ABSTRACT
To combat emerging coronaviruses, developing safe and efficient platforms to evaluate viral protease activities and the efficacy of protease inhibitors is a high priority. Here, we exploit a biosafety level 2 (BSL-2) chimeric Sindbis virus system to evaluate protease activities and the efficacy of inhibitors directed against the papain-like protease (PLpro) of severe acute respiratory syndrome coronavirus (SARS-CoV), a biosafety level 3 (BSL-3) pathogen. We engineered Sindbis virus to coexpress PLpro and a substrate, murine interferon-stimulated gene 15 (ISG15), and found that PLpro mediates removal of ISG15 (deISGylation) from cellular proteins. Mutation of the catalytic cysteine residue of PLpro or addition of a PLpro inhibitor blocked deISGylation in virus-infected cells. Thus, deISGylation is a marker of PLpro activity. Infection of alpha/beta interferon receptor knockout (IFNAR−/−) mice with these chimeric viruses revealed that PLpro deISGylation activity removed ISG15-mediated protection during viral infection. Importantly, administration of a PLpro inhibitor protected these mice from lethal infection, demonstrating the efficacy of a coronavirus protease inhibitor in a mouse model. However, this PLpro inhibitor was not sufficient to protect the mice from lethal infection with SARS-CoV MA15, suggesting that further optimization of the delivery and stability of PLpro inhibitors is needed. We extended the chimeric-virus platform to evaluate the papain-like protease/deISGylating activity of Middle East respiratory syndrome coronavirus (MERS-CoV) to provide a small-animal model to evaluate PLpro inhibitors of this recently emerged pathogen. This platform has the potential to be universally adaptable to other viral and cellular enzymes that have deISGylating activities.
IMPORTANCE Evaluating viral protease inhibitors in a small-animal model is a critical step in the path toward antiviral drug development. We modified a biosafety level 2 chimeric virus system to facilitate evaluation of inhibitors directed against highly pathogenic coronaviruses. We used this system to demonstrate the in vivo efficacy of an inhibitor of the papain-like protease of severe acute respiratory syndrome coronavirus. Furthermore, we demonstrate that the chimeric-virus system can be adapted to study the proteases of emerging human pathogens, such as Middle East respiratory syndrome coronavirus. This system provides an important tool to rapidly assess the efficacy of protease inhibitors targeting existing and emerging human pathogens, as well as other enzymes capable of removing ISG15 from cellular proteins.
INTRODUCTION
Emerging coronaviruses (CoVs) are now recognized for their life-threatening potential. The outbreak of severe acute respiratory syndrome coronavirus (SARS-CoV) that occurred a decade ago resulted in over 8,000 infected people, with 10% mortality (1). A recently emerged coronavirus, designated Middle East respiratory syndrome coronavirus (MERS-CoV), has infected over 837 people, with 291 deaths as of 24 July 2014 (http://www.who.int/csr/don/2014_07_23_mers/en/). Epidemiologic studies implicate animal reservoirs as the source for emerging coronaviruses. By identifying a SARS-like CoV from Chinese horseshoe bats and analyzing the mutations in the spike glycoprotein, first in intermediate hosts and then in humans, researchers were able to document the evolution of an emerging CoV (2). The footprint for the evolution of MERS-CoV is not yet clear. MERS-CoV has about 80% genome sequence identity to the bat coronaviruses HKU4 and HKU5 (3, 4). In addition, infectious MERS-CoV has been isolated from the respiratory tracts of young camels (5–7), and there is accumulating evidence that adult camels have specific antibodies to MERS-CoV, consistent with endemic infection in the camel population (8, 9). Currently, it is unclear if the human cases of MERS are from sporadic introduction from animal reservoirs with limited human-to-human transmission or if there is ongoing transmission of MERS-CoV in asymptomatic humans or intermediate hosts (10–12). It is clear that CoVs have zoonotic potential for crossing the species barrier and emerging in the human population to cause lethal disease.
Viral proteases are logical targets for antiviral drug development, and protease inhibitors have been identified to block the papain-like protease (PLpro) domain of SARS-CoV (13). PLpro is encoded in the viral replicase polyprotein and is critical for processing the polyprotein to generate a functional replicase complex. Structural and enzymatic studies revealed that PLpro is also a viral deubiquitinase (DUB) that can cleave ubiquitin (Ub) or ubiquitin-like molecules, such as interferon-stimulated gene 15 (ISG15), from substrate proteins (14–16). Moreover, the catalysis-dependent interferon antagonism of PLpro implies that it may be involved in evading host innate immunity (17, 18). High-throughput screening efforts led to the identification of small-molecule inhibitors directed against the viral papain-like protease domain, and synthetic medicinal chemistry and structure-activity relationship studies have produced compounds that inhibit replication of SARS-CoV in cell culture (13, 19). However, one of the challenges for preclinical antiviral drug development is the availability of a small-animal model for emerging CoVs. For SARS-CoV, infection of mouse-adapted strains in mice (20, 21) or transgenic mice expressing the receptor (angiotensin-converting enzyme 2 [ACE2]) (22, 23) may serve as model systems for evaluating the efficacy of therapeutics. However, these studies must be performed in biosafety level 3 (BSL-3) laboratories with select-agent status. For MERS-CoV, although rhesus macaques can be infected (24, 25), less expensive animal models, such as mice and hamsters, are not susceptible to natural infection (26, 27). Dipeptidyl peptidase 4 (DPP4) was identified as the receptor for MERS-CoV in human and bat cells (28). Recently, novel model systems were generated for MERS-CoV infection by infecting mice with recombinant adenovirus expressing the human DPP4 receptor, which renders them susceptible to infection under BSL-3 conditions (29), and by generating recombinant HKU5 expressing the SARS-CoV spike protein (30). Development of additional affordable and adaptable small-animal models is needed to evaluate antivirals against existing and potentially emerging coronaviruses.
The goal of our study was to develop a biosafety level 2 system to evaluate inhibitors of the papain-like proteases of highly pathogenic emerging coronaviruses, such as SARS-CoV and MERS-CoV. We were inspired by the fundamental work of Frias-Staheli et al., who first demonstrated that a chimeric Sindbis virus (SV) system could be used to evaluate the potential protease activity of a BSL-4 pathogen, Crimean Congo hemorrhagic fever virus (CCHFV) (31). This chimeric-virus system is based on the use of the positive-strand RNA virus SV, a BSL-2 pathogen that is rapidly cleared by the immune system after infection in mice. Lenschow and coworkers showed that the interferon response, particularly ISG15, is critical for efficient clearance of SV (32). Consequently, SV infection of interferon receptor knockout (IFNAR−/−) mice, which are unable to signal the induction of interferon-stimulated genes, results in a lethal infection. However, if the gene for ISG15 is inserted into and expressed by SV, then infection with the chimeric virus induces an antiviral state and mice are protected from lethal infection. To induce the antiviral state, ISG15 must be conjugated to host cell proteins, a process termed ISGylation (33). The removal of ISG15 by deISGylating enzymes, such as the L protease of CCHFV, results in the abrogation of the protection mediated by ISG15, and mice succumb to infection. Thus, deISGylating enzymes can be used to “toggle off” the effect of ISG15 in this system. Given that the PLpros of CoVs not only function to process the viral polyprotein, but also possess deISGylating activity (14, 34), we reasoned that the chimeric Sindbis-ISG15-protease system could be exploited as a surrogate system to evaluate enzymatic activity and inhibition of CoV PLpros. Here, we demonstrate the utility of the chimeric SIP system for evaluating the deISGylating activities of PLpros from SARS-CoV and MERS-CoV and the efficacy of a SARS-CoV PLpro inhibitor.
