ABSTRACT
Monocytic cells, including macrophages and dendritic cells, exist in different activation states that are critical to the regulation of antimicrobial immunity. Many pandemic viruses are monocytotropic, including porcine reproductive and respiratory syndrome virus (PRRSV), which directly infects subsets of monocytic cells and interferes with antiviral responses. To study antiviral responses in PRRSV-infected monocytic cells, we characterized inflammatory cytokine responses and genome-wide profiled signature genes to investigate response pathways in uninfected and PRRSV-infected monocytic cells at different activation states. Our findings showed suppressed interferon (IFN) production in macrophages in non-antiviral states and an arrest of lipid metabolic pathways in macrophages at antiviral states. Importantly, porcine monocytic cells at different activation states were susceptible to PRRSV and responded differently to viral infection. Based on Gene Ontology (GO) analysis, two approaches were used to potentiate antiviral activity: (i) pharmaceutical modulation of cellular lipid metabolism and (ii) in situ PRRSV replication-competent expression of interferon alpha (IFN-α). Both approaches significantly suppressed exogenous viral infection in monocytic cells. In particular, the engineered IFN-expressing PRRSV strain eliminated exogenous virus infection and sustained cell viability at 4 days postinfection in macrophages. These findings suggest an intricate interaction of viral infection with the activation status of porcine monocytic cells. An understanding and integration of antiviral infection with activation status of monocytic cells may provide a means of potentiating antiviral immunity.
IMPORTANCE Activation statuses of monocytic cells, including monocytes, macrophages (Mϕs), and dendritic cells (DCs), are critically important for antiviral immunity. Unfortunately, the activation status of porcine monocytic cells or how cell activation status functionally interacts with antiviral immunity remains largely unknown. This is a significant omission because many economically important porcine viruses are monocytotropic, including our focus, PRRSV, which alone causes nearly $800 million economic loss annually in the U.S. swine industries. PRRSV is ideal for deciphering how monocytic cell activation statuses interact with antiviral immunity, because it directly infects subsets of monocytic cells and subverts overall immune responses. In this study, we systematically investigate the activation status of porcine monocytic cells to determine the intricate interaction of viral infection with activation statuses and functionally regulate antiviral immunity within the framework of the activation paradigm. Our findings may provide a means of potentiating antiviral immunity and leading to novel vaccines for PRRS prevention.
INTRODUCTION
Monocytic cells, including blood monocytes (BMs), tissue macrophages (Mϕs), and dendritic cells (DCs), originate from common myeloid progenitor cells (1). After their origin, they circulate to locate throughout the body and specialize into a variety of activation statuses to functionally regulate defensive responses and immune homeostasis (1–5). The activation status of monocytic cells such as in Mϕs conventionally has been assigned as classical M1 and alternative M2 statuses, as well as other subtypes (2–4). For instance, classically activated (or M1 status) Mϕs develop in response to interferon gamma (IFN-γ) and bacterial products, such as lipopolysaccharides (LPS); the M2 status of those Mϕs alternatively activated by the Th2 cytokines interleukin-4 (IL-4) and IL-13 in response to parasitic infections is assigned to the M2a subclass. Accordingly, the other subclasses of M2 cells include M2b, obtained by triggering of Fcγ receptors plus the stimulation of Toll-like receptors (TLRs) in Mϕs, and M2c of deactivation programs elicited by immunosuppressive cytokines and hormones, such as IL-10, glucocorticoids (GCs), and transforming growth factor β (TGF-β) (2–4). Despite not being well studied, the M1/M2 activation paradigm is represented in both monocytes and DCs (1, 5–7). For example, human monocytes are divided based on the expression of CD16, with CD16+ monocytes representing M1 cells, which are more proinflammatory and microbicidal (5). A similar paradigm has been postulated for DCs, with type I DCs representing a subset inducing Th1 responses and type II DCs activating Th2 responses (8, 9). Nonetheless, the criteria for DC polarization and associated activation markers remain elusive in all species (1, 6, 7).
Monocytic cells at different activation statuses, as well characterized in Mϕs, functionally exert phenotypes to regulate inflammation, tissue repair, T- and B-cell proliferation, phagocytosis, and antimicrobial activity against bacteria and helminths (3–5). In addition, monocytic cells confer a cell-autonomous antiviral state induced upon viral infection or stimulation by viral mimics (10–13). Indeed, stimulation of type I IFN production and expression of IFN-stimulated genes (ISGs) to combat virus propagation are hallmarks of the antiviral state (10–15). Subsets of monocytic cells typically are among the major producers of type I IFNs (10–15), and recent studies have posited direct interaction between Mϕ polarization and viral infection (16–18). For example, HIV and respiratory syncytial virus (RSV) were shown to alter Mϕ activation statuses, thus affecting viral pathogenesis and host immune responses (16–18). In addition, most pandemic viral infections cause disease syndromes frequently complicated with coinfection from pathogens of other phyla (19–23). Thus, it is important to integrate the activation status with antiviral states linked to type I IFN production and action (16–19). Understanding the relationships among activation statuses and antiviral states not only extends the activation paradigm of monocytic cells but also integrates antiviral regulation into the scenario of immune responses, including inflammation, tissue repair, and overall antimicrobial activity (11, 17, 18, 21).
Many of the most economically important animal viruses are monocytotropic (21), including porcine reproductive and respiratory syndrome virus (PRRSV), the focus in this study. PRRSV is an enveloped positive-strand RNA virus that directly infects subsets of Mϕs and DCs and subverts immune responses in monocytic cells, making it ideal for deciphering how monocytic cell activation status interacts with antiviral immunity (21, 24–28). Monocytic cells are critical for providing early immune surveillance and bridging adaptive antiviral immunity (1–5, 7, 11). Direct infection of monocytic subsets of Mϕs and DCs by PRRSV inevitably leads to immune deviation and likely alters activation statuses (21). Few studies have focused on the activation status of porcine monocytic cells or how cell activation status modulates antiviral immunity (27, 28). Notably, in this study we focused on examining Gene Ontology (GO) analysis based on comparative transcriptomes revealed in macrophages at different activation statuses upon viral infection, rather than a transcriptomic comparison between infected and noninfected tissues/cells, which has been well documented in previous studies (29, 30). In this context, we used a multiplex cytokine assay and transcriptomic sequencing (RNA-Seq) analysis to profile gene response pathways in porcine monocytic cells polarized to typical activation statuses and antiviral states. We then showed that cells at different activation statuses (including antiviral states) reacted differently to PRRSV infection in terms of cytokine production and viral permissiveness. In addition, we investigated pharmaceutical and molecular approaches to promote antiviral immunity within the framework of cell activation statuses (2, 17, 18, 21) and showed that a lipid mediator and an IFN-expressing PRRSV vector successfully polarized porcine cells toward antiviral protection, which may facilitate novel adjuvant/vaccine approaches (21).
MATERIALS AND METHODS
Animals and isolation of primary cells.
Animal procedures and the isolation of alveolar macrophages and peripheral blood mononuclear cells (PBMCs) were conducted as described previously (29, 31–37). PBMCs were isolated using a 60% Ficoll-Paque Plus gradient (GE Healthcare, Piscataway, NJ). Monocytes were isolated from PBMCs with an anti-CD14 antibody (Ab), using magnetic beads conjugated with the corresponding secondary Ab (Miltenyi Biotec, Auburn, CA), and further phenotyped by a two-color immunofluorescence facilitated flow cytometric analysis (38). The monocyte-derived DCs (mDCs) were generated by culturing monocytes in the presence of IL-4 and granulocyte-macrophage colony-stimulating factor (GM-CSF) for 7 days as previously described (36, 37). Lavage fluids were centrifuged at 400 × g for 15 min to collect cells and further isolate Mϕs by plastic adherence (36, 37). Properties of both macrophages and mDCs were verified according to previously established procedures (35, 36). Cells were used immediately or cryopreserved in Recovery cell culture freezing medium (Invitrogen, Carlsbad, CA).
Cell polarization and cytokine phenotyping.
Mediators and conditions for polarization of porcine monocytic cells were applied as previously described (1–5, 29). In brief, Mϕs and DCs were stimulated with the mediators LPS, IFN-γ, IL-4, IL-10, IFN-α, and IFN-β at 20 ng/ml, and blood monocytes (BMs) were stimulated with either GM-CSF at 50 ng/ml or M-CSF at 100 ng/ml for 30 h. All mediators (purchased from R&D Systems, Minneapolis, MN, or Sigma-Aldrich, St. Louis, MO) were dissolved in 1× Dulbecco's phosphate-buffered saline (DPBS) (Invitrogen) containing 1% bovine serum albumin (BSA) (fraction V, cold ethanol precipitated; Sigma) and applied (1:100) to the cultured cells. For control cells, only BSA in DPBS was added to the cultures. Cells were then thoroughly washed and replenished with fresh medium for 16 h. Culture supernatants from an equal amount of cells from each treatment were collected, and secreted cytokines were measured using the SearchLight chemiluminescent multiplex assay (Aushon BioSystems, Billerica, MA) (24). Data were normalized as fold changes relative to the control cells and are presented as means of three independent replicates. Alternatively, polarized Mϕs and DCs were infected with the PRRSV strain incorporated with a red fluorescent protein (Ds-Red) for 16 h (29). The cells were then fixed with 4% formaldehyde in PBS, and PRRSV-positive cells and the expression of CD163 (a scavenger receptor critical to PRRSV cell entry) were examined using both two-color fluorescent flow cytometry and fluorescence microscopy. The antibody for CD163 detection was a fluorescein isothiocyanate (FITC)-conjugated mouse monoclonal antibody (MAb) (clone 2A10/11; AbD Serotec, Raleigh, NC) used directly for evaluation with flow cytometry or together with indirect labeling of the CD163 MAb by an Alexa Fluor 488-conjugated anti-mouse IgG (Invitrogen) for imaging by fluorescence microscopy. The CD163-positive cells in pictures were counted with ImageJ (http://imagej.nih.gov/ij/).
Genome-wide profiling marker genes related to macrophage activation statuses.
The expression of marker genes relative to each activation status and antiviral state was revealed using next-generation transcriptome sequencing (RNA-Seq) for genome-wide screening and confirmed family-wide using real-time reverse transcription-PCR (RT-PCR) assays. For RNA-Seq, equal quantities of primary alveolar macrophages were polarized for 30 h individually by the verified procedure described above and further infected for 5 h with a PRRSV strain as previously described (29). RNA preparation, RNA-Seq performance, and differentially expressed gene (DEG) analysis were done as previously described (29). In brief, 3 × 107 cells at each activation status (including the control DPBS mock stimulation) were pooled from three technical replicates representing cells obtained from four outbred pigs. Instead of applying the RNA-Seq procedure individually to each biological or technical replicate, replicates were pooled before proceeding to RNA-Seq analysis. In this case, the differentially expressed genes between two samples were assayed based on an algorithm as described previously (29). Functional classification of genes was conducted through Gene Ontology using the DAVID web tool (29, 30, 39). This method maps all DEGs to GO terms in the database (http://www.geneontology.org/), calculates gene numbers for every term, and then uses a hypergeometric test to find significantly enriched GO terms in DEGs compared with the genome background. The calculating formula is
where i is the number of successes, N is the number of all genes with GO annotation, n is the number of DEGs in N, M is the number of all genes that are annotated to the certain GO terms, and m is the number of DEGs in M. The calculated P value went through Bonferroni correction using the corrected P value of ≤0.05 as a threshold. GO terms fulfilling this condition were defined as significantly enriched GO terms in DEGs; this analysis recognizes the main biological functions that DEGs exercise. In addition, the expression of some genes was verified using real-time RT-PCR assays and a two-dimensional difference in gel electrophoresis (2D-DIGE) proteomic procedure (Applied Biomics, Inc., Hayward, CA) (28).
Antiviral/activation regulation with a lipid mediator, ToFA.
A cell-permeable inhibitor of acetyl coenzyme A (acetyl-CoA) carboxylase, 5-(tetradecyloxy)-2-furoic acid (ToFA) (Sigma-Aldrich) was added (5 μg/ml) to culture medium containing porcine BMs, Mϕs, and DCs for 30 h (40). Secretion of cytokines in the medium and expression of signature genes were analyzed as described above to determine the phenotype of the ToFA-induced activation status. The antiviral effects of ToFA treatment were evaluated by suppression of PRRSV replication and stimulated production of cytokines.
Construction of IFN-expressing PRRS viruses and infectivity analyses.
