ABSTRACT
Vegetatively propagated crop plants often suffer from infections with persistent RNA and DNA viruses. Such viruses appear to evade the plant defenses that normally restrict viral replication and spread. The major antiviral defense mechanism is based on RNA silencing generating viral short interfering RNAs (siRNAs) that can potentially repress viral genes posttranscriptionally through RNA cleavage and transcriptionally through DNA cytosine methylation. Here we examined the RNA silencing machinery of banana plants persistently infected with six pararetroviruses after many years of vegetative propagation. Using deep sequencing, we reconstructed consensus master genomes of the viruses and characterized virus-derived and endogenous small RNAs. Consistent with the presence of endogenous siRNAs that can potentially establish and maintain DNA methylation, the banana genomic DNA was extensively methylated in both healthy and virus-infected plants. A novel class of abundant 20-nucleotide (nt) endogenous small RNAs with 5′-terminal guanosine was identified. In all virus-infected plants, 21- to 24-nt viral siRNAs accumulated at relatively high levels (up to 22% of the total small RNA population) and covered the entire circular viral DNA genomes in both orientations. The hotspots of 21-nt and 22-nt siRNAs occurred within open reading frame (ORF) I and II and the 5′ portion of ORF III, while 24-nt siRNAs were more evenly distributed along the viral genome. Despite the presence of abundant viral siRNAs of different size classes, the viral DNA was largely free of cytosine methylation. Thus, the virus is able to evade siRNA-directed DNA methylation and thereby avoid transcriptional silencing. This evasion of silencing likely contributes to the persistence of pararetroviruses in banana plants.
IMPORTANCE We report that DNA pararetroviruses in Musa acuminata banana plants are able to evade DNA cytosine methylation and transcriptional gene silencing, despite being targeted by the host silencing machinery generating abundant 21- to 24-nucleotide short interfering RNAs. At the same time, the banana genomic DNA is extensively methylated in both healthy and virus-infected plants. Our findings shed light on the siRNA-generating gene silencing machinery of banana and provide a possible explanation why episomal pararetroviruses can persist in plants whereas true retroviruses with an obligatory genome-integration step in their replication cycle do not exist in plants.
INTRODUCTION
Viruses are often described as causing acute and persistent infections: acute virus infections cause severe disease symptoms and, in many cases, kill the host plant, whereas persistent virus infections normally cause mild symptoms and allow the host to recover from the disease; persistent virus infections can sometimes be beneficial for plant physiology (1). Depending on the environmental conditions and host plant species, the viral disease symptoms can oscillate from severe to nonvisible (recovery) and back. The molecular mechanisms underlying such oscillations in persistent virus infections are poorly understood, although an antiviral defense system based on RNA silencing has been implicated in plant recovery from DNA and RNA virus infections (2, 3).
RNA silencing, also known as RNA interference (RNAi), is an evolutionarily conserved sequence-specific mechanism which regulates gene expression and chromatin states and also defends against invasive nucleic acids such as transposons, transgenes, and viruses (4–7). It is induced by double-stranded RNA (dsRNA), which can be produced by DNA-dependent RNA polymerase II (Pol II) transcribing inverted repeats or certain genomic regions in both sense orientation and antisense orientation and, in some organisms, including plants, also by RNA-dependent RNA polymerase (RDR). In most eukaryotes, Dicer or Dicer-like (DCL) enzymes catalyze processing of dsRNA into small RNA (sRNA) duplexes, which are then sorted by Argonaute (AGO) family proteins. AGO forms the RNA-induced silencing complex (RISC) with one of the duplex strands. This single-stranded sRNA guides RISC to a complementary sequence in a target RNA. Following the complementary interaction, AGO catalyzes cleavage and/or translational repression of the target RNA, which results in posttranscriptional gene silencing (PTGS). In plants, fungi, and some animals, the RNA silencing machinery can also repress genes transcriptionally through DNA cytosine methylation and/or chromatin histone modification. In plants, transcriptional gene silencing (TGS) through sRNA-directed DNA methylation (RdDM) plays a key role in inactivation of transposons (8, 9). RdDM and TGS have also been implicated in defense against DNA geminiviruses (10) and pararetroviruses (11), although an involvement of RdDM is a matter of ongoing debate (12, 13).
Plants have evolved diverse sRNA-generating silencing pathways mediated by four distinct DCL enzymes. In Arabidopsis, DCL1 catalyzes processing of 21-to-22-nucleotide (nt) microRNAs (miRNAs) from hairpin structures of miRNA gene transcripts, while DCL4, DCL2, and DCL3 produce short interfering RNAs (siRNAs) of three major size classes (21 nt, 22 nt, and 24 nt, respectively) from perfect dsRNA precursors. DCL4 and DCL2 play a primary role in defense against RNA viruses by producing 21-nt and 22-nt viral siRNAs, respectively (14), whereas all the four DCLs generate DNA virus-derived 21-, 22-, and 24-nt siRNAs (15–18). Nuclear DCL3 generates highly abundant 24-nt viral siRNAs in Arabidopsis infected with the DNA geminivirus Cabbage leaf curl virus (CaLCuV) (17) and the pararetrovirus Cauliflower mosaic virus (CaMV) (16), which can potentially direct viral DNA methylation and TGS. However, CaMV-derived 24-nt siRNAs were not found to be associated with AGO4, the major effector protein in RdDM (16). This finding and other lines of evidence suggest that DNA viruses may evade siRNA-directed DNA methylation (13). The genetic requirements for viral siRNA biogenesis and antiviral defense have been investigated mostly in model plants such as Arabidopsis thaliana and Nicotiana benthamiana. Nonetheless, the antiviral silencing pathways appear to be conserved across the plant kingdom, since model and crop plants infected with various RNA and DNA viruses accumulate abundant viral siRNAs of the three major size classes (15–21).
However, very little is known about RNA silencing pathways and antiviral defenses in crop plants, including banana. Although the genome of Musa acuminata has been sequenced and conserved miRNAs have been identified in silico (22), banana RNA silencing genes and sRNA profiles have not been reported so far. Recently, RNAi transgenic banana plants which express inverted-repeat transgene-derived siRNAs cognate to Banana bunchy top virus (BBTV; a single-stranded DNA virus from the family Nanoviridae) were generated and shown to be resistant to BBTV infection (23). This suggests that the banana silencing machinery has the potential for antiviral defense.
Most cultivars of dessert bananas, plantains, and cooking bananas (Musa spp.) are derived from intra- or interspecific hybrids of the seedy banana species Musa acuminata (denoted the A genome) and Musa balbisiana (denoted the B genome). Banana streak disease was first described more than 50 years ago in Ivory Coast as the cause of characteristic leaf stripes on M. acuminata (24). Subsequently, the disease was recorded in most Musa-producing regions and in many Musa spp., and the causal agent was named banana streak virus (BSV) (25). BSV disease is transmitted by mealybugs (26, 27) but can also be spread by vegetative propagation of infected material. Significant variation in the severity of banana streak disease has been observed in different regions of the world, but the relative contributions of plant, virus, and environment to this variation are largely unknown (28). Banana streak Obino l'Ewai virus (BSOLV) from the triploid AAB plantain cv. Obino l'Ewai was the first sequenced BSV species associated with the disease (29). The circular 7.4-kb genome of BSOLV contains three consecutive open reading frames (ORFs), the first two encoding small (20.8-kDa and 14.5-kDa) proteins of unknown function and the third coding for a large (208-kDa) polyprotein consisting of a putative movement protein, an RNA binding coat protein (CP [analogous to retroviral Gag]), aspartyl proteinase, and a viral replicase (analogous to retroviral Pol) consisting of reverse transcriptase (RT) and RNase H domains. Based on the genome organization, protein functions, and other features, BSV is classified into genus Badnavirus (bacilliform DNA virus) of the family Caulimoviridae. This family comprises nine genera of plant pararetroviruses that replicate via reverse transcription of pregenomic RNA (pgRNA) and encapsidate open circular double-stranded (ds) genomic DNA into bacilliform or icosahedral virions (30, 31). According to an interpretation based mostly on extensive studies of CaMV from genus Caulimovirus (31, 32), the viral life cycle begins with a release of viral double-stranded DNA (dsDNA) from virions into the nucleus, where the gaps on both strands remaining after reverse transcription are repaired and the resulting covalently closed dsDNA associates with histones to form a minichromosome (episome). Pol II-mediated transcription of the circular minichromosome generates a capped and polyadenylated pgRNA with terminal repeats which is transported to the cytoplasm for translation of viral proteins and subsequently for reverse transcription. As demonstrated for CaMV and Rice tungro bacilliform virus (RTBV; genus Tungrovirus), translation of pgRNA is initiated by a shunt mechanism in which ribosomes bypass a long leader sequence containing multiple short ORFs (sORFs) and folding into a stable stem-loop structure (33–35), both features conserved in plant pararetroviruses (36). Several consecutive viral ORFs on polycistronic pgRNA are then translated by reinitiation (CaMV) (37) or leaky-scanning (RTBV) (38) mechanisms. The reverse transcription takes place in cytoplasmic inclusion bodies and presumably is initiated by CP-mediated packaging of pgRNA (39). The resulting open circular dsDNA can reenter the nucleus for the next round of transcription or be encapsidated into mature virions for movement within the host plant and for transmission to a new host plant by insect vectors. At late stages of pararetrovirus infection, plant cells accumulate high copy numbers of both circular covalently closed viral dsDNA in the nucleus and open circular dsDNA in the virions (31).
In contrast to retroviruses, pararetroviruses do not encode integrase and their life cycle does not require an integration step into the host genome. However, some plant pararetroviruses, including BSV, were able to get integrated into the host genomes, likely through a process of illegitimate recombination, and most of these integrants appear to be noninfectious relics of ancient infection events (40, 41). This is the case in the banana A genome, which contains multiple integrants of badnavirus-like sequences, but those contain only incomplete, highly rearranged, and fragmented genomes that exhibit distant similarity to the infectious BSV species described to date, limiting the possibility of giving rise to episomal virus infections (22, 40, 42, 43). In contrast, more recent BSV integrants in the B genome, including BSOLV (44), Banana streak Goldfinger virus (BSGFV) (45), Banana streak Imove virus (BSIMV) (41, 46), and, likely, Banana streak Mysore virus (BSMYV) (27), still retain infectivity. Taking the findings together, natural BSV infections of M. acuminata spp. and their intraspecific hybrids are possible only through insect transmission of a virus that had recombined out of the banana B genome in interspecific hybrids (e.g., BSOLV, BSGFV, BSIMV, or BSMYV) and/or of a nonintegrated virus persisting in Musa spp. or other hosts. Such persistent BSV species that seem to have no infectious counterparts in the genomes of Musa spp. include Banana streak Vietnam virus (BSVNV) (47) and Banana streak Cavendish virus (BSCAV) (48). The defense mechanisms induced in BSV-infected Musa spp. and viral counterdefense strategies have not been investigated so far.
