ABSTRACT
Mammalian genomes are replete with retrotransposable elements, including endogenous retroviruses. DNA methyltransferase 3-like (DNMT3L) is an epigenetic regulator expressed in prospermatogonia, growing oocytes, and embryonic stem (ES) cells. Here, we demonstrate that DNMT3L enhances the interaction of repressive epigenetic modifiers, including histone deacetylase 1 (HDAC1), SET domain, bifurcated 1 (SETDB1), DNA methyltransferase 3A (DNMT3A), and tripartite motif-containing protein 28 (TRIM28; also known as TIF1β and KAP1) in ES cells and orchestrates retroviral silencing activity with TRIM28 through mechanisms including, but not limited to, de novo DNA methylation. Ectopic expression of DNMT3L in somatic cells causes methylation-independent retroviral silencing activity by recruitment of the TRIM28/HDAC1/SETDB1/DNMT3A/DNMT3L complex to newly integrated Moloney murine leukemia virus (Mo-MuLV) proviral DNA. Concurrent with this recruitment, we also observed the accumulation of histone H3 lysine 9 trimethylation (H3K9me3) and heterochromatin protein 1 gamma (HP1γ), as well as reduced H3K9 and H3K27 acetylation at Mo-MuLV proviral sequences. Ectopic expression of DNMT3L in late-passage mouse embryonic fibroblasts (MEFs) recruited cytoplasmically localized HDAC1 to the nucleus. The formation of this epigenetic modifying complex requires interaction of DNMT3L with DNMT3A as well as with histone H3. In fetal testes at embryonic day 17.5, endogenous DNMT3L also enhanced the binding among TRIM28, DNMT3A, SETDB1, and HDAC1. We propose that DNMT3L may be involved in initiating a cascade of repressive epigenetic modifications by assisting in the preparation of a chromatin context that further attracts DNMT3A-DNMT3L binding and installs longer-term DNA methylation marks at newly integrated retroviruses.
IMPORTANCE Almost half of the mammalian genome is composed of endogenous retroviruses and other retrotransposable elements that threaten genomic integrity. These elements are usually subject to epigenetic silencing. We discovered that two epigenetic regulators that lack enzymatic activity, DNA methyltransferase 3-like (DNMT3L) and tripartite motif-containing protein 28 (TRIM28), collaborate with each other to impose retroviral silencing. In addition to modulating de novo DNA methylation, we found that by interacting with TRIM28, DNMT3L can attract various enzymes to form a DNMT3L-induced repressive complex to remove active marks and add repressive marks to histone proteins. Collectively, these results reveal a novel and pivotal function of DNMT3L in shaping the chromatin modifications necessary for retroviral and retrotransposon silencing.
INTRODUCTION
DNA methylation is important for the long-term silencing of certain endogenous retroviruses (ERVs) and retrotransposons, including intracisternal A particles (IAPs) (1–3). DNA methyltransferase 3-like (DNMT3L), which is highly expressed in germ and embryonic stem (ES) cells, is a known stimulator of de novo DNA methylation through its interaction with DNA methyltransferase 3A (DNMT3A) and DNA methyltransferase 3B (DNMT3B). DNMT3L binds to the N terminus of histone H3, which is unmethylated at lysine 4, to recruit the DNMT3L/DNMT3A/DNMT3A/DNMT3L complex (4, 5). DNMT3A and DNMT3B are required for methylation of endogenous repetitive elements (6) and to establish methylation marks on newly integrated Moloney murine leukemia virus (Mo-MuLV) proviral DNA in ES cells (7). DNMT3L is an important epigenetic regulator during germ cell development, with essential roles in the establishment of DNA methylation imprints in growing oocytes and retrotransposon silencing in prospermatogonia (8–10).
Germ line mutations in tripartite motif-containing protein 28 (TRIM28; also known as KAP1 or TIF1β) result in ERV activation and early embryonic lethality (11, 12). Apart from DNA methylation (13), ERVs are also silenced by enzymes that mediate posttranslational modifications of histones (14, 15). In ES cells, endogenous retrotransposon silencing requires epigenetic modifications, including the methylation of histone H3K9 and the demethylation of H3K4 (12, 14, 16–18). TRIM28 is a scaffold for epigenetic modifiers such as the histone methyltransferase SET domain, bifurcated 1 (SETDB1; also known as ESET) (19), heterochromatin protein 1 (HP1) (20), and the NuRD histone deacetylase (HDAC) complex (21). All of these are involved in transcriptional repression. TRIM28 is also essential for silencing endogenous retroviruses and euchromatic genes via the repressive histone modifier SETDB1 and HP1-mediated silencing machinery (12, 22–24).
Mouse embryonic cells repress the expression of endogenous retroelements as well as exogenous retroviruses. When responding to newly integrated Mo-MuLV proviral sequences in ES cells, TRIM28 and its interacting epigenetic modifiers target the primer binding site (PBS) sequence via zinc finger protein 809 (ZFP809) (25, 26). The Mo-MuLV PBS sequence is complementary to 18 nucleotides at the 3′ end of the host proline tRNA and is used as the primer for reverse transcription DNA synthesis (27). The PBS sequence is one of the retroviral repression targets (28–30). However, PBS-mediated repression can be abolished by a single G-to-A base pair mutation within the PBS sequence (the B2/PBSQ mutation) (28). Because TRIM28 interacts with various repressive modifiers, it is likely that TRIM28 facilitates repressive chromatin modifications by promoting the binding of its interacting proteins (including SETDB1, HDAC1, and HP1) and ZFP809 to the PBS sequence encoded within the 5′ end of the newly integrated Mo-MuLV proviral sequences.
It has been recently shown that DNMT3L and TRIM28 are required for de novo DNA methylation of proviral sequences in ES cells (23, 31). However, whether DNMT3L can interact with TRIM28 (or TRIM28-interacting epigenetic modifiers) and silence newly integrated proviral sequences by mechanisms other than de novo DNA methylation has not been clarified. To gain further insight into the various levels of epigenetic retroviral silencing activities mediated by DNMT3L and TRIM28, we performed functional analyses of the role of DNMT3L in different cell types with or without endogenous DNMT3L expression in silencing PBSpro-restricted or PBS-independent Mo-MuLV viruses as summarized in Table S1 in the supplemental material. Using a loss-of-function assay, we found that ES cells deficient for DNMT3L exhibit decreased retroviral silencing activity and reduced de novo methylation of newly integrated proviral sequences. DNMT3L and TRIM28 have synergistic effects on proviral silencing in ES cells, which can be partly explained by our immunoprecipitation results demonstrating that DNMT3L facilitated an interaction between TRIM28 and DNMT3A, SETDB1, and HDAC1 in ES cells. To further demonstrate the potential ability of DNMT3L in facilitating the binding between TRIM28 and various repressive epigenetic modifiers, we performed a gain-of-function experiment by ectopically expressing wild-type and mutant DNMT3L in somatic cells that normally lack expression of this protein. We confirmed the accumulation of a DNMT3L-dependent DNMT3A/TRIM28/HDAC1/SETDB1 complex on Mo-MuLV proviral sequences and the ability of this complex to install repressive chromatin modifications without triggering de novo DNA methylation in DNMT3L-overexpressing somatic cells. We discuss potential explanations for the differences between endogenous DNMT3L-mediated enhancement of retroviral silencing efficiency via both repressive histone modifications and de novo DNA methylation in ES cells and ectopic DNMT3L-induced repressive chromatin modification-mediated moderate retroviral silencing activity that lacks de novo DNA methylation in somatic cells.
MATERIALS AND METHODS
Ethics statement.
All animals used in this study were maintained in the Animal Center at the Institute of Biotechnology, National Taiwan University. Mice were kept under standard conditions. The animal use protocol has been reviewed and approved by the Institutional Animal Care and Use Committee. All efforts were made to minimize suffering.
Cell culture and sample preparation.
Dnmt3l-deficient ES cells of C57BL/6 and 129S4/SvJae genetic backgrounds were prepared as described previously (4, 10). The cells were maintained in high-glucose Dulbecco's modified Eagle medium (DMEM; Gibco-Invitrogen, Carlsbad, CA) supplemented with 15% fetal bovine serum (FBS), 2 mmol/liter of l-glutamine, MEM nonessential amino acids, 100 IU/ml of penicillin, 100 mg/ml of streptomycin, 0.12 mmol/liter of β-mercaptoethanol, and leukemia inhibitory factor (LIF). The medium for ES cells was changed daily, and the cultures were passaged every second day using a 1:3 ratio. Mouse embryonic fibroblasts (MEFs), 3T3 cells, and RAT2 cells were cultured in DMEM containing 10% FBS, 100 IU/ml of penicillin, and 100 mg/ml of streptomycin. The MEFs were passaged when the cells were approximately 70 to 80% confluent. Later-passage MEFs are defined as having been subjected to more than 5 passages.
Construction of expression vectors containing wild-type and mutant Dnmt3l.
cDNA from the mouse ES cell line D3 was used as the template for PCR amplification of wild-type Dnmt3l. The following PCR primers were used in these experiments: Dnmt3l-112F (CGATTACATCAATGGCTTCC) and Dnmt3l-1462R (GTGGAGGGAAGAGACCTTAG). We cloned the 1.35-kb PCR product into the TA vector (Invitrogen), and the sequence was verified with M13F and M13R primers. The Dnmt3l-TA vector was digested with EcoRI and then cloned into the EcoRI site of pIRES2-AcGFP1 (Clontech). The Dnmt3l-IRES-AcGFP fragments were digested by XhoI and NotI (NEB). The fragments were cloned into the SalI and NotI sites of preexcised pCAG-EGFP1 vector, eliminating the original enhanced green fluorescent protein (EGFP) fragment. To determine whether DNMT3L-mediated retroviral silencing activity requires interactions with histone tails and/or DNMT3A, we prepared 4 Dnmt3l mutant constructs abolishing these binding activities. The pCAG-Dnmt3l-IRES-AcGFP plasmid was used as the template for site-directed mutagenesis using the following primers: D124A F (TCCCTCTTCCTGTATGATGATGCTGGACACCAGAGTTACTGCACC), D124A R (CTGGTGTCCAGCATCATCATACAGGAAGAGGGACTC), I141W F (TGTTCCGGGGGTACCCTGTTCTGGTGTGAGAGCCCCGACTGTACC), I141W R (GCTCTCACACCAGAACAGGGTACCCCCGGAACAGC), F297A F (CGCTGTCCCGGCTGGTACATGGCCCAGTTCCACCGGATCCTGCAG), and F297A R (GTGGAACTGGGCCATGTACCAGCCGGGACAGCGAT). The C-terminal DNMT3L expression vector was generated by digesting pCAG-Dnmt3l-IRES-AcGFP with ScaI (NEB). The digested fragments were ligated using T4 DNA ligase (Promega).