We have constructed and produced a series of IFN-expressing PRRS viruses for evaluation of the effect of in situ-expressed IFNs associated with the virus replication. The previously constructed IFN-expressing constructs, successfully incorporated the intact coding regions of porcine IFN genes coding for IFN-α1, IFN-β, IFN-δ3, and IFN-ω5 and showed in situ production of IFN polypeptides in the virus-infected cells and inhibition of coinfected PRRSV corresponding to the subtype-specific anti-PRRSV activity (37). A question we confronted for molecular vaccine design, however, is how can sufficient PRRSV replication activity be achieved when expressing an IFN with high PRRSV antiviral activity? To address this problem, we inserted a leader linker in front of the IFN coding region that encodes a six-histidine tag, (His)6, plus a 10-residue polypeptide containing a C-terminal cleavage peptide (GKPILFFRLK) of cathepsin D (CSTD), a protease induced during PRRSV infection in Mϕs (Y. Sang, unpublished data). Two 36-nucleotide (nt) complement oligonucleotides containing codons optimized for encoding the cathepsin-cleavage peptide were synthesized (IDT DNA, Coralville, IA) with EcoRI cloning overhangs at their 5′ and 3′ termini. The synthetic inserts were ligated into a pHUE expression vector through EcoRI cloning site to fuse with the N-terminal vector sequence encoding the (His)6 tag and the C-terminal IFN-α6 coding region (previously cloned through the vector EcoRI and HindIII cloning sites). The cDNA flanking the region coding (His)6-GKPILFFRLK-IFN-α6 was amplified from authentic clones using a Phusion high-fidelity PCR (NEB), and two restriction enzyme digestion sites (AflII and MluI) were introduced into 5′ and 3′ ends of the coding region. The amplified coding region was purified and cloned into the expression cassette of the PRRSV infectious cDNA clone, pCMV-P129, following the previously described procedure (37). The rescue, titration, and infectivity analysis of progeny viruses were performed as described for other bioengineered PRRS viruses (37).
Data analyses.
The quality of RNA-Seq data was analyzed in terms of the proportion of gene-mapped reads and the saturation/randomness for genome-wide gene coverage using published procedures (29, 30, 39). Differentially expressed genes (DEGs) (genes with a false discovery rate [FDR] of ≤0.001, a fold change of >2) were profiled with an edgeR package (39). The gene expression data from real-time RT-PCR were normalized against threshold cycle (CT) values of internal reference genes and control samples (33, 34). Differences between groups were determined by Student's t test.
RESULTS
Polarization of monocytes, Mϕs, and mDCs in vitro and cytokine phenotyping.
Both forward and reverse approaches have been used to determine the activation statuses of porcine monocytic innate immune cells (2–5). In brief, primary tissue Mϕs, BMs, and DCs were polarized by well-established activation mediators and the expression of marker genes was analyzed at RNA and protein levels (forward). Confirmed signature genes then were used to classify the subsets of primary cells or specifically activated cells (reverse). To this end, we polarized BMs, alveolar Mϕs, and mDCs with LPS and IFN-γ for M1, IL-4 for M2a, IL-10 for M2c, and IFN-α/β for the antiviral state (Table 1) (2–5). Porcine monocytic cells were responsive to these mediators, and the corresponding cytokines produced elicited the proposed functional statuses. For example, LPS-/IFN-γ-induced M1 Mϕs (M1-LPS or M1-IFN-γ status) showed upregulation of proinflammatory cytokines, including IL-1β, IL-6, and IL-8; in contrast, IL-4-/IL-10-induced M2 cells (M2-IL-4 or M2-IL-10 status) displayed downregulation of these cytokines. Interestingly, Mϕs treated with type I IFNs to induce antiviral states had cytokine production patterns compatible with the activation paradigm. In particular, IFN-α-treated Mϕs were similar to M1-IFN-γ cells and IFN-β-treated Mϕs were similar to M2-IL-10 cells regarding the increase or decrease of cytokines. Blood monocytes primed with either GM-CSF or M-CSF showed dramatic increases in IL-1β, IL-8, tumor necrosis factor alpha (TNF-α), and IL-10; however, these two early-phase priming cytokines showed different responses in regulation of IL-6 and IL-8 (5). Monocyte-derived DCs (mDCs) showed a similar tendency, except they were less responsive in production of proinflammatory cytokines, such as IL-1β, IL-6, IL-12, and TNF-α; however, all had increased production of IL-8, indicating a high migration potential of mDCs (1, 6). In addition, IFN-β appeared more active in stimulating IL-1β production in mDCs than in alveolar Mϕs (Table 1).
TABLE 1.
Cytokine responses corresponding to porcine monocytic cell activation statusesa
| Cell type and mediator | Cytokine response (fold change)b |
|||||
|---|---|---|---|---|---|---|
| IL-1β | IL-6 | IL-8 | IL-12 | TNF-α | IL-10 | |
| Mϕs | ||||||
| LPS | 6.3 ± 0.6* | 0.9 ± 0.1 | 3.9 ± 0.4* | 1.0 ± 0.0 | 1.2 ± 0.1 | 1.0 ± 0.0 |
| IFN-γ | 2.0 ± 0.2* | 1.8 ± 0.2* | 1.4 ± 0.1 | 8.1 ± 0.8* | 1.0 ± 0.1 | 2.2 ± 0.2* |
| IL-4 | −1.8 ± 0.2* | −1.4 ± 0.2 | −2.3 ± 0.2* | 1.0 ± 0.0 | −1.1 ± 0.1 | 1.0 ± 0.0 |
| IL-10 | −3.0 ± 0.3* | 1.2 ± 0.2 | −5.0 ± 0.5* | 5.7 ± 0.5* | −1.4 ± 0.1 | 2.4 ± 0.2* |
| IFN-α | 1.6 ± 0.1 | 1.6 ± 0.2 | 1.7 ± 0.2* | 6.9 ± 0.7* | −1.5 ± 0.1 | 1.7 ± 0.2* |
| IFN-β | −1.5 ± 0.1 | 1.2 ± 0.1 | −4.6 ± 0.4* | 4.1 ± 0.4* | 1.2 ± 0.1 | 2.6 ± 0.2* |
| BMs | ||||||
| GM-CSF | 3.3 ± 0.1* | 2.1 ± 0.1* | 3.6 ± 0.0* | ND | 1.6 ± 0.4 | 6.9 ± 1.1* |
| M-CSF | 2.8 ± 0.1* | 1.4 ± 0.1 | 12.5 ± 0.1* | ND | 2.3 ± 0.6* | 7.1 ± 1.2* |
| mDCs | ||||||
| LPS | 1.8 ± 0.2* | 1.6 ± 0.2 | 3.3 ± 0.3* | ND | −2.7 ± 0.3* | −4.2 ± 0.4* |
| IFN-γ | 1.2 ± 0.1 | 1.1 ± 0.1 | 2.3 ± 0.2* | ND | −1.2 ± 0.1 | 1.0 ± 0.0 |
| IL-4 | −1.9 ± 0.2* | 2.3 ± 0.3* | 6.3 ± 0.6* | ND | −1.6 ± 0.2 | −1.4 ± 0.1 |
| IL-10 | −8.3 ± 0.8* | 1.0 ± 0.0 | −1.9 ± 0.2* | ND | −2.1 ± 0.4* | 2.1 ± 0.2* |
| IFN-α | 1.7 ± 0.2* | 1.0 ± 0.0 | 2.6 ± 0.2* | ND | −1.3 ± 0.1 | 1.0 ± 0.0 |
| IFN-β | 1.9 ± 0.2* | 1.0 ± 0.1 | 3.5 ± 0.3* | ND | −1.1 ± 0.1 | 1.1 ± 0.0 |
Cells were stimulated with the mediators (20 ng/ml each, except for GM-CSF at 50 ng/ml and M-CSF at 100 ng/ml in PBS containing 5% BSA) for 30 h and then thoroughly washed and replenished with fresh medium for 16 h. Culture supernatants were collected, and cytokine levels were measured.
Data are means ± SE from three replicates. Negative fold change values indicate a decrease. *, P < 0.05 relative to the control. ND, not determined.
Family-wide profile of cytokine and chemokine genes related to the activation status of porcine Mϕs upon PRRSV infection.
In addition to evaluating the secreted cytokines, we also used a nonbiased RNA-Seq procedure to profile the genome-wide response of the cytokine-cytokine receptor system in porcine Mϕs at different activation statuses. Serial assessments for quality control were conducted to verify that our RNA-Seq procedure and data met the criteria for genome-wide transcriptomic assays (29). As shown in Table 2 and Fig. S1 in the supplemental material, family-wide expression of cytokines, chemokines, and their receptors was detected. Gene expression of inflammatory cytokines, including IL-1β, IL-6, IL-8, IL-10, IL-12, and TNF-α, was consistent with the multiplex assays at the protein level, except for the highest expression of the IL-10 gene in Mϕs at the M1-LPS status (Table 1). In addition to these well-studied inflammatory cytokines, we also showed the status-specific expression of other cytokine-related genes at one or two statuses (Table 2; see Fig. S1). These novel cytokine- or chemokine-related gene markers included IL7, IL10RA (coding for IL-10 receptor alpha), IL15RA, and CCL21 in the M1-IFN-γ status, as well as IL4RA, IL6RA, IL10RB, CSF2RB, and CXCL4 particularly in the M2-IL-10 status. In general, we found 21 to 35 cytokine/chemokine-related genes significantly upregulated in each activation status: the M2-IL-10 status had the lowest number (i.e., 21), and the M1-LPS status had the highest number (i.e., 29). Notably, most chemokine genes were costimulated significantly in both M1-IFN-γ and IFN-α-activated antiviral (MaV-IFN-α) statuses. Some cytokine-related genes were specifically suppressed in certain activation statuses, including CCL19 and CCL22 in the IFN-γ-M1 status, CCL2 and CCL23 in the M2-IL-4 status, IL4RA and CCL21 in the M1-LPS status, and IL10RA, IL-18, and CCL8 in the MaV-IFN-α status. Overall, we identified 96 porcine transcripts of cytokines/chemokines and their receptors, and many of them, such as the genes of five porcine IL-17 receptors, were for the first time revealed family-wide at the transcriptional level (Table 2; see Fig. S1). These findings indicate that the RNA-Seq data could be exploited for identification of novel transcripts in addition to genome-wide quantitative analyses. Toll-like receptors (TLRs) are among the most important cellular receptors to recognize pathogen-associated molecular patterns and induce the production of immune effectors, including cytokines. Annotation of our RNA-Seq data revealed significant regulation of all porcine TLR paralogs except TLR5, which was specifically expressed on the basolateral surface of intestinal epithelial cells but not expressed in porcine alveolar macrophages (see Fig. S1C) (21). Corresponding to discoveries in murine macrophages, high expression of TLR2, TLR7, and TLR8 was found in cells activated at the M1-LPS, MaV-IFN-α, and M2-IL-10 statuses, respectively (Fig. 1A; see Fig. S1C). Constitutive expression of multiple TLRs (TLR2, TLR4, TLR6, and TLR8) was clearly associated with most activation statuses, indicating the immunocompetent potency of alveolar macrophages in response to diverse pathogenic stimuli (21). For confirmation, expression of genes of the IL-17, TLRs, and guanylate binding protein (GBP) families were also verified using an RT-PCR protocol (29) (Fig. 2A).
TABLE 2.
Cytokine and cytokine-receptor genes significantly regulated in macrophages at different activation statuses
| Status | Genes (total no.)a |
|
|---|---|---|
| Upregulated | Downregulated | |
| IFN-γ-M1 | IL1B, IL4RA, IL-6, IL-7, IL-8, IL10RA, IL12A, IL12B, IL12Rb1, IL13RA, IL-15, IL15RA, IL17RC, IL-18, IL18BP, IL23RA, IL27A, TNF, TNFAIP1, CSF2RA, CSF3, CCL2, CCL4, CCL5, CCL8, CCL20, CCL21, CCL23, CXCL9, CXCL10, CXCL11, CXCL16, CCR2L, CCR5, CCR7 (35) | IL1A, IL2RG, IL6RA, IL7RA, CSF1R, CSF2RB, CCL17, CCL19, CCL22, CCL24, CXCL4, CXCL14, CXCR2, CXCR4 (14) |
| IL-4-M2 | IL4I1, IL-7, IL7RA, IL-10, IL17RA, IL17RB, IL17RC, IL-18, IL23RA, IL27RA, CSF1R, CSF2RA, TGFB, CCL5, CCL8, CCL17, CCL22, CCL24, CXCL16, CCR7, CXCR2, CXCR2L(22) | IL1A, IL1B, IL2RG, IL-8, IL13RA, TNFAIP1, CSF3, CSF2RB, CCL2, CCL3, CCL4, CCL23, CXCL2, CXCL4, CXCL10, CXCL14, CCRL2, CCR5, CXCR4 (19) |
| LPS-M1 | IL1A, IL1B, IL1E, IL1RN, IL1R2, IL2RA, IL-5, IL-7, IL-8, IL-10, IL12A, IL12B, IL-18, IL18BP, IL-19, IL23P19, IL27A, IL27B, IL-33, TNF, TNFAIP1, CSF3, CCL2, CCL3, CCL4, CCL5, CCL8, CCL17, CCL20, CCL22, CCL23, CCL24, CXCL2, CCR7, CXCR2 (35) | IL2RG, IL4RA, IL6RA, IL7RA, IL11RA, IL13RA, IL15RA, IL-16, IL17RA, IL17RC, IL17RE, IL23RA, IL27RA, CSF1R, CSF2RA, CSF2RB, TGFB, CCL21, CXCL4, CXCL10, CXCL14, CXCL16, CCRL2, CCR5, CXCR4 (25) |
| IL-10-M2c | IL4RA, IL6RA, IL-7, IL7RA, IL-8, IL10RB, IL13RA, IL15RA, IL-18, IL18BP, IL21R, IL23RA, IL27RA, TNFAIP1, CSF1R, CSF2RB, CCL23, CXCL2, CXCL4, CXCL16, CXCR4 (21) | IL1A, IL1B, CSF2RA, CCL3, CCL4, CXCL10 (6) |
| IFNα-MaV | IL1RN, IL2RG, IL4I1, IL6RB, IL-7, IL7RA, IL-10, IL12Rb1, IL-15, IL15RA, IL17RA, IL18BP, IL21R, IL27A, IL27B, TNFAIP1, CSF2RA, CCL2, CCL4, CCL5, CCL23, CCL25, CXCL9, CXCL10, CXCL11, CXCL13, CXCL16, CCR2L, CCR5, CCR7 (30) | IL1A, IL1B, IL1R2, IL-8, IL10RA, IL10RB, IL13RA, IL17RC, IL-18, IL23RA, CSF3, CSF1R, CSF2RB, TGFB, CCL3, CCL8, CCL17, CCL24, CXCL2, CXCL4, CXCR1, CXCR2, CXCR4 (24) |
P (FDR) < 0.001 for a fold change of >2 for significant determination. Gene designations are according to the NCBI Gene database at http://www.ncbi.nlm.nih.gov/gene/. Boldface indicates genes up- or downregulated only in the indicated status but not in other statuses.