In this study, we characterized the sRNA-generating silencing machinery in persistently infected banana plants and examined whether or not episomal pararetroviruses are able to evade siRNA-directed DNA methylation. We used rolling circle amplification (RCA) and sequencing of viral DNA combined with sRNA deep sequencing to reconstruct the complete genomes and siRNA profiles for six episomal BSV species (BSOLV, BSGFV, BSIMV, BSMYV, BSVNV, and BSCAV) that persisted in M. acuminata triploid (AAA) banana plants over 10 years of vegetative propagation following transmission by mealybugs. We found that BSV infection induces multiple silencing pathways generating 21-, 22-, and 24-nt viral siRNAs which can potentially be associated with AGOs to target the viral genome. Despite the accumulation of 24-nt siRNAs covering the entire virus genome in sense and antisense orientations, the bulk of viral circular covalently closed dsDNA serving as the template for pgRNA transcription in the nucleus is largely not methylated. This implies that BSV evades RdDM and thereby accumulates multiple transcriptionally active minichromosomes in infected banana plants. Such evasion of viral DNA methylation and transcriptional silencing may contribute to persistence of episomal BSV in host plants.
MATERIALS AND METHODS
Banana plants and viruses.
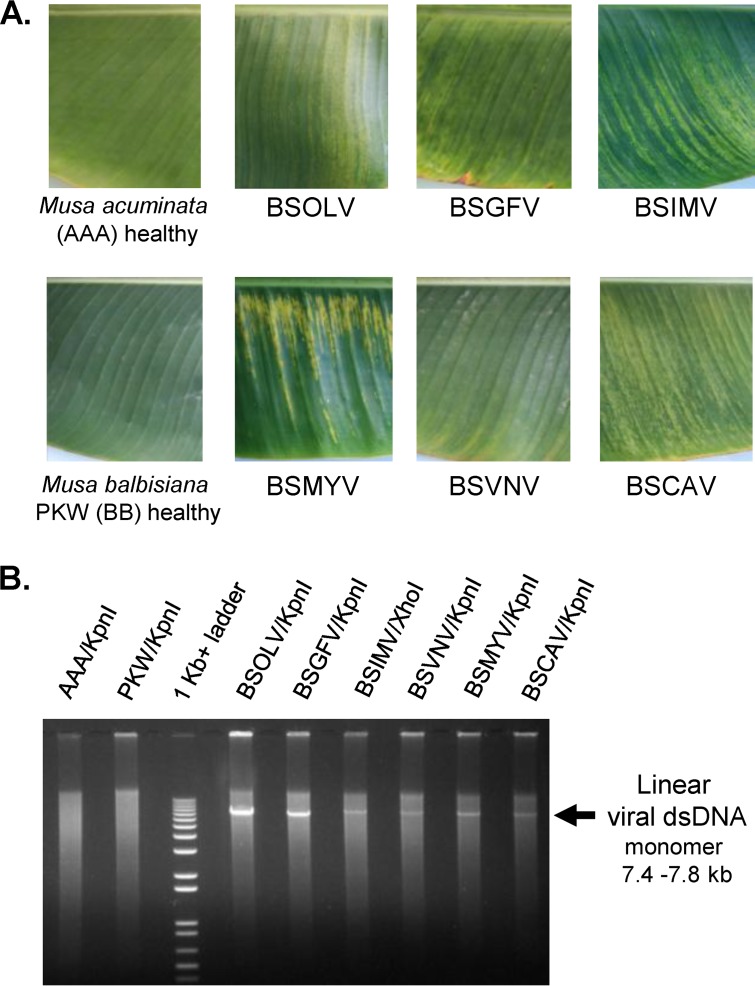
M. acuminata dwarf Cavendish plants (triploid AAA genome) were inoculated in 2000 by mealybugs (Planococcus citri) fed on fresh leaves of banana plants infected with a single BSV species each (Ben Lockhart, personal communication). Ben Lockhart's collection of the BSV-infected banana plants established in 1992 to 1993 was used as a source of inoculums for BSOLV, BSGFV, BSIMV, BSCAV, and BSMYV, while BSVNV came from plant ITC 1431 (M. acuminata siamea) (47). The resulting BSV-infected dwarf Cavendish collection was maintained at CIRAD (Montpellier, France) in a tropical greenhouse by vegetative propagation (i.e., growing of infected suckers) under the following conditions: 12 h of daylight with luminosity not exceeding 400 W/m2, 75% relative humidity, and temperatures of 26°C during the day and 24°C at night. The plants were regularly checked for BSV infection by immune capture-PCR as described earlier (49). The disease severity and virus load were oscillating depending on the season, with strong decreases in winter and intense multiplication in spring and summer. The leaves displaying the characteristic streak disease symptoms (Fig. 1A) were collected in November 2010. One of the two plants infected with BSGFV (the plant sample designated BPO-61 in Datasets S1 to S3 in the supplemental material) was found to be coinfected with BSCAV, which had accumulated much lower levels of viral DNA and viral siRNAs than BSGFV. Other plants proved to be infected with single BSV species.
FIG 1.
Leaf samples of BSV-infected and healthy control banana plants (A) and RCA analysis of viral DNA (B). Cropped pictures of the leaves from the healthy control M. acuminata cv. Cavendish (AAA) and M. balbisiana Pisang Klutuk Wulung (PKW) (BB) plants and the M. acuminata plants individually infected with BSOLV, BSGFV, BSIMV, BSMYV, BSVNV, and BSCAV are shown. Total DNA was taken for rolling circle amplification (RCA) reactions supplemented with a set of degenerate primers which are listed in Table S1 in the supplemental material. The resulting RCA products were digested with KpnI (except for BSIMV, where XhoI was used), separated on a 1% agarose gel, and stained with ethidium bromide (EtBr). As the DNA size marker, a 1-Kb+ ladder was used. The linear monomeric viral dsDNAs of expected sizes (from 7.4 to 7.8 kb) are indicated by arrows.
Total RNA and total DNA preparations.
The same banana leaf tissue was ground in liquid nitrogen and taken for preparation of total RNA and total DNA. Total RNA for both Illumina deep-sequencing (Fig. 2 and 3) and blot hybridization (Fig. 4) analyses of sRNAs was extracted from the banana leaves as described in reference 50. Briefly, 2 g of banana leaf tissue ground in liquid nitrogen was added to 10 ml of extraction buffer (100 mM Tris, 500 mM NaCl, 25 mM EDTA, 1.5% SDS, 2% polyvinylpyrrolidone [PVP], 0.7% 2-mercaptoethanol). The mixture was subjected to a vortex procedure, incubated at room temperature for 10 min, and centrifuged at 3,700 rpm for 15 min to pellet the debris. A one-third volume of precooled 5 M sodium acetate (pH 6) was added to the supernatant, mixed and incubated on ice for 30 min, and centrifuged at 10,000 rpm for 15 min at 4°C. The supernatant was treated with an equal volume of phenol-chloroform-isoamyl alcohol and centrifuged at 10,000 rpm for 10 min at 4°C. The supernatant was extracted with an equal volume of chloroform-isoamyl alcohol and centrifuged as described above. RNA in the aqueous phase was precipitated by addition of 2 to 3 volumes of cold ethanol, incubated at −70°C for 30 min, and pelleted by spinning at 10,000 rpm for 15 min at 4°C. RNA was dissolved in 500 μl of diethyl pyrocarbonate (DEPC)-treated water, extracted once again with chloroform-isoamyl alcohol, and precipitated with ethanol as described above. The RNA pellet was washed with 70% ethanol, air dried, dissolved in DEPC-treated water, and stored at −80°C until use.
FIG 2.
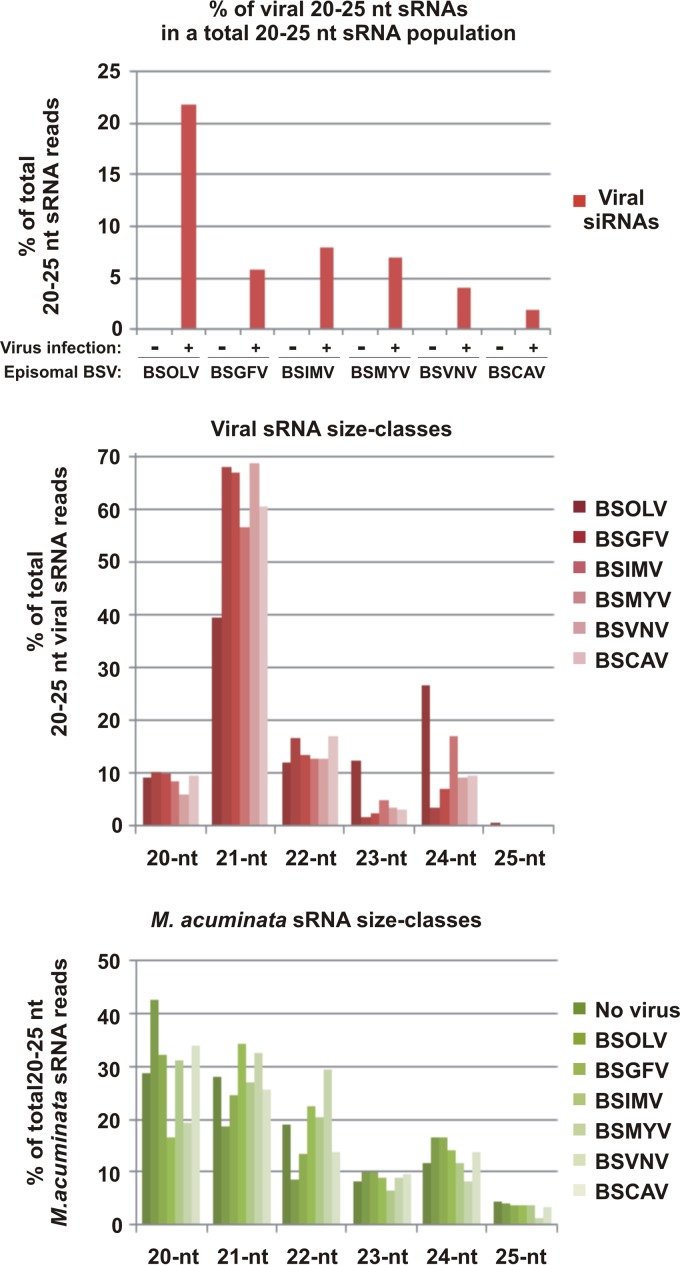
Characterization of viral and endogenous banana sRNAs in M. acuminata plants individually infected with six distinct BSV species and healthy control plants. The graphs show the percentages of 20-to-25-nt viral sRNAs in the pool of total (host and viral) 20-to-25-nt reads mapped to the M. acuminata and the genomes of the respective viruses (BSOLV, BSGFV, BSIMV, BSMYV, BSVNV, and BSCAV) with zero mismatches, the percentages of each size class of 20-to-25-nt viral sRNA reads mapped to each of the BSV species genomes with zero mismatches, and the percentages of each size class of 20-to-25-nt banana sRNA reads mapped to the genome of virus-free and BSV-infected plants with zero mismatches.
