ABSTRACT
Human polyomavirus 6 (HPyV6) and HPyV7 are commonly found on human skin. We have determined the X-ray structures of their major capsid protein, VP1, at resolutions of 1.8 and 1.7 Å, respectively. In polyomaviruses, VP1 commonly determines antigenicity as well as cell-surface receptor specificity, and the protein is therefore linked to attachment, tropism, and ultimately, viral pathogenicity. The structures of HPyV6 and HPyV7 VP1 reveal uniquely elongated loops that cover the bulk of the outer virion surfaces, obstructing a groove that binds sialylated glycan receptors in many other polyomaviruses. In support of this structural observation, interactions of VP1 with α2,3- and α2,6-linked sialic acids could not be detected in solution by nuclear magnetic resonance spectroscopy. Single-cell binding studies indicate that sialylated glycans are likely not required for initial attachment to cultured human cells. Our findings establish distinct antigenic properties of HPyV6 and HPyV7 capsids and indicate that these two viruses engage nonsialylated receptors.
IMPORTANCE Eleven new human polyomaviruses, including the skin viruses HPyV6 and HPyV7, have been identified during the last decade. In contrast to better-studied polyomaviruses, the routes of infection, cell tropism, and entry pathways of many of these new viruses remain largely mysterious. Our high-resolution X-ray structures of major capsid proteins VP1 from HPyV6 and from HPyV7 reveal critical differences in surface morphology from those of all other known polyomavirus structures. A groove that engages specific sialic acid-containing glycan receptors in related polyomaviruses is obstructed, and VP1 of HPyV6 and HPyV7 does not interact with sialylated compounds in solution or on cultured human cells. A comprehensive comparison with other structurally characterized polyomavirus VP1 proteins enhances our understanding of molecular determinants that underlie receptor specificity, antigenicity, and, ultimately, pathogenicity within the polyomavirus family and highlight the need for structure-based analysis to better define phylogenetic relationships within the growing polyomavirus family and perhaps also for other viruses.
INTRODUCTION
Polyomaviruses are a group of nonenveloped double-stranded DNA (dsDNA) viruses that were initially identified in mice (1) but have been found since then in birds and in several species of mammals, including humans (reviewed in references 2, 3, and 4). Due to recently developed techniques such as high-throughput sequencing and rolling-circle amplification, a number of new polyomaviruses, including 11 new human polyomaviruses, have been identified during the last decade (5–14). This expansion of the Polyomaviridae family led to its division into three genera, the ortho-, wuki-, and avipolyomaviruses (15). With high sequence homology and conserved overall architecture across genera, the family forms an attractive platform for analyzing determinants of cell entry, cell tropism, and host range as well as other factors that contribute to pathogenesis (4). Asymptomatic and latent infections with polyomaviruses are common in the healthy human population (16–18). Individuals with impaired immune responses due to organ transplantation, monoclonal antibody therapy, hematological diseases, or human immunodeficiency virus (HIV) infection are found to be especially susceptible to reactivation of polyomaviruses, which can lead to severe or fatal diseases (reviewed in references 4 and 19).
Cutaneous human polyomavirus 6 (HPyV6) and HPyV7 (8) are commonly shed from the skin together with the oncogenic Merkel cell polyomavirus (MCPyV) (8, 20). Although no human disease has been linked to HPyV6 and HPyV7 so far, initial studies indicate that persistent infections with both viruses are very common, resulting in seropositivity rates of 35% to 90% by adulthood (8, 17). Thus, an involvement of HPyV6 and/or HPyV7 in cutaneous tumors has to be considered (21–24). While the two viruses have the same tropism as MCPyV, they are more closely related in sequence to the Washington University and Karolinska Institute polyomaviruses (WUPyV and KIPyV, respectively), which were isolated from respiratory tract samples (5, 6). Hence, WUPyV, KIPyV, HPyV6, and HPyV7 have been classified together as wukipolyomaviruses (15).
The polyomavirus major capsid protein VP1 determines antigenicity and mediates attachment to host-cell receptors. It is well established that VP1 possesses a jelly-roll topology and assembles into 72 pentamers, which in turn form a T = 7d icosahedral capsid (25, 26). The VP1 pentamers associate with minor capsid proteins VP2 and VP3, which are located inside the capsid. The presence and roles of VP2 and VP3 seem to differ among polyomavirus species (27). All known structures of polyomavirus VP1 show extended and structurally variable surface loops that emanate from a conserved β-sheet core structure formed by strands B, I, D, and G and strands C, H, E, and F (25, 26, 28–35). These surface loops, named BC-, DE-, HI-, and EF-loops after the β-strands connected by them, are chiefly responsible for viral antigenicity. For each virus, they form a unique virus-host interaction platform that determines host range, cell tropism, viral spread, and pathogenicity. Engagement of sialylated glycan motifs during cell attachment and entry is a common feature of the better-studied orthopolyomaviruses (28–30, 32–36). In contrast, the routes of infection, transmission, cell tropism, receptor specificity, and entry pathways of wukipolyomaviruses remain largely mysterious. To provide an initial framework for investigating the molecular determinants of receptor specificity and tropism of HPyV6 and HPyV7, we determined high-resolution crystal structures of their recombinantly expressed VP1 proteins. While the core structures are highly conserved, the surface loops of HPyV6 and HPyV7 VP1 differ profoundly from those of related polyomavirus VP1 structures. Specific cell surface receptors remain to be unveiled for both viruses, but, interestingly, our findings indicate that sialylated glycans are likely not engaged during early infection steps. In support of the crystallographic analyses, interactions of VP1 with either α2,3- or α2,6-linked sialic acids could not be detected by flow cytometry cell binding studies and saturation transfer difference (STD) nuclear magnetic resonance (NMR) spectroscopy.
MATERIALS AND METHODS
DNA cloning and protein expression.
Following a strategy established for the expression of soluble, assembly-incompetent VP1 pentamers (36), DNAs coding for residues 20 to 291 of HPyV6 VP1 and residues 20 to 288 of HPyV7 VP1 (GenBank accession codes ADE45449 and ADE45474) were cloned into pET15b vectors (Novagen). Soluble pentamers were expressed and purified as described earlier (30, 33). Four nonnative residues (GSHM) are present at the N termini of both proteins after purification, and a nonnative glutamine forms the C terminus of HPyV7 VP1. For cell binding experiments, the JC polyomavirus (JCPyV) VP1 wild type and L54F mutant (residues 22 to 289) and murine polyomavirus VP1 (RA strain; residues 33 to 316) were expressed and purified accordingly (30, 37).
Crystallization, data collection, and structure determination.
