ABSTRACT
The noncoding regions (NCRs) of the eight-segmented viral RNAs (vRNAs) of influenza A virus consist of the highly conserved promoter region and the nonconserved segment-specific NCRs at both the 3′ and 5′ ends. The roles of the segment-specific NCRs of the eight segments have been extensively studied. However, the diversities in the same region of the two subtype-determinant hemagglutinin (HA) and neuraminidase (NA) segments have received little attention. In this study, we bioinformatically analyzed all available NCRs of HA and NA vRNAs of influenza A viruses and found that nucleotides in the segment-specific NCRs of HA and NA vRNAs are subtype specific and vary significantly in sequence and length at both the 3′ and 5′ ends among different subtypes. We then systematically studied the biological significance of the HA subtype-specific NCRs (HA ssNCRs) of the common HA subtypes (H1 to H7 and H9) in the context of the WSN (H1N1) reverse genetics system. We found that the HA ssNCRs play a critical role in HA vRNA virion incorporation. Upon HA vRNA incorporation, the 3′-end HA ssNCR plays a more critical role than the 5′-end HA ssNCR, and no stringent compatibility between the two ends is required. Furthermore, our data imply that, in addition to a particular nucleotide(s), the length of the HA ssNCR is involved in regulating HA vRNA incorporation efficiency. These results provide new insights into the HA segment virion incorporation that is critical for the emergence of epidemic and pandemic influenza A virus strains.
IMPORTANCE The nonconserved noncoding regions (NCRs) of the vRNAs of influenza A virus have been extensively studied, whereas the diversities in the nonconserved NCRs of the two subtype-determinant segments hemagglutinin (HA) and neuraminidase (NA) have received little attention. In this study, we bioinformatically analyzed all available NCRs of HA and NA vRNAs and discovered that the HA and NA vRNAs contain key subtype signatures in the NCRs. Our functional studies of the HA subtype-specific NCRs (HA ssNCRs) of the common HA subtypes in the context of WSN virus (H1N1) demonstrated that the HA ssNCR modulates virus replication efficiency by influencing HA segment virion incorporation. Moreover, we revealed important features of the HA ssNCR in determining HA vRNA incorporation efficiency. These data not only show new genetic characteristics of influenza A viruses, but also provide further evidence for understanding the selective genome packaging of influenza virus required for the emergence of epidemic and pandemic influenza virus strains.
INTRODUCTION
Influenza A viruses belong to the family Orthomyxoviridae of RNA viruses. Based on their two surface glycoproteins—hemagglutinin (HA) and neuraminidase (NA)—influenza A viruses are further divided into different subtypes (1). Among at least 16 HA and 9 NA subtypes, the subtypes H1 to H7 and H9 are commonly found in humans or birds. The high mutation rate of the virus and frequent reassortment among different influenza A virus strains pose great difficulties in preventing influenza epidemics and pandemics (2). Recent outbreaks of avian virus (H5N1 and H7N9) infection in humans further highlight the importance of understanding the molecular basis for influenza A virus genome packaging and reassortment, in particular for the two subtype-determinant HA and NA segments.
The influenza A virus genome (viral RNA [vRNA]) is composed of eight single-stranded negative-sense RNA segments, designated PB2, PB1, PA, HA, NP, NA, M, and NS. Each RNA segment is incorporated into virions as viral ribonucleoprotein complexes (vRNPs) (1). Within the vRNP complex, the 3′ and 5′ ends of each vRNA join together and bind to the viral heterotrimeric RNA-dependent RNA polymerase complex that is formed by PB1, PB2, and PA. The rest of the RNA wraps around a helical backbone formed by multiple copies of nucleoprotein (NP) (3, 4). The vRNP is the minimal unit for viral RNA transcription (vRNA→mRNA) and replication (vRNA↔cRNA) to occur in the host cell nucleus (1). The noncoding region (NCR) of each RNA segment is composed of the highly conserved promoter region followed by the nonconserved noncoding region at both the 3′ and 5′ ends (1, 5). The highly conserved viral RNA promoter is formed by 12 3′-terminal nucleotides (nt) and 13 5′-terminal nucleotides in a special secondary structure that is recognized by the viral RNA polymerase to initiate viral RNA transcription and replication (6, 7). The nonconserved regions are located between the promoter region and the start or stop codon, which are of variable lengths and nucleotide compositions at the 3′ and 5′ ends in different RNA segments. A stretch of uridines near the 5′-end nonconserved region is present in all eight RNA segments and serves as a polyadenylation site for viral mRNA polyadenylation (1, 8). Nucleotides in the nonconserved region are in most instances highly conserved in sequence and length for the same segment of different influenza A virus strains. Therefore, these regions are also called segment-specific NCRs (9).
It has been reported that the segment-specific NCRs play multiple roles in the life cycle of influenza A virus. Earlier studies indicated that mutations in the nonconserved NCRs significantly affected vRNA synthesis (10, 11). More importantly, the 3′ and 5′ NCRs, together with the terminal coding regions of each RNA segment, have been proposed to serve as packaging signals in the selective genome-packaging model (11–15). The cis-segment-specific packaging signals for each segment of H1N1 strain WSN or PR8 have been identified experimentally (16–25), although the precise mechanism is still poorly understood. Recently, it has been proposed that the packaging signal of the NP segment could be divided into two distinct signals, the segment-specific NCR, serving as the incorporation signal, and the terminal coding sequence, serving as the bundling signal (26).
Unlike the other six segments, the two subtype-determinant HA and NA segments have been previously shown to contain different nucleotide sequences of various lengths in the segment-specific NCRs (27), but this observation has received little attention. In this study, we performed systematic bioinformatics analyses of all available noncoding sequences of HA and NA RNA segments in the influenza virus databases. Interestingly, we discovered that HA and NA vRNAs contain key subtype-specific signatures in the nonconserved NCRs. We then systematically studied the functional significance of these HA subtype-specific NCRs (HA ssNCRs) within the WSN virus background. We demonstrated that the HA ssNCRs can significantly modulate virus replication efficiency by affecting HA segment virion incorporation. Moreover, we revealed important features of the HA ssNCR in determining HA vRNA incorporation efficiency. These data not only show new genetic characteristics of influenza A viruses, but also provide important insights into the roles of the nonconserved noncoding regions of HAs and NAs of influenza A viruses.
