ABSTRACT
Recent studies suggest that human endogenous retrovirus group K (HERV-K) provirus expression plays a role in the pathogenesis of HIV-1 infection. In particular, RNA from the HML-2 subgroup of HERV-K proviruses has been reported to be highly expressed at the cellular level and detectable in the plasma of HIV-1-infected patients, suggestive of virion production and, perhaps, replication. In this study, we developed an HML-2-specific quantitative-PCR assay that detects 51 of the 89 known HML-2 proviruses in the human genome. Plasma and peripheral blood mononuclear cells (PBMCs) from HIV-negative controls and HIV-1-infected patients were collected for analysis of HML-2 RNA expression. Contrary to previous reports, we did not detect high levels of HML-2 RNA in the plasma of HIV-1-infected patients, but we did observe a significant increase of HML-2 RNA in total PBMCs compared to HIV-negative controls. The level of HML-2 expression in PBMCs does not appear to be related to patient use of antiretrovirals or to HIV-1 plasma RNA, cellular RNA, or cellular DNA levels. To investigate the source of HML-2 RNA expression, patient PBMCs were sorted into CD3+ CD4+, CD3+ CD8+, CD3− CD14+, and CD3− CD20+ cell subsets and then analyzed for HML-2 RNA levels. No single cell subset was enriched for HML-2 RNA expression in HIV-1-infected patients, but there appears to be substantial variability in the level of HML-2 expression depending on the cell type.
IMPORTANCE Here, we report that human endogenous retrovirus group K (HERV-K) (HML-2) proviruses are expressed at significantly higher levels in peripheral blood mononuclear cells (PBMCs) from patients with HIV-1 infection than in those from uninfected individuals. However, contrary to previous reports, this expression did not lead to detectable virions in the plasma of these patients. In addition, we found that HML-2 proviruses were expressed in multiple blood cell types from HIV-1-infected individuals, and the magnitude of HML-2 expression was not related to HIV-1 disease markers in this patient cohort. These findings may have implications for HML-2-based therapies targeting HIV-1 infection.
INTRODUCTION
Retroviruses replicate by reverse transcribing their RNA genomes into double-stranded cDNA, which is subsequently integrated into the host genome. If a retrovirus infects a germ line cell and successfully integrates its cDNA to become a provirus, it is said to be endogenized and will be vertically transmitted to offspring in a Mendelian fashion (1). This process has occurred numerous times during the evolution of humans, with both truncated and full-length proviruses comprising roughly 8% of the genome, referred to collectively as human endogenous retrovirus (HERV) sequences (2).
Most HERV sequences have undergone extensive mutation after integration into the human genome. However, a few have maintained open reading frames (ORFs) for viral proteins and have the ability to form intact but noninfectious viral particles (2–6). One group of HERVs, named human endogenous retrovirus group K (HERV-K), includes many proviruses that have retained ORFs (5, 7). This group is made up of 11 subgroups named human MMTV-like (HML) to reflect their similarity to mouse mammary tumor virus (MMTV), a virus that causes mammary carcinoma in mice (2, 7–9). The HERV-K (HML-2) group includes the proviruses that have most recently integrated into the human ancestral genome. HML-2 proviruses are evolutionarily young compared to all other HERV sequences, with some having entered our genome less than 5 million years ago, after the human-chimpanzee split, and a few within the last few hundred thousand years (2). This subgroup is the only one that includes human-specific integration sites, at least 11 of which are insertionally polymorphic within the human population (7, 10–13). Furthermore, the subgroup contributes at least 89 full or partial proviral sequences to the human genome, many of which contain an ORF for gag, pro, pol, and/or env, genes essential for retroviral infectivity (7). While there is no conclusive proof that any endogenized HML-2 provirus is infective as is, two recombinant HML-2 viruses based on the most recent common ancestor of the human-specific proviruses, as inferred by phylogenetic analysis, have been shown to be weakly infectious (14, 15).
HERV-K was first associated with human disease after the discovery that a HERV-K provirus encodes the human teratocarcinoma-derived virus (HTDV) particles emanating from germ cell tumors (16–18). Since then, HERV-K expression has been observed in a variety of human diseases, most notably in different types of cancer and autoimmune disease (19–28). While these results are suggestive, conclusive evidence for a causal association of HERV-K with any human disease has yet to be obtained.
Recent studies have reported HERV-K (HML-2) expression during HIV-1 infection. Observations include the presence of HML-2-specific immune responses (29–32), detectable particle-associated HML-2 RNA in the plasma of HIV-1-infected individuals (33–36), and an increase in HML-2 RNA expression from cells infected with HIV-1 in vitro (37, 38), as well as from cells collected from HIV-1-infected patients (39). Additionally, HIV-1-infected patients on suppressive highly active antiretroviral therapy (HAART) exhibited lower plasma HML-2 RNA expression than those on a nonsuppressive regimen (33, 35), indicating a direct or indirect link between HIV-1 replication and HML-2 expression. It is not clear if the decrease in HML-2 expression is due to antiretroviral therapy (ART) administration, lack of HIV-1 replication, reduced immune activation and immune response to HIV-1 in the patients, or all three. Moreover, there is limited information about which HML-2 proviruses are expressed and which cell types could be producing them (36). Upregulation of HML-2 RNA in cells has been associated with HIV-1 Tat and Vif expression in cell culture models, though its relevance to in vivo HML-2 expression has not been explored (37). The cause(s) of HML-2 expression during HIV-1 infection remains to be clarified, as does the cell type(s) involved.
To address the remaining questions about how and where HML-2 is expressed during HIV infection, we studied plasma and peripheral blood mononuclear cells (PBMCs) from HIV-1 patients. To our surprise, and contrary to previous reports, we did not detect any HML-2 RNA in the plasma of the HIV-1-infected subjects in this study, although there was an increase in HML-2 DNA. However, we did detect an increase in HML-2 RNA in PBMCs collected from HIV-infected patients compared to uninfected controls. In further analysis of the cell source of HML-2 RNA, we were not able to identify a single cell type enriched for HML-2 expression over uninfected controls. In fact, all cell types tested were found to express HML-2, although at different levels.
MATERIALS AND METHODS
Sequence alignment.
An HML-2 provirus alignment containing 91 sequences was downloaded from a previous publication (7) and analyzed in BioEdit Sequence Alignment Editor (Ibis Biosciences, Carlsbad, CA). BioEdit was used to search for regions of high similarity for development of the HML-2 env-specific quantitative PCR (qPCR) and to determine which proviruses could be detected using the env-specific qPCR based on sequence identity to primers.
Quantification of HML-2 proviruses.
Human genomic DNA (Applied Biosystems TaqMan Human Control DNA; catalog number 4312660) was supplied at 10 ng/μl. Twofold serial dilutions of human gDNA were performed in 5 mM Tris-HCl (Invitrogen; catalog number 15568-025). The standard assumption of 3 pg DNA per haploid genome was used to estimate the number of genomes present in a known quantity of genomic DNA (gDNA). The gDNA (1.5 μl) was loaded into the HML-2 qPCR in triplicate. Plasmid DNA standards (Invitrogen; pcDNA 3.1; catalog number K4900-01) containing the HML-2 Env coding region from 7p22.1a (K108) were diluted to known quantities and used to estimate the numbers of HML-2 DNA copies in the different dilutions of human gDNA. Three runs of the assay were performed at different dilutions, and the results were used to estimate the number of HML-2 proviruses in human gDNA.
Clinical samples and ethics statement.
Plasma and PBMC samples were obtained at Tufts Medical Center (TMC), the NIH Clinical Center, and the University of Pittsburgh (UPitt). Human subject research was approved by the institution running each study, namely by the TMC IRB, the NIAID IRB, and the UPitt IRB. All participants provided written informed consent. Participants at NIH were all enrolled in clinical protocols (00-I-0110, 97-I-0082, and 95-I-0072) approved by the NIAID Institutional Review Board (FWA00005897) administered at the NIH Clinical Center in Bethesda, MD. Participants at NIH provided written consent for participation, for sample storage, and for sharing samples with collaborators outside the NIH. Patient characteristics are recorded in Tables 1 and 2 and Table S1 in the supplemental material. Two patients who provided plasma samples (Table 1) were hepatitis C virus (HCV) positive, one patient who provided PBMCs (Table 1) was herpes simplex virus positive, and three nonviremic patients who provided PBMCs (Table 2) were hepatitis C virus positive. Patients were recruited from TMC in 2012 and 2013 and confirmed to be off antiretroviral medication, 18 to 65 years of age, and free of confounding medical conditions, including cancer, schizophrenia, autoimmune disease, and pregnancy. Blood samples from TMC were drawn into BD Vacutainer CPT tubes with sodium citrate (Becton-Dickinson; catalog number 362760) to allow separation of plasma and PBMCs from the same blood draw. Samples were processed within 2 h of blood draw according to the manufacturer's instructions. Plasma was stored at −80°C in 1-ml aliquots, and PBMCs were frozen in 5% dimethyl sulfoxide (DMSO) in fetal bovine serum at 5 × 106 cells/ml and stored in liquid nitrogen until analysis. Blood samples from UPitt were drawn into BD Vacutainer K2EDTA tubes (Becton-Dickinson; catalog number 366643) with EDTA. Plasma was separated from whole blood by centrifugation at 400 × g followed by a second spin at 400 × g and was then stored at −80°C in 1.5-ml aliquots. PBMCs were isolated from the remaining (plasma-free) blood by Ficoll-Hypaque density gradient centrifugation. The PBMCs were frozen in 10% DMSO in fetal bovine serum at 5 × 106 cells/ml and stored in liquid nitrogen. Samples were shipped on dry ice and then transferred to liquid nitrogen storage until analysis. All uninfected control samples used were drawn at the NIH Clinical Center or UPitt from adult volunteers aged 18 to 65 with no known comorbidities. Control plasma and PBMC samples were stored as described above until they were thawed for analysis.
TABLE 1.