HPyV6 VP1 was concentrated to 6 mg/ml in 20 mM HEPES (pH 7.5)–150 mM NaCl–20 mM dithiothreitol (DTT), and crystals were obtained at 20°C using the sitting-drop vapor diffusion technique and a reservoir containing 100 mM Tris (pH 8.5)–200 mM NaSCN (sodium thiocyanate)–13.3% (wt/vol) polyethylene glycol (PEG) 3350. HPyV7 VP1 was crystallized using 7 mg/ml VP1–20 mM HEPES (pH 7.5)–150 mM NaCl and a reservoir containing 100 mM Na malonate (pH 4.5)–15% (wt/vol) PEG 3350 (hanging-drop vapor diffusion technique). Drops were set up with 1 μl protein solution and 1 μl reservoir solution in each case. Crystals were harvested into the respective reservoir solutions supplemented with 30% (vol/vol) glycerol prior to flash-freezing them in liquid nitrogen. Diffraction data were collected at beamlines ID14-4 at ESRF (Grenoble, France) (HPyV6) and X06DA at SLS (Villigen, Switzerland) (HPyV7). Data sets were processed with XDS (38), and structures were solved by molecular replacement with Phaser in CCP4 (39, 40). The WUPyV VP1 core structure (PDB accession no. 3S7X) (31) served as a search model to solve the HPyV7 VP1 structure, and the refined HPyV7 VP1 coordinates were then used to determine the structure of HPyV6 VP1. Rigid-body and simulated annealing refinement was in both cases carried out with Phenix (41), followed by alternating rounds of model building in Coot (42) and restrained refinement, including the translation-libration-screw method (43) and 5-fold noncrystallographic symmetry restraints with Refmac5 (44). Structural superpositions were done using secondary-structure matching (45) and the program Superpose in CCP4 (40). PyMOL (The PyMOL Molecular Graphics System, Version 1.3; Schrödinger, LLC) and the APBS tool plugin (46) were used to create structure figures.
Cell culture.
HeLa S3 and 293TT (47) cells were maintained in a humidified 37°C CO2 chamber in Dulbecco's modified Eagle medium (DMEM) supplemented with 1% penicillin-streptomycin, 6 mM l-glutamine, 1 mM sodium pyruvate, and 10% heat-inactivated fetal bovine serum (FBS). HeLa S3 cells were kindly provided by Katharina Rehn and Dirk Schwarzer (IFIB, Tübingen, Germany) and 293TT cells by Christopher C. Buck (National Cancer Institute, NIH).
Flow cytometry experiments.
VP1 pentamers were labeled with Alexa Fluor 488 C5 maleimide (Invitrogen). Proteins (1 mg/ml) were incubated in 20 mM HEPES (pH 7.5)–150 mM NaCl for 16 to 18 h with 80 mM DTT at 4°C. Excess DTT was removed using two 5-ml HiTrap desalting columns (GE Healthcare), and the dye (10 mM in 20 mM HEPES pH 7.0–150 mM NaCl) was added dropwise by gently mixing it into the protein solution (0.3 to 0.4 mg/ml in 20 mM HEPES pH 7.0–150 mM NaCl) to give a 8-fold molar excess of the dye. The reaction mixture was incubated for 18 h at 4°C. DTT (10 mM) was added, and the excess dye and DTT were removed by desalting (two 5-ml HiTrap desalting columns). The labeling efficiency was determined by UV light/visible light (UV/vis) absorption according to the manufacturer's protocol.
Cells (80% to 90% confluent) were detached nonenzymatically from flasks by incubation with Gibco enzyme-free cell dissociation buffer (Life Technologies) for 30 min at 37°C and were washed twice with phosphate-buffered saline (PBS). A total of 5 × 106 cells were suspended in 100 μl PBS. Cells were mock treated or pretreated with 0.2 U/ml neuraminidase (Clostridium perfringens neuraminidase type V; Sigma-Aldrich) at 37°C for 30 min and then washed 3 times with 500 μl PBS and pelleted after each wash at 200 × g (4 min; 4°C). Cells were incubated in 100 μl of labeled VP1 pentamer solution (50 μg/ml in PBS) on ice for 2 h with agitation every 15 to 20 min. HeLa S3 cells were washed twice in 500 μl PBS and fixed in 500 μl PBS containing 1% formaldehyde for 30 min. 293TT cells were washed twice in PBS and then suspended in 500 μl PBS for the measurement. DAPI (4′,6-diamidino-2-phenylindole) was added to 293TT cells to gate for live cells. Analysis was done using a BD FACSCanto (Becton, Dickinson and Company) flow cytometer equipped with a 488-nm excitation line. A total of 10,000 gated events were measured for each sample. Data were analyzed using FlowJo software (Tree Star, Inc.).
Saturation transfer difference NMR spectroscopy.
STD NMR spectra were recorded at 283 K using 3-mm-inner-diameter Match tubes (200-μl sample volume) and a Bruker AVIII-600 spectrometer equipped with a room temperature probe head and processed with TopSpin 3.0 (Bruker). Samples contained 1 mM α2,3-sialyllactose and α2,6-sialyllactose (Carbosynth) (each) and a 50 μM concentration of either HPyV6 or HPyV7 VP1. Proteins were buffer exchanged prior to NMR experiments in centrifugal concentrators to 20 mM potassium phosphate (pH 7.4)–150 mM NaCl in D2O. Off- and on-resonance irradiation frequencies were set to −80 ppm and 7.0 ppm, respectively. The irradiation power of the selective pulses was 57 Hz, the saturation time was 2 s, and the total relaxation delay was 3 s. A 50-ms continuous-wave spin-lock pulse with a strength of 3.2 kHz was employed to suppress residual protein signals. A total of 512 scans were recorded. A total of 10,000 points were collected, and spectra were multiplied with an exponential window function (line broadening, 1 Hz) prior to Fourier transformation. Spectra were referenced to 298 K using the α-d-Glc anomeric proton as an internal standard (48).
Protein structure accession numbers.
Coordinates and structure factor amplitudes have been deposited under accession numbers 4PCG (HPyV6) and 4PCH (HPyV7) with the RCSB Protein Data Bank (www.rcsb.org).
RESULTS
Overall structures of HPyV6 and HPyV7 VP1.
