dominant NP463-475 was then determined by HLA-class II antibodies (iii) and partial HLA matching of BLCLs (iv and v). (B) (i) The 13-mer peptides within 18-mer M1(91-108) and M1(97-114) were screened by ICS; the corresponding 18-mer results are shown by open bars. (ii) Several 13-mers and the two 18-mers were titrated. (iii to v) HLA restriction of the core 13-mer M194-106 was determined by HLA-class II antibodies (iii) and partial HLA matching of BLCLs (iv and v). (C) (i) The 13-mer peptides within 18-mer M1(97-114) and M1(103-120) were screened; 18-mer results are shown by open bars. (ii to v) Seven overlapping 13-mer peptides and two corresponding 18-mer peptides were titrated to compare their activities (ii), and HLA restriction of the core 13-mer M1105-117 was analyzed with HLA-class II antibodies (iii) and partially HLA matched BLCLs (iv and v). (D) (i) The 13-mer peptides within 18-mer NP97-114 and NP103-120 were screened as in panel A. (ii to iv) The core 13-mer peptide NP102-114 was identified by titration (ii), and its HLA restriction was determined with partial HLA matching of BLCLs (iii and iv).
FIG 6.
Identification of the core sequences and HLA restrictions of the immunodominant epitopes in donors 2, 3, and 4. (A) (i) The 13-mer peptides within 18-mer NP397-414 and NP403-420 observed in donor 2 were characterized. (ii to v) Four 13-mer peptides and two corresponding 18-mer peptides were titrated to compare their activities (ii), and HLA restriction of 13-mer NP404-416 was analyzed with HLA class II antibodies (iii) and partially HLA-matched BLCLs (iv and v). (B) (i) The 13-mer peptides within 18-mer M1(199-216) and M1(205-222) observed in donor 3 were screened as in panel A. (ii to iv) The most dominant 13-mer peptide was identified by titration (ii), and HLA restriction of M1209-221 was analyzed with partially HLA-matched BLCLs (iii and iv). (C) (i) The 13-mer peptides within 18-mer M1(127-144) observed in donor 4 were screened as in panel A. (ii) Four 13-mers and the corresponding 18-mer peptides were titrated to determine their activities. (iii to v) HLA restriction of 13-mer M1129-141 was analyzed with HLA class II antibodies (iii) and partially HLA-matched BLCLs (iv and v).
FIG 7.
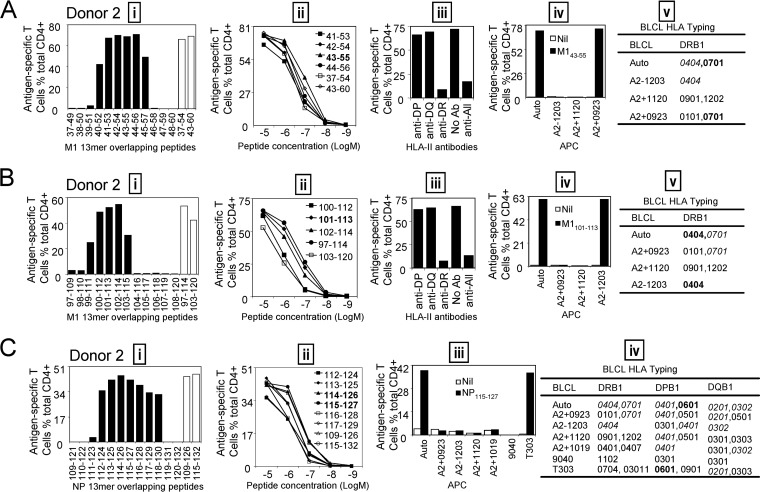
Identification of the core sequences and HLA restrictions of subdominant epitopes in donor 2. (A) (i) The 13-mer peptides within 18-mer M1(37-54) and M1(43-60) sequences were screened by ICS; the 18-mer results are shown by open bars. (ii) Several recognized 13-mers and the corresponding 18-mers were titrated. (iii to v) HLA restriction of the core 13-mer M143-55 was determined by HLA class II antibodies (iii) and partial HLA matching of BLCLs (iv and v). (B) (i) The 13-mer peptides within 18-mer M1(97-114) and M1(103-120) were screened; 18-mer results are shown by open bars. (ii to v) Three 13-mer overlapping peptides and two corresponding 18-mer peptides were titrated to compare their activities (ii), and HLA restriction of 13-mer M1101-113 was analyzed with HLA class II antibodies (iii) and partially HLA-matched BLCLs (iv and v). (C) (i) The 13-mer peptides within 18-mer NP109-126 and NP115-132 were screened as in panel A. (ii to iv) The 13-mer peptides NP114-126 and NP115-127 showed identical potentials when titrated, indicating that the core sequence is likely NP115-126 (ii), and HLA restriction of NP115-127 was determined by partial HLA matching of BLCLs (iii and iv).