FIG 1.
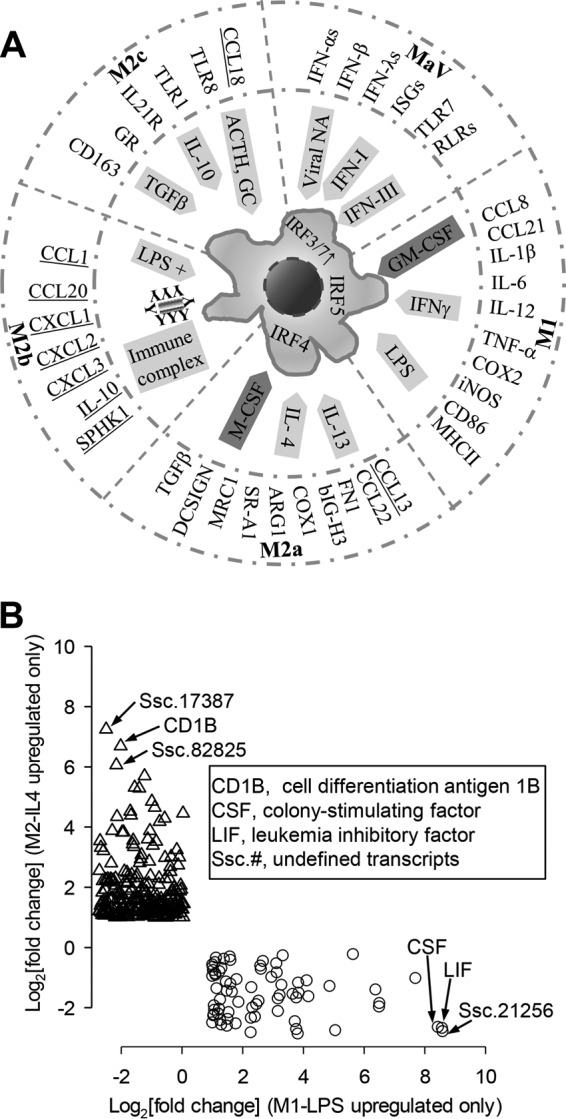
(A) Integration of antiviral states into the macrophage activation paradigm. Macrophages (Mϕs) are capable of displaying different functional phenotypes under the stimuli of specific mediators. Four conventional subtypes of Mϕ activation statuses (M1, M2a, M2b, and M2c) and antiviral states (MaV) are defined based on primary inductive mediators illustrated in the inner circle along with subtype-specific IRFs labeled in the cell. Mϕ polarization is associated with distinctive gene signatures selectively listed adjacent to the outer circle. As illustrated, most listed signature genes were identified and showed high expression corresponding to each activation status. The underlined M2b status was not tested, and porcine orthologs of CCL13 and CCL18 have not been identified thus far. (B) Illustration of potential signature genes profiled using RNA-Seq. Potential marker genes were grouped based on significant upregulation only in one activation status, such as LPS-stimulated (along the x axis) or IL-4-stimulated (along the y axis) cells. *, P (FDR) < 0.001; fold change, ≥2 for significant determination. Key abbreviations: ACTH, adrenocorticotropic hormone; ARG1, arginase 1; bIGH3, matrix-associated protein betaIG-H3; COX, cyclooxygenase; DCSIGN, DC-specific intercellular adhesion molecule-3-grabbing nonintegrin; FN1, fibronectin 1; GC, glucocorticoid; GR, GC receptor; IFN-I/III, type I or III interferon; iNOS, inducible nitric oxide synthase; IRF, IFN regulatory factor; ISG, IFN-stimulated genes; MRC1, mannose receptor C type-1; NA, nucleic acid; SPHK1, sphingosine kinase 1; RLR, RIG-I-like receptor; TLR, Toll-like receptor.
FIG 2.
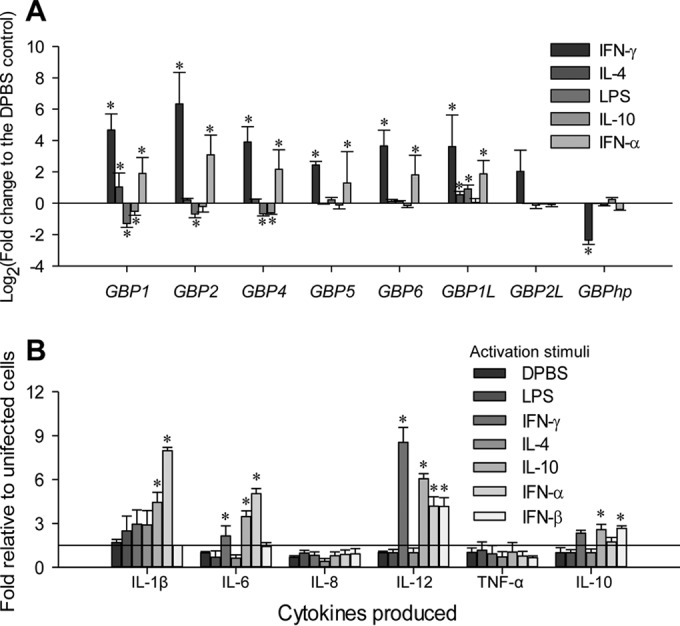
(A) Differential expression of the guanylate binding protein (GBP) family in polarized macrophages was verified with a real-time RT-PCR assay. (The y axis scale indicates fold change relative to M0-PBS status.) Aliquots of the cell RNA used for RNA-Seq were analyzed using a SYBR green-based real-time RT-PCR assay. CT values of the genes were normalized against CT values of a housekeeping gene (coding for glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) amplified from the same RNA samples to obtain 2−ΔCT, and the values of 2−ΔCT from infected cells were plotted against that from control cells to indicate up- or downregulation and to reflect the relative expression. *, P < 0.05; n = 3. Primers for RT-PCR detection are listed in Table S1 in the supplemental material. (B) Differential cytokine response to PRRSV infection in cells primed with different activation stimuli. Porcine alveolar Mϕs were treated as in Table 1 and infected with PRRSV (SDSU28983) for 16 h after 30 h of stimulation. Cytokine concentrations in culture supernatants from the infected cells are plotted against those from uninfected cells as in Table 1. Data are means ± standard errors (SE) from three replicates. *, P < 0.05 relative to the DPBS control.
Genome-wide profile of classic and species-specific signature genes associated with the activation status of porcine Mϕs upon PRRSV infection.
Phenotypic manifestation of activation statuses in monocytic cells has been associated with various exogenous stimuli and, in turn, endogenous expression of signature genes, including intracellular transcription factors, metabolic enzymes, and surface receptors, as well as secreted cytokine peptides (1–5). A series of typical signature genes have been characterized in macrophages at different activation statuses in humans and mice; however, their similarity or difference in pigs has not been systematically studied. Figure 1A shows signature genes pertinent to each well-characterized activation status in mice (1–5). Except for the M2b status, not evaluated herein, and genes CCL13 and CCL18, whose porcine orthologs have not been identified (41), most other listed signature genes were identified and showed high expression for each activation status. Notably, the antiviral state (MaV-IFN-α) integrated and fit very well in the paradigm of Mϕ activation statuses (Fig. 2B). For example, cells in the MaV-IFN-α state were primed primarily by type I or type III IFNs and underwent further maturation by TLRs or other receptors sensing viral molecular patterns (7, 10, 12, 15, 21). Signature genes characteristic of the antiviral state include those involved in the activation of IRF-3/IRF-7 and stimulation of antiviral ISGs (2, 7, 10, 12, 15). Based on our studies and those of others, we know that induction of the antiviral state is common in all monocytic cells (2, 10, 17, 18, 21), but whether subsets of monocytes and Mϕs are preset for antiviral responses, such as pDCs showing a natural propensity of IFN-α production, is not known (11, 24).
The unbiased RNA-Seq procedure also allowed us to profile species-specific signature genes pertaining to porcine macrophages at different activation statuses. For example, evaluation of potential signature genes identified 72 and 234 significant DEGs that were upregulated only at the LPS-M1 and IL-4-M2 statuses, respectively (Fig. 1B). These potential marker genes include some known genes, such as CD1B (coding for a major histocompatibility complex [MHC] protein mediating presentation of lipid antigens to T cells) and the genes coding for colony-stimulating factor (CSF) and leukemia inhibitory factor (LIF), in addition to transcripts with unknown identities (Fig. 1B). Most of these genes have not been studied for regulation of activation statuses and antiviral immunity.
PRRSV infection differentially altered cytokine production in macrophages at different activation statuses.
Because viruses such as HIV and RSV altered activation statuses in macrophages (16–19), we examined whether PRRSV infection interacts differentially with macrophages at different activation statuses. PRRSV infection induced differential cytokine responses in Mϕs polarized with different mediators (Fig. 2B). For example, MaV-IFN-α and M1-IFN-γ cells were more responsive than IFN-β-activated and M2-IL-4 cells in the production of proinflammatory cytokines, including IL-1β, IL-6, IL-8, IL-12, and TNF-α. In contrast, IFN-β-activated cells were most responsive in producing IL-10 upon virus infection. Although M2-IL-10 cells were intermediate, M2-IL-4 cells were rather inactive in cytokine production upon PRRSV infection (Fig. 2B). Interestingly, both IL-8 and TNF-α were not appreciably altered by PRRSV infection in macrophages at any activation status (Fig. 2B). Thus, porcine macrophages at different activation statuses responded heterogeneously to PRRSV infection with respect to cytokine production.
Macrophage and dendritic cells at different activation statuses differ in their permissiveness to PRRSV infection.