HIV-infected-patienta (NIH Clinical Center and TMC) characteristics for plasma and PBMCs analyzed in Fig. 2, 3, and 5
| Characteristic | Value |
|
|---|---|---|
| Plasma (n = 15) | PBMCs (n = 19) | |
| Demographic | ||
| Male [no. (%)] | 14 (93) | 18 (95) |
| 6 (40) | 9 (47) | |
| African-American [no. (%)] | 4 (27) | 3 (16) |
| Hispanic [no. (%)] | 5 (33) | 6 (32) |
| Asian, Pacific Islander [no. (%)] | 0 (0) | 1 (5) |
| Age (yr) [median (range)] | 42 (25.1–48.0) | 35.9 (19.5–51.9) |
| HIV | ||
| CD4 count [median (range)] | 523 (220–1,105) | 661 (210–1,105) |
| % CD4 cells [median (range)] | 24.8 (10.7–55.7) | 29 (9–55.7) |
| RNA viral load (log10 copies/ml) [median (range)] | 4.2 (1.95–5.6) | 4.11 (1.95–5.55) |
One viremic HIV-infected patient was off therapy during blood collection. All other patients were treatment naive and viremic.
TABLE 2.
| Characteristic | Value |
|
|---|---|---|
| Nonviremic patients (n = 10) | Viremic patients (n = 5) | |
| Demographic | ||
| Male [no. (%)] | 7 (70) | 5 (100) |
| White [no. (%)] | 3 (30) | 0 (0) |
| African-American [no. (%)] | 7 (70) | 4 (80) |
| Hispanic [no. (%)] | 0 (0) | 1 (20) |
| Asian, Pacific Islander [no. (%)] | 0 (0) | 0 (0) |
| Age (yr) [median (range)] | 51.5 (25–58) | 52 (29–53) |
| HIV | ||
| CD4 count [median (range)] | 617 (416–1,373) | 874 (244–1,091) |
| % CD4 cells [median (range)] | 31.1 (20.1–52.1) | 31.9 (9.3–47.4) |
| iSCAb viral load (log10 copies/ml) [median (range)] | NDc (<−0.2–5.31) | 3.44 (2.75–5.32) |
All nonviremic patients were on therapy. Viremic patients were either treatment naive, off therapy, or on therapy at the time of blood collection (see Table S1 in the supplemental material for treatment details).
iSCA is a modified SCA that detects a small region in the integrase portion of HIV-1 RNA.
ND, not determined. Four patients were below the limit of detection for iSCA.
RNA extraction from plasma.
Plasma RNA was extracted from HIV-infected and control patients according to a modified version of a previously published protocol (40). Plasma samples were thawed on ice the day of analysis, and a prespin at 2,500 × g was performed at room temperature for 15 min to pellet any cellular debris. Two hundred to 500 μl of plasma was diluted with an equal volume of Tris-buffered saline (Sigma; catalog number T5030), and virions were pelleted from the plasma at 21,000 × g for 1 h at 4°C. The pellet was resuspended in 50 μl of 5 mM Tris-HCl (Invitrogen; catalog number 15568-025) and treated with 20 mg/ml proteinase K (Ambion; catalog number AM2548) for 30 min at 55°C. Two hundred microliters of 6 M guanidinium isothiocyanate (Sigma; catalog number 50983) and 10 μl 20 mg/ml glycogen (Roche; catalog number 10901393001) were added and vortexed, and the mixture was incubated for 5 min at room temperature. Two hundred microliters of 100% isopropanol was added, and the sample was centrifuged at 21,000 × g at 4°C for 35 min. The pellet was washed with 500 μl 70% ethanol and centrifuged at 21,000 × g at 4°C for 15 min, and all ethanol was removed from the pellet through sequential spins and pipetting. The RNA pellet was air dried for 2 min and resuspended in buffer (965 μl 5 mM Tris-HCl, 25 μl RNasin or equivalent RNase-inhibiting enzyme, 10 μl 0.1 M dithiothreitol [DTT]). The RNA was left untreated or treated with 1 U of DNase (Ambion; Turbo DNA-free kit; catalog number AM1907) according to the manufacturer's instructions (as specified) for downstream analysis. For the HIV Roche TaqMan v2.0 assay, plasma HIV RNA was extracted using the automated Cobas AmpliPrep System v2.0 (Roche Molecular Diagnostics).
Nucleic acid extraction from cells.
Unsorted PBMCs were removed from liquid nitrogen storage and heated in a 37°C water bath until almost thawed. The cells were resuspended in 10 ml of phosphate-buffered saline (PBS) (Gibco; catalog number 14190) and centrifuged at 350 × g for 5 min at 4°C. The cell pellet was resuspended in 2 ml PBS, and 10 to 40 μl was used to obtain viable-cell counts using trypan blue and a hemocytometer. The cells were pelleted, and 1 ml of TRIzol (Ambion; catalog number 15596-026) was added to the cell pellet for lysis for 5 min at room temperature; 200 μl chloroform was added to the TRIzol solution and mixed vigorously for 15 s. Samples were incubated at room temperature for 3 min and then centrifuged at 12,000 × g for 15 min at 4°C; 400 μl of the aqueous phase was transferred into a new Eppendorf tube, and an equal volume of 70% ethanol was added. Samples were added to a downstream column purification kit (Ambion; PureLink RNA Mini; catalog number 1218301A) and treated according to the manufacturer's instructions. RNA was eluted into 60 μl of 5 mM Tris-HCl (Invitrogen; catalog number 15568-025) and treated with 2 U DNase for 1 h at 37°C, and the DNase was inactivated according to the manufacturer's instructions (Ambion; Turbo DNA-free kit; catalog number AM1907).
Sorted PBMCs were collected in PBS (Gibco; catalog number 14190) and kept on ice in a sorting tube (BD Biosciences; catalog number 352063) until they were transferred out of the sorting facility. The cells were pipetted into a new Eppendorf tube and centrifuged, and the RNA was extracted from the cell pellet according to the manufacturer's instructions (Qiagen; AllPrep RNA/DNA Mini; catalog number 80204). The column was eluted with 40 μl 5 mM Tris-HCl (Invitrogen; catalog number 15568-025), and the eluate was passed over the column twice to concentrate the RNA. The RNA was treated with 2 U DNase for 1 h at 37°C, and the DNase was inactivated as described above.
Nucleic acids were extracted from PBMCs at UPitt following a previously described protocol (41), with sonication during the lysis step and again after resuspension of nucleic acids.
Reverse transcription and quantitative PCR.
RNA was reverse transcribed using the HML-2 Env reverse primer for the plasma HML-2 virion analysis or with random hexamers (Invitrogen; catalog number N8080127) for the unsorted and sorted cell expression analyses to detect HML-2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts. All assays included both reverse transcription-positive (RT+) and RT-negative (RT−) wells to detect the presence of contaminating DNA in the RNA sample, and water RT+ and RT− controls were also prepared. Reverse transcription reactions were set up as recommended by the manufacturer (Invitrogen; Superscript III First Strand Synthesis; catalog number 18080-051) with the following cycling conditions: 50°C for 50 min, 85°C for 10 min, and 4°C hold. cDNA was used for downstream qPCRs. Reverse transcription for the HIV qPCR analysis was performed as described previously (40).
RNA standards were prepared for the HML-2 qPCR and GAPDH by cloning the amplicon sequence into a vector with a T7 promoter (Invitrogen; pcDNA 3.1 [catalog number K4900-01] or equivalent) and performing in vitro transcription (Ambion, MEGAscript T7; catalog number AM1334) to make copies of the qPCR amplicon. Standards for the HIV qPCR were either supplied by Mary Kearney (NCI Frederick) or prepared from a vector through in vitro transcription (IVT). IVT RNA was purified twice according to the manufacturer's instructions (Ambion; Megaclear; catalog number AM1908) and visualized on a denaturing gel to confirm the size of the IVT product. A known quantity of standard RNA was used in each assay to quantify the amount of RNA present in the test sample and was prepared in 10-fold serial dilutions and reverse transcribed at the same time as the test samples.
cDNA produced using an HML-2 gene-specific primer was detected using the HML-2 qPCR specific to the TM region of the env gene. The specificity of this qPCR for HML-2 env was verified by melting-curve analysis and sequencing of the amplified products. The primers (HML-2 env For, 5′ CTAACCATGTCCCAGTGATG 3′); HML-2 env Rev, 5′ GGAGACAGACTCATGAGCTTAGAA 3′) were used at a final concentration of 300 nM in a SYBR mastermix (Applied Biosystems; SYBR green PCR MM; catalog number 4309155). The HML-2 qPCR cycling conditions were as follows: 95°C for 10 min; 95°C for 15 s, 57°C for 15 s, 72°C for 45 s, and 74°C for 15 s; plate reading (45 cycles); and melting curve, 55°C to 95°C. Each sample and water RT+ and RT− well was analyzed in triplicate using the HML-2 qPCR. The GAPDH qPCR was performed as described for the HML-2 assay (GAPDH For, 5′ GTCAGTGGTGGACCTGACCT 3′; GAPDH Rev, 5′ TGCTGTAGCCAAATTCGTTG 3′). The GAPDH cycling conditions were as follows: 95°C for 10 min; 95°C for 15 s, 63.5°C for 15 s, 72°C for 45 s, and 82°C for 15 s; plate reading (45 cycles); and melting curve, 55°C to 95°C. HIV qPCR was performed as previously described (40).
For the Roche HIV TaqMan assay, plasma HIV RNA was reverse transcribed and quantitated using the automated Cobas TaqMan System v2.0 from Roche Molecular Diagnostics (quantitation limit, 20 cps/ml). Cellular HIV RNA and DNA were quantified using previously described protocols (41, 42). For DNA quantification, following estimation of the total nucleic acid concentration with a NanoDrop 1000 (Thermo Scientific), samples were diluted to a final concentration of <170 ng/μl to prevent inhibition of qPCR. Eight replicates from each sample were assayed for total HIV-1 DNA or RNA using published qPCR methods with normalization for cellular input (42). The 95% limit of detection (LOD) for the HIV-1 DNA or RNA was 5 copies per qPCR.