Cell transfection.
A total of 1 × 106 cells were plated in a 10-cm dish the day before transfection. DNMT3L expression plasmid and Lipofectamine 2000 (Invitrogen) were mixed in the appropriate proportions according to the manufacturer's protocol. Twenty-four hours after transfection, transfected cells were selected with G418. The final concentrations of G418 used for selection were 350 μg/ml for MEFs and 800 μg/ml for 3T3 cells. Untransfected cells did not survive G418 selection.
Retroviral preparation and infection.
The retroviral Mo-MuLV construct used in all of the studies contains a luciferase gene and a 42-bp insert within the U3 region of the Mo-MuLV long terminal repeat (LTR). The 42-bp insertion sequence is TTATACTCCTCCACACACCATCACTCACTCTTTCTCAATCCA. The luciferase reporter of Mo-MuLVLUC was used in place of the GFP of Mo-MuLVGFP that was constructed as described previously (31). 293T cells were transiently transfected with the retroviral packaging plasmids CMV-Intron, pMDG, and Mo-MuLVLUC using FuGene 6 (Roche) to produce replication-incompetent retroviruses Mo-MuLVLUC with PBS or Mo-MuLVLUC/PBSQ with the B2 mutation (PBSQ). The viral medium was harvested at 48 and 72 h after transfection and filtered through a 0.45-μm filter. To correct for variations in titers between virus preparations, we used as a standard RAT2 cells, which lack TRIM28-mediated repression activity (32). RAT2 cells infected with Mo-MuLVLUC and Mo-MuLVLUC/PBSQ had similar viral activities 2 days postinfection. The same batch of virus-containing medium was used for all experiments; the cells were supplemented with Polybrene (8 mg/ml) and incubated at 37°C for 6 h, after which the medium was replaced with fresh culture medium.
Luciferase activity assay.
A total of 2 × 105 cells were harvested after Mo-MuLVLUC or Mo-MuLVLUC/PBSQ infection for the desired number of days. A luciferase assay was performed using a ONE-GLO luciferase assay system (Promega) and a Centro XS3 LB 960 microplate luminometer (Berthold) according to the manufacturer's protocol. The Mo-MuLV luciferase activities were normalized to the total protein concentration quantified using a bicinchoninic acid (BCA) protein assay kit (Pierce), and the infected viral copy number was further determined through quantitative PCR (qPCR). The primer sequences for viral copy number determination were as follows: PBS forward, TGAGTGATTGACTACCCGTCAGC; PBS reverse, GTTCCGAACTCGTCAGTTCCAC; GAPDH forward, AGTGCCAGCCTCGTCCCGTAGACAAAATG; and GAPDH reverse, AAGTGGGCCCCGGCCTTCTCCAT.
DNA methylation analysis.
The DNA methylation mark mentioned in this work refers to methylation of carbon 5 of cytosine, in a CpG dinucleotide context. Experimentally, DNA methylation was examined by bisulfite conversion followed by Sanger sequencing. Bisulfite converts unmodified cytosine residues to uracil while keeping 5-methylcytosine unconverted. After PCR amplification, the uracil will be amplified as thymine and therefore differentiate unmethylated CG and methylated CG dinucleotide as TG and CG, respectively.
The ES cells, MEFs, and 3T3 cells transfected with control or DNMT3L expression vectors after various durations of Mo-MuLV infection were subjected to DNA extraction. The cells were harvested on different days postinfection. Cellular DNA was purified using a commercial kit (High Pure PCR template preparation kit; Roche). Genomic DNA (500 ng) was bisulfite converted using a commercial kit (EZ DNA Methylation Gold kit; Zymo Research) according to the manufacturer's protocol. Nested PCR was used to amplify the Mo-MuLV long terminal repeats. The PCR products were extracted from 1.5% agarose gels and purified using a gel extraction kit (Qiagen). The purified PCR fragments were cloned into pGEM-T vector systems and transferred into Escherichia coli for DNA sequencing. The analysis and statistical comparison of bisulfite data were performed using QUMA software (http://quma.cdb.riken.jp/) (33).
Oligonucleotides for PCR amplification of bisulfite-treated DNA.
Oligonucleotides used for PCR amplification of bisulfite-treated DNA consisted of LUCF1 (TTTTTTTATATATTATTATTTATTTTTTTT), LUCR1 (ATCAATCACTCAAAAAAAACCCTC), LUCF2 (TAGGGTTAAGAATAGATGGAATAGTTGA), and LUCR2 (AATAATCAATCACTCAAAAAAAACCC).
Antibodies.
For Western blotting, the following antibodies were used: anti-DNMT3L (13451; Cell Signaling), anti-DNMT3L polyclonal and anti-DNMT3A polyclonal antibodies (obtained from X. Cheng), anti-TRIM28 polyclonal (ab10484; Abcam), anti-HDAC1 polyclonal (ab7028; Abcam), anti-SETDB1 (ab12317; Abcam), anti-HP1γ (ab10480; Abcam), and anti-EZH2 (17-662; Millipore). For immunofluorescence staining, an anti-HDAC1 polyclonal (ab7028; Abcam) antibody was used.
Immunoprecipitation.
Protein from DNMT3L-expressing MEFs were extracted at 2 days posttransfection using NP-40. The same amounts (4 μg for each) of the anti-TRIM28 polyclonal antibody (ab10484; Abcam) or anti-HDAC1 (ab7028; Abcam) antibodies were used for immunoprecipitation by conjugating with Dynabeads protein G kit (Invitrogen). The anti-DNMT3L, anti-DNMT3A, anti-TRIM28, anti-HDAC1, anti-SETDB1, and anti-EZH2 antibodies described above were used for Western blot analysis of the immunoprecipitates. The cell lysates and antibodies were incubated overnight at 4°C. Other reagents were used according to the manufacturers' protocols.
ChIP.
Two million MEF cells were seeded in a 10-cm dish 24 h before transfection with plasmid DNA. The transfected MEFs were infected with the virus collected from 293T packaging cells, 2 days posttransfection. Two days after virus infection, 1 × 105 cells were harvested for one chromatin immunoprecipitation (ChIP) reaction using the LowCell# ChIP kit protein G (Diagenode). The following antibodies were used: normal mouse IgG (12-371B; Millipore); anti-TRIM28 (ab22553; Abcam), anti-HDAC1 (ab7028; Abcam), anti-SETDB1 (ab12317; Abcam), anti-histone H3K9ac (ab10812), anti-histone H3K27ac (ab4729; Abcam), and anti-HP1γ (ab10480; Abcam). All procedures were performed according to the manufacturers' protocols, and the molecular size of the sheared chromatin DNA was determined to be below 500 bp (data not shown). ChIP primer sets used for qPCR have been previously described (25). The relative recovery was quantified by qPCR and normalized to the input. The primer sequences for ERV qPCR are as follows: GAPDH F, ACC TTT AGC CTT GCC CTT T; GAPDH R, ACA TCA CCC CCA TCA CTC AT; ERV1 U3 F, CCC CAA ATG ACC GAG AAA TA; ERV1 U3 R, GCG GTT ACA GAA GCG AGA AG; MLV PBS CHIP F, GTA AAA ACT CCA CAC TCG GC; MLV PBS CHIP R, ACG ATT CGG ATG CAA ACA GC; IAP PBS CHIP F, CGT GAG AAC GCG TCG AAT AA; IAP PBS CHIP R, TTC TGG TTC TGG AAT GAG GG; MMERVK-10C_3854 F, CAA ATA GCC CTA CCA TAT GTC AG; MMERVK R, GTA TAC TTT CTT CTT CAG GTC CAC; MERVL-F, ATC TCC TGG CAC CTG GTA TG; MERVL-R, AGA AGA AGG CAT TTG CCA GA; MTA ChIP F, ATG TCT TGG GGA GGA CTG TG; and MTA ChIP R, AGC CCC AGC TAA CCA GAA CT.
Dnmt3l KO fetal testis preparation.
Mice heterozygous for the Dnmt3ltm1Enls mutant allele (10) were intercrossed to generate Dnmt3l knockout (KO) embryos and littermate controls. Fetal testes from embryonic day 17.5 (E17.5) Dnmt3l KO and wild-type littermates were collected for protein extraction and immunoprecipitation as described above.
RESULTS
DNMT3L and TRIM28 are required for efficient de novo DNA methylation of newly integrated Mo-MuLV proviral sequences in ES cells.