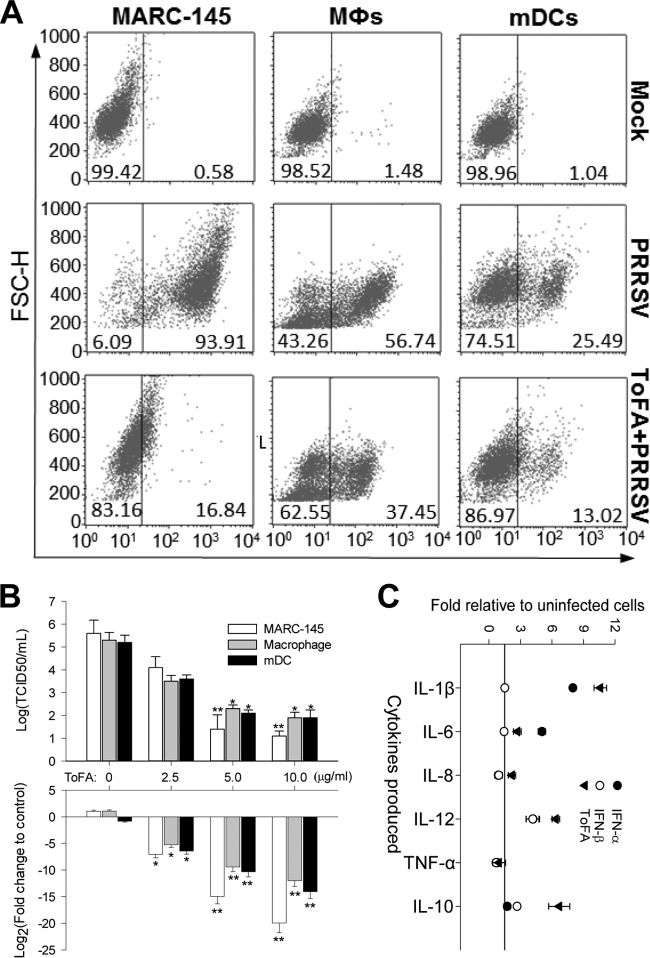
If PRRSV affects macrophage activation statuses via regulation of cytokine production, then does activation status of monocytic cells influence PRRSV infection? In pigs, subsets of lung Mϕs and mDCs are the primary cells supporting PRRSV infection, and PRRSV infections frequently promote anti-inflammatory cytokine IL-10 production but suppress proinflammatory cytokines, such as IL-12 (20–22). Using porcine monocytic cells polarized in vitro, we tested if PRRSV infection was altered in cells at different activation statuses. PRRSV infection was monitored using antibody to the PRRSV N protein and quantified by flow cytometry (33, 34). M2 polarization with IL-4 and IL-10 increased the cell population that was permissive to PRRSV infection by 10 to 20% in both Mϕs and mDCs (Fig. 3A). M1 polarization with LPS and IFN-γ showed a slight decrease in the population of permissive cells. Although both IFN-α and IFN-β have been shown to induce antiviral states (12–15), only IFN-α and not IFN-β significantly suppressed PRRSV infection in Mϕs (Fig. 3A and Table 3). IFN-α also inhibited PRRSV replication in cells showing a 10-fold decrease in fluorescence intensity of labeled PRRSV N proteins (Fig. 3A). In addition, we also monitored the expression of CD163, a scavenger receptor critical to PRRSV cell entry, and an M2-IL-10 cell marker (2, 26). Previous studies reported 60% to 95% CD163-positive cells in porcine alveolar Mϕs using indirect immunodetection of CD163 (26, 42). To study CD163 expression dynamics and correlate CD163 expression data detected using either direct or indirect immunodetection, we applied both detection procedures in alveolar Mϕs in different culture and activation states. As shown in Fig. 3B, while indirect immunodetection generally stained 60 to 80% of alveolar Mϕs to be CD163 positive, which is close to previous results (26, 42), the direct immunostaining using FITC-conjugated CD163 MAb detected only 5 to 15% of CD163high cells, which is consistent with our evaluation using the flow cytometric procedure in Fig. 3A and Table 3. Dynamically, primary alveolar Mϕs prior to culture had much less CD163-positive cells (2 to 5% by indirect immunostaining), the 30-h-culture condition dramatically elevated CD163-positive cells over 10-fold to 40 to 50% (42), and polarization mediators and PRRSV infection altered around 30% of overall CD163-positive cells. IL-10 significantly upregulated CD163 expression in mDCs (Fig. 3). GM-CSF and M-CSF, two colony-stimulating factors involved in priming the early phase of differentiation in monocytic cells, increased Mϕs and BMs that were permissive to PRRSV infection by approximately 20% (Table 3). In summary, porcine monocytic cells at different activation statuses not only respond but also are different in their permissiveness to PRRSV infection (Fig. 2B and Fig. 3). Because PRRSV infection may inevitably alter the cytokine response in porcine monocytic cells (Fig. 2B), it is likely that the virus may coopt the cell activation status to promote virus infection and exacerbate disease development in vivo.
FIG 3.
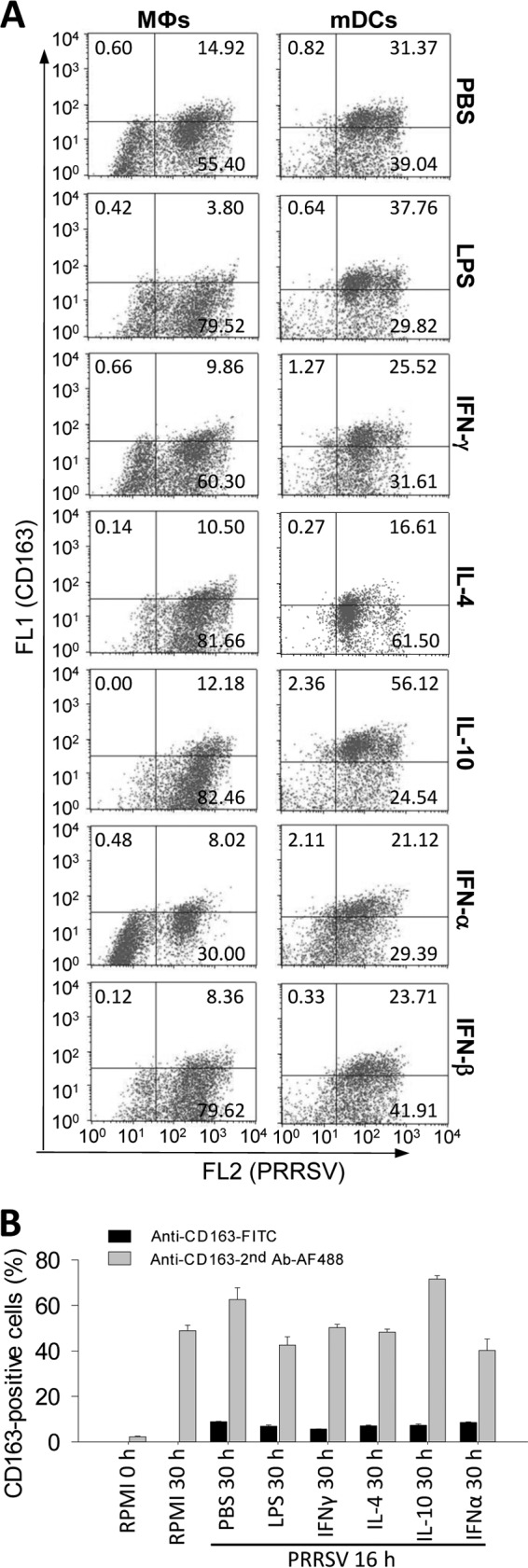
Effects of cell polarization on PRRSV infection. (A) Cells were treated with the indicated mediators as in Table 1, thoroughly washed, and infected with PRRSV for 16 h. Cells were collected and immunostained with specific MAbs for the PRRSV N protein (FL2, R-phycoerythrin) and CD163 (FL1, FITC), a scavenger receptor critical for PRRSV infection and a signature marker for M2-IL-10 activation. Positive cells were counted using flow cytometry. The data represent a set of four independent assays as presented in Table 3. (B) CD163-positive cells were detected using direct or indirect immunofluorescent approaches in alveolar macrophages under different culture and activation statuses. The CD163-positive cells in micrographs were counted with ImageJ (http://imagej.nih.gov/ij/).
TABLE 3.
Regulation of PRRSV infection and expression of CD163 by activation mediators
| Cell type and indicator | % of cells showing PRRSV-positive and high CD163 expressiona: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| PBS | LPS | IFN-γ | IL-4 | IL-10 | IFN-α | IFN-β | GM-CSF | M-CSF | |
| Mϕs | |||||||||
| Virus+ | 68.1 ± 5.4 | 80.3 ± 7.5 | 67.6 ± 6.3 | 88.8 ± 8.2* | 91.2 ± 8.5* | 36.6 ± 3.4* | 84.8 ± 8.0 | 89.1 ± 8.3* | 88.1 ± 8.2* |
| CD163high | 15.0 ± 1.4 | 4.1 ± 0.4* | 10.1 ± 1.0 | 10.2 ± 1.0 | 11.7 ± 1.1 | 8.2 ± 0.8* | 8.2 ± 0.8* | 10.4 ± 1.0 | 12.3 ± 1.2 |
| mDCs | |||||||||
| Virus+ | 69.5 ± 1.7 | 65.1 ± 6.1 | 55.0 ± 5.1 | 75.2 ± 7.0* | 77.7 ± 7.3* | 48.7 ± 4.5* | 63.2 ± 5.9 | ||
| CD163high | 31.0 ± 2.9 | 37.0 ± 3.5 | 25.8 ± 2.4 | 16.3 ± 1.5* | 56.3 ± 1.5* | 22.4 ± 2.1* | 23.2 ± 2.2* | ||
| BMs | |||||||||
| Virus+ | 38.8 ± 3.6 | 65.1 ± 6.1* | 55.9 ± 5.2* | ||||||
| CD163high | 5.1 ± 0.5 | 13.4 ± 1.3* | 10.2 ± 0.5* | ||||||
Percentages are of 5,000 cells (partial list from Fig. 3). Data are means ± SE from four replicates. *, P < 0.05 relative to the control (PBS).
GO analysis indicates that IFN signaling and lipid metabolism is significantly regulated in macrophages at different activation statuses.
For functional classification of DEGs from RNA-Seq data, we used Gene Ontology (GO) analysis to obtain expression patterns of DEGs annotated to given GO terms in the database (http://www.geneontology.org/). Collectively, DEGs of each activation status could be annotated to 493 to 1,970 GO terms; the most significant (correct, P < 0.05) GO terms enriched by the DEGs included those referring to immune system process, immune response, antigen processing and presentation, fatty acid/lipid metabolic process, and IFN signaling related to antiviral responses. Because deviation of the IFN system has been well demonstrated in PRRSV infection and is a potential target for antiviral regulation (7, 10, 12, 13, 15, 21), we first analyzed DEGs related to the IFN system, particularly type I IFN production and action in macrophages at differential activation statuses. In pigs, PRRSV strains exerted differential ability to inhibit the production and action of type I IFNs, particularly in IFN-α rather than IFN-β subtypes (21–25, 34). We previously showed that the swine genome contains at least 39 functional genes encoding 7 subtypes of type I IFNs, including multiple members of the IFNs IFN-α, IFN-δ, and IFN-ω as well as a single IFN-β gene (34). Emerging evidence shows that different subtypes of type I IFNs and even members of same subtype could be differentially responsive to viral infection (34, 43). Using RT-PCR assays, we showed that during the early phase (<6 h postinfection) of PRRSV infection in primary macrophages, the significant increase of IFN-β expression was accompanied by modest stimulation and even suppression of multiple IFN-α, IFN-δ, and IFN-ω subtypes (34; Y. Sang, data not shown). Here using RNA-Seq analysis, we show that except for IFN-β, which had a relatively higher expression than other IFN subtypes, isoform transcripts of the other subtypes were only slightly detectable, with few read counts and differential expression in cells at different activation statuses, compared to average RPKM (i.e., reads per kilobase transcript per million mapped reads) values of 10 to 40 of other cytokine genes, such as TNF (Fig. 4A). For example, IFN-α1, IFN-β, IFN-ω2, and IFN-ω6 were detected in cells at the M1-IFN-γ status, but IFN-α5, IFN-β, IFN-δ7, and IFN-ω4 were found in cells at the MaV-IFN-α status. In contrast, fewer isoforms of type I IFNs were detected in cells at the M1-LPS, M2-IL-4, and M2-IL-10 statuses. As expected, the highest expression of IFN-β was detected in cells at the M2-IL-10 regulatory status (Fig. 4A). Differential suppression of the IFN system (both type I and type II IFNs) during the early-phase PRRSV infection was also demonstrated using GO analysis, and the DEG cluster frequency of general cytokines was much higher than those of IFN production and response (Fig. 4B). Detail mapping of IFN receptors and other IFN-stimulated/induced genes (ISGs/IFIs) indicated that they retained higher expression in cells at the M1-IFN-γ and MaV-IFN-α statuses and were frequently suppressed at the M2-IL-4 status (Fig. 4C and D). For example, we analyzed IFN stimulation of the guanylate binding protein (GBP) family, which is located within the region of swine chromosome 4 (SSC4) and was recently genetically associated with pig anti-PRRSV immunity (44). Here we show that all porcine GBP genes, except an uncharacterized gene, GBP2L, were highly expressed in the macrophages upon both IFN-α and IFN-γ stimulation (Fig. 4D). Interestingly, we also detected a significant hypothetical transcript (GBPhp), clustered within porcine GBP region (Gene ID no. 100524372 in current swine genome assembly Sscrofa10.2), whose expression correlated negatively with IFN stimulation of porcine GBP genes (Fig. 4D). Thus, the general suppression of gene expression in the IFN system, particularly in IFN production, accompanied acute PRRSV infection in porcine macrophages, particularly at the M1-LPS and other M2 activation statuses (7, 10, 13, 15, 21).
FIG 4.