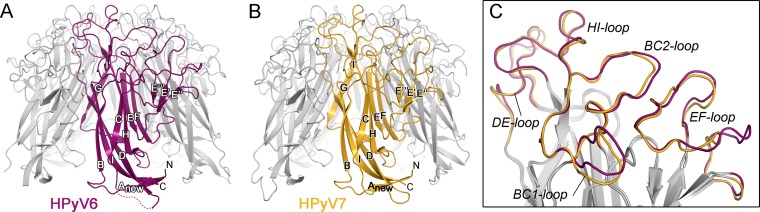
The HPyV6 and HPyV7 VP1 pentamer structures were solved at resolutions of 1.8 and 1.7 Å, respectively, and the refined structures have excellent statistics (Table 1). The final coordinates include residues 22 to 87 and 94 to 290 in the case of HPyV6 and residues 23 to 47, 55 to 85, and 96 to 286 of HPyV7 for all five VP1 chains in the asymmetric units. Each VP1 monomer within the pentamer adopts the iconic jelly-roll fold consisting of two apposed β-sheets (Fig. 1). The EF-loops fold into short three-stranded β-sheets (E′, E″, and E‴) and decorate the side of the pentamer. The I-strand is split into two parts named I and I′. As is typical for polyomavirus VP1 structures, rather poor electron density was observed for the CD-loops at the base of the HPyV6 and HPyV7 pentamers (25, 26, 28–32). This loop is flexible and assumes different conformations even in the context of the intact virion (25, 26, 28). The long BC-loop is divided for clarity into BC1- and BC2-loops that face in different directions (Fig. 1C). The BC1-loops of the HPyV7 VP1 pentamer have elevated mobility, and they have continuous electron density only when contacting the neighboring protomer within the crystal lattice. Thus, the final coordinates contain only three ordered BC1-loops per pentamer. In contrast, the BC1-loops of HPyV6 VP1 have good electron density and share similar conformations that are independent of the presence of crystal contacts. They could therefore be built for all five chains in the asymmetric unit.
TABLE 1.
Data collection and structural refinement statisticsa
| Parameter | HPyV6 VP1 | HPyV7 VP1 |
|---|---|---|
| Space group | C2 | C2 |
| Unit cell dimensions | ||
| a, b, c (Å) | 183.9, 89.4, 125.3 | 209.7, 86.4, 84.2 |
| α, β, γ (°) | 90.0, 131.3, 90.0 | 90.0, 92.1, 90.0 |
| Data collection | ||
| Resolution (Å) | 30.0–1.8 (1.85–1.80) | 30.0–1.70 (1.75–1.70) |
| No. of unique reflections | 140,450 (10,046) | 161,009 (9,962) |
| Redundancy | 4.1 (4.1) | 3.9 (3.9) |
| Rmeas (%) | 7.0 (56.8) | 6.7 (55.9) |
| I/σ | 14.8 (2.8) | 15.7 (2.4) |
| CC1/2 (%) | 99.8 (84.7) | 99.9 (75.4) |
| Completeness (%) | 99.2 (96.4) | 97.8 (82.1) |
| Wilson B-factor (Å2) | 28.9 | 26.0 |
| Refinement | ||
| Rwork/Rfree (%) | 14.8/17.4 | 16.9/19.6 |
| No. of atoms | ||
| Protein | 10,171 | 9,856 |
| Water | 1,070 | 990 |
| Average B-factor (Å2) | ||
| Protein | 28.7 | 24.8 |
| Water | 34.2 | 29.0 |
| RMSDs | ||
| Bond lengths (Å) | 0.009 | 0.010 |
| Bond angles (°) | 1.381 | 1.391 |
| Ramachandran plot (calculated using MolProbity Server) | ||
| Favored (%) | 95.9 | 97.5 |
| Allowed (%) | 4.1 | 2.4 |
| Outliers (%) | 0.0 | 0.1 |
Values for the highest-resolution bin are given in parentheses. CC1/2, correlation between intensities from random half-data sets. , where n is the number of observations of the reflection and <Ihkl,j> the intensity of symmetry (or Friedel)-related observations. , where Fobs and Fcalc are the observed and calculated structure factors, respectively. A total of 5% of the reflections were not used during structure refinement to calculate Rfree.
FIG 1.
Architecture of HPyV6 and HPyV7 VP1 pentamers. (A and B) Overall folds of HPyV6 (A) and HPyV7 (B) VP1 pentamers shown in a ribbon representation, with VP1 monomers highlighted in magenta and gold, respectively. A dashed line represents the missing HPyV6 CD-loop residues, which are defined only by rather poor electron density due to structural flexibility. (C) Closeup view of the surface loop architectures of HPyV6 and HPyV7 VP1 monomers. The loops are colored as described for panels A and B, respectively. VP1 monomer structures were superposed using the secondary-structure-matching (SSM) superposition tool (45) in the program Coot (42).
The Cα atoms of monomeric and pentameric HPyV6 and HPyV7 VP1 structures superpose with very low root mean square deviation (RMSD) values of 0.6 Å and 0.7 Å, respectively, reflecting their high sequence identity of 68% (49). RMSD values for individual residues exceed 1.5 Å only within the EF-loop, where three additional residues elongate the HPyV6 VP1 EF-loop somewhat so that it projects further away from the 5-fold axis (Fig. 1C).
In order to quantify the level of structural diversity of VP1 structures within the members of the Polyomaviridae family, the HPyV6 and HPyV7 VP1 coordinates were superposed with the most closely related WUPyV (PDB accession no. 3S7X) and KIPyV (PDB accession no. 3S7V) VP1 structures as well as the evolutionarily more distant MCPyV (PDB accession no. 4FMG) and simian virus 40 (SV40) (PDB accession no. 3BWQ) VP1 structures (15). Root mean square deviation (RMSD) values for superpositions of VP1 monomers from HPyV6 or HPyV7 onto their KIPyV and WUPyV counterparts are low (∼1.2 Å), in line with the classification of these four viruses into the wukipolyomavirus family. Superpositions of HPyV6 and HPyV7 VP1 onto VP1 monomers from the orthopolyomaviruses MCPyV and SV40 yield slightly higher RMSD values (1.4 to 1.7 Å). However, when entire VP1 pentamers are superposed, the RMSD differences all lie in a range from 1.3 Å (HPyV6-KIPyV) to 1.7 Å (HPyV6-SV40), demonstrating that the pentameric VP1 arrangements are similar across the wuki- and orthopolyomaviruses.
Organization of surface loops.