MATERIALS AND METHODS
Bioinformatics analysis of NCRs of HA and NA segments of influenza A viruses.
The DNA sequences of the HA and NA segments of all influenza A virus subtypes were obtained from the NCBI Influenza Virus Resource database (28) on 2 April 2013. Because motif 1 (AGC[A/G]AAAGCAGG) ([A/G] indicates a natural nucleotide variation of A or G at this position) and motif 2 (CCTTGTTTCTACT) are conserved at the 5′ and 3′ NCR, respectively, for all segments of type A influenza viruses, they were used as signatures for selecting NCR-containing sequences. We selected the NCR-containing sequences with either motif 1 or motif 2. The NCR-containing sequences were aligned using the software MUSCLE (29), with an additional manual check. After alignment, the positions with an 80% or higher ratio of gaps were removed. The NCR sequences for different type A influenza viruses were extracted and transformed into the negative-sense sequences. The NCR sequence logos showing the nucleotide frequency were constructed by the online Web server WebLogo (http://weblogo.berkeley.edu/logo.cgi) (30). The accession numbers for the sequences used here are available on request.
Cells and antibodies.
Madin-Darby canine kidney (MDCK) and 293T cells were purchased from the American Type Culture Collection (ATCC) (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C. Rabbit anti-β-actin monoclonal antibody (catalog no. 4970) was purchased from Cell Signaling Technology. Rabbit monoclonal anti-influenza virus HA (catalog no. 86001-RM01) was purchased from Sino Biological Inc. (China).
Plasmids.
The pcDNA plasmids of the RNP reconstitution system of influenza virus A/WSN/33/(H1N1) (pcDNA-PB2, pcDNA-PB1, pcDNA-PA, pcDNA-NP, pPOLI-HA, and pPOLI-sap-Rib) were kindly provided by Ervin Fodor (Oxford University, Oxford, United Kingdom). The eight plasmids of the influenza virus A/WSN/33 (H1N1) reverse genetic system (pHW2000-PB2, pHW2000-PB1, pHW2000-PA, pHW2000-HA, pHW2000-NP, pHW-2000NA, pHW2000-M, and pHW2000-NS) were previously described (31). Different HA ssNCR mutant pHW2000-HA plasmids were constructed by one- or two-step PCR with primers specific to HA subtype-specific NCR. The PCR products were then ligated into empty pHW2000 by BsmBI sites. The different HA ssNCR mutants in the pHW2000 plasmid were then subcloned into the pPOLI-HA plasmid by one-step PCR with universal promoter primers containing a BspQI site.
Reverse genetics.
A total of 0.5 μl (1 μg/μl) of each plasmid of the influenza virus A/WSN/33 (H1N1) reverse genetic system [pHW2000-PB2, -PB1, -PA, -HA(H1–H9), -NP, -NA, -M, and -NS] was mixed with 5 μl Lipofectamine 2000 and incubated at room temperature for 20 min. The mixtures were then added to a mixture of 106 293T and MDCK cells (at a ratio of 3:2) in a 6-well plate with the addition of 2 ml Opti-MEM (catalog no. 31985-070; Gibco). At 24 h posttransfection, the Opti-MEM medium was replaced with DMEM containing 0.5% FBS and 0.5 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma). When the cytopathic effect (CPE) reached about 50% of the total cells, the supernatants were harvested and the viruses were plaque purified. The purified viruses were passaged for two generations on MDCK cells and subjected to virus genome sequencing. The virus titers were then determined by plaque assay.
Virus genome sequencing.
The total RNA of each virus stock was extracted with a QIAamp Viral RNA minikit (Qiagen) and reverse transcribed using the SuperScript III first-strand synthesis system (Invitrogen) with a universal influenza A virus reverse transcription (RT) primer (a mixture of 5′-AGCAAAAGCAGG-3′ and 5′-AGCGAAAGCAGG-3′) (the natural nucleotide variation in the RT primers is underlined). The RT products were then amplified by PCR using universal HA NCR-specific primers containing a BsmBI site at the 5′ end (forward primer, 5′-GCGcgtctcCGGGAGCAAAAGCAGGGG-3′; reverse primer, 5′-GCGcgtctcCTATTAGTAGAAACAAGGGTG-3′) (underlined lowercase nucleotide sequence indicates the BsmBI cleavage site). Finally, the PCR products were ligated into the pHW2000 empty vector with a BsmBI site, followed by sequencing of the plasmid.
Growth curve analysis.
MDCK cells were infected with either wild-type (WT) WSN virus or HA NCR mutant viruses at a multiplicity of infection (MOI) of 0.001. At time points 0, 12, 24, 36, 48, 60, and 72 h postinfection (p.i.), the supernatants were collected. The virus titers were determined by plaque assay on MDCK cells. The growth curves shown are the average results of three independent experiments.
Silver staining of virion RNAs.
MDCK cells in 10-cm dishes were infected with wild-type WSN virus or HA NCR mutant viruses at a multiplicity of infection of 0.001. Viral supernatants were harvested at the time when the cells reached about 50% CPE and clarified by centrifugation at 2,000 rpm for 15 min. The supernatant was further centrifuged at 10,000 rpm for 30 min at 4°C. Then, the clarified supernatant was collected and loaded onto a 30% sucrose cushion and further centrifuged at 25,000 rpm for 2.5 h at 4°C. The RNA of the virus pellet was extracted using TRIzol reagent (Invitrogen) and separated on a 2.8% denaturing polyacrylamide gel containing 7 M urea. The gel was stained with a SilverQuest silver-staining kit (Invitrogen). The intensities of RNA bands were quantified using Image J software (NIH Image for Macintosh).
qPCR analysis of virion RNAs.
The quantitative-PCR (qPCR) analysis of packaged vRNAs has been described previously (21). Briefly, virion RNAs were extracted with TRIzol reagent (Invitrogen) from partially sucrose cushion-purified viruses. Approximately 20 ng vRNAs was reverse transcribed by SuperScript III (Invitrogen) with a universal primer (a mixture of 5′-AGCAAAAGCAGG-3′ and 5′-AGCGAAAGCAGG-3′). The RT product was then diluted 100-fold and used as the template for qPCR in a SYBR green-based method using the 7300 Real Time PCR System (Applied Biosystems). The WSN segment-specific PCR primers used to amplify each segment are available upon request. The lengths of the segment-specific qPCR products were kept between 130 and 155 bp. The relative concentrations of vRNAs were determined on the basis of an analysis of cycle threshold values.
