Abstract
The Ebola virus glycoprotein mucin-like domain (MLD) is implicated in Ebola virus cell entry and immune evasion. Using cryo-electron tomography of Ebola virus-like particles, we determined a three-dimensional structure for the full-length glycoprotein in a near-native state and compared it to that of a glycoprotein lacking the MLD. Our results, which show that the MLD is located at the apex and the sides of each glycoprotein monomer, provide a structural template for analysis of MLD function.
TEXT
The filoviruses Ebolavirus (EBOV) and Marburgvirus cause fatal hemorrhagic fevers in humans, with mortality rates approaching 90% (1, 2). Consequently, considerable effort is focused on developing vaccines and therapeutics to prevent and treat filoviral infections. All filoviruses display multiple copies of a single membrane-anchored glycoprotein (GP) projecting from their envelopes (reviewed in reference 3). Like most class I viral fusion proteins, GP is a trimer in which each monomer is a disulfide-bonded complex of a receptor binding subunit (GP1) and a fusion subunit (GP2) (reviewed in references 3 and 4). The structure of the trimeric ectodomain was described as a “chalice” consisting of a glycan cap, a head, and a base (5). The Ebola virus GP is the target of multiple neutralizing antibodies (Ab), several of which are effective in preventing the onset of disease in nonhuman primates when administered as part of a monoclonal Ab (MAb) cocktail 1 or 2 days after viral exposure (6, 7).
The mucin-like domain (MLD) is a highly glycosylated region spanning EBOV GP1 residues 313 to 501 (Fig. 1A). Although it is dispensable for EBOV infections in vitro (8, 9) and is not highly conserved (5), many functions have been attributed to the MLD. These include influencing GP structure (10), enhancing viral attachment to target cell surfaces (11, 12), protecting conserved regions of GP, such the receptor binding site, from antibody recognition (10, 13), and masking immune regulatory molecules, such as major histocompatibility complex 1 (MHC1), on infected cell surfaces (14, 15). In addition, many neutralizing antibodies, including two that comprise part of a promising therapeutic cocktail (7), are directed against the MLD (13, 16, 17). The goal of the present study was to provide structural information on the MLD of GP from EBOV as an aid to understanding the multiple functions of filoviral MLDs.
FIG 1.
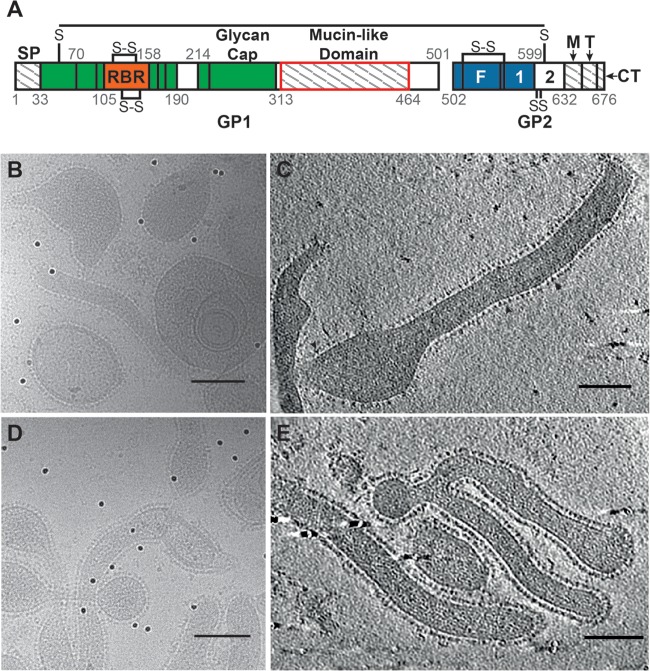
(A) A graphical representation of the Ebola virus GP sequence (adapted from references 5 and 24) shows segments present in our proteins but absent from the crystallized GP (regions with hatch marks), including the signal peptide (SP), MLD, MPER (M), and transmembrane region (T) and the cytoplasmic tail (CT). (Note that the SP is not present on the FL-GP or ΔMuc-GP VLPs, as it is cleaved off during GP biosynthesis). Domains present in the crystallized protein but not resolved in the crystal structure are shown in white and include HR2 (2) and a portion of the MLD. The segment missing from our ΔMuc-GP construct is outlined in red. Additional GP1 regions are colored green, except for the receptor binding region (RBR), which is orange. Additional GP2 regions, including the fusion peptide (F) and HR1 (1), are colored blue. (B and C) Projection image (B) and seven merged tomographic slices (C) of Ebola virus VLPs containing the FL-GP showing the various phenotypic appearances of the VLPs. GP spikes are clearly visible surrounding each particle, and the overall GP shape is visible in some well-separated GPs (indicated by arrowheads in panel C). (D and E) Projection image (D) and seven merged tomographic slices (E) showing Ebola virus VLPs containing ΔMuc-GPs. Scale bars in panels B to E represent 100 nm.
