ABSTRACT
Japanese encephalitis (JE) is an arthropod-borne disease associated with the majority of viral encephalitis cases in the Asia-Pacific region. The causative agent, Japanese encephalitis virus (JEV), has been phylogenetically divided into five genotypes. Recent surveillance data indicate that genotype I (GI) is gradually replacing genotype III (GIII) as the dominant genotype. To investigate the mechanism behind the genotype shift and the potential consequences in terms of vaccine efficacy, human cases, and virus dissemination, we collected (i) all full-length and partial JEV molecular sequences and (ii) associated genotype and host information comprising a data set of 873 sequences. We then examined differences between the two genotypes at the genetic and epidemiological level by investigating amino acid mutations, positive selection, and host range.
We found that although GI is dominant, it has fewer sites predicted to be under positive selection, a narrower host range, and significantly fewer human isolates. For the E protein, the sites under positive selection define a haplotype set for each genotype that shows striking differences in their composition and diversity, with GIII showing significantly more variety than GI. Our results suggest that GI has displaced GIII by achieving a replication cycle that is more efficient but is also more restricted in its host range.
IMPORTANCE Japanese encephalitis is an arthropod-borne disease associated with the majority of viral encephalitis cases in the Asia-Pacific region. The causative agent, Japanese encephalitis virus (JEV), has been divided into five genotypes based on sequence similarity. Recent data indicate that genotype I (GI) is gradually replacing genotype III (GIII) as the dominant genotype. Understanding the reasons behind this shift and the potential consequences in terms of vaccine efficacy, human cases, and virus dissemination is important for controlling the spread of the virus and reducing human fatalities. We collected all available full-length and partial JEV molecular sequences and associated genotype and host information. We then examined differences between the two genotypes at the genetic and epidemiological levels by investigating amino acid mutations, positive selection, and host range. Our results suggest that GI has displaced GIII by achieving a replication cycle that is more efficient but more restricted in host range.
INTRODUCTION
Japanese encephalitis (JE) is a zoonotic disease causing severe acute infections. Since it was first reported in Japan in 1924, JE cases have been identified in most Asian countries and especially in south Asia, southeast Asia, and east Asia (1). It has also been detected sporadically in parts of the western Pacific and northern Australia (2–4). JE is the leading cause of arbovirus encephalitis afflicting humans in terms of morbidity and mortality (5), with a recent study estimating the occurrence of ∼68,000 cases annually, resulting in between 10% and 50% fatalities (6, 7). Among the survivors, as many as 50% may end up with sequelae of irreversible neurological impairment or lingering mental symptoms (8, 9).
Japanese encephalitis virus (JEV), the infectious pathogen of JE, is a member of the genus Flavivirus in the family Flaviviridae (10). Similarly to other flaviviruses, JEV is an enveloped virus with a single-stranded positive-sense RNA genome that is approximately 11 kb in length. The 5′ capped genome molecule is flanked by a short 5′ untranslated region (UTR), a longer 3′ UTR, and a intervening single open reading frame (ORF) that encodes a polyprotein which is subsequently cleaved, by both viral and cellular proteases, into 10 functional proteins comprising 3 structural proteins (capsid [C], prematrix/matrix [prM/M], and envelope [E]) and 7 nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (11).
JEV is maintained in nature through cycles involving arthropods that transmit the virus via susceptible reservoir hosts. It has been confirmed that JEV can be transmitted through bites by a number of mosquito species, especially the rice-field-breeding Culex tritaeniorhynchus mosquito (12, 13). However, mosquitoes, pigs, and birds are the primary natural hosts, although a small number of isolates have been obtained from horses, bats, and midges.
Previous phylogenetic studies indicate that JEV can be classified into five genotypes based on nucleotide sequences of the E protein gene or the gene encoding the complete polyprotein (14), with most isolates classified as genotype I (GI) or genotype III (GIII) (15). GIII was previously dominant, but surveillance data from the last 20 years, in the form of nucleotide sequences from collected isolates, have revealed a gradual replacement of GIII by GI such that the latter is now dominant or cocirculating with GIII in many JEV regions of endemicity and epidemicity (16–19). In general, lineage replacement is a cause for concern as it can impact disease control measures, resulting in the irruption of new outbreaks (20, 21). Thus, understanding the reasons behind the current genotype replacement event can yield valuable insight into the future control and eradication of JE diseases.
In an earlier study, we observed that the displacement of GIII was accompanied by a corresponding order-of-magnitude increase in the relative genetic diversity of GI versus that of GIII (22). However, compared to many other viruses exhibiting multiple genotypes, there is relatively high conservation at the amino acid level within and between the JEV genotypes (with a nucleotide divergence of 11.6% and amino acid divergence of 2.8% between GI and GIII).
Previous epidemiology studies on JEV commonly focused on the E protein or on small numbers of genome sequences and, as such, did not permit a comprehensive comparative analysis of different lineages. However, a recent study of E protein gene sequences revealed that genotype I could be subdivided into two clades, GI-a and GI-b, with the majority of isolates placed in GI-b. Furthermore, viral multiplication and temperature sensitivity analyses revealed that the GI-b isolate had significantly higher infectivity titers in mosquito cells from 24 to 48 h postinfection, providing new insight into how GI emerged as the dominant lineage (23). Nevertheless, the recent publication of a large number of whole-genome sequences of JEV and the availability of a very large number of sequences from other genes make further examination of the GIII-to-GI displacement event worthwhile. In this study, we collected all available sequence data from all genomic regions and investigated the differences between the two genotypes not only from a genetic but also from an epidemiological perspective. Specifically, we investigated amino acid mutations, positive selection, and host range and considered the impact on the viral transmission cycle and ultimately on susceptible human populations in an attempt to identify determinants driving the observed lineage replacement.
MATERIALS AND METHODS
Phylogenetic analysis of JEV genome and E protein data sets.
Phylogenetic analyses were performed on a total of 121 full-length genome sequences and 570 complete E protein sequences (315 GI and 255 GIII) retrieved from the GenBank database (http://www.ncbi.nlm.nih.gov/). Vaccine or derivative strains, in vitro-cultured isolates, and isolates with sequences containing significant numbers of ambiguous nonstandard nucleotide or amino acid characters were excluded. The genome sequences were isolated from a variety of hosts (mosquitoes, midges, bats, pigs, horses, and humans) with isolation dates ranging from 1949 to 2009 and a geographic distribution spanning China, Japan, South Korea, Thailand, Viet Nam, India, and Taiwan. Although the earliest GIII strain (the Nakayama strain) was isolated in Japan in 1935, this sample was removed from the genome data set as it contains several ambiguous nucleotides. The E protein sequences from the genome data set were combined with the full-length E data set, and duplicate entries were removed. Only GI and GIII E sequences with minimal associated background information (genotype, year of isolation, and country of isolation) were retained to achieve a final data set of 347 GI and 275 GIII isolates. Background details of the genome and E sequences are available from us on request. Nucleotide and deduced amino acid sequences were aligned using ClustalW v2.0 (24) and manually edited using MEGA.
Phylogenetic analysis was conducted for the 10,299-nucleotide (nt) complete coding region and for the individual GI and GIII E protein gene data sets using the neighbor-joining (NJ) method and the maximum composite likelihood nucleotide substitution model implemented in MEGA v.5.05 (25). The robustness of phylograms was evaluated using 1,000 bootstrap replicates.
Mutation analysis and investigation of selection pressures.
To identify site mutations that characterized the differences between GI and GIII at the amino acid level, we generated a consensus alignment for the GIII sequences and compared the complete polyprotein sequences in order to identify sites with mutational differences between the two genotypes that were present in at least 95% of the GI isolates. The presence of positive selection was assessed using the Codeml program implemented in PAML software package version 4.0 (http://abacus.gene.ucl.ac.uk/software/paml.html) (26). We examined six different codon-based substitution models to estimate the ratio of nonsynonymous and synonymous nucleotide substitutions (ω = dN/dS) at each codon site: M0 (ratio of 1), M1a (nearly neutral), M2a (positive selection), M3 (discrete ω), M7 (ω values approximated by a β distribution), and M8 (ω values approximated by β distribution and a fraction of sites with ω of >1). As appropriate to each model, a maximum-likelihood approach was used to estimate ω and the proportion of sites in the alignment that fall into a specific predefined category. A Bayes empirical Bayes (BEB) approach was used to calculate the posterior probabilities that each codon belongs to a particular site class and for identifying sites under positive selection (27). Then, using the likelihood-ratio test (LRT), three independent pairwise comparisons (of M0 and M3, of M1a and M2a, and of M7 and M8) were performed to select the best-fitting models for the data. Amino acid sites on the E protein predicted to be under positive selection in each genotype (together with host, date, and location information) were compared for the GI and GIII sequences in turn to investigate correlations among these parameters. These relationships on the respective E protein gene trees were presented using the online iTOL software package (http://itol.embl.de/).
