ABSTRACT
The small hydrophobic (SH) protein is a 64-amino-acid polypeptide encoded by the human respiratory syncytial virus (hRSV). SH protein has a single α-helical transmembrane (TM) domain that forms pentameric ion channels. Herein, we report the first inhibitor of the SH protein channel, pyronin B, and we have mapped its binding site to a conserved surface of the RSV SH pentamer, at the C-terminal end of the transmembrane domain. The validity of the SH protein structural model used has been confirmed by using a bicellar membrane-mimicking environment. However, in bicelles the α-helical stretch of the TM domain extends up to His-51, and by comparison with previous models both His-22 and His-51 adopt an interhelical/lumenal orientation relative to the channel pore. Neither His residue was found to be essential for channel activity although His-51 protonation reduced channel activity at low pH, with His-22 adopting a more structural role. The latter results are in contrast with previous patch clamp data showing channel activation at low pH, which could not be reproduced in the present work. Overall, these results establish a solid ground for future drug development targeting this important viroporin.
IMPORTANCE The human respiratory syncytial virus (hRSV) is responsible for 64 million reported cases of infection and 160,000 deaths each year. Lack of adequate antivirals fuels the search for new targets for treatment. The small hydrophobic (SH) protein is a 64-amino-acid polypeptide encoded by hRSV and other paramyxoviruses, and its absence leads to viral attenuation in vivo and early apoptosis in infected cells. SH protein forms pentameric ion channels that may constitute novel drug targets, but no inhibitor for this channel activity has been reported so far. A small-molecule inhibitor, pyronin B, can reduce SH channel activity, and its likely binding site on the SH protein channel has been identified. Black lipid membrane (BLM) experiments confirm that protonation of both histidine residues reduces stability and channel activity. These results contrast with previous patch clamp data that showed low-pH activation, which we have not been able to reproduce.
INTRODUCTION
The human respiratory syncytial virus (hRSV) is an enveloped pneumovirus in the Paramyxoviridae family that causes lower respiratory tract disease in infants and in elderly and immunocompromised populations worldwide (1). Up to 64 million reported cases of hRSV infection and 160,000 deaths occur each year. Although the virus was identified almost half a century ago (2), there are still no vaccines or effective antiviral drugs available. hRSV can cause repeated reinfections throughout life, and its molecular mechanism of pathogenesis is not yet completely understood.
The hRSV genome comprises a nonsegmented negative-stranded RNA of ∼15 kb that transcribes 11 proteins, including the three membrane proteins F, G, and small hydrophobic (SH). Proteins F and G are key factors during virus attachment, fusion, and entry into host cells (3, 4). In contrast, the role of SH protein is less clear. RSV lacking the SH gene (RSVΔSH) was viable, caused formation of syncytia, and grew as well as the wild-type (WT) virus in cell culture (5–8). However, RSVΔSH virus replicated 10-fold less efficiently than the WT in the upper respiratory tract (7, 8). Also, RSVΔSH virus was attenuated in vivo by virus passage in mouse and chimpanzee models (5, 9). Overall, these results indicate involvement of SH protein in the pathogenesis of RSV infection.
Homologs of RSV SH protein are found in parainfluenza virus 5 (PIV5), mumps virus (MuV), and J paramyxovirus (JPV). In all of these systems, SH protein seems to block apoptosis in infected cells through inhibition of the tumor necrosis factor alpha (TNF-α) pathway (6, 10–12). It is thought that by delaying apoptosis, the virus may evade host inflammatory responses and the premature death of the host cells. Recent reports have also suggested that SH protein activates the NLRP3 inflammasome (13).
The SH protein is a type II integral membrane protein that is 64 (RSV subgroup A) or 65 (RSV subgroup B) amino acids long. In infected cells, most SH protein accumulates at the membranes of the Golgi complex, but it has also been detected in the endoplasmic reticulum and plasma membranes (14). During infection, the full-length unmodified form is the major species (15) although a truncated form (4.5 kDa) and glycosylated and phosphorylated forms have also been detected (16, 17). SH protein has a single predicted α-helical transmembrane (TM) domain (15) which is highly conserved (18, 19). The C- and N-terminal extramembrane domains are oriented lumenally/extracellularly and cytoplasmically, respectively.
Both the synthetic TM domain (residues 18 to 43) and full-length SH protein, which are responsible for channel activity, have been shown to form homopentamers in a variety of detergents (20, 21). The mutual orientation of monomers corresponding to the synthetic TM domain in lipidic membranes was determined using site-specific infrared dichroism (20), which showed His-22 to be in a lumenal, close to interhelical, orientation. This TM arrangement was confirmed by nuclear magnetic resonance (NMR) studies that reconstructed the pentameric α-helical bundle of the full-length protein in dodecylphosphocholine (DPC) micelles (21). In this micelle model, a single α-helical TM domain was flanked N-terminally by an α-helix and C-terminally by an extended β-hairpin.
The availability of SH channel inhibitors would help further understanding of the role of this channel activity in the viral life cycle of RSV and also have obvious therapeutic potential. However, thus far no compound has been reported that is able to inhibit SH protein channel activity. Here, we report one compound that shows partial inhibition and can constitute a starting point to obtain an effective channel inhibitor. The possible binding sites of this compound on the structure of the SH protein pentameric channel have been mapped using solution NMR in detergent micelles. To confirm that the structure of SH protein in detergent is representative of the structure present during channel activity assays in black lipid membranes (BLM), we have refined the structure of SH protein in a membrane-mimicking bicellar environment. Overall, the results presented establish a solid ground for future drug development targeting this important viroporin.
MATERIALS AND METHODS
Cloning of the SH gene and expression and purification of the His-MBP-SH construct.
The SH protein used in this work (strain S2 ts1C; GenBank accession number NP_044594.1) (22), was cloned, expressed, purified as wild type or mutant, and isotopically labeled essentially as described previously (21).
Liposome-based assay to screen for SH protein inhibitors.
Potassium flux into liposomes incorporating SH protein pentamers was measured by fluorescence changes of the dye Asante potassium green-2 (APG-2; TEFLabs, Austin, TX). The assay conditions, e.g., lipid, protein-to-lipid ratio, fluorophore, buffer type, and pH (internal and external), were first tested in larger volumes, and fluorescence was measured with a Cary eclipse fluorescence spectrophotometer. Once the conditions were optimized, the assay was implemented in a microplate reader (Tecan M200 Pro). First, 3 mg of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC; Avanti Polar Lipids) in chloroform was mixed with 26 μg of SH protein in methanol (molar ratio, 1,000:1) and dried under a N2 stream. After samples were dried overnight in a desiccator, 300 μl of internal buffer (25 mM tetraethylammonium [TEA]-HEPES, 0.5 mM EDTA, 100 mM TEA-SO4, 20 μM APG-2, pH 5.5) was added to obtain a lipid concentration of 10 mg/ml. The solution was vortexed for 10 min, sonicated, subjected to three freeze-thaw cycles, and extruded through a 200-nm polycarbonate membrane. Nonincorporated APG-2 was removed by passing the liposomes through an Econo-Pac 10DG (Bio-Rad) size exclusion column preequilibrated with internal buffer in the absence of APG-2. Liposome size was measured using a Brookhaven 90 Plus particle size analyzer. APG-2 fluorescence was measured from 150 μl of liposome suspension in a quartz cuvette (excitation wavelength [λex], 488 nm; emission wavelength [λem], 540 nm). The potassium concentration outside the vesicles was increased to 140 mM by addition of 560 mM KCl, 25 mM TEA-HEPES, and 0.5 mM EDTA, pH 7.5. Finally, 4 μl of 25% Triton X-100 (final 0.5%) was added.
For the microplate assay, the liposome fraction was diluted 3.5 times with 5 ml of internal buffer. An aliquot (75 μl) of the liposome suspension was placed in each well of a microplate (Greiner 96-well Fluotrac 200) containing 1 μl of compound dissolved in dimethyl sulfoxide (DMSO) from the National Cancer Institute (NCI). The final drug concentration was 10 μM. After a 15-min incubation, 25 μl of potassium buffer (560 mM KCl, 25 mM TEA-HEPES, 0.5 mM EDTA, pH 7.5) was added to obtain a final potassium concentration of 140 mM. Fluorescence of samples was read after a 5-min incubation. Finally, 4 μl of a solution of 25% Triton X-100 (to obtain a 0.5% final concentration) was added, and emission was measured after 5 min.
Plaque assay.