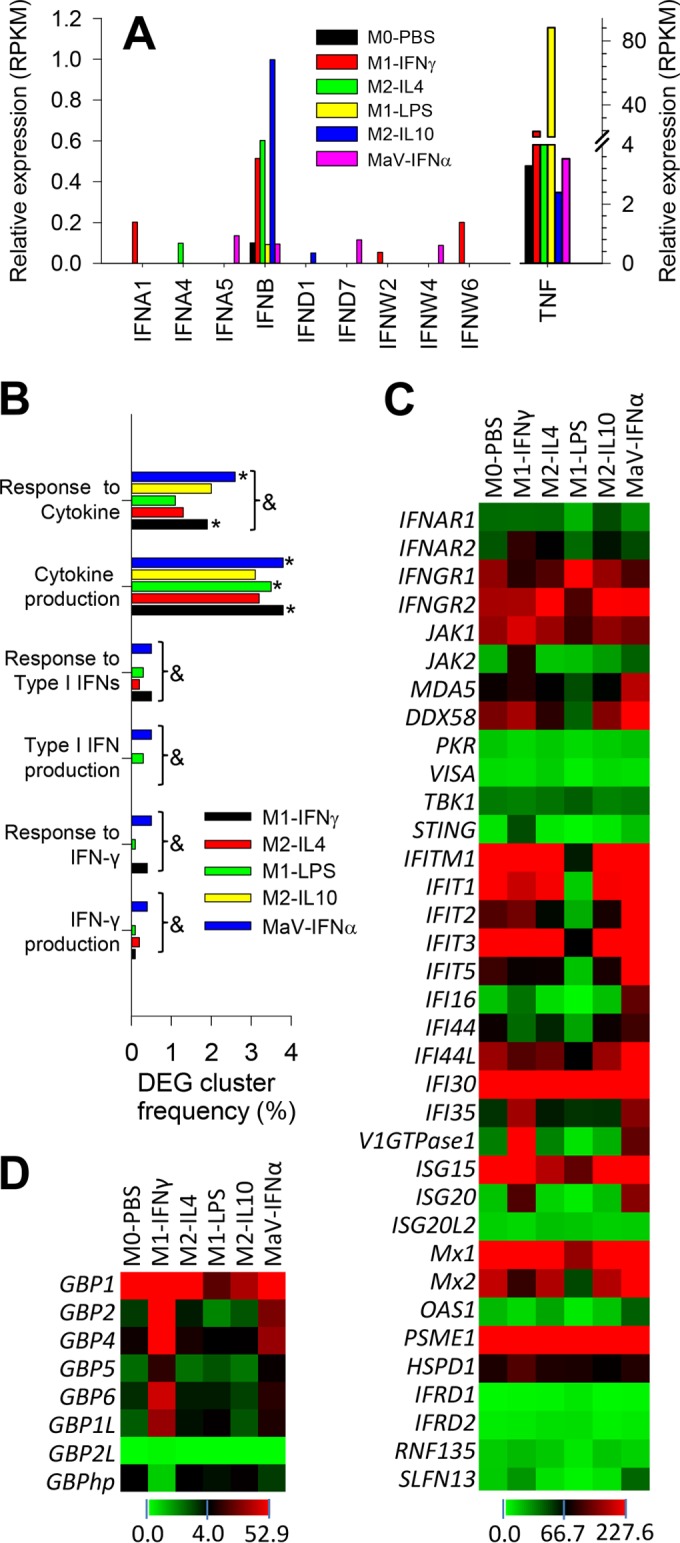
Quantitative expression and Gene Ontology (GO) analyses of interferon (IFN) production and action. (A) Quantitative expression analysis of type I IFN transcripts in PRRSV-infected macrophages at different activation statuses, showing suppressed type I IFN expression compared with the detection of TNF (tumor necrosis factor alpha). (B) GO analysis of DEGs clustered with IFN production and action compared with DEG cluster frequency to other cytokines, * and &, P < 0.05 compared with M0-PBS or among all statuses, respectively. (C) Heat map of the differential expression of genes encoding IFN receptors and IFN-stimulated/induced genes (ISGs/IFIs). (D) Family-wide analysis of the expression of genes encoding guanylate binding proteins (GBPs), a group of ISGs recently shown to be potentially associated with PRRSV pathogenesis (44). GBPhp is an unknown hypothetical gene that formed into a gene cluster with all other porcine GBP genes in chromosome 4 and had an expression pattern negatively correlated with most other porcine GBP genes. The color scale under each heat map illustrates the midpoint and range of RPKM values of the listed transcripts.
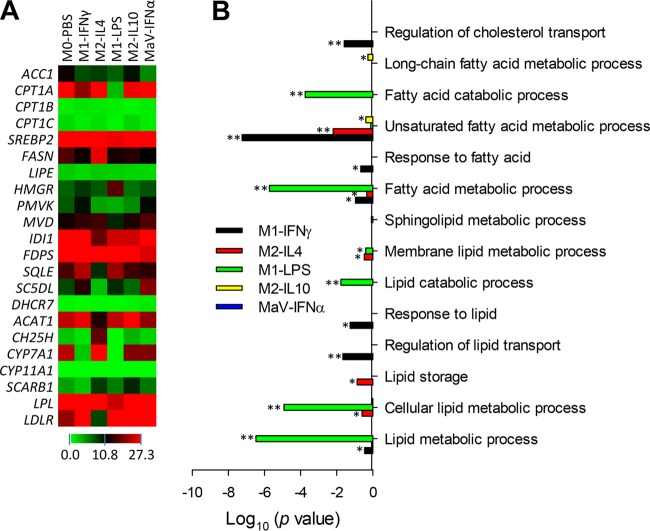
Using GO analyses, we determined lipid-related biological processes enriched by DEGs and their differential response in macrophages at different activation statuses upon PRRSV infection (Fig. 5 and Table 4). In general, DEGs associated with different activation statuses could be assigned to 14 to 56 GO terms pertinent to the processes of lipid/fatty acid biosynthesis, metabolism, and transport (Table 4). These total numbers of enriched GO terms were positively relevant to the DEG numbers filtered out at each activation status. For example, M1-LPS status and M2-IL-10 status had the highest (4,667) and lowest (153) number of DEGs and so were correspondingly enriched in lipid-related GO terms at the largest number, 56 and the smallest number, 14, respectively (Table 4). However, statistical analyses of the significance of DEGs enriched in these lipid-related biological processes indicated that despite comparable GO terms classified at the MaV-IFN-α state, no term was significantly enriched by DEGs (Table 4 and Fig. 5B). This finding indicated that lipid metabolic processes were generally arrested in macrophages at the MaV-IFN-α status. Further annotation of the key genes encoding enzymes catalyzing lipid metabolic processes indicated that most had relatively suppressed expression at the antiviral state (45). These suppressed key genes in lipid metabolism included FASN (encoding fatty acid synthase, the key protein for fatty acid synthesis), LIPE (encoding hormone-sensitive lipase, the key protein for lipolysis), HMGR (encoding 3-hydroxy-3-methylglutaryl-coenzyme-A reductase, the key protein for sterol synthesis), and particularly ACC1 (encoding acetyl-CoA carboxylase [ACC] alpha, the key protein for fatty acid oxidation) (Fig. 5A). Cholesterol metabolism was recently implicated in regulation of viral replication, and some cholesterol derivatives were shown to have direct antiviral activity (45, 46). As clearly shown in Fig. 5A, genes of most, if not all, key enzymes in cholesterol metabolism were significantly differentially regulated among cells at different activation statuses upon PRRSV infection.
FIG 5.
Heat map and gene ontology (GO) analyses of the biological processes related to lipid/fatty acid metabolism. (A) Differential expression of key genes in fatty acid oxidation, fatty acid synthesis, lipolysis, and sterol synthesis. (B) Significant biological processes of lipid/fatty acid metabolism enriched by DEGs in GO analysis. DEGs from the MaV-IFN-α state have no significantly enriched lipid metabolic pathways, implying an arrested state of lipid metabolism in macrophages in the MaV-IFN-α state (Table 3). *, P < 0.5; **, P < 0.05.
TABLE 4.
Numbers of DEG-enriched terms in GOa analysis of lipid/fatty acid metabolism
| Status | Total no. of GO terms | No. of significant GO terms at: |
|
|---|---|---|---|
| P < 0.5 | P < 0.05 | ||
| IFNγ-M1 | 49 | 7 | 3 |
| IL-4-M2 | 39 | 5 | 1 |
| LPS-M1 | 56 | 6 | 5 |
| IL-10-M2c | 14 | 2 | 0 |
| IFNα-MaV | 37 | 0 | 0 |
GO, Gene Ontology.
Modulation of the activation status of monocytic cells using a lipid mediator suppressing PRRSV infection.
For translational research based on profiling of gene response pathways, we are interested in developing a system to modulate the monocytic activation paradigm to aid in the management of PRRS (25, 47, 48). We began our investigation by targeting pathways that modulate inflammation, antiviral states, and T-cell proliferation. Because of the importance of lipid signaling in regulation of inflammation and overall immune responses, we first screened mediators that regulate lipid metabolism (49–51). Among multiple selected agonists and antagonists for enzymes in lipid metabolism, 5-(tetradecyloxy)-2-furoic acid (ToFA), a competitive inhibitor of acetyl-CoA carboxylase (ACC) (40), significantly suppressed PRRSV infection in all cells tested (Fig. 6A) with 90% effective concentration (EC90) and toxicity in terms of 50% cytotoxic concentration (CC50) at 6 and 25 μg/ml, respectively, in MARC-145 cells (a monkey cell line sensitive to PRRSV infection). ToFA at 5 μg/ml suppressed approximately 80% of PRRSV infection in MARC-145 cells and up to 50% in both porcine alveolar Mϕs and mDCs (Fig. 6A). Suppression of PRRSV infection in cells by ToFA treatment was also determined based on viral titers and viral replication for comparison of samples collected from ToFA-treated cells and the control cells. The dose-dependent assay showed that cells treated with ToFA at 5 to 10 μg/ml for 30 h significantly suppressed viral infection with respect to virus titers and viral genome replication (Fig. 6B). With respect to PRRSV-induced cytokine production, ToFA-treated Mϕs showed increased production of IL-1β, IL-8, IL-12, and IL-10, partially resembling that in IFN-α- or IFN-β-treated cells (Fig. 6C). ToFA-treated Mϕs also displayed the lowest response in IL-4 production. These data suggest that ToFA may have the potential to skew cells toward an antiviral/M1 status. Consistent with recent findings using mouse DCs (40), we also observed that ToFA-treated mDCs stimulate proliferation of blood T cells much better than the control cells (Y. Sang, unpublished data). These findings indicate that ToFA, through depletion of lipid content or alteration of lipid signaling (40), skews activation statuses by generally favoring antiviral or antigen-presenting processes, which may be similar to cells in the MaV-IFN-α state (Fig. 6).
FIG 6.
ToFA, a competitive inhibitor of acetyl-CoA carboxylase (ACC), suppresses PRRSV infection in cells and modulates the macrophage response to PRRSV infection with respect to cytokine production. (A and B) Cells (MARC-145, porcine alveolar macrophages, and monocyte-derived dendritic cell [mDCs]) were treated with ToFA (2.5 to 10 μg/ml diluted from a 5-mg/ml stock in dimethyl sulfoxide [DMSO]) or mock treatment (DMSO only for 30 h) and then infected with PRRSV at a multiplicity of infection (MOI) of 0.1 for 16 h for counting of PRRSV-positive cells (A), viral titration (upper part of panel B), and viral RNA detection (bottom part of panel B). Viral RNA was detected with a real-time RT-PCR assay to determine transcripts of PRRSV nucleocapsid (N) protein in total RNA (100 ng/20 μl PCR) purified from the infected cells. The quantitative PCR data were normalized against expression values (CT) of a housekeeping gene (coding for GAPDH) to obtain fold changes relative to the control cells. Viral titers were titrated in MARC-145 cells in 96-well plates using a serial dilution procedure as described previously (34). (C) Cells were stimulated with the mediators (IFN-α or IFN-β at 20 ng/ml and ToFA at 5 μg/ml) for 30 h and then washed and replenished with fresh medium containing PRRSV infected for 16 h before collection of medium for cytokine measurement as in Table 1. Data represent percentages of 10,000 cells counted by flow cytometry (A) or the mean ± SE from three replicates. *, P < 0.05, and **, P < 0.005, relative to the nontreated controls (B and C). The primers for RT-PCR detection are listed in Table S1 in the supplemental material.
Molecular modulation of the activation status of monocytic cells using IFN-expressing PRRSV to suppress coinfected viruses.