FIG 3.
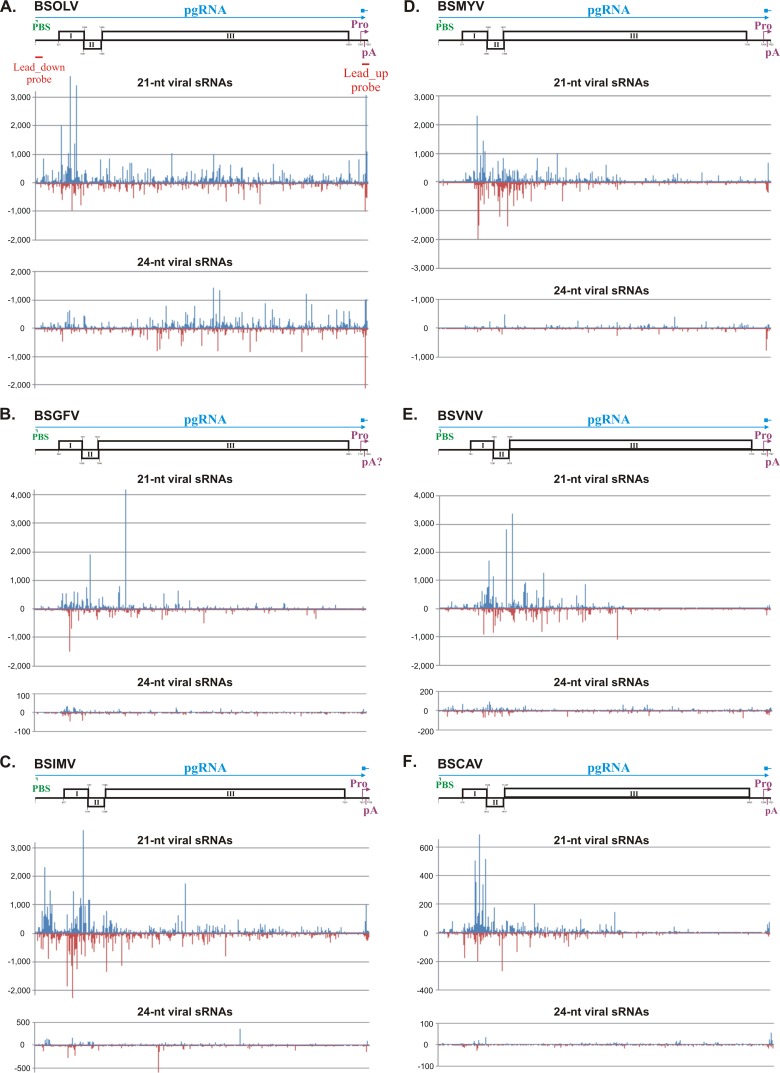
Single-nucleotide-resolution maps of 21-nt and 24-nt viral sRNAs from M. acuminata plants infected with six distinct episomal BSV species. The graphs plot the number of 21-nt and 24-nt viral sRNA reads at each nucleotide position of the reconstructed genome sequences of BSOLV (A), BSGFV (B), BSIMV (C), BSMYV (D), BSVNV (E), and BSCAV (F). Bars above the axis represent sense reads starting at each of the respective positions; those below the axis represent antisense reads ending at the respective positions (for the map details, see Dataset S3 in the supplemental material). The genome organization of each BSV species is shown schematically above the graph, starting with the Met-tRNA primer binding site (PBS) indicated in green and ending with the transcription start site indicated with a pink bent arrow (Pro), followed by the poly(A) signal (pA); the three ORFs (I, II, and III) are shown with boxes. The predicted pgRNA transcript is shown as a blue line (interrupted at the PBS). The positions of the probes used for sRNA blot hybridization (Lead-up and Lead-down; see Fig. 4) are indicated with red lines below the genome of BSOLV. Note that the putative BSOLV decoy region is located between the starts of transcription and reverse transcription, as indicated with the position of the “Lead-up probe.”
FIG 4.
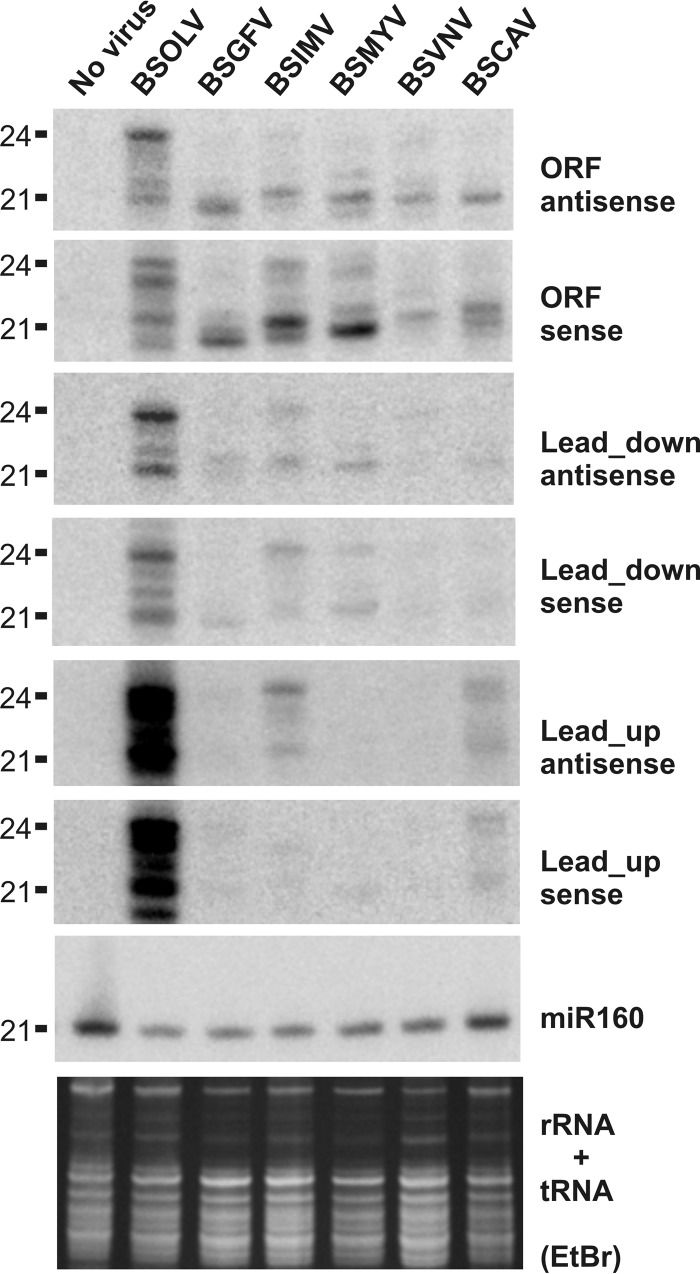
RNA blot hybridization analysis of viral and endogenous banana sRNAs in M. acuminata plants infected with six distinct BSV species. Total RNA samples from the healthy control M. acuminata plant and M. acuminata plants individually infected with BSOLV, BSGFV, BSIMV, BSMYV, BSVNV, and BSCAV were analyzed by RNA blot hybridization using 15% polyacrylamide gel electrophoresis (PAGE). The RNA blot membranes were successively hybridized with mixtures of 12 DNA oligonucleotide probes complementary to the respective viruses (2 probes per target region for each BSV species; for sequences and genome positions, see Table S1 in the supplemental material) and then to the evolutionary conserved miRNA (miR160). Positions of the BSV-specific probes Lead_up and Lead_down with respect of the reverse transcription start site are indicated in Fig. 3 for BSOLV. Ethidium bromide (EtBr) staining of ribosomal and transfer RNAs (rRNA + tRNA) is shown as a loading control. The 21-nt and 24-nt size markers are indicated. Note that sRNA mobility in 15% PAGE depends on the molecular weight: purine-rich sRNAs migrate more slowly by a ca. 0.5-to-1-nucleotide distance than pyrimidine-rich sRNAs of the same size.
Total DNA for both rolling circle amplification (Fig. 1B) and Southern blot hybridization (Fig. 5) analyses was isolated as described by Gawel and Jarret (51). In short, 100 mg of banana leaf tissues ground in liquid nitrogen was incubated with 500 μl of extraction buffer (100 mM Tris, 1.4 M NaCl, 20 mM EDTA, 2% mixed alkyl trimethyl ammonium bromide (MATAB; Sigma), 1% polyethylene glycol [PEG] 6000, 0.5% sodium sulfite) preheated up to 74°C. The slurry was mixed and incubated at 74°C for 20 min, and 500 μl of chloroform-isoamyl alcohol was added. The contents were mixed and centrifuged at 14,000 rpm for 15 min. DNA in the aqueous phase was precipitated with an equal volume of cold isopropanol and pelleted by centrifugation at 14,000 rpm for 30 min at 4°C. After a wash with 75% ethanol, the DNA pellet was dried by the use of a speed vacuum and dissolved in 100 μl of water.
FIG 5.