Visualization of sites within the JEV NS5 protein structure under positive selection.
Many GIII-to-GI mutations and sites under positive selection were located in the NS5 coding regions. In an attempt to investigate the significance of these mutations, the structure of the NS5 protein recently solved for Japanese encephalitis virus (4K6M) was downloaded from the protein database (http://www.pdb.org/pdb/home/home.do), and sites of interest were located on the structure and visualized using Pymol v1.5 (http://www.pymol.org/).
Investigation of host range and genotype shift.
To investigate the host range and variations in JEV GI and GIII isolates over time, a third data set (distinct from the genome and E protein data sets described above) was created. This data set comprised the full E sequences collected for the phylogenetic analysis combined with additional partial E, partial C, C-prM, and NS5 gene sequences (data available on request) that were also downloaded from GenBank. Derivative and genetically modified strains, as well as duplicate entries, were removed, and NJ trees were constructed to confirm the genotype of each sequence. GI and GIII sequences with minimal background information (genotype, year of isolation, and country of isolation) were selected to achieve a final data set of 893 isolates comprising 622 complete E protein gene sequences, 41 partial E and NS5 gene sequences, and 230 isolates for the partial C and C-prM sequences. The combined E/partial E/C-prM/NS5 data set was then used to classify the presence of GI and GIII isolates by year and country. Few isolates were obtained from horses, bats, midges, and birds, so only mosquito, pig, and human isolates were considered in the analysis. A 2-by-3 contingency table was generated based on the two genotypes and three hosts, and Pearson's chi-square test (χ2) implemented in the R statistical package (http://www.r-project.org) was performed to investigate the relationship between host distribution and genotype. Significance was assigned at P < 0.01.
RESULTS
Genome data set and phylogenetic analysis.
A summary of the GI and GIII genome sequences is available on request. The data corresponding to the NJ tree for the 121 whole-genome sequences for all JEV genotypes, sampled between 1949 and 2009, are available on request. The topology of the tree is consistent with previous reports (22) and is divided into five major groups corresponding to genotypes I, II, III, IV, and V. The majority of isolates belong to GI (54 sequences) and GIII (60 sequences). There are three and two subclades predicted for GI and GIII, respectively. For GIII, these subclades have strong bootstrap support; for GI, the bootstrap support is much weaker, but the strains in the two clades can be distinguished by the amino acid at site 2296 in the polyprotein (within NS4B), which is predicted to be under positive selection in both genotypes (see below). Schuh et al. (28) predicted the presence of two distinct G-I clades, G-Ia and G-Ib, on the basis of E protein data, but none of the G-Ia samples have full-length genome sequences available, so the tree represents only G-Ib isolates. Of the remaining genotypes, GV includes just two isolates; one was isolated in Malaysia in 1952 (29), and the other was collected recently (in 2009) in the Tibet autonomous region (30). There is a further report of an isolate from South Korea collected in 2010 (31), but no genome sequence is available. The GII isolates are from Australia and the south Pacific, and the single GIV isolate is from Indonesia. All of these GII and GIV isolates represent earlier strains collected 17 years ago. There are no recent isolates for either of these genotypes.
Mutations with amino acid properties.
The data with respect to the consensus amino acid changes in GIII versus GI are available on request. In terms of mutation density (i.e., number of mutations/number of sites for each coding region), the C protein is the most variable, with 5 changes across the 127 amino acids (aa) encoding the protein, followed by NS1, NS2A, and NS2B, with 1 mutation for every 40 amino acids, and NS3, NS4B, and NS5, with 1 mutation for every 80 amino acids. The most highly conserved proteins are E and NS4A.
Natural selection and adaptive evolution.
To investigate the extent of the selective pressure acting on the JEV genome, we estimated the ratio of nonsynonymous to synonymous substitutions across the entire polyprotein using six different codon substitution models implemented in the PAML software package to represent a range of possible scenarios. Details of the parameter estimates for each model are available on request. Table 1 shows the likelihood-ratio test (LRT) results for pairwise comparison of specific model pairs. The comparison of models M0 and M3 is a test of variable ω among sites, and the significant differences (supported by the χ2 test results) for both GI and GIII suggest that, as expected, both genotypes experience significant variation throughout their genomes. Comparison of models M1a and M2a is a test of neutral versus positive selection. Whereas there is a clear distinction between these two models for GIII, surprisingly, there is no significant difference estimated for GI (P < 0.05). A significant difference is achieved only in comparisons of the more complicated M7 and M8 models, which approximate site variation using a β distribution. But even in this case, although a small fraction of sites are predicted to be under positive selection for both genotypes, comparison of the respective β distributions reveals that, overall, GI is more strongly conserved than GIII.
TABLE 1.
Pairwise comparison of selected CODEML codon substitution models for genotype I and genotype IIIa
| Genotype | Model comparison |
||
|---|---|---|---|
| M0 vs M3 | M1 vs M2 | M7 vs M8 | |
| GI | 305.539 (0.000) | 5.062 (0.079) | 29.618 (0.000) |
| GIII | 616.530 (0.000) | 88.268 (0.000) | 68.798 (0.000) |
For each genotype and pairwise comparison, the first value represents the log likelihood determined by the comparison and the value in parentheses represents the estimated P value from the χ2 test.
Sites under positive selection.
A small number of individual sites under positive selection were detected in both genotypes. However, once again, there are notable differences (in terms of the numbers and distributions of these sites) between the predictions for GI and GIII (Table 2). For GIII, there are 11 sites predicted with a high posterior probability of having dN/dS of >1 (P of >95%) that are distributed throughout the genome. Conversely, only 5 sites are predicted for GI, and 4 of these sites are located within the NS4B and NS5 nonstructural proteins. Two sites, codon 417 and codon 2296 of the ORF (aa 123 in the E protein and aa 24 of NS4B), are shared by the two genotypes.
TABLE 2.
Sites predicted to shown positive selection in the genotype I and genotype III genome sequencesa
| Genotype | Polyprotein position [protein position] | Gene product |
|---|---|---|
| GI | 417 [123] (S/N)* | E |
| 2296 [24] (S/P)* | NS4b | |
| 2525 [253] (L/F) | NS4b | |
| 2956 [429] (D/G/S) | NS5 | |
| 3416 [889] (T/S) | NS5 | |
| GIII | 125 [125] (A/V) | C |
| 267 [140] (A/V/I) | PrM | |
| 417 [123] (S/R)* | E | |
| 503 [209] (R/K) | E | |
| 521 [227] (S/P) | E | |
| 702 [408] (S/L) | E | |
| 1673 [169] (D/E) | NS3 | |
| 2126 [3] (V/I) | NS4a | |
| 2280 [8] (K/R) | NS4b | |
| 2296 [24] (S/P/L)* | NS4b | |
| 2913 [386] (H/Y) | NS5 |
Asterisks indicate sites common to the two genotypes.
For site 417, a serine or arginine residue was observed in GIII, with the majority of isolates possessing a serine residue. The isolates containing an arginine residue are either from human cases isolated before 1965 or from bat isolates collected from Yunnan, China, between 1986 and 1997. For GI isolates, a serine or asparagine residue pair was observed. All the strains with an asparagine residue were isolated from mosquitoes in southeast China, with the exception of a single isolate from a human case in Thailand in 1985. This site is located in domain II of the E protein, and amino acid mutations within this domain can influence virulence by impacting the low-pH-mediated conformation transition associated with membrane fusion (32). It has also been reported that this site is correlated with the growth properties and pathogenicity of JEV; a serine-to-arginine mutation was shown to increase the growth rate in mouse neuroblastoma cells and pathogenicity in mice (33), suggesting a similar association with pathogenicity in GI. For site 521 (aa 227 of the E protein), it has been shown that Ser-227-Pro mutants can slightly retard the growth rate in Vero cells (33).