Primer extension assay and Western blot analysis.
Approximately 106 293T cells in a 6-well plate were transfected with four protein expression plasmids encoding PB1, PB2, PA, and NP of WSN virus, together with wild-type pPOLI HA or mutant pPOLI HA plasmid. At 24 h posttransfection, cells were harvested for either primer extension analysis or Western blot analysis. For primer extension analysis, total RNAs were extracted with TRIzol reagent. The levels of mRNA, cRNA, and vRNA of the HA template were analyzed by primer extension with two 32P-labled HA-specific primers: 5′-TACTCAACTGTCGCCAGTTC-3′ was used to detect vRNA, and 5′-GTCACTGCCACATTCTTCTC-3′ was used to detect mRNA and cRNA. Quantification of the RNA bands was performed by phosphorimager analysis using Image Quant TL software (GE). For Western blot analysis, the cells were lysed with Cytobuster lysis buffer (Novagen), followed by Western blotting with a monoclonal anti-HA antibody, with anti-β-actin antibody as an internal control.
Statistics.
Student's t test was used for two-group comparisons. P values of <0.05 and <0.01 were considered significant.
RESULTS
Bioinformatics analysis of the noncoding regions of influenza A virus HA and NA segments.
It has been shown that the two subtype-determinant segments, the HA and NA segments, of influenza A viruses contain diverse sequences among different subtypes in their nonconserved NCRs (27). In order to systematically compare the NCRs of all HA and NA segments, we obtained all available NCR sequences of HA and NA from the NCBI Influenza Virus Resource database (28) on 2 April 2013. The numbers of NCR sequences for the different HA and NA subtypes obtained varied significantly, because they correlate with the frequency at which each subtype occurred and was reported. These NCR sequences were then analyzed bioinformatically (29). The representative NCR logos of each HA or NA subtype were generated by the online Web server WebLogo (http://weblogo.berkeley.edu/logo.cgi) (30) (Fig. 1A and B). To validate these HA and NA ssNCR sequence logos, we also examined the NCR sequences from the literature, which were determined accurately by specific NCR-sequencing methods (27, 32–36), and compared them with the sequence logos generated from the bioinformatics analysis (Fig. 1C). The comparison further confirmed that these subtype-specific NCR logos were representative.
FIG 1.
Bioinformatics analysis of the noncoding regions of HA and NA segments of influenza A viruses. (A) NCR logos for 16 HA subtypes of influenza A viruses. The number in parentheses on the right of each HA subtype refers to the number of sequences used for analyzing the NCR sequence of the subtype. The NCR logos were sorted by the number of the NCR sequences analyzed (from large to small). The NCR logos of the common HA subtypes (≥75) that were used in our subsequent functional analysis are highlighted in red (H1 to H7 and H9). The height of the letter in the logo is in proportion to the frequency of a given nucleotide at that position. The black squares represent gaps in the sequence. The numbers above the logos refer to positions in the NCR sequence. The viral RNA promoter regions (12 3′-terminal nucleotides and 13 5′-terminal nucleotides) are shaded in gray. (B) NCR logos for 9 NA subtypes of influenza A viruses. They are presented in the same way as those in panel A. (C) Comparison between our bioinformatically analyzed sequence logos and accurately sequenced NCR sequences (see the text). The sequence logos under the lines are the NCR sequences reported by accurate NCR-sequencing methods. The sequence logos above the lines were derived from panels A and B. They are presented in the same way as for panel A.
Interestingly, the subtype-specific NCR logos show that the length and the sequence of the nucleotides in the nonconserved NCRs of all the different HA and NA subtypes are subtype specific, except for subtypes H10 and H12, which show the same nonconserved NCRs at both ends (Fig. 1A and B). It can be seen that the ssNCRs of HA and NA differ significantly in sequence and length (5 to 32 nt for the 3′ end and 10 to 33 nt for the 5′ end) at both ends. The relatively long HA and NA ssNCRs often contain variable or missing nucleotides at certain positions (Fig. 1A and B). We also noticed that all HA ssNCRs contain common HA-specific nucleotides—a 2-nt extension (CC) adjacent to the 3′ promoter region and a 3-nt extension (GUG)—except for the common U stretch next to the 5′ promoter region. In contrast, the length variation of the NA ssNCRs (6 to 10 nt for the 3′ end and 12 to 25 nt for the 5′ end) is not as large as that of HAs, and they do not show the common 2- to 3-nt extension at either end.
The HA ssNCRs play a role in determining influenza A virus replication efficiency.
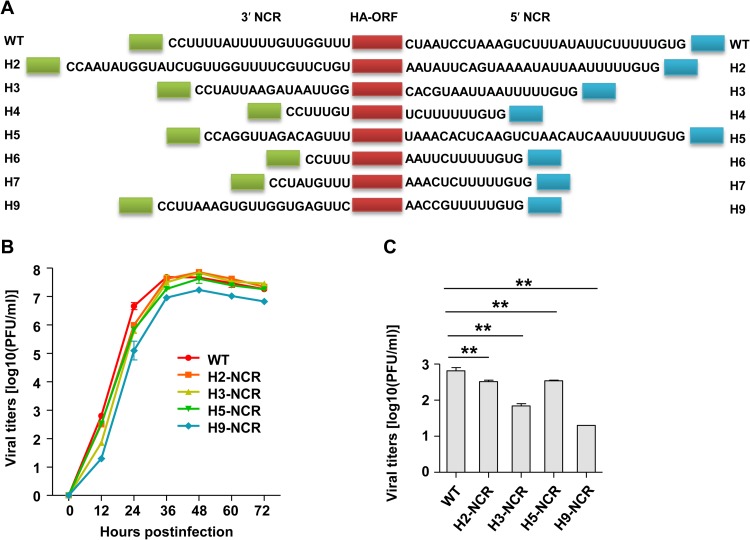
The discovery of the significantly varied HA ssNCRs of influenza A viruses prompted us to comparatively and systematically study the biological significance of the HA ssNCRs within the context of a virus. Using H1N1 WSN reverse genetics (31), we replaced the wild-type H1-NCR at both the 3′ and 5′ ends with the corresponding ssNCRs of the other common HA subtypes (H2- to H7- and H9-NCRs) individually in the PHW2000 HA plasmid and attempted to rescue the respective viruses (Fig. 2A). Interestingly, the wild-type WSN virus and four mutant viruses—H2-NCR, H3-NCR, H5-NCR, and H9-NCR—were successfully rescued, while the other three—H4-NCR, H6-NCR, and H7-NCR—were not viable. We noticed that the 3′ and 5′ segment-specific NCRs of H4, H6, and H7 are much shorter than those of H1, H2, H3, H5, and H9 at both ends (Fig. 1A and 2A).