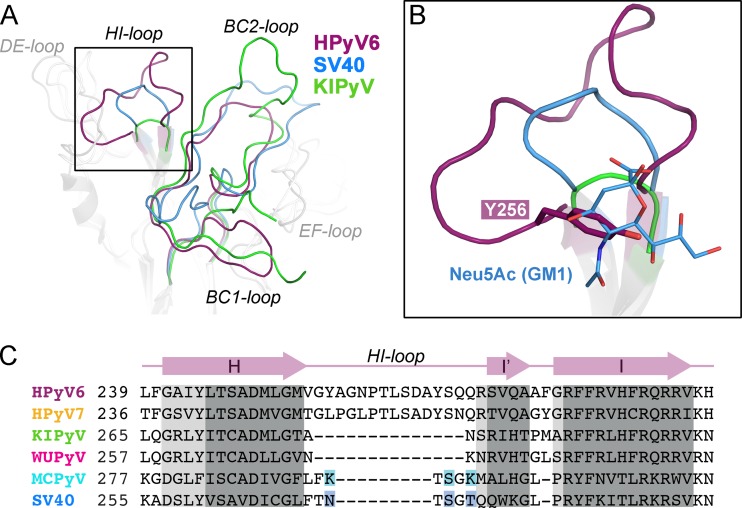
The top surface of the VP1 pentamer, which corresponds to the accessible surface of the virus, is almost entirely defined by the BC-, DE-, and HI-loops, and these loops endow each polyomavirus with a unique platform for specific interactions with individual receptors (Fig. 1C). Typically, this platform binds glycan receptors that terminate in sialic acid (Neu5Ac), but sequences and linkages of the recognized oligosaccharides differ among polyomaviruses, leading to specific interactions with a small subset of sialylated glycans in each case (28–30, 32–36). However, despite these differences, the location of the sialic acid binding site is conserved among the polyomaviruses for which VP1 structures have been available to date. The sialic acid binding site is typically located in a recessed area at the junction of the BC1-, BC2-, DE-, and HI-loops of a VP1 monomer, and additional contacts are contributed by the BC2- and DE-loops of counterclockwise (ccw) and clockwise (cw) neighboring monomers, respectively (28–30, 32–35). In order to assess the ability of the HPyV6 and HPyV7 surface loops to form such a sialic acid binding site, we compared the conformations and lengths of their surface-exposed loops with the equivalent loops in WUPyV and KIPyV, as well as in MCPyV and SV40. For clarity, only comparisons with the closely related KIPyV and the sialic acid-engaging SV40 polyomavirus are shown in Fig. 2. As the HPyV6 and HPyV7 loop structures are very similar (Fig. 1C), only HPyV6 VP1 is discussed and shown. The HI-loop of polyomavirus VP1 is only a short hairpin in all structures crystallized to date, and it typically forms a wall that closes the glycan-receptor binding site at one end. Strikingly, this loop is extended by 14 residues in HPyV6 compared to KIPyV and WUPyV and by 9 residues compared to SV40 and MCPyV (Fig. 2). Rather than projecting outward, the HPyV6 HI-loop folds on top of the pentamer and forms extensive contacts within itself and with the DE-loop (not shown in detail). To accommodate the extended HI-loop, the DE-loop of the ccw monomer is displaced toward the 5-fold symmetry axis. The BC2-loop of HPyV6 is severely truncated in comparison to other VP1 structures and lies flat on the VP1 surface (Fig. 2B). Taking these data together, the elongated HI-loops and the truncated BC2-loops of HPyV6 (and also HPyV7) lead to a profoundly altered surface loop network.
FIG 2.
Architecture of VP1 surface loops. Superposition and comparison of VP1 surface loop structures are shown for wukipolyomaviruses HPyV6 and KIPyV (PDB accession no. 3S7V) and orthopolyomavirus SV40 (PDB accession no. 3BWR). VP1 monomers were superposed using the secondary-structure-matching (SSM) superposition tool (45). (A) HI-, BC1-, and BC2-loops are highlighted in color in the overview. (B) Closeup view of the HI-loop receptor binding pocket. The sialic acid moiety (Neu5Ac) of the GM1 glycan in the SV40 VP1-GM1 pentasaccharide complex structure (PDB accession no. 3BWR) and the Y256 side chain of HPyV6 VP1 are shown in stick representation. The same view is taken for panels A and B. (C) Sequence alignment of the HI-loop region. Key residues interacting with the sialic acid moiety in the SV40-GM1 glycan and MCPyV VP1–α2,3-sialyllactosamine complex structures (PDB accession no. 4FMI) are highlighted in blue and cyan, respectively. Regions in which all VP1 structures align with root mean square deviation (RMSD) values of <1.0 Å (dark gray) and >1.5 Å (light gray) between Cα atoms are shaded.
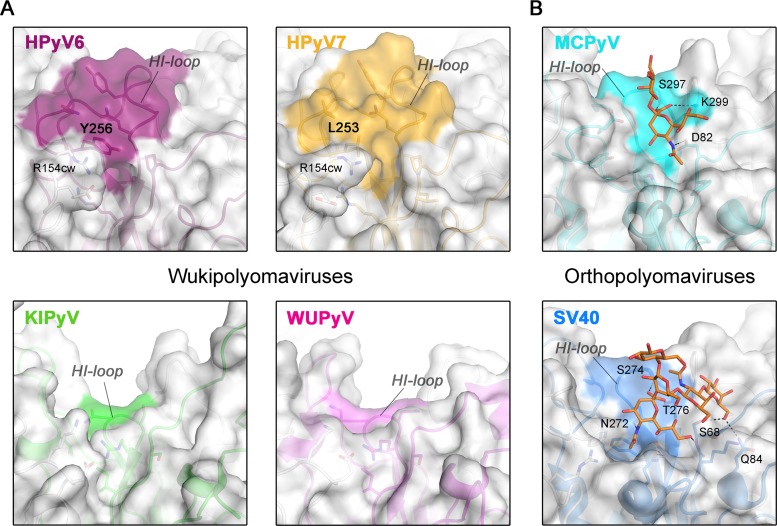
The HI-loop participates in the recognition of sialylated glycan receptors in all sialic-acid binding polyomaviruses whose structures have been determined to date (28–30, 32–35), by contributing parts of the shallow receptor binding groove on the protein surface and providing direct or water-mediated contacts with glycan components. The extended HI-loop in HPyV6 and HPyV7 produces a prominent, elevated ridge on the virion surface that partially covers the glycan binding site groove present in orthopolyomaviruses (Fig. 3). Thus, the conformation of the HI-loop appears to hinder the binding of sialic acids to the HPyV6 and HPyV7 capsids. In particular, HI-loop residues Y256 (HPyV6) and L253 (HPyV7) (Fig. 2B and 3A) project into the groove and would collide with a potential sialic acid ligand. The R154 side chains in the cw HPyV6 and HPyV7 EF-loops also help to close the binding site (Fig. 3A). The uniquely elongated HI-loops of HPyV6 and HPyV7 are not conserved in either KIPyV or WUPyV (Fig. 3A). In fact, these two viruses have unusually short HI-loops (see also Fig. 2C), and surface analysis shows that they possess a deep groove leading toward the central pore (Fig. 3A). Thus, HPyV6 and HPyV7 feature a remodeled surface structure compared to all other known VP1 structures, with likely consequences for receptor binding. The HPyV6 and HPyV7 genomes sequenced so far have revealed naturally occurring VP1 amino acid variations (8), which are mostly buried in the assembled virus (not shown) and thus are likely not critical for glycan receptor engagement. The only exceptions are HPyV7 residues 63 (threonine or proline), 153 (asparagine or aspartate), and 167 (serine or threonine), which are distributed across the BC2- and EF-loops and, therefore, could theoretically account for modulated receptor interactions.