There is growing evidence that indicates that NS4B plays a role in blocking interferon (IFN) signaling in West Nile virus (WNV) and dengue virus (DENV) infections (34). For site 24 in the NS4B protein, all the GIII isolates had a serine residue, with the exception of four human strains with a proline residue. In GI, however, there were approximately equal proportions of isolates with serine and proline residues. A previous study suggested that this site might be correlated with host preference in GI and that a Pro24 residue tends to be associated with human cases (35). However, our data set shows no significant association with hosts, with the proline residue occurring in mosquito, pig, and human isolates. In GI, these proline and serine residues clustered into separate clades in the estimated genome tree (data available on request), suggesting that the proline substitution occurred in a common ancestor and was inherited by present-day virus strains. In GIII, however, the four isolates with a Pro residue were dispersed throughout the three different clades. No clear pattern or functional significance was identified for the remaining GI-specific sites at codons 2525 (NS4B), 2956 (NS5), and 3416 (the C-terminal end of the NS5 RNA-dependent RNA polymerase [RdRp]).
Host adaptation.
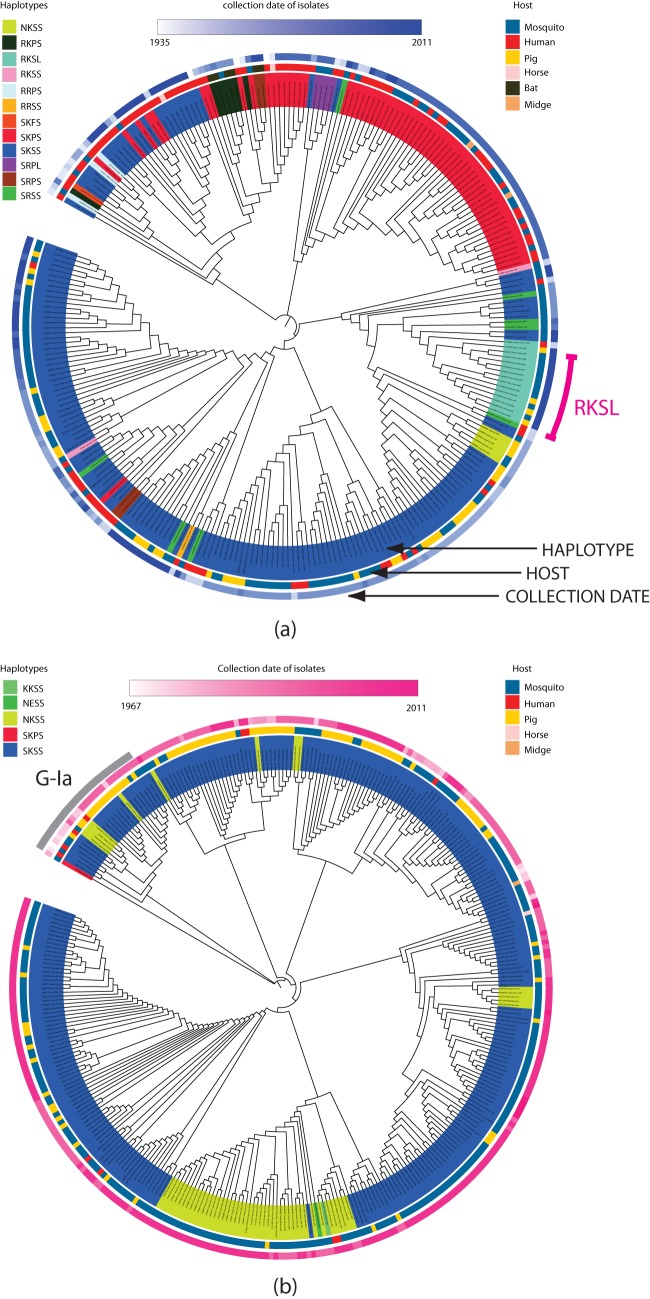
For the 11 sites predicted to be under positive selection in GIII, all of the bat samples (isolated from Yunnan province in China) have 100% identity, but no similar patterns exist for other hosts. For the sites located within the E protein (sites 417, 503, 521, and 702 in the polyprotein and sites 123, 209, 227, and 408 within the E protein), there are 12 haplotypes, with the SKSS and SKPS haplotypes predominant. Figure 1a shows the haplotypes mapped onto the estimated GIII E protein gene tree. Human and mosquito isolates show the greatest diversity, with 10 and 9 haplotypes, respectively (Table 3). Only three haplotypes, SKSS, NKSS, and SKSL, are present in porcine samples, suggesting a possible significance for sites 209K and 227S in this host. The other interesting feature in the tree is that RKSL appears to be an emerging haplotype on the east coast of China, as these isolates form a subclade with strong bootstrap support and the subclade is comprised of recent samples from mosquitoes and pigs (Fig. 1a).
FIG 1.
E protein gene tree for GIII and GI. The unrooted neighbor-joining E protein trees for GIII (a) and GI (b) were constructed from the combined E/genome data set alignments of 275 GIII and 347 GI isolates, respectively. Taxons are colored by haplotype as defined by amino acid sites 417, 503, 521, and 702 within the E protein that are predicted to be under positive selection in genotype III. The inner circle shows the hosts of isolates; the outer circle shows the isolation date of strains. (a) GIII displays greater diversity in both haplotypes and host range, with human and mosquito isolates defined by 10 and 9 haplotypes, respectively, and pigs associated with only 3 haplotypes. In addition, haplotype RKSL, marked at the bottom right of the tree, appears to be an emerging haplotype on the east coast of China, as these isolates form a subclade with strong bootstrap support, and the subclade is comprised of recent samples from mosquitoes and pigs. (b) GI has narrower host and haplotype ranges; most isolates were collected from mosquitoes and pigs, with fewer human case samples, and haplotypes SKSS and NKSS accounted for 99% of all isolates.
TABLE 3.
GI and GIII haplotypes for E protein by hosta
| Host | GIII result |
GI result |
||
|---|---|---|---|---|
| Haplotype | No. of samples | Haplotype | No. of samples | |
| Mosquito | SKSS | 79 | SKSS | 185 |
| SKPS | 25 | NKSS | 50 | |
| RKSL | 10 | KKSS | 1 | |
| SRSS | 5 | NESS | 1 | |
| SRPL | 3 | SKPS | 1 | |
| NKSS | 2 | |||
| RKSS | 2 | |||
| RRPS | 2 | |||
| RKPS | 1 | |||
| Human | SKSS | 45 | SKSS | 8 |
| SKPS | 38 | NKSS | 3 | |
| SRSS | 3 | |||
| RKPS | 2 | |||
| SRPS | 2 | |||
| RKSL | 1 | |||
| RRSS | 1 | |||
| SKFS | 1 | |||
| SRPL | 1 | |||
| RRPS | 1 | |||
| Pig | SKSS | 34 | SKSS | 91 |
| NKSS | 3 | NKSS | 5 | |
| RKSL | 4 | |||
| Midge | SKPS | 2 | SKSS | 1 |
| Horse | SKPS | 1 | SKSS | 1 |
| SKSS | 1 | |||
| Bat | SRPS | 2 | ||
| RKPS | 4 | |||
Haplotypes are defined by amino acid sites 417, 503, 521, and 702 within the E protein that are predicted to show positive selection in GIII.
Once again, the corresponding GI results are strikingly different. The GI isolates are defined by only five haplotypes, with SKSS and NKSS accounting for 99% of the isolates and only single occurrences of the remaining three haplotypes. Figure 1b shows the haplotypes mapped on to the GI E tree. The tree shows a classification of G-I isolates into G-Ia and G-Ib consistent with the findings of Schuh et al. (28). The majority of samples with NKSS sequences were recently (i.e., since 2003) collected from mosquitoes, with three earlier samples isolated from pigs. Thus, the GI isolates are defined by a range of haplotypes that is far narrower than that seen with the GIII isolates, although there is no distinct haplotype for the smaller G-Ia clade.