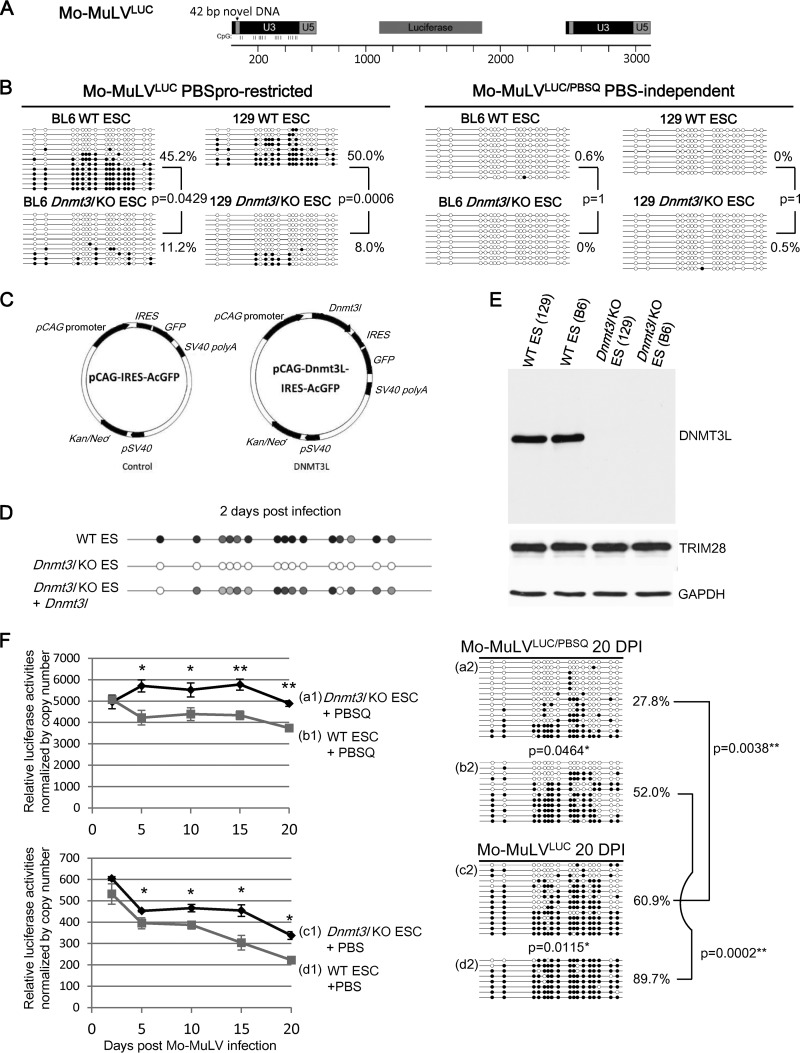
We infected wild-type and DNMT3L-deficient ES cells with Mo-MuLVLUC (PBSpro restricted; Fig. 1A), which can induce ZFP809-TRIM28-mediated retroviral silencing, and Mo-MuLVLUC/PBSQ (PBS-independent B2 mutation), which is insensitive to ZFP809-TRIM28-mediated silencing. At 2 days postinfection, DNA methylation at LTR sequences was present at very low levels (8.0 to 11.2%) in C57BL/6 and 129S4/SvJae Dnmt3l KO ES cell lines, compared with approximately 50% methylation of the same sequences in wild-type ES cells (Fig. 1B, left side). However, when we used PBS-independent Mo-MuLVLUC/PBSQ to infect wild-type and Dnmt3l KO ES cells of both the 129S4/SvJae and C57BL/6 genetic backgrounds, we did not detect any DNA methylation marks (Fig. 1B, right side). Transfecting Dnmt3l KO ES cells with wild-type Dnmt3l restored the efficiency of de novo methylation of proviral sequences (Fig. 1C and D). Wild-type and Dnmt3l KO ES cells expressed equal amounts of TRIM28 protein (Fig. 1E). These data indicate that both the DNMT3L and the ZFP809-TRIM28 pathways are important for initiating the de novo DNA methylation of newly introduced retroviral sequences.
FIG 1.
DNMT3L and the ZFP809-TRIM28 pathway are both required for epigenetic silencing of Mo-MuLV in ES cells. (A) Retroviral Mo-MuLV construct used in all studies containing a luciferase gene and a 42-bp insert within the U3 region of the Mo-MuLV long terminal repeat (LTR). This insert provides specific detection against preexisting and endogenous Mo-MuLV elements. The proviral DNA CpG sites for bisulfite sequencing are indicated. (B) ES cells lacking DNMT3L cannot add methylation marks to the LTR of proviral Mo-MuLV at 2 days postinfection. The PBSpro sequence from Mo-MuLV is also required for efficient de novo DNA methylation. DNA from wild-type (WT) or Dnmt3L KO ES cells was isolated at 2 days after Mo-MuLVLUC or Mo-MuLVLUC/PBSQ infection and analyzed by bisulfite sequencing using primers specific for the 42-bp insertion that were added to the LTRs of the Mo-MuLV constructs. The presence of a methylated or nonmethylated CpG within each sequence is indicated using a black or white circle, respectively. B6, C57BL/6 mice; 129, 129S4/SvJae mice. (C) Control and Dnmt3l expression vectors containing a neomycin resistance gene and IRES-GFP driven by a pCAG promoter. (D) Transfection of a Dnmt3l-expression vector rescues the methylation level in Dnmt3l KO ES cells. The bisulfite PCR product sequencing results are presented. The gray circles indicate the partially methylated CpG sites of mixed PCR fragments. (E) Protein expression levels of DNMT3L, TRIM28, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in wild-type and Dnmt3l KO ES cells. (F) DNMT3L and ZFP809-TRIM28 have a synergistic effect on the silencing of the proviral reporter and the methylation of the Mo-MuLV LTR DNAs. Wild-type (b1, b2, d1, and d2) and Dnmt3l KO (a1, a2, c1, and c2) 129S4/SvJae-derived ES cells were infected with either Mo-MuLVLUC (+PBS) (c1, c2, d1, and d2) or PBS-independent Mo-MuLVLUC/PBSQ (+PBSQ) (a1, a2, b1, and b2) viruses. We normalized the proviral luciferase activity to the relative virus copy numbers determined through quantitative PCR using primers specific to the PBS region of the exogenous Mo-MuLV as described previously (25) (Fig. 2D). The cells were cultured for the indicated periods, and luciferase activities were analyzed and normalized to the virus copy numbers revealed in panel F (left side). Data points are averages of three biological repeats; bars indicate SEMs. *, P < 0.05; **, P < 0.01. DNA methylation of the proviral LTRs 20 days postinfection is shown on the right side. DNA methylation percentage indicates the total number of black circles/total CpG sites. All P values were obtained by the nonparametric two-tailed Mann-Whitney test.
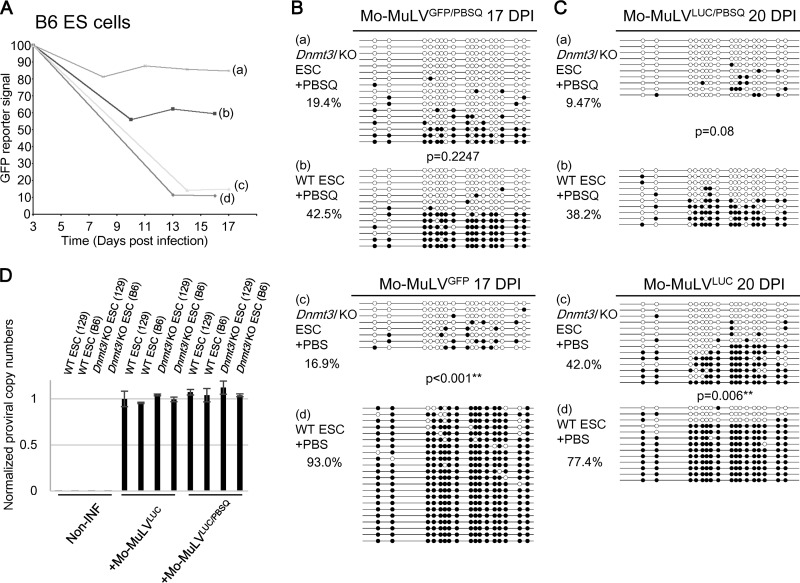
Analysis of proviral activity and DNA methylation status of Mo-MuLVLUC and Mo-MuLVLUC/PBSQ in wild-type and Dnmt3l KO ES cells 17 to 20 days after infection further revealed the intensity of DNMT3L-dependent and PBSpro-restricted retroviral silencing activity at the level of DNA methylation and beyond. Both DNMT3L and TRIM28 contributed to silencing of retrovirus (Fig. 1F and 2A). The luciferase activity measured among different samples (Fig. 1F) was normalized by copy number of proviral sequences (Fig. 2D). DNA methylation at Mo-MuLVLUC and Mo-MuLVGFP proviral sequences was significantly reduced in infected ES cells deficient for Dnmt3l (compare items c2 and d2 in Fig. 1F and items c and d in Fig. 2B and C). This decrease was associated with higher retroviral activity based on luciferase (Fig. 1F, items c1 and d1) and GFP reporter activity (Fig. 2A, items c and d). The mutant Mo-MuLVLUC/PBSQ and Mo-MuLVGFP/PBSQ proviral sequences, which cannot attract TRIM28 via ZFP809, accumulated significantly fewer DNA methylation marks than the ZFP809-TRIM28-accessible Mo-MuLVLUC and Mo-MuLVGFP sequences (Fig. 1F, items b2 and d2; Fig. 2B, items b and d; and Fig. 2C, items b and d). The PBS-independent Mo-MuLVPBSQ also retained approximately 10-fold-higher retroviral activity based on the luciferase (Fig. 1F, items b1 and d1) and GFP reporters used (Fig. 2A, items b and d), suggesting that TRIM28 is critical for retroviral silencing activity.
FIG 2.
DNMT3L- and ZFP809-TRIM28-mediated Mo-MuLV silencing in C57BL/6 background ES cells. (A) Wild-type and Dnmt3l KO ES cells in the C57BL/6 genetic background were sorted 3 days after Mo-MuLVGEP/PBS or Mo-MuLVGFP/PBSQ infection, and the GFP percentage was monitored. Wild-type (b and d) and Dnmt3l KO (a and c) C57BL/6-derived ES cells were infected with either Mo-MuLVGFP (c and d) or Mo-MuLVGFP/PBSQ (a and b) viruses for 3 days and then analyzed at the indicated time points by fluorescence-activated cell sorting (FACS) for GFP-positive ES cells. (B) DNA extracted and prepared for bisulfite analysis 17 days postinfection in the C57BL/6 genetic background. (C) Bisulfite sequencing of wild-type and Dnmt3l KO ES cells in the C57BL/6 genetic background after 20 days of Mo-MuLVLUC infection. DNA methylation percentage indicates the total number of black circles/total CpG sites. All P values were obtained by the nonparametric two-tailed Mann-Whitney test. **, P < 0.01. (D) qRT-PCR performed using primers specific to the PBS region of Mo-MuLV or internal control GAPDH after 2 days of Mo-MuLVLUC or Mo-MuLVLUC/PBSQ infection. Noninfected ES cells were used as a negative control. The values are averages of three biological repeats ± SEMs.