MATERIALS AND METHODS
Cells, virus, and plasmids.
A baby hamster kidney cell line (BHK-21) and Vero-E6 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% (vol/vol) fetal calf serum (FCS) and supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml). Stocks of the recombinant mouse-adapted SARS-CoV (rMA15) were propagated and titrated on Vero-E6 cells. The virus was cryopreserved at −80°C until use, as described below. pcDNA3–6×myc-mISG15 was kindly provided by Min-Jung Kim (Pohang University of Science and Technology, Pohang, Republic of Korea). pcDNA3-Ube1L, pcDNA3-UbcH8, and pcDNA3-Herc5 were kind gifts from Robert M. Krug (University of Texas). pcDNA-MERS-PLpro and its catalytic mutant (C1592A) were generated in our laboratory as described previously (35).
PLpro inhibitors 3e and 3h.
The synthesis and characterization of PLpro inhibitors 3e and 3h were described by Baez-Santos et al. (36). For mouse administration, inhibitor 3e was formulated with 5% dimethyl sulfoxide (DMSO), 25% polyethylene glycol 400 (PEG 400), and 70% phosphate-buffered saline (PBS) (vol/vol/vol).
SIP viruses.
The Sindbis virus vector dsTE12Q was kindly provided by Deborah Lenschow (Washington University, St. Louis, MO). To generate the chimeric Sindbis virus expressing ISG15 and PLpro, the DNA fragment of ISG15-IRES-PLpro comprising the murine ISG15 cDNA (nucleotides [nt] 1 to 465), the hepatitis C virus (HCV) internal ribosome entry site (IRES) (nt 40 to 371 of the genome of HCV genotype 2b), and PLpro in frame with a V5 epitope tag at the C terminus (available upon request) were generated by synthesis or two-step overlapping PCR and subsequently cloned into the BstEII restriction site of the dsTE12Q vector. The insert DNAs of each chimeric virus were generated as follows.
(i) SIP-SWT and SIP-SMT.
A DNA sequence comprising the ISG15-IRES-PLpro (amino acids 1599 to 1855 in pp1a of SARS-CoV) in frame with a V5 epitope tag was codon optimized, synthesized, and subcloned into the pUC57 vector (Genscript, NJ). A catalytically inactive mutant of PLpro (cysteine 1651 to alanine) was generated as described previously (18) by site-directed mutagenesis PCR using primers (available upon request). The DNAs of interest were cut from recombinant pUC57 plasmids, cloned into the TE12Q vector, and designated SIP-SWT and SIP-SMT (see below).
(ii) SIP-MWT and SIP-MMT.
A DNA sequence comprising ISG15-IRES-PLpro (amino acids 1483 to 1802 in pp1a of MERS-CoV) was generated by two-step overlapping PCR using primers (available upon request). Briefly, the fragment of ISG15-IRES was amplified from the recombinant pUC57 plasmid described above, and the fragment of MERS-CoV PLpro or its catalytic mutant in frame with the V5 epitope was amplified from the plasmids of pcDNA-MERS-PLpro or its catalytic mutant (cysteine 1592 to alanine), respectively. The fragment of ISG15-IRES-PLpro was generated by PCR amplification using primers (available upon request), cloned into the BstE II restriction site of the TE12Q vector, and designated SIP-MWT and SIP-MMT (see below).
All constructs were verified by sequencing and linearized by digestion with XhoI restriction enzyme. The viral RNA was synthesized by in vitro transcription reaction following the manufacturer's instructions (Ambion), and the RNA was subsequently electroporated into BHK-21 cells with 3 pulses under conditions of 850 V and 25 μF in a 0.4-cm cuvette cap (Bio-Rad). Viral supernatants were harvested at 16 to 24 h postelectroporation, and the titers were determined by standard plaque assay on the BHK-21 cells.
Western blotting.
Cell lysates were separated in an SDS-10% PAGE gel and transferred to a polyvinylidene difluoride (PVDF) membrane in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) for 1 h at 65 V. The membrane was blocked using 5% dried skim milk in TBST buffer (0.9% NaCl, 10 mM Tris-HCl, pH 7.5, 0.1% Tween 20) for 2 h at room temperature and subsequently incubated with primary antibodies overnight at 4°C. The mouse anti-myc tag monoclonal antibody (MBL) was used to detect myc-ISG15 and the ISGylated proteins. ISG15 was detected using rabbit anti-ISG15 polyclonal antibodies (a kind gift from the Lenschow laboratory). The expression of PLpro and β-actin was detected using mouse anti-V5 (Invitrogen) and anti-beta-actin (Ambion) monoclonal antibodies, respectively. Horseradish peroxidase (HRP)-conjugated goat anti-mouse (Southern Biotechnology) was used as the secondary antibody, detected by using Western Lighting Chemiluminescence Reagent Plus (PerkinElmer), and visualized using a FluoroChem E imager (ProteinSimple).
Viral growth kinetics.
To analyze the replication of SIP viruses, viral growth kinetics assays were performed. Briefly, 105 BHK-21 cells per well in a 24-well plate were infected with each SIP virus at a multiplicity of infection (MOI) of 5, and the cell supernatants were collected at the indicated time points. The viral titers of the supernatants were determined by standard plaque assay on the BHK-21 cells.
DeISGylation assay.
To determine the deISGylating activity of PLpro, BHK-21 cells in a 24-well plate were transfected with 0.25 μg pcDNA3-myc-ISG15 and 0.125 μg each of ISGylating enzyme expression plasmids (pcDNA3-Ube1L, pcDNA3-UbcH8, and pcDNA3-Herc5) by Lipofectamine 2000 following the manufacturer's instructions. At 6 h posttransfection, the medium was removed and replaced by mock or SIP virus inoculum at an MOI of 5. After 1 h of inoculation at 37°C, the inocula were replaced by fresh DMEM containing 1% FCS. Cell lysates were prepared at 18 h postinfection using 100 μl lysis buffer (4% SDS, 3% dithiothreitol [DTT], and 65 mM Tris, pH 6.8) and analyzed by Western blotting.
To determine the effects of PLpro inhibitors on deISGylating activity, BHK-21 cells were subjected to transfection and infection as described above, followed by the addition of fresh DMEM (1% FCS) containing the inhibitor at a final concentration of 50 μM or a serial dilution. After 17 h of treatment (18 h postinfection), cell lysates were prepared and analyzed by Western blotting as described above.
Infection of mice with SIP viruses.