Vero cells were placed in 12-well plates and cultured at 37°C overnight to reach about 90% confluence. Cells were infected with the RSV strain VR-1580 (ATCC) at a multiplicity of infection (MOI) of approximately 1. At 2 h postinfection, virus inoculum was removed, and 1 ml of fresh Dulbecco's modified Eagle's medium (DMEM) containing 2% fetal bovine serum (FBS) and 1 μl of DMSO, with and without different concentrations of pyronin B, was added. At 90 h postinfection, cultured supernatants were collected, and the 50% tissue culture infective doses (TCID50) of RSV were calculated by the Reed-Muench method (23). The experiment was repeated three times, and each sample was titrated twice.
Ion channel reconstitution and ionic current recording in black lipid membranes.
SH protein, wild type or mutant, was reconstituted in planar lipid bilayers formed by apposition of two monolayers prepared from a solution of a 3:1:1 mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)–1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)–1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phospho-l-serine (DOPS) (Avanti Polar Lipids) in pentane. Lipids were added on 70- to 90-μm-diameter orifices in the 15-μm-thick Teflon partition that separated two identical chambers (24, 25). The orifices were pretreated with a 1% solution of hexadecane in pentane. Aqueous solutions of 1 M KCl were buffered with 5 mM HEPES at pH 6. All measurements were performed at room temperature. After the addition of SH protein in one side of the chamber (here referred to as the cis side), a +100-mV voltage difference was applied to detect current. The electric potential was applied using Ag/AgCl electrodes in 2 M KCl–1.5% agarose bridges, assembled within standard 250-μl pipette tips. The potential was defined as positive when it was higher on the side of the protein addition (cis side), whereas the trans side was set to ground. An Axopatch 200 B amplifier (Molecular Devices, Sunnyvale, CA) in the voltage clamp mode was used to measure the current and the applied potential. The chamber and the head stage were isolated from external noise sources with a double metal screen (Amuneal Manufacturing Corp., Philadelphia, PA).
Experiments aimed to determine the conductance at different pHs were carried out with aqueous solutions of 1 M KCl buffered with 5 mM HEPES in 3:1:1 DOPC-DOPS-DOPE bilayers. The current amplitude recorded in approximately 30 independent experiments, each one lasting ∼200 s, was averaged. Then the data were fitted to a single Gaussian to obtain the mean value of conductance (G) as a function of pH.
The effect of the pyronin B on channel conductance was determined as follows: once a channel activity was recorded, different micromolar concentrations of inhibitor were added into the cis side (protein side addition), and the current amplitude in the presence of the inhibitor was analyzed. The dose-response curve at different pyronin B concentrations was built to obtain the effective Kd (dissociation constant).
Ion selectivity was evaluated by measuring the reversal potential (RP) under a 10-fold gradient concentration of KCl (500 mM/50 mM KCl, in planar bilayers of 3:1:1 DOPC-DOPS-DOPE). Ion selectivity experiments were also performed at basic (pH 7.5) and acidic (pH 3) pH to compare the channel selectivity and to evaluate a possible pH dependency related to His protonation.
Gel electrophoresis.
To perform electrophoresis in the presence of perfluorooctanoic (PFO) acid detergent, we modified Invitrogen's SDS-NuPAGE protocol by replacing SDS with PFO acid (26). Lyophilized peptide was dissolved in sample buffer containing 4% PFO acid and heated at 65°C for 5 min prior to loading. The gel was run at 80 V for 2 to 3 h with morpholineethanesulfonic acid (MES) running buffer containing 0.5% PFO acid. Blue native PAGE (BN-PAGE) was performed as described previously (27) using lyophilized peptide solubilized (0.1 mM) in sample buffer containing a 25 mM concentration of the chosen detergent.
NMR sample preparation.
The phospholipid bicelle system was composed of 1,2-dilauroyl-sn-glycero-3-phosphocholine (or dilauroylphosphatidylcholine [DLPC]) (Avanti Polar Lipids) and 1,2-dihexanoyl-sn-glycero-3-phosphocholine (or dihexanoylphosphatidylcholine [DHPC]) (Avanti Polar Lipids). Peptide-DLPC mixture in hexafluoroisopropanol (HFIP) was dried under a N2 stream followed by high vacuum. DHPC solution in appropriate buffer was added into the dried peptide-DLPC mixture, and the mixture was vortexed and sonicated. In this way, lyophilized SH peptides were reconstituted into DHPC-DLPC bicelles (9.5%, wt/vol; q (DLPC/DHPC molar ratio) = 0.3) at 0.7 mM (corresponding to approximately 1:200 peptide-to-lipid ratio) buffered with 20 mM sodium phosphate and 50 mM NaCl at pH 5.5.
Partial alignment of the SH protein/bicelle complexes relative to magnetic field was obtained by using stretched polyacrylamide hydrogels (28, 29). A 4.2% polyacrylamide gel was polymerized in a gel chamber having an inner diameter of 5.4 mm. After polymerization was complete, gels were washed in H2O overnight and then twice with sample buffer containing 20 mM sodium phosphate and 50 mM NaCl at pH 5.5. The gels were then completely dried at room temperature. The protein solution containing SH protein/bicelle complex was soaked into the dried gels over 2 days to ensure complete rehydration. The hydrated 4.2% gel was then radically compressed into an open-ended tube (inner diameter, 4.2 mm) using a gel press assembly (New Era Enterprise, Inc.).
NMR spectroscopy.
NMR experiments were performed at 40°C (313 K) using Bruker Avance-II 700 and 600 NMR spectrometers equipped with cryogenic TXI probes (Bruker BioSpin). Sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) was used as the internal reference for 1H nuclei. The chemical shifts of 13C and 15N nuclei were calculated from the 1H chemical shifts. The NMR data were processed using TopSpin, version 3.1 (Bruker, Billerica, MA) and analyzed using CARA (www.nmr.ch). Sequence-specific assignment of backbone 1HN, 15N, 13C′, and 13Cα was achieved by using two-dimensional (2D) [1H-15N] TROSY-HSQC (transverse relaxation optimized spectroscopy–heteronuclear single-quantum coherence) and three-dimensional (3D) HNCO, HN(CA)CO, HNCA, HN(CO)CA, and HNCACB experiments on a 15N/13C-labeled SH protein. Side chain resonances were assigned utilizing 3D 15N-resolved nuclear Overhauser effect spectroscopy (NOESY)-HSQC (mixing time, 150 ms) HCC(CO)NH, and 13C-resolved NOESY-HSQC (120-ms mixing time). 1H-15N heteronuclear steady-state NOEs were obtained by recording spectra with and without 4-s 1H presaturation at 700 MHz to determine backbone flexibility.
The titration experiment with the drug was performed with 15N-labeled SH protein in DPC micelles (21). Chemical shift perturbations were monitored using [1H-15N] TROSY-HSQC spectra where the magnitude of the perturbation was calculated using the following formula: CSP (ppm) = √ΔδH2 + (0.24 · ΔδN)2, where CSP is chemical shift perturbation and δH and δN are chemical shift increments of proton and nitrogen, respectively.
Structure calculation.
NOE distance restraints were obtained from 15N NOESY-HSQC (mixing time, 150 ms) and 13C NOESY-HSQC (mixing time, 120 ms) spectra. Backbone dihedral angle restraints (φ and ψ) were derived from 13C′, 13Cα, 13Cβ, 1Hα, and 1Hβ chemical shift values using TALOS+ (30). The short-range and medium-range NOE connectivities were used to establish the sequence-specific 1H NMR assignment and to identify elements of the regular secondary structure. Structure calculations were started from 100 random conformers, using the standard simulated annealing protocol in CYANA, version 3.0 (31, 32). Seven cycles of NOE assignment and structure reconstruction resulted in a bundle of 20 conformers. CNS, version 1.3 (33, 34), was used to refine the structure using the standard simulated annealing protocol. A total of 20 structures were calculated, and the structure statistics are summarized in Tables 1and 2.
TABLE 1.
Structure statistics for the selected 20 structures corresponding to the N-terminal extramembrane domain of SH protein
| Parameter | Value for the parameter |
|---|---|
| NMR restraints (no.) | |
| Total unambiguous distance restraints | 228 |
| Intraresidual | 91 |
| Sequential (| i − j | = 1)a | 58 |
| Short-range (| i − j | < = 1) | 149 |
| Medium (2 ≤ | i − j | ≤ 4) | 61 |
| Long range (| i − j | ≥ 5) | 18 |
| Dihedral angle restraints | 8 |
| Hydrogen bond restraintsb | 6 |
| RDC restraintsc | 9 |
| RMSD from the avg atomic coordinates (residues 1–14 [Å])d | |
| Backbone atoms | 0.33 ± 0.13 |
| All heavy atoms | 0.93 ± 0.23 |
| Ramachandran analysis (%) | |
| Residues in most favored regions | 72.9 |
| Residues in additional allowed regions | 26.2 |
| Residues in generously allowed regions | 0.8 |
| Residues in disallowed regions | 0.0 |
i and j are the positions in the sequence of two given residues.