Flow cytometry and analysis.
PBMCs collected from 10 HIV-infected patients (Table 2) were thawed in a 37°C water bath until just icy and washed twice in buffer (PBS without Mg2+ or Ca2+, 2 mM EDTA, 0.1% bovine serum albumin [BSA]). The cells were resuspended in buffer and stained with antibodies to CD3 (clone UCHT1), CD4 (clone RPA-T4), CD8 (clone RPA-T8), CD14 (clone MΦP9), and CD20 (clone 2H7); antibodies and their isotype controls were procured from BD Biosciences (San Jose, CA) and BioLegend (San Diego, CA). Additionally, the cells were stained with LIVE/DEAD (L/D) stain (Molecular Probes; L/D Fixable Aqua Dead Cell Stain kit; catalog number L34957) or propidium iodide to allow sorting of live cells only, preincubated with an Fc receptor-blocking solution to prevent nonspecific binding of antibodies (BioLegend; Human TruStain FcX; catalog number 422302), and passed through a 40-μm mesh strainer prior to sorting (BD Biosciences; catalog number 352235) to minimize clumping. Samples were sorted on a BD Influx (BD Biosciences; 405 nm, 488 nm, and 635 nm) using the Spigot software package. All cells were gated on absence of L/D stain and size (pulse width × forward scatter [FSC] and FSC × side scatter [SSC]). Cells were sorted into lineages based on expression of the following markers: CD3+ CD4+ (CD4+ T cells), CD3+ CD8+ (CD8+ T cells), CD3− CD20+ (B cells), and CD3− CD14+ (monocytes). The cells were sorted into PBS and kept on ice until RNA extraction. All flow cytometry analysis postsort was performed using Summit version 4.3 build 2445 (Beckman Coulter, Fullerton, CA).
Statistical analysis.
All statistical analyses were performed using GraphPad (San Diego, CA) Prism software. The data were analyzed using the specified nonparametric tests due to the small sample size and uncertainty about normal distribution in the population. Correlation analyses were performed as either linear correlations or nonlinear correlations based on the data sets and are indicated for each analysis. P values and R2 values are noted where applicable, and a P value of <0.05 defined statistical significance.
RESULTS
Detection of HML-2 proviruses.
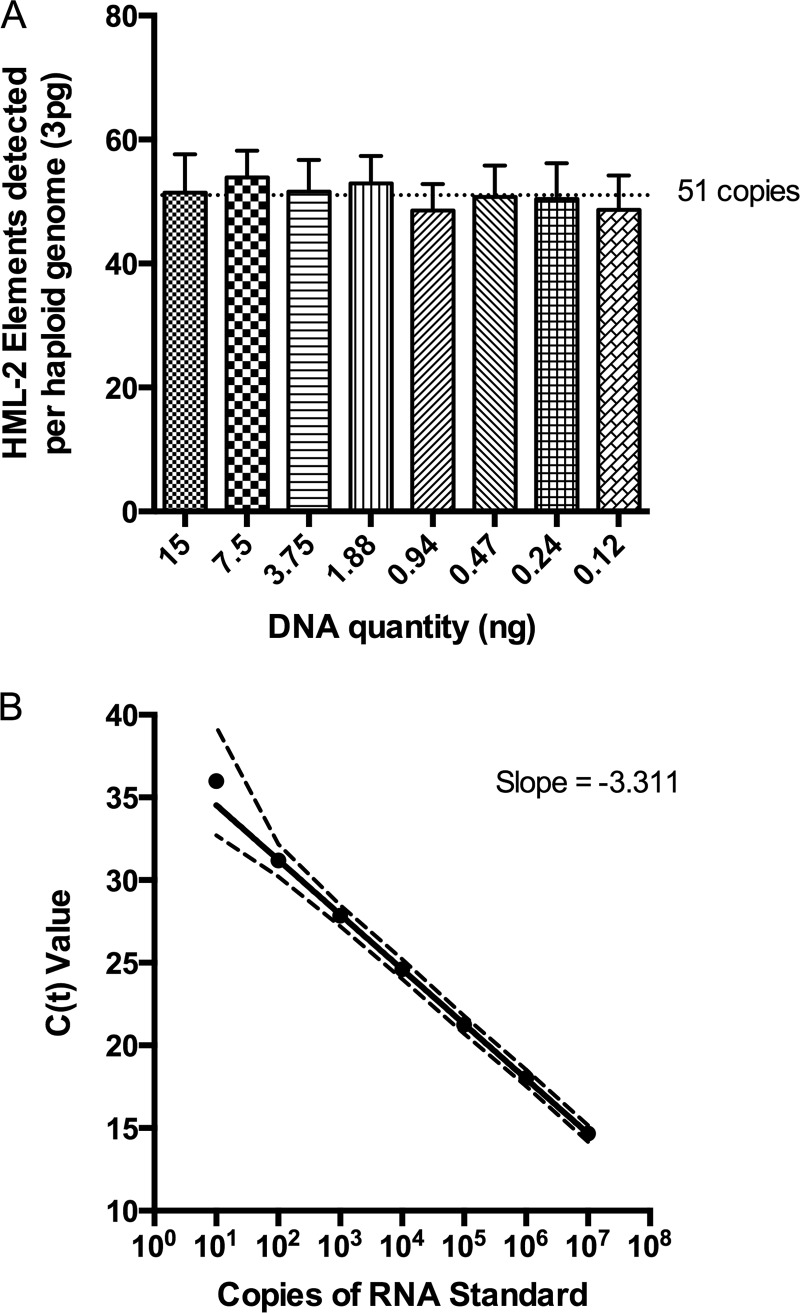
The HML-2 subgroup of HERV-K comprises at least 89 proviruses per haploid human genome (7), many of which have accumulated inactivating mutations or deletions. To design a qPCR assay capable of detecting the majority of these proviruses, a master alignment of all known elements was prepared and searched for regions of highest sequence conservation. Importantly, HML-2 proviruses can be categorized into type 1 or type 2 (2). Type 1 HML-2 proviruses have a characteristic 292-bp deletion in the SU region of the envelope gene and are associated with the production of an alternative accessory protein named Np9 (43), whereas those type 2 proviruses that retain a full coding sequence produce the accessory protein Rec (44). A 119-bp sequence located in the TM region of env was chosen as the target of our qPCR assay due to its high sequence conservation among both type 1 and type 2 HML-2 proviruses. Based on primer sequence similarity to individual proviruses, an estimated 67 proviruses out of the 91 included in the alignment should be detected using the env qPCR assay, including both recent and ancient integrations (data not shown). The number of proviruses detected experimentally, 51 copies per haploid genome (Fig. 1A), was similar to, although slightly lower than, this prediction. All other primer locations would detect a similar number, since many of them are missing substantial portions of their genomes. The identity of the individual proviruses detected using the env qPCR assay was not investigated in detail, though the env primers were confirmed to detect K111 proviruses, known to be highly repeated due to postintegration duplication (reference 45 and data not shown). Known quantities of RNA standards, based on the K108 (7p22.1a) env sequence, were created to quantitate HML-2 env RNA signal from cell and plasma specimens. Assay performance between multiple runs of the standards showed high repeatability, high PCR efficiency, and a linear dynamic range of 107 down to 10 copies (Fig. 1B).
FIG 1.
Quantitative-PCR detection of HML-2 proviruses. (A) The number of HML-2 proviruses detected per haploid genome using env qPCR was determined using dilutions of human genomic DNA at known concentrations (using the estimate of 6 pg DNA/cell and 3 pg DNA/haploid genome) and determining the copy numbers of HML-2 DNA at these different dilutions using plasmid DNA standards containing the 7p22.1a env sequence. The mean and standard deviation are plotted for each dilution, where the mean is the average of replicate wells from 3 assays. (B) RNA standard serial dilutions were reverse transcribed in duplicate, and the cDNA was assayed using SYBR green env qPCR. Mean cycle threshold values (CT; y axis) and the 95% confidence intervals (CI) are plotted for each dilution of the RNA standard (x axis). CT values are compiled from multiple assays (n = 41 for 10 to 106 copies and n = 26 for 107 copies). The slope of the line is −3.311, corresponding to a PCR efficiency of 100.46%.
Lack of HML-2 RNA detection in plasma from HIV-infected patients.
Prior reports (33–36) presented data in support of the idea that HML-2 virions were released into the blood of HIV-1-infected patients. To see if we could confirm this result, plasma was collected from two different centers and tested for the presence of HML-2 virions. Deidentified plasma from HIV-1-infected patients (n = 9) was collected from the archives at the NIH Clinical Center. Additional patients (n = 6) were recruited in a clinical study ongoing at Tufts Medical Center specifically for HIV-1-infected patients naive to therapy or off ART for >2 months. All samples tested were from patients off ART at the time of collection to exclude any possible effect of ART on HML-2 detection (33, 34). Sample characteristics are detailed in Table 1.