IFNAR−/− mice on a C57BL/6 background were initially obtained from Deborah Lenschow (Washington University, St. Louis, MO). The mice were bred and maintained at Loyola University, Chicago, IL, in accordance with all federal and Loyola University Chicago Institutional Animal Care and Use Committee (IACUC) guidelines. Seven- to 8-week-old male IFNAR−/− mice were infected subcutaneously in the left hind footpad with 6 × 106 PFU of SIP virus diluted in 25 μl DMEM, followed by daily body weight monitoring. When the weight loss of an infected mouse was more than 25% of the initial body weight, the mouse was humanely euthanized. The survival rate was calculated by counting the dead or euthanized mice and analyzed by log-rank test with GraphPad Prism software.
To determine if the PLpro inhibitor can block the function of PLpro in vivo, we tested PLpro 3e inhibitor in mice infected with SIP viruses. For each injection, 50 μg per dose was administered intraperitoneally (i.p.) at 0 and 2 days postinfection (p.i.). The weight loss of the mice was monitored daily, and the survival rate was analyzed as described above.
Infection of mice with SARS-CoV MA15 virus.
All viral and animal experiments were performed in a class II biological safety cabinet in a certified biosafety level 3 laboratory containing redundant exhaust fans, and workers wore personal protective equipment, including Tyvek suits, hoods, and high-efficiency particle arrestor-filtered powered air-purifying respirators (PAPRs).
Eight-week-old BALB/c mice were purchased from Harlan Laboratories and housed in accordance with all University of North Carolina (UNC)—Chapel Hill IACUC guidelines. The mice were administered either a single dose of 20 μg poly(I·C) inranasally (i.n.) or 50 μg per dose of inhibitor 3e or vehicle i.p. The mice were anesthetized with a mixture of ketamine-xylazine and were infected i.n. either with 2 × 104 PFU of the rMA15 virus or with PBS in a dose of 50 μl. Infected animals were monitored daily for weight loss and sacrificed upon approaching 80% of their starting body weight or manifesting severe clinical symptoms, according to the IACUC guidelines. The lung tissues of infected mice were collected at 4 days p.i., and the viral titers were determined by plaque assay on Vero-E6 cells.
The DNA sequences comprising the ISG15-IRES-PLpro (amino acids 1599 to 1855 in pp1a of SARS-CoV and amino acids 1483 to 1802 in pp1a of MERS-CoV) appear in GenBank under accession numbers NP_828849 and AFS88944, respectively.
RESULTS
Exploiting chimeric Sindbis virus to evaluate CoV PLpro activity.
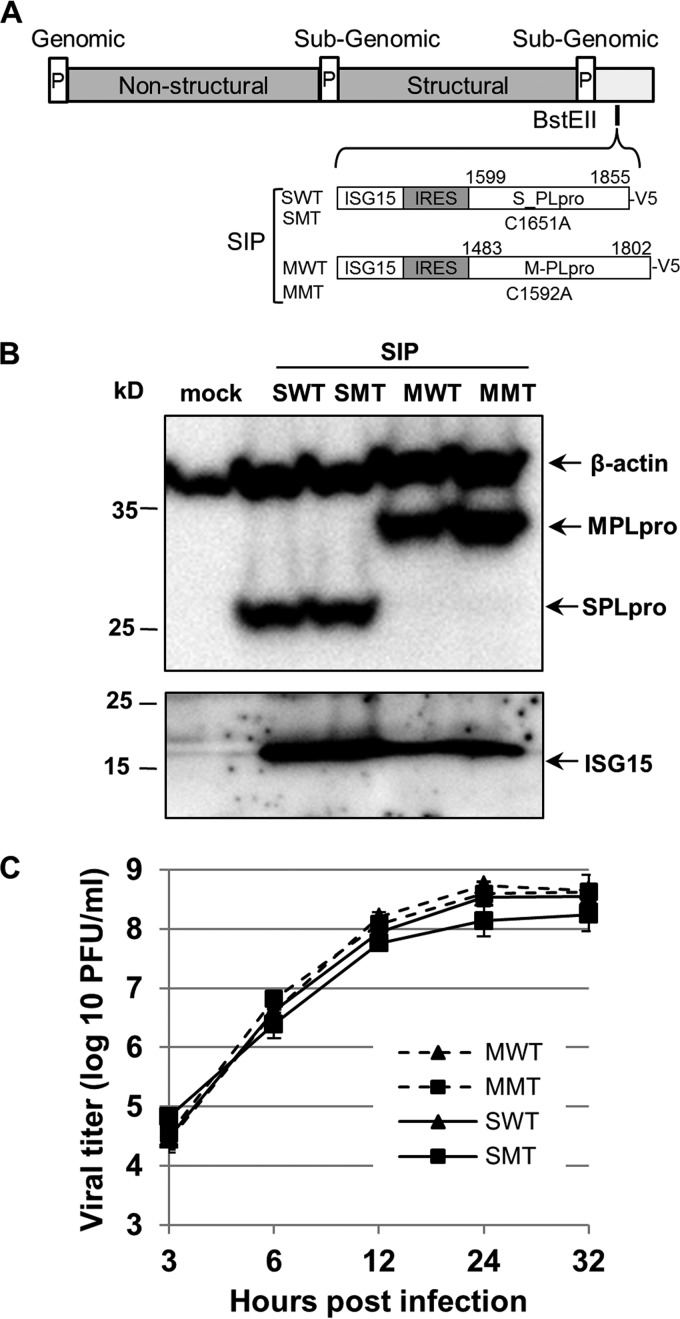
The goal of our study was to establish a BSL-2 mouse model system to assess the function of CoV PLpros and to evaluate PLpro inhibitors in vivo. To do this, we modified a chimeric Sindbis virus that coexpresses ISG15 and a viral operational taxonomic unit (OTU) domain deISGylating enzyme (31). To modify the system to allow expression of the larger ubiquitin-specific protease (USP) family enzyme from SARS-CoV, we replaced the original encephalomyocarditis virus (EMCV) IRES with the IRES from HCV. A dicistronic DNA fragment containing ISG15 cDNA followed by the HCV IRES and subsequent PLpro coding region was synthesized and inserted into the BstEII site of the recombinant Sindbis genome, namely, Sindbis-ISG15-IRES-PLpro (SIP). Four chimeric Sindbis viruses expressing either wild-type (WT) or catalytic cysteine mutant (MT) PLpros from SARS-CoV and MERS-CoV were generated and named SIP-SWT, SIP-SMT, SIP-MWT, and SIP-MMT, respectively (Fig. 1A). The expression of ISG15 and PLpro was detected by immunoblotting lysates prepared from virus-infected cells (Fig. 1B). Analysis of the viral growth kinetics revealed that the four SIP viruses replicate with similar kinetics and to high titer in BHK-21 cells (Fig. 1C). Furthermore, these viruses were stable upon passage in BHK-21 cells, which was in contrast to chimeric viruses containing larger insertions that had an EMCV IRES (data not shown).
FIG 1.
Generation of chimeric SIP viruses. (A) Schematic diagram of chimeric Sindbis viruses expressing ISG15, HCV IRES, and SARS-CoV PLpro (S_PLpro) or MERS-CoV PLpro (M-PLpro). (B) Detection of PLpro and ISG15 from cells infected with SIP viruses. BHK-21 cells were mock infected or infected with SIP viruses as indicated at an MOI of 5, lysates were prepared at 18 h postinfection, and proteins were detected by immunoblotting. (C) Replication kinetics of SIP viruses. BHK-21 cells were infected with SIP viruses as indicated at an MOI of 5, and supernatants were collected at the indicated time points. The viral titers are representative of three independent experiments. The error bars represent standard deviations (SD).
