ABSTRACT
Viral protease inhibitors are remarkably effective at blocking the replication of viruses such as human immunodeficiency virus and hepatitis C virus, but they inevitably lead to the selection of inhibitor-resistant mutants, which may contribute to ongoing disease. Protease inhibitors blocking the replication of coronavirus (CoV), including the causative agents of severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), provide a promising foundation for the development of anticoronaviral therapeutics. However, the selection and consequences of inhibitor-resistant CoVs are unknown. In this study, we exploited the model coronavirus, mouse hepatitis virus (MHV), to investigate the genotype and phenotype of MHV quasispecies selected for resistance to a broad-spectrum CoV 3C-like protease (3CLpro) inhibitor. Clonal sequencing identified single or double mutations within the 3CLpro coding sequence of inhibitor-resistant virus. Using reverse genetics to generate isogenic viruses with mutant 3CLpros, we found that viruses encoding double-mutant 3CLpros are fully resistant to the inhibitor and exhibit a significant delay in proteolytic processing of the viral replicase polyprotein. The inhibitor-resistant viruses also exhibited postponed and reduced production of infectious virus particles. Biochemical analysis verified double-mutant 3CLpro enzyme as impaired for protease activity and exhibiting reduced sensitivity to the inhibitor and revealed a delayed kinetics of inhibitor hydrolysis and activity restoration. Furthermore, the inhibitor-resistant virus was shown to be highly attenuated in mice. Our study provides the first insight into the pathogenicity and mechanism of 3CLpro inhibitor-resistant CoV mutants, revealing a low genetic barrier but high fitness cost of resistance.
IMPORTANCE RNA viruses are infamous for their ability to evolve in response to selective pressure, such as the presence of antiviral drugs. For coronaviruses such as the causative agent of Middle East respiratory syndrome (MERS), protease inhibitors have been developed and shown to block virus replication, but the consequences of selection of inhibitor-resistant mutants have not been studied. Here, we report the low genetic barrier and relatively high deleterious consequences of CoV resistance to a 3CLpro protease inhibitor in a coronavirus model system, mouse hepatitis virus (MHV). We found that although mutations that confer resistance arise quickly, the resistant viruses replicate slowly and do not cause lethal disease in mice. Overall, our study provides the first analysis of the low barrier but high cost of resistance to a CoV 3CLpro inhibitor, which will facilitate the further development of protease inhibitors as anti-coronavirus therapeutics.
INTRODUCTION
Treatment of viral infections with antiviral drugs leads to selection within the quasispecies and the amplification of drug-resistant mutants (1–3). The pathogenicity of drug-resistant mutants is a primary concern for the implementation of antiviral therapies. The virulence of drug-resistant mutants of human immunodeficiency virus type 1 (HIV-1) was the major factor contributing to the failure of single-drug antiretroviral trials (4). In contrast, acyclovir-resistant mutants of herpes simplex virus with viral thymidine kinase deficiency are attenuated in immunocompetent individuals (5), which allows for effective single-drug therapy. Investigating the pathogenicity of drug-selected viral quasispecies is important for understanding viral pathogenesis and informative for antiviral drug design and therapeutic approaches.
Coronaviruses (CoVs) are a large family of RNA viruses that cause illness in animals, including humans, with symptoms ranging from common colds to severe and fatal respiratory or gastrointestinal infection. Emerging coronaviruses have become a significant threat to human health. The most infamous CoV, severe acute respiratory syndrome coronavirus (SARS-CoV), caused the outbreak of 2002-2003 with more than 8,000 infected people and a 10% mortality rate (6). A recently emerged coronavirus detected in Saudi Arabia (7), designated Middle East respiratory syndrome coronavirus (MERS-CoV) (8), has infected at least 536 people, with 145 deaths as of 7 May 2014 (9). Besides SARS-CoV and MERS-CoV causing severe respiratory syndrome, four other human coronaviruses are associated with mild to moderate respiratory diseases, including human CoV 229E (HCoV-229E) (10), HCoV-OC43 (11), HCoV-NL63 (12, 13), and HCoV-HKU1 (14). These endemic human coronaviruses are recognized to cause primarily upper respiratory tract infection and occasionally lower respiratory tract disease in elderly, newborn, and immunocompromised individuals (15). Important for antiviral therapy, analysis of respiratory samples from SARS patients showed that peak viral titers occurred 10 days after the onset of fever, indicating a potential “window” period for antiviral therapy (16). Efforts are under way to identify specific antiviral inhibitors of SARS-CoV and MERS-CoV that target viral entry or replication (reviewed in reference 17).
Coronaviruses contain the largest known RNA genome, which ranges in size from 27 to 32 kb for different CoVs and encodes a replicase polyprotein that is processed by viral proteases, the papain-like protease (PLP) and the 3C-like protease (3CLpro, also known as the main protease). The PLP domain within nonstructural protein 3 (nsp3) cleaves the replicase polyprotein to generate nsp1 to nsp3 (18), while 3CLpro (nsp5) mediates the cleavage of nsp4 to nsp16 (19). Because of their essential role in viral replication, both proteases are considered attractive targets for antiviral therapeutics. Numerous protease inhibitors have been synthesized and identified to inhibit protease enzymatic activity and block CoV replication in cell culture (20–26). As 3CLpro is the main protease and structurally conserved among CoVs (27–31), the 3CLpro protease inhibitors have been intensively studied (20, 21, 23, 24, 27, 28, 31–34). However, the probability of developing resistance (the genetic barrier) and the effect of resistance on the replication capacity (the relative viral fitness) have not been investigated for coronaviruses.
In the present study, we exploited the murine coronavirus, mouse hepatitis virus (MHV), as a model system to study the phenotype, genotype, and pathogenicity of viruses resistant to a broad-spectrum 3CLpro inhibitor, GRL-001. GRL-001 is a 5-chloropyridyl ester-derived compound which has been shown to inhibit 3CLpro enzymatic activity of SARS-CoV and MERS-CoV (24, 35) and to block the replication of SARS-CoV, MERS-CoV, and bat coronavirus HKU5 (24, 36). Therefore, GRL-001 is a potential lead compound for developing anticoronavirus therapeutics. We selected for inhibitor-resistant MHV by serially passaging the virus in the presence of GRL-001 and then evaluated the replication and pathogenicity of isogenic viruses generated using reverse genetics. This study represents the first investigation of the 3CLpro enzymatic activity and pathogenicity of a protease inhibitor-resistant virus and illustrates the low genetic barrier but high fitness cost of resistance.
MATERIALS AND METHODS
Cells and virus.
Delayed brain tumor (DBT) cells (37) and baby hamster kidney 21 cells expressing the MHV receptor (BHK-MHVR) were used for all experiments. The DBT cells were grown in modified Eagle's medium (MEM) supplemented with 10% tryptose phosphate broth (TPB) media, 5% heat-inactivated fetal calf serum (FCS), 2% penicillin-streptomycin, and 2% glutamine. The BHK-MHVR medium was Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% heat-inactivated FCS and G418 (0.8 mg/ml; HyClone) to maintain selection for MHVR expression. Wild-type (WT) MHV strain A59 (GenBank accession no. AY910861) generated by reverse genetics was the parental strain used for inhibitor selection and mouse infection studies.
Selection of inhibitor-resistant mutants.
In order to isolate inhibitor-resistant mutants, the WT MHV strain was serially passaged on DBT cells in the presence of increasing concentrations of GRL-001. Briefly, confluent DBT cell monolayers were infected with WT virus at a multiplicity of infection (MOI) of 0.1 and subsequently incubated for 24 h at 37°C in postinfection medium (MEM with 100 IU/ml of penicillin, 100 μg/ml of streptomycin, and 5% FCS) supplemented with GRL-001 at 1× the 50% effective concentration (EC50). GRL-001 concentrations were increased to a concentration of 2× the EC50 of GRL-001 during the 3rd and 4th passages. Aliquots of final-passage viruses were subjected to plaque assay, and the viral plaques were isolated for further propagation. The RNA of plaque-purified virus was extracted, and the nsp5 gene of these virus clones was sequenced with primers (available upon request).
Recovery of infectious clone 3CLpro mutant MHVs.
To generate the 3CLpro mutant MHV, nucleotide changes were introduced into the C fragment of the MHV reverse genetics system as described previously by Yount et al. (38) by site-directed mutagenesis PCR with primers (available upon request). The C fragment with mutations was verified by sequencing. Viral RNA generated from an in vitro transcription reaction by using ligated genomic cDNA fragments was electroporated into BHK-MHVR cells. Cell supernatant was collected as viral stock when electroporated cells demonstrated abundant cytopathic effects. All infectious clones of 3CLpro mutant virus were plaque purified and propagated on DBT cells. 3CLpro mutations of mutant virus was verified by RT-PCR amplification and sequencing of nsp5.
Antiviral-activity assay.
DBT cells at a density of approximately 1 × 104 cells per well in a 96-well plate were either mock infected with serum-free MEM or infected with an MOI of 1 in 100 μl of serum-free MEM and incubated for 1 h at 37°C. The viral inoculum was removed after 1 h of incubation, and then 100 μl of MEM, supplemented with 5% FCS and the GRL-001 inhibitor at final concentrations ranging from 3.125 to 50 μM, was added. Cells were then incubated for 48 h at 37°C with 5% CO2. All controls and each inhibitor concentration were set up in triplicate, and the antiviral-activity assays were performed independently on at least two separate occasions. Cell viability was determined approximately at 24 h after infection using the Cell-Titer Glo luminescent cell viability assay (Promega). The EC50s were determined using a nonlinear regression program with Prism 5 software (GraphPad, La Jolla, CA).
Plaque size comparison and viral growth kinetics.