For donor 1, the immunodominant CD4+ T cells responding to the NP457-474and NP463-480 18-mer peptides (Fig. 4A) recognized six 13-mer peptides (Fig. 5A, i). Titration of these six 13-mers showed that NP463-475 was the most potent core epitope sequence (Fig. 5A, ii). To determine the restricting HLA molecule for NP463-475, a class II antibody-blocking assay was conducted. The anti-DR antibody efficiently blocked T cell activation to peptide NP463-475, whereas the anti-DP and anti-DQ antibodies did not (Fig. 5A, iii). To further confirm the HLA-DR restriction of NP463-475, a panel of BLCLs with different DR alleles (Fig. 5A, iv and v) were used as APCs after being pulsed with NP463-475 to stimulate the peptide-specific T cell line. Autologous BLCLs and BLCL A2+1120 both expressing HLA-DRB1*0901, efficiently activated peptide-specific T cells. In contrast, BLCLs 9053 and 9088 did not express HLA-DRB1*0901 and failed to present the peptide (Fig. 5A, iv and v). Therefore, the NP463-475-specific immunodominant CD4+ T cell response in donor 1 was restricted to HLA-DRB1*0901. Using a similar approach, three comparable dominant epitopes—M194-106, restricted to HLA-DRB1*1302 (Fig. 5B); M1105-117, restricted to HLA-DRB1*0901 (Fig. 5C); and NP102-114, restricted to DPB1*0101 (Fig. 5D)—were identified from donor 1.
For donor 2, the immunodominant CD4+ T cells responding to the NP397-414and NP403-420 18-mer peptides (Fig. 4B) recognized seven 13-mer peptides (Fig. 6A, i). Peptide titration (Fig. 6A, ii) indicated that NP404-416 was the most potent and likely minimal epitope, as NP403-415 and NP405-417 were not as potent. Antibody blocking assays showed that NP404-416 was also HLA-DR restricted (Fig. 6A, iii). Furthermore, using the peptide-pulsed BLCLs A2−1203, we showed that the peptide is presented by HLA-DRB1*0404 (Fig. 6A, iv and v).
For donor 3, M1(205-222) 18-mer-specific immunodominant CD4+ T cells also recognized six 13-mer peptides within the corresponding 18-mer sequence (Fig. 6B, i). As shown in the peptide titration assay (Fig. 6B, ii), three neighboring 13-mer peptides, M1(208-220), M1(209-221), and M1(210-222), induced almost the same responses at different peptide concentrations, indicating that all three of these 13-mer peptides contained the epitope core sequence. Thus, the core sequence of this immunodominant epitope was deduced to be an 11-mer, M1210-220. Six peptide-pulsed BLCLs were used directly to identify the HLA restriction. Only BLCL 9040, which shared DPB1*0301 and DQB1*0301 with an autologous BLCL, induced a comparable response, while the other three BLCLs, A2+1019, A2+1120, and T258, sharing DQB1*0301, failed to activate this epitope-specific CD4+ T cell response. Therefore, the peptide is presented by HLA-DPB1*0301 (Fig. 6B, iii and iv).
For donor 4, 13-mer mapping and titration showed that the core sequence of this immunodominant epitope was M1129-141 (Fig. 6C, i and ii). An antibody-blocking assay (Fig. 6C, iii) and BLCL peptide presentation (Fig. 6C, iv and v) showed that M1129-141 was presented by HLA-DRB1*0101.
Ex vivo assessment of epitope-specific memory CD4+ T cell precursor frequencies.
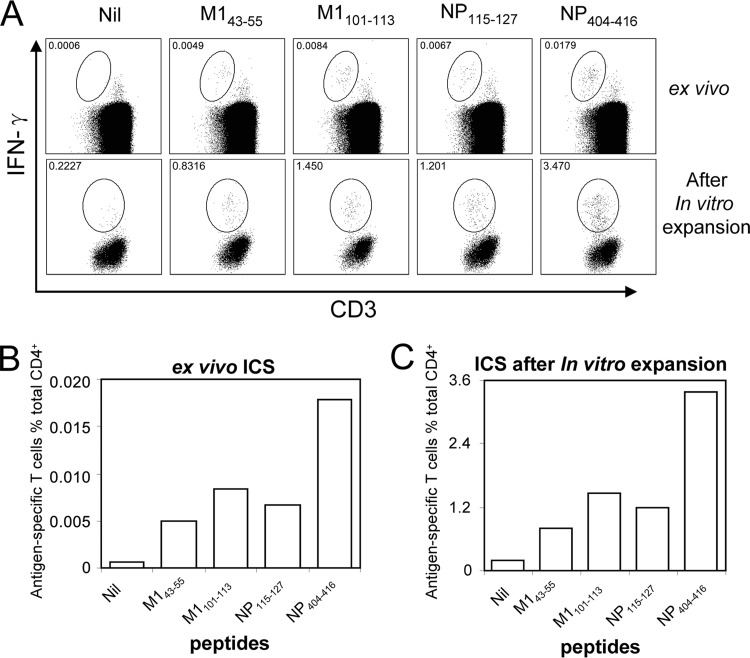
The immunodominant CD4+ T cell responses were identified by in vitro-expanded IAV-specific T cell lines. The immunodominance hierarchy might have been changed by the expansion procedure. To assess that possibility, some subdominant epitopes were also characterized in detail for their epitope core sequences and HLA restrictions. After that, ex vivo assessments of these epitope-specific memory CD4+ T cell precursor frequencies were conducted to confirm the immunodominance hierarchy (ranking). Using the same approach described above, three subdominant epitopes—M143-55, restricted to HLA-DRB1*0701 (Fig. 7A); M1101-113, restricted to HLA-DRB1*0404 (Fig. 7B); and NP115-127, restricted to DPB1*0601 (Fig. 7C)—were characterized from donor 2. Then, PBMCs were directly stimulated ex vivo with the three above-mentioned subdominant peptides and the immunodominant NP404-416. As shown in Fig. 8A, the epitope-specific CD4+ T cells, no matter whether subdominant or dominant, are calculated to have made an approximately 180-fold (170- to 194-fold) expansion in this assay. The immunodominance hierarchies were similar when tested ex vivo (Fig. 8B) and after in vitro expansion (Fig. 8C), indicating that a 10-day in vitro culture period did not significantly alter the immunodominance hierarchy.