FIG 2.
Effects of the HA ssNCRs on influenza A virus replication efficiency. (A) Schematic representation of recombinant WSN HA vRNAs with subtype-specific NCR substitutions at both the 3′ and 5′ ends. The green and blue boxes represent the 12 and 13 terminal promoter nucleotides at the 3′ and 5′ ends, respectively. The red boxes represent ORFs of WSN virus HA. The nucleotides shown between the promoter nucleotides and the ORFs are the subtype-specific NCRs of the indicated HA subtypes. (B) Growth curves of rescued viruses in MDCK cells. WT and mutant WSN viruses were plaque purified and passaged for two generations. MDCK cells were infected with virus at an MOI of 0.001. At 0 h, 12 h, 24 h, 48 h, 60 h, and 72 h after infection, the virus titer in the supernatant was determined by plaque assay of the MDCK cells. The error bars indicate the standard deviations of three independent experiments. H4-NCR, H6-NCR, and H7-NCR were not able to be rescued. (C) Virus titers were determined at 12 h p.i. **, P < 0.01; two-tailed Student's t test.
To further characterize these viruses, we then plaque purified each virus and passaged it for two generations. The full genomes of the passaged viruses were sequenced to ensure the sequences of recombinant HA vRNAs were correct and that no mutations occurred in other segments. The growth kinetics of these viruses were then examined by plaque assay (Fig. 2B). It can be seen that all the mutant viruses showed growth kinetics different from that of the WT virus. Especially at earlier time points postinfection (12 and 24 h), the four mutant viruses (H2-NCR, H3-NCR, H5-NCR, and H9-NCR) showed lower replication efficiencies than the WT virus (Fig. 2C). In particular, the H9-NCR virus showed about 1- to 1.5-log-unit reductions at all time points examined. We did not observe any differences in the sizes of the plaques among the WT and the mutant viruses. Together, these results suggested that the HA ssNCR plays a role in determining virus replication efficiency.
The HA ssNCRs do not significantly affect viral RNA synthesis and protein expression.
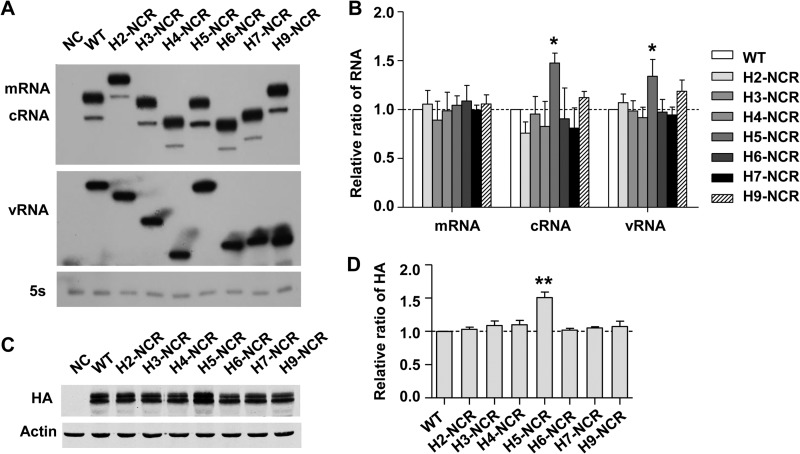
Since the nonconserved NCR could be part of cis-acting signals for viral RNA transcription, replication, and translation, we first examined the effects of all HA ssNCR substitutions on viral RNA synthesis and protein expression in an RNP reconstitution system derived from the WSN virus (37). The RNA-expressing plasmids pPOLI-H1NCR-HA, pPOLI-H2NCR-HA, pPOLI-H3NCR-HA, pPOLI-H4NCR-HA, pPOLI-H5NCR-HA, pPOLI-H6NCR-HA, pPOLI-H7NCR-HA, and pPOLI-H9NCR-HA were individually cotransfected with pcDNA-PB1, pcDNA-PB2, pcDNA-PA, and pcDNA-NP in 293T cells. At 24 h posttransfection, the total RNAs were harvested and the levels of three species of viral RNAs (vRNA, mRNA, and cRNA) were detected by primer extension analysis (37). As shown in Fig. 3A and B, all of the viral RNA templates showed comparable viral RNA synthesis efficiencies, although subtle differences were observed, especially for the cRNA and vRNA levels of H5-NCR, which were significantly higher than those of the others. To examine whether these HA ssNCRs affected viral protein expression at the translational level, the cells were harvested and lysed for Western blot analysis with a monoclonal anti-HA antibody. We found that the levels of the HA protein were generally comparable, except for H5-NCR (Fig. 3C and D). Intriguingly, H5-NCR showed a higher level of HA protein expression, whereas its mRNA level (Fig. 3A) was not significantly higher than those of the others, implying that the NCR of H5 might modulate its vRNA and cRNA synthesis at the transcriptional level and its protein expression at the translational level. In general, these results suggested that the reductions in viral replication efficiencies were not due to the effects of HA ssNCRs on viral RNA synthesis and viral protein translation.
FIG 3.
Effects of HA ssNCRs on viral RNA synthesis and protein expression in an RNP reconstitution system derived from the WSN virus. (A) Effects of HA ssNCRs on viral RNA synthesis. 293T cells were transfected with four protein expression plasmids (pcDNA-PA, -PB1, -PB2, and -NP), together with a viral RNA expression plasmid (pPOLI-HA) in which the H1 ssNCR was replaced with other HA ssNCRs. The levels of mRNA, cRNA, and vRNA were detected by primer extension analyses. NC was the negative control, with only pPOLI-HA transfected. (B) Statistical analysis of the viral RNAs in panel A. The values of viral RNAs in the transfected 293T cells were standardized to the 5S rRNA level and then normalized to the levels of viral RNAs in the NC. The data represent the means and standard deviations (SD) of three independent experiments. (C) Effects of HA ssNCRs on viral protein expression. 293T cells were transfected as indicated for panel A. The levels of HA protein were analyzed by Western blotting. (D) Statistical analysis of HA protein expression levels in panel C. *, P < 0.05; **, P < 0.01; two-tailed Student's t test. The data represent the means and SD of three independent experiments.