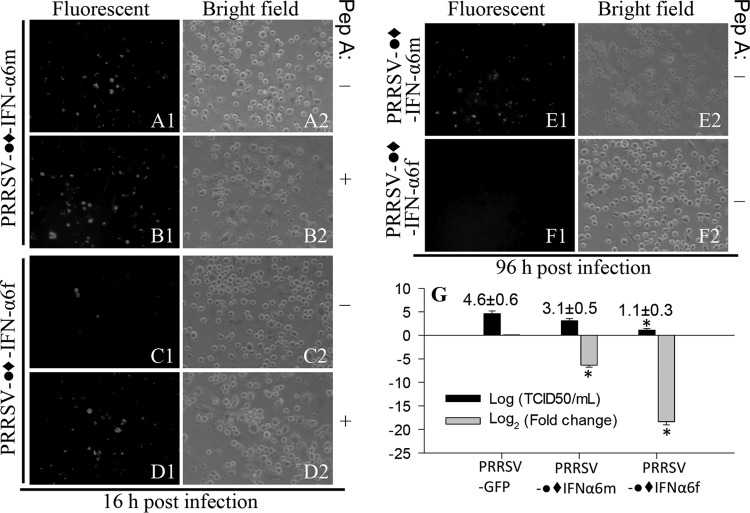
To potentiate antiviral immunity within the context of the porcine monocytic cell activation paradigm, a molecular tool was needed. One molecular tool that we developed is an IFN-expressing PRRSV infectious cDNA clone (37, 52). Using this PRRSV cDNA vector, we have stably expressed immune effectors, including multiple type I IFNs (37). The production of type I IFNs is fundamental to antiviral immunity and is suppressed in PRRSV infection (Fig. 4) (21–24). We reasoned that PRRSV replication-mediated IFN-α production in situ would reverse IFN subversion by wild-type viruses. We believe that this construct is a prototype for molecular vaccines to simultaneously optimize an antiviral state and to present viral antigens. However, because pigs have at least 39 functional type I IFN genes with diverse antiviral activity (34), which one should be used? We chose the gene coding for IFN-α6 as our first candidate, because it has high antiviral activity against PRRSV (34). Another question we confronted is how can sufficient PRRSV replication activity be achieved when expressing an IFN with high PRRSV antiviral activity? We approached this problem by inserting a leader linker in front of the IFN coding region that encodes a leader linker containing a C-terminal cleavage site of an endogenous protease expressed in Mϕs (Fig. 7A) (53). Using the designed construct, we were able to express and release active IFN peptides as well as rescue sufficient virions in MARC-145 cells (Fig. 7B and C) (37). In porcine Mϕs, virus replication-competent expression of the full-length peptide (with signal peptide, IFN-α6f), but not the mature IFN-α6 peptide (without signal peptide, IFN-α6m), efficiently inhibited the infection of coinfected PRRSV (Fig. 8). This was not unexpected because IFN peptides need the signal peptide included in the full-length construct for correct processing and secretion extracellularly to act on IFN receptors (IFNAR1/2) located on the plasma membrane (12, 15). This finding indicates that the correct partition of the expressed IFN by the signal peptide is essential for IFN antiviral activity. This was further confirmed when virus expression resumed after a protease inhibitor (peptide A) was added, likely by competitively inhibiting the release of the IFN peptide from the linker. Macrophages infected with the virus expressing the full-length IFN-α6 were able to clear the virus infection and were protected from virus-induced cytolysis for 96 h of coinfection. In contrast, cells infected with the virus expressing the linked mature peptide of IFN-α6 could not prevent virus replication and cytolysis (Fig. 8E and F). Quantification of virus titers and viral genome transcripts also indicated efficient replication of the IFN-expressing PRRSV, particularly in MARC cells (virus titers of 10 to 103 50% tissue culture infective doses [TCID50/ml]) (Fig. 7C) and significant suppression of PRRSV infection by the replication-competent expression of the full-length porcine antiviral IFN-α6 (Fig. 8G).
FIG 7.
Conditional expression of antiviral interferon (IFN) using an infectious PRRSV cDNA clone. (A) Schematic of the vector promoter and the DNA encoding the viral genome. The open reading frame (ORF) of an antiviral IFN was inserted in the viral ORF1b/ORF2 junction region using two restriction enzymes (AflII and MluI). The expressed IFN peptide has a leader of a His-tagged peptide containing a C-terminal protease cleavage site. This design allows rescue of high-titer recombinant virus in MARC-145 cells and IFN peptide release by the protease highly expressed in inflamed Mϕs. CMV, cytomegalovirus. (B) A Western blot shows successful expression and processing of the His-tagged IFN-α6 peptides. Cell lysates from MARC-145 cells infected with IFN-α6-expressing PRRSV were treated (+) with 5 U of the protease and blotted with both anti-His tag (1:1,000) and anti-IFN-α (1:200) antibodies (R&D). Note the loss of His tag and reduction of ∼2 kDa of the protein band in the protease-treated samples. The image on the right shows merged His tag (red) and PRRSV (green) immunofluorescence showing colocalization (yellow) of IFN-α6-PRRSV in the infected MARC-145 cells. (C) Propagation rate of IFN-engineered viruses. Values in parentheses are titers (log TCID50/ml) of passage 3 (P3) viruses. Growth kinetics of the recombinant viruses were determined or titrated in MARC-145 cells. Culture supernatants from cells transfected with infectious clones were harvested at 5 days posttransfection and are designated “P1.” The P1 virus was used to inoculate fresh MARC-145 cells to collect P2 and then P3 at an interval of 4 to 5 days between successive passages. Each passage virus was titrated, aliquoted, and stored at −80°C until use. Growth curves of the rescued viruses from MARC-145 or alveolar macrophages were evaluated by inoculating MARC-145 cells with P3 viruses at an MOI of 0.1.
FIG 8.
Antiviral activity of a PRRSV cDNA infectious clone expressing porcine IFN-α6. The expression of the full length form of IFN-α6 (IFN-α6f, with signal peptide) instead of the mature form (IFN-α6m, without signal peptide), protects alveolar Mϕs from infection and cytolysis by a coinfected PRRSV. The symbol ●◆ indicates a His-tagged linker of an endogenous protease cleavage site. Fluorescent images show coinfected green fluorescent protein (GFP)-labeled PRRSV, and bright-field images show cell cytolysis. (A1 to B2) Mϕs coinfected with the IFN-α6m-expressing PRRSV and the GFP-labeled virus with (B) or without (A) addition of a protease inhibitor (Pep. A). (C1 to D2) Same as in panels A and B but with IFN-α6f-expressed PRRSV showing suppression of coinfected GFP-PRRSV. (E1 to F2) IFN-α6f-expressing PRRSV protects cells from infection and cytolysis after 96 h of coinfection with GFP-PRRSV. The data represent three repeats with similar results. (G) Suppression of PRRSV infection in macrophages by conditional expression of full-length IFN-α6. Virus-containing medium and RNA were prepared from macrophages infected by GFP-PRRSV and IFN-expressing PRRSV for 96 h. The total viruses were titrated in MARC-145 cells to estimate TCID50 values, and the transcripts of PRRSV nucleocapsid (N) protein in total RNA (100 ng/20 μl PCR) purified from the infected cells were detected using a real-time RT-PCR to calculate fold decreases relative to the GFP-PRRSV-only control. *, P < 0.05 relative to the GFP-PRRSV-only control. The primers for RT-PCR detection are listed in Table S1 in the supplemental material.
DISCUSSION
Our studies here integrate antiviral regulation into the paradigm of activation statuses in porcine monocytic cells, a group of immune cells that not only are critical for overall immune responses but also are targeted by numerous economically devastating viruses to evade antiviral immunity (16–19, 21). These monocytotropic viruses include many of the world's most dreaded porcine viruses, such as African swine fever virus (ASFV), classical swine fever virus (CSFV), porcine circovirus 2 (PCV2), foot-and-mouth disease virus (FMDV), pseudorabies virus (PrV), swine influenza virus (SIV), and the focus of our studies, PRRSV (19, 21, 24, 36). These viruses collectively cause the majority of swine viral infections and significant global economic losses. Because of the lack of an effective vaccine, PRRSV has devastated swine herds worldwide for more than 20 years with an estimated loss nearing $800 million annually in the United States (44). Furthermore, this porcine virus evolves rapidly to form novel, highly pathogenic strains, causing new pandemic threats, such as the highly pathogenic PRRSV first seen in China in 2006 (30, 44).
Activation statuses of monocytic cells regulate inflammation, tissue repair, antimicrobial activity, and T- and B-cell responses (1–5). Here we integrate antiviral regulation within the activation paradigm of monocytic cells. Studies have shown that PRRSV directly infects subgroups of Mϕs and DCs and diverts immune responses in these cells through suppression of type I IFN production/action and impairment of antigen presentation (20–26). Therefore, understanding the activation status of monocytic cells and their interaction with PRRSV infection is essential not only to porcine immunology but also to determine the immunological etiology underlying PRRS (20–22, 25, 47, 48) and providing a framework for immunometabolic modulation to potentiate antiviral immunity in vivo (21, 48). Following standard procedures established in our laboratory, we defined that the macrophage population typically comprises >90% pulmonary lavage cells and showed a functional macrophage phenotype with respect to cell morphology and bacterial and phagocytic activity (35). The mDCs were differentiated following a standard protocol, which has been well determined for their morphology and surface phenotype of major histocompatibility complex class I-positive (MHC-I+), MHC-II+, and CD11c+ cells as well as low expression of CD16, CD14, and CD80/86 (36). Porcine blood monocytes were recently grouped based on the surface expression of CD14, CD163, and swine leukocyte antigen DR (SLA-DR) (38); clearly, monocytes used in this study contain predominant CD14+ CD163low cells (>90%) and few CD14+ CD163high (6%) cells (see Fig. S2 in the supplemental material). In addition, we determined the secreted cytokine profiles of these monocytic cells at basal and stimulated statuses, which further define their cell phenotypes pertaining to functional property. Furthermore, macrophages and mDCs used in this study showed 70 to 94% permissiveness to PRRSV infection, which indicates their macrophage and mDC phenotypes because these monocytic subsets are predominant cell types known to support PRRSV infection (26).
The activation status of porcine monocytic cells has not been systematically studied compared to the well-characterized paradigm in humans and mice, although a few previous studies have shown that the origin and differentiation of these cell lineages affect susceptibility to PRRSV (27, 28). Using primary monocytic cells isolated from the lungs and blood of naive piglets, we showed that porcine monocytic cells could be polarized into different activation statuses with typical mediators characterized in other species. Although some cell-specific responses were noted, such as higher production of IL-8 in activated monocytes and diminished production of IL-12 in mDCs, in general, secretion of typical cytokines correlated with their phenotypes (Table 1). Although studies of activation statuses in monocytes and DCs are limited, the M1and M2 activation paradigm appears to be active in both monocytes and DCs (6–9). In particular, we showed here that the antiviral states induced by type I IFNs, especially IFN-α, fit the paradigm of activation statuses and generally elicited more M1 statuses than M2 statuses. Interestingly, the cytokine profile of macrophages responding to IFN-β stimulation was more similar to the IL-10 regulatory status, indicating that IFN-β may have less antiviral but more immunomodulatory activity than IFN-α in macrophages (15) (Table 1 and Fig. 1A and 2B).
To study antiviral responses in cells at different activation statuses, we infected porcine macrophages with a PRRSV strain and investigated the early gene response (5 h postinfection) in cells polarized in vitro. We showed that during interaction with the virus in this early phase, macrophages kept their original polarization and cell integrity without any observed loss of viability, which allowed us to investigate early immunometabolic responses prior to cellular exhaustion by viral replication and release. Using PRRSV-infected macrophages at different activation statuses, we verified the expression of most cytokines and their receptors at the transcriptomic level. In addition, novel status-specific identifiers, such as the IL10RA and CCL21 genes, at the M1-IFN-γ status were identified, which might reflect novel signature genes (Table 2; see Fig. S1 in the supplemental material). Similarly, the signature expression of chemokines and their receptors as well as TLRs was also characteristically correlated with different activation statuses (Table 2; see Fig. S1 in the supplemental material). Of the key genes characterized for each activation status in macrophages, we showed most if not all typical signature genes specific to each activation status that have been characterized in other species were conserved in pigs. In particular, the marker genes related to the onset of antiviral states fit well into the paradigm of macrophage activation statuses; together they can be incorporated into a framework linking antiviral regulation to immune homeostasis in monocytic cells (Fig. 1A).
To reveal genes critical to status-specific antiviral and immune-homeostatic regulation, we compared response genes or gene response pathways among the PRRSV-infected macrophages at different activation statuses. In this context, we genome-wide profiled as potential signature genes those that were significantly upregulated (Fig. 1B) or downregulated in each activation status. Detailed annotation of those potential signature genes revealed some common signature genes (such as LIF and CD1B for the M1 and M2 statuses, respectively) characterized in other species and novel discoveries, including many undefined noncoding RNAs (Fig. 1B). However, most of these genes have not been studied for regulation of activation statuses and antiviral immunity. Therefore, our collection of potential signature genes pertinent to PRRSV-infected macrophages at each activation status provides a rich resource for targeting molecular regulation of the antiviral response in the paradigm of activation statuses.