FIG 8.
In vitro expansion did not significantly alter the immunodominance hierarchy. (A) M143-55, M1101-113, NP115-127, and NP404-416 were used to stimulate PBMC samples ex vivo or IAV-specific T cell lines after 10-day in vitro expansion from donor 2 in ICS assays (gated on CD3- and CD4-double-positive cells). Epitope-specific cells were gated out from the background based on their downregulated CD3 expression. (B) The immunodominance hierarchy determined ex vivo shown as antigen-specific T cell numbers among 106 CD4+ T cells. (C) The immunodominance hierarchy for the same T cell specificities after 10-day in vitro expansion shown as percentages of antigen-specific T cells among total CD4+ T cells.
DISCUSSION
In this study, we evaluated the immunodominant CD4+ T cell responses to IAV in PBMCs collected from healthy individuals using a systematic approach (Fig. 1). We demonstrated that M1 and NP are the immunodominant antigens targeted by IAV-specific CD4+ T cells (Fig. 2). We also definitively characterized 10 epitopes derived from these antigens using synthetic overlapping peptides (Fig. 4, 5, 6, and 7). A few previously reported epitopes had their immunodominant status confirmed, although some of them were found to be restricted to different HLA-II molecules. Moreover, several novel immunodominant epitopes were defined.
Multiple previous studies (9–11) used synthetic overlapping peptides to stimulate PBMCs in an ex vivo enzyme-linked immunospot (ELISPOT) system. Due to limited PBMC availability, peptide pools and/or 2- or even 3-dimensional peptide matrix systems had to be utilized (9–11). Although many responses were identified, they might have been significantly underestimated by such an approach due to peptide competition and/or limited serum protease capacity required for trimming so many long peptides at high concentration (18, 19). Most importantly, the immunodominance hierarchy at the single-epitope and HLA levels remained unclear. To be able to identify the most immunodominant antigens and then their antigenic epitopes systematically, a short in vitro T cell expansion method was developed to raise the frequency of IAV-specific T cells in an unbiased fashion. Considering the IAV protein composition differences between IAV virions and those within the IAV-infected cells and also considering that soluble antigen is processed and presented by class II molecules most efficiently, we used lysates from IAV-infected P815 cells to mimic the IAV protein composition and abundance derived from IAV-infected cells. The lysate-expanded IAV-specific CD4+ T cell lines were then assessed by lysates containing individual IAV proteins encoded by individual rVVs to reveal the immunodominant IAV protein antigens responded to by the multispecificity T cell lines. We demonstrated that our novel systematic approach is not only reliable, but also accurate, for the following three reasons. First, these rVVs are known to have similar infectious capacities, as shown in our previous report (7), and the target IAV proteins were also effectively produced in the infected cells, judging by both vaccinia virus-derived proteins (recognized by antibody TW2.3 [15]) and the encoded IAV proteins (shown by anti HA [16]) (Fig. 3A). Second, we compared our home-made IAV protein-containing lysates with commercial purified recombinant IAV proteins at different concentrations, and rVV-infected P815 cell lysates stimulated an immunodominance hierarchy similar to that stimulated by the purified IAV proteins (Fig. 3B), indicating that a significant bias that might confound the identification of dominant antigen was unlikely. Finally, to make sure that the in vitro expansion did not significantly alter the immunodominance hierarchy, ex vivo ICS was further conducted to confirm that the initial frequencies of IAV-specific dominant and subdominant CD4+ T cell precursors show the same hierarchy as those after in vitro expansion (Fig. 8). Importantly, as the in vitro expansion increased the total cell number significantly, our approach allowed us to screen every overlapping peptide within the dominant proteins, which would not be possible using PBMCs ex vivo. The overall immunodominance hierarchies were demonstrated first at the antigen level and then at the single-epitope level in the present study (Fig. 2 and 4).
The IAV-specific CD4+ T cell responses identified here mainly focused on M1 and NP (Fig. 2). This profile was consistent with a report from Lee et al. (10). PB1-specific CD4+ T cell responses were also observed in more than half (5/8) of the investigated donors. However, most of these responses were subdominant, although Assarsson et al. found that PB1 was a major target for both CD4+ and CD8+ T cell responses (11). In the study by Assarsson et al., the epitope peptides were selected according to algorithm prediction based only on DR supertypes. Their study led to the confirmation of a single DR-restricted NP peptide, NP330-344 (11). The discrepancies between their study and ours may have contributed to the different observed outcomes for the immunodominant IAV antigens. More importantly, HA, which was considered one of the most abundant IAV antigens and the most dominant target of IAV-specific CD4+ T cells, judging by the number of identified CD4+ T cell epitopes listed in the IEDB (>50% of the total listed epitopes) (http://www.iedb.org/), did not induce significant responses in our study, which was the opposite of the results of a previous study, in which peptide pools were used (9). Similarly, NA, the other major surface IAV protein, did not stimulate dominant IAV-specific CD4+ T cell responses. Therefore, IAV-specific CD4+ T cell responses mainly targeted internal proteins rather than surface proteins, and M1 and NP were the most dominant targets.