Visualization of the sites under selection in the NS5 protein structure.
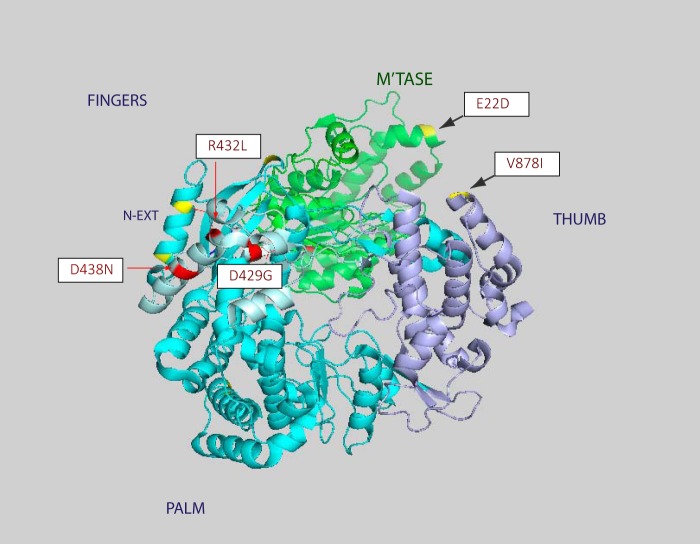
Given the number of GI-to-GIII mutations and the number of sites under positive selection that are located within the RdRp catalytic domain of the NS5 protein, we next attempted to investigate their locations within the folded protein structure for this domain. The NS5 protein comprises a methyltransferase (MTase) at the N-terminal and an RdRp at the C-terminal end. The recently solved crystal structure of the full-length JEV NS5 protein (36) was downloaded from the Protein Database (PDB code 4K6M), and a schematic representation is shown in Fig. 2, with the GI-to-GIII mutations and sites under positive selection marked.
FIG 2.
Locations of sites under positive selection within the NS5 three-dimensional (3D) structure. The solved structure shows the RdRp structure at the front of the image and the MTase structure behind. The JEV structure follows the standard flavivirus RdRp right-hand conformation consisting of fingers, palm, and thumb characteristic of the flavivirus polymerase. The locations of the four positive-charge changes on the “fingertip” region of the finger subdomain that occur between the GI and GIII NS5 sequences are marked red. Two of these mutations, sites 429 and 432, produce compensatory charge mutations of +1 and −1, respectively. The remaining mutations are marked yellow. See Table 2 and the main text for full details.
The JEV NS5 architecture is characteristic of the recognized flavivirus polymerase structure, with a right-hand conformation consisting of fingers, palm, and thumb (37), and shows significant structural homology to the corresponding architecture in both WNV and DV (36). The majority of GI-to-GIII mutations are located in the region coding for RdRp, but there is a single mutation in the MTase region (E22D), a site that has been shown to affect replication efficiency in dengue virus 4 (DV4) (38). Of the remaining mutations, there are two in the N-terminal extension (K280R and K287R) and one each in the palm (A372V), middle finger (E588G), and thumb (V878I) regions. It is interesting that the four remaining mutations correspond to charge changes in close proximity within the NS5 structure, located in the fingertip region of the finger subdomain (37). Two of the mutations, at sites 429 and 432, are charge complementary, producing charge changes of +1 and −1, respectively. These two sites are in close proximity to motif F (aa 453 to 476) in JEV, which is thought to provide a binding site for incoming nucleoside triphosphates (NTPs) entering through the tunnel from the back of RdRp molecule (39). Two Zn2+ binding sites have been identified in the RdRp structure, one of which is coordinated by sites E440, H444, C449, and C452 in JEV. Site D438N (associated with a charge change of +1 in G) is in the proximity of this zinc binding site and has been shown to be related to replication efficiency in DV2 (38).
Host preference of genotypes.
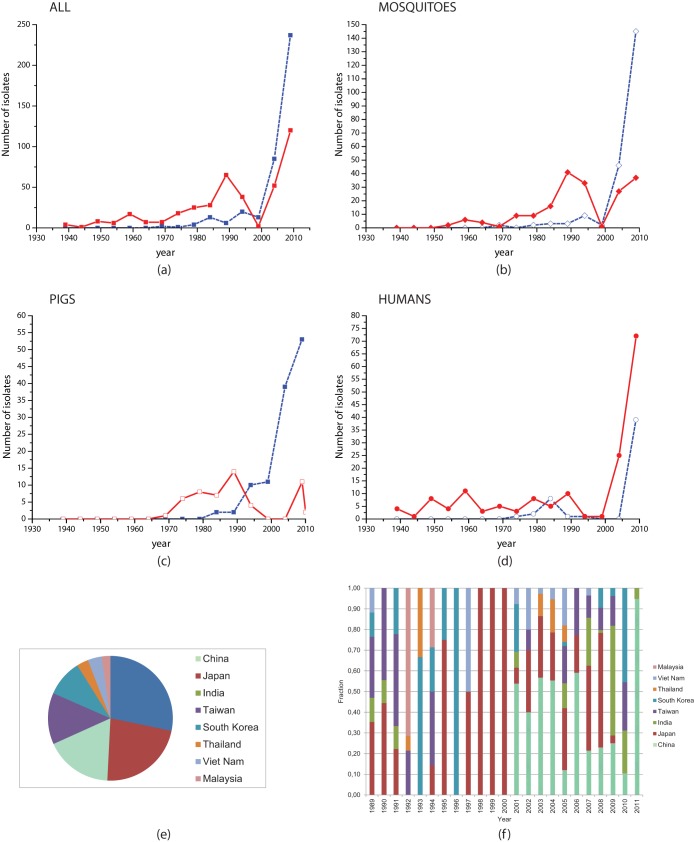
The combined E/partial E/C-prM/NS5 data set comprising 454 GI strains and 439 GIII isolates was used to investigate the host preference of the two major genotypes. Hosts comprised mosquitoes, midges, humans, and pigs, with a small number of isolates collected from horse, bats, and birds. The majority of strains were isolated from mosquitoes, with 278 and 197 GI and GIII isolates, respectively. Figure 3 shows the variations in the numbers of GI and GIII isolates collected over time. Figure 3a shows the variations for all isolates and highlights how GIII began to be displaced by GI as the dominant genotype in the middle of the 1990s. The variations for mosquitoes (Fig. 3b) and pigs (Fig. 3c) reflect this displacement pattern. However, Fig. 3d shows that the majority of human cases remain associated with GIII. Furthermore, it seems that, overall, GI has a much narrower host preference (Table 4). To determine whether these observed differences are significant, we constructed a contingency table to analyze the relationship among hosts and genotypes. Statistical analysis revealed a highly significant difference (Pearson's chi-square test, P value = 2.2 × 10−16); i.e., the host composition of GI is significantly different from the host composition of GIII. Panels e and f of Fig. 3 show the geographical composition of the samples collected between 1990 to 2011, the time range over which the genotype shift primarily occurred. China, Taiwan, Japan, South Korea, and Thailand are all well represented across the time interval, suggesting that there is no evidence of sampling bias for any specific country. The full temporal-spatial details of the isolates are available on request.
FIG 3.
Change in GI versus GIII host shift over time. Data represent variations over time in the number of collected JEV isolates for GI and GIII for all samples and for sample subsets. The red solid line represents GIII, and the blue dashed line represents GI. (a) Variations for all isolates, highlighting how GIII was displaced by GI as the dominant genotype in the middle of the 1990s. (b and c) Variations over time for mosquitoes and pigs, respectively, demonstrating similar displacement patterns. (d) Variations over time for human isolates. Despite GIII being displaced as the dominant genotype, the majority of human cases remain associated with GIII. (e and f) Geographical composition of samples collected over the time period (1990 to 2011) during which genotype replacement appears to have occurred. Panel e represents the sum over the entire period; panel f shows the variation by year. There is no region in which samples appear to have been preferentially sampled, and many regions have been continually represented over the time course.
TABLE 4.