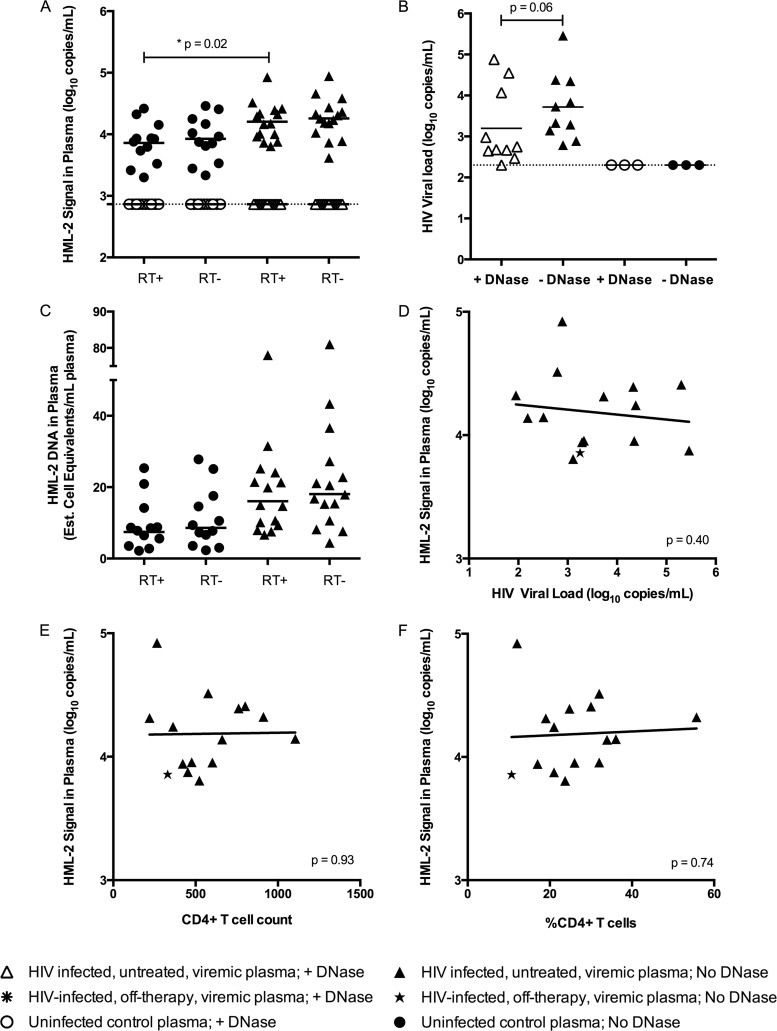
RNA was extracted from pelleted plasma and treated with recombinant DNase I to degrade any contaminating genomic DNA that might have led to false HML-2 RNA signal in the env qPCR assay. DNase-treated RNA was reverse transcribed and analyzed for virus expression using env qPCR or a modified HIV single-copy assay (SCA) (40). A previously published report estimated that ∼95% of patients infected with HIV-1 have detectable HML-2 RNA in their plasma (35) and, moreover, that extremely high levels of HML-2 RNA are present in the plasma of those patients, ranging from to 103 to 1010 copies per ml (33, 34, 36, 45). Contrary to these reports, we could not detect HML-2 RNA in the plasma of any HIV-1-infected patient tested (Fig. 2A). It is possible that the DNase used contained contaminating RNases or that the DNase was not properly inactivated, leading to a reduction in HML-2 detection. Therefore, a comparison between DNase-treated and untreated RNAs was performed with and without reverse transcriptase to determine if an HML-2 RNA signal could be measured in the absence of DNase. However, even without DNase treatment, an HML-2 RNA signal was not detected, and the RT+ and RT− wells yielded equivalent copy values, suggestive of only DNA template being present (Fig. 2A). Additionally, the cellular GAPDH gene could be detected when the untreated RT− RNA was used as a qPCR template but not when DNase-treated RT− RNA was used, consistent with the presence of a contaminating DNA template (data not shown).
FIG 2.
Absence of HML-2 virions in plasma from HIV-1-infected patients. (A) Plasma from HIV-1-infected viremic patients and uninfected controls was centrifuged at 21,000 × g to pellet virions. RNA extraction was followed by DNase treatment (+ DNase) or no treatment (No DNase). RNA was reverse transcribed (RT+), along with no-RT controls (RT−). RT+ and RT− wells were analyzed in triplicate using env qPCR. The dotted line represents the limit of detection (730 copies), and the geometric mean is plotted for each group of samples (*, P = 0.02; Mann-Whitney test). (B) The same samples used for HML-2 detection were analyzed using a modified HIV SCA qPCR. The dotted line represents the limit of detection (200 copies), and the geometric means are plotted for both HIV-1-infected patient groups (P = 0.06; Wilcoxon signed-rank test). (C) The amounts of HML-2 DNA in plasma from healthy controls and HIV-1-infected subjects were estimated for the RT+ and RT− wells shown in panel A. A conversion factor between RNA and DNA signals in the env qPCR was determined by running RNA and DNA standards simultaneously. The cell number was calculated from the resulting DNA copy number (102 copies of HML-2 DNA/cell). Means are plotted for each group. (D to F) Level of HML-2 RNA plotted against the level of HIV-1 viremia (D), the CD4+ T-cell count (E), and the percentage of CD4+ T cells (F). The P values for linear regressions are shown in the lower right-hand corners.
Our failure to detect HML-2 RNA was not due to improper extraction from the plasma samples, since HIV RNA was detected in the clinical specimens using the same RNA preparations that were used for the HML-2 assay (Fig. 2B). Additionally, HIV was detected with or without DNase treatment of viral RNA, although there was an ∼4-fold reduction in RNA levels after DNase treatment (Fig. 2B). In addition, the HML-2 RNA extraction and detection procedures were validated using supernatants from the teratocarcinoma cell line Tera-1, which is known to produce HML-2 virions (data not shown) (16–18, 46). In case the extraction and detection of HML-2 virions failed for unknown reasons, previously published methods for both were also used as controls on a subset of clinical samples. Similar to what is shown in Fig. 2A, these alternative methods did not lead to the detection of HIV RNA (data not shown). Thus, in the group of patients tested, we could find no evidence for HML-2 virions in plasma.
In contrast to the lack of HML-2 RNA detection in plasma, our experiments did show a significant difference in the levels of HML-2 DNA in the plasma of HIV-1-infected patients compared to controls (Mann-Whitney test; P = 0.02) (Fig. 2A). Based on the estimated number of copies of HML-2 proviral DNA detected per haploid genome (Fig. 1A) and the difference in signal intensities from HML-2 RNA versus DNA in the env qPCR assay (not shown), the control plasma DNA level was calculated to be equivalent to ∼11 lysed cells per ml, whereas the DNA levels from HIV-1-infected patients were about 2-fold higher, with an estimated ∼23 lysed cells per ml of plasma (Fig. 2C). The source of the cell DNA is unknown, but it could be related to immune surveillance and/or infected-cell killing during active HIV replication in the absence of ART. However, the level of HML-2 DNA signal in the plasma was not strongly associated with HIV RNA levels, the number of CD4+ T cells/μl, or the percentage of CD4 T cells in the HIV-infected patients (Fig. 2D to F).
HML-2 RNA detection in PBMCs from HIV-1-infected patients.
Although HML-2 virions were not detected in the plasma of the tested HIV-1-infected or control patients, it was possible that HML-2 was actively transcribed in PBMCs (39). As with the plasma samples, PBMCs were obtained from archived samples from the NIH Clinical Center (n = 13) and also from an ongoing clinical study at Tufts Medical Center (n = 6), where all patients were verified to be off ART. Sample characteristics are listed in Table 1. PBMCs were assessed for viability, and 1 million to 2 million cells were used for total RNA extraction, followed by DNase treatment, reverse transcription, and analysis for the total copy number using qPCR assays for env and the reference GAPDH transcripts.
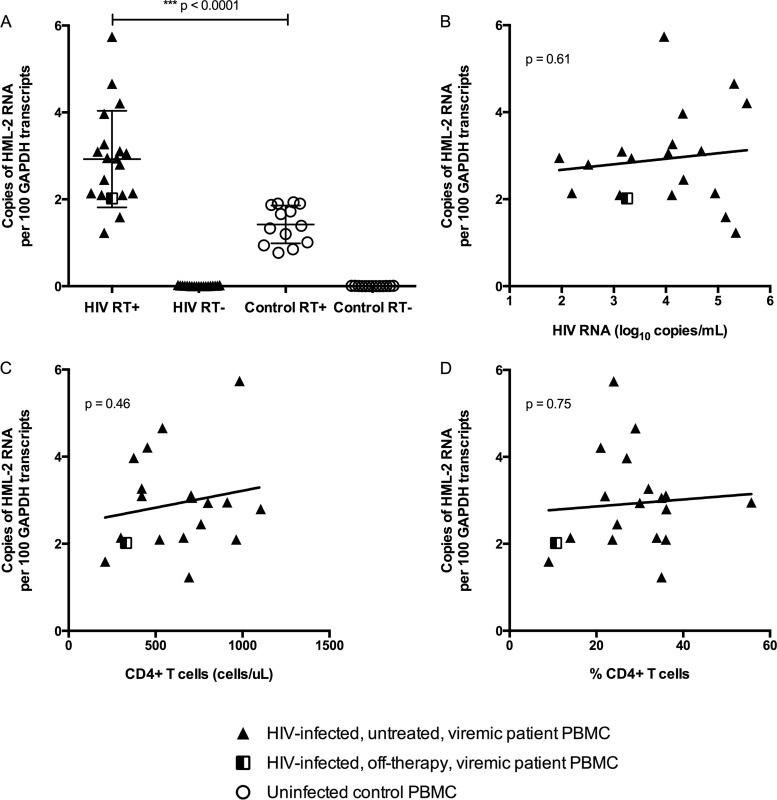
PBMCs isolated from HIV-infected patients showed significant upregulation in HML-2 transcription relative to GAPDH and compared to uninfected controls (Mann-Whitney test; P < 0.0001) (Fig. 3A). The extent of HML-2 transcription was not significantly associated with any of the tested HIV-1 disease markers, including plasma HIV RNA levels, the number of CD4+ T cells/μl, or the percentage of CD4 T cells (Fig. 3B to D). To more directly test the correlation of HML-2 expression and HIV replication and to determine if HIV replication is necessary for the apparent upregulation of HML-2 transcription, a blinded study was performed to assess HML-2 RNA levels in PBMCs from 15 HIV-1 patients on therapy, 10 without viremia (<75 copies of RNA/ml) and 5 with detectable viremia, compared to uninfected controls (n = 4) (Table 2). Of the 5 patients with viremia, 3 were off ART and 2 were on failing ART regimens (see Table S1 in the supplemental material).
FIG 3.