One of the key factors in determining IAV-specific CD4+ T cell immunodominance is protein abundance (20). M1 and NP were two of the most abundant proteins contained in IAV particles (21), which may explain why they are so frequently targeted by IAV-specific CD4+ T cells. However, HA is as abundant as M1 and NP (21), and it is therefore unclear why HA does not induce comparable responses. We believe that some antigen-processing and presentation preference may have contributed to such a biased outcome. For example, increasing evidence shows that antigens produced endogenously by APCs can also be presented on major histocompatibility complex (MHC) class II molecules (22–25), including an epitope from IAV M1 (26). If this kind of antigen presentation exists widely during IAV infection for unknown reasons, it is relatively easy to understand why CD4+ T cell responses, just like CD8+ T cell responses, may also focus on the same dominant proteins, M1 and NP. However, such a biased antigen presentation during IAV infection remains to be established. Furthermore, as HA proteins are much more polymorphic than NP and M1, it is also possible that other H1 HA sequences may stimulate responses larger than that stimulated by the H1 from PR8. It is also possible that most identified HA-derived epitopes are relatively subdominant.
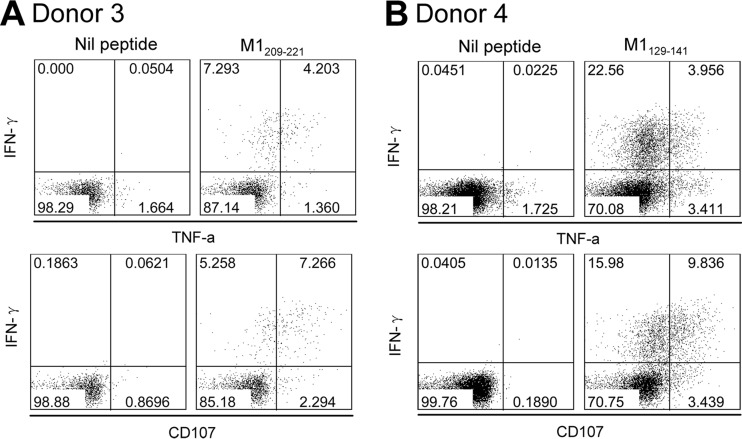
The primary role of CD4+ T cells is to provide help for B cells to produce neutralizing antibodies specific to surface viral HA and NA (3). However, increasing evidence shows that CD4+ T cells act as more than just helpers during IAV infection (4). They also can provide direct protective responses against IAV infection through helper-independent mechanisms (5, 6, 27, 28). IAV-specific CD4+ T cells may secrete cytokines to recruit innate and adaptive immune cells to the infection site and create an antiviral microenvironment, they may activate tissue-resident APCs to specifically enhance immunity, and some may even kill infected cells directly, like CD8+ T cells (6, 29). The IAV-specific T cells in this study showed a Th1 cell phenotype, and their major products, IFN-γ and TNF-α (Fig. 9), could exert powerful anti-influenza virus effects (30, 31). We also observed that the IAV-specific T cells presented cytotoxic characteristics by expressing the degranulation marker CD107a (6, 17) (Fig. 9). In such a scenario, focusing their responses on more conserved internal proteins would enable the cellular immunity to better handle invading IAV by antigen mutations to their surface proteins, which may back up the resultant ineffective humoral immunity.
FIG 9.
Functional assessment of IAV-specific CD4+ T cells. In vitro-expanded immunodominant epitopes, M1209-221 in donor 3 (A) and M1129-141 in donor 4 (B) specific T cell lines, were stimulated with the corresponding peptides. IFN-γ, TNF-α, and CD107a expression was detected in an ICS assay.
Many IAV-derived CD4+ T cell epitopes have been identified and indexed in the IEDB. As shown in Table 1, some epitopes identified in the present study were reported previously. However, their immunodominance status and HLA restrictions were often undetermined (6, 9). Up to six different HLA class II alleles could be coexpressed in the same subject, and they may all have the ability to present IAV epitopes (8). Our present data demonstrated that, in the same subject (donors 1 and 2), at least three different HLA class II alleles presented IAV epitopes (Fig. 5, 6, and 7). Epitope prediction is generally not accurate for immunodominant CD8+ T cell epitopes (7) and is likely less so for immunodominant CD4+ T cell epitopes (32) due to the fact that class II binding is more promiscuous (33, 34). Using the online prediction tool of the IEDB (35, 36), only two epitopes identified in our study (M1105–117/DRB1*0901 and M1129-141/DRB1*0101) had relatively low percentile ranks, which means high binding affinity. The rest were either not at all predictable or predicted with a low binding affinity (Table 1). Our systematic approach is likely efficient and effective, as demonstrated by deriving the exact sequences of two previously reported epitopes (M1209-221 and NP404-416) while confirming their immunodominant status (Table 1). Interestingly, one of the two, M1209-221, although reported to be restricted to DRB1*0404 and DR1101 (37), was found in our study to be presented by DPB1*0301 (donor 3) (Fig. 6B, iii and iv). Donor 2, donor 5, and donor 6 in our study indeed expressed DRB1*0404; however, the M1209-221-specific response was either not detected or detected as a minor response (Fig. 4). These results indicate that many CD4+ T cell epitopes may be presented by multiple HLA molecules, and their immunodominance status will naturally be HLA allele dependent. We also identified three novel epitopes, M1129-141, NP102-114, and NP463-475. Among them, two were immunodominant (Table 1). All of the epitopes identified here were defined to their most potent core sequences by 13-mer overlapping peptides and peptide titration.