PLpros expressed by chimeric Sindbis virus have deISGylating activity.
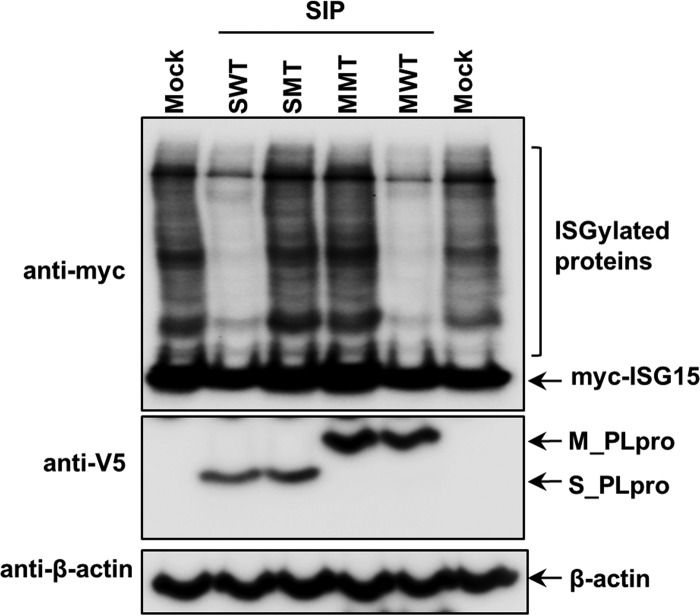
To determine whether PLpros expressed from Sindbis virus are able to cleave ISG15, BHK-21 cells were transfected with plasmids expressing ISGylation substrates and enzymes (myc-tagged ISG15, Ube1L, UbcH8, and Herc5) and subsequently infected with SIP-WT or SIP-MT viruses. Western blot results showed that cellular proteins were ISGylated in mock-infected cells transfected with ISGylation machinery plasmids. In contrast, the level of ISGylated proteins was significantly reduced in cells infected with SIP-WT, but not SIP-MT, virus (Fig. 2). The decreased level of ISGylated proteins in SIP-WT-infected cells is not due to virus-induced cell death or lack of host translation in the context of a Sindbis virus infection, as the SIP-MT virus grows to levels similar to those of the SIP-WT virus (Fig. 1C), and cellular proteins are ISGylated in SIP-MT-infected cells. These results indicate that both SARS-CoV PLpro and MERS-CoV PLpro exhibit broad deISGylating activity, even when expressed in the context of Sindbis virus infection. These results extend the work of Frias-Staheli et al. (31) and show that viral USP-type enzymes, like the viral OTU-type enzyme used in their study, can function as deISGylating proteases in the context of Sindbis virus infection.
FIG 2.
PLpros expressed by chimeric Sindbis virus have deISGylating activity. BHK-21 cells were transfected with ISGylating machinery expression plasmids (myc-ISG15, Ube1L, UbcH8, and Herc5) and subsequently infected with SIP viruses as indicated. The cell lysates were probed with anti-myc to detect ISGylated proteins and unconjugated ISG15. The expression of PLpro and beta-actin as a loading control was detected with anti-V5 and anti-beta-actin antibodies, respectively.
PLpro disrupts ISG15-mediated protection in mice.
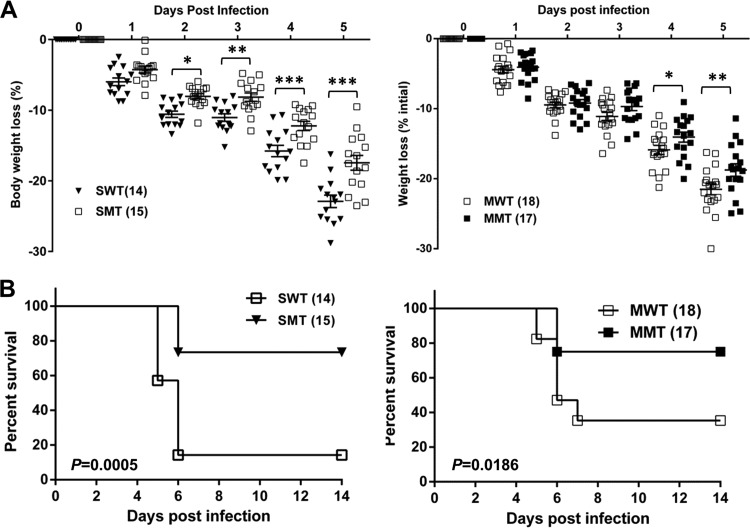
To determine whether coexpression of PLpro removes the protective effect of ISG15 during Sindbis virus infection, we infected IFNAR−/− mice with SIP-SWT or SIP-SMT virus and monitored for weight loss and survival. Mice exhibiting greater than 25% weight loss were humanely euthanized in accordance with animal care guidelines. As expected, the mice infected with SIP-SWT virus lost more body weight than mice infected with SIP-SMT virus at days 2 to 5 p.i. (Fig. 3A). SIP-SWT infection results in over 80% mortality, which is significantly higher than the mortality observed in mice infected with the SIP-SMT virus (26.7%) (P = 0.0005) (Fig. 3B). Moreover, we found that the SIP virus expressing wild-type PLpro from MERS-CoV (SIP-MWT), but not the catalytic inactive cysteine mutant of MERS-CoV PLpro (SIP-MMT), is able to inhibit the ISG15 function. The weight loss at 4 to 5 days p.i. and the mortality of mice infected with SIP-MWT were significantly greater than those of SIP-MMT-infected mice (Fig. 3C and D). These results indicate that PLpros of SARS-CoV and MERS-CoV are capable of disrupting the ISG15-mediated protection in IFNAR−/− mice, suggesting its role in antagonizing the innate immune response. In our system, the SIP virus-infected mice approached maximum weight loss at 5 to 6 days p.i. and then either recovered or succumbed to infection, whereas in Frias-Staheli's work, the majority of chimeric-virus-infected mice succumbed to infection at 8 to 12 days p.i. The differential survival times may due to the different genetic backgrounds of the mouse strains (129/SV/Pas in their work) or the expression of different proteases. Next, we wanted to determine if this chimeric Sindbis virus-mouse system could be used for evaluating small-molecule inhibitors directed against the PLpro domain.
FIG 3.
PLpros inhibit the ISG15-mediated antiviral effect in IFNAR−/− mice. Seven- to 8-week-old male IFNAR−/− mice were injected in the footpad with the WT (SWT or MWT) or MT (SMT or MMT) SIP viruses at 6 × 106 PFU and monitored for weight loss. The data are pooled from three independent experiments. The numbers of mice per group are indicated in parentheses. The statistical differences in body weight loss (A) and survival rate (B) were analyzed using Prism software with the two-way analysis of variance (ANOVA) test and the log-rank test, respectively. The error bars represent standard errors of the mean (SEM). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
An inhibitor of SARS-CoV PLpro specifically blocks deISGylating activity.