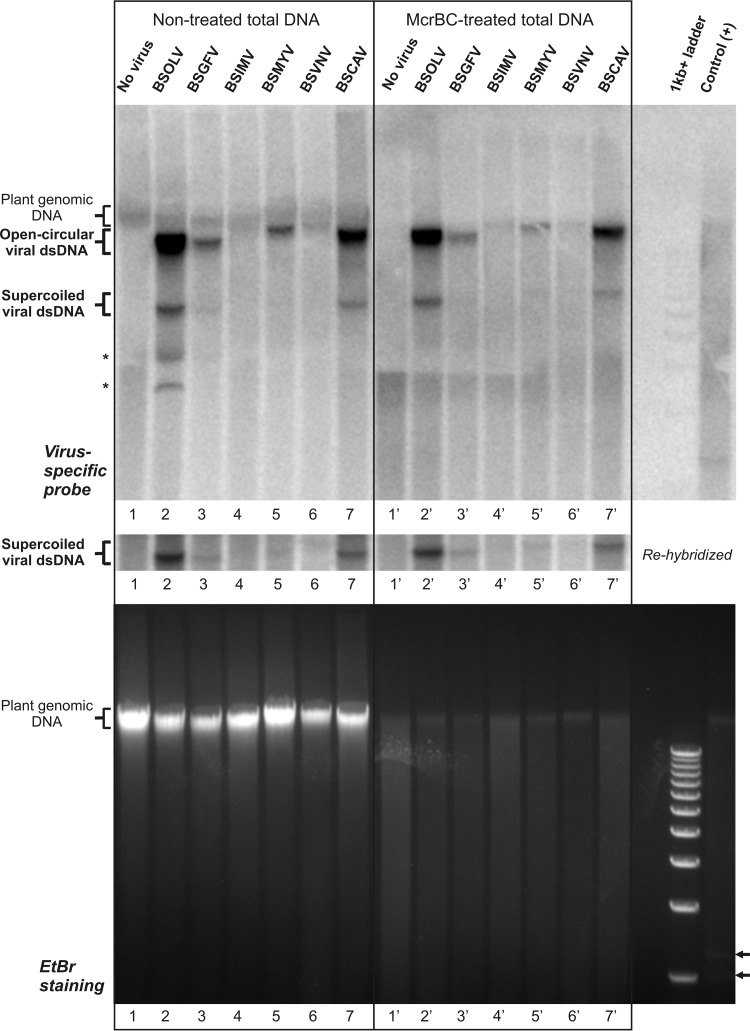
Analysis of relative accumulations of viral DNA and methylation statuses of the supercoiled and open circular forms of viral dsDNA using Southern blot hybridization. Total DNA samples from the healthy control M. acuminata plant and M. acuminata plants individually infected with BSOLV, BSGFV, BSIMV, BSMYV, BSVNV, and BSCAV were treated with the methylation-dependent enzyme McrBC and then analyzed by Southern blotting hybridization using 1% agarose gel electrophoresis as described in Materials and Methods. As a “nontreated” control, aliquots of the total DNA samples were incubated in parallel in the same buffer but without enzyme (left side of the blot). As a positive control [Control (+)], a methylated plasmid DNA was spiked in total DNA from the M. balbisiana plant (the digestion products are indicated by arrowheads in lower panel). As a DNA size marker, a 1-Kb+ ladder was used. Following separation, the gel was stained with EtBr (lower panel) and then the DNA was transferred to the membrane for hybridization with a mixture of the BSV-specific probes (see Materials and Methods). The hybridized membrane image is shown in the upper panel, with the positions of plant genomic DNA, viral dsDNA forms (open circular and supercoiled), and two McrBC-sensitive bands (asterisks) indicated. The membrane was rehybridized at 35°C to enhance the hybridization signal of the supercoiled form of viral DNA (cropped image in the middle panel).
RCA and Southern blot hybridization.
Total DNA was taken for rolling circle amplification (RCA) using an illustra TempliPhi amplification kit (GE Healthcare Life Sciences) and a set of degenerate primers (see Table S1 in the supplemental material) according to the protocol described in reference 48. Briefly, ∼25 ng of total DNA and 1 μl of degenerate primer mix (4.16 pmol/μl of each primer; see Table S1) were added to 5 μl of sample buffer from the kit and the mixture was heated to 95°C for 3 min. The mixture was cooled on ice, 5 μl of reaction buffer from the kit (premixed with 0.2 μl of bacteriophage Phi29 DNA polymerase) was added, and the reaction was allowed to proceed for 18 h at 30°C. The reaction was stopped by incubation at 65°C for 10 min, and an aliquot of the RCA products was digested with KpnI (except for BSIMV, where XhoI was used) and separated on 1% agarose gel (Fig. 1B).
For McrBC (New England BioLabs) treatment and subsequent Southern blot hybridization, 10 μg total DNA was taken and digested with 30 U of McrBC enzyme overnight at 37°C as recommended by the manufacturer. As a positive control for McrBC analysis, 0.5 μg of methylated plasmid (with one McrBC site; supplied by the manufacturer) was spiked in 10 μg of total DNA isolated from the M. balbisiana plant (see Fig. 5; arrows indicate the two products of McrBC digestion of the methylated plasmid). The nontreated total DNA samples represented in Fig. 5 were incubated in parallel under the same conditions as the McrBC-treated total DNA samples but without the McrBC enzyme.
Following the treatment with or without McrBC, the DNA of each total reaction mixture was separated in one 1% agarose gel in 1× TNE buffer (40 mM Tris-acetate, 20 mM sodium acetate, 2 mM EDTA, pH 7.5), stained with EtBr (Fig. 5, lower part), and then transferred onto a Hybond N+ membrane (Amersham). The membrane was hybridized overnight at 45°C in UltraHyb-oligo buffer (Ambion) with a mixture of probes specific for each BSV. Two 40-nt (or 41-nt) oligonucleotides of same GC content and melting point (see Table S1 in the supplemental material) were designed for each of the six BSV species, and all the 12 oligonucleotides were pooled and end labeled with P32 by the use of polynucleotide kinase for hybridization. After 16 h of hybridization, the blot was washed two times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% SDS for 30 min at 45°C and the signal was detected after 20 h to 5 days of exposure to a phosphor screen using a Molecular Imager (Typhoon FLA 7000; GE Healthcare Life Sciences). For repeated hybridizations, the membrane was stripped with 0.5× SSC–0.5% SDS for 30 min at 80°C and then with 0.1× SSC–0.5% SDS for 30 min at 80°C. To enhance the signal of the viral supercoiled DNA form, the membrane was stripped and rehybridized at 35°C. Note that, due to low titers of viral DNA, the level of supercoiled DNA for two of the six viruses analyzed (namely, BSIMV and BSVNV) was below the detection threshold.
Illumina deep-sequencing and bioinformatic analysis of viral nucleic acids.
The RCA products were fragmented by the use of a Bioruptor with settings for 350-bp fragments. The DNA fragments were separated using a 2% agarose gel, extracted, and sequenced following the Illumina protocols using an Illumina Hi-Seq 2000 Genome Analyzer (GA) (a 1-by-50-bp run in a v3 flow cell) and a TruSeq SBS kit (v3). After the removal of adaptors, the data sets of 50 reads were used for de novo reconstruction of BSV genomes and identification of single nucleotide polymorphisms (SNPs) and indels (insertions/deletions). To reconstruct viral genomes, the reads were assembled into contigs using Velvet 1.2.07 (https://www.ebi.ac.uk/∼zerbino/velvet/) (52) followed by Oases 0.2.08 (http://www.ebi.ac.uk/∼zerbino/oases/) (53). Oases contigs were merged using the Seqman module of the Lasergene DNASTAR 8.1.2 Core Suite (DNAStar, Madison, WI). SNP/indel calling and correction of errors in the viral genomes were done using Integrative Genomics Viewer (IGV; www.broadinstitute.org/igv) (54) with redundant and nonredundant reads. The reconstructed virus genomes and SNPs were further verified using the data sets of sRNA reads obtained by deep sequencing (see below).
For sRNA deep sequencing, cDNA libraries of 19-to-30-nt RNA fractions of the total RNA samples were prepared as described previously (16). The libraries were sequenced on a Hi-Seq 2000 GA using a TruSeq kit (v5). After the adaptor sequences were trimmed, the data sets of reads were mapped to the reference genome sequences of the DH-Pahang subspecies malaccensis Musa acuminata plant (22) and the six reconstructed BSV species using a Burrows-Wheeler Alignment Tool (BWA version 0.5.9) (55) with zero mismatches to each reference sequence. The results of the bioinformatics analysis of the mapped reads are summarized in Fig. 2 and 3 and Datasets S1 to S3 in the supplemental material. Reads mapping to several positions on the reference genome were attributed at random to one of them. To account for the circular BSV genome, the first 50 bases of the viral sequence were added to its 3′ end. For each reference genome or sequence and each sRNA size class (20 to 25 nt), we counted the total number of reads, the numbers of reads in the forward and reverse orientations, and the numbers of reads starting with A, C, G, and T (see Dataset S2). The single-base resolution maps of 20-, 21-, 22-, 23-, 24-, and 25-nt viral sRNAs (Fig. 3; see also Dataset S3) were generated by a map tool, MISIS (http://www.fasteris.com/apps/) (56). In these maps, for each position on the sequence (starting from the 5′ end of the reference sequence), the numbers of matches starting at this position in the forward (first base of the read) and reverse (last base of the read) orientations for each read length are given. Note that the reads mapped to the last 50 bases of the extended viral sequence were added to the reads mapped to the first 50 bases. By default, MISIS generates two maps, one with zero mismatches to the reference genome (see Dataset S3) and another with up to two mismatches. The comparison of the two maps was informative for initial identification of SNPs in the reference sequence, which could then be confirmed using the IGV tool described above.
The sequence analyses of the reconstructed BSV genomes (see Sequence Analysis S1 in the supplemental material) were performed using the following tools: NCBI nucleotide BLAST (blastn), the NCBI ORF finder with a protein BLAST option (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), the EMBOSS Needle for pairwise sequence alignment (http://www.ebi.ac.uk/Tools/psa/emboss_needle/nucleotide.html), ClustalW 2.0.12 for multiple alignment (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::clustalw-multialign), and MFold for prediction of RNA secondary structure (http://mfold.rna.albany.edu/?q = mfold/RNA-Folding-Form).
sRNA blot hybridization analysis.
sRNA blot hybridization analysis was performed as described by Blevins et al. (18) using the short DNA oligonucleotide probes listed in Table S1 in the supplemental material. For each of the several successive hybridizations shown in Fig. 4, a mixture of 12 p32-labeled DNA oligonucleotides (six pairs specific for the corresponding region of each virus) was used as a probe (see Table S1).
Nucleotide sequence accession numbers.
The NCBI accession numbers for the Montpellier (MP) isolates are as follows: for BSOLV-MP, KJ013506; for BSGFV-MP, KJ013507; for BSIMV-MP, KJ013508; for BSMYV-MP, KJ013509; for BSVNV-MP, KJ013510; and for BSCAV-MP, KJ013511.
RESULTS AND DISCUSSION
De novo reconstruction of consensus master genomes of six BSV species causing individual persistent infections in banana plants.