TABLE 1.
Epitopes identified in this study and previously reported epitopes containing the same sequences
| Identified epitope | Donor | IDD/SDDa | HLA restriction | Rank (percentile)b | Reported epitope(s) | Reference | HLA restriction | Method |
|---|---|---|---|---|---|---|---|---|
| M143-55 | 2 | SDD | DRB1*0701 | 4.6 | M141-60 | 39 | DPB1*04:01 | Tetramer |
| M141-60 | 37 | DRB1*07:01c | Tetramer | |||||
| M141-60 | 40 | DRB1*11:01 | Tetramer | |||||
| M143-59 | 9 | Undetermined | ELISPOT | |||||
| M141-60 | 41 | DRB1*12:01 | Tetramer | |||||
| M141-60 | 42 | DRB1*01:03 | Tetramer and ELISPOT | |||||
| M141-60 | 42 | DPB1*04:01 | Tetramer and ELISPOT | |||||
| M194-106 | 1 | SDD | DRB1*1302 | 63.6 | M189-108 | 40 | DRB1*13:01 | Tetramer |
| M1101-113 | 2 | SDD | DRB1*0404 | 4.5 | M197-116 | 37 | DRB1*07:01 | Tetramer |
| M197-116 | 37 | DRB1*04:04 | Tetramer | |||||
| M197-116 | 37 | DRB1*11:01 | Tetramer | |||||
| M1101-113 | IEDB | DRB1*04:01 | Tetramer | |||||
| M197-116 | 40 | DRB1*13:01 | Tetramer | |||||
| M197-116 | 40 | DRB1*11:01 | Tetramer | |||||
| M1101-113 | 43 | DRB1*04:01 | [3H]thymidine and ELISPOT | |||||
| M197-113 | 9 | DR | ELISPOT | |||||
| M197-116 | 44 | DRB1*04:01 | Cytometric bead array | |||||
| M1100-114 | 45 | DR | ELISPOT | |||||
| M197-114 | 6 | undetermined | ELISPOT | |||||
| M197-116 | 42 | DRB1*01:03 | Tetramer and ELISPOT | |||||
| M1105-117 | 1 | SDD | DRB1* 0901 | 1.08 | M1105-117 | IEDB | DRB1*07:01 | Tetramer |
| M1105-124 | 42 | DRB1*01:03 | Tetramer and ELISPOT | |||||
| M1129-141 | 4 | IDD | DRB1*0101 | 0.6 | Not reported previously | |||
| M1209-221 | 3 | IDD | DPB1*0301 | NA | M1209-228 | 37 | DRB1*04:04 | Tetramer |
| M1209-228 | 37 | DRB1*11:01 | Tetramer | |||||
| NP102-114 | 1 | SDD | DPB1*0101 | 6.1 | Not reported previously | |||
| NP115-127 | 2 | SDD | DPB1*0601 | NA | NP113-132 | IEDB | DRB1*03:01 | Tetramer |
| NP115-131 | 9 | undetermined | ELISPOT | |||||
| NP404-416 | 2 | IDD | DRB1*0404 | 6.9 | NP401-420 | 46 | DRB1*14:01 | Tetramer |
| NP401-420 | 44 | DRB1*04:01 | Cytometric bead array | |||||
| NP404-417 | IEDB | DRB1*04:01 | Tetramer | |||||
| NP404-417 | IEDB | DRB1*04:04 | Tetramer | |||||
| NP404-417 | IEDB | DRB1*01:01 | Tetramer | |||||
| NP401-420 | IEDB | DRB1*15:01 | Tetramer | |||||
| NP404-416 | IEDB | DRB1*15:01 | Tetramer | |||||
| NP463-475 | 1 | IDD | DRB1*0901 | 4 | Not reported previously |
Considering the future use of these epitopes in vaccine design, it is obviously important to assess their sequence conservation with the pandemic and seasonal IAVs. Interestingly, plenty of variants were found (Table 2), although the internal viral proteins were considered to be more conserved than surface proteins. We also noticed that immunodominant epitopes seemed to have been subjected to greater selective pressure than subdominant ones, if the number of such variants is a direct indication. For three of the PR8-derived immunodominant epitopes, M1209-221, NP404-416, and especially NP463-475, a perfect sequence match can rarely be found in the IAV strains that have emerged in the last decade in Australia, while the subdominant epitopes, such as M143-55 and M1101-113, showed higher levels of conservation. Although IAV infections are resolved quickly in each infected individual, it is entirely possible that such epitope variants can be generated due to both the immune pressure and the lack of an error-proof nature of the IAV polymerase. However, the mechanisms associated with how such mutations are accumulated in newer strains at the population level are poorly understood (38). Furthermore, the cost of mutations to the viral fitness has not been studied. It is not clear at this stage whether the T cells specifically recognizing those epitopes derived from PR8 could recognize the variant peptides derived from the newer strains listed in Table 2. To improve coverage of future T cell-based vaccines, one possible strategy could be to integrate multiple NP and M1 proteins encoded by various circulating strains, which may induce and/or boost the immunodominant T cells specific for the covered epitope variants.
TABLE 2.