Backbone hydrogen bonds of α-helix are applied for regions confirmed to be α-helix according to local NOE pattern.
RDC, residual dipolar coupling.
Statistics are calculated and averaged over an ensemble of the 20 structures with lowest target function from CYANA.
TABLE 2.
Structure statistics for the selected 20 structures of the C-terminal extramembrane domain of SH protein
| Parameter | Value for the parameter |
|---|---|
| NMR restraints (no.) | |
| Total unambiguous distance restraints | 514 |
| Intraresidual | 165 |
| Sequential (| i− j| = 1)a | 146 |
| Short-range (| i− j | < = 1) | 311 |
| Medium (2 ≤ | i− j | ≤ 4) | 96 |
| Long range (| i − j | ≥ 5) | 107 |
| Dihedral angle restraints | 34 |
| Hydrogen bond restraintsb | 8 |
| RDC restraintsc | 21 |
| RMSD from the avg atomic coordinates (residues 38–64 [Å])d | |
| Backbone atoms | 0.44 ± 0.17 |
| All heavy atoms | 0.94 ± 0.15 |
| Ramachandran analysis (%) | |
| Residues in most favored regions | 59.4 |
| Residues in additional allowed regions | 35.8 |
| Residues in generously allowed regions | 4.8 |
| Residues in disallowed regions | 0.0 |
i and j are the positions in the sequence of two given residues.
Backbone hydrogen bonds of α-helix are applied for regions confirmed to be α-helix according to local NOE pattern.
RDC, residual dipolar coupling.
Statistics are calculated and averaged over an ensemble of the 20 structures with lowest target function from CYANA.
Cell culture and transient transfection.
Standard procedures were utilized for cell culture and transfection, as described by Dossena et al. (35), with modifications. HEK293 Phoenix cells were cultured in minimum essential Eagle medium (MEM; Sigma, Austria) supplemented with 10% fetal bovine serum (FBS; Lonza), 2 mM l-glutamine, 100 μg/ml penicillin, 100 U/ml streptomycin, and 1 mM pyruvic acid (sodium salt). The cells were maintained at 37°C in 5% CO2 and 95% air at 100% humidity. Subcultures were routinely established every second to third day by seeding the cells into 100-mm-diameter petri dishes following trypsin/EDTA treatment. For patch clamp experiments, HEK293 Phoenix cells were seeded into 30-mm-diameter petri dishes, grown overnight, and transfected with 3 μg of plasmid DNA by the calcium phosphate coprecipitation method. Medium was changed at 8 h posttransfection, and the cells were seeded on glass coverslips (diameter, 10 mm) at 32 h posttransfection. Electrophysiology measurements were performed at 48 to 58 h posttransfection.
Electrophysiological recordings.
HEK293 Phoenix cells were transfected with the bicistronic mammalian expression vector pIRES-AcGFP1 (Clontech) (where IRES is internal ribosome entry site and AcGFP1 is Aequorea coerulescens monomeric green fluorescent protein) coding either for AcGFP only or for both AcGFP and the full-length SH expressed as separate proteins. Control experiments were conducted in cells transfected with the pIRES-AcGFP1 vector. Single cells expressing AcGFP were selected by fluorescence microscopy and voltage clamped using the whole-cell patch clamp technique. The resistance of the glass pipettes was 3 to 8 MΩ when they were filled with the pipette solution (135 mM potassium gluconate, 10 mM KCl, 10 mM HEPES, 0.5 mM EGTA, 2 mM Mg++-ATP, pH 7.3 [adjusted with KOH]). The pH 7.4 bath solution was composed of 124 mM NaCl, 3.5 mM KCl, 0.01 mM NaH2PO4, 26.2 mM NaHCO3, 1.3 mM MgSO4, 2.5 mM CaCl2, and 10 mM d-(+)-glucose, pH 7.4, while the pH 5.5 bath solution was composed of 124 mM NaCl, 3.5 mM KCl, 0.01 mM NaH2PO4, 0.3 mM NaHCO3, 25.9 mM sodium gluconate, 1.3 mM MgSO4, 2.5 mM CaCl2, and 10 mM d-(+)-glucose. Both bath solutions were constantly bubbled with 95% O2 and 5% CO2 to maintain their respective pHs. The Henderson-Hasselbalch equation was used to calculate the amount of bicarbonate required to achieve the respective pH in the presence of 5% CO2. Fast exchange of the pH 7.4 bath solution with the acidic bath solution was accomplished using a perfusion system with a flow rate of 5 ml/min and a bath volume of ∼300 μl. All experiments were carried out at room temperature. For data acquisition, an EPC-8 (HEKA Elektronik, Germany) amplifier controlled by a Macintosh computer running the Patch Master (HEKA Elektronik, Germany) software was used. Access resistance as well as fast and slow capacitance were compensated and monitored throughout the recordings. All current measurements were filtered at 5 kHz and digitized at 50 kHz. The cells were held at 0 mV, and step pulses of 400-ms duration were applied from 0 mV to +40 mV every 20 s to monitor current changes over time. To establish the current-to-voltage (I/V) relationship, step pulses of 500-ms duration were applied every 5 min from −120 mV to +100 mV in 20-mV increments from a holding potential of 0 mV. For data analysis, Fit Master (HEKA Elektronik, Germany) and Excel (Microsoft, USA) software were used. The current values were normalized to the membrane capacity to obtain the current density. Membrane potentials were corrected for liquid junction potentials, and membrane currents were leakage subtracted.
RESULTS
Inhibition of SH protein channel activity by pyronin B.
Since no SH protein channel inhibitors have been reported to date, a liposome-based fluorescence assay was used to screen for compounds that can inhibit SH channel activity (see Materials and Methods). For this assay, four microplates from the NCI library containing 80 compounds each (10 μM) were tested against SH protein reconstituted in liposomes. Of these compounds, five showed significant inhibition of channel activity (>50%): NSC526417, NSC81189, NSC78627, NSC77827, and NSC44690 (results not shown).
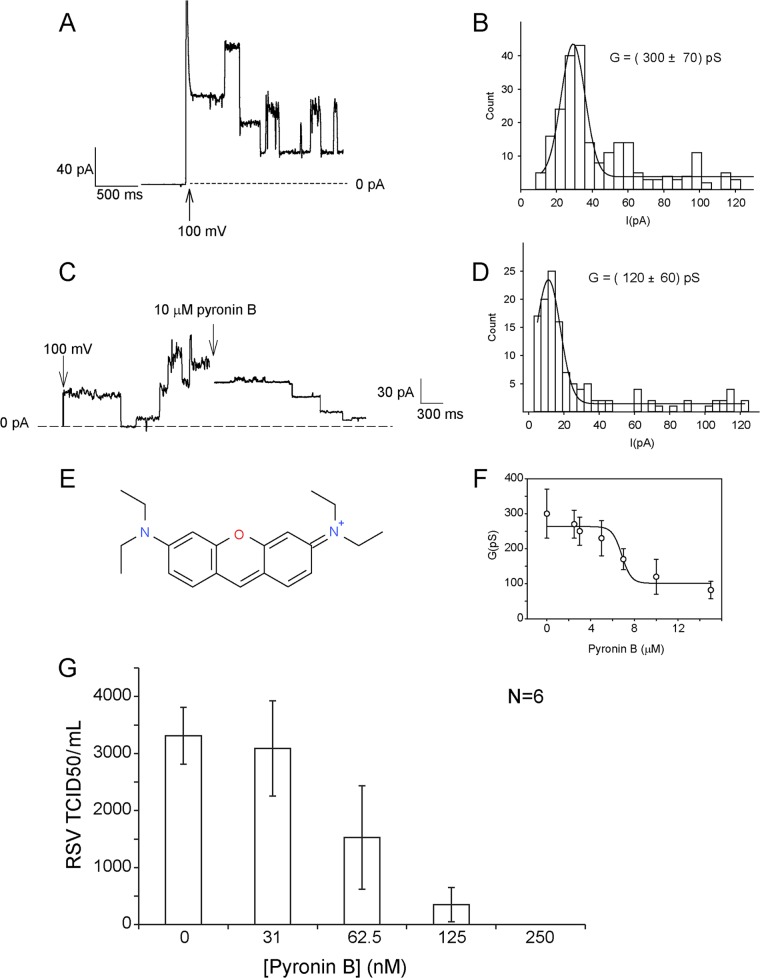
These compounds were then retested against purified SH protein reconstituted in planar lipid bilayers (BLM). The most potent of these was NSC44690 (pyronin B). A concentration of 10 μM pyronin B led to a ∼60% inhibition of channel activity; conductance changed from 300 ± 70 pS under control conditions (Fig. 1A and B) to 120 ± 60 pS in the presence of pyronin B (Fig. 1C and D). The structure of pyronin B is shown in Fig. 1E. The binding affinity (Kd) of the compound was obtained from the dose-response curve and determined to be around 6.8 μM (Fig. 1F). The effect of pyronin B was tested on RSV replication in Vero cells (Fig. 1G), where tissue culture infectivity (50% infective dose, TCID50) was zero at a drug concentration of 0.25 μM.