To begin the investigation of persistent viral infections in M. acuminata, we first reconstructed consensus master genomes of BSOLV, BSGFV, BSIMV, BSMYV, BSVNV, and BSCAV from the leaf samples of individual banana plants, which had been inoculated with single BSV species by insect transmission and then propagated vegetatively for 10 years. Each sample corresponded to the third leaf before the last emerged leaf that showed characteristic banana streak disease symptoms. Total DNA and RNA were extracted from the leaf tissues of six distinct virus-infected plants and of virus-free plants of M. acuminata triploid (AAA) cultivars and seedy M. balbisiana (BB) as controls (Fig. 1A). To amplify each viral circular genomic DNA, total DNA was subjected to rolling circle amplification (RCA) supplemented with degenerate primers specific for all the six investigated BSV species (designed based on their full-length reference genomes from NCBI GenBank; see Table S1 in the supplemental material). The resulting RCA products were analyzed by the use of restriction endonucleases. As expected, the amplified viral genomic DNA from infected plants was monomerized by single cutters and, after gel separation, appeared as a single band of linear dsDNA of genome size; no band of corresponding size was detected in the control plants (Fig. 1B). To reconstruct consensus master genomes and identify their potential variants in the viral quasispecies, the undigested RCA products were then deep sequenced using Illumina technology and the complete circular viral genomes were reconstructed de novo from the sequencing reads using bioinformatic tools (see Materials and Methods). The reconstructed genomes were validated by SNP (single nucleotide polymorphism) and indel (insertion/deletion) calling with redundant reads. In each case, several SNPs were identified (see Dataset S1 in the supplemental material), highlighting the quasispecies nature of persistent BSV species which, like all viruses, exist in “clouds” of microvariants deviating from a consensus master genome (57). Interestingly, most of the SNPs in all the BSV species were found to be “silent”: SNPs in the intergenic region did not affect conserved cis-acting elements (see below), while most of SNPs in the coding sequences did not change the encoded amino acids (most of them occurred in the wobble position of codons) and only a few SNPs resulted in amino acid change. This suggests that the majority of SNPs belong to presumably viable variants of the viral genomes. To account for potential errors of the RCA method, the master genomes and SNPs were further verified by deep sequencing and bioinformatic analysis of viral siRNAs (see Dataset S1 and below) using a siRomics approach, proven to be applicable for de novo reconstruction of consensus master genomes of RNA and DNA viruses (58), and were deposited to NCBI GenBank as isolates of the previously identified BSVs (see Materials and Methods).
Sequence analysis revealed that the reconstructed viral genomes of our BSV isolates share 93.8% to 99.7% nucleotide identities with the GenBank reference genomes (see Sequence Analysis S1A in the supplemental material). For BSOLV, BSGFV, BSIMV, BSMYV, and BSVNV, which share 99.7%, 99.6%, 99.2%, 99.7%, and 98.9% identity, respectively, the differences are rather minor (except for BSVNV; see below) and include several single nucleotide substitutions, some of which result in amino acid substitutions of the viral ORF products, and a few short indels in the intergenic region (see Sequence Analysis S1A). Some or all these differences may represent the mutations that each original virus genome had accumulated following transmission to a new host and adaptation to a changing environment during the long persistence in vegetatively propagated host plants. Consistent with this hypothesis, de novo reconstruction of a BSGFV master genome from a second persistently infected and independently propagated banana plant revealed 10 single nucleotide substitutions (see Dataset S1 in the supplemental material). Interestingly, all these substitutions were present as variants deviating from the consensus master genome in one or both BSGFV-infected plants (see Dataset S1), indicating the ongoing evolution of the viral quasispecies. Among other alterations, our isolate of BSVNV has two 1-nt deletions in ORF I, which shift the frame and thereby elongate the ORF I product by 31 amino acids (see Sequence Analysis S1A). However, the elongated ORF I overlaps with ORF II, similarly to other BSVs (see Sequence Analysis S1B), and encodes a protein similar to those encoded by its homologs from other BSVs. This suggests that the previously described BSVNV genome (47) may contain cloning or sequencing errors that resulted in the truncation of ORF I. This hypothesis remains to be validated through generation of an infectious clone of BSVNV. In the case of BSCAV, the two isolates share 93.8% nucleotide identity and have much more substantial differences in the intergenic regions and the amino acid contents of three ORFs (see Sequence Analysis S1A), which classifies our isolate as a new strain of the virus.
Further sequence analysis revealed that the reconstructed genomes preserve the cis elements predicted to regulate transcription and translation of BSV pgRNA, which include the Pol II promoter and terminator elements and the elements driving ribosome shunting and leaky scanning. For the promoter and terminator elements, the only exception is BSGFV, for which both of our isolates and the GenBank reference sequence lack a CCAAT-box upstream of the TATA box and a recognizable poly(A) signal between the transcription start site and the Met-tRNA primer binding site (see Sequence Analysis S1 in the supplemental material). In all the BSV species, the ribosome shunt configuration is preserved in the pgRNA leader region, despite the drastic differences in the lengths and nucleotide compositions of the leader sequence (see Sequence Analysis S1B). Similarly to other plant pararetroviruses (36), this configuration comprises (i) a short ORF (sORF 1) terminating at a short distance (6 or 7 nt) in front of a large stem-loop structure, (ii) a stable bottom helix of the structure, and (iii) an unstructured landing site downstream of the structure followed by the ORF I start codon (AUG or CUG) (see Sequence Analysis S1B). The shapes and stabilities of the stem-loop structure, which is bypassed by shunting ribosomes, differ between the BSV species and also differ from those in CaMV and RTBV (shown schematically in Sequence Analysis S1B), for which the ribosome shunting mechanism has been dissected (reference 35 and references therein). Remarkably, and consistent with the shunt model, the only primary sequences which were found to be nearly identical in all the six BSV leaders include (i) the Met-tRNA primer binding site, (ii) sORF 1 (of 18 nt), and (iii) a 22-nt conserved motif just downstream of sORF 1 (see Sequence Analysis S1A). sORF 1 and the downstream motif likely constitute a takeoff site for shunting ribosomes. The primary leader sequence downstream of the takeoff site elements is not conserved and contains variable numbers of sORFs (see Sequence Analysis S1A), possibly because the function of this sequence is to form a secondary structure that brings the takeoff site in close proximity to the unstructured landing site just upstream of the ORF I start codon. Furthermore, a top section of the stem-loop structure in all the BSV leaders contains a purine-rich sequence which may serve as a packaging signal, in accordance with a model for CP-mediated packaging of CaMV pgRNA (39).
Consistent with the leaky-scanning model for initiation of translation of ORF II and ORF III, the ORF I and ORF II sequences are devoid of AUG triplets, except for suboptimal start codons of the ORFs I and II themselves. ORF I contains a few sORFs only in BSCAV and BSOLV (see Sequence Analysis S1B in the supplemental material). These sORFs, however, are not expected to block downstream translation. Thus, an artificial sORF introduced in ORF II of RTBV decreased leaky-scanning-mediated translation initiation of ORF III only slightly, showing that ribosomes can reinitiate translation following a short translational event (38).
In summary, the reconstructed genomes of six episomal BSV species encode a complete set of (potentially functional) viral proteins and, with the exception of BSGFV, possess all the conserved cis elements known to be required for viral replication and gene expression. This indicates that they represent infectious master genomes. The lack of recognizable CCAAT-box and poly(A) signal in the two BSGFV isolates and the GenBank reference sequence implies that alternative mechanisms of pgRNA transcription regulation evolved in this virus.
Persistent BSV infection induces multiple siRNA-generating silencing pathways targeting the entire virus genome.
To examine the activity of sRNA-generating silencing machinery in persistently infected banana plants, a fraction of 19-to-30-nt sRNAs from the BSV-infected and noninfected banana plants was deep sequenced using Illumina technology and analyzed by mapping of sRNA reads to the banana genome and the reconstructed BSV genomes (see Materials and Methods). The sRNA mapping and counting results are detailed in Datasets S2 and S3 in the supplemental material and summarized in Fig. 2 and 3. We restricted our analysis to sRNAs ranging in size from 20 to 25 nt, the size range known to correspond to functional plant miRNAs and siRNAs. The size profile of sRNAs matching the banana genome with zero mismatches was found to be interesting, as the relative accumulation of 20-nt sRNAs was comparable to that of 21-nt sRNAs, whereas 22-nt and 24-nt sRNAs represented the third- and fourth-most-abundant classes, respectively (Fig. 2; see also Dataset S2). In contrast, the endogenous sRNA profiles in other monocot and dicot plants, e.g., rice (59), maize (60), poplar (61), and Arabidopsis (16, 17, 62), differ in that the 24-nt class (mostly populated with heterochromatic siRNAs) is predominant, followed in most cases by the 21-nt class (populated with miRNAs and secondary-phase siRNAs). The analysis of 5′ nucleotide identities of the banana sRNAs revealed strong biases in each of the four major classes, with 5′ G dominating in 20-nt sRNAs (82%), 5′ U in 21-nt sRNAs (71%) and 22-nt sRNAs (74%), and 5′ A in 24-nt sRNAs (54%). These biases suggest that sRNAs of different size classes are sorted by different AGO family proteins based in most cases on the 5′ nucleotide as established in A. thaliana (63–65). The predominance of 20-nt 5′-G sRNAs in banana suggests that they represent a novel class of plant sRNAs, the biogenesis and function of which remain to be investigated. Our preliminary results indicated that most of the 20-nt 5′-G sRNAs are derived from so-called “unanchored” sequences of the recently released M. acuminata DH Pahang genome (22) which together constitute one-third of the banana genome and cannot be anchored to any of the 11 chromosomes.