Differences in host composition for genotype I and III isolates
| Host | No. of isolates with indicated genotype |
|
|---|---|---|
| GIII | GI | |
| Mosquito | 197 | 278 |
| Pig | 54 | 120 |
| Human | 175 | 54 |
| Horse | 4 | 1 |
| Bat | 6 | 0 |
| Midge | 2 | 1 |
| Bird | 1 | 0 |
| Total | 439 | 454 |
The two genotypes also showed notable differences in the composition of vector hosts (Table 5). Of the 279 GI isolates and 199 GIII vector isolates, 218 and 128 isolates were able to be resolved to the group or subgroup level. For both genotypes, the dominant species is Culex tritaeniorhynchus, which accounts for 94% (n = 204) of isolates in GI but for only 74% (n = 95) in GIII. The difference also extends to the genus-group-subgroup range of the two genotypes. For GI, the remaining 6% of isolates (n = 13) span 9 genera, groups, or subgroups; for GIII, the remaining 24% of isolates (n = 31) span 14 different genera, groups, or subgroups. However, the classification of subgroups within Culex families is based on morphological characteristics and is somewhat subjective (40, 41), so the significance of these differences is unclear.
TABLE 5.
Comparison of the range of vector isolates for GI and GIIIa
| Subgroup and species | GI |
GIII |
||
|---|---|---|---|---|
| No. of isolates | % isolates | No. of isolates | % isolates | |
| Mosquitoes | ||||
| Aedes vexans | 2 | 1.56 | ||
| Aedes lineatopennis | 1 | 0.78 | ||
| Anopheles minimus | 1 | 0.78 | ||
| Anopheles sinensis | 1 | 0.46 | 3 | 2.34 |
| Armigeres spp. | 2 | 0.92 | 4 | 3.13 |
| Armigeres subalbatus | 3 | 2.34 | ||
| Culex modestus | 1 | 0.46 | ||
| Culex whitmorei | 1 | 0.78 | ||
| Culex epidesmus | 1 | 0.78 | ||
| Culex gelidus | 1 | 0.46 | ||
| Culex pipiens | 1 | 0.46 | ||
| Culex fuscocephalus | 1 | 0.78 | ||
| Culex pipiens pallens | 3 | 1.38 | ||
| Culex quinquefasciatus | 1 | 0.46 | 2 | 1.56 |
| Culex theileri | 3 | 2.34 | ||
| Culex vishnui | 1 | 0.46 | 5 | 3.91 |
| Culex annulus | 2 | 1.56 | ||
| Culex pseudovishnui | 2 | 0.92 | 2 | 2.11 |
| Culex tritaeniorhynchus | 204 | 93.58 | 95 | 74.22 |
| Midges | ||||
| Culicoides | 2 | 1.56 | ||
| Lasiohelea taiwana Shiraki | 1 | 0.46 | ||
| Total | 218 | 100.00 | 128 | 100.00 |
GI had the narrower host range, with almost all isolates collected from Culex tritaeniorhynchus. For GIII, more than 25% of isolates were from other subgenera or Culex subgroups.
DISCUSSION
In this work, we consolidated all sequence data collected from across the JEV incidence region to try and identify differences between “before” and “after” GIII-to-GI shift events. In this way, we created a comprehensive data set describing the impact on JEV in the Asia-Pacific region over the course of the last 30 years. Previous studies have considered the phylogeographic events associated with this genotype shift and compared the differences in genetic diversity by investigating a single protein. Here, by collecting and consolidating all publicly available samples and their associated background data, we have also obtained a perspective on the differences that exist between these two genotypes among the primary hosts and reservoirs in the JEV replication cycle.
Previous studies have shown that the evolution of JEV is driven predominantly by strong purifying selection, and few sites have been predicted to be under positive selection. However, those analyses were conducted on individual genes (E or prM-C) or on a small number of genome sequences (42–44). Here, we reexamined this issue by the use of a much larger data set and, for the first time, investigated the two genotypes separately. Overall, our results are consistent with those from previous studies with respect to predicting a consistent phylogeny, estimating low ratios of nonsynonymous substitutions to synonymous substitutions (dN/dS) in both genotypes, and identifying a small number of individual sites under positive selection. However, the differences in the specific predictions for the individual genotypes are surprising. Typically, an emerging strain is considered to have gained some adaptive advantage over other strains with respect to host or environment and this is reflected in the identification of positive selection within its viral genome (45). Based on this, we would have expected to identify stronger effects in GI as the emerging lineage. However, while 11 sites were identified as evolving under diversifying selective pressure in GIII, only 5 sites were identified in GI. While similar differences in the numbers of sites for GI and GIII in the predictions for the E protein were reported in a recent publication by Schuh et al. (23), there are discrepancies between these two studies in the identified sites. However, similar inconsistencies were also observed between their predictions based on the fast, unconstrained Bayesian approximation (FUBAR) and fixed-effects-likelihood (FEL) methods. These differences are likely a consequence of the highly conserved nature of the JEV genome (compared to, for example, that of HIV) and make it difficult to apply complex site variation models to predict sites under positive selection. What is significant among all these results is that there are consistently fewer sites predicted for GI than for GIII. Similarly, while GIII was predicted to show positive rather than neutral selection, there was insufficient support to allow making this distinction for GI.
Analysis of full-length genome sequences revealed that the differences between the two lineages can be represented by only 30 amino changes within the 3,432 amino acids comprising the polyprotein, corresponding to a difference of less than 1%. While the significance of the set of identified GI-to-GIII site mutations and sites predicted to be under positive selection needs to be established, the high similarity of the two lineages means that this set represents a relatively small number of sites, and it seems that they would be worthwhile candidates for experimental studies, as many of them may be associated with host adaptation or play relevant functional roles in the virus life cycle.
Further differences were revealed when we considered the host composition of the more comprehensive set of E/partial E/C-prM/NS5 sequences. The change in the ratio of GI isolates to GIII isolates from pig and mosquito is consistent with the gradual emergence of GI, but, unexpectedly, the majority of human isolates are still GIII. Moreover, these host differences have been shown to be statistically significant with very strong support. The differences in host composition also extended to the vector host, with the GIII mosquito isolates collected from a wider range of mosquitoes and GI isolates almost exclusively restricted to Culex tritaeniorhynchus.
The highly conserved nature of the JEV genome is reflected in the predicted trees for the genome and the respective E protein gene trees for GI and GIII. Whereas the genome tree predicted two and three subclades, respectively, for GI and GIII, these classifications were not reflected in the tree generated from the shorter E protein gene sequences. However, mapping of the sites predicted to be under positive selection in this protein showed strong clustering by haplotype and, once again, showed distinct differences that highlighted the greater sequence variation within the GIII isolates.
Given the similarities of the two genotypes at the nucleotide and amino acid levels, it is remarkable that GI has displaced GIII so effectively in so many regions. This is in stark contrast to, for example, DENV, which exists as multiple genotypes within each of the four serotypes and exhibits far more complex genotype and serotype emergence patterns (for examples, see references 46 and 47). However, notable distinctions exist between DENV and JEV in terms of their respective host ranges and viral life cycles. The DENV transmission cycle involves humans acting as reservoirs and mosquitoes serving as vectors (48). The predominant vector is Aedes aegypti, with secondary contributions from Aedes albopictus and sporadic reports of outbreaks associated with Aedes hensilli, although only sporadic cases of dengue hemorrhagic fever (DHF) have been associated with Aedes albopictus outbreaks (49, 50). In contrast, in the enzootic JEV cycle, a number of mosquito species and other arthropod insects act as vectors, swine serve as an amplifying host (51), and wading ardeid water birds (e.g., herons and egrets) act as virus reservoirs. While each of these hosts play an important role in the maintenance of JEV in nature (13), humans are considered a dead-end host, developing insufficient viremia to infect feeding mosquitoes (52). Of all the vector hosts, Culex tritaeniorhynchus has been demonstrated to be the most efficient and prevalent (13). However, other Culex species (e.g., Culex annulirostris, Culex vishnui, Culex bitaeniorhynchus, and Culex pipiens) (52–54) and other mosquitoes of different genera (e.g., Armigeres, Aedes, Anopheles, and Mansonia) have demonstrated secondary roles and have been identified as potential viral transmitters (55, 56).