To compare the plaque sizes of WT and mutant viruses, a plaque assay was performed on DBT cells. Briefly, DBT cells in 6-well plates were infected with series diluted viral stock for 1 h at 37°C, followed by overlaying with a 0.4% Noble agar-MEM mixture. Plates were incubated at 37°C for 48 h and fixed by 4% formaldehyde solution for 1 h. Viral plaques were visualized by staining with crystal violet and photographed. The area of plaques was calculated by using Photoshop CS software (Adobe). To analyze the growth kinetics of mutant virus, DBT cells were infected with an MOI of 0.1 at 37°C for 1 h and the medium was replaced with fresh MEM containing 2% FCS. Cell supernatant was harvested at various time points and titrated on DBT cells as described above. The average titer of each time point was calculated from three plaque assays.
Radiolabeling of newly synthesized proteins and RIPAs.
DBT cells were infected with WT or double-mutant virus at an MOI of 1 and incubated at 37°C for 1 h. Newly synthesized proteins were metabolically labeled with 50 μCi/ml of [35S]-translabeled methionine (ICN, Costa Mesa, CA) for 30 min, followed by chase at 30-min intervals. At the time of harvesting, radioactively labeled cells were washed with phosphate-buffered saline (PBS), and cell lysates were prepared by scraping the cells in lysis buffer A (4% SDS, 3% dithiothreitol [DTT], 40% glycerol, and 0.065 M Tris [pH 6.8]) (39). The lysates were either used directly for immunoprecipitation assays or stored at −70°C for future studies. Radiolabeled cell lysate was diluted in 1.0 ml of radioimmunoprecipitation assay (RIPA) buffer (0.5% Triton X-100, 0.1% SDS, 300 mM NaCl, 4 mM EDTA, and 50 mM Tris-HCl [pH 7.4]) (39) and subjected to immunoprecipitation with anti-nsp5 or -nsp8 rabbit polyclonal antibodies (40) and protein G magnetic beads (Millipore). The immunoprecipitated products were eluted with 2× Laemmli sample buffer (Bio-Rad), incubated at 37°C for 30 min, and analyzed by electrophoresis on a 10% or 5 to 12.5% gradient polyacrylamide gel containing 0.1% SDS. Following electrophoresis, the gel was fixed in 25% methanol–10% acetic acid, enhanced with Amplify solution (Amersham Biosciences) for 60 min, dried, and exposed to Kodak X-ray film.
Mouse experiments.
Four-week-old C57BL/6 mice purchased from The Jackson Laboratory were intracranially inoculated with 600, 1,200, or 3,000 PFU of WT or mutant MHV. Fourteen-week-old IFNAR−/− (C57BL/6 background) mice were initially obtained from Deborah Lenschow (Washington University in St. Louis), bred, and maintained at Loyola University Chicago in accordance with all federal and university guidelines. IFNAR−/− mice were infected intraperitoneally with 50 PFU of WT or mutant MHV. Infected mice were monitored for body weight daily and euthanized when the weight loss was over 25% according to the protocol. Graphs of survival rate and weight loss were generated by Prism 5 software. The statistical analyses of survival rate and weight loss were conducted with log rank test and 2-way analysis of variance (ANOVA) test, respectively.
In vitro assays for enzymatic activity of 3CLpro.
The enzymatic efficiencies (kapp) of both 3CLpro-WT and 3CLpro-T26I/D65G were determined at ambient temperature and 37°C. The total concentration of the UIVT3 substrate was varied to give final concentrations of 0.125, 0.25, 0.5, 1.0, 1.5, and 2.0 μM. After separate incubations of the assay buffer and UIVT3 substrate at the appropriate temperature for 20 min, 3CLpro was added to the wells containing assay buffer, yielding a final 3CLpro concentration of 100 nM. The plates were further incubated at the appropriate temperature for 5 min, after which the reaction was initiated by the addition of 20 μl of the appropriate concentration of substrate. The fluorescence intensity of the reaction was then measured over time as relative fluorescence units (RFUs) for a period of 11 min, using an excitation wavelength of 485 nm and a bandwidth of 20 nm and monitoring emission at 528 nm and a bandwidth of 20 nm using a BioTek Synergy H1 multimode microplate reader. The initial reaction rates (Vi) were determined by calculating the initial slope of the progress curve, which was then converted to the amount of product (μM) produced per minute using the experimentally determined value of the fluorescence extinction coefficient for UIVT3. Plotting Vi/[enzyme] versus [UIVT3] gave a linear correlation, the slope of which was taken to be kapp. These values were determined in the nonlinear regression program SigmaPlot.
Determination of IC50 of GRL-001 for WT and T26I/D65G MHV 3CLpro.
The 50% inhibitory concentrations (IC50s) for both WT and T26I/D65G MHV 3CLpros were determined at ambient temperature (25°C) and 37°C. The GRL-001 inhibitor was tested at concentrations of 0.1, 0.25, 0.5, 1, 2.5, 5, 10, and 25 μM. The inhibitor was added to 100 nM enzyme in assay buffer, and the enzyme-inhibitor mixture was incubated for 20 min at the appropriate temperature. The reaction was initiated by the addition of 2 μM UIVT3 substrate. The fluorescence intensity of the reaction was then measured over 20 min. The inhibition of the 3CLpro enzymes by GRL-001 was monitored by following the change in RFUs over time, using the initial slope of the progress curve to determine the initial rate. The percent inhibition of the 3CLpro enzymes was then plotted as a function of inhibitor concentration. IC50s were determined for both WT and T26I/D65G at both ambient temperature and 37°C using the nonlinear regression program SigmaPlot.
Esterase activities of WT and T26I/D65G MHV 3CLpro toward GRL-001 inhibitor.
To determine the qualitative rate of hydrolysis of GRL-001 by both WT and T26I/D65G MHV 3CLpro enzymes at ambient temperature and 37°C, the enzymes were incubated with two equivalents of the inhibitor at the appropriate temperature and their activities were tested over the course of 6 h. At each time point, 10 μl of enzyme-inhibitor stock reaction mixture was added to 70 μl of assay buffer in triplicate and the reaction was initiated by the addition of 20 μl of 5 μM UIVT3 substrate, resulting in a final concentrations of 100 nM, 200 nM, and 1 μM for 3CLpro, GRL-001, and UIVT3, respectively. The fluorescence intensity of the reaction was then measured at specific time points as RFUs for a period of 5 min, using an excitation wavelength of 485/20 nm and monitoring emission at 528/20 nm using a BioTek Synergy H1 multimode microplate reader. The time course of the GRL-001 hydrolysis reaction (and subsequent reactivation of the enzyme) was followed by monitoring the change in RFUs over time; the initial slope of the progress curve was then converted to the amount of product (μM) produced per minute using the experimentally determined fluorescence extinction coefficient for the UIVT3 substrate. The raw RFU values were corrected for background fluorescence. This value (μM UIVT3 hydrolysis product produced per minute) was then taken as a percentage of the uninhibited enzyme value and plotted over time.
Sequence alignments and modeling of MHV nsp5 structures.
The amino acid sequences of crystalized 3CLpros were retrieved from the Protein Data Bank (PDB) (HKU1, 3D23; SARS-CoV, 2V6N; HKU4, 2YNA; transmissible gastroenteritis virus [TGEV], 1LVO; 229E, 1P9S; NL63, 3TLO; and infectious bronchitis virus [IBV], 2Q6D) or GenBank for those without crystal structures (MHV-A59, NP_740610; OC43, NP_937947; MERS-CoV, AFY13306; and HKU5, YP_001039961). Sequences were aligned by the MUSCLE (multiple-sequence comparison by log expectation) algorithm. The X-ray crystal structures of SARS-CoV 3CLpro (PDB identification number [PDB ID], 2V6N) and HCoV-HKU1 3CLpro (PDB ID, 3D23) were used as a structural model of comparison (23, 29). Structural alignment and annotations were generated using PyMol (DeLano Scientific).
RESULTS
Selection of MHV-A59 inhibitor-resistant viruses and identification of residues associated with resistance.