The HA ssNCR substitutions significantly affect virion incorporation of the HA segment per se.
According to the influenza virus selective genome-packaging model, the HA ssNCR is the key part of the packaging signals. We investigated whether the HA ssNCR substitutions could differentially affect virus genome packaging. The silver staining of the virion RNAs obtained from wild-type or HA ssNCR substitution viruses showed that, in contrast to the other seven segments, only the HA vRNA content in the virions was significantly reduced in the four HA ssNCR substitution viruses when they were compared with that of the WT WSN virus (Fig. 4A). The quantification of the band intensities by densitometry showed that relative HA/NP ratios correlated well with the virus growth properties (Fig. 4B).
FIG 4.
Effects of HA ssNCRs on virus selective genome packaging. (A) Analysis of the packaging efficiency of each individual vRNA with silver-staining gel. Wild-type (H1-NCR) and mutant (H2-NCR, H3-NCR, H5-NCR, and H9-NCR) viruses were amplified on MDCK cells and purified by ultracentrifugation through a 30% sucrose cushion. Then, the virion vRNAs were extracted with TRIzol reagent, analyzed on a 2.8% denaturing polyacrylamide gel, and visualized by silver staining. (B) Statistical analysis of the HA vRNA packaging efficiency in panel A. The band intensities were quantified using Image J software. The HA/NP vRNA ratio in the WT virus was set as 1. The relative HA/NP vRNA ratios in mutant viruses were calculated (means and SD of three independent experiments; *, P < 0.05; **, P < 0.01; two-tailed Student's t test). (C) Analysis of the packaging efficiency of each individual vRNA segment by RT-qPCR. The NP vRNA was used as an internal reference. The relative ratio of each vRNA segment versus NP vRNA was calculated. The standard deviations were derived from three independent virus preparations, with vRNA levels quantified in duplicate. *, P < 0.05; **, P < 0.01; two-tailed Student's t test.
It has been reported that the packaging signal in one segment could also affect the incorporation of other segments in the proposed selective-packaging model (12, 13). In order to accurately examine whether the substitutions of these ssNCRs in the WSN virus background might affect the packaging efficiency of other segments, we further analyzed the virion RNAs with RT-qPCR (21). The incorporation efficiency of each vRNA was calculated as the ratio (as a percentage) of the level of each vRNA to the level of NP vRNA (Fig. 4C). In general, the RT-qPCR results were consistent with the results from the RNA-staining experiment, showing that the HA ssNCR substitutions resulted in the reduction of HA virion incorporation at different levels and did not significantly reduce the incorporation efficiency of other segments. Moreover, we observed that the reduction of HA incorporation by the HA ssNCR substitutions at both ends could lead to an increase of PB1, and in particular PA, vRNA incorporation in the context of WSN virus (Fig. 4C). Taken together, these data indicated that the HA ssNCR plays a critical role in determining HA segment virion incorporation.
Single-end HA ssNCR substitutions at either the 3′ or the 5′ end result in differential effects on HA vRNA virion incorporation.
In the configuration of the vRNPs, the partial complementary nucleotides in the terminal promoter regions at the 3′ and the 5′ ends join together to form a special secondary structure to be bound by the polymerase complex (7, 9, 38). It can be speculated that the HA ssNCRs at the two ends, adjacent to the promoter region, could also be brought together and might be in close contact with each other. Thus, we were interested to test whether compatibility is required between the HA ssNCRs at the 3′ and the 5′ ends in regulating HA segment incorporation. To this end, we generated a series of PHW2000 HA single-end substitution constructs with one end of the H1-NCR unchanged but the other end replaced with the corresponding NCRs of other subtype-specific NCRs (H2- to H7- and H9-NCRs) (Fig. 5A and B).
FIG 5.
Effects of single-end HA ssNCR substitutions on virus replication efficiency. (A and B) Schematic representation of recombinant WSN HA vRNAs with subtype-specific NCR substitutions at either the 3′ end (A) or the 5′ end (B). The green and blue boxes represent the terminal 12 and 13 promoter nucleotides at the 3′ and 5′ ends, respectively. The HA ssNCR at either the 3′ end or the 5′ end are shown. The HA ssNCRs were sorted by the lengths of the NCR sequences. (C and D) Growth curves of the recombinant viruses with 3′-end (C) and 5′-end (D) HA ssNCR substitutions. The recombinant WSN viruses were rescued and purified as described in the legend to Fig. 2B. The error bars indicate the standard deviations of three independent experiments.
We first tried to rescue these single-end HA ssNCR substitution viruses. Surprisingly, all 14 recombinant viruses were successfully rescued, including the three subtypes that could not be rescued previously with the full HA ssNCR substitutions (H4, H6, and H7) (Fig. 2A and B). To further characterize these rescued viruses, the viral growth kinetics was first examined. Interestingly, the two sets of recombinant viruses showed quite distinct growth properties (Fig. 5C and D). In general, in comparison with the WT viruses, the 3′-ssNCR substitution viruses showed more drastic reductions (maximum 3 to 5 log units) in growth levels than the 5′-ssNCR substitution viruses.
Among the 3′-end ssNCR substitution viruses (Fig. 5C), the H3–3′-NCR and H9–3′-NCR viruses could replicate to a level close to that of the WT virus (≤1 log unit), while the H2–3′-NCR, H5–3′-NCR, H4–3′-NCR, and H6–3′-NCR viruses showed 2- to 3.5-log-unit reductions from the WT growth level. The H7-3′-NCR virus showed the greatest reduction (4 log units) in virus replication efficiency. It is interesting that the lengths of the H3 (17 nt) and H9 (21 nt) 3′ ssNCRs were similar to that of the WT H1 3′ ssNCR (20 nt). The other mutant viruses with either a longer 3′ ssNCR (H2) or shorter 3′ ssNCRs (H4, H6, and H7) attenuated virus growth to various extents. An exception was the H5–3′-NCR virus, which showed quite significant reduction from WT virus growth levels, but the length of its 3′ ssNCR is only 1 nucleotide shorter than that of the WT H1 3′ ssNCR. Together, these results suggested that the range of lengths for the HA 3′ ssNCR might be important in determining virus replication efficiency, while particular nucleotides of the 3′ ssNCR within a certain length range are also important.