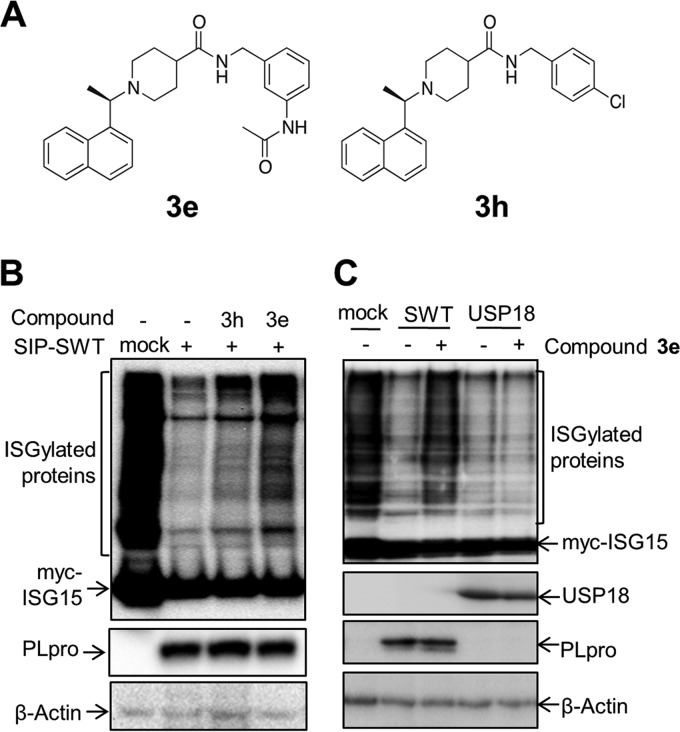
We reported the identification of small-molecule compounds that block PLpro activity in vitro and block the replication of SARS-CoV in cell culture (13, 19). We recently designed and evaluated the biological activity of a second generation of SARS-CoV PLpro inhibitors in vitro (36). One of these compounds, 3e, inhibits SARS-CoV PLpro with a potency of 390 nM and has an antiviral potency of 8.3 μM against SARS-CoV in Vero-E6 cells. Importantly, the compound is not cytotoxic, does not bind to human serum albumin, and has increased metabolic stability compared to other compounds evaluated (36). Therefore, to determine if compound 3e blocked PLpro activity in the context of the chimeric SIP virus, we exploited the deISGylation assay in cell culture. We found that the cells treated with compound 3e, but not the control compound 3h (Fig. 4A), a structural homolog of 3e with a higher 50% inhibitory concentration (IC50) (600 nM) and no antiviral activity (36), showed an increase in the level of ISGylated proteins in SIP-WT virus-infected cells (Fig. 4B). This indicates that the deISGylating activity of PLpro was blocked by compound 3e. We also found that the deISGylating activity of PLpro was inhibited by compound 3e in a dose-dependent manner (data not shown). To assess the specificity of compound 3e, we tested the activity of a cellular deISGylating enzyme, USP18 (also known as Ubp43), in the presence of 3e. Western blot results revealed that the level of ISGylated proteins in USP18-transfected cells was significantly decreased compared to the control, and there was no observable change in the level of ISGylation in the transfected cells treated with compound 3e (Fig. 4C). This result indicates that compound 3e does not inhibit the deISGylating activity of USP18. Taken together, these results suggest that compound 3e specifically blocks SARS-CoV PLpro deISGylating activity in cell culture during replication of the SIP virus.
FIG 4.
Compound 3e blocks the deISGylating activity of SARS-CoV PLpro in cell culture. (A) Structures of tested compounds. (B) BHK-21 cells were transfected with ISGylating machinery plasmids and subsequently infected with SIP-SWT virus as indicated, followed by addition of 50 μM compound 3e or 3h. (C) BHK-21 cells cotransfected with ISGylating machinery plasmids and USP18 expression plasmids were treated with 50 μM compound 3e or DMSO. Cell lysates were immunoblotted with anti-myc antibody for detecting the ISGylated proteins. Expression of PLpro, USP18, and beta-actin as a loading control was detected with anti-V5, anti-hemagglutinin (HA), and anti-beta-actin antibodies, respectively.
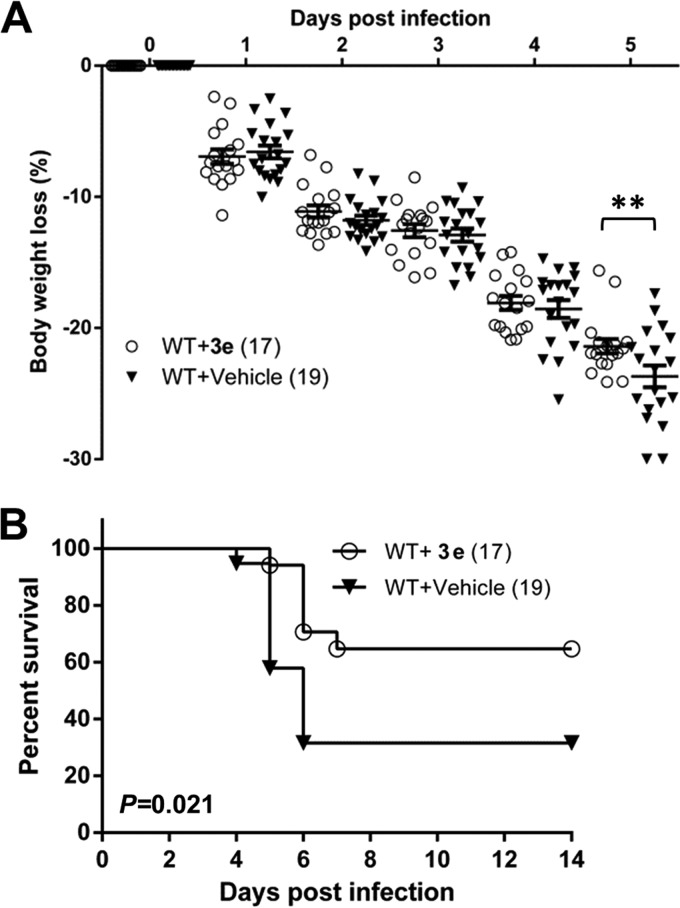
SARS-CoV PLpro inhibitor 3e protects mice from lethal SIP virus infection.
To determine whether the PLpro inhibitor was effective in a small-animal model, we injected IFNAR−/− mice subcutaneously with SIP-WT and administered compound 3e (50 μg/dose) i.p. at 0 and 2 days p.i. Although we found that there was no significant difference in weight loss between 3e- and vehicle-treated mice at early time points, except at 5 days p.i. (P < 0.01), the majority of the 3e-treated-mice (64.7%) recovered from infection before approaching the weight loss leading to euthanasia. In contrast, SIP-WT-infected mice administered the vehicle control exhibited significantly higher mortality (31.6% survival rate; P = 0.021) (Fig. 5). The effect of compound 3e was specific to SARS-CoV PLpro activity, as it had no significant effect on the pathogenesis of Sindbis virus infection of IFNAR−/− mice and there was no evidence of weight loss in mice injected with compound 3e alone (data not shown). These results demonstrate the protective effect of compound 3e during SIP-WT virus infection.
FIG 5.