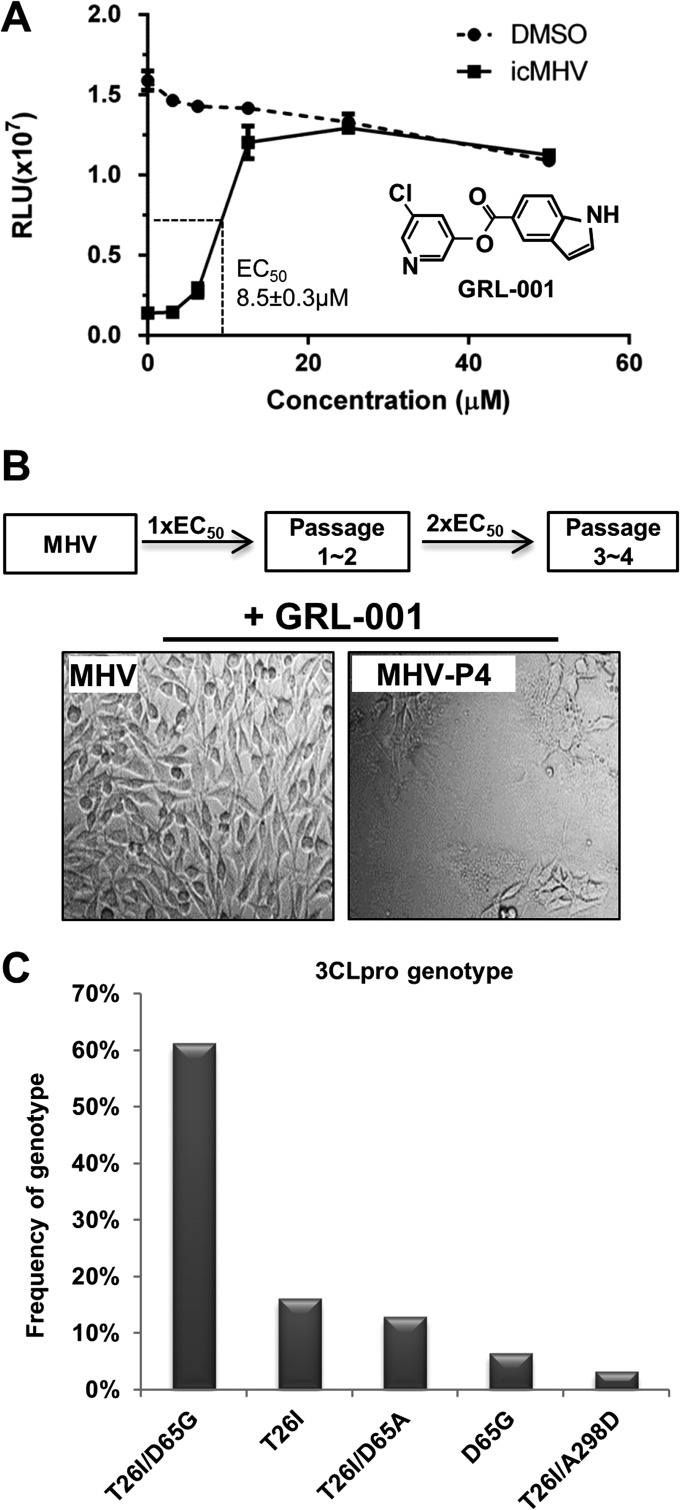
To study the properties of inhibitor resistance in coronaviruses, 3CLpro inhibitor GRL-001 (also termed CE-5) was used to select for inhibitor-resistant MHVs. We found that the EC50 of GRL-001 for MHV-A59 was 8.5 ± 0.3 μM (Fig. 1A). To obtain inhibitor-resistant viruses, wild-type (WT) MHV was serially passaged in the presence of GRL-001 at a concentration of 1× the EC50 for two passages and further passaged in the presence of GRL-001 at a concentration of 2× the EC50 for two additional passages. We observed cytopathic effect (syncytium formation) in virus-infected cells after four passages (Fig. 1B), consistent with the presence of inhibitor-resistant viruses that had been selected and amplified during the passaging process. The supernatant collected from serial passage 4 was designated inhibitor-resistant passage 4 (MHV-P4).
FIG 1.
Treatment with 3CLpro inhibitor GRL-001 selects for resistant strains of MHV-A59. (A) Chemical structure of GRL-001 and dose-dependent inhibition of MHV-induced cell death. DBT cells were infected with infectious clone WT MHV (icMHV) at an MOI of 0.1 and treated with GRL-001 at serial concentrations. Cell viability was determined by cell titer Glo assay (Promega). The EC50s were determined using nonlinear regression program with Prism 5 software. DMSO, dimethyl sulfoxide. (B) Scheme for selection of inhibitor-resistant viruses and evidence of selection for viruses that induce cytopathic effect in the presence of GRL-001. (C) Frequency of genotypes detected in plaque-purified MHV isolates. RNA was isolated from 31 randomly isolated plaques and the 3CLpro region was amplified and sequenced.
To identify the mutation(s) associated with GRL-001 resistance, viruses within the MHV-P4 supernatant were plaque purified and the viral RNA isolated from individual plaques was subjected to reverse transcription-PCR (RT-PCR) amplification of the 3CLpro region. Sequence analysis of 31 plaque isolates revealed single nucleotide changes at multiple genome sites of 10285 (ACA→AUA), 10402 (GAU→GGU or GCU), and 11101 (GCU→GAU), which led to single- or double-mutant genotypes within 3CLpro: single mutants with changes of threonine 26 to isoleucine (T26I) or aspartic acid 65 to glycine or alanine (D65G or D65A) and T26I/D65G, T26I/D65A, or T26I/A298D double mutants. We found that the T26I/D65G double mutant was the most frequently identified genotype, with 61% (19/31) of the plaque-purified isolates showing these mutations (Fig. 1C). These results demonstrate that only one nucleotide change is sufficient to gain amino acid substitutions at 3CLpro T26, D65, and/or A298, which are likely to confer resistance to the inhibitor.
3CLpro mutant viruses are resistant to GRL-001.
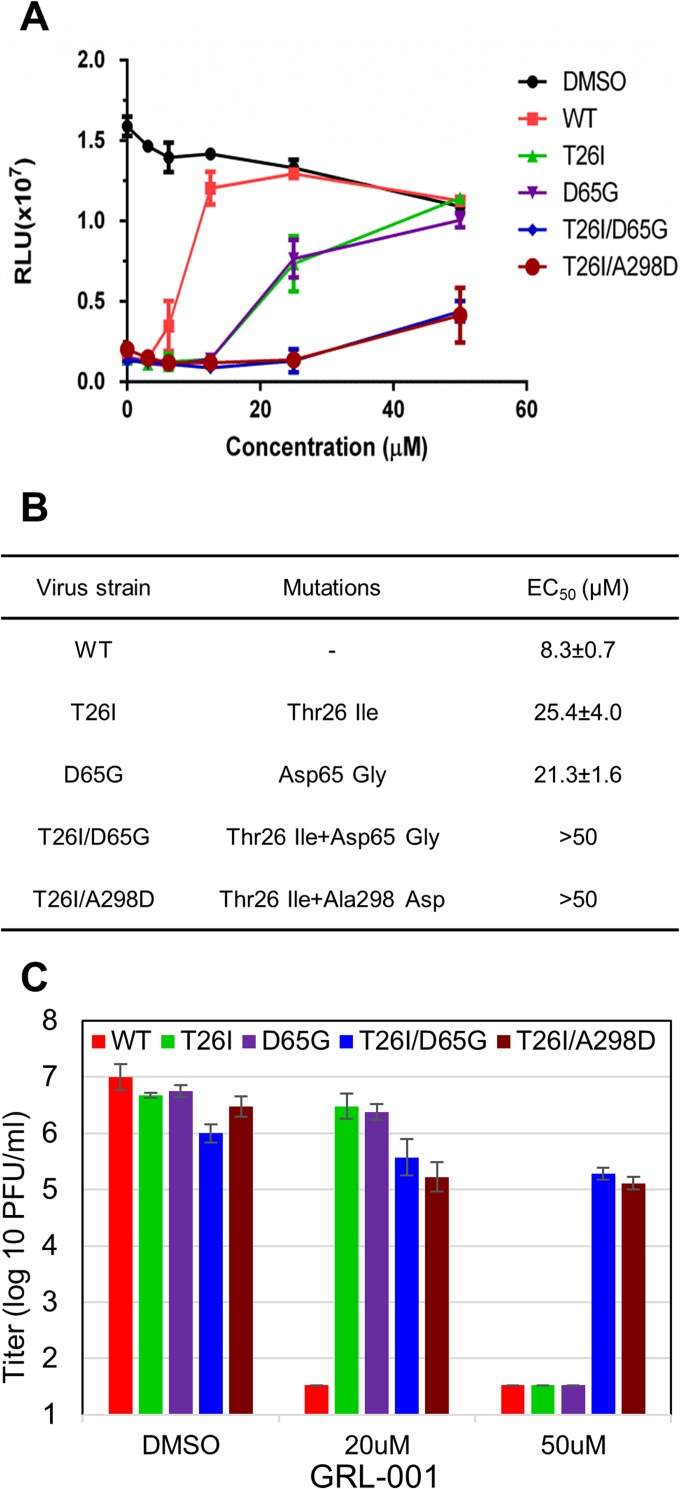
To determine if the mutations detected in the plaque-purified isolates are responsible for resistance to GRL-001, we used a site-directed mutagenesis approach and MHV reverse genetics to engineer isogenic isolates of MHV-A59 with mutations in 3CLpro (38). Engineered MHV isolates encoding 3CLpro substitutions were generated and designated MHV-T26I, MHV-D65G, MHV-T26I/D65G, and MHV-T26I/A298D. These mutant viruses were subjected to plaque purification, and engineered mutations were verified by sequence analysis of nsp5. The sensitivity of these mutant viruses to GRL-001 was analyzed using an antiviral-activity assay, and the EC50 was determined for each genotype (Fig. 2). Compared to WT virus, we found that the single-mutant viruses T26I and D65G were partially resistant, with EC50s of 25.4 ± 4.0 μM and 21.3 ± 1.6 μM, respectively. In contrast, the double-mutant viruses (T26I/D65G and T26I/A298D) induced significant cytopathic effects in the presence of high concentrations GRL-001 (50 μM), indicating that double-mutant viruses have much greater resistance to GRL-001 (EC50 > 50 μM) than single-mutant viruses (Fig. 2A and B). We further determined the effect of GRL-001 on the production of infectious virus (Fig. 2C). The plaque assay results show that the production of WT MHV infectious particles is significantly reduced in the presence of the inhibitor at 20 μM or 50 μM. The replication of the single-mutant viruses was not significantly affected at 20 μM GRL-001 but was dramatically reduced at 50 μM. As expected, the double-mutant viruses replicated to a high titer (∼105 PFU/ml) even in the presence of 50 μM GRL-001. Taken together, the accumulation of these drug-resistant double mutants within 4 passages is consistent with a low genetic barrier for CoV to gain resistance to this 3CLpro inhibitor.
FIG 2.
MHVs with two substitutions in 3CLpro are resistant to GRL-001. (A) Antiviral-activity assay reveals range of sensitivities to GRL-001. DBT cells were infected with virus at an MOI of 1, followed by addition of GRL-001 at the indicated concentrations. Cells were evaluated for viability using the cell titer Glo assay at 24 h postinfection. (B) EC50s of WT and 3CLpro mutant viruses were calculated based on the results of the antiviral-activity assay. (C) Production of WT and mutant viruses produced in the presence of 3CLpro inhibitor GRL-001. DBT cells were incubated with viral inoculum for 1 h, and then the medium was replaced with fresh medium containing DMSO or GRL-001 inhibitor at 20 μM or 50 μM and cells were incubated for 17 h. The infectious virus titer in the supernatant was determined by plaque assay on DBT cells.