In contrast, among the series of 5′ HA ssNCR mutant viruses (Fig. 5D), except for the H9–5′-NCR substitution virus, which showed about 2-log-unit attenuation in virus growth, the other five (H5-5′ NCR, H2–5′ NCR, H3–5′ NCR, H4–5′ NCR, H6-5′ NCR, and H7-5′ NCR) showed relatively minor effects on virus growth. We speculated that the H9-5′ NCR might contain a particular unfavorable element at the 5′ end for WSN virus replication, which may also account for the full H9-NCR substitution virus having the lowest replication efficiency (Fig. 2A to C). Interestingly, the replication efficiencies of the other five mutant viruses showed a tendency for the relative replication levels to be correlated with their relative lengths. Considering the quite significant length and sequence variations among the five 5′ NCRs and relatively minor reductions in virus replication levels (in comparison with those of the series of 3′-NCR substitution viruses), we concluded that a stringent compatibility between the two ends is not required and that the length of the HA 5′ ssNCR may play a significant role in determining virus replication efficiency.
We went on to examine whether these HA NCR single-end substitutions could have differential effects on viral RNA synthesis and protein expression in the RNP reconstitution system (Fig. 6A to H). In comparison with the wild-type template, both the 3′-NCR substitutions and the 5′-NCR substitutions did not show any differential effects on viral synthesis and protein expression. Furthermore, this implied that the differential effects previously observed for the full H5-NCR substitution (Fig. 3) were due to the presence of both the H5-3′ NCR and the H5-5′ NCR simultaneously.
FIG 6.
Effects of single-end substitutions of HA ssNCR on viral RNA synthesis and protein expression in the WSN RNP reconstitution system. The experiments and statistical analysis were performed as described in the legend to Fig. 3, except that the RNA-expressing plasmid pPOLI-HA was replaced individually with the PHW-HA plasmids containing single-end HA ssNCR substitutions. (A and E) Effects of the 3′-end (A) and 5′-end (E) HA ssNCR substitutions on viral RNA synthesis. (B and F) Statistical analysis of the viral RNAs in panels A and E, respectively. (C and G) Effects of the 3′-end (C) and 5′-end (G) HA ssNCR substitutions on viral protein expression. (D and H) Statistical analysis of the HA protein expression levels in panels C and G, respectively. *, P < 0.05; two-tailed Student's t test. The data represent the means and SD of three independent experiments.
We then further examined the effects of these single-end HA ssNCR substitutions on the virus genome-packaging efficiency with both the silver staining and the RT-qPCR analysis. Consistent with what we observed in virus growth, the series of 3′-NCR substitution viruses showed more drastic defects on HA vRNA incorporation than the series of 5′-ssNCR substitution viruses in general in the silver-staining analysis (Fig. 7A and C). The extents of reduction also correlated well with the virus growth levels for both sets of mutant viruses (Fig. 7B and D). Furthermore, our RT-qPCR results were also consistent with these results (data not shown)
FIG 7.
Effects of single-end HA ssNCR substitutions on virus genome packaging. (A) Analysis of the vRNA genome-packaging efficiencies of the wild-type virus and the HA 3′-end substitution viruses (H2–3′ NCR, H3-3′ NCR, H4-3′ NCR, H5-3′ NCR, H6-3′ NCR, H7-3′ NCR, and H9-3′ NCR) with silver-staining gel. The experiments were performed as described in the legend to Fig. 4A. (B) Statistical analysis of the HA vRNA packaging efficiencies in panel A. The HA/NP vRNA ratio in the WT virus was set as 1. The relative HA/NP vRNA ratios in mutant viruses were calculated (means and SD of three independent experiments; *, P < 0.05; **, P < 0.01; two-tailed Student's t test). (C) Analysis of the vRNA genome-packaging efficiencies of the wild-type virus and the HA 5′-end substitution viruses (H2–5′ NCR, H3–5′ NCR, H4–5′ NCR, H5-5′ NCR, H6-5′ NCR, H7-5′ NCR, and H9-5′ NCR) with silver-staining gel. (D) Statistical analysis of the HA vRNA packaging efficiencies in panel C. The analyses were performed as described in the legends to panels A and B. The standard deviations were calculated based on three independent experiments with different virus preparations.
Taken together, these data further confirmed that the HA ssNCR is critical in determining HA vRNA incorporation. Regarding the properties of different HA ssNCRs in regulating HA virion incorporation, we concluded that (i) the HA 3′ ssNCR plays a more important role than the HA 5′ ssNCR, (ii) no stringent compatibility is required between the 3′ and 5′ ends of HA ssNCRs, and (iii) in addition to a particular nucleotide(s), the lengths of the HA ssNCRs at both ends may play an important role in determining HA vRNA incorporation efficiency.
DISCUSSION
In this study, we performed, for the first time, a bioinformatics analysis of all the variable noncoding regions of HAs and NAs. Interestingly, we revealed that the HA and NA segments contain key subtype-specific signatures in the nonconserved NCRs. To our knowledge, the result of our bioinformatics analysis is the first systematic description of subtype genetic characteristics in the noncoding region of influenza A viruses. Among the relatively long HA and NA subtype-specific NCRs, the nucleotides at certain positions were variable or missing (Fig. 1A and B). Although we could not exclude the possibility that these variations might be due to the relatively high error rates often occurring during NCR sequencing, considering the large numbers of NCRs analyzed for these common subtypes and the consistent variations or missing nucleotides observed at these positions, it is more likely that most of these variations reflect natural flexibilities of the subtype-specific NCRs that underwent functional selection during long-term virus evolution. Moreover, we discovered that all HAs contain a pair of HA-specific nucleotides in the nonconserved NCRs at both ends, CC at positions 13 and 14 from the 3′ end and GUG at positions 14 to 16 from the 5′ end. The presence of these highly conserved HA-specific nucleotides implies that they may play a critical role during influenza A virus replication. Further studies on the role of these HA-specific nucleotides are under way.