HML-2 RNA expression in PBMCs from HIV-1-infected patients. (A) HML-2 RNA was quantitated in PBMCs collected from viremic HIV-1-infected subjects, as well as uninfected controls. Relative expression of HML-2 was assessed for each sample in triplicate by comparing the HML-2 copy number to the reference GAPDH gene copy number. RT− controls are shown for all samples. The data are reported as copies of HML-2 RNA per 100 copies of GAPDH RNA (***, P < 0.0001; Mann-Whitney test). The mean and standard deviation are plotted for each group. (B to D) Extent of HML-2 upregulation in PBMCs plotted against HIV plasma RNA (B), CD4 T cells/μl (C), or the percentage of CD4 T cells (D). The P values for linear regression are shown in the upper left-hand corners.
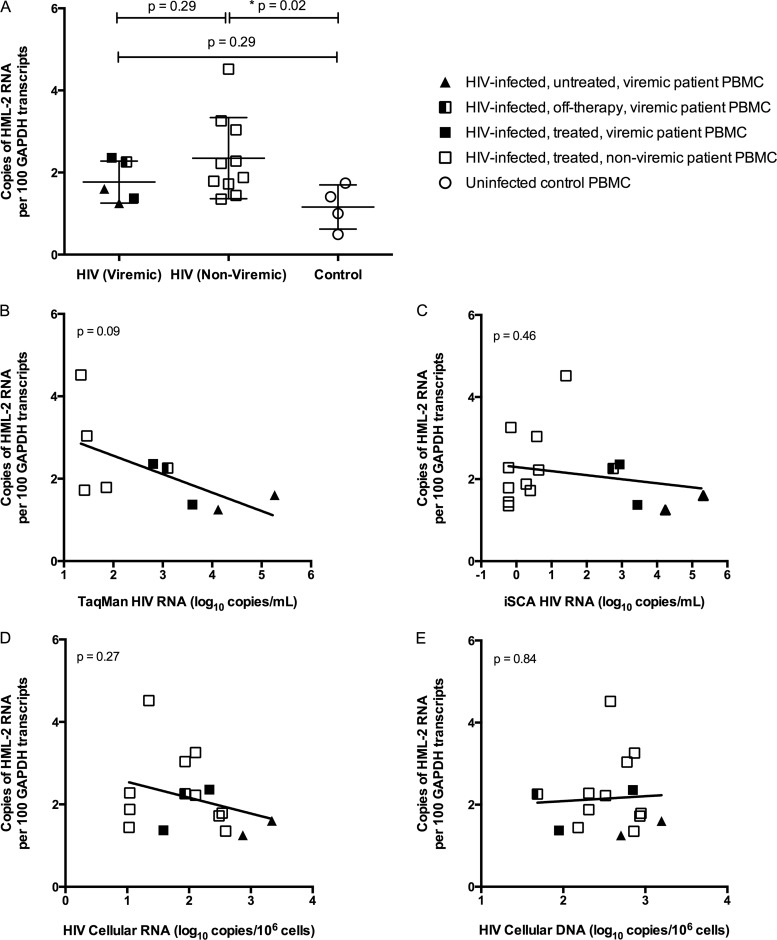
Compared to the uninfected controls, there was upregulation in HML-2 RNA in both groups of patients on ART, but to a significant level only in the aviremic (Mann-Whitney test; P = 0.02) and not the viremic (Mann-Whitney test; P = 0.29) patients (Fig. 4A). The lack of statistical significance for the viremic group could have been due to the smaller sample size (n = 5). The magnitudes of HML-2 upregulation in HIV-infected patients compared to uninfected controls were similar in viremic patients and in those with suppressed viremia on ART (Fig. 3A and data not shown). Thus, the use of antiretrovirals and lack of HIV replication did not appear to have a major effect on HML-2 RNA transcription (Fig. 4B to D). In addition, there was no association of HML-2 RNA with HIV DNA levels (Fig. 4E).
FIG 4.
HML-2 RNA in PBMCs from HIV-1-infected patients on antiretroviral therapy. (A) HML-2 RNA was quantitated in PBMCs collected from HIV-1-infected subjects on antiretroviral therapy without detectable viremia; from patients with viremia who were either on therapy (treated), currently off therapy, or naive to therapy (untreated); and from uninfected controls. Note that the open symbols indicate samples with clinically undetectable HIV RNA (<75 copies). RT− wells were below the limit of detection (data not shown). The data are reported as copies of HML-2 RNA per 100 copies of GAPDH RNA (*, P = 0.02; Mann-Whitney test). The mean and standard deviation are plotted for each group. (B to E) Level of HML-2 RNA expression in PBMCs plotted against HIV plasma RNA measured by TaqMan assay (B), HIV plasma RNA measured by iSCA assay (C), cellular HIV RNA (D), or cellular HIV DNA (E). The P values for linear regression are shown in the upper left-hand corners.
Cell-type-specific expression of HML-2 RNA in PBMCs.
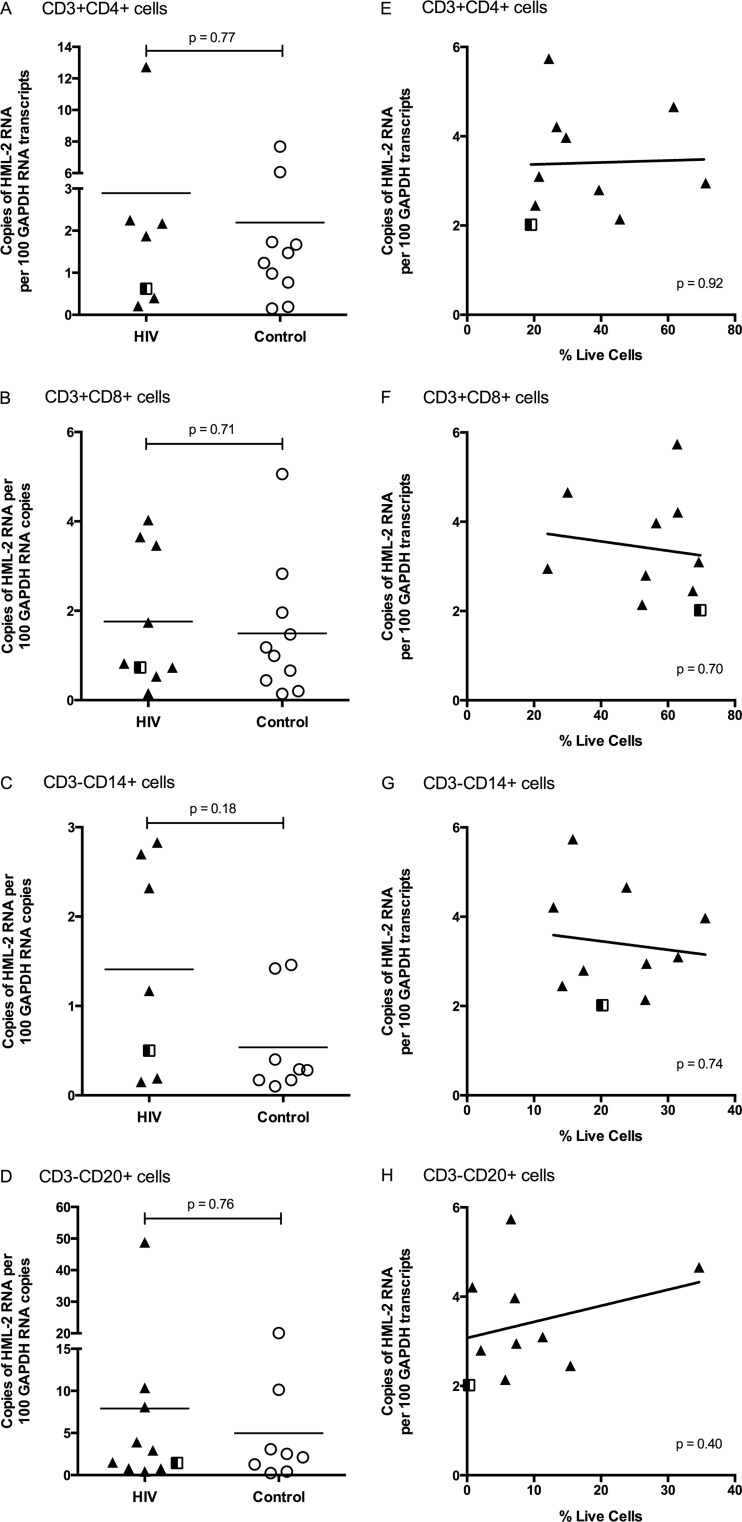
We next investigated the cell source of the HML-2 RNA expression to begin to elucidate the mechanism(s) governing HML-2 provirus activation in HIV-infected patients. Attempts to isolate HML-2-expressing cells using a commercially available antibody to HERV-K Env were unsuccessful; therefore, PBMCs were sorted by cell type and then assayed for HML-2 RNA. Specifically, PBMCs isolated from patients described in Table 1 (NIH Clinical Center and TMC only) were subjected to live cell sorting into these major PBMC subsets: CD4+ T cells (CD3+ CD4+), CD8+ T cells (CD3+ CD8+), B cells (CD3− CD20+), and monocytes (CD3− CD14+). HML-2 RNA levels were determined as described for total PBMCs.
Consistent with the lack of correlation of HML-2 RNA levels with HIV replication, sorted CD4+ T cells were not enriched for HML-2 RNA compared to other cell subsets (Fig. 5A). Interestingly, no cell type was significantly enriched for HML-2 RNA transcription; in fact, all PBMC subsets tested showed detectable HML-2 expression in both the HIV-infected and uninfected populations, although to different extents (Fig. 5A to D). This result is consistent with previous assessments showing HML-2 expression in blood cells (47, 48). For all cell types, the HIV-infected patients exhibited a slightly higher level of HML-2 expression, with the greatest difference in monocytes (Mann-Whitney test; P = 0.18) (Fig. 5C). However, no significant correlation was found between the percentage of monocytes in PBMCs and HML-2 RNA upregulation in total PBMCs (P = 0.56) (Fig. 5G). Indeed, in no cell type did the difference reach statistical significance, even though there was a significant difference when all PBMCs were analyzed (Fig. 3A).