FIG 3.
Surface structures of VP1 pentamers. Closeup views of VP1 pentamer top-surface regions that are involved in sialic acid engagement in the case of MCPyV and SV40 are shown. Equivalent surface sections are shown in surface and cartoon representations for VP1 pentamers from HPyV6, HPyV7, and KIPyV (PDB accession no. 3S7V) and WUPyV (PDB accession no. 3S7X) (A) and from SV40 (PDB accession no. 3BWR) and MCPyV (PDB accession no. 4FMI) (B). HI-loop residues are highlighted on the surface representations according to the colors assigned to the respective viruses. Carbohydrates (Neu5Ac and Gal of α2,3-sialyllactosamine and GM1 pentasaccharide) in panel B are shown in stick representations (colored by atom type; carbons in orange, oxygen in red, and nitrogen in blue), and glycan-protein contacts (hydrogen bonding and salt bridges) are shown as dashed lines for MCPyV and SV40 VP1.
HPyV6 and HPyV7 VP1 do not bind sialic acids.
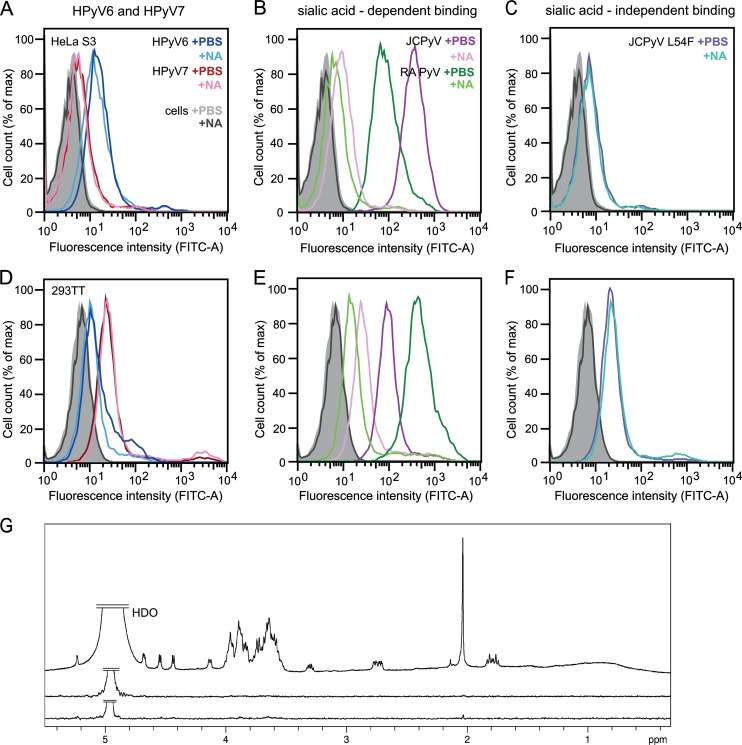
Unassembled recombinant VP1 pentamers are useful tools to study cell attachment and entry and cellular trafficking of polyomaviruses (30, 50). To investigate whether HPyV6 and HPyV7 engage sialic acids on cell surfaces during early steps of infection, we analyzed binding of their VP1 pentamers to two cultured human cell lines, HeLa S3 and 293TT, by flow cytometry (Fig. 4). Prior to single-cell binding experiments with Alexa Fluor 488-conjugated VP1 pentamers, cells were mock treated or incubated with Clostridium perfringens neuraminidase type V to remove terminal α2,3-, α2,6-, and α2,8-linked sialic acids from the cell surface. JCPyV and murine polyomavirus (RA strain) VP1 pentamers bind to both cell lines in a neuraminidase-sensitive manner (30, 51) (Fig. 4B and E), in line with their known use of sialylated receptors for attachment. In contrast, HPyV6 and HPyV7 VP1 attachment to both cell lines is not affected by enzymatic removal of sialic acids (Fig. 4A and D). The measured fluorescence signals for HPyV6 and HPyV7 VP1 pentamers are also similar to those seen with the control representing neuraminidase-insensitive binding, the JCPyV VP1 L54F mutant (Fig. 4C and F). This mutant has a disrupted VP1 sialic acid binding site and no longer engages sialylated receptors (37).
FIG 4.
HPyV6 and HPyV7 VP1 do not engage sialic acids. (A to F) Cell binding analysis. (G) Saturation transfer difference (STD) NMR spectroscopy of HPyV6 and HPyV7 VP1 pentamers with α2,3- and α2,8-sialyllactose. (A to F) HeLa S3 (A to C) and 293TT (D to F) cells were subjected to mock treatment (PBS) or were pretreated with 0.2 U/ml Clostridium perfringens neuraminidase (NA), washed, and then incubated with Alexa Fluor 488-conjugated VP1 pentamers. VP1 pentamer binding was then analyzed by flow cytometry. Histograms represent the fluorescence intensity of Alexa Fluor 488 for 10,000 gated events in each case. Data for cells alone are colored gray and black for mock- and NA-treated cells, respectively. Three independent experiments were performed, and results of a typical experiment are presented. (B and E) JCPyV and murine polyomavirus (RA strain) VP1 pentamers are included as positive controls for neuraminidase-sensitive attachment (30, 51). (C and F) JCPyV L54F is a VP1 mutant with an abolished sialic acid binding site (37) and was used to test for sialic acid-independent cell binding. FITC, fluorescein isothiocyanate; max, maximum. (G) From top to bottom: 1H reference spectrum of 50 μM HPyV7 VP1 with 1 mM α2,3- and α2,6-sialyllactose each; STD NMR difference spectrum recorded with the same sample; STD NMR difference spectrum of 50 μM HPyV6 VP1 with 1 mM (each) α2,3- and α2,8-sialyllactose. No significant saturation transfer to either capsid protein was observed. HDO peaks were truncated for clarity.
To probe for interactions in solution and to identify ligand atoms in contact with VP1, we analyzed binding of the HPyV6 and HPyV7 VP1 pentamers to the sialylated model compounds α2,3- and α2,6-linked sialyllactose by STD NMR spectroscopy (52). STD NMR spectroscopy has been successfully used to define, for example, the interactions of MCPyV and BKPyV VP1 with specific glycan receptor motifs (32, 33). No significant magnetization transfer was observed with either type of sialyllactose from HPyV6 or HPyV7 VP1, suggesting that neither protein interacts with simple α2,3- or α2,6-linked sialylated oligosaccharides (Fig. 4G). These findings are consistent with the crystal structure analysis, and they are also in agreement with experiments employing crystal soaking and glycan array screening, neither of which revealed interactions with tested sialylated compounds (data not shown).