Compound 3e blocks PLpro from disrupting ISG15-mediated protection in IFNAR−/− mice. IFNAR−/− mice were infected subcutaneously with SIP-SWT virus at 0 days p.i. and administered 50 μg per mouse of compound 3e or vehicle only i.p. at 0 and 2 days p.i. The mice were monitored for body weight loss (A) and survival (B). The data are pooled from three independent experiments. Total mouse numbers per group are indicated in parentheses. The statistical differences in weight loss and survival were analyzed with Prism software using the 2-way ANOVA test and the log-rank test, respectively. The error bars represent SEM. **, P < 0.01.
Evaluation of inhibitor 3e in SARS-CoV MA15-infected mice.
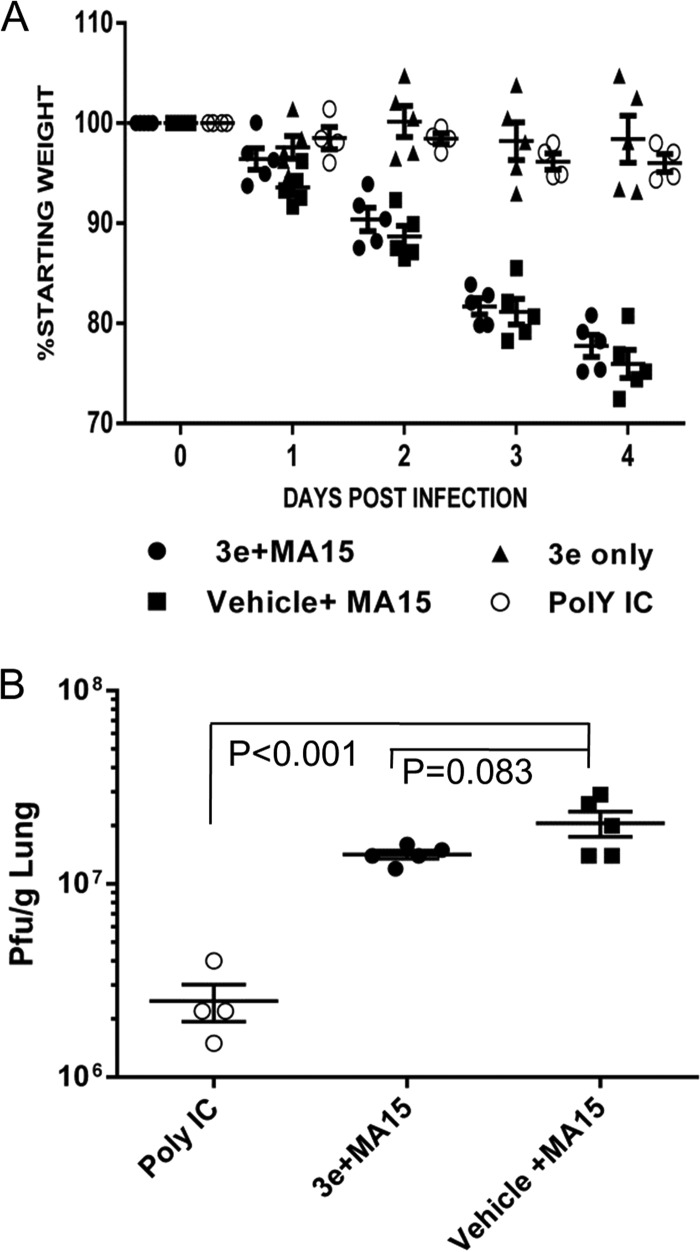
To extend our studies to a respiratory tract model system, we evaluated 3e using the established SARS-CoV MA15 intranasal-infection model (20). Wild-type BALB/c mice were pretreated intranasally with a 20-μg single dose of poly(I·C) as a positive control or 1 dose of compound 3e (50 μg/dose) 4 h prior to infection. Overall, the mice were administered 50 μg/dose of 3e twice a day on days 0 (4 h postinfection), 1, and 2 postinfection, the mice were monitored for weight loss through day 4 postinfection, and virus titers in the lungs were assessed on day 4 postinfection. Mice treated with compound 3e were not protected from virus-induced weight loss or virus replication, whereas the poly(I·C)-treated mice were completely protected from weight loss, and the virus titers on day 4 showed significant reduction in virus replication compared to the untreated-virus control (Fig. 6). The results suggest that although protease inhibitor 3e was capable of protecting the majority of mice from lethal systemic SIP virus infection, the inhibitor is either not sufficiently stable or bioavailable in the respiratory tract to reduce the replication and pathogenesis of a respiratory tract infection with SARS-CoV MA15 in mice.
FIG 6.
Evaluation of PLpro inhibitor 3e in the SARS-CoV MA15 mouse model. (A) Five 8-week-old BALB/c mice from each group were administered either a single dose of 20 μg poly(I·C) (Poly IC) i.n. or 50 μg per dose of 3e or vehicle i.p. at 4 h prior to infection i.n. with 2 × 104 PFU strain MA15 virus. The infected mice were further treated with 3e or vehicle twice a day at 0, 1, and 2 days p.i. The mice were monitored for weight loss and mortality. (B) The viral titer in the lung was determined at 4 days p.i. The statistical differences in weight loss and titer were analyzed with Prism software using the t test. The error bars represent SEM.
DISCUSSION
This work establishes a chimeric Sindbis virus-mouse model system for assessing the deISGylating activity of SARS-CoV and MERS-CoV PLpros and for evaluating SARS-CoV PLpro inhibitors in BSL-2 containment. To develop this model system, we exploited a chimeric Sindbis virus system pioneered by Frias-Staheli and coworkers, who first showed that viral proteases with deISGylating activity could remove the protective antiviral state induced by ISG15 (31). We extended their studies by (i) modifying the chimeric virus to express either SARS-CoV PLpro or MERS-CoV PLpro under translational control of the HCV IRES (this shortened IRES enabled the insertion of larger CoV USP-like proteases) and (ii) showing that SARS-CoV PLpro inhibitors could be evaluated in SIP virus-infected IFNAR−/− mice. This is the first demonstration of the efficacy and specificity of an inhibitor that targets a viral papain-like cysteine protease (PLpro) in a virus-infected animal. This is important because there are over 100 cellular DUBs, and previously, it was unclear if the PLpro inhibitor was sufficiently specific to alter protease/deISGylating activity in an infected animal. In general, this chimeric Sindbis virus-protease system enables the study of enzymes with deISGylating activity and establishes a BSL-2 model that can be used to evaluate the efficacy of small-molecule inhibitors of existing and emerging coronaviruses in vivo.
We hypothesize that the multifunctionality of CoV PLpros as proteases and deubiquitinating and deISGylating enzymes is important in viral pathogenesis, especially in antagonizing the innate immune response. ISG15 functions as an antiviral molecule through ISGylation of host substrates and by eliciting cytokine activity (36). Mice lacking ISG15 are more susceptible to lethal infection with Sindbis virus, herpesvirus, and influenza virus (32, 37). The role of ISG15 in CoV pathogenesis is not yet clear. Ma and coworkers showed that murine coronavirus infection of USP18-deficient mice, where ISGylation levels are high, resulted in lower viral titers and prolonged survival compared to wild-type mice, suggesting that high levels of ISGylation may delay CoV replication and pathogenesis (38). However, USP18 also mediates ISGylation-independent dendritic cell maturation (39), and thus, the loss of USP18 function may affect the kinetics of the immune response to viral infection. In the present study, we directly showed that CoV PLpros are capable of disrupting the protective effect of ISG15 in vivo, suggesting that PLpros have evolved an ISGylation antagonism mechanism to promote viral replication.