Inhibitor-resistant virus exhibits delayed replicase processing and production of progeny virus.
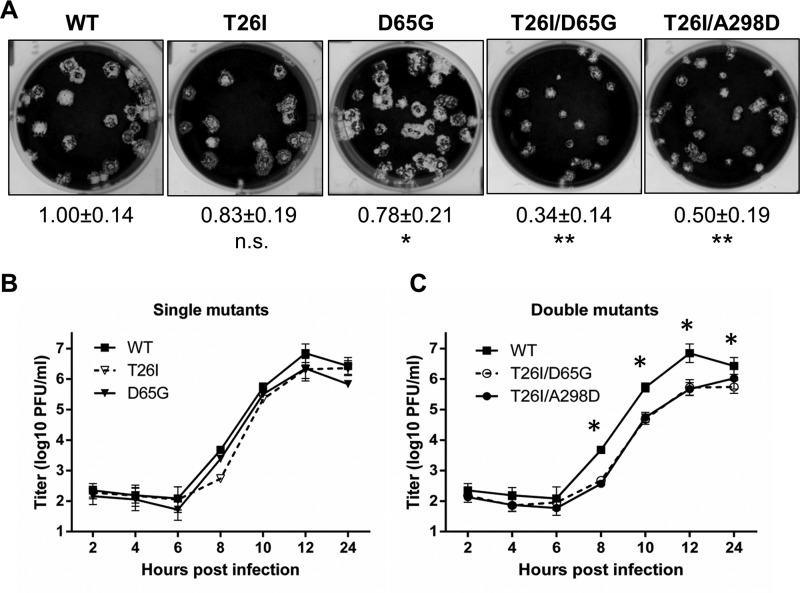
To evaluate the impact of the mutations associated with resistance to the 3CLpro inhibitor GRL-001 on coronavirus replication (fitness cost), we compared the plaque size, replication kinetics, and polyprotein processing of the engineered viruses. We found that the plaques generated by single-mutant viruses were smaller than that of WT virus (∼20% less), and double-mutant viruses generated plaques that were significantly smaller than the plaques generated by WT virus (∼50% less) (Fig. 3A). To evaluate replication kinetics, we infected cells with a multiplicity of infection (MOI) of 0.1 and harvested supernatant for plaque assay over a time course of 24 h. We found that single-mutant viruses replicated with kinetics similar to those of WT virus. In contrast, the double-mutant viruses replicated with a significant delay and to a lower titer at 12 h postinfection (Fig. 3B) compared to WT virus. In addition, we observed similar results for cells infected with a plaque-purified isolate of resistant MHV, which harbors T26I and A298D mutations (data not shown). These results indicate that viruses containing 3CLpro GRL-001 resistance double mutations are impaired for replication in cell culture.
FIG 3.
Inhibitor-resistant virus exhibits deficiencies in viral replication. (A) Representative plaques formed by WT and 3CLpro mutant MHVs at 48 h postinfection at 37°C. The relative areas of at least 12 single plaques of each virus were measured by Photoshop CS software to determine average area. n.s., not significant; *, P < 0.01; **, P < 0.001. (B and C) Growth kinetics of WT and 3CLpro mutant viruses in DBT cells infected at an MOI of 0.1. The error bars represent SD from the results of three plaque assays.*, P < 0.01.
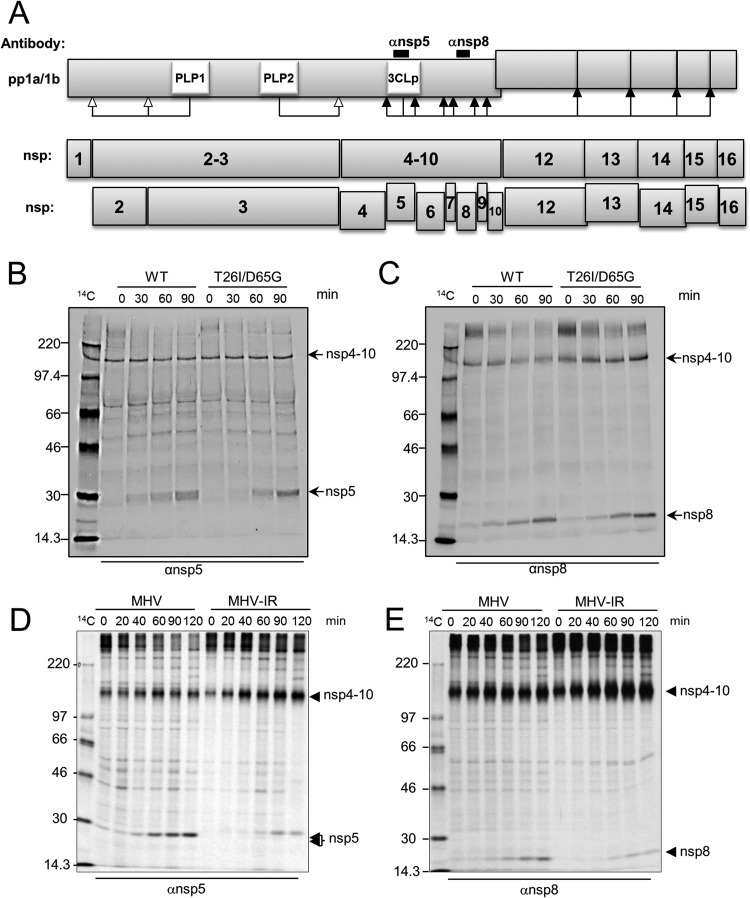
We hypothesized that the delay in virus replication observed in the double-mutant viruses was due to a delay in 3CLpro-mediated proteolytic processing of the replicase polyprotein. To test this hypothesis, we performed radiolabeling and pulse-chase studies to evaluate the precursor-product relationships in cells infected with WT and double-mutant viruses. The processing of the MHV replicase polyprotein is mediated by papain-like proteases to generate nsp1, nsp2, nsp3, and the p150 intermediate and by 3CLpro to process the p150 intermediate to generate nsp4 to nsp10 from ORF1a and nsp12 to nsp16 from ORF1b (Fig. 4A). Previous studies have shown that the precursors and products of the MHV replicase polyprotein can be immunoprecipitated by cognate antisera (39). To identify products generated during replication, we infected DBT cells with either WT or MHV-T26I/D65G and pulse-labeled newly synthesized proteins with [35S]methionine for 30 min from 4.5 h postinfection. Following the pulse, the [35S]methionine was removed and chased by the addition of excess unlabeled methionine in the medium. Cells were harvested and lysates prepared and subjected to immunoprecipitation with antibodies directed against nsp5 and nsp8. The products of the immunoprecipitation were analyzed by electrophoresis and visualized by autoradiography. We observed a delay in the accumulation of nsp5 and nsp8 in MHV-T26I/D65G-infected cells (Fig. 4B and C). In WT-infected cells, the nsp5 and nsp8 products are detected 30 min into the chase, whereas in the cells infected with the double-mutant virus, the nsp5 and nsp8 products were detected at 60 min into the chase. In addition, we observed a similar pattern of processing in cells infected with a plaque-purified isolate of resistant MHV, which harbors T26I and A298D mutations (Fig. 4D and E). Therefore, viruses resistant to GRL-001 are delayed in 3CLpro-mediated processing during virus replication. Taken together, these data suggest that although the genetic barrier of MHV resistance to the inhibitor GRL-001 is low, the fitness cost is high.
FIG 4.
Inhibitor-resistant viruses have delayed replicase processing. (A) Schematic diagram of 3CLpro-mediated replicase processing. (B to E) Comparison of proteolytic processing of WT and resistant viruses (infectious clone MHV-T26I/D65G [B and C] and plaque-purified isolate of inhibitor-resistant MHV [MHV-IR] that harbors T26I and A298D mutations [D and E]) by pulse-chase analysis. DBT cells were infected with WT or resistant viruses, and newly synthesized proteins were pulse-labeled with [35S]Met for 20 or 30 min at 4.5 h postinfection. Labeling medium was removed and replaced with medium containing excess unlabeled methionine and cysteine, and cells were harvested at 30-min intervals during the chase period. Cell lysates were subjected to immunoprecipitation with antisera for nsp5 (B and D) and nsp8 (C and E), and the products were analyzed by 10% SDS-PAGE and visualized by autoradiography.
GRL-001 protease inhibitor resistant virus is highly attenuated in mice.