Unique features of the virion surface.
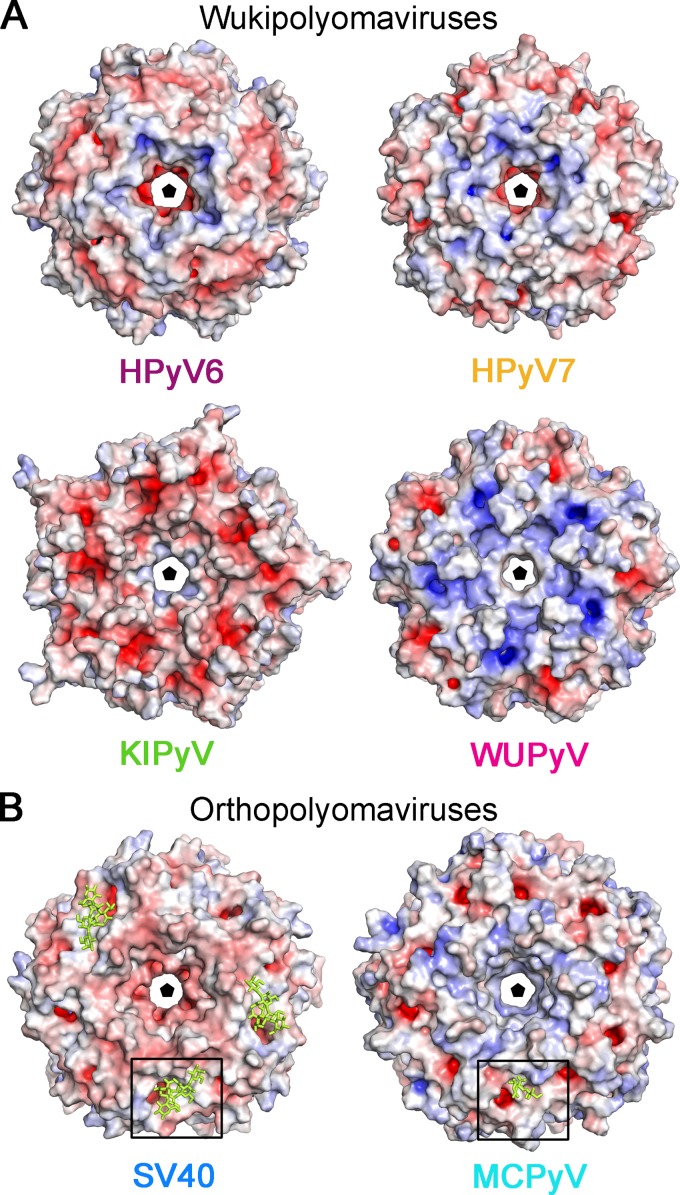
In order to obtain some initial clues about the molecular determinants of the receptor specificities and antigenic properties of HPyV6 and HPyV7 VP1, we examined their electrostatic surface potentials and compared these with the electrostatic surface potentials of other polyomavirus VP1 pentamers (Fig. 5). Consistent with their structural conservation, the electrostatic potentials for HPyV6 and HPyV7 VP1 are very similar (Fig. 5A). Both pentamers possess negatively charged patches within the central pore, which are surrounded by concentric positively and negatively charged rings around the 5-fold axis. The similarities in the distribution of surface charges suggest that the two viruses pursue similar strategies for cell surface attachment and perhaps also for viral entry.
FIG 5.
Electrostatic surface potentials of VP1 pentamer from the wuki- and orthopolyomavirus genera. Overall surface representations of HPyV6, HPyV7, and KIPyV (PDB accession no. 3S7V) and WUPyV (PDB accession no. 3S7X) (A) and SV40 VP1 (PDB accession no. 3BWR) and MCPyV (PDB accession no. 4FMI) (B) pentamers are colored according to electrostatic potential (calculated using APBS tool 2.1; 46), with blue and red corresponding to +7 kT and −7 kT, respectively. Views are equivalent in panels A and B and are shown from the top—the outer surface of the virion—along the 5-fold axis of the pentamer. Carbohydrates (GM1 pentasaccharide and Neu5Ac and Gal of α2,3-sialyllactosamine) in panel B are shown in yellow stick representations, and the glycan binding site is highlighted for clarity with a box.
The electrostatic surface potentials of the remaining wukipolyomavirus family members, WUPyV and KIPyV, differ and are also distinct from those of HPyV6 and HPyV7 (Fig. 5A). Whereas the WUPyV VP1 pentamer surface is mostly electropositive, the corresponding KIPyV residues have a more negative potential. Interestingly, the surface potentials of HPyV6 and HPyV7 VP1 are more similar to that of MCPyV (compare Fig. 5A and B) and feature a negatively charged area, a central pore around the 5-fold axis, surrounded by positively and negatively charged patches (Fig. 5A and B).
DISCUSSION
We have determined the high-resolution structures of VP1 pentamers of the recently identified human polyomaviruses HPyV6 and HPyV7. Our structure analysis reveals essential features and critical differences in surface morphology that are likely important for antigenicity and receptor engagement of these two skin viruses. The region that accommodates sialic acid receptors in other polyomaviruses is obstructed, and consistent with this, cell binding analysis and STD NMR spectroscopy (Fig. 4) as well as experiments employing crystal soaking and glycan array screening (data not shown) did not yield indications of binding of sialic acid by HPyV6 or by HPyV7.
A new sialic acid binding site different in its overall location on the VP1 pentamer from the polyomavirus sialic acid binding sites structurally characterized so far (28–30, 32–35) is possible but, in the light of our experiments, rather unlikely. The findings of our structure-function analysis suggest instead that HPyV6 and HPyV7 do not engage sialylated glycans on cell surfaces during attachment and entry and likely recognize a different receptor type. It is still possible, however, that HPyV6 and HPyV7 bind sialic acids in a different location that is present only in the fully assembled virus, such as in canyons between adjacent pentamers, and not in the free VP1 pentamer.
The analysis of electrostatic surface potentials has in some cases helped to identify potential receptor binding regions in viral proteins, for example, in sialic acid binding adeno- and rotaviruses (53, 54). We note that it is important to keep in mind that sialic acid binding sites need not always display a positive electrostatic potential, as hydrogen bonds rather than salt bridges mediate contacts with the sialic acid carboxylate in at least some polyomaviruses (55) (see also Fig. 5B). As HPyV6, HPyV7, and MCPyV are all shed from skin (8), this similarity might indicate a conserved strategy for receptor engagement. MCPyV is unique among the members of the polyomavirus family because of its sequential engagement of negatively charged glycosaminoglycan (GAG) and sialylated receptors (56). GAG binding sites on proteins are typically lysine/arginine-rich (57, 58); however, the binding site for GAGs on MCPyV VP1 is not known. The electrostatic surface potentials of HPyV6 and HPyV7 VP1 are indeed similar to that of MCPyV, with potential consequences for receptor specificity, including engagement of GAGs, but the typical shallow depression that harbors sialic acid binding sites on MCPyV (32) and other polyomavirus VP1s is clearly not evident in our HPyV6 and HPyV7 VP1 structures. Thus, only a few conclusions concerning specific receptor engagement for HPyV6 and HPyV7 are possible until their receptor class has been identified.