FIG 5.
HML-2 RNA expression in different cell types. (A to D) PBMCs from viremic HIV-infected patients and healthy controls were stained with antibodies specific to CD3, CD4, CD8, CD14, and CD20. Live cells that were CD3+ CD4+, CD3+ CD8+, CD3− CD14+, and CD3− CD20+ were sorted for each patient and analyzed for HML-2 RNA expression. (A) CD3+ CD4+ T cells. (B) CD3+ CD8+ T cells. (C) CD3− CD14+ monocytes. (D) CD3− CD20+ B cells. The P values for Mann-Whitney t tests are shown. (E to H) Correlation between the percentage of PBMCs (after size and LIVE/DEAD gating) and HML-2 upregulation in total unsorted PBMCs, including CD3+ CD4+ T cells (E), CD3+ CD8+ T cells (F), CD3− CD14+ monocytes (G), and CD3− CD20+ B cells (H). The P values for linear regression are shown in the lower right-hand corners. See the legend to Fig. 3 for symbol definitions.
Taken together, our results imply that increased HML-2 expression in HIV-infected patient PBMCs is due to an indirect effect of HIV-1 infection. Furthermore, HIV-1 infection does not result in the release of a large amount of HML-2 virions in the plasma of patients represented in our cohort, as measured by our assay, contrary to publications showing HML-2 RNA detection at 103 copies/ml and higher (33, 34, 36, 45).
DISCUSSION
The HML-2 group of endogenous retroviruses is of great interest due to their recent integration into the human germ line, their sequence preservation, and the association of their expression with human diseases (2). Previous reports investigating HML-2 activity during HIV-1 infection have described detectable HML-2 virions in patient plasma, often at levels higher than those of HIV (33–36, 45); HML-2 RNA upregulation in patient CD4+ and CD8+ T cells (39); and the presence of cytotoxic immune responses specific to HML-2 antigens (29–31). The observation of HML-2 RNA and protein expression during HIV-1 infection has led to a proposal for development of a vaccination strategy to eliminate HIV-1-infected cells, which may be targeted by their ability to display HML-2 antigens (32). However, the mechanisms governing HML-2 expression are unclear and, given the large number of proviruses, likely to be very complex, and the cells expressing HML-2 RNA and protein are largely undefined.
In this study, we investigated HML-2 expression in a cohort of HIV-infected patients to determine the cell source of HML-2 expression and the relationship of HML-2 expression to HIV disease status. Using a qPCR assay capable of detecting RNA from more than half of the known HML-2 elements (Fig. 1A), we observed, contrary to previous reports, that HML-2 virions were not detectable in patient plasma during untreated HIV-1 infection (Fig. 2A). The discrepancy between this and previous results was not due to differences in methodology, since our finding was confirmed using a previously published extraction and detection method with a subset of the clinical samples (Fig. 2A and data not shown). Similarly, a recent study using deep-sequencing techniques did not uncover an increase in HERV RNA sequences from the plasma of HIV-1 subtype B subjects compared to uninfected controls (49). It is possible that virion production was occurring below our limit of detection using the env qPCR, as we did not achieve single-copy sensitivity.
The presence of HML-2 virions, at any level, would be highly interesting. Though no HML-2 provirus has been shown to be infectious as is, studies on reconstituted HERV-K (HML-2) viruses suggest that low-level infectivity can be acquired through a few recombination events between different proviruses (14, 15). Studies with higher sensitivity should be performed to determine if such events can occur and if they are correlated with HIV disease progression. Another, less likely, possibility to explain our finding is that HML-2 virion production occurs in a patient population not captured in our study.
There are limited clinical data profiling the exact comorbidities and ethnicities of study subjects reported to show HML-2 virions in their plasma. Our patient population included varied ethnic backgrounds, and most individuals lacked comorbidities associated with HIV, like HCV or human T cell leukemia virus (HTLV) infection. The two patients in our plasma sample cohort (Table 1) infected with HCV did not show virion production (Fig. 2A). While HTLV Tax was associated with HERV transcription in one study (50), HTLV infection was not associated with HML-2-specific immune responses, differing from HIV (51). Infection with various herpesviruses has been associated with an increase in HERV transcription (52–54), although its impact on eliciting virions in vivo is unknown.
During this analysis, we found that HIV-1-infected patient plasma showed a significant increase in extracellular DNA compared to uninfected individuals (Fig. 2A). Plasma DNA, specifically mitochondrial DNA, has been observed in HIV-1-infected patients previously, though not at a level significantly different from controls (55). The finding of extracellular DNA in the plasma of HIV-1 patients could help explain the discrepancies between our results and those from previous publications. If extracted plasma RNA was not treated with DNase and adequate reverse transcriptase controls were not run, our assay could have led to the erroneous detection of HML-2 RNA in patient plasma (Fig. 2A). However, the signal would have derived solely from extracellular DNA present in the plasma of HIV-1 patients and not from HML-2 RNA. This is possible because HML-2 proviruses are present at a high copy number in the human genome, with at least 89 proviruses present per haploid genome (7). Furthermore, levels of DNA in patient plasma could also be affected by sample handling, i.e., if blood samples were not immediately processed, increasing amounts of DNA could be present in patient plasma samples and provide more template for erroneous HML-2 RNA detection. Finally, quantitation of contaminating genomic DNA with an RNA-based standard could further inflate HML-2 signal due to the less than 100% efficiency of the RT step used for the standard curve. Therefore, a combination of the above factors could lead to the false detection of high levels of HML-2 RNA and possibly explain why previous publications have shown HML-2 RNA in HIV-1 patient plasma.
Extracellular double-stranded DNA (dsDNA) could potentially function as a damage-associated molecular pattern if taken up by cells and detected by a dsDNA receptor in the endosome or cytosol, leading to immune activation (56). In this study, association of plasma DNA with immune activation markers could not be assessed. Signaling due to extracellular DNA could potentially represent an additional pathway to chronic immune activation commonly seen in HIV-1-infected individuals (57). An additional study examining the levels of plasma DNA in patients on antiretroviral therapy would also be informative, as chronic activation is present, albeit at a lower level, in patients on long-term therapy and is associated with decreased longevity, cardiovascular disease, and metabolic syndrome (58).
A significant upregulation in HML-2 RNA was assessed in PBMCs from HIV-1-infected patients in the absence or presence of antiretroviral therapy (P < 0.0001 [Fig. 3A] and P = 0.02 [Fig. 4A]). On average, the level of expression was about 2-fold higher than that seen in uninfected controls, although some patients showed levels of HML-2 transcription equivalent to those of HIV-negative subjects. This result could signify that HML-2 upregulation is not a universal phenomenon or perhaps that the cell source of expression was not highly abundant in the patients with no measurable increase in transcription. The initial analysis was performed using unsorted PBMCs (Fig. 3A), so cell subset frequency could have affected the overall measured expression levels. To assess this possibility, total PBMCs from HIV-infected and uninfected patients were sorted into CD4+ and CD8+ T cell subsets, in addition to B cells and monocytes. These cell types represent the major constituents of PBMCs, although smaller populations, including dendritic cells and NK cells, could potentially represent sources of HML-2 expression. When HML-2 RNA upregulation was assessed in sorted cells, we saw no significant difference in expression in any subset from HIV patients compared to controls, although each subset individually showed a small increase in patients compared to controls, with the greatest difference in monocytes, a cell type that is not a target for HIV infection in vivo (59). The lack of enrichment in the CD4+ T cell population is consistent with HML-2 RNA expression having no clear relationship to HIV replication (Fig. 4), while the lack of enrichment in other cell populations could exemplify the indirect mechanism regulating HML-2 expression in HIV-1-infected patients. The differences in the magnitude of HML-2 transcription in cell subsets could be due to cell-specific transcription factors, methylation patterns, or other epigenetic changes, which are believed to control endogenous retrovirus expression in differentiated tissues (60, 61). Based on these results, it appears that differential expression from multiple cell sources may lead to the 2-fold difference in HML-2 expression in HIV-infected versus control individuals.
It is important to point out that our results do not exclude the possibility of significant upregulation of expression of HML-2 in HIV-infected cells. On average, less than 0.2% of CD4 cells are infected with HIV during chronic infection (62), so even a 10-fold upregulation of HML-2 RNA would lead to only a 2% increase in the total cell population. To answer this question directly, it will be necessary to sort either HIV- or HML-2-expressing cells from patient samples, a daunting task.
In this study, no significant correlation was observed between HIV-1 disease markers, including the level of viremia and intracellular HIV RNA and DNA, and HML-2 expression in patient PBMCs (Fig. 3B to D and 4), and no enrichment was seen in HIV-1 target CD4+ T cells, as mentioned above (Fig. 5A). In addition, there was no effect of antiretroviral therapy on HML-2 transcription (Fig. 4A). The lack of effect of HIV replication or antiretrovirals is interesting, as they have been reported to have positive and negative effects, respectively, on HML-2 virion production in HIV-1-infected patients (33, 35). In vitro, the accessory proteins Tat and Vif have been reported to positively influence HML-2 RNA and protein expression (32, 37). The difference between the in vitro and in vivo results may be due to the higher levels of infection and therefore protein production seen during in vitro infections, or potentially a noncanonical function the accessory proteins assume under in vitro conditions.
Immune activation in HIV-1 infection remains a possible indirect mechanism that could lead to HML-2 expression. It is known that other endogenous retroviruses, including porcine endogenous retroviruses (PERV), murine leukemia virus (MLV), and MMTV, exhibit increased expression after treatment with mitogens or immune-activating agents (63–66). Although we did not see a difference in HML-2 upregulation between patients on or off antiretroviral therapy (Fig. 4A), which are associated with different levels of immune activation (57), immune activation markers were not specifically assessed in these patient groups. A previous study reported a negative correlation between immune activation (CD38+ HLA− DR+) in CD4+ and CD8+ T cells from HIV-1-infected patients and HML-2 expression in vivo (39), though a positive effect of stimulating agents like phorbol myristate acetate (PMA)/ionomycin, phytohemagglutinin (PHA), and interleukin 2 (IL-2) has been reported in vitro (37, 38). This point remains to be clarified with additional studies.