Conservation of peptide sequences within H1N1 and H3N2 viruses
| Peptide | Donor | IDD/SDDa | Peptide sequenceb | Frequency (%) of peptide variants |
|
|---|---|---|---|---|---|
| H1N1 | H3N2 | ||||
| M143-55 | 2 | SDD | MEWLKTRPILSPLc | 100 | 96 |
| ·······S····· | 4 | ||||
| M194-106 | 1 | SDD | DKAVKLYRKLKREc | 76 | |
| ·R··········· | 40 | 24 | |||
| ·R·····K····· | 55 | ||||
| GR·····K····· | 5 | ||||
| M1101-113 | 2 | SDD | RKLKREITFHGAKc | 35 | 100 |
| K············ | 60 | ||||
| ········L···· | 5 | ||||
| M1105-117 | 1 | SDD | REITFHGAKEISLc | 4 | |
| ···········A· | 30 | 92 | |||
| ··········V·· | 55 | ||||
| ··········VA· | 10 | 4 | |||
| ····L······A· | 5 | ||||
| M1129-141 | 4 | IDD | GLIYNRMGAVTTEc | 35 | 88 |
| ········T···· | 55 | 8 | |||
| ········T·A·· | 5 | ||||
| ········T··I· | 5 | ||||
| ·········I··· | 4 | ||||
| M1209-221 | 3 | IDD | ARQMVQAMRTIGTc | 10 | 12 |
| T····H······· | 50 | ||||
| ·········A··· | 30 | 32 | |||
| T···IH······· | 5 | ||||
| ··H······A··· | 5 | ||||
| ··········V·· | 36 | ||||
| ·········AV·· | 16 | ||||
| ··R······AV·· | 4 | ||||
| NP102-114 | 1 | SDD | GKWMRELILYDKEc | 64 | 4 |
| R············ | 2 | ||||
| ·······V····· | 2 | 60 | |||
| ···V···V····· | 29 | ||||
| ···V········· | 2 | ||||
| ·R·····V····· | 31 | ||||
| ·······V····K | 1 | ||||
| ·······V····G | 1 | ||||
| ·R·V···V····· | 1 | ||||
| ·····G·V····· | 1 | ||||
| NP115-127 | 2 | SDD | EIRRIWRQANNGDc | 31 | 19 |
| ············E | 4 | 79 | |||
| ·V··········· | 9 | ||||
| ····V·······E | 56 | ||||
| ··········S·E | 1 | ||||
| NP404-416 | 2 | IDD | GQISIQPTFSVQRc | 16 | 3 |
| ····T········ | 22 | ||||
| ····V········ | 62 | 58 | |||
| ··T·V········ | 31 | ||||
| ····V··A····· | 8 | ||||
| NP463-475 | 1 | IDD | VFELSDEKAASPIc | 4 | 1 |
| ·········TN·· | 62 | 88 | |||
| ·········TN·V | 4 | ||||
| ·······R··N·· | 2 | ||||
| ·······R·TN·· | 31 | ||||
| ··········N·· | 7 | ||||
IDD, immunodominant determinant; SDD, subdominant determinant.
Australian H1N1 (n = 20 for M1; n = 45 for NP) and H3N2 (n = 25 for M1; n = 72 for NP) sequences included the full-length sequences of viruses available from the NCBI influenza virus resource database (accessed on 25 April 2014). The search criteria were Australia, M1/NP, and H1N1/H3N2; identical sequences were represented by the oldest sequence in the group, full length only.
Peptide sequence was identified and used in this study.
In conclusion, using a systematic screening approach to evaluate immunodominant CD4+ T cell responses to IAV in healthy donors led to the demonstration that IAV-specific CD4+ T cell responses focus on M1 and NP. Detailed characterization of such T cell responses led to the discovery of 10 IAV M1 and NP CD4+ T cell epitopes. Although some of them have been previously reported, their immunodominance statuses were confirmed in this study. Moreover, several novel immunodominant epitopes were identified. However, variants of these epitopes existed widely, indicating greater immune selective pressure on the immunodominant epitopes than on the subdominant ones. To fully appreciate immunity to IAV infection and its interaction with the human host or human population, antigen specificity, especially the immunodominance hierarchy of CD4+ T cell responses and their selective pressure on IAV adaptation, will need to be studied in greater detail in the future.
ACKNOWLEDGMENTS
L.C. is the recipient of a China Scholarship. W.C. is an NHMRC Senior Research Fellow (603104). This project was partly supported by NHMRC Program Grant 567122 (W.C.) and the Chinese National Key Technology R&D Program (2014BAI15B00 and 2014BAI15B01).