Our analysis of the VP1 pentamer surface properties, such as electrostatic distributions and loop morphology, suggests that a structural-biology approach can improve the phylogenetic polyomavirus classification that is based entirely on sequence similarity (15). In terms of surface loop length and structure, HPyV6 and HPyV7 differ significantly from the other structurally characterized members of the wukipolyomavirus genus, WUPyV and KIPyV. While the latter two viruses possess strongly charged VP1 pentamers, the electrostatic surfaces of the former two resemble that of the orthopolyomavirus MCPyV more closely. Our analysis thus demonstrates the need for structure-based comparisons to inform understanding of potential receptor binding sites and conserved regions of antigenicity, two characteristics that are largely determined by the electrostatics and loop structure of VP1. It has been shown that, in some cases, structural analysis even allows the identification of evolutionary relationships that are not revealed by sequence analysis (59–61).
Our comprehensive analysis and comparison of VP1 pentamers from different polyomavirus genera lead us to propose that the observed structural diversity explains differences and similarities in the tissue tropism of wukipolyomaviruses compared with other polyomaviruses. Cellular and tissue distributions of receptors and coreceptors are important determinants of viral tropism. The establishment of experimental models and cell culture systems to propagate HPyV6 and HPyV7 as well as other newly identified polyomaviruses is clearly required. Such models should enable functional studies of receptor engagement and cell entry mechanisms of these viruses that are widely circulating within the human population.
ACKNOWLEDGMENTS
We thank members of the Stehle group for critical discussions and the staff at beamlines X06DA of the Swiss Light Source (Villigen, Switzerland) and ID14-4 at ESRF (Grenoble, France) for beam time and assistance with data collection. We acknowledge Remco Sprangers (Max Planck Institute for Developmental Biology, Tübingen, Germany) for assistance in recording the NMR data and the staff of the Flow Cytometry Core Facility, University of Tübingen, Tübingen, Germany, for technical support.
This work was supported by contract research “Glykobiologie/Glykomik” of the Baden-Württemberg Stiftung (T.S.). R.L.G. was supported by NIH grant CA37667.
Footnotes
Published ahead of print 9 July 2014
REFERENCES
- 1.Gross L. 1953. Neck tumors, or leukemia, developing in adult C3H mice following inoculation, in early infancy, with filtered (Berkefeld N), or centrifugated (144,000 X g), Ak-leukemic extracts. Cancer 6:948–958. [DOI] [PubMed] [Google Scholar]
- 2.DeCaprio JA, Garcea RL. 2013. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 11:264–276. 10.1038/nrmicro2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feltkamp MC, Kazem S, van der Meijden E, Lauber C, Gorbalenya AE. 2013. From Stockholm to Malawi: recent developments in studying human polyomaviruses. J. Gen. Virol. 94:482–496. 10.1099/vir.0.048462-0 [DOI] [PubMed] [Google Scholar]
- 4.Dalianis T, Hirsch HH. 2013. Human polyomaviruses in disease and cancer. Virology 437:63–72. 10.1016/j.virol.2012.12.015 [DOI] [PubMed] [Google Scholar]
- 5.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MAA, Dalianis T, Ramqvist T, Andersson B. 2007. Identification of a third human polyomavirus. J. Virol. 81:4130–4136. 10.1128/JVI.00028-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3:e64. 10.1371/journal.ppat.0030064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. 10.1126/science.1152586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515. 10.1016/j.chom.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 6:e1001024. 10.1371/journal.ppat.1001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, Hengel H, Ehlers B. 2011. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J. Virol. 85:4586–4590. 10.1128/JVI.02602-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2012. Identification of MW polyomavirus, a novel polyomavirus in human stool. J. Virol. 86:10321–10326. 10.1128/JVI.01210-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2013. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 436:295–303. 10.1016/j.virol.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B. 2013. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS One 8:e58021. 10.1371/journal.pone.0058021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra N, Pereira M, Rhodes RH, An P, Pipas JM, Jain K, Kapoor A, Briese T, Faust PL, Lipkin WI. 1 May 2014. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J. Infect. Dis. 10.1093/infdis/jiu250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johne R, Buck CB, Allander T, Atwood WJ, Garcea RL, Imperiale MJ, Major EO, Ramqvist T, Norkin LC. 2011. Taxonomical developments in the family Polyomaviridae. Arch. Virol. 156:1627–1634. 10.1007/s00705-011-1008-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kean JM, Rao S, Wang M, Garcea RL. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog. 5:e1000363. 10.1371/journal.ppat.1000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Meijden E, Bialasiewicz S, Rockett RJ, Tozer SJ, Sloots TP, Feltkamp MC. 2013. Different serologic behavior of MCPyV, TSPyV, HPyV6, HPyV7 and HPyV9 polyomaviruses found on the skin. PLoS One 8:e81078. 10.1371/journal.pone.0081078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moens U, Van Ghelue M, Song XB, Ehlers B. 2013. Serological cross-reactivity between human polyomaviruses. Rev. Med. Virol. 23:250–264. 10.1002/rmv.1747 [DOI] [PubMed] [Google Scholar]
- 19.De Gascun CF, Carr MJ. 2013. Human polyomavirus reactivation: disease pathogenesis and treatment approaches. Clin. Dev. Immunol. 2013:373579. 10.1155/2013/373579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wieland U, Silling S, Hellmich M, Potthoff A, Pfister H, Kreuter A. 13 January 2014. Human polyomaviruses 6, 7, 9, 10 and Trichodysplasia spinulosa-associated polyomavirus in HIV-infected men. J. Gen. Virol. 10.1099/vir.0.061259-0 [DOI] [PubMed] [Google Scholar]
- 21.Schrama D, Buck CB, Houben R, Becker JC. 2012. No evidence for association of HPyV6 or HPyV7 with different skin cancers. J. Invest. Dermatol. 132:239–241. 10.1038/jid.2011.261 [DOI] [PubMed] [Google Scholar]
- 22.Duncavage EJ, Pfeifer JD. 2011. Human polyomaviruses 6 and 7 are not detectable in Merkel cell polyomavirus-negative Merkel cell carcinoma. J. Cutan. Pathol. 38:790–796. 10.1111/j.1600-0560.2011.01765.x [DOI] [PubMed] [Google Scholar]
- 23.Scola N, Wieland U, Silling S, Altmeyer P, Stucker M, Kreuter A. 2012. Prevalence of human polyomaviruses in common and rare types of non-Merkel cell carcinoma skin cancer. Br. J. Dermatol. 167:1315–1320. 10.1111/j.1365-2133.2012.11141.x [DOI] [PubMed] [Google Scholar]