FIG 1.
Ionic current and inhibition of SH channel activity. (A) Ionic current through SH protein in a 3:1:1 DOPC-DOPS-DOPE planar membrane in 1 M KCl at pH 6. (B) Histogram of current amplitude. (C) Example of original recording of channel activity before and after addition (arrow) of 10 μM compound NSC44690 (pyronin B) under the same conditions as described in panel A. (D) Corresponding current amplitude histogram in the presence of pyronin B. (E) Structure of pyronin B. (F) Dose-response curve obtained in the presence of 0, 2.5, 3, 5, 7, 10, and 15 μM pyronin B. The conductance values were fitted using a standard sigmoidal dose-response curve to obtain a Kd of ∼ 6.8 μM. (G) Effect of increasing concentration of pyronin B on RSV replication in Vero cells, calculated as the 50% tissue culture infective dose (TCID50).
Binding site of pyronin B on the SH protein pentameric surface.
Binding of pyronin B to the SH protein was monitored with a series of [1H-15N] TROSY-HSQC spectra recorded in the absence and presence of pyronin B at different concentrations. When a 4.8 mM concentration of the drug was added to 0.6 mM uniformly 15N-labeled SH protein, backbone resonances of several residues in SH protein were significantly perturbed (Fig. 2A), and large (≥0.07 ppm) chemical shift perturbations (CSPs) were observed at both ends of the TM domain. At the N-terminal end, residues more affected were Ile-6 and Ile-21 (Fig. 2B). At the C-terminal end, the residue most affected was Ala-39 (Fig. 2B) and a group of nearby residues, i.e., Ile-38, Ile-40, Leu 41, and Lys-43 (Fig. 2C), located at the C-terminal end of the TM domain. Interestingly, most of these juxtamembrane residues (residues 38 to 43) form a conserved motif in the RSV SH protein, A39ILNKL43 (Fig. 2D), which suggests that this is a critical region for SH protein, where emergence of resistance mutations would be difficult for the virus. By comparison, residues at positions 6 and 21 are not conserved. Determination of intermolecular nuclear Overhauser effects (NOEs) for the drug-protein complex was not possible, probably due to weak interaction.
FIG 2.
Pyronin B binding significantly perturbed the chemical shifts of SH oligomer. (A) Superposition of TROSY-HSQC spectra of uniformly 15N-labeled SH protein (monomer concentration, 0.6 mM) in the absence (blue) and presence (red) of 4.8 mM pyronin B. Peaks that undergo significant shifts upon complex formation are highlighted. (B) Selected regions in TROSY-HSQC spectrum in the presence of pyronin B concentrations increasing from 0 (red) to 0.3 (pink), 1.2 (yellow), 2.4 (green), and 4.8 mM (blue). (C) Chemical shift perturbation (CSP, ppm) of the backbone amide resonances of 0.6 mM 15N-labeled SH protein upon titration with 4.8 mM pyronin B. The missing and overlapping residues were eliminated from the analysis. (D) Result from a Clustal X alignment of >40 variants of RSV SH protein where identity (star) and similarity (dot) are indicated.
Docking of pyronin B to the SH protein pentameric surface.
To provide more insight into the nature of the pyronin B binding sites, docking studies of pyronin B to the SH protein were performed, using as a model for docking the SH pentameric structure obtained in DPC micelles (21). The Patchdock server (36, 37) performed first a blind docking search such that pyronin B could explore the entire surface of SH protein. The ten best structures were selected according to the geometric shape complementarity score. Interestingly, in nine of these structures, pyronin B was located near the residues that showed the largest chemical shift perturbation, i.e., Ile-6, Ile-21, and Ala-39 (Fig. 3). Thus, the following search was restricted to regions close to these residues and led to the prediction of two possible binding sites at both ends of the TM domain. The N-terminal one is formed by Phe-14 and Ile-21 in one monomer (i+1) and Ile-6 of the previous monomer (the ith monomer) (Fig. 3A and B). The C-terminal one is formed by residues Ile-40, Leu-41, and Lys-43 in one monomer (i+1) and Ile-38 and Ala-39 of the previous monomer (i) (Fig. 3A to C). An analysis of the druggable pockets on the SH pentamer surface by the software DogSiteScorer (38) also identified these same regions (Fig. 3D).
FIG 3.
Mapping of pyronin B binding to SH protein. (A) The CSP values in the presence of pyronin B are mapped onto the structure of SH protein, with residues showing larger (CSP of ≥0.07 ppm) and smaller (CSP of ≥0.04 ppm) shifts represented in red and yellow, respectively. (B and C) Model of the two pyronin B binding sites on the SH pentamer at the N-terminal (B) and C-terminal (C) ends of the TM domain (dotted circles), where residues with a CSP of ≥0.07 ppm are labeled in red. Only two monomers of SH protein pentamer (i and i + 1) are shown for simplicity. (D) Druggable pockets (green mesh) predicted by DoGSiteScorer (38), an automated pocket detection and druggability assessment tool. For comparison, the main residues that showed largest NMR chemical shift changes are shown in red. Two monomers have been removed from the pentamer for clarity.
SH mutants abolish inhibition by pyronin B.
To more specifically delineate the site(s) of inhibition by pyronin B, we tested the effect of the drug on (i) a truncated form of SH protein, encompassing only its TM domain (SH-TM, residues 18 to 43) and (ii) full-length mutants at the two proposed binding sites, i.e., at the N or the C termini of the TM domain. The SH-TM should contain an intact binding site at the C-terminal juxtamembrane domain (residues 38 to 43), whereas most residues identified at the N-terminal end are absent. The SH-TM (20) and the four single mutants of the full-length SH protein tested were able to form pentamers: A39S, I21F, and I21Y in PFO gels (Fig. 4A) and H22F in blue native gels (Fig. 4B). All of them also showed channel activity although in I21F this was reduced to about 50% (Fig. 4C). The effect of pyronin B on SH-TM was similar to full-length SH protein (40 and 60% inhibition, respectively) at 10 μM, suggesting that the intact C-terminal end of the TM domain (residues 38 to 41) is sufficient for inhibition. Indeed, the conservative mutation A39S in that region almost completely prevented inhibition (∼10% inhibition).
FIG 4.
Channel conductance of SH protein mutants and effect of pyronin B on channel activity. (A) PFO gel electrophoresis of several full-length SH mutants at the two proposed binding sites of pyronin B, loaded with 3 μg (left) and 12 μg (right). Arrowheads indicate the main oligomers. (B) Blue native gel electrophoreses in DHPC and C14-betaine of His-Phe mutants. Arrowheads indicate the bands corresponding to pentamers. (C) Conductance of the wild type (WT), transmembrane domain (SH-TM), and single SH mutants in the absence of inhibitor (control) or in the presence of 10 μM pyronin B. (D) Representative ionic current through mutant A39S in a 3:1:1 DOPC-DOPE-DOPS planar membrane at pH 6 before and after the addition of 10 μM pyronin B.
Surprisingly, the three other mutants also showed resistance to inhibition by pyronin B (∼20% inhibition), despite the intact C-terminal binding site. These results can be rationalized by assuming a destabilization of the C-terminal binding site in the presence of these more disruptive mutations at the N-terminal end of the TM. I21F and I21Y represent more bulky substitutions, whereas H22F affects His-22, which was suggested to have a structural role (21). Although H22F does not affect pentamerization (Fig. 4B), H22A completely disrupted pentamerization, in contrast to H51A (21). Thus, cross talk between the two ends of the TM may lead to a modification of the pyronin B binding site at the C-terminal region, without affecting channel activity.
Secondary structure of SH protein reconstituted in bicelles.
The comparison between the inhibitory effect of pyronin B (BLM) and its binding to SH protein (NMR) was performed using lipid membrane and detergent micelle environments, respectively. However, examples exist in the literature of the better suitability of bicelles over micelles in studying membrane protein structure. For example, in α integrins (39, 40), the presence of a reverse loop in the juxtamembrane cytoplasmic region was observed in a bicellar or membrane environment but not in detergent micelles or organic solvents (41–43). Therefore, to confirm that proposed binding sites of pyronin B to SH protein represent those present in membranes, we analyzed the structure of SH protein in a membrane-mimicking bicellar environment.