The Ebola virus GP crystal structure was obtained using a truncated protein that lacks both the MLD and the transmembrane domain (5) (Fig. 1A) and gives only a partial representation of the GP structure. To localize the MLD on Ebola virus GP in as near a native state as possible, we produced entry-competent virus-like particles (VLPs), as described previously (18), and performed cryo-electron tomography and subtomogram averaging of GP spikes using previously described methods (19).
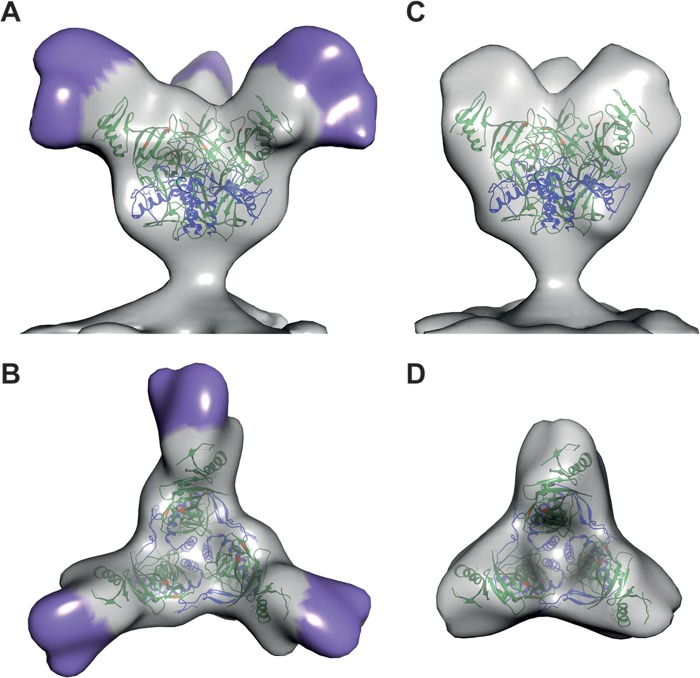
We imaged Ebola virus VLPs that express GPs in either the full-length (FL-GP) (Fig. 1B and C) or MLD-deleted (ΔMuc-GP) state (Fig. 1D and E), with the goal of identifying density corresponding to the MLD. GP spikes are continuous on the VLP surface in both particle types and are visible in two-dimensional (2D) projection images (Fig. 1B and D) and in merged tomographic slices (Fig. 1C and E). From 32 tomograms of Ebola virus FL-GP VLPs, 5,298 potential GP spikes were picked using an automated spike-picking program (20), which was modified to recognize spike density on the surface of both spherical and cylindrical VLPs. Potential spike particles were then subjected to subtomogram averaging for density map generation. The resulting FL-GP map shows a chalice-shaped, trimeric structure, as expected (5) (Fig. 2A and B). Compared to the crystal structure, extra density is visible at the apex and sides of each GP1 monomer (Fig. 2A and B, indicated by purple shading), suggesting that this is the location of the MLD. Excess density is also visible near the base of the map, which likely corresponds to the membrane-proximal external region (MPER) and the HR2 region of GP2, which were absent from or not visualized in the crystallized protein (5) (Fig. 1A).
FIG 2.