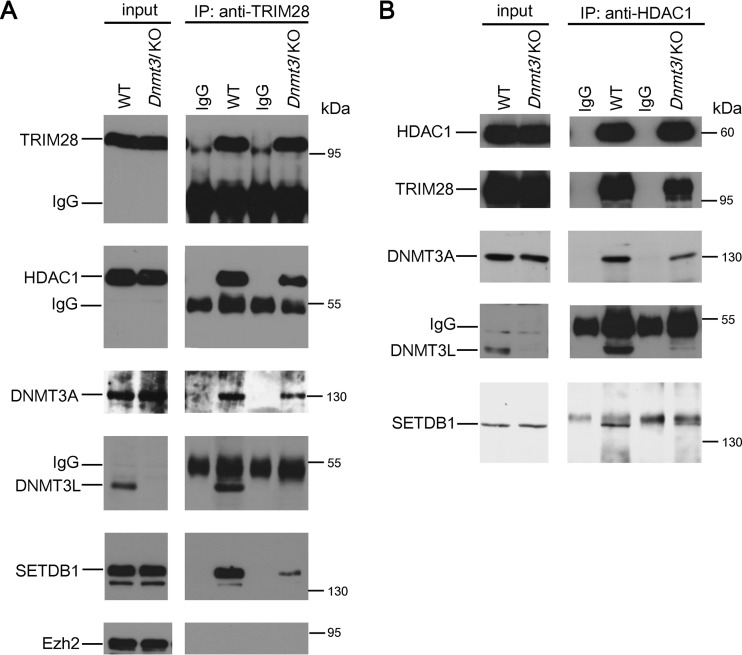
ZFP809-TRIM28-induced proviral silencing was dramatically greater than the silencing effect induced by DNMT3L (Fig. 1F and 2A). The strong ZFP809-TRIM28-induced silencing effect suggests that TRIM28 may have recruited substantially more silencers that masked the modest silencing effect induced by DNMT3L in ES cells. Indeed, DNMT3L-induced modest retroviral silencing activity (Fig. 1F, items a1 and b1, P < 0.01) was not completely dependent on DNA methylation (Fig. 1F, items a2 and b2, P = 0.0464 in the 129S4/SvJae genetic background; Fig. 2B, items a and b, P = 0.22; and Fig. 2C, items a and b, P = 0.08 in the C57BL/6 genetic background). These results suggest that DNMT3L may trigger proviral silencing activities beyond its ability to facilitate de novo DNA methylation activity by DNMT3A. To clarify whether DNMT3L can induce the interaction between enzymes capable of mediating posttranslational modifications of histone proteins in ES cells, we performed immunoprecipitation experiments. These revealed that DNMT3L is associated in a complex with HDAC1, TRIM28, DNMT3A, and SETDB1 (Fig. 3). Interestingly, we observed reduced association of DNMT3A and SETDB1 in the absence of DNMT3L. The polycomb repressor complex component EZH2, a reputed DNMT3L-interacting modifier which is also involved in HIV silencing (34, 35), was absent from the TRIM28 complex (Fig. 3A). These results indicate that TRIM28 can synergistically act with DNMT3L to affect DNA methylation-dependent and -independent pathways that are responsible for repressing Mo-MuLV retroviral activity.
FIG 3.
DNMT3L facilitated the formation of the DNMT3A/SETDB1/HDAC1 protein complex in ES cells 2 days after Mo-MuLVLUC infection. Equal amounts of control IgG and anti-TRIM28 (A) or anti-HDAC1 (B) immunoprecipitates from wild-type and Dnmt3l KO ES cell extracts were analyzed by Western blotting using anti-TRIM28, anti-DNMT3L, anti-DNMT3A, anti-SETDB1, anti-HDAC1, and EZH2 antibodies, as indicated.
Ectopic Dnmt3L induces modest retroviral silencing activity in later-passage MEFs.
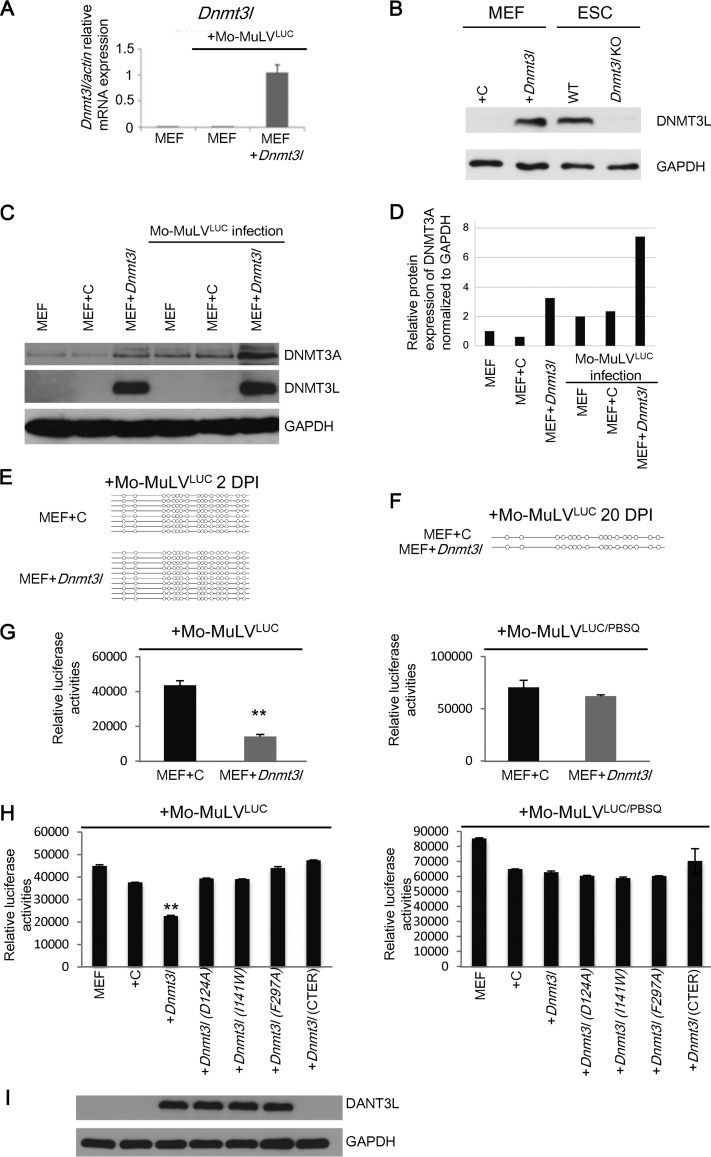
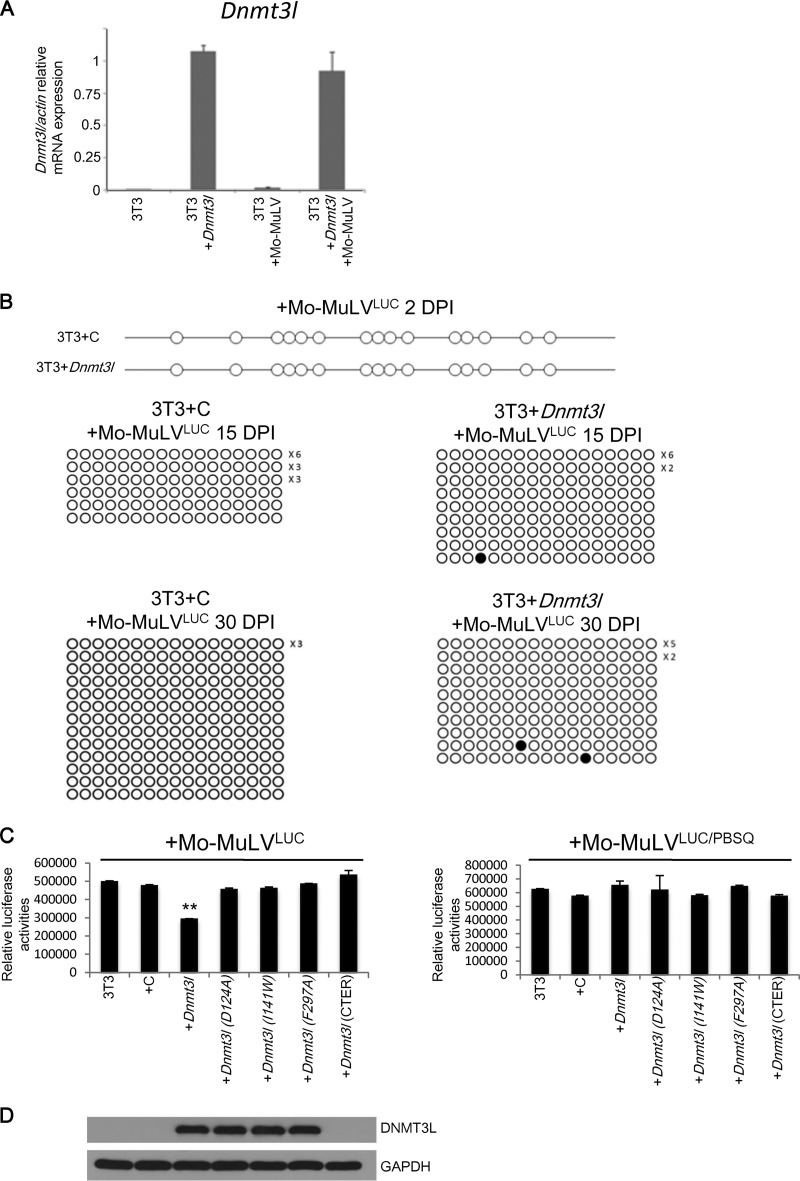
We observed that DNMT3L and TRIM28 may cooperate in retroviral silencing activity through other machineries beyond de novo DNA methylation in ES cells. To further clarify the involvement of DNMT3L in DNA methylation and chromatin modification in retroviral silencing, we transfected control and DNMT3L overexpression vectors (Fig. 1C) into MEFs and then infected the cells with Mo-MuLVLUC. Dnmt3l mRNA and protein expression levels in the Dnmt3l-transfected MEFs were confirmed through quantitative reverse transcription-PCR (qRT-PCR) and immunoblotting for DNMT3L, respectively (Fig. 4A and B). DNMT3A expression was upregulated approximately 2-fold after DNMT3L overexpression or 5-fold after DNMT3L overexpression following Mo-MuLV infection (Fig. 4C and D). However, we did not detect DNMT3L-induced methylation on the LTR regions of Mo-MuLV proviral DNA within 20 days postinfection (Fig. 4E and F). Surprisingly, DNMT3L-expressing MEFs repressed Mo-MuLVLUC retroviral activities compared with MEFs transfected with control vectors (Fig. 4G, left side), although ectopic DNMT3L did not silence the PBS-independent viral construct Mo-MuLVLUC/PBSQ in MEFs (Fig. 4G, right side), suggesting that the modest silencing induced by DNMT3L is PBSpro sequence dependent. To examine the generality of this repression mechanism, we repeated these experiments with 3T3 cells. Again, even with high Dnmt3l RNA expression (Fig. 5A), we did not detect DNMT3L-induced methylation on the LTR regions of Mo-MuLV proviruses 30 days after infection of 3T3 cells (Fig. 5B). However, a modest decrease in retroviral activity was observed in DNMT3L-expressing 3T3 cells infected with Mo-MuLVLUC compared with cells transfected with control vectors (Fig. 5C, left side). These results demonstrate that DNMT3L can exert retroviral silencing activity via a DNA methylation-independent mechanism.