The contrasting transmission cycles of these two viruses suggest a possible explanation for the differences in our GI and GIII results and the recent displacement of GIII. Our results indicate that GIII shows greater diversity than GI in both sequence variation and host range. In particular, the narrow haplotype range of the GI E protein is associated with a narrower vector range, suggesting that the GI strain has been optimized for transmission to and from Culex tritaeniorhynchus and pigs. This hypothesis is supported by the results from an experimental study by Schuh et al. (23), which showed that the GI-b clade demonstrates an increased in vitro multiplicative ability compared to other sublineages, possibly resulting in increased numbers of mosquito→avian→transmission cycles. Our results support these findings and also suggest a more efficient transmission pattern for the virus (from vector to amplifying host and back to vector). This was achieved by narrowing the host range of the GI-b lineage with the trade-off of reduced numbers of infected human hosts (evidenced by reduced numbers of human cases after the genotype switch). However, since humans are a dead-end host, this does not impact the infection efficiency of the virus.
While our results imply a reduced human risk in regions where GI is dominant, JEV will continue to present a serious threat to public health. The close proximity of pig breeding farms or feral pig populations to human residential areas increases the risks of exposure of human populations to JEV, resulting in significant levels of spillover to humans (57–59). This link was demonstrated in Taiwan by an order-of-magnitude drop in the incident rate in nonvaccinated infants between 1966 and 1980 that coincided with the urbanization of many rural areas (60). In many regions of endemicity, there has been a switch to periurban agricultural methods in recent years in response to growing requirements for pork production that has led to increases in populations of pigs which, in their role as amplifying hosts, may have drastically altered the dynamics of the transmission mode (59, 61, 62). Given the variations in JEV GIII vectors in terms of resting locations (63) and feeding preferences (64–66) as well as seasonal variations in populations (67, 68), the virus would be well positioned to adapt to changes in the infection landscape.
However, there have been two recent reports of human outbreaks associated with GI. South Korea reported JE fatalities (7 of 26 cases) in 2010, and recently published sequences of strains isolated from mosquitoes in this outbreak confirmed that the cases were associated exclusively with GI isolates (69), but no human sequences have been published. Another report from an outbreak in India showed greater numbers of human cases associated with GI, but GIII cases remained dominant (41 of 68 sequences submitted to GenBank [data available on request]). Only the C-prM sequence was published for the majority of the isolates, and the full-length E sequence is available for only two of the GI strains (70). However, they are placed in a distinct clade in our estimated E protein gene tree and contain distinct amino acid mutations that are not present in any other GI isolates, suggesting the possible emergence of another lineage.
While it is tempting to dismiss these differences as merely a consequence of sampling biases, there is no evidence of any striking geographical bias in the samples; many geographically distinct regions, including Japan, China, India, Taiwan, South Korea, and Thailand, have been well represented over the period in which the genotype shift has occurred (Fig. 3e and f), and there is strong statistical support for all these results. Importantly, these are the same data that are used as evidence for genotype replacement. Nevertheless, this highlights not only the need for more surveillance data but also the benefits of some kind of central repository for this information. Having access to a comprehensive data source describing the impact of JEV as well as details of environment and effective control measures could permit more insightful analyses that extend beyond standard phylogenetic investigations. This has already been demonstrated in two recent reports, which investigated the link between environment and the incidence of JEV infection in Nepal (71) and estimated the distribution of JEV in Asia (72) by considering environment and vector distribution based on the integration and analysis of data obtained from WorldClim (http://www.worldclim.org/bioclim) and MosquitoMap (73). Complementing current vaccination and mosquito control programs with the use of novel analytical techniques applied to comprehensive data collections can lead to a more effective approach for the continued control and eradication of JEV.
ACKNOWLEDGMENTS
This work was supported by the 973 program (2012CB721102) and Chinese Academy of Science project (KSCX2-EW-Z-3).
We thank Junko Kuno for assistance in obtaining background information on Japanese JEV data sets, Yoo Hyosoon and Cho Sung-il for assistance in obtaining the background on Korean data sets, and Jan Hawkwind for helpful comments on the structure and style of the manuscript.
Footnotes
Published ahead of print 23 July 2014
REFERENCES
- 1.Erlanger TE, Weiss S, Keiser J, Utzinger J, Wiedenmayer K. 2009. Past, present, and future of Japanese encephalitis. Emerg. Infect. Dis. 15:1–7. 10.3201/eid1501.080311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanna JN, Ritchie SA, Phillips DA, Shield J, Bailey MC, Mackenzie JS, Poidinger M, McCall BJ, Mills PJ. 1996. An outbreak of Japanese encephalitis in the Torres Strait, Australia, 1995. Med. J. Aust. 165:256–260 [DOI] [PubMed] [Google Scholar]
- 3.Hanna JN, Ritchie SA, Phillips DA, Lee JM, Hills SL, van den Hurk AF, Pyke AT, Johansen CA, Mackenzie JS. 1999. Japanese encephalitis in north Queensland, Australia, 1998. Med. J. Aust. 170:533–536 [DOI] [PubMed] [Google Scholar]
- 4.Paul WS, Moore PS, Karabatsos N, Flood SP, Yamada S, Jackson T, Tsai TF. 1993. Outbreak of Japanese encephalitis on the island of Saipan, 1990. J. Infect. Dis. 167:1053–1058. 10.1093/infdis/167.5.1053 [DOI] [PubMed] [Google Scholar]
- 5.Igarashi A. 1992. Epidemiology and control of Japanese encephalitis. World Health Stat. Q. 45:299–305 [PubMed] [Google Scholar]
- 6.Tsai TF. 2000. New initiatives for the control of Japanese encephalitis by vaccination: minutes of a WHO/CVI meeting, Bangkok, Thailand, 13–15 October 1998. Vaccine 18(Suppl 2):S1–S25. 10.1016/S0264-410X(00)00037-2 [DOI] [PubMed] [Google Scholar]
- 7.Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, Hombach JM, Marfin AA, Solomon T, Tsai TF, Tsu VD, Ginsburg AS. 2011. Estimated global incidence of Japanese encephalitis: a systematic review. Bull. World Health Organ. 89:766–774, 774A–774E. 10.2471/BLT.10.085233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monath TP. 2002. Japanese encephalitis vaccines: current vaccines and future prospects. Curr. Top. Microbiol. Immunol. 267:105–138. 10.1007/978-3-642-59403-8_6 [DOI] [PubMed] [Google Scholar]
- 9.Sarkari NB, Thacker AK, Barthwal SP, Mishra VK, Prapann S, Srivastava D, Sarkari M. 2012. Japanese encephalitis (JE) part II: 14 years' follow-up of survivors. J. Neurol. 259:58–69. 10.1007/s00415-011-6131-9 [DOI] [PubMed] [Google Scholar]
- 10.Lindenbach BD, Rice CM. 2001. Flaviviridae: the viruses and their replication, p 991–1041 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 11.Unni SK, Ruzek D, Chhatbar C, Mishra R, Johri MK, Singh SK. 2011. Japanese encephalitis virus: from genome to infectome. Microbes Infect. 13:312–321. 10.1016/j.micinf.2011.01.002 [DOI] [PubMed] [Google Scholar]