Characterization of signature genes for macrophage activation statuses also allowed us to demonstrate the consequence of interaction between PRRSV and Mϕ activation statuses (Fig. 2B and Fig. 3). PRRSV infection induced differential cytokine responses in macrophages at different activation statuses, indicating that the potential of PRRSV to repolarize monocytic cells and the population sizes of macrophages at different activation statuses were factors in determining Mϕ-initiated immune responses (16–18). Notably, the high cytokine responses of IL-1β, IL-6, and IL-12 were found at the MaV-IFN-α or M1-IFN-γ status, and a moderate response was found at the M2-IL-10 status, whereas, macrophages at the M2-IL-4 and M1-LPS statuses, which mimic parasitic and bacterial infections, respectively, were the least responsive to PRRSV infection (Fig. 2B). Conversely, we showed that the magnitude of cytokine response was somewhat reciprocal to cell permissiveness toward the PRRSV. For example, both M2-IL-4 and M1-LPS had a higher portion of PRRSV-infected cells than the two IFN-stimulated statuses. However, monocytic cells at the M2-IL-10 status had the highest ratio of permissive cells, which was perhaps because of their anti-inflammatory state and higher expression of the scavenger receptor CD163, a confirmed cellular receptor for PRRSV for infection (26). Interestingly, IFN-β at the same dose of IFN-α stimulated IL-10 production (Fig. 2B) and, in turn, had little suppressive effect on PRRSV infection in macrophages (Fig. 3A and Table 3). The regulation of IFN-β in IL-10 production was recently shown in both macrophages and DCs, indicating the critical role of IFN-β in regulation of immune homeostasis in addition to its antiviral stimulation (54, 55). Although CD163 has been shown to be a cellular receptor capable of mediating PRRSV infection, its expression level in porcine monocytic cells was not always proportionally correlated with PRRSV permissiveness. For example, MaV-IFN-α macrophages had 2-fold more CD163-positive cells but were only half as permissive to PRRSV as the M1-LPS macrophages (Table 3). Indeed, annotation of our RNA-Seq data showed that CD163 had a constitutive expression level (the lowest RPKM of 113.4 of the M1-LPS-M1 compared with average RPKM values of most genes of 10 to 40), indicating that expression of CD163 in porcine monocytic cells is not a limiting factor (but could be an additive factor, such as at the M2-IL-10 status) for PRRSV infection (26). In porcine macrophages, CD163-specific antibody blocked up to 75% of PRRSV infection, whereas a combination of both antibodies to CD163 and sialoadhesin completely blocked the infection (42). Notably, our two-color fluorescence facilitated flow cytometric analyses detected highly positive cells pertaining to the expression levels of PRRSV N protein and the directly immunolabeling CD163; therefore, some portion of the “negative”-staining cells should be critically determined as CD163low cells. Although we know of no study exactly evaluating how many CD163 molecules per cell are sufficient to mediate PRRSV entrance, we interpret that a low threshold is better than a high one relative to pathogenic success of the virus. Thus, it is reasonable that a PRRSV infection ratio is higher than the CD163high cell population, as shown in Table 3 (26, 42).
In addition to genome-wide profiling of signature genes, Gene Ontology (GO) analysis of the DEGs identified multiple biological processes significantly and differentially altered by PRRSV infection in macrophages at different activation statuses. In this context, two biological processes, the IFN system (IFN production and action) and the lipid metabolic process, were analyzed to target antiviral regulation. Suppression of the IFN system typically has been associated with PRRSV infection. Nonstructural proteins (Nsp) of PRRSV, such as Nsp1, inhibit IRF3 and ISGF3 nuclear translocation, which result in suppression of both type I IFN production and induction of ISGs in host cells (21, 24, 25). Furthermore, differential suppression of IFN-α rather than IFN-β subtypes of porcine type I IFNs by PRRSV infection was shown in mDCs (21, 24, 25). Compared with other cytokine genes, many fewer reads pertinent to IFN transcripts were detected in our RNA-Seq data. For example, RPKM values for all IFN subtypes were <1.0, except for IFN-β at the M2-IL-10 status, whereas other cytokine genes generally had RPKM values averaging 10 to 40 (Fig. 4A). Clearly, general suppression of type I IFN production by PRRSV in macrophages could be ascribed to the low detection of IFN transcripts. In addition, an AU-rich element (ARE)-mediated mRNA decay could contribute the low detection of IFN transcripts (56). Similar to their orthologs in humans, mRNAs of most porcine type I IFNs contain an ARE in the 3′ untranslated region, and ARE-mediated mRNA decay is a critical mechanism in posttranscriptional control of the IFN level (56). In this context, we observed that genes coding for several cellular proteins, including the KHSRP gene (coding for KH-type splicing regulatory protein) and BRF2 (coding for butyrate response factor), which control ARE-mediated mRNA decay (56), were stimulated in most PRRSV-infected cells (unpublished data). Determination of whether PRRSV has a mechanism to suppress IFN production through the ARE-mediated mRNA decay mechanism should be pursued. Although the low read number of IFN transcripts did not attain a significant threshold between the cells at different activation statuses, comparison of IFN levels detected among different activation statuses demonstrated that IFN gene expression was different in cells at different activation statuses in terms of IFN subtypes and expression level. This was indicated by the finding that more subtypes and higher IFN expression were detected at both MaV-IFN-α and M1-IFN-γ statuses. Using current RNA-Seq analysis, it is difficult to estimate if PRRSV infection inhibits IFN induction of ISGs; however, expression of most ISGs was much higher in PRRSV-infected macrophages at the MaV-IFN-α status, implying that polarization of monocytic cells using exogenously delivered IFN is feasible for overcoming PRRSV suppression of IFN (37, 57). In addition, macrophages at the M1-LPS status were inactive in terms of induction of most ISG expression upon PRRSV infection, indicating that bacterial coinfection could be a major factor that exacerbates the viral pathogenicity in PRRS (20, 25).
Multiple biological processes in lipid/fatty acid metabolism were significantly enriched by DEGs in GO analyses. In general, DEGs involved in most lipid biological processes could be detected in cells at all activation statuses, except M2-IL-10, which had a limited number of 153 significant DEGs and enriched the fewest GO terms. However, when a hypergeometric test was used to find significantly enriched GO terms in DEGs, we observed several lipid metabolic processes that were significantly altered in cells at all activation statuses, except MaV-IFN-α (Table 4). This likely indicates an arrest of lipid metabolism in the antiviral state induced by IFN-α or that modulation of cellular lipid metabolism may provide a means to simulate antiviral states (21, 45). Profiling of key genes involved in lipid metabolism revealed that the expression of genes encoding some critical enzymes, such as ACC1, FASN, LIPE and HMGR, was suppressed, particularly in the antiviral state (Fig. 5 and Table 4). Therefore, modulation of the activity of these enzymes may provide another approach to antiviral regulation (45, 46, 50, 51).
Based on RNA-Seq analyses, we validated our observations of gene expression with authentic antiviral regulation in cells. First, we tested a series of agonists or antagonists in regulation of lipid metabolic processes. ToFA, a competitive inhibitor of acetyl-CoA carboxylase (ACC), showed significant activity in suppressing PRRSV infection in all tested cells (Fig. 6A and B). We also showed that ToFA at a physiological dosage elicited porcine macrophages to produce proinflammatory cytokines (including IL-1β, IL-8, TNF-α, and IL-12) as well as the anti-inflammatory cytokine IL-10, intermediately as seen in the antiviral state induced by IFN-α and IFN-β (Fig. 6C). Acetyl-CoA carboxylase serves as a key enzyme in cellular lipid metabolism to catalyze malonyl-CoA production for new fatty acid synthesis, meanwhile inhibiting the beta-oxidation of fatty acids in the mitochondria. Because lipid metabolic status is essential to fuel cell activation and some lipid metabolites also directly intervene in virus-host interaction, suppression of ACC by ToFA thus may affect viral infection directly via the virus infection process (such as viral fusion and budding processes) (37) and indirectly through regulation of cell antiviral status. Further lipidomic profiling will determine lipid metabolites altered by ToFA treatment in monocytic cells, However, the present study indicates that ToFA treatment may skew porcine macrophages to an antiviral state, similar to what happens in cells stimulated with type I IFNs with respect to inflammatory cytokine production (Fig. 6C) and suppression of lipid metabolism (Fig. 5 and Table 4). In addition, ToFA treatment was shown to enhance DC processing of antigens for activation of T cells in mice (40); a similar mechanism should be considered for vaccine design and validation in pigs. These findings indicate that an activation status with virostatic potency could be induced through modulation of cellular lipid metabolism (40, 45, 50, 51).
Exogenous application of type I IFNs, particularly the IFN-α subtype, has been used successfully for treatment of several viral diseases (12–15). Several studies reported that suppression of type I IFN production was a key mechanism that contributed to success of PRRSV field isolates and that some attenuated strains showed less suppression and even stimulation of IFN production (21, 24, 25, 47, 48). Based on mechanistic studies of the interaction between PRRSV and the porcine IFN system, we and others have proposed reversing type I IFN suppression at infection sites to combat PRRSV infection (21, 48, 57). For example, in two reports, IFN-α was introduced either intramuscularly as a cDNA adjuvant component (58) or as an adenoviral expression construct (57); although the former showed little synergistic effect for vaccine protection, the timely virus vector-mediated IFN-α expression alone successfully protected against PRRSV challenge (57, 58). We therefore suggest that a type I IFN expressed by a replication-competent PRRSV cDNA infectious clone may be an ideal means for reversing PRRSV suppression of type IFN production (21, 37). We were able to express different subtypes of type I IFNs, including IFN-α1, IFN-α6, IFN-δ3, and IFN-ω5, with a PRRSV cDNA infection clone and showed that the replication-competent PRRSV-IFN constructs expressing the IFN-α subtype induced antiviral protection in cells (37). To obtain sufficient PRRSV replication while expressing an IFN with high PRRSV antiviral activity, we inserted a leader linker in front of the IFN coding region. This design allowed for rescue of enough virions in MARC-145 cells and the release of active IFN peptides in porcine Mϕs to overcome the virus suppression of IFN production (Fig. 7C and Fig. 8). This construct can be a prototype for molecular vaccines to optimize an antiviral state and to simultaneously present viral antigens.
Several studies have reported genome-wide gene response pathways directly in porcine tissues or cell collections (30, 44). Our use of RNA-Seq to profile signature genes and responsive pathways associated with PRRSV infection in subsets of monocytic cells not only confirmed previous identifications and revealed novel gene response pathways but also examined their involvement in antiviral regulation through pharmaceutical and molecular manipulation. Our efforts now will be to introduce a modified lipid nanoparticle (LNP) procedure (59) to deliver both ToFA (as an adjuvant) and the PRRSV-IFN construct as a vaccine platform in pigs to further validate the vaccine effect of ToFA and the PRRSV-IFN expression construct in vivo.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by USDA AFRI NIFA/DHS 2010-39559-21860 and USDA NIFA 2013-67015-21236. We thank the Department of Anatomy and Physiology COBRE facility cores (funded by NIH P20-RR017686) for technical and equipment support.
We thank Barbara Lutjemeier and Danielle Goodband for excellent technical support, Wenjing Sang and Andrew Schade for help in RNA sample preparation, and Samantha Lyman and Wyatt Brichalli for help in RNA-Seq data analyses.
Footnotes
Published ahead of print 23 July 2014
This article is contribution no. 14-243-J from the Kansas Agricultural Experiment Station.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01714-14.