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1. Tamura S, Kurata T. 2004. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn. J. Infect. Dis. 57:236–247 [PubMed] [Google Scholar]
- 2. Hamada H, Bassity E, Flies A, Strutt TM, Garcia-Hernandez MDL, McKinstry KK, Zou T, Swain SL, Dutton RW. 2013. Multiple redundant effector mechanisms of CD8+ T cells protect against influenza infection. J. Immunol. 190:296–306. 10.4049/jimmunol.1200571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun J, Braciale TJ. 2013. Role of T cell immunity in recovery from influenza virus infection. Curr. Opin. Virol. 3:425–429. 10.1016/j.coviro.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swain SL, McKinstry KK, Strutt TM. 2012. Expanding roles for CD4(+) T cells in immunity to viruses. Nat. Rev. Immunol. 12:136–148. 10.1038/nri3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McKinstry KK, Strutt TM, Kuang Y, Brown DM, Sell S, Dutton RW, Swain SL. 2012. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J. Clin. Invest. 122:2847–2856. 10.1172/JCI63689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, Xu XN. 2012. Preexisting influenza-specific CD4(+) T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 18:274–280. 10.1038/nm.2612 [DOI] [PubMed] [Google Scholar]
- 7. Wu C, Zanker D, Valkenburg S, Tan B, Kedzierska K, Zou QM, Doherty PC, Chen W. 2011. Systematic identification of immunodominant CD8+ T-cell responses to influenza A virus in HLA-A2 individuals. Proc. Natl. Acad. Sci. U. S. A. 108:9178–9183. 10.1073/pnas.1105624108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grant E, Wu C, Chan KF, Eckle S, Bharadwaj M, Zou QM, Kedzierska K, Chen W. 2013. Nucleoprotein of influenza A virus is a major target of immunodominant CD8(+) T-cell responses. Immunol. Cell Biol. 91:184–194. 10.1038/icb.2012.78 [DOI] [PubMed] [Google Scholar]
- 9. Babon JA, Cruz J, Orphin L, Pazoles P, Co MD, Ennis FA, Terajima M. 2009. Genome-wide screening of human T-cell epitopes in influenza A virus reveals a broad spectrum of CD4(+) T-cell responses to internal proteins, hemagglutinins, and neuraminidases. Hum. Immunol. 70:711–721. 10.1016/j.humimm.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee LY, Ha DO, Simmons LAC, de Jong MD, Chau NV, Schumacher R, Peng YC, McMichael AJ, Farrar JJ, Smith GL, Townsend AR, Askonas BA, Rowland-Jones S, Dong T. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118:3478–3490. 10.1172/JCI32460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Assarsson E, Bui HH, Sidney J, Zhang Q, Glenn J, Oseroff C, Mbawuike IN, Alexander J, Newman MJ, Grey H, Sette A. 2008. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 82:12241–12251. 10.1128/JVI.01563-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217. 10.1038/nm0297-212 [DOI] [PubMed] [Google Scholar]
- 13. Goulder PJ, Sewell AK, Lalloo DG, Price DA, Whelan JA, Evans J, Taylor GP, Luzzi G, Giangrande P, Phillips RE, McMichael AJ. 1997. Patterns of immunodominance in HIV-1-specific cytotoxic T lymphocyte responses in two human histocompatibility leukocyte antigens (HLA)-identical siblings with HLA-A*0201 are influenced by epitope mutation. J. Exp. Med. 185:1423–1433. 10.1084/jem.185.8.1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen L, Li B, Yang WC, He JL, Li NY, Hu J, He YF, Yu S, Zhao Z, Luo P, Zhang JY, Li HB, Zeng M, Lu DS, Li BS, Guo H, Yang SM, Guo G, Mao XH, Chen W, Wu C, Zou QM. 2013. A dominant CD4+ T-cell response to Helicobacter pylori reduces risk for gastric disease in humans. Gastroenterology 144:591–600. 10.1053/j.gastro.2012.12.002 [DOI] [PubMed] [Google Scholar]
- 15. Yuwen H, Cox JH, Yewdell JW, Bennink JR, Moss B. 1993. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology 195:732–744. 10.1006/viro.1993.1424 [DOI] [PubMed] [Google Scholar]
- 16. Gerhard W, Yewdell J, Frankel ME, Webster R. 1981. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290:713–717. 10.1038/290713a0 [DOI] [PubMed] [Google Scholar]
- 17. Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. 2003. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 281:65–78. 10.1016/S0022-1759(03)00265-5 [DOI] [PubMed] [Google Scholar]
- 18. Pala P, Bodmer HC, Pemberton RM, Cerottini JC, Maryanski JL, Askonas BA. 1988. Competition between unrelated peptides recognized by H-2-Kd restricted T cells. J. Immunol. 141:2289–2294 [PubMed] [Google Scholar]
- 19. Kozlowski S, Corr M, Shirai M, Boyd LF, Pendleton CD, Berzofsky JA, Margulies DH. 1993. Multiple pathways are involved in the extracellular processing of MHC class I-restricted peptides. J. Immunol. 151:4033–4044 [PubMed] [Google Scholar]
- 20. Sant AJ, Chaves FA, Krafcik FR, Lazarski CA, Menges P, Richards K, Weaver JM. 2007. Immunodominance in CD4 T-cell responses: implications for immune responses to influenza virus and for vaccine design. Expert Rev. Vaccines 6:357–368. 10.1586/14760584.6.3.357 [DOI] [PubMed] [Google Scholar]
- 21. Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. 2008. Cellular proteins in influenza virus particles. PLoS Pathog. 4:e1000085. 10.1371/journal.ppat.1000085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li P, Gregg JL, Wang N, Zhou D, O'Donnell P, Blum JS, Crotzer VL. 2005. Compartmentalization of class II antigen presentation: contribution of cytoplasmic and endosomal processing. Immunol. Rev. 207:206–217. 10.1111/j.0105-2896.2005.00297.x [DOI] [PubMed] [Google Scholar]
- 23. Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. 2005. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307:593–596. 10.1126/science.1104904 [DOI] [PubMed] [Google Scholar]