Following the discovery of the presence of subtype-specific NCRs, we systematically and comparatively studied the biological significance of the common HA subtype-specific NCRs (H1 to H7 and H9) within the influenza A/WSN/33 virus background. We found that the HA ssNCR modulates virus replication efficiency significantly by exerting its influence on HA vRNA incorporation. According to the selective-packaging model, the eight segments of influenza A viruses are selectively packaged into virions, probably by intersegment interactions (13, 24, 39, 40). The PB2 vRNA has been proposed to play a primary role in the packaging of the full set of genomes (25, 41). Marsh et al. previously reported that the absence of the HA vRNA in delta HA virus led to reduction in the packaging of other vRNA segments. They also identified a key region of the H1 open reading frame (ORF) that is critical for efficient packaging of the H1 HA segment (21). In this study, we found that the substitution of HA subtype NCRs mainly affected HA vRNA incorporation and did not significantly reduce the incorporation efficiencies of the other seven segments; rather, it significantly increased PB1 and PA segment incorporation into the HA ssNCR substitution viruses. Taken together, these facts may suggest that nucleotides in the nonconserved NCR play a critical role during virion selective incorporation and that nucleotides in the coding region are mainly responsible for the intersegment interactions. This conclusion is also consistent with a recent proposal that the noncoding region of the NP segment acts as a virion incorporation signal, while the terminal coding region serves as an intersegment bundling signal (26).
In addition, considering that different HA and NA subtypes contain diverse subtype-specific NCRs and their significantly varied coding sequences, the packaging signals, composed of the NCR and the terminal coding nucleotides, may also vary significantly among different HA and NA subtypes. Therefore, apart from previously identified packaging signals for H1N1 WSN or PR8 virus, it is necessary to further identify the packaging signals for each HA and NA subtype, particularly for the common HA subtypes.
It has been previously reported that the 3′ end of the NA vRNA coding region played a more important role than the 5′ coding region in determining NA vRNA virion incorporation (24). Here, we found that 3′-end HA ssNCR substitution viruses showed more drastic reductions in HA vRNA incorporation than 5′-end HA ssNCR substitution viruses. In addition, our observations that the full H4-, H6-, and H7-ssNCR substitutions failed to produce viable viruses whereas the 5′ single-end replacement of the H4, H6, and H7 viruses with the unchanged WT H1 3′ ssNCR showed quite significant growth capacities further support the notion that the 3′-end HA ssNCR plays a more critical role than the 5′-end HA ssNCR in HA vRNA incorporation. Moreover, the significant replication capability obtained with the series of 5′ single-end substitution viruses suggested that stringent compatibility is not required between the two HA ssNCR ends. Taken together, these data support the idea that the 3′-end packaging signals play a more important role than the 5′-end packaging signals during the selective genome packaging.
It has been reported that the lengths and sequences of the coding nucleotides within the cis-acting packaging signal for each segment are equally important (22, 39). Here, we found that the lengths of the noncoding regions at both ends might play a more significant role than specific sequences. This observation is consistent with the conclusion from a linker-scanning mutagenesis study that the incorporation of influenza virus genome segments does not absolutely require specific sequences in the packaging signals (42). However, both we and the authors of that study could not exclude the possibility that a specific residue at a particular position(s) is preferred. Therefore, further investigation of the mechanisms by which the lengths of the subtype-specific NCRs exert their effects on HA vRNA incorporation is necessary.
In summary, we report here for the first time that the nonconserved NCRs of HA and NA segments are subtype specific. This key viral genetic information not only can further facilitate influenza A virus genome subtyping, but also can provide a framework for studying subtype differences of influenza A viruses in terms of virus genome packaging, sequence coevolution, reassortment, and host specificity. Furthermore, our functional study indicated that the HA ssNCRs play a key role in determining HA vRNA virion incorporation. We also revealed certain characteristics of the HA ssNCRs in determining HA segment incorporation efficiency. These results provide new insights into the selective genome packaging of influenza A virus. A better understanding of the HA and NA vRNA-packaging efficiencies in the different virus backgrounds would significantly facilitate preparedness for and prevention of new emerging influenza A virus epidemics and pandemics.
ACKNOWLEDGMENTS
We thank George Brownlee and Ervin Fodor (University of Oxford, Oxford, United Kingdom) for their valuable advice and discussion.
This work was supported by grants from the Chinese Science and Technology Key Project (grant numbers 2013ZX10004601 and 2013ZX10004611), the National Natural Science Foundation of China (grant number 31070152) to T.D, the Biomedical Supercomputing Center Project of Hunan University to T.J. (531106011004), and the Young Teacher's Development Plan of Hunan University to Y.P. (531107040720).