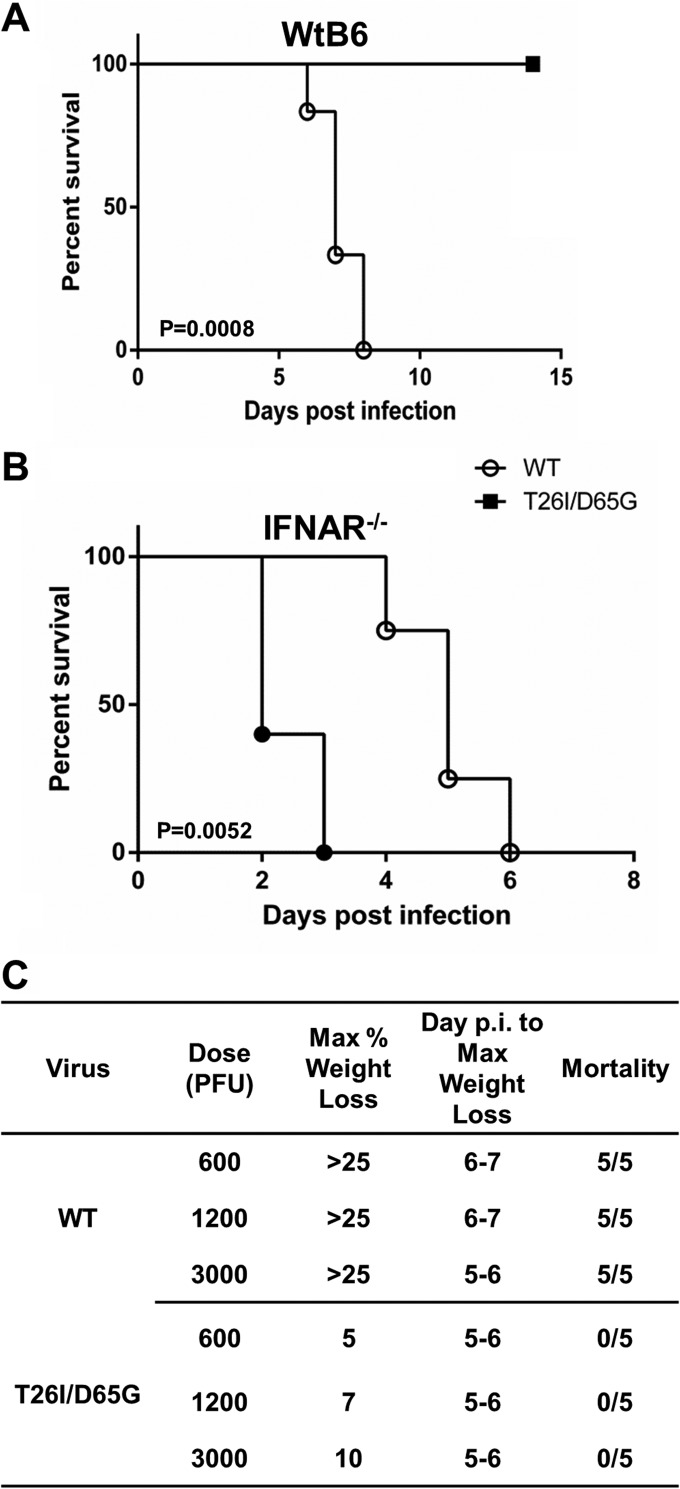
Our results demonstrating the delayed processing and replication kinetics associated with resistance to GRL-001 led us to hypothesize that this virus might be attenuated in vivo. To test this hypothesis, we infected wild-type C57BL/6 (WtB6) mice intracranially with WT MHV or MHV-T26I/D65G at 600, 1,200, and 3,000 PFU per mouse and analyzed weight loss to evaluate pathogenicity. Mice reaching 25% weight loss were euthanized according to our protocol. We found that mice infected with 600 PFU of WT MHV succumbed to infection by day 7. In contrast, mice infected with up to 3,000 PFU of MHV-T26I/D65G showed transient weight loss but never succumbed to infection (Fig. 5A). The estimated 50% lethal dose (LD50) of MHV-T26I/D65G is greater than 3,000 PFU, which indicates the GRL-001-resistant virus is greatly attenuated compared to WT virus (Fig. 5C). These results indicate that treatment with the 3CLpro inhibitor GRL-001 selects for viruses that exhibit delayed processing and replication kinetics and which are highly attenuated in animals.
FIG 5.
MHV-T26I/D65G inhibitor-resistant virus is highly attenuated. (A) C57BL/6 (WtB6) mice succumb to wild-type MHV (WT) but survive infection with mutant MHV (T26I/D65G). Four-week-old wild-type C57BL/6 mice were intracranially infected with 600 PFU of the WT (n = 5) or MHV-T26I/D65G mutant (n = 5). (B) Pathogenesis of MHV-T26I/D65G is delayed compared to that of WT MHV in highly susceptible type I IFN receptor knockout (IFNAR−/−) mice. Fourteen-week-old IFNAR−/− mice were intraperitoneally inoculated with 50 PFU of the WT (n = 6) or MHV-T26I/D65G (n = 6). Body weight loss was monitored daily. The statistical differences in survival were analyzed by Prism 5 software using the log rank test. (C) Morbidity and mortality in C57BL/6 mice following intracranial administration of WT and MHV-T26I/D65G. p.i., postinfection.
Type I interferon (IFN) signaling is crucial for controlling MHV infection, and type I IFN receptor knockout (IFNAR−/−) mice are exquisitely sensitive to MHV infection (41). To further evaluate the pathogenicity of MHV-T26I/D65G and minimize the contribution of host immune response to the attenuation of the virus, we inoculated IFNAR−/− mice intraperitoneally with 50 PFU of WT MHV or MHV-T26I/D65G and monitored for weight loss. We found that mice infected with WT virus succumbed to infection by day 3. In contrast, MHV-T26I/D65G-infected mice survived significantly longer than WT MHV-infected mice (P = 0.0052) (Fig. 5B). These results further document the attenuation of MHV-T26I/D65G in the highly sensitive IFNAR−/− mouse model.
3CLpro-T26I/D65G exhibits reduced enzymatic activity and sensitivity to GRL-001.
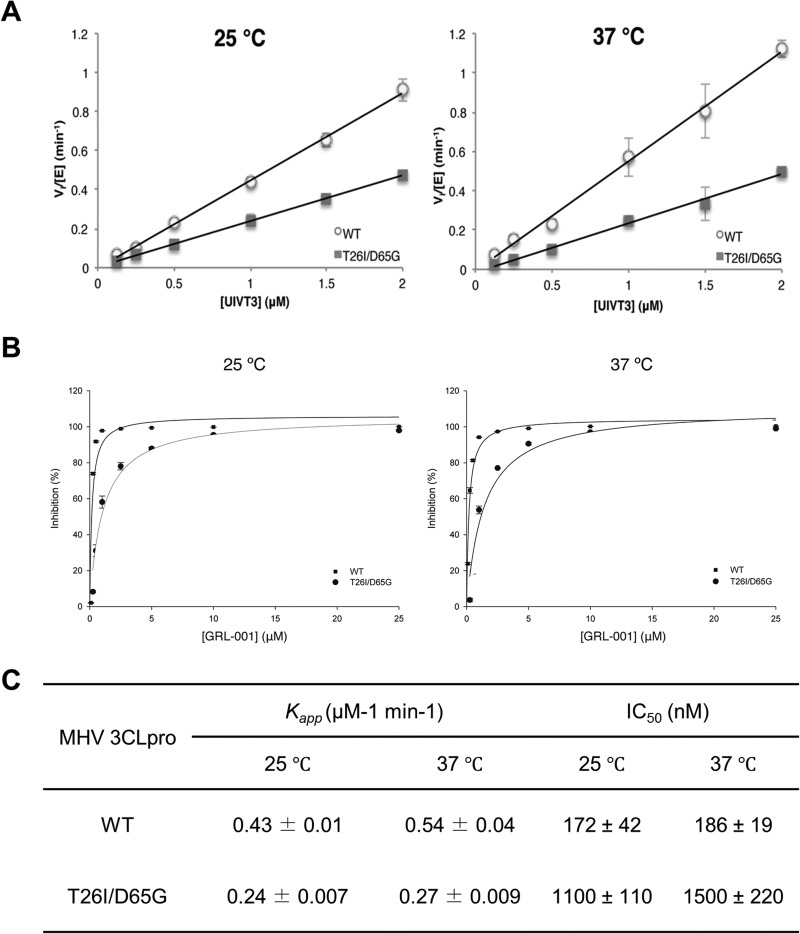
To further characterize 3CLpro resistance, WT and T26I/D65G mutant forms of 3CLpro were expressed in Escherichia coli and purified for in vitro analysis. The apparent rate constants for enzymatic catalysis (kapp) for both WT and 3CLpro-T26I/D65G were determined at ambient temperature (25°C) and 37°C (Fig. 6A) by determining the initial rate of the reaction using a fluorescence resonance energy transfer (FRET)-based substrate termed UIVT3, which contains a 3CLpro consensus cleavage site. The initial rate of the reaction was determined as a function of UIVT3 substrate by monitoring the fluorescence intensity over time. The slopes of the best-fit lines in Fig. 6A are kapp. We found that 3CLpro-T26I/D65G exhibits 50% less activity than the WT, with kapps of 0.43 ± 0.01 μM−1 min−1 for 3CLpro-WT and 0.24 ± 0.009 μM−1 min−1 for 3CLpro-T26I/D65G at 25°C and 0.54 ± 0.04 μM−1 min−1 for 3CLpro-WT and 0.27 ± 0.009 μM−1 min−1 for 3CLpro-T26I/D65G at 37°C (Fig. 6C). These data indicate that the T26I/D65G mutant is less efficient at catalyzing hydrolysis of the UIVT3 substrate, which correlates with impaired trans-cleavage activity of 3CLpro. Thus, the decreased catalytic activity of the T26I/D65G mutant likely contributes to the delayed polyprotein processing observed in the pulse-chase analysis.
FIG 6.
T26I/D65G MHV 3CLpro has reduced enzymatic efficiency and is not efficiently blocked by GRL-001. The enzymatic efficiency (kapp) (A) and inhibition by GRL-001 (IC50) (B) of both WT and T26I/D65G 3CLpro were determined at ambient temperature (25°C) and 37°C. (A) The initial reaction rates (Vi) were determined by calculating the initial slope of the progress curve, which was then converted to the amount of product (μM) produced per minute using the experimentally determined value of the fluorescence extinction coefficient for UIVT3. Plotting Vi/[E] versus [UIVT3] gave a linear correlation, the slope of which was taken to be the kapp. (B) The inhibition of the 3CLpro by GRL-001 was monitored by following the change in RFUs over time, using the initial slope of the progress curve to determine the initial rate. The percent inhibition of the 3CLpro enzymes was then plotted as a function of inhibitor concentration. (C) kapp and IC50s were determined using the nonlinear regression program SigmaPlot.