FIG 4.
DNMT3L-induced retroviral silencing activity depends on PBSpro sequence and functional DNMT3L harboring proper binding activities to the N-terminal tail of histone H3 and DNMT3A. (A) The average relative Dnmt3l/actin ratio (±SEM) was determined through qRT-PCR using primers specific for the Dnmt3l gene and normalized to the actin gene in MEFs. (B) Dnmt3L-overexpressing MEFs have protein expression levels similar to those in ES cells, as confirmed by Western blot analysis using an anti-DNMT3L antibody or an anti-GAPDH antibody as a control. (C) DNMT3A protein expression is upregulated by DNMT3L expression in MEFs or by Mo-MuLV infection. Protein expression of DNMT3A, DNMT3L, and GAPDH in MEFs overexpressing DNMT3L and/or infected with Mo-MuLVLUC is shown. (D) Quantification of the results of DNMT3A expression normalized to GAPDH. (E and F) Bisulfite sequencing indicated that DNMT3L-expressing MEFs did not methylate proviral DNA at 2 days (E) or 20 days (F) postinfection. (G) DNMT3L-induced retroviral silencing is PBSpro restricted. Control and DNMT3L-expressing MEFs were infected with Mo-MuLVLUC (left) and Mo-MuLVLUC/PBSQ (right). Relative mean luciferase activities ± SEMs normalized to the total protein amounts were calculated 2 days posttransfection following 2 days of Mo-MuLV infection. (H) Mutated DNMT3L expression constructs consisting of Dnmt3lD124A, Dnmt3lI141w, Dnmt3lF297A, and the C-terminus-only domain did not induce retroviral silencing. Control and DNMT3L-expressing MEFs were infected with Mo-MuLVLUC (left) or Mo-MuLVLUC/PBSQ (right). Relative mean luciferase activities ± SEMs normalized by total protein were calculated 2 days after transfection following 2 days of Mo-MuLV infection. Student's t tests were used for statistical analysis: **, P < 0.01. (I) Protein expression of DNMT3L and GAPDH in MEFs overexpressing wild-type and mutant DNMT3L.
FIG 5.
DNMT3L induces retroviral silencing activity in 3T3 cells. (A) Relative mRNA expression level of 3T3 cells transfected with Dnmt3l expression construct. (B) Dnmt3l-transfected 3T3 cells lack the DNA methylation mark-adding ability at 2 to 30 days postinfection. (C) DNMT3L mutations reduce the DNMT3L-induced silencing activity of 3T3 cells. Expression constructs carrying wild-type or mutant DNMT3L (D124A, I141W, and F297A and truncated DNMT3L without the ADD domain) were transfected into 3T3 cells. Relative mean luciferase activities ± SEMs normalized by total protein were calculated 2 days after transfection following 2 days of Mo-MuLV infection. Student's t tests were used for statistical analysis. **, P < 0.01. (D) Protein expression of DNMT3L and GAPDH in MEFs overexpressing wild-type and mutant DNMT3L.
We next probed which DNMT3L interactions are important for silencing activity. DNMT3L has previously been shown to interact with histone H3 via the N-terminal ADD domain. Interaction with DNMT3A is mediated by the C-terminal methyltransferase folds. To disrupt histone H3 binding, we engineered 3 mutant proteins: two with D124A and I141W mutations and a C-terminus-only protein (4). To disrupt DNMT3A-DNMT3L heterodimer formation, we engineered a mutation at F261A (36). None of these four mutant forms were capable of inducing retroviral silencing when ectopically expressed in MEFs (Fig. 4H and I) or 3T3 cells (Fig. 5C and D).
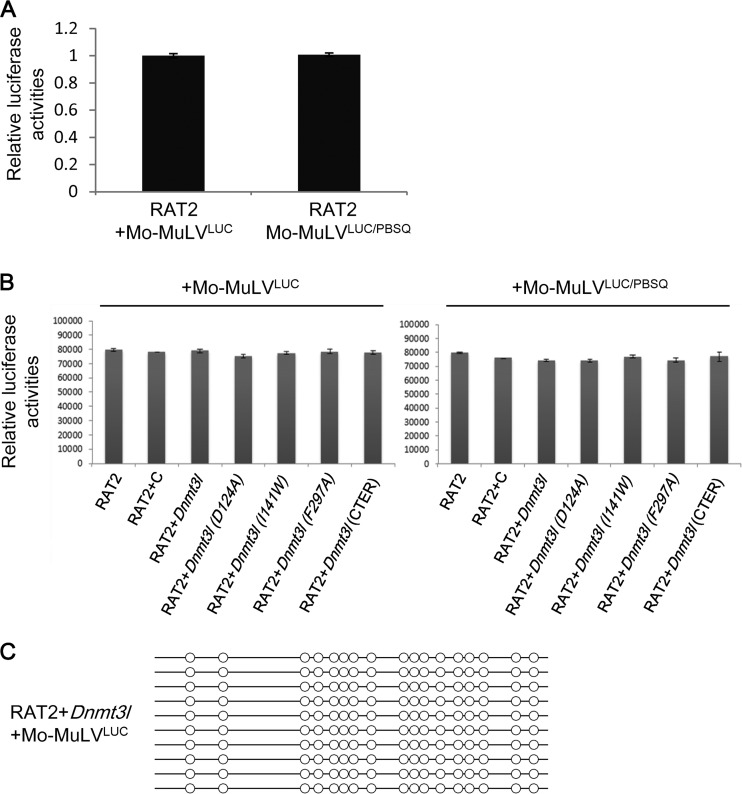
RAT2 cells have previously been reported to lack TRIM28-mediated retroviral repression. Using this cell line as a negative control, we found that there were indeed no differences in luciferase activity between RAT2 cells with or without the expression of wild-type or mutant DNMT3L (Fig. 6A and B). Furthermore, despite the ectopic presence of Dnmt3L protein, there was no de novo DNA methylation of the proviral LTR in DNMT3L-expressing RAT2 cells 20 days postinfection (Fig. 6C). Collectively, these results indicate that DNMT3L-induced, PBSpro-restricted retroviral silencing requires interactions between DNMT3L, histone H3, and DNMT3A.
FIG 6.
Mo-MuLVLUC and Mo-MuLVLUC/PBSQ have the same infection titers. (A) RAT2 cells lack PBS-mediated silencing activity (32) and were used to normalize the titer of Mo-MuLV/LUC and Mo-MuLV/LUC/PBSQ. (B) The relative luciferase activity showed no infection activity differences between Mo-MuLVLUC and Mo-MuLVLUC/PBSQ in wild-type RAT2 cells or RAT2 cells transfected with either the wild type or the indicated mutant DNMT3L expression constructs. (C) DNMT3L did not induce the DNA methylation of the proviral LTR sequence in RAT2 cells 20 days postinfection.
Ectopic DNMT3L induces the formation of a repressive chromatin modifier complex to introduce repressive histone modifications at Mo-MuLV proviral DNA and ERVs in DNMT3L-expressing MEFs.
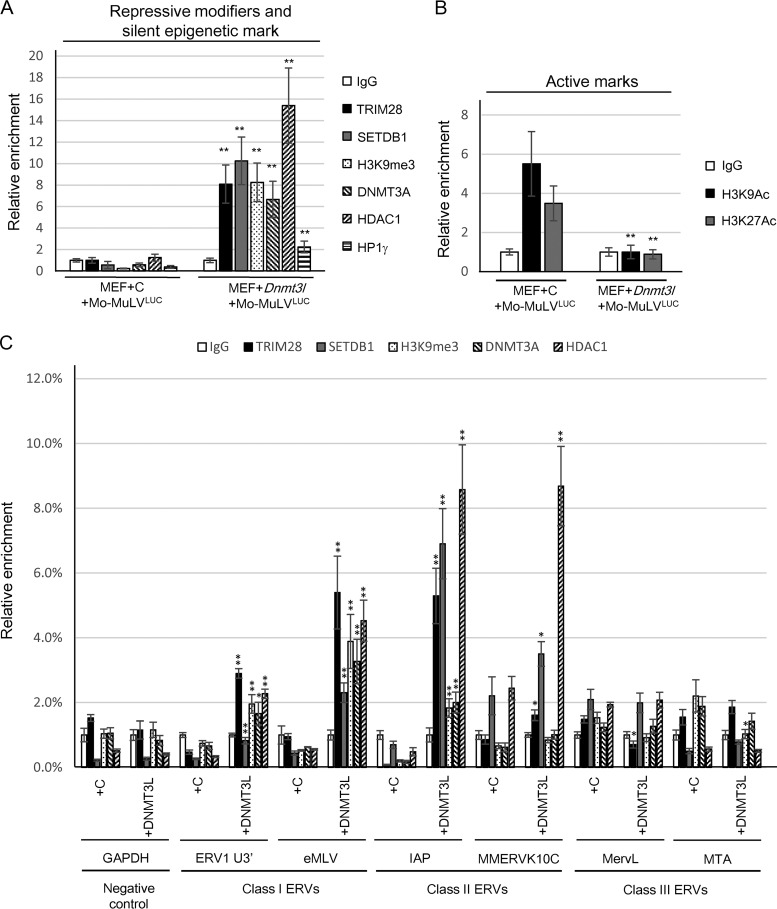
We next sought to determine whether DNMT3L-induced retroviral silencing correlated with a change in the chromatin state of the newly integrated proviral DNA. Chromatin immunoprecipitation (ChIP) was performed on DNMT3L-expressing MEFs following infection with Mo-MuLVLUC or Mo-MuLVLUC/PBSQ. We observed that ectopic DNMT3L induced the enrichment of TRIM28, HDAC1, DNMT3A, and SETDB1 (with modest amounts of HP1γ) at the Mo-MuLV LTR (Fig. 7A) and triggered H3K9me3 accumulation. This enrichment of DNMT3L-associated modifiers was not observed in cells transfected with control plasmid. Our results indicate that DNMT3L can induce retroviral silencing via recruitment of TRIM28, SETDB1, H3K9me3, and HDAC1 to the Mo-MuLV LTR. The opposite effect was observed for H3K9ac and H3K27ac modifications that are typically associated with transcriptionally active chromatin (Fig. 7B). The presence of more modifications was associated with transcriptional silencing, and fewer marks associated with transcriptional activity were observed after MEFs were transfected with Dnmt3l. We then asked whether DNMT3L-mediated enrichment of these chromatin modifiers could also occur at preexisting ERVs. ChIP analysis showed significant enrichment of TRIM28, SETDB1, and HDAC1 mainly for the class I and class II ERVs (Fig. 7C). Our data are consistent with previous observations showing that IAP elements (a class II ERV) are upregulated by deletion of Trim28 in mouse ES cells and in early embryos (12). Thus, ectopic DNMT3L may rescue some of the ES cell-specific TRIM28-mediated ERV silencing in MEFs.