A major advantage of the SIP virus system is that we were able to study the PLpros of pathogenic CoVs in mice in a BSL-2 environment. This is particularly important because of the limited number of small-animal models currently available for the study of MERS-CoV PLpro inhibitors (29, 30). MERS-CoV enters cells by interaction with DPP4, and both human and bat DPP4s are functional receptors (28). In contrast, mice and rats are resistant to infection (26, 27), likely because of differences in the portion of DPP4 that interacts with the receptor binding domain of the spike glycoprotein of MERS-CoV (40, 41). The development of mouse-adapted strains of MERS-CoV and the generation of transgenic mice expressing the human DPP4 receptor are aimed at providing critical tools needed for understanding pathogenesis and evaluating candidate vaccines, but this work will be performed in BSL-3 containment. In contrast, the SIP virus mouse model is used in BSL-2 containment, and experiments are cost-effective for evaluating the efficacy and toxicity of PLpro inhibitors. In addition, we envision expanding the SIP virus system to identify broad-spectrum PLpro inhibitors that block a wide array of human and bat PLpro activities and protect mice from lethal viral infection. Using the SIP system, we can evaluate the efficacy of an inhibitor of a novel bat PLpro without the need for developing a transgenic-mouse model expressing the receptor for the novel virus. An important caveat of the SIP virus system is that it is a systemic infection and lethality is due to transmission of the virus through the central nervous system (32). Thus, it is a very sensitive system for evaluating protease inhibitors, and we demonstrate the efficacy of protease inhibitor 3e, which was previously shown to block replication of SARS-CoV in cell culture (36). However, as we report here, protease inhibitors must also be evaluated in the context of a respiratory tract infection, such as with mouse-adapted SARS-CoV MA15 (20, 21). We found that the protease inhibitor 3e was not effective at blocking replication and pathogenesis of SARS-CoV MA15 in the respiratory tracts of infected mice. This lack of efficacy may be due to the relative instability of 3e (36) or limitations of delivery onto the mucosal surfaces of the respiratory tract, the site of natural infection. Further work is required to optimize PLpro inhibitors for bioavailability, stability and appropriate delivery to block replication and pathogenesis of coronavirus.
Papain-like proteases (PLpros or PLPs) are conserved in all coronaviruses, and the goal of identifying a broad-spectrum inhibitor would be to inhibit existing human and potential emerging CoVs. Several CoV PLPs have been identified as deISGylating enzymes, such as the PLpro domain of SARS-CoV (14) and the PLP2 domain of human coronavirus (HCoV) NL63 (18). The SIP system described here could be extended to study the function of the PLpro/PLP2 domain of endemic HCoVs, such as HCoV NL63, HCoV HKU1, HCoV 229E, and HCoV OC43. HCoV NL63 generally causes mild upper respiratory tract disease in adults, but it can cause more severe respiratory disease in young children (42, 43). HCoV-HKU1 has been associated with pneumonia in the elderly (44). Currently, there are no animal models for evaluating inhibitors to HCoV NL63 or HCoV HKU1. Other PLPs, including those of bat coronaviruses, are postulated to act as deISGylating enzymes, as well, since they share conserved catalytic elements and are predicted to recognize similar cleavage sites in the viral polyprotein. We are currently developing SIP viruses expressing the PLP2 domain of HCoV NL63 and other CoVs with the long-term goal of identifying broad-spectrum PLpro inhibitors that could block replication of existing and emerging coronaviruses.
Overall, this study provides evidence of CoV PLpro deISGylating activity in the context of viral infection and establishes a BSL-2 mouse model for evaluating PLpro inhibitors. The current studies are designed to facilitate antiviral drug development for existing and emerging coronavirus infections and are a forerunner of the development of similar platforms aimed at testing inhibitors against other deISGylating enzymes in vivo.
ACKNOWLEDGMENTS
We thank Jonathan E. Snyder and Richard Kuhn at Purdue University for providing reagents. We thank Tom Gallagher at Loyola University, Chicago, IL, for his helpful comments on the manuscript. We also thank Andy Kilianski in the Baker laboratory for his technical support and discussions.
This work was supported by the National Institutes of Health (NIAID-R01AI085089 to S.C.B. and A.D.M.) and NIAID U19 AI 107810 to R.S.B. A.M.M was supported by an Arthur J. Schmitt fellowship (Loyola University, Chicago, IL).
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1. Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439–450. 10.1038/nrmicro2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Song H-D, Tu C-C, Zhang G-W, Wang S-Y, Zheng K, Lei L-C, Chen Q-X, Gao Y-W, Zhou H-Q, Xiang H, Zheng H-J, Chern S-WW, Cheng F, Pan C-M, Xuan H, Chen S-J, Luo H-M, Zhou D-H, Liu Y-F, He J-F, Qin P-Z, Li L-H, Ren Y-Q, Liang W-J, Yu Y-D, Anderson L, Wang M, Xu R-H, Wu X-W, Zheng H-Y, Chen J-D, Liang G, Gao Y, Liao M, Fang L, Jiang L-Y, Li H, Chen F, Di B, He L-J, Lin J-Y, Tong S, Kong X, Du L, Hao P, Tang H, Bernini A, Yu X-J, Spiga O, Guo Z-M, Pan H-Y, He W-Z, Manuguerra J-C, Fontanet A, Danchin A, Niccolai N, Li Y-X, Wu C-I, Zhao G-P. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U. S. A. 102:2430–2435. 10.1073/pnas.0409608102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 4. Van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus ADME, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RAM. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3:e00473–12. 10.1128/mBio.00473-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Memish ZA, Cotten M, Meyer B, Watson SJ, Alsahafi AJ, Al Rabeeah AA, Corman VM, Sieberg A, Makhdoom HQ, Assiri A, Al Masri M, Aldabbagh S, Bosch B-J, Beer M, Müller MA, Kellam P, Drosten C. 2014. Human Infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg. Infect. Dis. 20:1012–1015. 10.3201/eid2006.140402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hemida MG, Chu DKW, Poon LLM, Perera RAPM, Alhammadi MA, Ng H-Y, Siu LY, Guan Y, Alnaeem A, Peiris M. 2014. MERS coronavirus in dromedary camel herd, Saudi Arabia. Infect. Dis. 20:1231–1234. 10.3201/eid2007.140571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM, Madani TA. 2014. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 370:2499–2505. 10.1056/NEJMoa1401505 [DOI] [PubMed] [Google Scholar]
- 8. Perera RA, Wang P, Gomaa MR, El-Shesheny R, Kandeil A, Bagato O, Siu LY, Shehata MM, Kayed AS, Moatasim Y, Li M, Poon LL, Guan Y, Webby RJ, Ali MA, Peiris JS, Kayali G. 2013. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 18:pii=20574 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20574 [DOI] [PubMed] [Google Scholar]
- 9. Reusken CBEM, Haagmans BL, Müller MA, Gutierrez C, Godeke G-J, Meyer B, Muth D, Raj VS, Smits-De Vries L, Corman VM, Drexler J-F, Smits SL, El Tahir YE, De Sousa R, van Beek J, Nowotny N, van Maanen K, Hidalgo-Hermoso E, Bosch B-J, Rottier P, Osterhaus A, Gortázar-Schmidt C, Drosten C, Koopmans MPG. 2013. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 13:859–866. 10.1016/S1473-3099(13)70164-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. 2013. Family cluster of Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 368:2487–2494. 10.1056/NEJMoa1303729 [DOI] [PubMed] [Google Scholar]