(A) Isosurface representation of the density map of FL-GP from the surface of Ebola virus VLPs shown as a side view. The density assigned to the MLD is indicated by purple shading. (B) The top view of the FL-GP map is shown with MLD indicated in purple. (C) Isosurface representation, shown in side view, of the density map of a GP spike from Ebola virus ΔMuc VLPs. (D) The top view of the ΔMuc-GP map is shown. In all panels, crystal coordinates (Protein Data Bank [PDB] 3CSY) are colored with the GP1 region in green and the GP2 region in blue. Residues important for binding to NPC1 are colored orange. All X-ray coordinates were fitted using the University of California, San Francisco (UCSF), Chimera software package (25).
To generate a density map of ΔMuc-GP for comparison, 6,304 potential GP spikes were manually picked from 29 tomograms of the ΔMuc-GP VLPs and subjected to subtomogram averaging. The ΔMuc-GP density map appears truncated compared to the FL-GP map (Fig. 2C and D), as expected, based on the reduced mass of the ΔMuc-GP protein (Fig. 1A). The ΔMuc-GP map extends ∼13 nm from the membrane compared to ∼14 nm for the FL-GP (Fig. 3A). The striking difference between the two maps in the density measurements is seen when the ΔMuc-GP map is superimposed with the FL-GP map (Fig. 3A). Density missing from the ΔMuc-GP map (and, therefore, attributable to the MLD) protrudes from the top and sides of each monomer in the spike (Fig. 3A, mesh).
FIG 3.
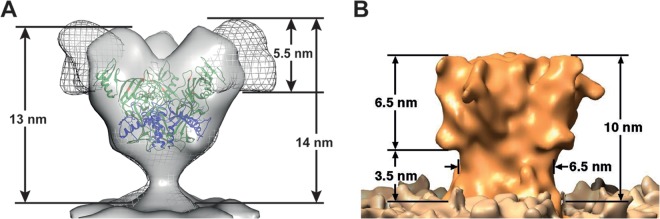
(A) A superposition of the ΔMuc-GP map (solid gray) with the FL-GP map (gray mesh) is shown. Heights of the FL and ΔMuc-GP maps (measured from the membrane to the tallest point on the map) are shown. The proposed location of the MLD corresponds to the mesh density in the FL GP map. (B) A map for Ebola virus GP, previously reported by Beniac and coworkers (21), which does not resemble the results we present here, is shown for comparison.
An alternative structure for FL-GP, also determined by cryo-electron tomography, was reported previously by Beniac and coworkers (21) (Fig. 3B). That study used a combination of Ebola virus VLPs and Ebola virus fixed with paraformaldehyde (21). In that map, very little extra density was seen extending beyond the space filled by the crystal structure, and those authors hypothesized that the MLD must fill the chalice (21). Their conclusions are in contrast to the results we present here regarding the localization of the MLD (Fig. 3), with the differences likely due to different starting materials (as described above) as well as different methods. Our map was obtained using unfixed Ebola virus VLPs and a single, proven method of cryo-EM tomography with subtomogram averaging (19). The Beniac map was derived using a combination of different Ebola virus samples and processing methods, including combining subtomogram images of Ebola virus VLPs with images of Ebola virus fixed in paraformaldehyde (21).
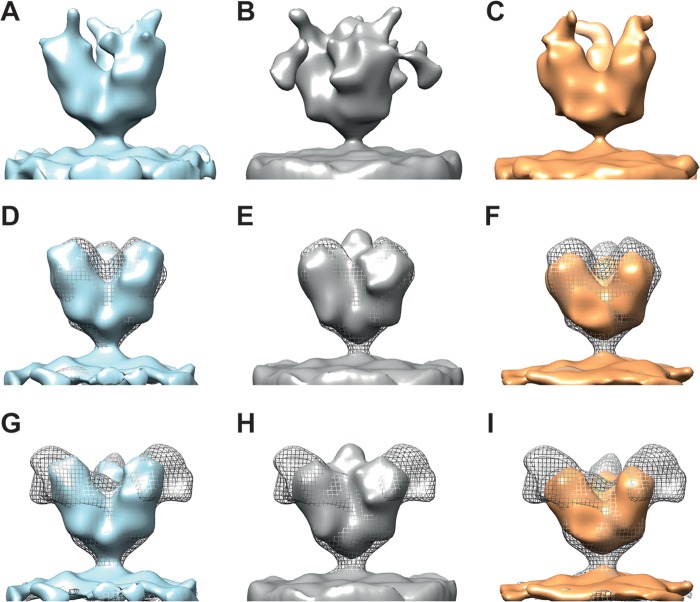