REFERENCES
- 1.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. 2010. Development of monocytes, macrophages, and dendritic cells. Science 327:656–661. 10.1126/science.1178331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mosser DM, Edwards JP. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8:958–969. 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon S, Martinez FO. 2010. Alternative activation of macrophages: mechanism and functions. Immunity 32:593–604. 10.1016/j.immuni.2010.05.007 [DOI] [PubMed] [Google Scholar]
- 4.Martinez FO, Sica A, Mantovani A, Locati M. 2008. Macrophage activation and polarization. Front. Biosci. 13:453–461. 10.2741/2692 [DOI] [PubMed] [Google Scholar]
- 5.Martinez FO. 2009. The transcriptome of human monocyte subsets begins to emerge. J. Biol. 8:99. 10.1186/jbiol206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid MA, Kingston D, Boddupalli S, Manz MG. 2010. Instructive cytokine signals in dendritic cell lineage commitment. Immunol. Rev. 234:32–44. 10.1111/j.0105-2896.2009.00877.x [DOI] [PubMed] [Google Scholar]
- 7.Tailor P, Tamura T, Ozato K. 2006. IRF family proteins and type I interferon induction in dendritic cells. Cell Res. 16:134–140. 10.1038/sj.cr.7310018 [DOI] [PubMed] [Google Scholar]
- 8.Kaliński P, Hilkens CM, Wierenga EA, Kapsenberg ML. 1999. T-cell priming by type-1 and type-2 polarized dendritic cells: the concept of a third signal. Immunol. Today 20:561–567. 10.1016/S0167-5699(99)01547-9 [DOI] [PubMed] [Google Scholar]
- 9.Skalova K, Mollova K, Michalek J. 2010. Human myeloid dendritic cells for cancer therapy: does maturation matter? Vaccine 28:5153–5160. 10.1016/j.vaccine.2010.05.042 [DOI] [PubMed] [Google Scholar]
- 10.Kumagai Y, Takeuchi O, Kato H, Kumar H, Matsui K, Morii E, Aozasa K, Kawai T, Akira S. 2007. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27:240–252. 10.1016/j.immuni.2007.07.013 [DOI] [PubMed] [Google Scholar]
- 11.Gilliet M, Cao W, Liu YJ. 2008. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 8:594–606. 10.1038/nri2358 [DOI] [PubMed] [Google Scholar]
- 12.Kaisho T. 2008. Type I interferon production by nucleic acid-stimulated dendritic cells. Front. Biosci. 13:6034–6042. 10.2741/3134 [DOI] [PubMed] [Google Scholar]
- 13.Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. 2012. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 36:166–174. 10.1016/j.immuni.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1:519–525. 10.1016/j.coviro.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González-Navajas JM, Lee J, David M, Raz E. 2012. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 12:125–135. 10.1038/nri3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cassol E, Cassetta L, Rizzi C, Alfano M, Poli G. 2009. M1 and M2a polarization of human monocyte-derived macrophages inhibits HIV-1 replication by distinct mechanisms. J. Immunol. 182:6237–6246. 10.4049/jimmunol.0803447 [DOI] [PubMed] [Google Scholar]
- 17.Herbein G, Varin A. 2010. The macrophage in HIV-1 infection: from activation to deactivation? Retrovirology 7:33. 10.1186/1742-4690-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirey KA, Pletneva LM, Puche AC, Keegan AD, Prince GA, Blanco JC, Vogel SN. 2010. Control of RSV-induced lung injury by alternatively activated macrophages is IL-4R alpha, TLR4-, and IFN-beta-dependent. Mucosal Immunol. 3:291–300. 10.1038/mi.2010.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen WH, Toapanta FR, Shirey KA, Zhang L, Giannelou A, Page C, Frieman MB, Vogel SN, Cross AS. 2012. Potential role for alternatively activated macrophages in the secondary bacterial infection during recovery from influenza. Immunol. Lett. 141:227–234. 10.1016/j.imlet.2011.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molitor TW, Bautista EM, Choi CS. 1997. Immunity to PRRSV: double-edged sword. Vet. Microbiol. 55:265–276. 10.1016/S0378-1135(96)01327-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sang Y, Rowland RR, Blecha F. 2011. Interaction between innate immunity and porcine reproductive and respiratory syndrome virus. Anim. Health Res. Rev. 12:149–167. 10.1017/S1466252311000144 [DOI] [PubMed] [Google Scholar]
- 22.Thacker EL. 2001. Immunology of the porcine respiratory disease complex. Vet. Clin. North Am. Food Anim. Pract. 17:551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walson JL, John-Stewart G. 2007. Treatment of helminth co-infection in individuals with HIV-1: A systematic review of the literature. PLoS Negl. Trop. Dis. 1:e102. 10.1371/journal.pntd.0000102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calzada-Nova G, Schnitzlein WM, Husmann RJ, Zuckermann FA. 2011. North American porcine reproductive and respiratory viruses inhibit type I interferon production by plasmacytoid dendritic cells. J. Virol. 85:2703–2713. 10.1128/JVI.01616-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darwich L, Díaz I, Mateu E. 2010. Certainties, doubts and hypotheses in porcine reproductive and respiratory syndrome virus immunobiology. Virus Res. 154:123–132. 10.1016/j.virusres.2010.07.017 [DOI] [PubMed] [Google Scholar]
- 26.Welch SK, Calvert JG. 2010. A brief review of CD163 and its role in PRRSV infection. Virus Res. 154:98–103. 10.1016/j.virusres.2010.07.018 [DOI] [PubMed] [Google Scholar]
- 27.Charerntantanakul W, Platt R, Roth JA. 2006. Effects of porcine reproductive and respiratory syndrome virus-infected antigen-presenting cells on T cell activation and antiviral cytokine production. Viral Immunol. 19:646–661. 10.1089/vim.2006.19.646 [DOI] [PubMed] [Google Scholar]
- 28.Duan X, Nauwynck HJ, Pensaert MB. 1997. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch. Virol. 142:2483–2497. 10.1007/s007050050256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sang Y, Brichalli W, Rowland RR, Blecha F. 2014. Genome-wide analysis of signature genes for antiviral regulation in PRRSV-infected porcine macrophages at different activation statuses. PLoS One 9:e87613. 10.1371/journal.pone.0087613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao S, Jia J, Mo D, Wang Q, Qin L, He Z, Zhao X, Huang Y, Li A, Yu J, Niu Y, Liu X, Chen Y. 2010. Understanding PRRSV infection in porcine lung based on genome-wide transcriptome response identified by deep sequencing. PLoS One 5:e11377. 10.1371/journal.pone.0011377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DY, Kaiser TJ, Horlen K, Keith ML, Taylor LP, Jolie R, Calvert JG, Rowland RR. 2009. Insertion and deletion in a non-essential region of the nonstructural protein 2 (nsp2) of porcine reproductive and respiratory syndrome (PRRS) virus: effects on virulence and immunogenicity. Virus Genes 38:118–128. 10.1007/s11262-008-0303-4 [DOI] [PubMed] [Google Scholar]
- 32.Rowland RR. 2010. The interaction between PRRSV and the late gestation pig fetus. Virus Res. 154:114–122. 10.1016/j.virusres.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sang Y, Ross CR, Rowland RR, Blecha F. 2008. Toll-like receptor 3 activation decreases porcine arterivirus infection. Viral Immunol. 21:303–313. 10.1089/vim.2008.0042 [DOI] [PubMed] [Google Scholar]
- 34.Sang Y, Rowland RR, Hesse RA, Blecha F. 2010. Differential expression and activity of the porcine type I interferon family. Physiol. Genomics 42:248–258. 10.1152/physiolgenomics.00198.2009 [DOI] [PubMed] [Google Scholar]
- 35.Chitko-McKown CG, Chapes SK, Brown RE, Phillips RM, McKown RD, Blecha F. 1991. Porcine alveolar and pulmonary intravascular macrophages: comparison of immune functions. J. Leukoc. Biol. 50:364–372 [DOI] [PubMed] [Google Scholar]
- 36.Loving CL, Brockmeier SL, Sacco RE. 2007. Differential type I interferon activation and susceptibility of dendritic cell populations to porcine arterivirus. Immunology 120:217–229. 10.1111/j.1365-2567.2006.02493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sang Y, Shi J, Sang W, Rowland RR, Blecha F. 2012. Replication-competent recombinant porcine reproductive and respiratory syndrome (PRRS) viruses expressing indicator proteins and antiviral cytokines. Viruses 4:102–116. 10.3390/v4010102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez C, Revilla C, Alvarez B, Chamorro S, Correa C, Domenech N, Alonso F, Ezquerra A, Domínguez J. 2007. Phenotypic and functional characterization of porcine granulocyte developmental stages using two new markers. Dev. Comp. Immunol. 31:296–306. 10.1016/j.dci.2006.06.002 [DOI] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herber DL, Cao W, Nefedova Y, Novitskiy SV, Nagaraj S, Tyurin VA, Corzo A, Cho HI, Celis E, Lennox B, Knight SC, Padhya T, McCaffrey TV, McCaffrey JC, Antonia S, Fishman M, Ferris RL, Kagan VE, Gabrilovich DI. 2010. Lipid accumulation and dendritic cell dysfunction in cancer. Nat. Med. 16:880–886. 10.1038/nm.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dawson HD, Loveland JE, Pascal G, Gilbert JG, Uenishi H, Mann KM, Sang Y, Zhang J, Carvalho-Silva D, Hunt T, Hardy M, Hu Z, Zhao SH, Anselmo A, Shinkai H, Chen C, Badaoui B, Berman D, Amid C, Kay M, Lloyd D, Snow C, Morozumi T, Cheng RP, Bystrom M, Kapetanovic R, Schwartz JC, Kataria R, Astley M, Fritz E, Steward C, Thomas M, Wilming L, Toki D, Archibald AL, Bed'Hom B, Beraldi D, Huang TH, Ait-Ali T, Blecha F, Botti S, Freeman TC, Giuffra E, Hume DA, Lunney JK, Murtaugh MP, Reecy JM, Harrow JL, Rogel-Gaillard C, Tuggle CK. 2013. Structural and functional annotation of the porcine immunome. BMC Genomics 14:332. 10.1186/1471-2164-14-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. 2008. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 89:2943–2953. 10.1099/vir.0.2008/005009-0 [DOI] [PubMed] [Google Scholar]
- 43.Fung KY, Mangan NE, Cumming H, Horvat JC, Mayall JR, Stifter SA, De Weerd N, Roisman LC, Rossjohn J, Robertson SA, Schjenken JE, Parker B, Gargett CE, Nguyen HP, Carr DJ, Hansbro PM, Hertzog PJ. 2013. Interferon-ε protects the female reproductive tract from viral and bacterial infection. Science 339:1088–1092. 10.1126/science.1233321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowland RR, Lunney J, Dekkers J. 2012. Control of porcine reproductive and respiratory syndrome (PRRS) through genetic improvements in disease resistance and tolerance. Front. Genet. 3:260. 10.3389/fgene.2012.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanc M, Hsieh WY, Robertson KA, Watterson S, Shui G, Lacaze P, Khondoker M, Dickinson P, Sing G, Rodríguez-Martín S, Phelan P, Forster T, Strobl B, Müller M, Riemersma R, Osborne T, Wenk MR, Angulo A, Ghazal P. 2011. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol. 9:e1000598. 10.1371/journal.pbio.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, Freiberg AN, Su L, Lee B, Cheng G. 2013. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38:92–105. 10.1016/j.immuni.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang YW, Meng XJ. 2010. Novel strategies and approaches to develop the next generation of vaccines against porcine reproductive and respiratory syndrome virus (PRRSV). Virus Res. 154:141–149. 10.1016/j.virusres.2010.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimman TG, Cornelissen LA, Moormann RJ, Rebel JM, Stockhofe-Zurwieden N. 2009. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 27:3704–3718. 10.1016/j.vaccine.2009.04.022 [DOI] [PubMed] [Google Scholar]
- 49.Huang YH, Sauer K. 2010. Lipid signaling in T-cell development and function. Cold Spring Harb. Perspect. Biol. 2:a002428. 10.1101/cshperspect.a002428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serhan CN. 2010. Novel lipid mediators and resolution mechanisms in acute inflammation: to resolve or not? Am. J. Pathol. 177:1576–1591. 10.2353/ajpath.2010.100322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teijaro JR, Walsh KB, Cahalan S, Fremgen DM, Roberts E, Scott F, Martinborough E, Peach R, Oldstone MB, Rosen H. 2011. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell 146:980–991. 10.1016/j.cell.2011.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoo D, Welch SK, Lee C, Calvert JG. 2004. Infectious cDNA clones of porcine reproductive and respiratory syndrome virus and their potential as vaccine vectors. Vet. Immunol. Immunopathol. 102:143–154. 10.1016/j.vetimm.2004.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shpacovitch V, Feld M, Bunnett NW, Steinhoff M. 2007. Protease-activated receptors: novel PARtners in innate immunity. Trends Immunol. 28:541–550. 10.1016/j.it.2007.09.001 [DOI] [PubMed] [Google Scholar]
- 54.Saraiva M, O'Garra A. 2010. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10:170–181. 10.1038/nri2711 [DOI] [PubMed] [Google Scholar]
- 55.de Weerd NA, Vivian JP, Nguyen TK, Mangan NE, Gould JA, Braniff SJ, Zaker-Tabrizi L, Fung KY, Forster SC, Beddoe T, Reid HH, Rossjohn J, Hertzog PJ. 2013. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 14:901–907. 10.1038/ni.2667 [DOI] [PubMed] [Google Scholar]
- 56.Lin WJ, Zheng X, Lin CC, Tsao J, Zhu X, Cody JJ, Coleman JM, Gherzi R, Luo M, Townes TM, Parker JN, Chen CY. 2011. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol. Cell. Biol. 31:3196–3207. 10.1128/MCB.05073-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brockmeier SL, Loving CL, Nelson EA, Miller LC, Nicholson TL, Register KB, Grubman MJ, Brough DE, Kehrli ME., Jr 2012. The presence of alpha interferon at the time of infection alters the innate and adaptive immune responses to porcine reproductive and respiratory syndrome virus. Clin. Vaccine Immunol. 19:508–514. 10.1128/CVI.05490-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Charerntantanakul W, Platt R, Johnson W, Roof M, Vaughn E, Roth JA. 2006. Immune responses and protection by vaccine and various vaccine adjuvant candidates to virulent porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 109:99–115. 10.1016/j.vetimm.2005.07.026 [DOI] [PubMed] [Google Scholar]
- 59.Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, Cu Y, Beard CW, Brito LA, Krucker T, O'Hagan DT, Singh M, Mason PW, Valiante NM, Dormitzer PR, Barnett SW, Rappuoli R, Ulmer JB, Mandl CW. 2012. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U. S. A. 109:14604–14609. 10.1073/pnas.1209367109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.