- 11. Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DAT, Alabdullatif ZN, Assad M, Almulhim A, Makhdoom H, Madani H, Alhakeem R, Al-Tawfiq JA, Cotten M, Watson SJ, Kellam P, Zumla AI, Memish ZA. 2013. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 369:407–416. 10.1056/NEJMoa1306742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cotten M, Watson SJ, Kellam P, Al-Rabeeah AA, Makhdoom HQ, Assiri A, Al-Tawfiq JA, Alhakeem RF, Madani H, AlRabiah FA, Al Hajjar S, Al-Nassir WN, Albarrak A, Flemban H, Balkhy HH, Alsubaie S, Palser AL, Gall A, Bashford-Rogers R, Rambaut A, Zumla AI, Memish ZA. 2013. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet 382:1993–2002. 10.1016/S0140-6736(13)61887-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson ME, Baker SC, Ghosh AK, Mesecar AD. 2008. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 105:16119–16124. 10.1073/pnas.0805240105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Ménard R. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199–15208. 10.1128/JVI.79.24.15199-15208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189–15198. 10.1128/JVI.79.24.15189-15198.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, Mesecar AD. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 103:5717–5722. 10.1073/pnas.0510851103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 83:6689–6705. 10.1128/JVI.02220-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clementz M, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84:4619–4629. 10.1128/JVI.02406-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghosh AK, Takayama J, Rao KV, Ratia K, Chaudhuri R, Mulhearn DC, Lee H, Nichols DB, Baliji S, Baker SC, Johnson ME, Mesecar AD. 2010. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation. J. Med. Chem. 53:4968–4979. 10.1021/jm1004489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roberts A, Deming D, Paddock CD, Cheng A, Yount B, Vogel L, Herman BD, Sheahan T, Heise M, Genrich GL, Zaki SR, Baric R, Subbarao K. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3:e5. 10.1371/journal.ppat.0030005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Day CW, Baric R, Cai SX, Frieman M, Kumaki Y, Morrey JD, Smee DF, Barnard DL. 2009. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology 395:210–222. 10.1016/j.virol.2009.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81:813–821. 10.1128/JVI.02012-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tseng C-TK, Huang C, Newman P, Wang N, Narayanan K, Watts DM, Makino S, Packard MM, Zaki SR, Chan T-S, Peters CJ. 2007. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human angiotensin-converting enzyme 2 virus receptor. J. Virol. 81:1162–1173. 10.1128/JVI.01702-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Munster VJ, de Wit E, Feldmann H. 2013. Pneumonia from human coronavirus in a macaque model. N. Engl. J. Med. 368:1560–1562. 10.1056/NEJMc1215691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Wit E, Rasmussen AL, Falzarano D, Bushmaker T, Feldmann F, Brining DL, Fischer ER, Martellaro C, Okumura A, Chang J, Scott D, Benecke AG, Katze MG, Feldmann H, Munster VJ. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 110:16598–16603. 10.1073/pnas.1310744110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coleman CM, Matthews KL, Goicochea L, Frieman MB. 2013. Wild type and innate immune deficient mice are not susceptible to the Middle East Respiratory Syndrome Coronavirus. J. Gen. Virol. 95:408–412. 10.1099/vir.0.060640-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Wit E, Prescott J, Baseler L, Bushmaker T, Thomas T, Lackemeyer MG, Martellaro C, Milne-Price S, Haddock E, Haagmans BL, Feldmann H, Munster VJ. 2013. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS One 8:e69127. 10.1371/journal.pone.0069127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM, Thiel V, Drosten C, Rottier PJM, Osterhaus ADME, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ, Baric RS, Enjuanes L, Gallagher T, McCray PB, Perlman S. 2014. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U. S. A. 111:4970–4975. 10.1073/pnas.1323279111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Agnihothram S, Yount BL, Donaldson EF, Huynh J, Menachery VD, Gralinski LE, Graham RL, Becker MM, Tomar S, Scobey TD, Osswald HL, Whitmore A, Gopal R, Ghosh AK, Mesecar A, Zambon M, Heise M, Denison MR, Baric RS. 2014. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. mBio 5:e00047–14. 10.1128/mBio.00047-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, García-Sastre A, Virgin HW. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404– 416. 10.1016/j.chom.2007.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, Guin AKO, Schmidt RE, Levine B, Virgin HW., IV 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974–13983. 10.1128/JVI.79.22.13974-13983.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Skaug B, Chen ZJ. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143:187–190. 10.1016/j.cell.2010.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, Shanks J, Sterner DE, Bernal A, Mattern MR, Wilkinson KD, Butt TR. 2008. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 17:1035–1043. 10.1110/ps.083450408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kilianski A, Mielech A, Deng X, Baker SC. 2013. Assessing activity and inhibition of MERS-CoV papain-like and 3C-like proteases using luciferase-based biosensors. J. Virol. 87:11955–11962. 10.1128/JVI.02105-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baez-Santos YM, Barraza SJ, Wilson MW, Agius M, Mielech AM, Davis NM, Baker SC, Larsen SD, Mesecar AD. 2014. X-Ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J. Med. Chem. 57:2393–2412. 10.1021/jm401712t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, Garcı A, Leib DA, Pekosz A, Knobeloch K, Horak I, Whiting HV., IV 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U. S. A. 104:1371–1376. 10.1073/pnas.0607038104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma X-Z, Bartczak A, Zhang J, He W, Shalev I, Smil D, Chen L, Phillips J, Feld JJ, Selzner N, Levy G, McGilvray I. 2014. Protein interferon-stimulated gene 15 conjugation delays but does not overcome coronavirus proliferation in a model of fulminant hepatitis. J. Virol. 88:6195–6204. 10.1128/JVI.03801-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cong X-L, Lo M-C, Reuter BA, Yan M, Fan J-B, Zhang D-E. 2012. Usp18 promotes conventional CD11b+ dendritic cell development. J. Immunol. 188:4776–4781. 10.4049/jimmunol.1101609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. 2013. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231. 10.1038/nature12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Y, Rajashankar KR, Yang Y, Agnihothram SS, Liu C, Lin Y-L, Baric RS, Li F. 2013. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J. Virol. 87:10777–10783. 10.1128/JVI.01756-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fouchier RAM, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH, Osterhaus ADME. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 101:6212–6216. 10.1073/pnas.0400762101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJM, Wolthers KC, Wertheim-van Dillen PME, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat. Med. 10:368–373. 10.1038/nm1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Woo PCY, Lau SKP, Chu C, Chan K, Tsoi H, Huang Y, Wong BHL, Poon RWS, Cai JJ, Luk W, Poon LLM, Wong SSY, Guan Y, Peiris JSM, Yuen K. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884–895. 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]