As the env qPCR measures total HML-2 signal and does not distinguish between the proviruses expressed, it is possible that individuals express distinct proviruses and the expression of only some of these was associated with HIV disease markers. It will be beneficial to explore which HML-2 polymorphic loci are expressed, as this information could indicate different disease outcomes based on inherited polymorphisms or provirus expression patterns. In addition, different HML-2 proviruses have different ORFs that dictate which retroviral proteins are expressed in vivo (7). Likewise, investigation into whether type 1 or type 2 proviruses are expressed differently during HIV-1 infection could elucidate new cellular functions for the accessory proteins Rec and Np9, as their expression is associated with malignancy (43, 67, 68).
The results of this study confirm the increased expression of HML-2 RNA in PBMCs from HIV-1-infected patients; however, they do not support the claim that HML-2 virions are present in blood. In addition, our observations of HML-2 RNA expression in sorted PBMCs show that all the major cell types in the blood express HML-2 and possibly protein, consistent with a previous analysis using whole blood (48). Based on these observations, it appears that the use of HML-2 expression as a way to target HIV infection carries a significant risk of off-target effects. Our data suggest that HML-2 protein may be expressed in CD8+ T cells and B cells. Thus, targeting HML-2 epitopes may affect these cells and weaken the cytotoxic and humoral arms of an individual's immune response to HIV-1 infection (69). HML-2 expression has also been detected in embryonic stem cells, presenting an additional cause for concern (70). The mode of HML-2 activation in cells remains an open issue; however, neither the levels of HIV replication nor antiretroviral therapy appears to affect its expression. Our study reveals the complexity of HML-2 RNA expression during HIV-1 infection and the need for additional studies to clarify its effects on HIV-1 pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank all the patient volunteers who participated in this study. We also thank Mary Kearney, Liz Anderson, and Ann Wiegand at NCI Frederick and Elizabeth Fyne at the University of Pittsburgh for providing reagents and coordinating sample shipments and Michael Jordan, Jose Caro, Maureen K. Ward, and Kimeto Bailey at Tufts Medical Center for their efforts in patient recruitment and sample collection.
This work was supported by research grant R37 CA 089441 from the National Cancer Institute and by Leidos contract 25XS119 to J.M. through the National Cancer Institute. N.B. was a Howard Hughes Medical Institute (HHMI) Med into Grad Scholar supported in part by a grant to Tufts University from HHMI through the Med into Grad Initiative and the recipient of a travel award from the Tufts/Brown/Life Span Center for AIDS Research. J.M.C. was a Research Professor of the American Cancer Society with support from the F. M. Kirby Foundation.
Footnotes
Published ahead of print 23 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01623-14.
REFERENCES
- 1.Boeke JD, Stoye JS. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p 343–435 In Coffin JM, Hughes SH, Varmus HE. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 2.Bannert N, Kurth R. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 7:149–173. 10.1146/annurev.genom.7.080505.115700 [DOI] [PubMed] [Google Scholar]
- 3.Dewannieux M, Blaise S, Heidmann T. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 79:15573–15577. 10.1128/JVI.79.24.15573-15577.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boller K, Schonfeld K, Lischer S, Fischer N, Hoffmann A, Kurth R, Tonjes RR. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89:567–572. 10.1099/vir.0.83534-0 [DOI] [PubMed] [Google Scholar]
- 5.Löwer R, Löwer J, Kurth R. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 93:5177–5184. 10.1073/pnas.93.11.5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. 2004. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl. Acad. Sci. U. S. A. 101:1731–1736. 10.1073/pnas.0305763101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90. 10.1186/1742-4690-8-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bittner JJ. 1936. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science 84:162. [DOI] [PubMed] [Google Scholar]
- 9.Jackson RB, Little CC. 1933. The existence of non-chromosomal influence in the incidence of mammary tumors in mice. Science 78:465–466. 10.1126/science.78.2029.465 [DOI] [PubMed] [Google Scholar]
- 10.Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861–868. 10.1016/S0960-9822(99)80390-X [DOI] [PubMed] [Google Scholar]
- 11.Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 79:12507–12514. 10.1128/JVI.79.19.12507-12514.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes JF, Coffin JM. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. U. S. A. 101:1668–1672. 10.1073/pnas.0307885100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531–1535. 10.1016/S0960-9822(01)00455-9 [DOI] [PubMed] [Google Scholar]
- 14.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548–1556. 10.1101/gr.5565706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YN, Bieniasz PD. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3:e10. 10.1371/journal.ppat.0030010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bronson DL, Fraley EE, Fogh J, Kalter SS. 1979. Induction of retrovirus particles in human testicular tumor (Tera-1) cell cultures: an electron microscopic study. J. Natl. Cancer Inst. 63:337–339 [PubMed] [Google Scholar]
- 17.Lower R, Lower J, Frank H, Harzmann R, Kurth R. 1984. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J. Gen. Virol. 65:887–898. 10.1099/0022-1317-65-5-887 [DOI] [PubMed] [Google Scholar]
- 18.Ruprecht K, Ferreira H, Flockerzi A, Wahl S, Sauter M, Mayer J, Mueller-Lantzsch N. 2008. Human endogenous retrovirus family HERV-K(HML-2) RNA transcripts are selectively packaged into retroviral particles produced by the human germ cell tumor line Tera-1 and originate mainly from a provirus on chromosome 22q11.21. J. Virol. 82:10008–10016. 10.1128/JVI.01016-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, Strong TV. 2001. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin. Cancer Res. 7:1553–1560 [PubMed] [Google Scholar]
- 20.Frank O, Verbeke C, Schwarz N, Mayer J, Fabarius A, Hehlmann R, Leib-Mosch C, Seifarth W. 2008. Variable transcriptional activity of endogenous retroviruses in human breast cancer. J. Virol. 82:1808–1818. 10.1128/JVI.02115-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seifarth W, Skladny H, Krieg-Schneider F, Reichert A, Hehlmann R, Leib-Mosch C. 1995. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J. Virol. 69:6408–6416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, Fodinger D, Seppele H, Schanab O, Magin-Lachmann C, Lower R, Jansen B, Pehamberger H, Wolff K. 2003. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 63:8735–8741 [PubMed] [Google Scholar]
- 23.Buscher K, Hahn S, Hofmann M, Trefzer U, Ozel M, Sterry W, Lower J, Lower R, Kurth R, Denner J. 2006. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 16:223–234. 10.1097/01.cmr.0000215031.07941.ca [DOI] [PubMed] [Google Scholar]
- 24.Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. 2007. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int. J. Cancer 120:81–90. 10.1002/ijc.22256 [DOI] [PubMed] [Google Scholar]
- 25.Contreras-Galindo R, Kaplan MH, Leissner P, Verjat T, Ferlenghi I, Bagnoli F, Giusti F, Dosik MH, Hayes DF, Gitlin SD, Markovitz DM. 2008. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J. Virol. 82:9329–9336. 10.1128/JVI.00646-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehlhardt S, Seifert M, Schneider J, Ojak A, Zang KD, Mehraein Y. 2006. Human endogenous retrovirus HERV-K(HML-2) Rec expression and transcriptional activities in normal and rheumatoid arthritis synovia. J. Rheumatol. 33:16–23 [PubMed] [Google Scholar]
- 27.Reynier F, Verjat T, Turrel F, Imbert PE, Marotte H, Mougin B, Miossec P. 2009. Increase in human endogenous retrovirus HERV-K (HML-2) viral load in active rheumatoid arthritis. Scand. J. Immunol. 70:295–299. 10.1111/j.1365-3083.2009.02271.x [DOI] [PubMed] [Google Scholar]
- 28.Sicat J, Sutkowski N, Huber BT. 2005. Expression of human endogenous retrovirus HERV-K18 superantigen is elevated in juvenile rheumatoid arthritis. J. Rheumatol. 32:1821–1831 [PubMed] [Google Scholar]
- 29.Garrison KE, Jones RB, Meiklejohn DA, Anwar N, Ndhlovu LC, Chapman JM, Erickson AL, Agrawal A, Spotts G, Hecht FM, Rakoff-Nahoum S, Lenz J, Ostrowski MA, Nixon DF. 2007. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 3:e165. 10.1371/journal.ppat.0030165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tandon R, SenGupta D, Ndhlovu LC, Vieira RG, Jones RB, York VA, Vieira VA, Sharp ER, Wiznia AA, Ostrowski MA, Rosenberg MG, Nixon DF. 2011. Identification of human endogenous retrovirus-specific T cell responses in vertically HIV-1-infected subjects. J. Virol. 85:11526–11531. 10.1128/JVI.05418-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.SenGupta D, Tandon R, Vieira RG, Ndhlovu LC, Lown-Hecht R, Ormsby CE, Loh L, Jones RB, Garrison KE, Martin JN, York VA, Spotts G, Reyes-Teran G, Ostrowski MA, Hecht FM, Deeks SG, Nixon DF. 2011. Strong human endogenous retrovirus-specific T cell responses are associated with control of HIV-1 in chronic infection. J. Virol. 85:6977–6985. 10.1128/JVI.00179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones RB, Garrison KE, Mujib S, Mihajlovic V, Aidarus N, Hunter DV, Martin E, John VM, Zhan W, Faruk NF, Gyenes G, Sheppard NC, Priumboom-Brees IM, Goodwin DA, Chen L, Rieger M, Muscat-King S, Loudon PT, Stanley C, Holditch SJ, Wong JC, Clayton K, Duan E, Song H, Xu Y, SenGupta D, Tandon R, Sacha JB, Brockman MA, Benko E, Kovacs C, Nixon DF, Ostrowski MA. 2012. HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J. Clin. Invest. 122:4473–4489. 10.1172/JCI64560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Contreras-Galindo R, Almodovar-Camacho S, Gonzalez-Ramirez S, Lorenzo E, Yamamura Y. 2007. Comparative longitudinal studies of HERV-K and HIV-1 RNA titers in HIV-1-infected patients receiving successful versus unsuccessful highly active antiretroviral therapy. AIDS Res. Hum. Retrovir. 23:1083–1086. 10.1089/aid.2007.0054 [DOI] [PubMed] [Google Scholar]