Reconstitution of SH protein in dihexanoylphosphatidylcholine-dilauroylphosphatidylcholine (DHPC-DLPC) bicelles resulted in well resolved 1H-15N TROSY-HSQC spectra (Fig. 5A). However, residues 16 to 37, which form the core of the TM domain (21), were missing from the spectrum. This problem was also found for bicellar compositions 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)-DHPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)-DHPC, 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC)-DHPC, and DLPC-DHPC when spectra were collected at 313 K, pH 5.5, and the protein concentration was 0.3 mM (data not shown). This broadening may be caused by the TM domain being more structured and has been reported previously for similar viral membrane proteins reconstituted in bicelles (44). Nevertheless, the TM ends, juxtamembrane, and extramembrane parts of the SH protein were still observable.
FIG 5.
Spectral resolution and secondary structure of SH extramembrane domains in bicelles. (A) [1H-15N] TROSY-HSQC of SH protein in DHPC-DLPC bicelles. The cross-peaks are labeled by one-letter code and residue number. The Trp-15 indole group is shown in the inset. (B) Comparison of 13Cα secondary chemical shifts, i.e., the difference between experimental and tabulated random-coil 13Cα chemical shifts, of SH protein in DPC micelles (red), DHPC-DLPC bicelles (green), and DHPC-DMPC bicelles (blue). Regions that exhibit significant 13Cα secondary shift differences between bicelle- and micelle-embedded SH protein are indicated by a box. (C and D) Secondary structure of SH protein and NOE connectivity. Sequential and medium-ranged NOE connectivity for extramembrane N-terminal (residues 1 to 14) (C) and C-terminal (residues 38 to 64) (D) domains, displayed as bands under the respective residues. (E) Heteronuclear 1H-15N NOE (HNOE) experiment.
The secondary 13Cα chemical shifts are highly correlated to secondary structure (45, 46). Therefore, the shifts found previously in DPC micelles (21) and those found here in DHPC-DLPC bicelles were compared (Fig. 5B). Only small differences were present at the N-terminal extramembrane fragment, where residues 1 to 7 were close to random-coil values, and the rest were more α-helical. Similarly, the region proposed as the pyronin B binding site (residues 38 to 43) showed identical Cα chemical shifts.
However, dramatic differences were observed in the C-terminal juxtamembrane fragment Leu-44 to Asn-48 (Fig. 5B, box). NOE connectivities (Fig. 5C and D) show that this fragment is α-helical in bicelles, in contrast with the β-structure previously found in DPC micelles (21). The dynamic properties of the extramembrane domains of SH protein were studied using a [1H-15N] steady-state heteronuclear NOE (HNOE) experiment (Fig. 5E). The majority of the HNOE values correspond to a well-folded structure (residues 5 to 14 and 38 to 60).
Three-dimensional model of SH protein monomer in bicelles.
The restraints shown in Tables 1 and 2 were used in the calculations to obtain a total of 20 structures for the N- and C-terminal extramembrane domains of SH protein (root mean square deviation [RMSD] for backbone atoms of 0.33 and 0.44 Å, respectively). The N-terminal domain (residues 5 to 14) forms an α-helix (Fig. 6A) similar to that observed in DPC micelles (21). However, in the C-terminal domain (residues 38 to 64), the TM α-helix extends up to His-51 (Fig. 6B), in contrast with the β-strand-like loop structure present in micelles. Comparison between these two models (Fig. 6C) shows that Pro-58 is located at the tip of a C-terminal loop formed by the non-α-helical residues after His-51. Due to that extended helical region, the total α-helical content of SH protein is increased from ∼53% in DPC micelles to ∼60% in bicelles, which is almost identical to the percentage obtained using Fourier transform infrared spectroscopy (FTIR) of SH protein reconstituted in DMPC membranes (21).
FIG 6.
Structural model of SH protein in bicelles. Ensemble of 20 low-energy structures of N-terminal (A) and C-terminal (B) extramembrane domains of SH protein calculated using the NMR restraints summarized in Tables 1 and 2 are shown. The side chains are shown in line representation. (C) Overlay of the structures obtained in DHPC-DLPC bicelles (blue) and DPC micelles (red) (21). Some side chains are indicated for comparison of the two structures. (D) Superposition of the helical stretches of the two models. (E) Lumenal view of the proposed SH protein pentamer structure and orientation of the His-51 side chain. (F) Orientation of His-22 in the pentameric oligomer. (G) Possible contacts of His-22 with Phe-10 and Ile-21, both in a neighboring monomer.
A top-down view of the helices superimposed in the two models (Fig. 6D) shows the relative orientation of the two His residues and indicates that residues His-51 and Lys-43 are aligned. To assess the orientation of His-51 relative to the channel lumen, the SymmDock server (37, 47) was used to generate a pentameric oligomer of the C-terminal extramembrane domain using the mutual orientation of the TM domains obtained in DPC micelles (21). This is justified since in the latter model these mutual orientations were also very similar to the ones obtained using infrared dichroism data of synthetic SH-TM in lipidic membranes (20). Further, similar approaches were followed to obtain the pentameric TM model of severe acute respiratory syndrome-corona virus (SARS-CoV) E protein lipid environments that were confirmed in DPC micelles by solution NMR (48, 49). In that case, our reported TM-TM orientations were consistent with mutations that recovered channel activity in infected rats (50).
With this TM orientation, His-51 side chains face the lumen of the channel (Fig. 6E) although Leu-44 has even a more evident lumenal position, and His-22 is slightly skewed toward the other helix-helix interface (Fig. 6D). This orientation for the His-22 side chain is obviously retained from the model obtained in micelles (21). The good alignment of His-51 and Lys-43 (see above) allows visualization of the relative orientation of the two His residues (Fig. 6F). Possible contacts of His-22 with Phe-10 and Ile-21, at a different monomer, may stabilize the pentameric form (Fig. 6G).
Given this proposed lumenal/interfacial orientation of the two His residues, we examined their contribution to channel activity and selectivity using black lipid membranes (BLM) and purified SH protein.
Characterization of SH protein channel activity.
Purified WT SH protein, two single mutants, H22F and H51F, and one double mutant, H22F H51F, were tested in BLM. All of these forms were able to form pentameric oligomers (Fig. 4B) and showed channel activity (Fig. 7A to D). When a concentration gradient was used, the reversal potential (RP), i.e., the applied transmembrane voltage that yields zero current, provided an estimate of channel selectivity for anions or cations. At pH 7.5, the RP of all the forms tested showed mild but clear cationic selectivity (Fig. 7E to H). In contrast, at pH 3 all of these forms showed mild anionic selectivity (data not shown). A summary of the averaged results is shown in Table 3.
FIG 7.

SH channel activity measurements. (A to D) Original current recordings obtained with a 500/50 mM KCl gradient, pH 7.5, showing that a current is observed for WT and mutant SH proteins at 0 mV. (E to H) Data from the current-to-voltage relationships corresponding to panels A to D showing the reversal potential (RP) of each SH variant. (I and J) Cation (P+/P−) selectivity at 7.5 (I) and anion (P−/P+) selectivity and at pH 3 (J) for each SH variant, shown as a function of the difference of global charge with respect to the WT. (K) Channel conductance G (pS) for the WT SH protein at different pHs in 1 M KCl and 3:1:1 DOPC-DOPS-DOPE. The experimental points were fitted by a sigmoidal curve. Error bars represent standard errors of the means (n = 6).
TABLE 3.
Reversal potential of SH protein (WT and mutants)a
| Protein | Characterization of channel activity at: |
||||
|---|---|---|---|---|---|
| pH 7.5 |
pH 3 |
||||
| RP (mV) | P+ /P− | RP (mV) | P− /P+ | Chargeb | |
| WT | −27.3 ± 2.2 | 3.9 ± 0.1 | 40 ± 3 | 8.9 ± 0.4 | 0 |
| H51F | −25 ± 3 | 3.5 ± 0.1 | 37 ± 3 | 7.1 ± 0.3 | −1 |
| H22F | −14 ± 1.5 | 2.00 ± 0.05 | 39.4 ± 2.0 | 8.4 ± 0.4 | −1 |
| H51F H22F | −21.6 ± 2.0 | 2.9 ± 0.1 | 36 ± 3 | 6.7 ± 0.3 | −2 |
Results are from artificial membranes in the presence of a 10-fold KCl concentration gradient (see Materials and Methods). Each reversal potential (RP) value is the average of a minimum of 10 experiments.
The charge value represents the charge versus that of the WT SH protein at pH 3.
At pH 7.5, H22F, H22F, and the double mutant H22F H51F (Fig. 7I) showed lower cation selectivity, P+/P− (where P+ and P− refer to cation and anion permeability, respectively). Since the net charge of SH protein at this pH should be similar for all of these variants given the pKa of His residues of ∼6.5 (51), the differences observed in RP are likely to reflect structural changes induced by the H22F mutation. These changes, however, do not affect the pentameric structure (Fig. 4B), but in the case of H22F, the mutation may destabilize pyronin binding (Fig. 4C).