To directly test if the mutations in 3CLpro confer resistance to GRL-001, we determined the IC50s of GRL-001 for 3CLpro-WT and 3CLpro-T26I/D65G at ambient temperature and 37°C (Fig. 6B). The inhibition of the 3CLpro by GRL-001 as a function of GRL-001 concentration was determined, and the data were fit to a dose-response curve to obtain the IC50s. We observed that the IC50s of GRL-001 for WT and T26I/D65G 3CLpro were 172 ± 42 nM and 1,100 ± 110 nM, respectively, at 25°C and 186 ± 19 nM and 1,500 ± 220 nM, respectively, at 37°C (Fig. 6C). These results demonstrate that 3CLpro-T26I/D65G is about 6 to ∼8 times more resistant to GRL-001, which is in accordance with the insensitivity of MHV-T26I/D65G to this inhibitor (Fig. 2).
Analysis of the mechanism of resistance of 3CLpro-T26I/D65G to GRL-001.
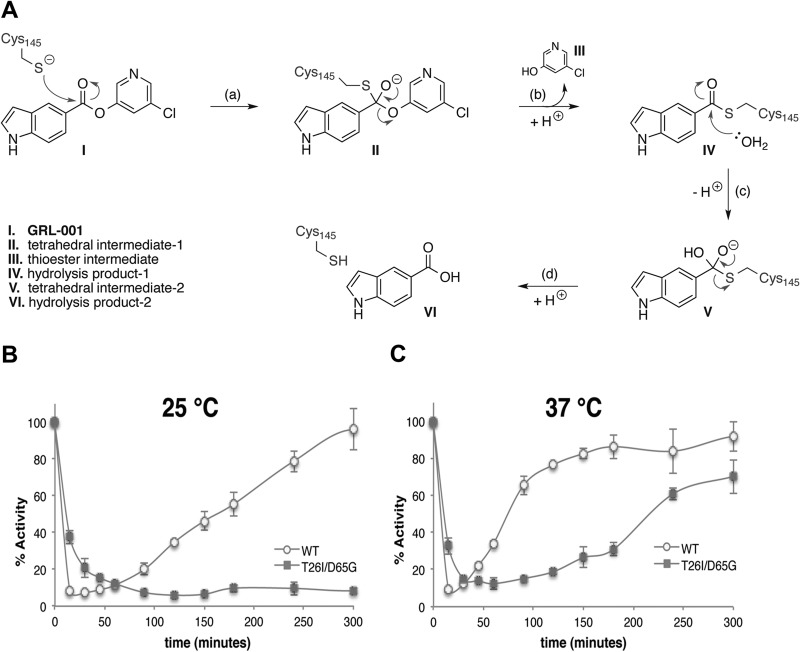
GRL-001 functions as a competitive substrate of 3CLpro and therefore forms various covalent intermediates by reaction with the catalytic cysteine (Cys145) throughout the hydrolysis process. Upon completion of inhibitor hydrolysis, the GRL-001 hydrolysis products are liberated from the 3CLpro active site and the free thiolate of the catalytic cysteine is regenerated. A detailed mechanism of GRL-001 hydrolysis is shown in Fig. 7A. By monitoring the “rebound” or restoration of the enzymatic activity of 3CLpro-WT and 3CLpro-T26I/D65G after a completed reaction cycle (release of item VI in Fig. 7A), we could evaluate the efficiency of GRL-001 hydrolysis and activity restoration. Purified 3CLpro-WT and 3CLpro-T26I/D65G were incubated with two equivalents of GRL-001 at the appropriate temperature, and their activities were measured over the course of 6 h. The UIVT3 substrate was added at specific time points, and the initial rates were measured to assess the percent remaining activity of inhibited versus uninhibited enzyme (or free enzyme). We found that at 25°C, 3CLpro-WT exhibited a rapid reduction of activity at early time points and enhanced activity toward UIVT3 substrate hydrolysis at late time points, indicating efficient initial inhibition by GRL-001 followed by relatively fast restoration of activity of WT enzyme. In contrast, 3CLpro-T26I/D65G was not inhibited as efficiently by GRL-001 at early time points. Interestingly, enzymatic activity was not restored over the time course of 6 h, indicating that in the absence of coincubation with the UIVT3 substrate, GRL-001 is an effective inhibitor of 3CLpro-T26I/D65G (Fig. 7B). At 37°C, 3CLpro-T26I/D65G exhibited some evidence of activity restoration, but it was again substantially slower than 3CLpro-WT (Fig. 7C). These results demonstrate that in the absence of coincubation with the UIVT3 substrate, 3CLpro-T26I/D65G possesses less esterase activity toward GRL-001 and exhibits delayed restoration of enzymatic activity relative to 3CLpro-WT. These results are consistent with a change in a rate-limiting step(s) associated with the acetylation and deacetylation steps of the 3CLpro-catalyzed reaction via introduction of the T26I/D65G double mutant. The rate of the acetylation (steps a and b in Fig. 7A) appears to decrease slightly upon mutation, whereas the rate of deacetylation (steps c and d in Fig. 7A) is substantially decreased.
FIG 7.
Esterase activities of WT and T26I/D65G MHV 3CLpro toward GRL-001 inhibitor. (A) Mechanism of GRL-001 hydrolysis catalyzed by MHV 3CLpro where the catalytic cysteine (Cys145) is indicated, the inhibitor (GRL-001) is shown in bold type, and intermediates and hydrolysis products are labeled with Roman numerals and identified in the bottom left corner. (B) GRL-001 hydrolysis time point assay at 25°C, where the restoration of enzymatic activity correlates to the enzymatic rate of hydrolysis of GRL-001. WT and T26I/D65G MHV 3CLpro enzymes were incubated in a 1:2 enzyme/GRL-001 ratio at the appropriate temperature, and enzymatic activity was monitored by measuring the fluorescence intensity of the reaction after addition of the UIVT3 substrate at each time point and determined as a percentage of the appropriate uninhibited enzyme at each time point. Note that in the absence of coincubation with both the UIVT3 substrate and GRL-001, T26I/D65G MHV 3CLpro is substantially slower at GRL-001 hydrolysis than with the WT MHV 3CLpro enzyme. (C) GRL-001 hydrolysis time point assay at 37°C. Note the enhanced rates of both the WT and T26I/D65G MHV 3CLpro toward GRL-001 hydrolysis.
DISCUSSION
Our understanding of the potential outcomes after treatment with viral protease inhibitors benefits from the years of study of protease inhibitors directed against HIV and hepatitis C virus (HCV). The simultaneous use of three distinct inhibitors to HIV (termed highly active antiretroviral therapy [HAART]) maximizes the therapeutic benefit and reduces the incidence of resistant strains (1). Studies using protease inhibitors directed against HCV NS3/4A protease, termed direct-acting antivirals (DAAs), have evaluated the genetic barrier and replicative cost of resistance (reviewed in reference 2). Initial studies of HCV protease inhibitors using viral replicons suggested that although the genetic barrier to resistance is low (i.e., resistant mutants are viable and are rapidly selected), the cost of resistance was high, with reduced levels of replication (42–44). Recent studies using two antivirals (one protease inhibitor and one RNA polymerase inhibitor) suggest that for HCV, double-drug therapy may be sufficient to “cure” HCV infection (45–47). Indeed, the use of DAAs for HCV has revolutionized the treatment of HCV-infected patients. For infections with coronaviruses, such as the recently emerged MERS-CoV, it is unclear if single-, double-, or triple-drug therapy may be required to provide therapeutic benefit. Because coronaviruses encode multiple proteases, such as 3CLpro and PLP, and multiple enzymatic activities, such as helicase and polymerase, there are certainly sufficient distinct targets for therapeutic development.
Low genetic barrier but high cost of resistance to 3CLpro inhibitor GRL-001.
During the evaluation of resistance to protease inhibitors, it is important to determine if there is any genetic barrier to resistance, i.e., whether the inhibitor-resistant virus is viable. For MHV, we identified a quasispecies of resistant mutants within four passages in cell culture (Fig. 1). This suggests that the barrier to the development of resistance to GRL-001 is low, as resistant mutants accumulate rapidly under selective pressure. The identification of single and double mutants provided the opportunity to analyze the effect of each site for the ability to confer resistance to the inhibitor. Interestingly, two mutations within 3CLpro were required to confer full resistance to the inhibitor (Fig. 2). The fact that these double mutants were the predominant population within 4 virus passages illustrates the low genetic barrier to resistance. The next issue is then to determine the “cost” of resistance for virus replication and pathogenesis. We found that the inhibitor-resistant viruses were delayed in replication, proteolytic processing, and production of infectious virus (Fig. 3). An important consequence of this defective replication is that the inhibitor-resistant virus is highly attenuated in mice (Fig. 4). This indicates that resistance to GRL-001 comes at the cost of replication efficiency and pathogenesis. These studies were performed with virus selected in cell culture in the presence of GRL-001. Studies are in progress to develop analogs of GRL-001 that can be administered in animal studies. It will be important to determine if similar or distinct viruses are selected in mice treated with analogs of GRL-001 and if the inhibitor-resistant viruses contribute to pathogenesis.
Modeling “resistance” onto 3CLpro.