FIG 7.
DNMT3L can recruit epigenetic modifiers to induce repressive histone modifications on Mo-MuLV LTR and ERVs in MEFs. (A) DNMT3L induces TRIM28, SETDB1, DNMT3A, HDAC1, and HP1γ binding to proviral PBS sequence and increases H3K9me3 accumulation on the associated chromatin. Results of a chromatin immunoprecipitation (ChIP) assay of TRIM28, SETDB1, H3K9me3, DNMT3A, HDAC1, and HP1γ at the viral PBS sequence of control and DNMT3L-overexpressing MEFs after 2 days of Mo-MuLVLUC infection are shown. (B) DNMT3L decreases the active H3K9ac and H3K27ac marks on the proviral PBS sequence. (C) DNMT3L induces TRIM28, SETDB1, and HDAC1 binding to endogenous IAPs. Results of a ChIP assay of TRIM28, SETDB1, H3K9me3, DNMT3A, and HDAC1 at the ERVs are shown. ChIP using IgG antibody was used for background control. qRT-PCR was performed using sequence-specific primers ERV1 U3′ and endogenous MLV (class I), IAP and MMERVK10C (class II), and MervL and MTA (class III) as described previously (52). Primers specific for GAPDH served a negative control. Each ChIP experiment shows the mean enrichment ± the SEM from three biological repeats. Relative enrichment values (percentage of input) in all ChIP experiments were normalized to the total input of the samples. The relative enrichment values were compared statistically between control (+C) and DNMT3L-expressing (+DNMT3L) MEFs in each ERV primer set. Student's t tests were used for statistical analysis. *, P < 0.05; **, P < 0.01.
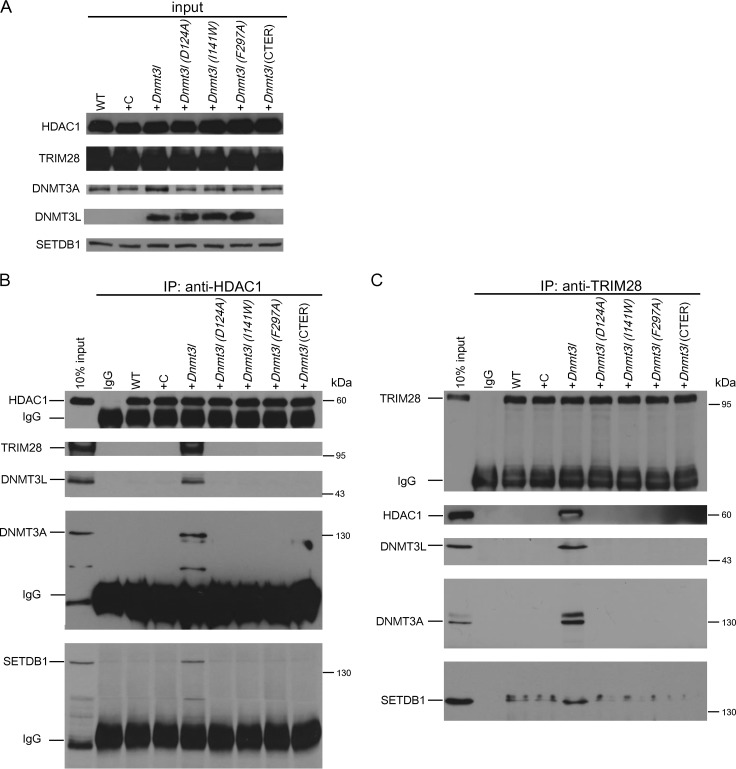
We next tested whether DNMT3L-induced chromatin modification was dependent on the formation of a repressive modifier complex. We performed immunoprecipitation assays using an anti-TRIM28 antibody and an anti-HDAC1 antibody to investigate the protein complex in MEFs after transfection for one passage. The total inputs of HDAC1, TRIM28, DNMT3A, DNMT3L, and SETDB1 are shown in Fig. 8A. Only MEFs transfected with wild-type DNMT3L induced the formation of a complex containing HDAC1, TRIM28, DNMT3A, DNMT3L, and SETDB1, which immunoprecipitated with HDAC1 (Fig. 8B). Similar results were obtained using an anti-TRIM28 antibody as an alternative means of purifying this complex (Fig. 8C). The point mutations D124A, I141W, and F297A and the truncated DNMT3L abolished the formation of this protein complex (Fig. 8B and C), indicating that the interaction between DNMT3L and histone tails and the binding between DNMT3A and DNMT3L are both required for DNMT3L-induced epigenetic modifier complex formation.
FIG 8.
Ectopic DNMT3L induces the formation of a repressive chromatin modifier complex in DNMT3L-expressing MEFs. (A) Western blot analysis of HDAC1, TRIM28, DNMT3A, DNMT3L, and SETDB1 in a MEF lysate input fraction used for immunoprecipitation. (B) DNMT3L induces formation of a HDAC1/DNMTA/SETDB1/TRIM28-containing complex in MEFs. MEFs were transfected with a control vector (+C) or a wild-type Dnmt3l vector (+Dnmt3l) or the indicated mutant expression construct for one passage. The same amounts of control IgG and anti-HDAC1 immunoprecipitates from MEF extracts were analyzed by Western blot assays using the antibodies indicated. (C) Western blot using antibodies indicated on material enriched following immunoprecipitation using anti-TRIM28 antibody.
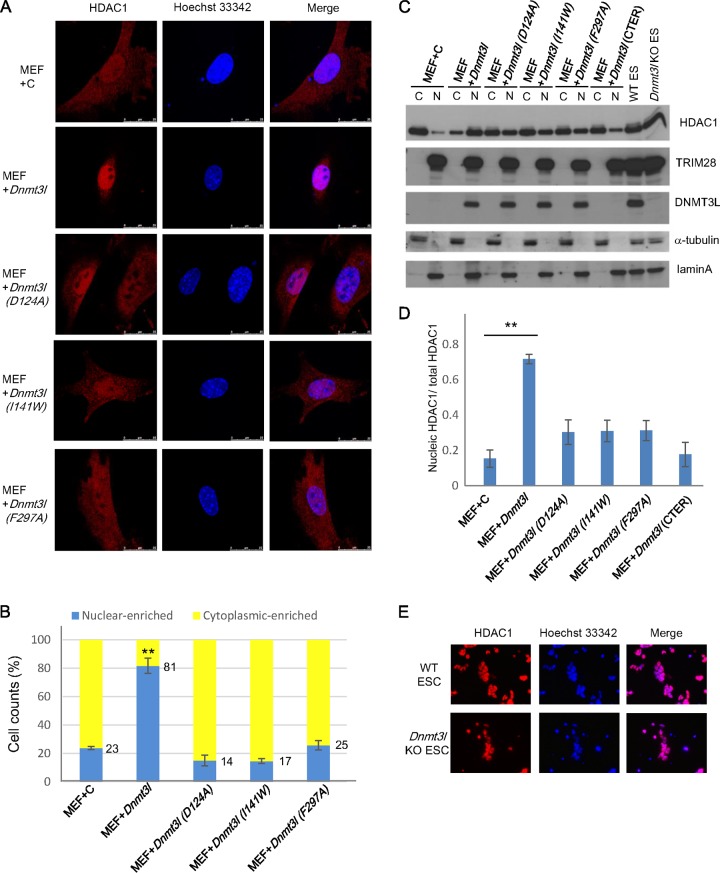
DNMT3L induces HDAC1 translocation into the nucleus.
It has previously been reported that DNMT3L interacts with and increases the deacetylase activity of HDAC1 in vitro (37). TRIM28 mediates gene silencing (20, 26) by recruiting the NuRD histone deacetylase complex to target promoters (21). Our data showed that DNMT3L induced the formation of a repressive protein complex (Fig. 8). Additionally, our chromatin immunoprecipitation data revealed that ectopic expression of full-length DNMT3L results in the recruitment of HDAC1 to newly integrated proviral LTRs and some endogenous retroviruses (Fig. 7A and C). Based on these findings, we investigated whether DNMT3L can recruit HDAC1 into the nucleus. Expression of DNMT3L in later-passage (beyond passage 5 [P5]) MEFs resulted in the translocation of HDAC1 from the cytosol to the nucleus; this was abolished if Dnmt3L was unable to interact with either histone H3 or Dnmt3A (Fig. 9A and B). These data indicate that the interactions between DNMT3L and histone tails and DNMT3A are necessary for recruiting HDAC1 to the nucleus. The translocation of HDAC1 to the nucleus was further confirmed by examining its distribution in the cytosolic and nuclear fractions. An immunoblot of HDAC1 protein expression (Fig. 9C) and the accompanying quantification (Fig. 9D) revealed that significantly increased HDAC1 translocation to the nucleus happened only when the fully functional wild-type DNMT3L was ectopically expressed in MEFs. Similarly, significantly increased HDAC1 translocation to the nucleus was found in DNMT3L-expressing 3T3 cells compared with mutant DNMT3L-transfected 3T3 cells (data not shown).