- 24.Kreuter A, Silling S, Dewan M, Stucker M, Wieland U. 2011. Evaluation of 4 recently discovered human polyomaviruses in primary cutaneous B-cell and T-cell lymphoma. Arch. Dermatol. 147:1449–1451. 10.1001/archdermatol.2011.330 [DOI] [PubMed] [Google Scholar]
- 25.Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354:278–284. 10.1038/354278a0 [DOI] [PubMed] [Google Scholar]
- 26.Stehle T, Gamblin SJ, Yan Y, Harrison SC. 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure 4:165–182. 10.1016/S0969-2126(96)00020-2 [DOI] [PubMed] [Google Scholar]
- 27.Schowalter RM, Buck CB. 2013. The Merkel cell polyomavirus minor capsid protein. PLoS Pathog. 9:e1003558. 10.1371/journal.ppat.1003558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stehle T, Yan Y, Benjamin TL, Harrison SC. 1994. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369:160–163. 10.1038/369160a0 [DOI] [PubMed] [Google Scholar]
- 29.Neu U, Woellner K, Gauglitz G, Stehle T. 2008. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc. Natl. Acad. Sci. U. S. A. 105:5219–5224. 10.1073/pnas.0710301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neu U, Maginnis MS, Palma AS, Stroh LJ, Nelson CD, Feizi T, Atwood WJ, Stehle T. 2010. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe 8:309–319. 10.1016/j.chom.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neu U, Wang J, Macejak D, Garcea RL, Stehle T. 2011. Structures of the major capsid proteins of the human Karolinska Institutet and Washington University polyomaviruses. J. Virol. 85:7384–7392. 10.1128/JVI.00382-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neu U, Hengel H, Blaum BS, Schowalter RM, Macejak D, Gilbert M, Wakarchuk WW, Imamura A, Ando H, Kiso M, Arnberg N, Garcea RL, Peters T, Buck CB, Stehle T. 2012. Structures of Merkel cell polyomavirus VP1 complexes define a sialic acid binding site required for infection. PLoS Pathog. 8:e1002738. 10.1371/journal.ppat.1002738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neu U, Allen SA, Blaum BS, Liu Y, Frank M, Palma AS, Stroh LJ, Feizi T, Peters T, Atwood WJ, Stehle T. 2013. A structure-guided mutation in the major capsid protein retargets BK polyomavirus. PLoS Pathog. 9:e1003688. 10.1371/journal.ppat.1003688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neu U, Khan ZM, Schuch B, Palma AS, Liu Y, Pawlita M, Feizi T, Stehle T. 2013. Structures of B-lymphotropic polyomavirus VP1 in complex with oligosaccharide ligands. PLoS Pathog. 9:e1003714. 10.1371/journal.ppat.1003714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan ZM, Liu Y, Neu U, Gilbert M, Ehlers B, Feizi T, Stehle T. 19 March 2014. Crystallographic and glycan microarray analysis of human polyomavirus 9 VP1 identifies N-glycolyl neuraminic acid as a receptor candidate. J. Virol. 10.1128/JVI.03455-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stehle T, Harrison SC. 1997. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 16:5139–5148. 10.1093/emboj/16.16.5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maginnis MS, Stroh LJ, Gee GV, O'Hara BA, Derdowski A, Stehle T, Atwood WJ. 2013. Progressive multifocal leukoencephalopathy-associated mutations in the JC polyomavirus capsid disrupt lactoseries tetrasaccharide c binding. mBio 4:e00247-13. 10.1128/mBio.00247-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afonine PV, Grosse-Kunstleve RW, Adams PD. 2005. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr. D Biol. Crystallogr. 61:850–855. 10.1107/S0907444905007894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Painter J, Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450. 10.1107/S0907444906005270 [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 45.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256–2268. 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- 46.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2004. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 78:751–757. 10.1128/JVI.78.2.751-757.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC. 1995. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal. Chem. 67:793–811. 10.1021/ac00101a004 [DOI] [PubMed] [Google Scholar]
- 49.Goujon M, McWilliam H, Li WZ, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–W699. 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson CD, Derdowski A, Maginnis MS, O'Hara BA, Atwood WJ. 2012. The VP1 subunit of JC polyomavirus recapitulates early events in viral trafficking and is a novel tool to study polyomavirus entry. Virology 428:30–40. 10.1016/j.virol.2012.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bauer PH, Cui C, Liu WR, Stehle T, Harrison SC, DeCaprio JA, Benjamin TL. 1999. Discrimination between sialic acid-containing receptors and pseudoreceptors regulates polyomavirus spread in the mouse. J. Virol. 73:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayer M, Meyer B. 1999. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem. Int. Ed. 38:1784–1788. [DOI] [PubMed] [Google Scholar]
- 53.Dormitzer PR, Sun ZY, Wagner G, Harrison SC. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885–897. 10.1093/emboj/21.5.885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burmeister WP, Guilligay D, Cusack S, Wadell G, Arnberg N. 2004. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J. Virol. 78:7727–7736. 10.1128/JVI.78.14.7727-7736.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neu U, Bauer J, Stehle T. 2011. Viruses and sialic acids: rules of engagement. Curr. Opin. Struct. Biol. 21:610–618. 10.1016/j.sbi.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schowalter RM, Pastrana DV, Buck CB. 2011. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 7:e1002161. 10.1371/journal.ppat.1002161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Q, Lerch TF, Meyer NL, Chapman MS. 2011. Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6). Virology 420:10–19. 10.1016/j.virol.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kern A, Schmidt K, Leder C, Muller OJ, Wobus CE, Bettinger K, Von der Lieth CW, King JA, Kleinschmidt JA. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072–11081. 10.1128/JVI.77.20.11072-11081.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bahar MW, Graham SC, Stuart DI, Grimes JM. 2011. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure 19:1011–1020. 10.1016/j.str.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ravantti J, Bamford D, Stuart DI. 2013. Automatic comparison and classification of protein structures. J. Struct. Biol. 183:47–56. 10.1016/j.jsb.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 61.Rissanen I, Grimes JM, Pawlowski A, Mantynen S, Harlos K, Bamford JK, Stuart DI. 2013. Bacteriophage P23–77 capsid protein structures reveal the archetype of an ancient branch from a major virus lineage. Structure 21:718–726. 10.1016/j.str.2013.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]