At pH 3, anion selectivity (P−/P+) was somewhat correlated with the expected global charge (Fig. 7J and Table 3). At this pH, both His residues should be protonated, and therefore the net charge relative to the WT depends on the number of His residues present. Anion selectivity was higher for WT, lower for the single mutants, and even lower for the H22F H51F double mutant. In this case, the absence of His-22 (H22F) led to a behavior similar to that of the WT, suggesting a small contribution of this residue to ion selectivity. In contrast, mutants where His-51 was absent (H51F and H22F H51F) produced a significant reduction in P−/P+, suggesting a more important involvement of His-51 in modulating channel selectivity.
These results are somewhat consistent with the respective orientation of these two side chains in the pentameric channel, with His-51 being more exposed to the lumen and His-22 being more rotated toward the helix-helix interface and/or making stabilizing contacts with neighboring monomers (Fig. 6F and G). These data are also consistent with the observed electrophoretic pattern of His mutants (21); mutant H22A showed aberrant mobility and/or aggregation, suggesting a structural role. In contrast, H51A showed mobility similar to that of the WT form. Finally, the overall conductance was pH dependent and consistent with the titration of histidine residues (Fig. 7K), with less conductance at lower pH, where both His residues are protonated. A dramatic reduction of channel activity at lower pH was also observed previously for synthetic SH-TM (residues 18 to 43) in planar lipid bilayers (20). In SH-TM, only His-22 was present, suggesting that protonation of this histidine alone can reduce or block channel activity.
Patch clamp studies of transfected HEK293 Phoenix cells.
The results obtained by functional reconstitution of the TM domain (20) or the full-length SH protein (Fig. 7) in BLM show (i) that channel activity is reduced at low pH and (ii) that His-to-Phe mutants are channel active. This is in stark contrast with previous patch clamp results (21), which showed channel activation at low pH and no channel activity for a H22F H51F double mutant. Therefore, we tried to reproduce those original patch clamp experiments, where HEK293 Phoenix cells were transiently transfected with either the pIRES-AcGFP1 vector carrying the SH protein or with the control vector pIRES-AcGFP1. Transfected cells were initially kept in a pH 7.4 bath solution. After the seal was realized and the whole-cell configuration was obtained, channel activity was monitored in a freshly bubbled (95% O2, 5% CO2) pH 7.4 bath.
The kinetics of the elicited currents (Fig. 8A) resemble those of outwardly rectifying and voltage-dependent potassium currents of the Kv channel family (52) previously observed in these cells (53). These channels are stimulated at positive potentials and inactive at negative potentials; therefore, the currents measured at negative potentials (e.g., at −80 mV) were considered leakage currents and were subtracted from the measured currents. Comparing currents from control and SH-transfected cells without leakage subtraction did not reveal an SH-dependent current at pH 7.4 (Fig. 8A) or 5.5 (not shown).
FIG 8.
Patch clamp assays in SH-overexpressing cells. (A) Representative original tracings of currents recorded by the patch clamp technique in whole-cell configuration, leakage subtracted and obtained at pH 7.4 in control (left) and SH-transfected (right) cells. Currents were elicited with voltage increments of 20 mV from −120 to +100 mV applied from a holding potential of 0 mV (upper right inset). (B and C) Current density-to-voltage relationships measured in a pH 7.4 bath solution (B) and at 10 min following replacement of the extracellular solution with an acidic, pH 5.5, bath (C).
Current values representing the maximum activation of channels (Fig. 8A, rectangle) were averaged and plotted in the current density-to-voltage relationships determined at pH 7.4 (Fig. 8B) and pH 5.5 (Fig. 8C). At pH 7.4, no differences were observed between currents measured in cells overexpressing SH protein and in control cells (Fig. 8A and B). Next, replacing the extracellular solution with a freshly bubbled acidic (pH 5.5) bath was performed every 5 min. The switch to an acidic environment did not cause any obvious change of the current density-to-voltage relationship over a period of 10 min (Fig. 8C).
The hypothesis that the measured currents herein are due to endogenous potassium-selective channels is supported by a separate experiment (data not shown) that shows that the experimental RP was close to the equilibrium potential of potassium predicted by the Nernst law. Thus, the ion channel activity of SH protein, if any, may have been masked by such current. No significant difference was observed in RPs between control and SH-expressing cells at pH 7.4 and 5.5. This observation points to the conclusion that SH expression, or pH, did not modify the ion selectivity of the measured current; i.e., the patch clamp experiments revealed only an endogenous current whose characteristics did not change following SH expression.
In another set of patch clamp experiments, the exact same cell line (HEK293) used in previous studies (21) was analyzed. In these experiments, the currents measured in SH-expressing cells were lower, not higher, than those of the control (data not shown). This effect could have been caused by cellular stress induced by SH expression, reducing the activity, expression, or membrane trafficking of the endogenous potassium channels. A similarly reduced channel activity was observed previously after expression of another viroporin, SARS-CoV E, in HEK-293T cells (54) and was attributed to an indirect influence of the viroporin on other cellular proteins involved in ion transport. However, in that case, SARS-CoV E was not detected in plasma membrane. In contrast, both expression and plasma membrane localization of FLAG-tagged SH protein was shown in HEK293 cells (21).
Thus, the possibility that the full-length nontagged version of SH protein used in patch clamp experiments was not expressed or did not accumulate at plasma membranes was tested by attempting to observe channel activity in HEK293 Phoenix cells overexpressing a FLAG-tagged (C- or N-terminal) SH protein or a C- and N-terminally truncated SH protein that encompassed the TM domain. None of these conditions produced channel activity that could be attributed to SH protein in a whole-cell configuration (data not shown). We conclude that previous patch clamp data that showed low-pH activation of SH protein cannot be reproduced.
DISCUSSION
Inhibition of SH protein by pyronin B.
Small-molecule inhibitors can be used to probe the role of SH protein channel activity during the cell cycle. Although with modest Kd (∼6.8 μM), pyronin B compares favorably with the 16 μM for amantadine inhibition of influenza virus M2 protein (55), the ∼10 μM reported for hexamethylene amiloride (HMA) inhibition of SARS-CoV E (56), 50 to 100 μM for rimantadine inhibition of HCV p7 (57), or the >100 μM HMA for p7 inhibition (58). Pyronin B was found to bind SH protein mostly at the lipid-facing side of the TM α-helices and not into the pore lumen. This mechanism of inhibition is thus probably allosteric, similar to that proposed for rimantadine binding to the TM domain of M2 (57) or rimantadine to hepatitis C virus (HCV) p7 (57). In fact, the proposed binding site for pyronin B, encompassing residues 38 to 43, is totally conserved in RSV SH proteins, which suggests that inhibition would be more difficult to overcome by compensatory mutations. By reference to the pentameric model of SH protein in DPC micelles, the proposed binding site is located at the narrowest region of the channel lumen (Fig. 9A and B). Inhibition is more easily rationalized here than near the N-terminal end (cytoplasmic) of the TM, where the channel lumen opens significantly.
FIG 9.
Model of the SH protein channel. Schematic drawing representing a sectional view of the SH protein channel at pH 7.5 (A) and 3.0 (B). (C) FASTA multiple alignment of SH proteins and homologs in human RSV (subgroups A and B), bovine RSV, parainfluenza virus 5 (PIV5), and mumps virus (MuV). The TM domain (purple) predicted using TMHMM (64) and the C-terminal extramembrane His residues (blue) are highlighted.
Effect of bicelles on juxtamembrane domains of membrane proteins.
The juxtamembrane regions in membrane proteins are sensitive to the hydrophobic environment used experimentally, e.g., micelles versus bicelles. This has been shown for the HIV envelope protein (59), integrin transmembrane domains (39, 40), or BtuB (a 22-strand β-barrel protein) (60). We have shown herein that significant structural differences are observed for SH protein when it is reconstituted in bicelles or micelles. In this case, these effects are unlikely to derive from the different lengths of hydrophobic tails in the phospholipids or detergents used to reconstitute the protein. Indeed, both DLPC and DPC hydrophobic tails, used in bicelles and micelles, respectively, have 12 carbon atoms. In addition, the secondary 13Cα chemical shifts from SH protein reconstituted in bicelles of different hydrophobic tail lengths, e.g., DHPC-DLPC versus DHPC-DMPC, were almost identical. All of these data suggest that it is the curvature of the micelle surface that may dramatically affect the structure at the juxtamembrane region.
The role of histidine residues in SH protein channel activity.