Analysis of sRNAs matching the consensus master genomes of BSV species with zero mismatches revealed that the viral sRNAs constitute a significant fraction of the total 20-to-25-nt sRNAs in BSV-infected banana plants (∼2% for BSCAV, ∼4% for BSVNV, ∼6% for BSGFV, ∼7% for BSMYV, 8% for BSIMV, and 22% for BSOLV) but not in healthy banana plants (Fig. 2; see also Dataset S2 in the supplemental material). Given the much smaller size of the virus genome (7.3 to 7.8 Kb) compared to the banana genome (∼473 Mb), banana plants produce large amounts of virus genome-derived siRNAs, especially in the case of BSOLV. The differences in the relative accumulations of viral siRNAs may reflect different proportions of the infected cells in the analyzed leaf tissues or different stages of virus replication (with different copy numbers of the viral DNA per cell) in the oscillating persistent infections. Thus, deep sequencing of sRNAs from the second plant infected with BSGFV revealed lower levels of viral siRNAs (∼2% of total 20 to 25 sRNAs; see Dataset S2). The size profile of viral sRNAs differs substantially from that of the endogenous banana sRNAs: 21-nt viral sRNAs are predominant, followed by the 22-nt class (BSGFV, BSIMV, BSVNV, and BSCAV) or the 24-nt class (BSOLV and BSMYV), whereas the 20-nt class is only the third or the fourth most abundant (Fig. 2). The relative accumulation of 25-nt viral sRNAs is negligible. Interestingly, viral 20-nt sRNAs do not exhibit any strong bias to 5′ G as observed for endogenous 20-nt sRNAs (see Dataset S2), suggesting that the banana 20-nt 5′-G sRNA pathway is not involved in targeting BSV. For most BSV species, viral siRNAs of the 21-nt and 22-nt classes exhibit bias to 5′ U, albeit the bias is less pronounced than that seen with endogenous sRNAs of these classes, while viral 24-nt sRNAs are dominated by abundant 5′ A and 5′ U species (see Dataset S2). Taking the data together, viral sRNAs can potentially be associated with multiple AGOs because of the high diversity of sRNA species in each size class (see below). The predominance of 21-nt viral sRNAs together with the underrepresentation of 24-nt viral sRNAs in BSGFV, BSIMV, BSVNV, and BSCAV infections suggested that these viruses induce mostly PTGS pathways in banana plants. These presumptive PTGS pathways of Musa acuminata would be analogous to the Arabidopsis thaliana DCL4- and DCL2-dependent pathways that generate 21-nt and 22-nt siRNAs, respectively, and play a primary role in defense against RNA viruses (14, 18). In BSOLV and BSMYV infections, however, the presence of relatively high levels of 24-nt viral sRNAs implies the involvement of a nuclear TGS pathway in defense against episomal BSVs in banana plants. In Arabidopsis, the DCL3-dependent nuclear pathway generates 24-nt siRNAs and thereby contributes together with the PTGS pathways to defense against DNA viruses, as established for the geminivirus CaLCuV and the pararetrovirus CaMV (15–18).
Analysis of single-nucleotide-resolution maps of virus-derived sRNAs revealed that nonredundant 20-to-25-nt sRNA species cover the entire circular virus genome without gaps (see the “total” column in Dataset S3 in the supplemental material). A few strand-specific gaps of more than 24 nt in the 5′ and 3′ nucleotide coverage were observed only in BSCAV, which leaves less than 100 nt not covered with the viral reads on each strand, but all these small gaps were covered on the opposite stand (see the “total_forward” and “total_reverse” columns in Dataset S3). Strand-specific gaps in sRNA coverage were not observed for other BSV species. Since BSCAV has the lowest number of total reads (47,473), whereas other viruses have much higher numbers (ranging from 134,023 reads in BSIMV to 658,896 reads in BSOLV; see Dataset S2), it could be expected that deeper sequencing of BSCAV sRNAs would close the gaps. In conclusion, the entire circular genome of BSV is covered with sense and antisense precursors of viral sRNAs. Similar findings have been reported for CaLCuV (17), grapevine geminivirus (58), and CaMV (58). For CaLCuV and other geminiviruses that transcribe circular viral DNA bidirectionally, these findings have implicated Pol II in production of sense and antisense transcripts forming dsRNA precursors of viral siRNAs (13). Indeed, the genetic evidence using CaLCuV-infected Arabidopsis mutants for silencing-related RNA polymerases ruled out involvement of Pol IV, Pol V, RDR1, RDR2, or RDR6 in the biogenesis of viral siRNAs (17, 18). In CaMV and other pararetroviruses with monodirectional Pol II transcription, the mechanism of dsRNA production remains to be investigated. For CaMV, genetic evidence indicates that Pol IV, Pol V, RDR1, RDR2, and RDR6 are not required for viral siRNA biogenesis (16, 18), suggesting that Pol II might be involved (13).
Analysis of redundant sRNA reads revealed that viral sRNA hotspots are unevenly distributed along the genome (see Fig. 3 for 21-nt and 24-nt sRNAs and Dataset S3 in the supplemental material for all the size classes). In most BSVs (except BSOLV), the hotspots of 21-nt and 22-nt viral sRNAs of both sense and antisense polarity tend to concentrate within ORF I, ORF II, and the 5′ portion of ORF III, while the 3′ half of ORF III and the promoter portion of the intergenic region upstream of the transcription start site are relatively poorly covered with 21-nt or 22-nt viral sRNAs. In contrast, 24-nt viral sRNA hotspots are more evenly distributed along the viral genomes (Fig. 3). This suggests that, in the hot spot regions, 21-nt and 22-nt viral sRNAs are produced from the more abundant dsRNA precursors which may preferentially be amplified by an RDR activity. In BSOLV, 24-nt siRNAs are of exceptionally high abundance and their hot spots are slightly depleted from the ORF I-to-ORF II region that generates the most abundant 21-nt and 22-nt viral sRNAs (Fig. 3). This implies that 24-nt and 21-to-22-nt viral sRNAs may be processed from different dsRNA precursors.
In CaMV, the pgRNA leader region between the start site for Pol II transcription and the primer binding site for reverse transcription generates the most abundant viral siRNAs of the three major size classes and both polarities (16). This region was proposed to produce a highly abundant dsRNA decoy that diverts components of the plant silencing machinery from the upstream promoter region and the downstream coding regions (13, 16). In all the BSV species, the primer binding site is located much closer to the transcription start site than in CaMV, and therefore much of the pgRNA leader sequence which contains the ribosome shunt elements and forms the large stem-loop structure is located downstream of it (see Sequence Analysis S1 in the supplemental material). The structured leader region is populated with the hotspots of 21/22-nt and 24-nt viral sRNAs only in BSIMV, while the corresponding region in other BSV species is a relatively poor source of sRNAs (Fig. 3). The latter finding indicates that a stem-loop secondary structure per se may not be a major determinant for massive production of viral sRNAs. Moreover, the high abundance of viral sRNA of both polarities implies that viral sRNA duplexes are produced from perfect dsRNA precursors rather than from the structured regions of pgRNA. In BSOLV, a “decoy” region between the transcription and reverse transcription starts spawns highly abundant 21-nt and 24-nt viral sRNAs (Fig. 3A; Lead_up). This and most other findings from the deep sequencing analysis were validated by sRNA blot hybridization analysis (Fig. 4; see Lead_up probes). Thus, BSOLV may have evolved a CaMV-like decoy strategy of silencing evasion (16), which may explain why the accumulation of BSOLV DNA is higher than that of other species (Fig. 5). However, such an evasion mechanism appears to be less efficient in persistently infected banana plants in diverting the siRNA-generating machinery from other regions of the viral genome (Fig. 3A and 4) than the CaMV decoy mechanism in Arabidopsis plants that exhibit severe CaMV disease with no recovery (16).
We noted that BSV infection alters the relative accumulations of endogenous banana sRNAs, but no clear tendency for either increased or decreased accumulation of any particular sRNA size class was observed (Fig. 2). Interestingly, BSOLV, whose sRNA size class profile resembles that of CaMV (the two species produce comparable amounts of 21-nt and 24-nt viral siRNAs), has a CaMV-like impact on the endogenous sRNA profile, boosting endogenous 24-nt sRNA production at the expense of 21-nt sRNAs (16). The biological significance of this alteration remains to be investigated.
In summary, the RNA silencing machinery of banana plants infected with BSV generates abundant 21-to-22-nt and 24-nt viral sRNAs, possibly in the cytoplasm and the nucleus, respectively. Based on the characteristic sizes, distributions along the entire genome in both orientations, and 5′ nucleotide biases, these viral sRNAs can be classified as typical siRNAs that have the potential to bind distinct AGO proteins and target viral nucleic acids for posttranscriptional and transcriptional silencing, respectively. The prevalence of 21-nt viral siRNAs with their hotspots in a 5′ portion of pgRNA in most BSV infections is reminiscent of the siRNA profile of an Evade retrotransposon activated in certain epigenetic mutants of Arabidopsis (8). Highly abundant 21-nt siRNAs derived from this retrotransposon are generated by the RDR6- and DCL4-dependent mechanism and loaded onto AGO1 and AGO2. However, these siRNAs are not effective in silencing the retrotransposon transcript, because PTGS is evaded via Gag protein-mediated packaging of the transcript into virus-like particles (8). By analogy, pararetroviral CP/Gag that encapsidates pgRNA for reverse transcription might also protect the pgRNA from viral siRNA-directed cleavage and/or translational repression. Whether or not BSV and other badnaviruses have evolved specialized suppressors of silencing remains to be investigated.
Viral circular dsDNA evades cytosine methylation in persistently infected banana plants.
To address whether or not the viral siRNAs accumulating in BSV-infected banana plants direct methylation of viral dsDNA, we exploited the cleavage activity of the McrBC methylation-dependent enzyme. This enzyme recognizes 5-methylcytosines (a hallmark of plant DNA methylation) in an RmC context (where R = A or G) and cleaves between two recognition sites separated by ∼30 to ∼3,000 bp at distance of 25 to 30 bp from one of the two sites. A circular plasmid DNA containing a single RmC site on one strand could also be linearized by McrBC (66). Thus, any circular viral dsDNA that contains at least one 5′ methylcytosine in an RmC context should be digested by McrBC. We found that each of the six BSV genomes contains numerous McrBC recognition sites. These sites can potentially be targets for methylation, presumably by a banana RdDM pathway.
Total DNA extracted from the banana leaf samples was treated overnight with McrBC and then separated on a 1% agarose gel along with total DNA aliquots of the same samples treated in parallel under the same conditions but without McrBC. As a control, a methylated plasmid was mixed with total banana DNA and the mixture was treated with McrBC. Ethidium bromide (EtBr) staining revealed that the methylated plasmid was fully digested, yielding two fragments as expected. The banana genomic DNA in the control reaction and all the other reactions was almost fully digested by McrBC (Fig. 5; compare the McrBC-treated and nontreated sample results). This indicates that McrBC enzymatic activity is not inhibited by total banana genomic DNA and that the bulk of banana DNA is extensively methylated. The smear of McrBC digestion products (Fig. 5) suggests that the cytosine methylation sites are scattered along the banana genome.