To further verify the MLD location on FL-GP, we attempted to determine the structure of FL-GP bound to the mouse antibody, 14G7, which binds MLD residues 477 to 493 (22). 14G7 is protective in 90% of Ebola virus-challenged mice when administered 1 day postchallenge (13), making this region an intriguing therapeutic target. Ebola virus VLPs were incubated with 14G7 antibody on ice for 30 to 60 min before electron microscopy (EM) grids were made and imaged. Although 14G7 binding to the FL-GP was apparent by an enzyme-linked immunosorbent assay (ELISA) (data not shown), we were unable to resolve a reproducible map containing density attributable to the anti-MLD antibody. The map classes obtained from these data show noisy density data extending from the top and sides of the maps (Fig. 4A to C). However, when the contours of the maps were adjusted so that only the strongest density was shown, the resulting maps were equivalent in size to the ΔMuc-GP map (Fig. 4D to F), indicating that neither the MLD nor the anti-MLD antibody was resolved. The lack of an MLD on these maps was verified by superimposing each map onto the FL-GP map (Fig. 4G to I). The missing MLD and antibody density is potentially a result of increased flexibility of the mucin domain upon antibody binding, making it probable that the MLD region is smeared with 3D averaging.
FIG 4.
(A to C) Isosurface representations of three individual map classes that resulted from data collected on FL Ebola virus VLP samples incubated with the 14G7 anti-MLD antibody. All maps are shown in side view. In general, the maps are noisy, with weak regions of densities extending from the top and/or sides of the apex of the spike. (D, E, and F) Maps from panels A, B, and C, respectively, are shown with contours adjusted so that only the strongest regions of density are visualized; the maps in panels D, E, and F are shown in solid blue, gray, and orange, respectively, to match the colors of the corresponding maps in panels A, B, and C. Maps obtained from antibody-bound spikes (contoured to show only the strongest regions of density) are superimposed with the ΔMuc-GP map (shown in gray mesh) for size and feature comparisons. (G to I) Solid-color maps for antibody-bound spikes shown as described for panels D to F but superimposed with the FL-GP map (shown in gray mesh).
Our results structurally define the MLD of the EBOV GP as a region surrounding the apex and sides of each GP1 monomer. This finding is significant due to its spatial relationship to epitopes that are important for cell entry and infection (23). The GP epitope bound by the Ebola virus receptor, NPC1, is buried in the center of the protein (residues shown in orange in Fig. 2) and would not be easily accessible to neutralizing antibodies due to the mass of the MLD and glycans covering the apex. Our maps visually demonstrate how the MLD shields these vulnerable epitopes from neutralization.
The FL-GP and ΔMuc-GP maps were deposited in the Electron Microscopy Data Bank (EMDB; http://www.emdatabank.org/) under the EMDB accession numbers 6003 and 6004, respectively, for the full-length and ΔMuc-GP maps.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant AI103601 to J.M.W.
We thank Steve Fellini and Susan Chacko and their colleagues for continued support with use of the Biowulf cluster for computing at NIH, Gene Olinger for supplying the 14G7 antibody, and Audray Harris and Joel Meyerson for aiding in sample preparation. We thank Mario Borgnia and Oleg Kuybeda for computational support.