The funnel-like architecture adopted by the TM domain in SH protein (21) has been observed in other viroporins, e.g., influenza virus M2 protein (61), SARS-CoV E protein (48), and HCV p7 (57). All of these viral channels contain narrow regions that probably serve as selectivity filters. In SH protein, this narrower region (Ser-29 to Cys-45) (21) is lined with hydrophobic side chains (Ile-32, Ile-36, Ile-40, and Leu-44), and Ile-36 defines the narrowest point in the channel lumen (Fig. 9A and B). This figure shows that in our present refined model, obtained in bicelles, His-22 is located at the largest opening of this funnel, whereas His-51 is at the tip of the smallest opening. While both His residues are accessible to the lumen of the channel, our structural data point to a stabilizing role for His-22, whereas His-51 is more important for channel activity, consistent with its location in a tighter region of the channel. While at pH 7.5 the channel is mildly selective for cations, at pH 3, His-51 protonation may slow cation transport due to an electrostatic effect (Fig. 9B). Nevertheless, ion selectivity in SH protein is poor, as also observed in other viroporins, e.g., SARS-CoV E protein (62) or HCV p7 (58), and it may be a hallmark of incomplete specialization of these channels.
An indication of the relative importance of these two His residues can also be obtained by analyzing their conservation in close homologs (Fig. 9C). This comparison is justified since PIV5 SH protein could be replaced by the SH protein from MuV or RSV SH in strains A2 or B1 to produce a similar phenotype (6, 12). These data suggest a role of a membrane-permeabilizing pentameric structure common to these species. Alignment of the homologous sequences of RSV SH protein, bovine RSV, PIV5, and MuV shows that there is at least one His residue near the position equivalent to RSV SH His-51, i.e., several residues after the TM domain. In fact, in these other sequences a C-terminal His residue is present with a 3- to 4-residue periodicity, expected for α-helices. Therefore, by comparison with the orientation of hRSV SH His-51, these His residues should also have a lumenal orientation. In contrast, the equivalent residue to His-22 is replaced with Phe in these homologous sequences, supporting a more structural role for this residue.
Finally, based on these results, it can only be speculated that SH channel activity can be strongly regulated by lower pH when present in native biological membranes and the unknown conditions present in the cell. The intravesicular pH drops along the endocytic pathway from pH 6.0 to 6.5 in early endosomes to pH 4.5 to 5.5 in late endosomes and lysosomes (63), whereas in the Golgi lumen the pH is only one unit below that of the cytoplasm. These small pH changes, together with the low pK measured in the present paper and the relatively low effect of pH on both conductance and selectivity, do not suggest that pH plays a major role in modulating channel activity during the life cycle of the virus. However, mutation of these His residues in the context of the infected cell is an interesting avenue for future experiments, especially since SH homologs have a conserved His residue at or near the equivalent position to His-51 in hRSV and since that current was abolished when SH-TM was exposed to a pH of 4 (20).
ACKNOWLEDGMENTS
J.T. acknowledges the funding of the National Research Foundation (grant NRF-CRP4-2008-02) and Tier 1 grant RG 51/13.
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1. Dowell SF, Anderson LJ, Gary HE, Jr, Erdman DD, Plouffe JF, File TM, Jr, Marston BJ, Breiman RF. 1996. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J. Infect. Dis. 174:456–462. 10.1093/infdis/174.3.456 [DOI] [PubMed] [Google Scholar]
- 2. Blount RE, Jr, Morris JA, Savage RE. 1956. Recovery of cytopathogenic agent from chimpanzees with coryza. Proc. Soc. Exp. Biol. Med. 92:544–549. 10.3181/00379727-92-22538 [DOI] [PubMed] [Google Scholar]
- 3. Krusat T, Streckert HJ. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247–1254. 10.1007/s007050050156 [DOI] [PubMed] [Google Scholar]
- 4. Lamb RA. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1–11. 10.1006/viro.1993.1561 [DOI] [PubMed] [Google Scholar]
- 5. Bukreyev A, Whitehead SS, Murphy BR, Collins PL. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973–8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fuentes S, Tran KC, Luthra P, Teng MN, He B. 2007. Function of the respiratory syncytial virus small hydrophobic protein. J. Virol. 81:8361–8366. 10.1128/JVI.02717-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin H, Zhou H, Cheng X, Tang R, Munoz M, Nguyen N. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210–218. 10.1006/viro.2000.0393 [DOI] [PubMed] [Google Scholar]
- 8. Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, Harris DO, Randolph VB, Udem SA, Murphy BR, Sidhu MS. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. U. S. A. 94:13961–13966. 10.1073/pnas.94.25.13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whitehead SS, Bukreyev A, Teng MN, Firestone CY, St Claire M, Elkins WR, Collins PL, Murphy BR. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73:3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z, Xu J, Patel J, Fuentes S, Lin YA, Anderson D, Sakamoto K, Wang LF, He BA. 2011. Function of the small hydrophobic protein of J paramyxovirus. J. Virol. 85:32–42. 10.1128/JVI.01673-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin Y, Bright AC, Rothermel TA, He B. 2003. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J. Virol. 77:3371–3383. 10.1128/JVI.77.6.3371-3383.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilson RL, Fuentes SM, Wang P, Taddeo EC, Klatt A, Henderson AJ, He B. 2006. Function of small hydrophobic proteins of paramyxovirus. J. Virol. 80:1700–1709. 10.1128/JVI.80.4.1700-1709.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. 2013. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax 68:66–75. 10.1136/thoraxjnl-2012-202182 [DOI] [PubMed] [Google Scholar]
- 14. Rixon HW, Brown G, Aitken J, McDonald T, Graham S, Sugrue RJ. 2004. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J. Gen. Virol. 85:1153–1165. 10.1099/vir.0.19769-0 [DOI] [PubMed] [Google Scholar]
- 15. Collins PL, Mottet G. 1993. Membrane orientation and oligomerization of the small hydrophobic protein of human respiratory syncytial virus. J. Gen. Virol. 74:1445–1450. 10.1099/0022-1317-74-7-1445 [DOI] [PubMed] [Google Scholar]
- 16. Olmsted RA, Collins PL. 1989. The 1A protein of respiratory syncytial virus is an integral membrane protein present as multiple, structurally distinct species. J. Virol. 63:2019–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rixon HW, Brown G, Murray JT, Sugrue RJ. 2005. The respiratory syncytial virus small hydrophobic protein is phosphorylated via a mitogen-activated protein kinase p38-dependent tyrosine kinase activity during virus infection. J. Gen. Virol. 86:375–384. 10.1099/vir.0.80563-0 [DOI] [PubMed] [Google Scholar]
- 18. Chen MD, Vazquez M, Buonocore L, Kahn JS. 2000. Conservation of the respiratory syncytial virus SH gene. J. Infect. Dis. 182:1228–1233. 10.1086/315829 [DOI] [PubMed] [Google Scholar]
- 19. Collins PL, Olmsted RA, Johnson PR. 1990. The small hydrophobic protein of human respiratory syncytial virus: comparison between antigenic subgroups A and B. J. Gen. Virol. 71:1571–1576. 10.1099/0022-1317-71-7-1571 [DOI] [PubMed] [Google Scholar]
- 20. Gan SW, Ng L, Lin X, Gong X, Torres J. 2008. Structure and ion channel activity of the human respiratory syncytial virus (hRSV) small hydrophobic protein transmembrane domain. Protein Sci. 17:813–820. 10.1110/ps.073366208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gan SW, Tan E, Lin X, Yu D, Wang J, Tan GM-Y, Vararattanavech A, Yeo CY, Soon CH, Soong TW, Pervushin K, Torres J. 2012. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J. Biol. Chem. 287:24671–24689. 10.1074/jbc.M111.332791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tolley KP, Marriott AC, Simpson A, Plows DJ, Matthews DA, Longhurst SJ, Evans JE, Johnson JL, Cane PA, Randolph VB, Easton AJ, Pringle CR. 1996. Identification of mutations contributing to the reduced virulence of a modified strain of respiratory syncytial virus. Vaccine 14:1637–1646 [DOI] [PubMed] [Google Scholar]