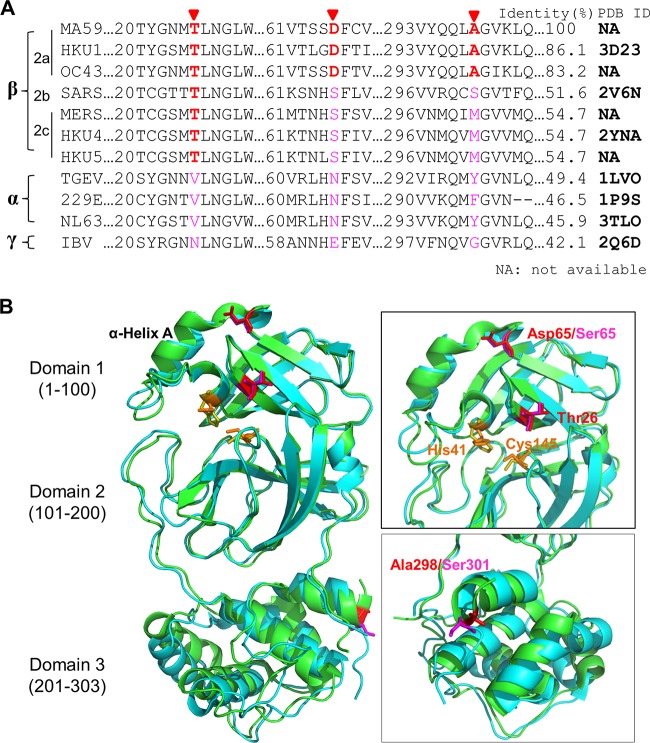
Structural studies would facilitate our understanding of the mechanism of resistance to GRL-001. However, the structure of MHV 3CLpro has not yet been solved. Structures are available for seven different coronavirus 3CLpro domains (27–31) in the Protein Data Bank (PDB). Even though the amino acid identities of these seven 3CLpros range from 40 to 80% (Fig. 8A), the structures are highly conserved. The structures of SARS-CoV 3CLpro bound with inhibitors have been intensively investigated, providing substantial information of enzyme folding, maturation, and inhibitor design (23, 27, 28, 33, 48, 49). Zhao et al. reported the crystal structure of HKU1 3CLpro (29), which is most similar in amino acid sequence to MHV. We further modeled the resistance residues T26, D65, and A298 onto the structures of HKU1 and SARS-CoV 3CLpros (Fig. 8B). The modification of these sites suggests that the T26I substitution may affect the binding of the substrate and inhibitor within the active site (27, 29). The fact that threonine at this position is highly conserved among the beta-coronaviruses (Fig. 8A) suggests that there may be limited flexibility in this position for optimal protease activity. Our in vitro protease activity data indicate that the T26I/D65G substitutions decrease enzymatic efficiency and reduce the off-rate of the substrate. We speculate that the side chain of residues D65 or S65 may affect the local conformation surrounding α-helix A of 3CLpro domain 1 (amino acids [aa] 1 to 100) and thereby the conformation at the catalytic site (Fig. 8B). Structural studies are needed to fully evaluate this issue. Determining if these two mutations confer resistance to GRL-001 in other emerging CoVs needs to be investigated.
FIG 8.
Context of sites that confer resistance to 3CLpro inhibitor GRL-001. (A) Alignment of 3CLpro amino acid sequence from selected CoVs with MUSCLE algorithm. Residues of 3CLpro associated with resistance in MHV (T26, D65, and A298) and corresponding sites among selected CoVs are highlighted. The Protein Data Bank identification numbers (PDB ID) of available structures of 3CLpros are listed. (B) Structural alignment of SARS-CoV 3CLpro (cyan, 2V6N) and HKU1 3CLpro (green, 3D23) to model the position identified in MHV 3CLpro that confers resistance to GRL-001. Catalytic residues Cys145 and His41 are labeled and colored (SARS-CoV 3CLpro, orange; HKU1 3CLpro, olive), and resistance-associated residues are labeled and colored (SARS-CoV 3CLpro, red, and HKU1 3CLpro, magenta).
The role of A298D substitution in conferring resistance to GRL-001 is currently unclear. A298 of MHV 3CLpro is located at the tail of domain 3 (aa 201 to 303) and is not conserved among coronaviruses. However, the α-helix tail of D3 has been shown to be critical for catalytic activity and enzyme dimerization (49–52). The substitution of A298D may alter the conformation of at the α-helix tail and thereby impair catalysis. In the present study, the double-mutant MHV (T26I/A298D) has an increased EC50 compared to that of the T26I single-mutant MHV, suggesting that the A298D mutation also affects inhibitor binding. Overall, our results support a model whereby an altered interaction with substrate and inhibitor likely contributes to both resistance to GRL-001 and less efficient proteolytic processing of the replicase polyprotein.
Implications for therapeutic potential of 3CLpro inhibitors.
This study is the first to describe the genotypes and pathogenesis of an inhibitor-resistant coronavirus. We show that the inhibitor-resistant virus is attenuated both in cell culture and in infected mice. To further advance the field, more structural information is needed to fully define the inhibitor binding site and to determine the molecular basis of drug resistance. Structural studies of HCV NS3/4A inhibitor-resistant mutants revealed the unique molecular basis of resistance to distinct inhibitors and emphasized how inhibitor binding simultaneously interfered with the recognition of the viral substrates (53, 54). These studies revealed that drug resistance mutations were frequently associated with residues at the edge of the substrate/inhibitor binding pocket. Mutations at the edges of the substrate binding pocket may be tolerated, although virus replication may be attenuated. Mutations that confer resistance by altering the binding pocket itself may be nonviable because of an inability to interact with the viral polyprotein substrate. Therefore, efforts directed at defining the substrate binding site and generating inhibitors that directly compete with substrate binding without extending outside the binding pocket may be the most efficacious.
Our studies have important implications for the development of therapeutics against existing and emerging coronaviruses in humans. Recently, additional broad-spectrum coronavirus inhibitors have been identified by screening existing drugs for their ability to block the replication of MERS-CoV, SARS-CoV, and other CoVs (55–57). Three compounds, imatinib mesylate, gemcitabine hydrochloride, and chlorpromazine hydrochloride, were identified in two independent studies (55, 57). Currently the targets for these compounds in CoV-infected cells are unclear, and these drugs may impact host cell rather than viral targets. Regarding viral targets, further study of protease inhibitors and protease inhibitor-resistant mutants selected in cell culture and studies of drug selection in animals are needed to determine if similar or distinct mutants are selected and if the viruses selected in animals are as attenuated as the viruses selected in cell culture.
ACKNOWLEDGMENTS
We thank Sakshi Tomar and Andrew Kilianski for their technical assistance.
This research was supported by grants from the National Institutes of Health (AI085089 to S.C.B. and A.D.M. and AI026603 to A.D.M.) and in part by the Walther Cancer Foundation to A.D.M.
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1. Siliciano JD, Siliciano RF. 2013. Recent trends in HIV-1 drug resistance. Curr. Opin. Virol. 3:487–494. 10.1016/j.coviro.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Halfon P, Locarnini S. 2011. Hepatitis C virus resistance to protease inhibitors. J. Hepatol. 55:192–206. 10.1016/j.jhep.2011.01.011 [DOI] [PubMed] [Google Scholar]
- 3. Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solórzano A, García-Sastre A, Palese P, Bouvier NM. 2013. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat. Commun. 4:2854. 10.1038/ncomms3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Menéndez-Arias L. 2013. Molecular basis of human immunodeficiency virus type 1 drug resistance: overview and recent developments. Antiviral Res. 98:93–120. 10.1016/j.antiviral.2013.01.007 [DOI] [PubMed] [Google Scholar]
- 5. Piret J, Boivin G. 2011. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob. Agents Chemother. 55:459–472. 10.1128/AAC.00615-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peiris JSM, Guan Y, Yuen KY. 2004. Severe acute respiratory syndrome. Nat. Med. 10:S88–S97. 10.1038/nm1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RM. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 8. De Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RAM, Galiano M, Gorbalenya AE, Memish ZA, Perlman S, Poon LLM, Snijder EJ, Stephens GM, Woo PCY, Zaki AM, Zambon M, Ziebuhr J. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 87:7790–7792. 10.1128/JVI.01244-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2014. First confirmed cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities—May 2014. MMWR Morb. Mortal. Wkly. Rep. 63:431–436 [PMC free article] [PubMed] [Google Scholar]
- 10. Becker WB, McIntosh K, Dees JH, Chanock RM. 1967. Morphogenesis of avian infectious bronchitis virus and a related human virus (strain 229E). J. Virol. 1:1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McIntosh K, Becker WB, Chanock RM. 1967. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. U. S. A. 58:2268–2273. 10.1073/pnas.58.6.2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJM, Wolthers KC, Wertheim-van Dillen PME, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat. Med. 10:368–373. 10.1038/nm1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fouchier RAM, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH, Osterhaus ADME. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 101:6212–6216. 10.1073/pnas.0400762101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woo PCY, Lau SKP, Chu C, Chan K, Tsoi H, Huang Y, Wong BHL, Poon RWS, Cai JJ, Luk W, Poon LLM, Wong SSY, Guan Y, Peiris JSM, Yuen K. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884–895. 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garbino J, Crespo S, Aubert J-D, Rochat T, Ninet B, Deffernez C, Wunderli W, Pache J-C, Soccal PM, Kaiser L. 2006. A prospective hospital-based study of the clinical impact of non-severe acute respiratory syndrome (non-SARS)-related human coronavirus infection. Clin. Infect. Dis. 43:1009–1015. 10.1086/507898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peiris J, Chu C, Cheng V, Chan K, Hung I, Poon L, Law K, Tang B, Hon T, Chan C, Chan K, Ng J, Zheng B, Ng W, Lai R, Guan Y, Yuen K. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767–1772. 10.1016/S0140-6736(03)13412-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kilianski A, Baker SC. 2014. Cell-based antiviral screening against coronaviruses: developing virus-specific and broad-spectrum inhibitors. Antiviral Res. 101:105–112. 10.1016/j.antiviral.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mielech AM, Chen Y, Mesecar AD, Baker SC. 2014. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 10.1016/j.virusres.2014.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853–879 [DOI] [PubMed] [Google Scholar]