Footnotes
Published ahead of print 9 July 2014
REFERENCES
- 1.Hartman AL, Towner JS, Nichol ST. 2010. Ebola and Marburg hemorrhagic fever. Clin. Lab Med. 30:161–177. 10.1016/j.cll.2009.12.001 [DOI] [PubMed] [Google Scholar]
- 2.Feldmann H, Geisbert TW. 2011. Ebola haemorrhagic fever. Lancet 377:849–862. 10.1016/S0140-6736(10)60667-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JE, Saphire EO. 2009. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 4:621–635. 10.2217/fvl.09.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219. 10.1080/10409230802058320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JE, Saphire EO. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182. 10.1038/nature07082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu X, Audet J, Wong G, Pillet S, Bello A, Cabral T, Strong JE, Plummer F, Corbett CR, Alimonti JB, Kobinger GP. 2012. Successful treatment of Ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci. Transl. Med. 4:138ra81. 10.1126/scitranslmed.3003876 [DOI] [PubMed] [Google Scholar]
- 7.Olinger GG, Jr, Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, Pauly M, Whaley KJ, Lear CM, Biggins JE, Scully C, Hensley L, Zeitlin L. 2012. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 109:18030–18035. 10.1073/pnas.1213709109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaletsky RL, Simmons G, Bates P. 2007. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 81:13378–13384. 10.1128/JVI.01170-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174–4178. 10.1128/JVI.80.8.4174-4178.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennemann NJ, Rhein BA, Ndungo E, Chandran K, Qiu X, Maury W. 2014. Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. mBio 5:e00862-13. 10.1128/mBio.00862-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marzi A Möller P, Hanna SL, Harrer T, Eisemann J, Steinkasserer A, Becker S, Baribaud F, Pöhlmann S. 2007. Analysis of the interaction of Ebola virus glycoprotein with DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin) and its homologue DC-SIGNR. J. Infect. Dis. 196(Suppl 2):S237–S246. 10.1086/520607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuno K, Nakayama E, Noyori O, Marzi A, Ebihara H, Irimura T, Feldmann H, Takada A. 2010. C-type lectins do not act as functional receptors for filovirus entry into cells. Biochem. Biophys. Res. Commun. 403:144–148. 10.1016/j.bbrc.2010.10.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. 2000. Epitopes involved in antibody-mediated protection from Ebola virus. Science 287:1664–1666. 10.1126/science.287.5458.1664 [DOI] [PubMed] [Google Scholar]
- 14.Reynard O, Borowiak M, Volchkova VA, Delpeut S, Mateo M, Volchkov VE. 2009. Ebolavirus glycoprotein GP masks both its own epitopes and the presence of cellular surface proteins. J. Virol. 83:9596–9601. 10.1128/JVI.00784-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francica JR, Varela-Rohena A, Medvec A, Plesa G, Riley JL, Bates P. 2010. Steric shielding of surface epitopes and impaired immune recognition induced by the Ebola virus glycoprotein. PLoS Pathog. 6:e1001098. 10.1371/journal.ppat.1001098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez O, Tantral L, Mulherkar N, Chandran K, Basler CF. 2011. Impact of Ebola mucin-like domain on antiglycoprotein antibody responses induced by Ebola virus-like particles. J. Infect. Dis. 204(Suppl 3):S825–S832. 10.1093/infdis/jir295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowling W, Thompson E, Badger C, Mellquist JL, Garrison AR, Smith JM, Paragas J, Hogan RJ, Schmaljohn C. 2007. Influences of glycosylation on antigenicity, immunogenicity, and protective efficacy of Ebola virus GP DNA vaccines. J. Virol. 81:1821–1837. 10.1128/JVI.02098-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoemaker CJ, Schornberg KL, Delos SE, Scully C, Pajouhesh H, Olinger GG, Johansen LM, White JM. 2013. Multiple cationic amphiphiles induce a Niemann-Pick C phenotype and inhibit Ebola virus entry and infection. PLoS One 8:e56265. 10.1371/journal.pone.0056265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8:e1002797. 10.1371/journal.ppat.1002797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6:e1001249. 10.1371/journal.ppat.1001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beniac DR, Melito PL, Devarennes SL, Hiebert SL, Rabb MJ, Lamboo LL, Jones SM, Booth TF. 2012. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS One 7:e29608. 10.1371/journal.pone.0029608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olal D, Kuehne AI, Bale S, Halfmann P, Hashiguchi T, Fusco ML, Lee JE, King LB, Kawaoka Y, Dye JM, Jr, Saphire EO. 2012. Structure of an antibody in complex with its mucin domain linear epitope that is protective against Ebola virus. J. Virol. 86:2809–2816. 10.1128/JVI.05549-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, Ruthel G, Pfeffer SR, Dye JM, Whelan SP, Brummelkamp TR, Chandran K. 2012. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 31:1947–1960. 10.1038/emboj.2012.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dube D, Brecher MB, Delos SE, Rose SC, Park EW, Schornberg KL, Kuhn JH, White JM. 2009. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J. Virol. 83:2883–2891. 10.1128/JVI.01956-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]