- 34.Contreras-Galindo R, Gonzalez M, Almodovar-Camacho S, Gonzalez-Ramirez S, Lorenzo E, Yamamura Y. 2006. A new real-time-RT-PCR for quantitation of human endogenous retroviruses type K (HERV-K) RNA load in plasma samples: increased HERV-K RNA titers in HIV-1 patients with HAART non-suppressive regimens. J. Virol. Methods 136:51–57. 10.1016/j.jviromet.2006.03.029 [DOI] [PubMed] [Google Scholar]
- 35.Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, Yamamura Y. 2006. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res. Hum. Retrovir. 22:979–984. 10.1089/aid.2006.22.979 [DOI] [PubMed] [Google Scholar]
- 36.Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, Markovitz DM. 2012. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J. Virol. 86:262–276. 10.1128/JVI.00602-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Hernandez MJ, Swanson MD, Contreras-Galindo R, Cookinham S, King SR, Noel RJ, Jr, Kaplan MH, Markovitz DM. 2012. Expression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J. Virol. 86:7790–7805. 10.1128/JVI.07215-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Contreras-Galindo R, Lopez P, Velez R, Yamamura Y. 2007. HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS Res. Hum. Retrovir. 23:116–122. 10.1089/aid.2006.0117 [DOI] [PubMed] [Google Scholar]
- 39.Ormsby CE, Sengupta D, Tandon R, Deeks SG, Martin JN, Jones RB, Ostrowski MA, Garrison KE, Vazquez-Perez JA, Reyes-Teran G, Nixon DF. 2012. Human endogenous retrovirus expression is inversely associated with chronic immune activation in HIV-1 infection. PLoS One 7:e41021. 10.1371/journal.pone.0041021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mens H, Kearney M, Wiegand A, Spindler J, Maldarelli F, Mellors JW, Coffin JM. 2011. Amplifying and quantifying HIV-1 RNA in HIV infected individuals with viral loads below the limit of detection by standard clinical assays. J. Vis. Exp. 55:e2960. 10.3791/2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cillo AR, Sobolewski MD, Bosch RJ, Fyne E, Piatak M, Jr, Coffin JM, Mellors JW. 2014. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 111:7078–7083. 10.1073/pnas.1402873111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cillo AR, Krishnan A, Mitsuyasu RT, McMahon DK, Li S, Rossi JJ, Zaia JA, Mellors JW. 2013. Plasma viremia and cellular HIV-1 DNA persist despite autologous hematopoietic stem cell transplantation for HIV-related lymphoma. J. Acquir. Immune Defic. Syndr. 63:438–441. 10.1097/QAI.0b013e31828e6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armbruester V, Sauter M, Krautkraemer E, Meese E, Kleiman A, Best B, Roemer K, Mueller-Lantzsch N. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 8:1800–1807 [PubMed] [Google Scholar]
- 44.Löwer R, Tönjes RR, Korbmacher C, Kurth R, Löwer J. 1995. Identification of a rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Contreras-Galindo R, Kaplan MH, He S, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Kappes F, Dube D, Chan SM, Robinson D, Meng F, Dai M, Gitlin SD, Chinnaiyan AM, Omenn GS, Markovitz DM. 2013. HIV infection reveals widespread expansion of novel centromeric human endogenous retroviruses. Genome Res. 23:1505–1513. 10.1101/gr.144303.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bieda K, Hoffmann A, Boller K. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82:591–596 [DOI] [PubMed] [Google Scholar]
- 47.Seifarth W, Spiess B, Zeilfelder U, Speth C, Hehlmann R, Leib-Mosch C. 2003. Assessment of retroviral activity using a universal retrovirus chip. J. Virol. Methods 112:79–91. 10.1016/S0166-0934(03)00194-0 [DOI] [PubMed] [Google Scholar]
- 48.Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mosch C. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 79:341–352. 10.1128/JVI.79.1.341-352.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L, Deng X, Linsuwanon P, Bangsberg D, Bwana MB, Hunt P, Martin JN, Deeks SG, Delwart E. 2013. AIDS alters the commensal plasma virome. J. Virol. 87:10912–10915. 10.1128/JVI.01839-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toufaily C, Landry S, Leib-Mosch C, Rassart E, Barbeau B. 2011. Activation of LTRs from different human endogenous retrovirus (HERV) families by the HTLV-1 tax protein and T-cell activators. Viruses 3:2146–2159. 10.3390/v3112146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones RB, Leal FE, Hasenkrug AM, Segurado AC, Nixon DF, Ostrowski MA, Kallas EG. 2013. Human endogenous retrovirus K(HML-2) Gag and Env specific T-cell responses are not detected in HTLV-I-infected subjects using standard peptide screening methods. J. Negat. Results Biomed. 12:3. 10.1186/1477-5751-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sutkowski N, Conrad B, Thorley-Lawson DA, Huber BT. 2001. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 15:579–589. 10.1016/S1074-7613(01)00210-2 [DOI] [PubMed] [Google Scholar]
- 53.Tai AK, Luka J, Ablashi D, Huber BT. 2009. HHV-6A infection induces expression of HERV-K18-encoded superantigen. J. Clin. Virol. 46:47–48. 10.1016/j.jcv.2009.05.019 [DOI] [PubMed] [Google Scholar]
- 54.Kwun HJ, Han HJ, Lee WJ, Kim HS, Jang KL. 2002. Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate early protein 0. Virus Res. 86:93–100. 10.1016/S0168-1702(02)00058-8 [DOI] [PubMed] [Google Scholar]
- 55.Lauring AS, Lee TH, Martin JN, Hunt PW, Deeks SG, Busch M. 2012. Lack of evidence for mtDNA as a biomarker of innate immune activation in HIV infection. PLoS One 7:e50486. 10.1371/journal.pone.0050486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paludan SR, Bowie AG. 2013. Immune sensing of DNA. Immunity 38:870–880. 10.1016/j.immuni.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.d'Ettorre G, Paiardini M, Ceccarelli G, Silvestri G, Vullo V. 2011. HIV-associated immune activation: from bench to bedside. AIDS Res. Hum. Retrovir. 27:355–364. 10.1089/aid.2010.0342 [DOI] [PubMed] [Google Scholar]
- 58.Plaeger SF, Collins BS, Musib R, Deeks SG, Read S, Embry A. 2012. Immune activation in the pathogenesis of treated chronic HIV disease: a workshop summary. AIDS Res. Hum. Retrovir. 28:469–477. 10.1089/AID.2011.0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, Epling L, Tan A, Ho T, Lemey P, Shao W, Hunt PW, Somsouk M, Wylie W, Douek DC, Loeb L, Custer J, Hoh R, Poole L, Deeks SG, Hecht F, Palmer S. 2013. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc. Natl. Acad. Sci. U. S. A. 110:E4987–E4996. 10.1073/pnas.1308313110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manghera M, Douville RN. 2013. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology 10:16. 10.1186/1742-4690-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maksakova IA, Mager DL, Reiss D. 2008. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell. Mol. Life Sci. 65:3329–3347. 10.1007/s00018-008-8494-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Josefsson L, King MS, Makitalo B, Brannstrom J, Shao W, Maldarelli F, Kearney MF, Hu WS, Chen J, Gaines H, Mellors JW, Albert J, Coffin JM, Palmer SE. 2011. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc. Natl. Acad. Sci. U. S. A. 108:11199–11204. 10.1073/pnas.1107729108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tacke SJ, Specke V, Denner J. 2003. Differences in release and determination of subtype of porcine endogenous retroviruses produced by stimulated normal pig blood cells. Intervirology 46:17–24. 10.1159/000068120 [DOI] [PubMed] [Google Scholar]
- 64.Moroni C, Schumann G. 1978. Mitogen induction of murine C-type viruses. IV. Effects of lipoprotein E. coli, pokeweed mitogen and dextran sulphate. J. Gen. Virol. 38:497–503 [DOI] [PubMed] [Google Scholar]
- 65.Greenberger JS, Phillips SM, Stephenson JR, Aaronson SA. 1975. Induction of mouse type-C RNA virus by lipopolysaccharide. J. Immunol. 115:317–320 [PubMed] [Google Scholar]
- 66.King LB, Corley RB. 1990. Lipopolysaccharide and dexamethasone induce mouse mammary tumor proviral gene expression and differentiation in B lymphocytes through distinct regulatory pathways. Mol. Cell. Biol. 10:4211–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galli UM, Sauter M, Lecher B, Maurer S, Herbst H, Roemer K, Mueller-Lantzsch N. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 24:3223–3228. 10.1038/sj.onc.1208543 [DOI] [PubMed] [Google Scholar]
- 68.Gross H, Barth S, Pfuhl T, Willnecker V, Spurk A, Gurtsevitch V, Sauter M, Hu B, Noessner E, Mueller-Lantzsch N, Kremmer E, Grasser FA. 2011. The NP9 protein encoded by the human endogenous retrovirus HERV-K(HML-2) negatively regulates gene activation of the Epstein-Barr virus nuclear antigen 2 (EBNA2). Int. J. Cancer 129:1105–1115. 10.1002/ijc.25760 [DOI] [PubMed] [Google Scholar]
- 69.Walker BD, Yu XG. 2013. Unravelling the mechanisms of durable control of HIV-1. Nat. Rev. Immunol. 13:487–498. 10.1038/nri3478 [DOI] [PubMed] [Google Scholar]
- 70.Fuchs NV, Loewer S, Daley GQ, Izsvak Z, Lower J, Lower R. 2013. Human endogenous retrovirus K (HML-2) RNA and protein expression is a marker for human embryonic and induced pluripotent stem cells. Retrovirology 10:115. 10.1186/1742-4690-10-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.