FIG 9.
DNMT3L induces HDAC1 translocation to the nucleus in later-passage MEFs. The subcellular localization of HDAC1 in wild-type and Dnmt3l-transfected MEFs at passage 8 is shown. (A) Control and Dnmt3L-, Dnmt3LD124A-, Dnmt3LI141w-, and Dnmt3LF297A-transfected MEFs were immunostained with an anti-HDAC1 antibody and Hoechst 33342 and analyzed by confocal microscopy. Red, HDAC1; blue, Hoechst 33342. Scale bar, 2.5 μm. (B) Cell counts of HDAC1 subcellular localization. Random microscopic fields of cells from three biological repeats were used for counting the cells with nucleus-enriched or cytoplasm-enriched HDAC1 distribution. Blue bars, percentage of cells with predominantly nuclear localized HDAC1; yellow bars, percentage of cells with predominantly cytoplasmic localized HDAC1. Total cell counts for each group are 111 for +C, 132 for +Dnmt3l, 120 for +Dnmt3l (D124A), 106 for +Dnmt3l (I141W), and 108 for +Dnmt3l (F297A). The values reflect the average cell count percentages from three biological repeats ± SEMs. Student's t test was used for statistical analysis. **, P < 0.01. (C) Western blot analysis of protein expression in the cytosol or nucleus using anti-HDAC1, anti-TRIM28, anti-DNMT3L, anti-alpha-tubulin (control for cytoplasmic signal), and lamin A (control for nuclear signal) antibodies. (D) Quantification of immunoblotting results using ImageJ software. The mean nuclear HDAC1 signal ± SEM from three biological repeats was normalized to the total HDAC1 signal. Student's t test was used for statistical analysis. **, P < 0.01. (E) Wild-type and Dnmt3l KO ES cells were immunostained with an anti-HDAC1 antibody and Hoechst 33342 and analyzed by confocal microscopy. Red, HDAC1; blue, Hoechst 33342.
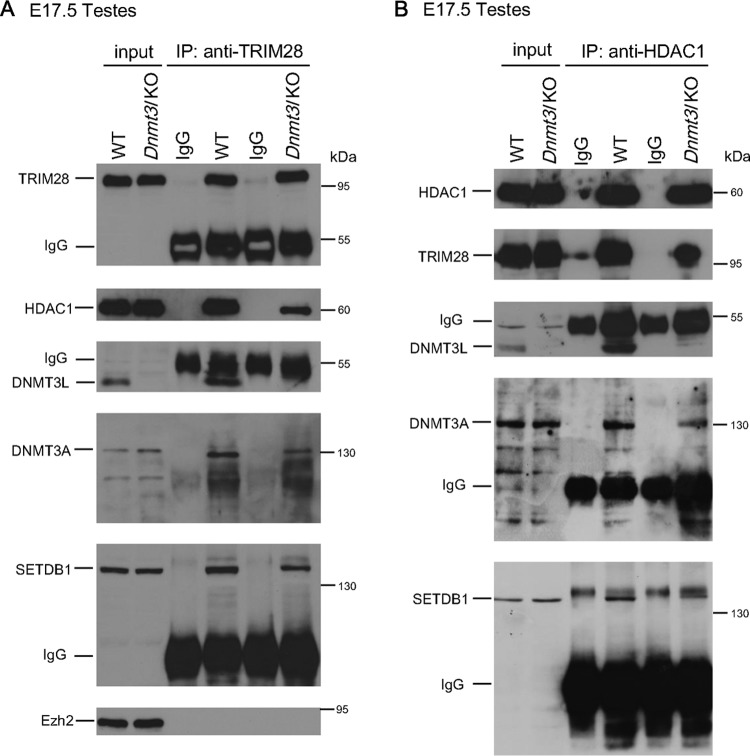
DNMT3L-repressive chromatin modifier complex is present in fetal gonads.
DNMT3L is normally expressed in male gonads from E14 onwards (38). We examined whether the DNMT3L-associated repressor complex observed in our gain-of-function model is also present in the gonads. We performed immunoprecipitation using anti-TRIM28 or anti-HDAC1 antibody to analyze associated proteins in E17.5 gonads. Both proteins were observed in complex with DNMT3A, DNMT3L, and SETDB1. In the absence of DNMT3L, we observed a decrease in the interaction among SETDB1, DNMT3A, HDAC1, and TRIM28 compared with that in gonads from wild-type littermates (Fig. 10). Similar to the results from other cell types, we also did not observe any enrichment for Ezh2 when immunoprecipitating for TRIM28. These data support the existence of this corepressor complex in cells that normally express DNMT3L and suggest that it is of biological relevance during gametogenesis.
FIG 10.
DNMT3L facilitates the formation of the protein complex containing DNMT3A, SETDB1, and HDAC1 in E17.5 gonads. Immunoprecipitation was performed with 50 μg of total protein from wild-type or Dnmt3l KO E17.5 gonads. The same amount of protein from control IgG and anti-TRIM28 (A) or anti-HDAC1 (B) immunoprecipitates from wild-type and Dnmt3l KO gonad cell extracts was analyzed through Western blot assays using anti-TRIM28, anti-DNMT3L, anti-DNMT3A, anti-SETDB1, anti-HDAC1, and anti-EZH2 antibodies as indicated.
DISCUSSION
Methylation-independent retroviral silencing by ectopic DNMT3L in somatic cells.
Using a combination of various cells types that normally or ectopically express DNMT3L, we have further defined the different levels of epigenetic mechanisms by which DNMT3L functions to repress retroviral activity (summarized in Table S2 in the supplemental material). Ectopic expression of DNMT3L triggered the formation of a repressive epigenetic modifying complex consisting of DNMT3L, DNMT3A, TRIM28, HDAC1, and SETDB1 that targets newly integrated Mo-MuLV proviral sequences and introduces repressive chromatin modifications in somatic cells without triggering de novo DNA methylation activity.
Although several studies have shown that DNA methylation and histone modifications coordinately silence retroviral sequences (15, 23, 39–43), the importance of DNMT3L in coordinating histone modifications for proviral silencing has not been previously described. DNMT3L-induced retroviral silencing activity in somatic cells depends on the TRIM28-associated silencing pathway. Our observation that formation of a DNMT3L-induced DNMT3L/DNMT3A/TRIM28/HDAC1/SETDB1 complex in MEFs is abrogated by expression of mutant DNMT3L proteins unable to interact with histone H3 tails highlights the importance of the chromatin targeting. We propose that the entire DNMT3L-induced epigenetic silencing complex is built upon the Mo-MuLV proviral DNA. Although MEFs and 3T3 cells do not have detectable ZFP809 protein expression, in the presence of DNMT3L, TRIM28 may still be recruited to Mo-MuLV sequences through another ZFP protein. In addition, as the binding between DNMT3L and histone proteins is essential for the formation of TRIM28-included epigenetic modification complex formation, DNMT3L may also help TRIM28 to anchor to the chromatin with Mo-MuLV sequences.
Expression of a DNMT3L mutant unable to interact with DNMT3A also failed to trigger DNMT3L-induced assembly of the repressive chromatin-modifying complex. Both DNMT3A and DNMT3L interact with TRIM28 and HDAC1 (37, 44–46); the former attracts SETDB1 to introduce H3K9 methylation and further recruits HP1γ (24, 47). In addition, DNMT3A may also interact with SETDB1 directly and mediate gene silencing (23, 48). We propose that Dnmt3L serves as a platform to amplify recruitment of this silencing complex at newly integrated proviral sequences.
Among the chromatin modifiers that DNMT3L recruits to the Mo-MuLV sequences in MEFs, HDAC1 is the most significantly enriched. Previous studies indicate that DNMT3L can directly interact with HDAC1 (37). Our immunofluorescence and nucleus/cytosol immunoblot analyses demonstrated that DNMT3L overexpression can change the subcellular localization of HDAC1 from the cytosol to the nucleus in late-passage MEFs, in which a significant amount of HDAC1 is located in the cytoplasm. A similar DNMT3L-induced HDAC1 translocation from the cytoplasm to the nucleus is found in 3T3 cells. This effect may partly explain how DNMT3L increases the recruitment of HDAC1 to the TRIM28 silencing complex. In contrast, in ES cells (Fig. 9E) or MEFs at lower passages, when HDAC1 is predominantly localized to the nucleus, DNMT3L does not further influence the subcellular localization of HDAC1 significantly. The fact that HDAC1 is already localized to the nucleus in ES cells may account for their inherent ability to suppress retroviral activity (49, 50).
DNMT3L and TRIM28 synergistically silence proviral sequences in ES cells through multiple mechanisms.
Although our initial observations were made by overexpressing DNMT3L in somatic and transformed cell lines, the associated repressive chromatin-modifying complex also exists in more physiologically relevant cell types. In mutant ES cells and fetal testes, the absence of DNMT3L reduces the interaction between TRIM28 and repressive epigenetic modifiers, including DNMT3A, HDAC1, and SETDB1. DNMT3L may therefore help to initiate a repressive epigenetic silencing cascade by adding repressive histone marks to attract more repressive epigenetic modifiers and eventually lead to the de novo DNA methylation of newly infected retroviral sequences in ES cells.
In ES cells, both DNMT3L and TRIM28 are important for facilitating efficient de novo DNA methylation. Our gain-of-function study with MEFs indicated that DNMT3L enhances the recruitment of a repressive chromatin-modifying complex to Mo-MuLV proviral sequences to administer repressive histone modifications (summarized in Table S2 in the supplemental material). The decreased SETDB1/HDAC1/TRIM28 complex formation in Dnmt3l-deficient ES cells may significantly reduce the speed and strength of the retroviral silencing activity at the chromatin level. DNMT3L is a crucial regulator of DNA methylation establishment and is thought to specifically enhance DNMT3A-mediated methylation by a number of mechanisms. DNMT3L has been shown to form heterotetramers with DNMT3A, which can halt the homo-oligomerization of DNMT3A and release DNMT3A from heterochromatin regions (51).