The methylation status of viral DNA was then evaluated by Southern blotting hybridization using a mixture of six pairs of DNA oligonucleotide probes with equal annealing temperatures, each pair being designed to hybridize specifically to one of the six BSV species. This experimental design allows simultaneous detection of major forms of viral DNA of all the six BSV species and measurement of their relative levels of accumulation. The results revealed two major forms of circular viral dsDNA, the more abundant open circular dsDNA and the less abundant covalently closed (supercoiled) dsDNA, before and after McrBC treatment. Compared to the banana genomic DNA, both major forms of viral DNA appeared to be resistant to McrBC (Fig. 5, upper panel). This indicates that the major fraction of viral genomic DNA is not methylated. More specifically, the resistance to McrBC digestion of the supercoiled form of viral dsDNA, which is clearly detectable in BSOLV, BSCAV, BSGFV, and BSMYV (Fig. 5, cropped image in the middle panel), suggests that viral minichromosomes accumulating in the nucleus for Pol II-mediated transcription of pgRNA are largely not methylated. We cannot exclude the possibility that a fraction of the episomal DNA is methylated. In fact, some reduction in open circular dsDNA levels after the enzymatic treatment was observed (Fig. 5). However, this form of viral DNA is packaged in the virions following reverse transcription in the cytoplasm and cannot be methylated by the nuclear RdDM machinery. A potential nonspecific activity of McrBC during the overnight incubation may account for minor loss of viral DNA. Since no substantial decrease in the ratio of the supercoiled form to the open circular form was observed (Fig. 5), this argues for little (if any) methylation of the supercoiled DNA. It is difficult to evaluate the methylation status of episomal DNA in BSIMV and BSVNV, for which the supercoiled form was barely detectable even under low-stringency hybridization and washing conditions (cropped portion in the middle panel of Fig. 5). We expect, however, that if these two low-titer viruses were to replicate more actively and accumulate supercoiled DNA at detectable levels, this DNA could also be largely unmethylated, like the supercoiled DNA for the four higher-titer viruses.
The Southern blot hybridization analysis combined with the viral sRNA profiles shows that the relative accumulation of viral DNA does not correlate with the relative abundance of viral siRNAs. Indeed, BSOLV, spawning siRNAs that were 10 times more abundant than those spawned by BSCAV (Fig. 2), accumulated viral DNA at an abundance only three times greater than that seen with BSCAV. The other four BSV species produced larger amounts of viral siRNAs than BSCAV and accumulated smaller amounts of viral DNA than BSCAV (Fig. 5), but these amounts of DNA differed dramatically, unlike the amounts of viral siRNAs, which were comparable. Finally, BSVNV, with barely detectable levels of viral DNA, accumulated viral siRNAs that were only five times less abundant than those seen with BSOLV, which accumulated the largest amounts of DNA. This implies that viral siRNAs do not necessarily restrict accumulation of viral DNA, although the total abundance of viral siRNAs may not reflect the proportion of viral siRNAs incorporated into functional RISCs. Moreover, the fact that similar ratios of the supercoiled form to the open circular form were observed in BSOLV and BSCAV, which produced the largest and the smallest amounts of siRNAs, respectively, indicates that viral siRNAs do not influence the distribution of viral DNA between the cytoplasm (populated with the open circular form) and the nucleus (populated mostly with the supercoiled form). Therefore, the efficiency of viral siRNA production (or the relative abundance of 24-nt viral siRNAs) does not appear to affect the viral replication cycle, in which the nucleus supplies viral pgRNA to the cytoplasm for reverse transcription whereas the cytoplasm supplies the open circular viral DNA to the nucleus for repair (i.e., production of covalently closed, supercoiled DNA) and subsequent Pol II-mediated transcription of pgRNA on the covalently closed DNA (reviewed in reference 13).
In the case of BSOLV (but not other BSVs), two additional bands of viral DNA were detected which migrated faster than the supercoiled form, and both bands disappeared after the McrBC treatment (Fig. 5; asterisks). Note that these bands cannot be an artifact of DNA overload, since equal amounts of total DNA were taken for treatment in the presence and absence of McrBC followed by Southern blotting hybridization. The nature of these bands and why these are the only apparently methylated forms of viral DNA remain to be investigated. In geminiviruses, the only form of viral DNA which contains a substantial proportion of methylated cytosines is heterogeneous linear dsDNA, a byproduct of recombination-dependent replication (12, 13). Pararetroviruses are not known to exploit this replication mechanism.
In conclusion, most of the circular covalently closed viral dsDNA in BSV-infected M. acuminata plants is nonmethylated. Thus, multiple BSV minichromosomes appear to evade siRNA-directed DNA methylation in the nucleus and thereby retain the potential for active Pol II transcription. One can argue that 24-nt viral siRNA levels are not sufficient to exert DNA methylation. However, the relatively highly abundant 24-nt viral siRNAs accumulating in BSOLV-infected plants, which cover the entire virus genome in both orientations, also fail to direct methylation of viral DNA. The mechanism of RdDM evasion by episomal BSV remains to be investigated. In Arabidopsis, the establishment and maintenance of cytosine methylation at the RdDM loci require Pol V and Pol IV, respectively. Pol V generates a scaffold transcript and recruits de novo methyltransferase through interaction with 24-nt siRNA-AGO4 complexes, while Pol IV and RDR2 together generate dsRNA precursors of 24-nt siRNAs (reviewed in reference 13). Genetic evidence indicates that Pol V, Pol IV, and RDR2 are not required for the biogenesis of 24-nt viral siRNAs in CaMV-infected Arabidopsis (16, 18), which effectively uncouples the viral siRNAs from the RdDM machinery (13). This may explain the lack of detectable cytosine methylation of episomal viral DNA in CaMV-infected turnip plants (11) or in kohlrabi plants that recovered from CaMV disease symptoms (2). Note, however, that those earlier studies of CaMV made use of methylation-sensitive enzymes, which would report cytosine methylation only at the respective enzyme recognition sites. Investigation of genetic requirements for viral siRNA biogenesis in banana plants is currently difficult, because no information on the banana RNA silencing genes and no respective gene mutant lines are available. Our analysis of the viral and endogenous sRNA profiles and the finding that the banana genome is extensively methylated suggest the existence of conserved nuclear and cytoplasmic components of RNA silencing machinery that can mediate both PTGS and RdDM/TGS in banana, similarly to other plant species. Our finding that the banana sRNA population contains a dominant 20-nt 5′-G RNA class which is absent in the viral siRNA population raises the intriguing possibility that those sRNAs might be indirectly involved in the establishment or maintenance of DNA cytosine methylation in banana plants.
The findings reported here may also be relevant for further understanding and better control of the banana streak disease during industrial banana cultivation, but it will be important to investigate the banana silencing machinery and BSV infections under open field conditions and in other banana genotypes.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by the Swiss National Science Foundation (31003A_143882/1 to M.M.P.), the European Commission Marie Curie fellowship (PIIF-237493-SUPRA to R.R.), the European Cooperation in Science and Technology (COST) action FA0806 (SER no. C09.0176 to L.F. and M.M.P.), and the CIRAD (Ph.D. grant to P.-O.D).
We thank Thomas Boller for supporting research of the M.M.P. group at the University of Basel and Ben Lockhart, who performed the mealybug-mediated BSV infection of all the banana plants used in this study.
R.R., M.M.P., M.-L.I.-C., M.C., and L.F. designed the research, R.R., J.S., M.C., P.-O.D., and N.L. performed the research, J.S., M.M.P., R.R., M.C., and M.-L.I.-C. analyzed the data, and M.M.P. wrote the paper.
Footnotes
Published ahead of print 23 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01496-14.
REFERENCES
- 1.Roossinck MJ. 2013. Plant virus ecology. PLoS Pathog. 9:e1003304. 10.1371/journal.ppat.1003304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Covey SN, Al-Kaff NS, Lángara A, Turner DS. 1997. Plants combat infection by gene silencing. Nature 385:781–782. 10.1038/385781a0 [DOI] [Google Scholar]
- 3.Ratcliff FG, MacFarlane SA, Baulcombe DC. 1999. Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11:1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10:94–108. 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11:204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding SW, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426. 10.1016/j.cell.2007.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marí-Ordóñez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O. 2013. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45:1029–1039. 10.1038/ng.2703 [DOI] [PubMed] [Google Scholar]
- 9.Panda K, Slotkin RK. 2013. Proposed mechanism for the initiation of transposable element silencing by the RDR6-directed DNA methylation pathway. Plant Signal. Behav. 8:e25206. 10.4161/psb.25206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raja P, Wolf JN, Bisaro DM. 2010. RNA silencing directed against geminiviruses: post-transcriptional and epigenetic components. Biochim. Biophys. Acta 1799:337–351. 10.1016/j.bbagrm.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 11.Al-Kaff NS, Covey SN, Kreike MM, Page AM, Pinder R, Dale PJ. 1998. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113–2115. 10.1126/science.279.5359.2113 [DOI] [PubMed] [Google Scholar]
- 12.Paprotka T, Deuschle K, Metzler V, Jeske H. 2011. Conformation-selective methylation of geminivirus DNA. J. Virol. 85:12001–12012. 10.1128/JVI.05567-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pooggin MM. 2013. How can plant DNA viruses evade siRNA-directed DNA methylation and silencing? Int. J. Mol. Sci. 14:15233–15259. 10.3390/ijms140815233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68–71. 10.1126/science.1128214 [DOI] [PubMed] [Google Scholar]