- 12.Scherer WF, Buescher EL, Flemings MB, Noguchi A, Scanlon J. 1959. Ecologic studies of Japanese encephalitis virus in Japan. III. Mosquito factors. Zootropism and vertical flight of Culex tritaeniorhynchus with observations on variations in collections from animal-baited traps in different habitats. Am. J. Trop. Med. Hyg. 8:665–677 [PubMed] [Google Scholar]
- 13.van den Hurk AF, Ritchie SA, Mackenzie JS. 2009. Ecology and geographical expansion of Japanese encephalitis virus. Annu. Rev. Entomol. 54:17–35. 10.1146/annurev.ento.54.110807.090510 [DOI] [PubMed] [Google Scholar]
- 14.Zheng Y, Li M, Wang H, Liang G. 8 March 2012. Japanese encephalitis and Japanese encephalitis virus in mainland China. Rev. Med. Virol. 10.1002/rmv.1710 [DOI] [PubMed] [Google Scholar]
- 15.Solomon T, Ni H, Beasley DW, Ekkelenkamp M, Cardosa MJ, Barrett AD. 2003. Origin and evolution of Japanese encephalitis virus in southeast Asia. J. Virol. 77:3091–3098. 10.1128/JVI.77.5.3091-3098.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun SM, Cho JE, Ju YR, Kim SY, Ryou J, Han MG, Choi WY, Jeong YE. 2010. Molecular epidemiology of Japanese encephalitis virus circulating in South Korea, 1983–2005. Virol. J. 7:127. 10.1186/1743-422X-7-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nitatpattana N, Dubot-Peres A, Gouilh MA, Souris M, Barbazan P, Yoksan S, de Lamballerie X, Gonzalez JP. 2008. Change in Japanese encephalitis virus distribution, Thailand. Emerg. Infect. Dis. 14:1762–1765. 10.3201/eid1411.080542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nga PT, del Carmen Parquet M, Cuong VD, Ma SP, Hasebe F, Inoue S, Makino Y, Takagi M, Nam VS, Morita K. 2004. Shift in Japanese encephalitis virus (JEV) genotype circulating in northern Vietnam: implications for frequent introductions of JEV from Southeast Asia to East Asia. J. Gen. Virol. 85:1625–1631. 10.1099/vir.0.79797-0 [DOI] [PubMed] [Google Scholar]
- 19.Wang HY, Takasaki T, Fu SH, Sun XH, Zhang HL, Wang ZX, Hao ZY, Zhang JK, Tang Q, Kotaki A, Tajima S, Liang XF, Yang WZ, Kurane I, Liang GD. 2007. Molecular epidemiological analysis of Japanese encephalitis virus in China. J. Gen. Virol. 88:885–894. 10.1099/vir.0.82185-0 [DOI] [PubMed] [Google Scholar]
- 20.Huang K, Zhu H, Fan X, Wang J, Cheung CL, Duan L, Hong W, Liu Y, Li L, Smith DK, Chen H, Webster RG, Webby RJ, Peiris M, Guan Y. 2012. Establishment and lineage replacement of H6 influenza viruses in domestic ducks in southern China. J. Virol. 86:6075–6083. 10.1128/JVI.06389-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vu TT, Holmes EC, Duong V, Nguyen TQ, Tran TH, Quail M, Churcher C, Parkhill J, Cardosa J, Farrar J, Wills B, Lennon NJ, Birren BW, Buchy P, Henn MR, Simmons CP. 2010. Emergence of the Asian 1 genotype of dengue virus serotype 2 in Viet Nam: in vivo fitness advantage and lineage replacement in South-East Asia. PLoS Negl. Trop. Dis. 4:e757. 10.1371/journal.pntd.0000757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X-L, Liu H, Wang H-Y, Fu S-H, Liu H-Z, Zhang H-L, Li M-H, Gao X-Y, Wang J-L, Sun X-H, Lu X-J, Zhai Y-G, Meng W-S, He Y, Wang H-Q, Han N, Wei B, Wu Y-G, Feng Y, Yang D-J, Wang L-H, Tang Q, Xia G, Kurane I, Rayner S, Liang G-D. 2011. Emergence of genotype I of Japanese encephalitis virus as the dominant genotype in Asia. J. Virol. 85:9847–9853. 10.1128/JVI.00825-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuh AJ, Ward MJ, Leigh Brown AJ, Barrett AD. 5 February 2014. Dynamics of the emergence and establishment of a newly dominant genotype of Japanese encephalitis virus throughout Asia. J. Virol. 10.1128/JVI.02686-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 00:2.3:2.3.1–2.3.22. 10.1002/0471250953.bi0203s00 [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 26.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Wong WS, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22:1107–1118. 10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]
- 28.Schuh AJ, Ward MJ, Brown AJ, Barrett AD. 2013. Phylogeography of Japanese encephalitis virus: genotype is associated with climate. PLoS Negl. Trop. Dis. 7:e2411. 10.1371/journal.pntd.0002411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammed MA, Galbraith SE, Radford AD, Dove W, Takasaki T, Kurane I, Solomon T. 2011. Molecular phylogenetic and evolutionary analyses of Muar strain of Japanese encephalitis virus reveal it is the missing fifth genotype. Infect. Genet. Evol. 11:855–862. 10.1016/j.meegid.2011.01.020 [DOI] [PubMed] [Google Scholar]
- 30.Li MH, Fu SH, Chen WX, Wang HY, Guo YH, Liu QY, Li YX, Luo HM, Da W, Duo Ji DZ, Ye XM, Liang GD. 2011. Genotype v Japanese encephalitis virus is emerging. PLoS Negl. Trop. Dis. 5:e1231. 10.1371/journal.pntd.0001231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takhampunya R, Kim HC, Tippayachai B, Kengluecha A, Klein TA, Lee WJ, Grieco J, Evans BP. 2011. Emergence of Japanese encephalitis virus genotype V in the Republic of Korea. Virol. J. 8:449. 10.1186/1743-422X-8-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee E, Hall RA, Lobigs M. 2004. Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and West Nile viruses. J. Virol. 78:8271–8280. 10.1128/JVI.78.15.8271-8280.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajima S, Nerome R, Nukui Y, Kato F, Takasaki T, Kurane I. 2010. A single mutation in the Japanese encephalitis virus E protein (S123R) increases its growth rate in mouse neuroblastoma cells and its pathogenicity in mice. Virology 396:298–304. 10.1016/j.virol.2009.10.035 [DOI] [PubMed] [Google Scholar]
- 34.Evans JD, Seeger C. 2007. Differential effects of mutations in NS4B on West Nile virus replication and inhibition of interferon signaling. J. Virol. 81:11809–11816. 10.1128/JVI.00791-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muñoz-Jordán JL, Laurent-Rolle M, Ashour J, Martínez-Sobrido L, Ashok M, Lipkin WI, García-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004–8013. 10.1128/JVI.79.13.8004-8013.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu G, Gong P. 2013. Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 9:e1003549. 10.1371/journal.ppat.1003549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. 2007. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 81:4753–4765. 10.1128/JVI.02283-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanley KA, Lee JJ, Blaney JE, Jr, Murphy BR, Whitehead SS. 2002. Paired charge-to-alanine mutagenesis of dengue virus type 4 NS5 generates mutants with temperature-sensitive, host range, and mouse attenuation phenotypes. J. Virol. 76:525–531. 10.1128/JVI.76.2.525-531.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235–240. 10.1038/35065653 [DOI] [PubMed] [Google Scholar]
- 40.Harbach RE. 2007. The Culicidae (Diptera): a review of taxonomy, classification and phylogeny. Zootaxa 1668:48 [Google Scholar]
- 41.Harbach RE. 2011. Classification within the cosmopolitan genus Culex (Diptera: Culicidae): the foundation for molecular systematics and phylogenetic research. Acta Trop. 120:1–14. 10.1016/j.actatropica.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 42.Carney J, Daly JM, Nisalak A, Solomon T. 2012. Recombination and positive selection identified in complete genome sequences of Japanese encephalitis virus. Arch. Virol. 157:75–83. 10.1007/s00705-011-1143-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito M, Taira K, Itokazu K, Mori N. 2007. Recent change of the antigenicity and genotype of Japanese encephalitis viruses distributed on Okinawa Island, Japan. Am. J. Trop. Med. Hyg. 77:737–746 [PubMed] [Google Scholar]