- 23. Yamada Y, Liu DX. 2009. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 83:8744–8758. 10.1128/JVI.00613-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bezrukov SM, Vodyanoy I. 1993. Probing alamethicin channels with water-soluble polymers—effect on conductance of channel states. Biophys. J. 64:16–25. 10.1016/S0006-3495(93)81336-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montal M, Mueller P. 1972. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. U. S. A. 69:3561–3566. 10.1073/pnas.69.12.3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Y, Surya W, Claudine S, Torres J. 2014. Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J. Biol. Chem. 289:12535–12549. 10.1074/jbc.M114.560094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gan SW, Vararattanavech A, Nordin N, Eshaghi S, Torres J. 2011. A cost-effective method for simultaneous homo-oligomeric size determination and monodispersity conditions for membrane proteins. Anal. Biochem. 416:100–106. 10.1016/j.ab.2011.05.007 [DOI] [PubMed] [Google Scholar]
- 28. Tycko R, Blanco FJ, Ishii Y. 2000. Alignment of biopolymers in strained gels: a new way to create detectable dipole-dipole couplings in high-resolution biomolecular NMR. J. Am. Chem. Soc. 122:9340–9341. 10.1021/ja002133q [DOI] [Google Scholar]
- 29. Ulmer TS, Ramirez BE, Delaglio F, Bax A. 2003. Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy. J. Am. Chem. Soc. 125:9179–9191. 10.1021/ja0350684 [DOI] [PubMed] [Google Scholar]
- 30. Cornilescu G, Delaglio F, Bax A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289–302. 10.1023/A:1008392405740 [DOI] [PubMed] [Google Scholar]
- 31. Guntert P, Mumenthaler C, Wuthrich K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283–298. 10.1006/jmbi.1997.1284 [DOI] [PubMed] [Google Scholar]
- 32. Herrmann T, Guntert P, Wuthrich K. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319:209–227. 10.1016/S0022-2836(02)00241-3 [DOI] [PubMed] [Google Scholar]
- 33. Brunger AT. 2007. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2:2728–2733. 10.1038/nprot.2007.406 [DOI] [PubMed] [Google Scholar]
- 34. Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921. 10.1107/S0907444998003254 [DOI] [PubMed] [Google Scholar]
- 35. Dossena S, Gandini R, Tamma G, Vezzoli V, Nofziger C, Tamplenizza M, Salvioni E, Bernardinelli E, Meyer G, Valenti G, Wolf-Watz M, Furst J, Paulmichl M. 2011. The molecular and functional interaction between ICln and HSPC038 proteins modulates the regulation of cell volume. J. Biol. Chem. 286:40659–40670. 10.1074/jbc.M111.260430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duhovny D, Nussinov R, Wolfson HJ. 2002. Efficient unbound docking of rigid molecules. Lecture Notes Comput. Sci. 2452:185–200. 10.1007/3-540-45784-4_14 [DOI] [Google Scholar]
- 37. Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. 2005. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33:W363–W367. 10.1093/nar/gki481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Volkamer A, Kuhn D, Rippmann F, Rarey M. 2012. DoGSiteScorer: a web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics 28:2074–2075. 10.1093/bioinformatics/bts310 [DOI] [PubMed] [Google Scholar]
- 39. Lau TL, Kim C, Ginsberg MH, Ulmer TS. 2009. The structure of the integrin αIIbβ3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 28:1351–1361. 10.1038/emboj.2009.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Surya W, Li Y, Millet O, Diercks T, Torres J. 2013. Transmembrane and juxtamembrane structure of αL integrin in bicelles. PLoS One 8:e74281. 10.1371/journal.pone.0074281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bhunia A, Tang XY, Mohanram H, Tan SM, Bhattacharjya S. 2009. NMR solution conformations and interactions of integrin αLβ2 cytoplasmic tails. J. Biol. Chem. 284:3873–3884. 10.1074/jbc.M807236200 [DOI] [PubMed] [Google Scholar]
- 42. Lai C, Liu X, Tian C, Wu F. 2013. Integrin α1 has a long helix, extending from the transmembrane region to the cytoplasmic tail in detergent micelles. PLoS One 8:e62954. 10.1371/journal.pone.0062954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vinogradova O, Haas T, Plow EF, Qin J. 2000. A structural basis for integrin activation by the cytoplasmic tail of the alpha(IIb)-subunit. Proc. Natl. Acad. Sci. U. S. A. 97:1450–1455. 10.1073/pnas.040548197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Son WS, Park SH, Nothnagel HJ, Lu GJ, Wang Y, Zhang H, Cook GA, Howell SC, Opella SJ. 2012. “q-Titration” of long-chain and short-chain lipids differentiates between structured and mobile residues of membrane proteins studied in bicelles by solution NMR spectroscopy. J. Magn. Reson. 214:111–118. 10.1016/j.jmr.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spera S, Bax A. 1991. Empirical correlation between protein backbone conformation and C.alpha. and C.beta. 13C nuclear magnetic resonance chemical shifts. J. Am. Chem. Soc. 113:5490–5492. 10.1021/ja00014a071 [DOI] [Google Scholar]
- 46. Xu X-P, Case D. 2001. Automated prediction of 15N, 13Cα, 13Cβ and 13C′ chemical shifts in proteins using a density functional database. J. Biomol. NMR 21:321–333. 10.1023/A:1013324104681 [DOI] [PubMed] [Google Scholar]
- 47. Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. 2005. Geometry-based flexible and symmetric protein docking. Proteins 60:224–231. 10.1002/prot.20562 [DOI] [PubMed] [Google Scholar]
- 48. Pervushin K, Tan E, Parthasarathy K, Xin L, Jiang FL, Yu D, Vararattanavech A, Soong TW, Liu DX, Torres J. 2009. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 5:e1000511. 10.1371/journal.ppat.1000511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Torres J, Parthasarathy K, Lin X, Saravanan R, Kukol A, Liu DX. 2006. Model of a putative pore: the pentameric alpha-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 91:938–947. 10.1529/biophysj.105.080119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nieto-Torres JL, Dediego ML, Verdia-Baguena C, Jimenez-Guardeno JM, Regla-Nava JA, Fernandez-Delgado R, Castano-Rodriguez C, Alcaraz A, Torres J, Aguilella VM, Enjuanes L. 2014. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 10:e1004077. 10.1371/journal.ppat.1004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cymes GD, Ni Y, Grosman C. 2005. Probing ion-channel pores one proton at a time. Nature 438:975–980. 10.1038/nature04293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Paulmichl M, Nasmith P, Hellmiss R, Reed K, Boyle WA, Nerbonne JM, Peralta EG, Clapham DE. 1991. Cloning and expression of a rat cardiac delayed rectifier potassium channel. Proc. Natl. Acad. Sci. U. S. A. 88:7892–7895. 10.1073/pnas.88.17.7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dossena S, Maccagni A, Vezzoli V, Bazzini C, Garavaglia ML, Meyer G, Furst J, Ritter M, Fugazzola L, Persani L, Zorowka P, Storelli C, Beck-Peccoz P, Botta G, Paulmichl M. 2005. The expression of wild-type pendrin (SLC26A4) in human embryonic kidney (HEK 293 Phoenix) cells leads to the activation of cationic currents. Eur. J. Endocrinol. 153:693–699. 10.1530/eje.1.02018 [DOI] [PubMed] [Google Scholar]
- 54. Nieto-Torres JL, DeDiego ML, Alvarez E, Jimenez-Guardeno JM, Regla-Nava JA, Llorente M, Kremer L, Shuo S, Enjuanes L. 2011. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology 415:69–82. 10.1016/j.virol.2011.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jing X, Ma C, Ohigashi Y, Oliveira FA, Jardetzky TS, Pinto LH, Lamb RA. 2008. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc. Natl. Acad. Sci. U. S. A. 105:10967–10972. 10.1073/pnas.0804958105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilson L, Gage P, Ewart G. 2006. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353:294–306. 10.1016/j.virol.2006.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. OuYang B, Xie S, Berardi MJ, Zhao X, Dev J, Yu W, Sun B, Chou JJ. 2013. Unusual architecture of the p7 channel from hepatitis C virus. Nature 498:521–525. 10.1038/nature12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Premkumar A, Wilson L, Ewart GD, Gage PW. 2004. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 557:99–103. 10.1016/S0014-5793(03)01453-4 [DOI] [PubMed] [Google Scholar]
- 59. Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. 2002. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. J. Am. Chem. Soc. 124:2450–2451. 10.1021/ja017875d [DOI] [PubMed] [Google Scholar]
- 60. Fanucci GE, Lee JY, Cafiso DS. 2003. Membrane mimetic environments alter the conformation of the outer membrane protein BtuB. J. Am. Chem. Soc. 125:13932–13933. 10.1021/ja0376442 [DOI] [PubMed] [Google Scholar]
- 61. Schnell JR, Chou JJ. 2008. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451:591–595. 10.1038/nature06531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Verdia-Baguena C, Nieto-Torres JL, Alcaraz A, DeDiego ML, Torres J, Aguilella VM, Enjuanes L. 2012. Coronavirus E protein forms ion channels with functionally and structurally involved membrane lipids. Virology 432:485–494. 10.1016/j.virol.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sorkin A, von Zastrow M. 2002. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 3:600–614. 10.1038/nrm883 [DOI] [PubMed] [Google Scholar]
- 64. Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]