Footnotes
Published ahead of print 23 July 2014
REFERENCES
- 1.Palese P, Shaw M. 2013. Orthomyxoviridae, p 1151–1185 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed), Fields virology, 6th ed. Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 2.Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931–939. 10.1038/nature08157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J. 2012. The structure of native influenza virion ribonucleoproteins. Science 338:1634–1637. 10.1126/science.1228172 [DOI] [PubMed] [Google Scholar]
- 4.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. 2012. Organization of the influenza virus replication machinery. Science 338:1631–1634. 10.1126/science.1227270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fodor E. 2013. The RNA polymerase of influenza A virus: mechanisms of viral transcription and replication. Acta Virol. 57:113–122. 10.4149/av_2013_02_113 [DOI] [PubMed] [Google Scholar]
- 6.Fodor E, Brownlee GG. 2002. Influenza virus replication, p 1–29 In Potter CW. (ed), Influenza. Elsevier Science, New York, NY [Google Scholar]
- 7.Robertson JS. 1979. 5′ and 3′ terminal nucleotide sequences of the RNA genome segments of influenza virus. Nucleic Acids Res. 6:3745–3757. 10.1093/nar/6.12.3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poon LL, Pritlove DC, Fodor E, Brownlee GG. 1999. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 73:3473–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desselberger U, Racaniello VR, Zazra JJ, Palese P. 1980. The 3′ and 5′ terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene 8:315–328. 10.1016/0378-1119(80)90007-4 [DOI] [PubMed] [Google Scholar]
- 10.Bergmann M, Muster T. 1996. Mutations in the nonconserved noncoding sequences of the influenza A virus segments affect viral vRNA formation. Virus Res. 44:23–31. 10.1016/0168-1702(96)01335-4 [DOI] [PubMed] [Google Scholar]
- 11.Zheng H, Palese P, Garcia-Sastre A. 1996. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication. Virology 217:242–251. 10.1006/viro.1996.0111 [DOI] [PubMed] [Google Scholar]
- 12.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. 2010. Genome packaging in influenza A virus. J. Gen. Virol. 91:313–328. 10.1099/vir.0.017608-0 [DOI] [PubMed] [Google Scholar]
- 13.Noda T, Kawaoka Y. 2012. Packaging of influenza virus genome: robustness of selection. Proc. Natl. Acad. Sci. U. S. A. 109:8797–8798. 10.1073/pnas.1206736109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou YY, Heaton NS, Gao Q, Palese P, Singer RH, Lionnet T. 2013. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog. 9:e1003358. 10.1371/journal.ppat.1003358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou YY, Vafabakhsh R, Doganay S, Gao Q, Ha T, Palese P. 2012. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc. Natl. Acad. Sci. U. S. A. 109:9101–9106. 10.1073/pnas.1206069109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozawa M, Fujii K, Muramoto Y, Yamada S, Yamayoshi S, Takada A, Goto H, Horimoto T, Kawaoka Y. 2007. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 81:30–41. 10.1128/JVI.01434-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dos Santos Afonso E, Escriou N, Leclercq I, van der Werf S, Naffakh N. 2005. The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology 341:34–46. 10.1016/j.virol.2005.06.040 [DOI] [PubMed] [Google Scholar]
- 18.Giannecchini S, Wise HM, Digard P, Clausi V, Del Poggetto E, Vesco L, Puzelli S, Donatelli I, Azzi A. 2011. Packaging signals in the 5′-ends of influenza virus PA, PB1, and PB2 genes as potential targets to develop nucleic-acid based antiviral molecules. Antiviral Res. 92:64–72. 10.1016/j.antiviral.2011.06.013 [DOI] [PubMed] [Google Scholar]
- 19.Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. 2008. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 82:11869–11879. 10.1128/JVI.01634-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Y, Hong Y, Parslow TG. 2005. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 79:10348–10355. 10.1128/JVI.79.16.10348-10355.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh GA, Hatami R, Palese P. 2007. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 81:9727–9736. 10.1128/JVI.01144-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozawa M, Maeda J, Iwatsuki-Horimoto K, Watanabe S, Goto H, Horimoto T, Kawaoka Y. 2009. Nucleotide sequence requirements at the 5′ end of the influenza A virus M RNA segment for efficient virus replication. J. Virol. 83:3384–3388. 10.1128/JVI.02513-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii K, Fujii Y, Noda T, Muramoto Y, Watanabe T, Takada A, Goto H, Horimoto T, Kawaoka Y. 2005. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 79:3766–3774. 10.1128/JVI.79.6.3766-3774.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y. 2003. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. U. S. A. 100:2002–2007. 10.1073/pnas.0437772100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. 2006. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 80:2318–2325. 10.1128/JVI.80.5.2318-2325.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto H, Muramoto Y, Noda T, Kawaoka Y. 2013. The genome-packaging signal of the influenza A virus genome comprises a genome incorporation signal and a genome-bundling signal. J. Virol. 87:11316–11322. 10.1128/JVI.01301-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Lee CW. 2009. Sequencing and mutational analysis of the non-coding regions of influenza A virus. Vet. Microbiol. 135:239–247. 10.1016/j.vetmic.2008.09.067 [DOI] [PubMed] [Google Scholar]
- 28.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601. 10.1128/JVI.02005-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann E, Krauss S, Perez D, Webby R, Webster RG. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165–3170. 10.1016/S0264-410X(02)00268-2 [DOI] [PubMed] [Google Scholar]
- 32.Wang R, Taubenberger JK. 2014. Characterization of the noncoding regions of the 1918 influenza A H1N1 virus. J. Virol. 88:1815–1818. 10.1128/JVI.03098-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, Xiao Y, Taubenberger JK. 2014. Rapid sequencing of influenza A virus vRNA, cRNA and mRNA non-coding regions. J. Virol. Methods 195:26–33. 10.1016/j.jviromet.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park SJ, Kang BK, Jeoung HY, Moon HJ, Hong M, Na W, Park BK, Poo H, Kim JK, An DJ, Song DS. 2013. Complete genome sequence of a canine-origin H3N2 feline influenza virus isolated from domestic cats in South Korea. Genome Announc. 1:e0025312. 10.1128/genomeA.00253-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SJ, Park BK, Song DS, Poo H. 2012. Complete genome sequence of a mammalian species-infectious and -pathogenic H6N5 avian influenza virus without evidence of adaptation. J. Virol. 86:12459–12460. 10.1128/JVI.02301-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su S, Cao N, Chen J, Zhao F, Li H, Zhao M, Wang Y, Huang Z, Yuan L, Wang H, Zhang G, Li S. 2012. Complete genome sequence of an avian-origin H3N2 canine influenza A virus isolated in farmed dogs in southern China. J. Virol. 86:10238. 10.1128/JVI.01601-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989–9001. 10.1128/JVI.76.18.8989-9001.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. 1987. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl. Acad. Sci. U. S. A. 84:8140–8144. 10.1073/pnas.84.22.8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y, Huang T, Ly H, Parslow TG, Liang Y. 2008. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J. Virol. 82:229–236. 10.1128/JVI.01541-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fournier E, Moules V, Essere B, Paillart JC, Sirbat JD, Isel C, Cavalier A, Rolland JP, Thomas D, Lina B, Marquet R. 2012. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 40:2197–2209. 10.1093/nar/gkr985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsh GA, Rabadan R, Levine AJ, Palese P. 2008. Highly conserved regions of influenza A virus polymerase gene segments are critical for efficient viral RNA packaging. J. Virol. 82:2295–2304. 10.1128/JVI.02267-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujii K, Ozawa M, Iwatsuki-Horimoto K, Horimoto T, Kawaoka Y. 2009. Incorporation of influenza A virus genome segments does not absolutely require wild-type sequences. J. Gen. Virol. 90:1734–1740. 10.1099/vir.0.010355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]