- 24. Schmid D, Pypaert M, Munz C. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 26:79–92. 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Munz C. 2012. Antigen processing for MHC class II presentation via autophagy. Front. Immunol. 3:9. 10.3389/fimmu.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menendez-Benito V, Neefjes J. 2007. Autophagy in MHC class II presentation: sampling from within. Immunity 26:1–3. 10.1016/j.immuni.2007.01.005 [DOI] [PubMed] [Google Scholar]
- 27. Teijaro JR, Verhoeven D, Page CA, Turner D, Farber DL. 2010. Memory CD4 T cells direct protective responses to influenza virus in the lungs through helper-independent mechanisms. J. Virol. 84:9217–9226. 10.1128/JVI.01069-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown DM, Lee S, Garcia-Hernandez MDL, Swain SL. 2012. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J. Virol. 86:6792–6803. 10.1128/JVI.07172-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soghoian DZ, Streeck H. 2010. Cytolytic CD4(+) T cells in viral immunity. Expert Rev. Vaccines 9:1453–1463. 10.1586/erv.10.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bot A, Bot S, Bona CA. 1998. Protective role of gamma interferon during the recall response to influenza virus. J. Virol. 72:6637–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seo SH, Webster RG. 2002. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J. Virol. 76:1071–1076. 10.1128/JVI.76.3.1071-1076.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chaves FA, Lee AH, Nayak JL, Richards KA, Sant AJ. 2012. The utility and limitations of current Web-available algorithms to predict peptides recognized by CD4 T cells in response to pathogen infection. J. Immunol. 188:4235–4248. 10.4049/jimmunol.1103640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doolan DL, Southwood S, Chesnut R, Appella E, Gomez E, Richards A, Higashimoto YI, Maewal A, Sidney J, Gramzinski RA, Mason C, Koech D, Hoffman SL, Sette A. 2000. HLA-DR-promiscuous T cell epitopes from Plasmodium falciparum pre-erythrocytic-stage antigens restricted by multiple HLA class II alleles. J. Immunol. 165:1123–1137. 10.4049/jimmunol.165.2.1123 [DOI] [PubMed] [Google Scholar]
- 34. Hammer J, Valsasnini P, Tolba K, Bolin D, Higelin J, Takacs B, Sinigaglia F. 1993. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell 74:197–203. 10.1016/0092-8674(93)90306-B [DOI] [PubMed] [Google Scholar]
- 35. Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. 2008. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 4:e1000048. 10.1371/journal.pcbi.1000048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. 2010. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics 11:568. 10.1186/1471-2105-11-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. 2008. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J. Immunol. 180:1758–1768. 10.4049/jimmunol.180.3.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gog JR, Rimmelzwaan GF, Osterhaus AD, Grenfell BT. 2003. Population dynamics of rapid fixation in cytotoxic T lymphocyte escape mutants of influenza A. Proc. Natl. Acad. Sci. U. S. A. 100:11143–11147. 10.1073/pnas.1830296100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang J, Huston L, Berger D, Danke NA, Liu AW, Disis ML, Kwok WW. 2005. Expression of HLA-DP0401 molecules for identification of DP0401 restricted antigen specific T cells. J. Clin. Immunol. 25:428–436. 10.1007/s10875-005-6095-6 [DOI] [PubMed] [Google Scholar]
- 40. Faner R, James E, Huston L, Pujol-Borrel R, Kwok WW, Juan M. 2010. Reassessing the role of HLA-DRB3 T-cell responses: evidence for significant expression and complementary antigen presentation. Eur. J. Immunol. 40:91–102. 10.1002/eji.200939225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chow IT, James EA, Tan V, Moustakas AK, Papadopoulos GK, Kwok WW. 2012. DRB1*12:01 presents a unique subset of epitopes by preferring aromatics in pocket 9. Mol. Immunol. 50:26–34. 10.1016/j.molimm.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang J, James EA, Huston L, Danke NA, Liu AW, Kwok WW. 2006. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clin. Immunol. 120:21–32. 10.1016/j.clim.2006.03.008 [DOI] [PubMed] [Google Scholar]
- 43. Ge X, Tan V, Bollyky PL, Standifer NE, James EA, Kwok WW. 2010. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J. Virol. 84:3312–3319. 10.1128/JVI.02226-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ge X, Gebe JA, Bollyky PL, James EA, Yang J, Stern LJ, Kwok WW. 2010. Peptide-MHC cellular microarray with innovative data analysis system for simultaneously detecting multiple CD4 T-cell responses. PLoS One 5:e11355. 10.1371/journal.pone.0011355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alexander J, Bilsel P, del Guercio MF, Stewart S, Marinkovic-Petrovic A, Southwood S, Crimi C, Vang L, Walker L, Ishioka G, Chitnis V, Sette A, Assarsson E, Hannaman D, Botten J, Newman MJ. 2010. Universal influenza DNA vaccine encoding conserved CD4+ T cell epitopes protects against lethal viral challenge in HLA-DR transgenic mice. Vaccine 28:664–672. 10.1016/j.vaccine.2009.10.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. James EA, Moustakas AK, Berger D, Huston L, Papadopoulos GK, Kwok WW. 2008. Definition of the peptide binding motif within DRB1*1401 restricted epitopes by peptide competition and structural modeling. Mol. Immunol. 45:2651–2659. 10.1016/j.molimm.2007.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. 2008. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 4:e1000048. 10.1371/journal.pcbi.1000048 [DOI] [PMC free article] [PubMed] [Google Scholar]