Under the histone recognition hypothesis (31) and from the data described herein, we propose that in the absence of DNMT3L, fewer repressive histone modifiers are recruited to chromatin-containing newly integrated proviral sequences. The relative open chromatin signature, in turn, further reduces DNMT3A binding and hinders the acquisition of DNA methylation. This may explain how DNMT3L facilitates effective de novo DNA methylation on Mo-MuLV proviral sequences in ES cells. In addition, our observation that TRIM28 is found in complex with DNMT3A and DNMT3L may explain the necessity of TRIM28 for efficient and effective de novo methylation and repression of newly integrated Mo-MuLVs. The DNMT3L-induced PBSpro-restricted epigenetic silencing activity at both the DNA methylation and chromatin modification levels may also account for the observation that endogenous retrotransposons are hypomethylated in Dnmt3l-deficient ES cells (31).
DNMT3L-induced retroviral silencing does not rely on DNA methylation of proviruses in somatic cells.
While endogenous DNMT3L can enhance the efficiency of retroviral silencing activity by facilitating the recruitment of repressive histone modifiers and accelerating de novo DNA methylation in ES cells, ectopic expression of DNMT3L in somatic cells is only sufficient to initiate moderate retroviral silencing activity via repressive histone modifications. Although DNMT3A is also involved in the DNMT3L-induced repressive epigenetic modification complex, we did not detect de novo methylation on the Mo-MuLV sequences 20 days after infection in MEFs or 30 days after infection in 3T3 cells. There are at least two possible reasons why ectopic DNMT3L was not sufficient to trigger de novo methylation on Mo-MuLV sequences in somatic cells. First, compared with ES cells, the MEFs and 3T3 cells may still lack components that are critical for facilitating the de novo methylation of newly infected Mo-MuLV sequences, despite the ectopic expression of DNMT3L. Second, as was suggested recently for ES cells, DNMT3L may serve as a dominant negative inhibitor of DNMT3A by competing for the same binding sites on the H3K27 methyltransferase EZH2 (35); ectopic DNMT3L may therefore protect certain promoters from DNMT3A-dependent de novo DNA methylation in differentiated cells. One could speculate whether DNMT3L also inhibits DNMT3A from binding to a repressive chromatin modifier(s) on Mo-MuLV sequences through a similar mechanism. However, this situation is unlikely because the interaction between DNMT3A and DNMT3L is necessary for the formation of the repressive epigenetic modifier complex, and the ectopically expressed DNMT3L in fact attracted more DNMT3A to Mo-MuLV LTR regions. We therefore favor the first hypothesis, that a critical accessory component for methylating the Mo-MuLV DNA sequences in ES cells is still missing in DNMT3L-expressing somatic cells. Further experiments contrasting DNMT3L-interacting proteins and potential regulatory RNAs between ES cells and the DNMT3L-overexpressing somatic cells may reveal the missing components necessary for introducing DNA methylation marks on newly infected Mo-MuLV proviral sequences.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate insightful discussions and comments from Deborah Bourc'his (Institute Curie, France) and the members of the S.-P.L. and T.H.B. laboratories.
This work was supported by the National Science Council (grants NSC 99-2314-B-002 −108 -MY3 and NSC 102-2321-B-002-031) and the National Taiwan University (grant 103R7602D3).
S.-P.L., T.H.B., and T.-H.K. conceived and designed the experiments; T.-H.K., H.-F.L., D.W., S.K.T.O., and C.-Y.C. performed the experiments; T.-H.K. and S.-P.L. analyzed and interpreted the data; and X.Z., X.C., K.H., S.P.G., and H.-C.K. contributed reagents, materials, and analysis tools. All authors were involved in discussing the implications of the findings; T.-H.K., S.-P.L., S.K.T.O., and T.H.B. wrote the paper.
Footnotes
Published ahead of print 2 July 2014
This article is dedicated to the memory of Daniel Wolf.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01176-14.
REFERENCES
- 1.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23:5594–5605. 10.1128/MCB.23.16.5594-5605.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaudet F, Rideout WM, III, Meissner A, Dausman J, Leonhardt H, Jaenisch R. 2004. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol. Cell. Biol. 24:1640–1648. 10.1128/MCB.24.4.1640-1648.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh CP, Chaillet JR, Bestor TH. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116–117. 10.1038/2413 [DOI] [PubMed] [Google Scholar]
- 4.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714–717. 10.1038/nature05987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, Jeltsch A. 2010. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 38:4246–4253. 10.1093/nar/gkq147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang GG, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22:480–491. 10.1128/MCB.22.2.480-491.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- 8.Bourc'his D, Bestor TH. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431:96–99. 10.1038/nature02886 [DOI] [PubMed] [Google Scholar]
- 9.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536–2539. 10.1126/science.1065848 [DOI] [PubMed] [Google Scholar]
- 10.Hata K, Okano M, Lei H, Li E. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983–1993 [DOI] [PubMed] [Google Scholar]
- 11.Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. 2000. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development 127:2955–2963 [DOI] [PubMed] [Google Scholar]
- 12.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463:237–240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- 13.Jähner D, Stuhlmann H, Stewart CL, Harbers K, Löhler J, Simon I, Jaenisch R. 1982. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 298:623–628. 10.1038/298623a0 [DOI] [PubMed] [Google Scholar]
- 14.Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, Lorincz MC. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8:676–687. 10.1016/j.stem.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pannell D, Osborne CS, Yao SY, Sukonnik T, Pasceri P, Karaiskakis A, Okano M, Li E, Lipshitz HD, Ellis J. 2000. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J. 19:5884–5894. 10.1093/emboj/19.21.5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, Pfaff SL. 2011. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 25:594–607. 10.1101/gad.2008511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464:927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- 18.Schlesinger S, Goff SP. 2013. Silencing of proviruses in embryonic cells: efficiency, stability and chromatin modifications. EMBO Rep. 14:73–79. 10.1038/embor.2012.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Gene Dev. 16:919–932. 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sripathy SP, Stevens J, Schultz DC. 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 26:8623–8638. 10.1128/MCB.00487-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultz DC, Friedman JR, Rauscher FJ., III 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 15:428–443. 10.1101/gad.869501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. 1999. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18:6385–6395. 10.1093/emboj/18.22.6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. 2013. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140:519–529. 10.1242/dev.087585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ., III 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19:4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57. 10.1016/j.cell.2007.07.026 [DOI] [PubMed] [Google Scholar]
- 26.Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458:1201–1204. 10.1038/nature07844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada F, Peters GG, Dahlberg JE. 1979. The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J. Biol. Chem. 254:10979–10985 [PubMed] [Google Scholar]
- 28.Barklis E, Mulligan RC, Jaenisch R. 1986. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell 47:391–399. 10.1016/0092-8674(86)90596-9 [DOI] [PubMed] [Google Scholar]
- 29.Feuer G, Taketo M, Hanecak RC, Fan H. 1989. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J. Virol. 63:2317–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loh TP, Sievert LL, Scott RW. 1990. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonal carcinoma cells. Mol. Cell. Biol. 10:4045–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, Goff SP, Bestor TH. 2010. Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin 3:17. 10.1186/1756-8935-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolf D, Cammas F, Losson R, Goff SP. 2008. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J. Virol. 82:4675–4679. 10.1128/JVI.02445-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumaki Y, Oda M, Okano M. 2008. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 36:W170–W175. 10.1093/nar/gkn294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman J, Cho WK, Chu CK, Keedy KS, Archin NM, Margolis DM, Karn J. 2011. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 85:9078–9089. 10.1128/JVI.00836-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, Matarese F, Stunnenberg HG, Oliviero S. 2013. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell 155:121–134. 10.1016/j.cell.2013.08.056 [DOI] [PubMed] [Google Scholar]
- 36.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. 2007. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449:248–251. 10.1038/nature06146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deplus R, Brenner C, Burgers WA, Putmans P, Kouzarides T, de Launoit Y, Fuks F. 2002. Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res. 30:3831–3838. 10.1093/nar/gkf509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakai Y, Suetake I, Shinozaki F, Yamashina S, Tajima S. 2004. Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr. Patterns 5:231–237. 10.1016/j.modgep.2004.07.011 [DOI] [PubMed] [Google Scholar]
- 39.Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554. 10.1371/journal.ppat.1000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. 2000. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol. Cell. Biol. 20:7419–7426. 10.1128/MCB.20.20.7419-7426.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golding MC, Zhang L, Mann MR. 2010. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell 6:457–467. 10.1016/j.stem.2010.03.014 [DOI] [PubMed] [Google Scholar]
- 42.He J, Yang Q, Chang LJ. 2005. Dynamic DNA methylation and histone modifications contribute to lentiviral transgene silencing in murine embryonic carcinoma cells. J. Virol. 79:13497–13508. 10.1128/JVI.79.21.13497-13508.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung DC, Lorincz MC. 2012. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem. Sci. 37:127–133. 10.1016/j.tibs.2011.11.006 [DOI] [PubMed] [Google Scholar]
- 44.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20:2536–2544. 10.1093/emboj/20.10.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, Trono D. 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 44:361–372. 10.1016/j.molcel.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, Li X. 2012. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J. Biol. Chem. 287:2107–2118. 10.1074/jbc.M111.322644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120. 10.1038/35065132 [DOI] [PubMed] [Google Scholar]
- 48.Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD, Pfeifer GP. 2006. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J. Biol. Chem. 281:19489–19500. 10.1074/jbc.M513249200 [DOI] [PubMed] [Google Scholar]
- 49.Haas DL, Lutzko C, Logan AC, Cho GJ, Skelton D, Jin Yu X, Pepper KA, Kohn DB. 2003. The Moloney murine leukemia virus repressor binding site represses expression in murine and human hematopoietic stem cells. J. Virol. 77:9439–9450. 10.1128/JVI.77.17.9439-9450.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowe HM, Trono D. 2011. Dynamic control of endogenous retroviruses during development. Virology 411:273–287. 10.1016/j.virol.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 51.Jurkowska RZ, Rajavelu A, Anspach N, Urbanke C, Jankevicius G, Ragozin S, Nellen W, Jeltsch A. 2011. Oligomerization and binding of the Dnmt3a DNA methyltransferase to parallel DNA molecules: heterochromatic localization and role of Dnmt3L. J. Biol. Chem. 286:24200–24207. 10.1074/jbc.M111.254987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlesinger S, Lee AH, Wang GZ, Green L, Goff SP. 2013. Proviral silencing in embryonic cells is regulated by Yin Yang 1. Cell Rep. 4:50–58. 10.1016/j.celrep.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.