- 15.Akbergenov R, Si-Ammour A, Blevins T, Amin I, Kutter C, Vanderschuren H, Zhang P, Gruissem W, Meins F, Jr, Hohn T, Pooggin MM. 2006. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 34:462–471. 10.1093/nar/gkj447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blevins T, Rajeswaran R, Aregger M, Borah BK, Schepetilnikov M, Baerlocher L, Farinelli L, Meins F, Jr, Hohn T, Pooggin MM. 2011. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 39:5003–5014. 10.1093/nar/gkr119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aregger M, Borah BK, Seguin J, Rajeswaran R, Gubaeva EG, Zvereva AS, Windels D, Vazquez F, Blevins T, Farinelli L, Pooggin MM. 2012. Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 8:e1002941. 10.1371/journal.ppat.1002941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Jr, Hohn T, Pooggin MM. 2006. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34:6233–6246. 10.1093/nar/gkl886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donaire L, Wang Y, Gonzalez-Ibeas D, Mayer KF, Aranda MA, Llave C. 2009. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392:203–214. 10.1016/j.virol.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 20.Pantaleo V, Saldarelli P, Miozzi L, Giampetruzzi A, Gisel A, Moxon S, Dalmay T, Bisztray G, Burgyan J. 2010. Deep sequencing analysis of viral short RNAs from an infected Pinot Noir grapevine. Virology 408:49–56. 10.1016/j.virol.2010.09.001 [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Wang Y, Guo W, Xie Y, Xie Q, Fan L, Zhou X. 2011. Characterization of small interfering RNAs derived from the geminivirus/betasatellite complex using deep sequencing. PLoS One 6:e16928. 10.1371/journal.pone.0016928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, Da Silva C, Jabbari K, Cardi C, Poulain J, Souquet M, Labadie K, Jourda C, Lengellé J, Rodier-Goud M, Alberti A, Bernard M, Correa M, Ayyampalayam S, Mckain MR, Leebens-Mack J, Burgess D, Freeling M, Mbéguié-A-Mbéguié D, Chabannes M, Wicker T, Panaud O, Barbosa J, Hribova E, Heslop-Harrison P, Habas R, Rivallan R, Francois P, Poiron C, Kilian A, Burthia D, Jenny C, Bakry F, Brown S, Guignon V, Kema G, Dita M, Waalwijk C, Joseph S, Dievart A, et al. 2012. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–217. 10.1038/nature11241 [DOI] [PubMed] [Google Scholar]
- 23.Shekhawat UK, Ganapathi TR, Hadapad AB. 2012. Transgenic banana plants expressing small interfering RNAs targeted against viral replication initiation gene display high-level resistance to banana bunchy top virus infection. J. Gen. Virol. 93:1804–1813. 10.1099/vir.0.041871-0 [DOI] [PubMed] [Google Scholar]
- 24.Yot-Dauthy D, Bové JM. 1966. Mosaïque du bananier. Identification et purification de diverses souches du virus. Fruit 21:449–465 [Google Scholar]
- 25.Lockhart BEL. 1986. Purification and serology of a bacilliform virus associated with banana streak disease. Phytopathology 76:995–999. 10.1094/Phyto-76-995 [DOI] [Google Scholar]
- 26.Kubiriba J, Legg JP, Tushemereirwe W, Adipala E. 2001. Vector transmission of Banana streak virus in the screenhouse in Uganda. Ann. Appl. Biol. 139:37–43. 10.1111/j.1744-7348.2001.tb00128.x [DOI] [Google Scholar]
- 27.Geering AD, Pooggin MM, Olszewski NE, Lockhart BE, Thomas JE. 2005. Characterisation of Banana streak Mysore virus and evidence that its DNA is integrated in the B genome of cultivated Musa. Arch. Virol. 150:787–796. 10.1007/s00705-004-0471-z [DOI] [PubMed] [Google Scholar]
- 28.Geering ADW, Thomas JE. 2002. Banana streak virus. Web descriptions of plant viruses, description no. 390. Assoc. Appl. Biol. http://www.dpvweb.net/dpv/showdpv.php?dpvno=390
- 29.Harper G, Hull R. 1998. Cloning and sequence analysis of banana streak virus DNA. Virus Genes 17:271–278. 10.1023/A:1008021921849 [DOI] [PubMed] [Google Scholar]
- 30.Hull R. 2007. Caulimoviridae (plant pararetroviruses). eLS, John Wiley & Sons Ltd., Chichester, United Kingdom: http://www.els.net. 10.1002/9780470015902.a0000746.pub2 [DOI] [Google Scholar]
- 31.Hohn T, Rothnie H. 2013. Plant pararetroviruses: replication and expression. Curr. Opin. Virol. 3:621–628. 10.1016/j.coviro.2013.08.013 [DOI] [PubMed] [Google Scholar]
- 32.Hohn T. 2013. Plant pararetroviruses: interactions of cauliflower mosaic virus with plants and insects. Curr. Opin. Virol. 3:629–638. 10.1016/j.coviro.2013.08.014 [DOI] [PubMed] [Google Scholar]
- 33.Fütterer J, Kiss-László Z, Hohn T. 1993. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 73:789–802. 10.1016/0092-8674(93)90257-Q [DOI] [PubMed] [Google Scholar]
- 34.Pooggin MM, Ryabova LA, He X, Fütterer J, Hohn T. 2006. Mechanism of ribosome shunting in Rice tungro bacilliform pararetrovirus. RNA 12:841–850. 10.1261/rna.2285806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pooggin MM, Fütterer J, Hohn T. 2008. Cross-species functionality of pararetroviral elements driving ribosome shunting. PLoS One 3:e1650. 10.1371/journal.pone.0001650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pooggin MM, Fütterer J, Skryabin KG, Hohn T. 1999. A short open reading frame terminating in front of a stable hairpin is the conserved feature in pregenomic RNA leaders of plant pararetroviruses. J. Gen. Virol. 80:2217–2228 [DOI] [PubMed] [Google Scholar]
- 37.Ryabova LA, Pooggin MM, Hohn T. 2002. Viral strategies of translation initiation: ribosomal shunt and reinitiation. Prog. Nucleic Acid Res. Mol. Biol. 72:1–39. 10.1016/S0079-6603(02)72066-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fütterer J, Rothnie HM, Hohn T, Potrykus I. 1997. Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning. J. Virol. 71:7984–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerra-Peraza O, de Tapia M, Hohn T, Hemmings-Mieszczak M. 2000. Interaction of the cauliflower mosaic virus coat protein with the pregenomic RNA leader. J. Virol. 74:2067–2072. 10.1128/JVI.74.5.2067-2072.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geering AD, Olszewski NE, Harper G, Lockhart BE, Hull R, Thomas JE. 2005. Banana contains a diverse array of endogenous badnaviruses. J. Gen. Virol. 86:511–520. 10.1099/vir.0.80261-0 [DOI] [PubMed] [Google Scholar]
- 41.Chabannes M, Iskra-Caruana ML. 2013. Endogenous pararetroviruses–a reservoir of virus infection in plants. Curr. Opin. Virol. 3:615–620. 10.1016/j.coviro.2013.08.012 [DOI] [PubMed] [Google Scholar]
- 42.Geering AD, Olszewski NE, Dahal G, Thomas JE, Lockhart BE. 2001. Analysis of the distribution and structure of integrated Banana streak virus DNA in a range of Musa cultivars. Mol. Plant Pathol. 2:207–213. 10.1046/j.1464-6722.2001.00071.x [DOI] [PubMed] [Google Scholar]
- 43.Gayral P, Iskra-Caruana ML. 2009. Phylogeny of Banana Streak Virus reveals recent and repetitive endogenization in the genome of its banana host (Musa sp.). J. Mol. Evol. 69:65–80. 10.1007/s00239-009-9253-2 [DOI] [PubMed] [Google Scholar]
- 44.Chabannes M, Baurens FC, Duroy PO, Bocs S, Vernerey MS, Rodier-Goud M, Barbe V, Gayral P, Iskra-Caruana ML. 2013. Three infectious viral species lying in wait in the banana genome. J. Virol. 87:8624–8637. 10.1128/JVI.00899-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gayral P, Noa-Carrazana JC, Lescot M, Lheureux F, Lockhart BE, Matsumoto T, Piffanelli P, Iskra-Caruana ML. 2008. A single Banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus. J. Virol. 82:6697–6710. 10.1128/JVI.00212-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geering AD, Parry JN, Thomas JE. 2011. Complete genome sequence of a novel badnavirus, banana streak IM virus. Arch. Virol. 156:733–737. 10.1007/s00705-011-0946-7 [DOI] [PubMed] [Google Scholar]
- 47.Lheureux F, Laboureau N, Muller E, Lockhart BE, Iskra-Caruana ML. 2007. Molecular characterization of banana streak acuminata Vietnam virus isolated from Musa acuminata siamea (banana cultivar). Arch. Virol. 152:1409–1416. 10.1007/s00705-007-0946-9 [DOI] [PubMed] [Google Scholar]
- 48.James AP, Geijskes RJ, Dale JL, Harding RM. 2011. Molecular characterisation of six badnavirus species associated with leaf streak disease of banana in East Africa. Ann. Appl. Biol. 158:346–353. 10.1111/j.1744-7348.2011.00466.x [DOI] [Google Scholar]
- 49.Le Provost G, Iskra-Caruana ML, Acina I, Teycheney PY. 2006. Improved detection of episomal Banana streak viruses by multiplex immunocapture PCR. J. Virol. Methods 137:7–13. 10.1016/j.jviromet.2006.05.021 [DOI] [PubMed] [Google Scholar]
- 50.Liu JJ, Goh CJ, Loh CS, Liu P, Pua EC. 1998. A method for isolation of total RNA from fruit tissues of banana. Plant Mol. Biol. Rep. 16:1–6. 10.1023/A:1017158311412 [DOI] [Google Scholar]
- 51.Gawel NJ, Jarret RL. 1991. Chloroplast DNA restriction fragment length polymorphisms (RFLPs) in Musa species. Theor. Appl. Genet. 81:783–786 [DOI] [PubMed] [Google Scholar]
- 52.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz MH, Zerbino DR, Vingron M, Birney E. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092. 10.1093/bioinformatics/bts094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14:178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seguin J, Otten P, Baerlocher L, Farinelli L, Pooggin MM. 2014. MISIS: a bioinformatics tool to view and analyze maps of small RNAs derived from viruses and genomic loci generating multiple small RNAs. J. Virol. Methods 195:120–122. 10.1016/j.jviromet.2013.10.013 [DOI] [PubMed] [Google Scholar]
- 57.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76:159–216. 10.1128/MMBR.05023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seguin J, Rajeswaran R, Malpica-López N, Martin RR, Kasschau K, Dolja VV, Otten P, Farinelli L, Pooggin MM. 2014. De novo reconstruction of consensus master genomes of plant RNA and DNA viruses from siRNAs. PLoS One 9:e88513. 10.1371/journal.pone.0088513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ. 2011. Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23:4185–4207. 10.1105/tpc.111.089045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiao Y, Song W, Zhang M, Lai J. 2011. Identification of novel maize miRNAs by measuring the precision of precursor processing. BMC Plant Biol. 11:141. 10.1186/1471-2229-11-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klevebring D, Street NR, Fahlgren N, Kasschau KD, Carrington JC, Lundeberg J, Jansson S. 2009. Genome-wide profiling of populus small RNAs. BMC Genomics 10:620. 10.1186/1471-2164-10-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. 2007. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5:e57. 10.1371/journal.pbio.0050057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, Chen S, Hannon GJ, Qi Y. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116–127. 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. 2008. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133:128–141. 10.1016/j.cell.2008.02.033 [DOI] [PubMed] [Google Scholar]
- 65.Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. 2010. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22:321–334. 10.1105/tpc.109.072199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Panne D, Raleigh EA, Bickle TA. 1999. The McrBC endonuclease translocates DNA in a reaction dependent on GTP hydrolysis. J. Mol. Biol. 290:49–60. 10.1006/jmbi.1999.2894 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.