- 44.Schuh AJ, Tesh RB, Barrett AD. 2011. Genetic characterization of Japanese encephalitis virus genotype II strains isolated from 1951 to 1978. J. Gen. Virol. 92:516–527. 10.1099/vir.0.027110-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pond SL, Frost SD, Grossman Z, Gravenor MB, Richman DD, Brown AJ. 2006. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput. Biol. 2:e62. 10.1371/journal.pcbi.0020062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennett SN, Drummond AJ, Kapan DD, Suchard MA, Munoz-Jordan JL, Pybus OG, Holmes EC, Gubler DJ. 2010. Epidemic dynamics revealed in dengue evolution. Mol. Biol. Evol. 27:811–818. 10.1093/molbev/msp285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambrechts L, Fansiri T, Pongsiri A, Thaisomboonsuk B, Klungthong C, Richardson JH, Ponlawat A, Jarman RG, Scott TW. 2012. Dengue-1 virus clade replacement in Thailand associated with enhanced mosquito transmission. J. Virol. 86:1853–1861. 10.1128/JVI.06458-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasilakis N, Weaver SC. 2008. The history and evolution of human dengue emergence. Adv. Virus Res. 72:1–76. 10.1016/S0065-3527(08)00401-6 [DOI] [PubMed] [Google Scholar]
- 49.Jansen CC, Beebe NW. 2010. The dengue vector Aedes aegypti: what comes next. Microbes Infect. 12:272–279. 10.1016/j.micinf.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 50.Lambrechts L, Scott TW, Gubler DJ. 2010. Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLoS Negl. Trop. Dis. 4:e646. 10.1371/journal.pntd.0000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nidaira M, Taira K, Okano S, Shinzato T, Morikawa T, Tokumine M, Asato Y, Tada Y, Miyagi K, Matsuda S, Itokazu K, Kudaka J, Nakamura M, Tamanaha K. 2009. Survey of Japanese encephalitis virus in pigs on Miyako, Ishigaki, Kume, and Yonaguni Islands in Okinawa, Japan. Jpn. J. Infect. Dis. 62:220–224 [PubMed] [Google Scholar]
- 52.Endy TP, Nisalak A. 2002. Japanese encephalitis virus: ecology and epidemiology. Curr. Top. Microbiol. Immunol. 267:11–48. 10.1007/978-3-642-59403-8_2 [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharya S, Chakraborty SK, Chakraborty S, Ghosh KK, Palit A, Mukherjee KK, Chakraborty MS, Tandon N, Hati AK. 1986. Density of Culex vishnui and appearance of JE antibody in sentinel chicks and wild birds in relation to Japanese encephalitis cases. Trop. Geogr. Med. 38:46–50 [PubMed] [Google Scholar]
- 54.Buescher EL, Scherer WF, Rosenberg MZ, Gresser I, Hardy JL, Bullock HR. 1959. Ecologic studies of Japanese encephalitis virus in Japan. II. Mosquito infection. Am. J. Trop. Med. Hyg. 8:651–664 [DOI] [PubMed] [Google Scholar]
- 55.Chen WJ, Dong CF, Chiou LY, Chuang WL. 2000. Potential role of Armigeres subalbatus (Diptera: Culicidae) in the transmission of Japanese encephalitis virus in the absence of rice culture on Liu-chiu islet, Taiwan. J. Med. Entomol. 37:108–113. 10.1603/0022-2585-37.1.108 [DOI] [PubMed] [Google Scholar]
- 56.Weng MH, Lien JC, Wang YM, Lin CC, Lin HC, Chin C. 1999. Isolation of Japanese encephalitis virus from mosquitoes collected in northern Taiwan between 1995 and 1996. J. Microbiol. Immunol. Infect. 32:9–13 [PubMed] [Google Scholar]
- 57.Scherer WF, Moyer JT, Izumi T, Gresser I, McCown J. 1959. Ecologic studies of Japanese encephalitis virus in Japan. VI. Swine infection. Am. J. Trop. Med. Hyg. 8:698–706 [DOI] [PubMed] [Google Scholar]
- 58.Kim HC, Klein TA, Takhampunya R, Evans BP, Mingmongkolchai S, Kengluecha A, Grieco J, Masuoka P, Kim MS, Chong ST, Lee JK, Lee WJ. 2011. Japanese encephalitis virus in culicine mosquitoes (Diptera: Culicidae) collected at Daeseongdong, a village in the demilitarized zone of the Republic of Korea. J. Med. Entomol. 48:1250–1256. 10.1603/ME11091 [DOI] [PubMed] [Google Scholar]
- 59.Lindahl J, Chirico J, Boqvist S, Thu HT, Magnusson U. 2012. Occurrence of Japanese encephalitis virus mosquito vectors in relation to urban pig holdings. Am. J. Trop. Med. Hyg. 87:1076–1082. 10.4269/ajtmh.2012.12-0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu YC, Huang YS, Chien LJ, Lin TL, Yueh YY, Tseng WL, Chang KJ, Wang GR. 1999. The epidemiology of Japanese encephalitis on Taiwan during 1966–1997. Am. J. Trop. Med. Hyg. 61:78–84 [DOI] [PubMed] [Google Scholar]
- 61.Hedlund A, Witter E, An BX. 2003. Assessment of N, P and K management by nutrient balances and flows on peri-urban smallholder farms in southern Vietnam. Eur. J. Agron. 20:71–87. 10.1016/S1161-0301(03)00076-5 [DOI] [Google Scholar]
- 62.Vagneron I. 2007. Economic appraisal of profitability and sustainability of peri-urban agriculture in Bangkok. Ecol. Econ. 61:516–529. 10.1016/j.ecolecon.2006.04.006 [DOI] [Google Scholar]
- 63.Das BP, Lal S, Saxena VK. 2004. Outdoor resting preference of Culex tritaeniorhynchus, the vector of Japanese encephalitis in Warangal and Karim Nagar districts, Andhra Pradesh. J. Vector Borne Dis. 41:32–36 [PubMed] [Google Scholar]
- 64.Mitchell CJ, Chen PS, Boreham PF. 1973. Host-feeding patterns and behaviour of 4 Culex species in an endemic area of Japanese encephalitis. Bull. World Health Organ. 49:293–299 [PMC free article] [PubMed] [Google Scholar]
- 65.Arunachalam N, Samuel PP, Hiriyan J, Rajendran R, Dash AP. 2005. Short report: observations on the multiple feeding behavior of Culex tritaeniorhynchus (Diptera: culicidae), the vector of Japanese encephalitis in Kerala in southern India. Am. J. Trop. Med. Hyg. 72:198–200 [PubMed] [Google Scholar]
- 66.Philip Samuel P, Arunachalam N, Hiriyan J, Tyagi BK. 2008. Host feeding pattern of Japanese encephalitis virus vector mosquitoes (Diptera: Culicidae) from Kuttanadu, Kerala, India. J. Med. Entomol. 45:927–932. 10.1603/0022-2585(2008)45[927:HFPOJE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 67.Self LS, Shin HK, Kim KH, Lee KW, Chow CY, Hong HK. 1973. Ecological studies on Culex tritaeniorhynchus as a vector of Japanese encephalitis. Bull. World Health Organ. 49:41–47 [PMC free article] [PubMed] [Google Scholar]
- 68.Nitatpattana N, Apiwathnasorn C, Barbazan P, Leemingsawat S, Yoksan S, Gonzalez JP. 2005. First isolation of Japanese encephalitis from Culex quinquefasciatus in Thailand. Southeast Asian J. Trop. Med. Public Health 36:875–878 [PubMed] [Google Scholar]
- 69.Seo HJ, Kim HC, Klein TA, Ramey AM, Lee JH, Kyung SG, Park JY, Cho YS, Cho IS, Yeh JY. 2013. Molecular detection and genotyping of Japanese encephalitis virus in mosquitoes during a 2010 outbreak in the Republic of Korea. PLoS One 8:e55165. 10.1371/journal.pone.0055165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sarkar A, Taraphdar D, Mukhopadhyay SK, Chakrabarti S, Chatterjee S. 2012. Molecular evidence for the occurrence of Japanese encephalitis virus genotype I and III infection associated with acute encephalitis in patients of West Bengal, India, 2010. Virol. J. 9:271. 10.1186/1743-422X-9-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Impoinvil DE, Solomon T, Schluter WW, Rayamajhi A, Bichha RP, Shakya G, Caminade C, Baylis M. 2011. The spatial heterogeneity between Japanese encephalitis incidence distribution and environmental variables in Nepal. PLoS One 6:e22192. 10.1371/journal.pone.0022192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miller RH, Masuoka P, Klein TA, Kim HC, Somer T, Grieco J. 2012. Ecological niche modeling to estimate the distribution of Japanese encephalitis virus in Asia. PLoS Negl. Trop. Dis. 6:e1678. 10.1371/journal.pntd.0001678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Foley DH, Wilkerson RC, Birney I, Harrison S, Christensen J, Rueda LM. 2010. MosquitoMap and the Mal-area calculator: new Web tools to relate mosquito species distribution with vector borne disease. Int. J. Health Geogr. 9:11. 10.1186/1476-072X-9-11 [DOI] [PMC free article] [PubMed] [Google Scholar]