- 20. Ghosh AK, Xi K, Ratia K, Santarsiero BD, Fu W, Harcourt BH, Rota PA, Baker SC, Johnson ME, Mesecar AD. 2005. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 48:6767–6771. 10.1021/jm050548m [DOI] [PubMed] [Google Scholar]
- 21. Ghosh AK, Xi K, Grum-Tokars V, Xu X, Ratia K, Fu W, Houser KV, Baker SC, Johnson ME, Mesecar AD. 2007. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 17:5876–5880. 10.1016/j.bmcl.2007.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramajayam R, Tan K-P, Liang P-H. 2011. Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem. Soc. Trans. 39:1371–1375. 10.1042/BST0391371 [DOI] [PubMed] [Google Scholar]
- 23. Verschueren KHG, Pumpor K, Aneml̈ler S, Chen S, Mesters JR, Hilgenfeld R. 2008. A structural view of the inactivation of the SARS coronavirus main proteinase by benzotriazole esters. Chem. Biol. 15:597–606. 10.1016/j.chembiol.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghosh AK, Gong G, Grum-Tokars V, Mulhearn DC, Baker SC, Coughlin M, Prabhakar BS, Sleeman K, Johnson ME, Mesecar AD. 2008. Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 18:5684–5688. 10.1016/j.bmcl.2008.08.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson ME, Baker SC, Ghosh AK, Mesecar AD. 2008. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 105:16119–16124. 10.1073/pnas.0805240105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosh AK, Takayama J, Rao KV, Ratia K, Chaudhuri R, Mulhearn DC, Lee H, Nichols DB, Baliji S, Baker SC, Johnson ME, Mesecar AD. 2010. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation. J. Med. Chem. 53:4968–4979. 10.1021/jm1004489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xue X, Yu H, Yang H, Xue F, Wu Z, Shen W, Li J, Zhou Z, Ding Y, Zhao Q, Zhang XC, Liao M, Bartlam M, Rao Z. 2008. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 82:2515–2527. 10.1128/JVI.02114-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 100:13190–13195. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao Q, Li S, Xue F, Zou Y, Chen C, Bartlam M, Rao Z. 2008. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J. Virol. 82:8647–8655. 10.1128/JVI.00298-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 21:3213–3224. 10.1093/emboj/cdf327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- 32. Chuck C-P, Chen C, Ke Z, Wan DC-C, Chow H-F, Wong K-B. 2013. Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases. Eur. J. Med. Chem. 59:1–6. 10.1016/j.ejmech.2012.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacobs J, Grum-Tokars V, Zhou Y, Turlington M, Saldanha SA, Chase P, Eggler A, Dawson ES, Baez-Santos YM, Tomar S, Mielech AM, Baker SC, Lindsley CW, Hodder P, Mesecar A, Stauffer SR. 2013. Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro. J. Med. Chem. 56:534–546. 10.1021/jm301580n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turlington M, Chun A, Tomar S, Eggler A, Grum-Tokars V, Jacobs J, Daniels JS, Dawson E, Saldanha A, Chase P, Baez-Santos YM, Lindsley CW, Hodder P, Mesecar AD, Stauffer SR. 2013. Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 23:6172–6177. 10.1016/j.bmcl.2013.08.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kilianski A, Mielech A, Deng X, Baker SC. 2013. Assessing activity and inhibition of MERS-CoV papain-like and 3C-like proteases using luciferase-based biosensors. J. Virol. 87:11955–11962. 10.1128/JVI.02105-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Agnihothram S, Yount BL, Donaldson EF, Huynh J, Menachery VD, Gralinski LE, Graham RL, Becker MM, Tomar S, Scobey TD, Osswald HL, Whitmore A, Gopal R, Ghosh AK, Mesecar A, Zambon M, Heise M, Denison MR, Baric RS. 2014. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. mBio 5:e00047–14. 10.1128/mBio.00047-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hirano N, Fujiwara K, Hino S, Matumoto M. 1974. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch. Gesamte Virusforsch. 44:298–302. 10.1007/BF01240618 [DOI] [PubMed] [Google Scholar]
- 38. Yount B, Denison MR, Weiss SR, Ralph S, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065–11078. 10.1128/JVI.76.21.11065-11078.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schiller JJ, Kanjanahaluethai A, Baker SC. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288–302. 10.1006/viro.1997.9010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697–3708. 10.1128/JVI.76.8.3697-3708.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cervantes-Barragan L, Züst R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, Ludewig B. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E, Zeuzem S, Sarrazin C. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718. 10.1002/hep.23192 [DOI] [PubMed] [Google Scholar]
- 43. Rong L, Dahari H, Ribeiro RM, Perelson AS. 2010. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci. Transl. Med. 2:30ra32. 10.1126/scitranslmed.3000544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kieffer TL, Kwong AD, Picchio GR. 2010. Viral resistance to specifically targeted antiviral therapies for hepatitis C (STAT-Cs). J. Antimicrob. Chemother. 65:202–212. 10.1093/jac/dkp388 [DOI] [PubMed] [Google Scholar]
- 45. Ferenci P, Bernstein D, Lalezari J, Cohen D, Luo Y, Cooper C, Tam E, Marinho RT, Tsai N, Nyberg A, Box TD, Younes Z, Enayati P, Green S, Baruch Y, Bhandari BR, Caruntu FA, Sepe T, Chulanov V, Janczewska E, Rizzardini G, Gervain J, Planas R, Moreno C, Hassanein T, Xie W, King M, Podsadecki T, Reddy KR. 2014. ABT-450/r-ombitasvir and dasabuvir with or without ribavirin for HCV. N. Engl. J. Med. 10.1056/NEJMoa1402338 [DOI] [PubMed] [Google Scholar]
- 46. Afdhal N, Zeuzem S, Kwo P, Chojkier M, Gitlin N, Puoti M, Romero-Gomez M, Zarski J-P, Agarwal K, Buggisch P, Foster GR, Bräu N, Buti M, Jacobson IM, Subramanian GM, Ding X, Mo H, Yang JC, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Mangia A, Marcellin P. 2014. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N. Engl. J. Med. 10.1056/NEJMoa1402454 [DOI] [PubMed] [Google Scholar]
- 47. Afdhal N, Reddy KR, Nelson DR, Lawitz E, Gordon SC, Schiff E, Nahass R, Ghalib R, Gitlin N, Herring R, Lalezari J, Younes ZH, Pockros PJ, Di Bisceglie AM, Arora S, Subramanian GM, Zhu Y, Dvory-Sobol H, Yang JC, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Sulkowski M, Kwo P. 2014. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N. Engl. J. Med. 370:1483–1493. 10.1056/NEJMoa1316366 [DOI] [PubMed] [Google Scholar]
- 48. Goetz DH, Choe Y, Hansell E, Chen YT, McDowell M, Jonsson CB, Roush WR, McKerrow J, Craik CS. 2007. Substrate specificity profiling and identification of a new class of inhibitor for the major protease of the SARS coronavirus. Biochemistry 46:8744–8752. 10.1021/bi0621415 [DOI] [PubMed] [Google Scholar]
- 49. Kang X, Zhong N, Zou P, Zhang S, Jin C, Xia B. 2012. Foldon unfolding mediates the interconversion between M(pro)-C monomer and 3D domain-swapped dimer. Proc. Natl. Acad. Sci. U. S. A. 109:14900–14905. 10.1073/pnas.1205241109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chou C-Y, Chang H-C, Hsu W-C, Lin T-Z, Lin C-H, Chang G-G. 2004. Quaternary structure of the severe acute respiratory syndrome (SARS) coronavirus main protease. Biochemistry 43:14958–14970. 10.1021/bi0490237 [DOI] [PubMed] [Google Scholar]
- 51. Hsu W-C, Chang H-C, Chou C-Y, Tsai P-J, Lin P-I, Chang G-G. 2005. Critical assessment of important regions in the subunit association and catalytic action of the severe acute respiratory syndrome coronavirus main protease. J. Biol. Chem. 280:22741–22748. 10.1074/jbc.M502556200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xia B, Kang X. 2011. Activation and maturation of SARS-CoV main protease. Protein Cell 2:282–290. 10.1007/s13238-011-1034-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Romano KP, Ali A, Aydin C, Soumana D, Ozen A, Deveau LM, Silver C, Cao H, Newton A, Petropoulos CJ, Huang W, Schiffer CA. 2012. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog. 8:e1002832. 10.1371/journal.ppat.1002832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Romano KP, Ali A, Royer WE, Schiffer CA. 2010. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc. Natl. Acad. Sci. U. S. A. 107:20986–20991. 10.1073/pnas.1006370107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. De Wilde AH, Jochmans D, Posthuma CC, Zevenhoven-Dobbe JC, van Nieuwkoop S, Bestebroer TM, van den Hoogen BG, Neyts J, Snijder EJ. 19 May 2014. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 10.1128/AAC.03011-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adedeji AO, Singh K, Kassim A, Coleman CM, Elliott R, Weiss SR, Frieman MB, Sarafianos SG. 19 May 2014. Evaluation of SSYA10-001 as a replication inhibitor of SARS, MHV and MERS coronaviruses. Antimicrob. Agents Chemother. 10.1128/AAC.02994-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dyall J, Coleman CM, Hart BJ, Venkataraman T, Holbrook MR, Kindrachuk J, Johnson RF, Olinger GG, Jahrling PB, Laidlaw M, Johansen LM, Lear CM, Glass PJ, Hensley LE, Frieman MB. 19 May 2014. Repurposing of clinically developed drugs for treatment of Middle East respiratory coronavirus infection. Antimicrob. Agents Chemother. 10.1128/AAC.03036-14 [DOI] [PMC free article] [PubMed] [Google Scholar]