ABSTRACT
The hepatitis C virus (HCV) envelope glycoprotein E1E2 complex is a candidate vaccine antigen. Previous immunization studies of E1E2 have yielded various results on its ability to induce virus-neutralizing antibodies in animal models and humans. The murine model has become a vital tool for HCV research owing to the development of humanized mice susceptible to HCV infection. In this study, we investigated the antibody responses of mice immunized with E1E2 and a novel soluble form of E1E2 (sE1E2) by a DNA prime and protein boost strategy. The results showed that sE1E2 elicited higher antibody titers and a greater breadth of reactivity than the wild-type cell-associated E1E2. However, immune sera elicited by either immunogen were only weakly neutralizing. In order to understand the contrasting results of binding and serum neutralizing activities, epitopes targeted by the polyclonal antibody responses were mapped and monoclonal antibodies (MAbs) were generated. The results showed that the majority of serum antibodies were directed to the E1 region 211 to 250 and the E2 regions 421 to 469, 512 to 539, 568 to 609, and 638 to 651, instead of the well-known immunodominant E2 hypervariable region 1 (HVR1). Unexpectedly, in MAb analysis, ∼12% of MAbs isolated were specific to the conserved E2 antigenic site 412 to 423, and 85% of them cross-neutralized multiple HCV isolates. The epitopes recognized by these MAbs are similar but distinct from the previously reported HCV1 and AP33 broadly neutralizing epitopes. In conclusion, E1E2 can prime B cells specific to conserved neutralizing epitopes, but the levels of serum neutralizing antibodies elicited are insufficient for effective virus neutralization. The sE1E2 constructs described in this study can be a useful template for rational antigen engineering.
IMPORTANCE Hepatitis C virus infects 2 to 3% of the world's population and is a leading cause of liver failures and the need for liver transplantation. The virus envelope glycoprotein complex E1E2 produced by detergent extraction of cells overexpressing the protein was evaluated in a phase I clinical trial but failed to induce neutralizing antibodies in most subjects. In this study, we designed a novel form of E1E2 which is secreted from cells and is soluble and compared it to wild-type E1E2 by DNA immunization of mice. The results showed that this new E1E2 is more immunogenic than wild-type E1E2. Detailed mapping of the antibody responses revealed that antibodies to the conserved E2 antigenic site 412 to 423 were elicited but the serum concentrations were too low to neutralize the virus effectively. This soluble E1E2 provides a new reagent for studying HCV and for rational vaccine design.
INTRODUCTION
Hepatitis C virus (HCV) is a leading cause of liver cirrhosis and hepatocellular carcinoma in developed countries, with an estimated 170 million people being infected worldwide (1, 2). Of particular concern in the United States is the increasing number of cases in the 15- to 24-year-old age group, while the national rate of symptomatic HCV infection declined and began to level off in 2006 (3, 4). Standard-of-care therapy consists of pegylated alpha interferon, ribavirin, and a direct-acting antiviral (DAA), boceprevir (5, 6) or teleprevir (7), which is partially effective but has significant side effects. New DAAs are on the horizon and show great promise in replacing the interferon-based treatment in the foreseeable future (8). However, it is uncertain if affordable treatment will eventually be available to the majority of patients due to the high drug costs. To combat this global public health problem, it is imperative that more affordable drugs, as well as a broadly effective HCV vaccine to prevent new infections, be developed.
Although vaccines and therapeutic antibodies have been successfully developed to protect at-risk populations against many viral diseases, so far they have not been successful for HCV. The extreme genetic diversity of circulating HCV is a major roadblock to an HCV vaccine. The sequences of HCV isolates from different genotypes can differ by as much as 35% (9). Consequently, any given vaccine based on a single isolate is unlikely to be effective. To overcome this challenge, a broadly effective vaccine must target conserved B or T cell epitopes. To study conserved B cell epitopes, we and others have isolated murine, rat, and human monoclonal antibodies (MAbs) that can cross neutralize diverse HCV isolates (10–19). The majority of cross-neutralizing MAbs have been found to neutralize HCV by blocking the viral envelope glycoprotein E2 from binding to the HCV receptor/entry factor CD81. These MAbs include MAbs HCV1, AP33, 3/11, and HC-33 binding to the E2 antigenic site from 412 to 423 (10, 18, 20) and a group of MAbs recognizing a cluster of overlapping discontinuous epitopes forming antigenic region 3 (AR3) of the E1E2 complex (12, 16). AR3 has been mapped to the E2 regions from amino acids (aa) 396 to 424, 436 to 447, and 523 to 540 and to E2 residues S424, G523, G530, and D535 by competition enzyme-linked immunosorbent assay (ELISA) and alanine scanning mutagenesis (16). Of note, these E2 residues are also critical for E2 binding to CD81 (21). MAbs similar to AR3 antibodies recognizing epitopes with some variations in the contacting residues have also been identified by others (13–15). Another interesting murine MAb, H77.39, was reported to be able to block E2 binding to both CD81 and another HCV receptor, the scavenger receptor class B1 (SR-B1), and it was reported that E2 residues N415 and N417 are involved in the interactions (22). For E1, the human MAbs H-111 (23) and IGH526 (17) were reported to neutralize HCV by binding to the E1 antigenic sites from aa 192 to 202 and 313 to 327, respectively. Another important conserved neutralizing epitope is present on the quaternary structure of the E1E2 complex, defined by the MAb AR4A (12). The binding of MAb AR4A depends on both E1 and E2, and mutations in the E1 region from aa 201 to 206, the E2 region from aa 657 to 659, and E2 aa 692 that disrupt E1E2 complex formation abolish binding of this MAb. Protection against heterologous HCV isolated by the AR3- and AR4-specific MAbs and MAb HCV1 has been demonstrated in the humanized mouse and chimpanzee model, respectively (12, 24). These new findings underscore the fact that, despite the extreme genetic diversity, conserved B cell epitopes are present on HCV for rational vaccine design.
The E1E2 glycoprotein complex has been evaluated as a vaccine candidate in a phase I clinical trial (25–27). Early immunization studies in the chimpanzee model provided proof that HCV infection can be prevented and controlled by antibodies to E1E2 (28). However, protection was achieved only for autologous virus challenge and not heterologous virus challenge. Immunizations with E1E2 formulated in MF59 adjuvant for the elicitation of neutralizing antibodies (NAbs) have produced various results, ranging from good immunogenicity in guinea pigs to modest immunogenicity in mice and humans (25, 26, 29, 30). E1E2 had also been presented on virus-like particles (VLPs) (31) and retroviral particles (32) or delivered by recombinant measles viruses (33), where modest levels of cross-NAbs were reported in some of the studies. Overall, the E1E2 complex is an attractive vaccine candidate, although improvement of its ability to elicit broadly NAbs (bNAbs) is required for it to be useful as a broadly effective HCV vaccine. Currently, only limited information on the specificities of antibody responses following immunization with the glycoproteins is available. A better understanding of the antigenic and immunogenic properties of E1E2 will provide useful information for improving E1E2 as a vaccine antigen. The mouse model was utilized in this study because several humanized mouse models for HCV infection have now been developed (34–38), whereas the chimpanzee model is no longer available for academic research. Consequently, the humanized mouse models will likely be used in most preclinical HCV vaccine studies; therefore, a detailed characterization of murine antibody responses to HCV antigens is warranted.
In this study, we took advantage of recent advances in HCV antibody studies and investigated the possibility of designing a soluble form of the E1E2 complex that maintains the quaternary fold of the complex. This complex will be useful as a template for engineering immunogens to focus B cell responses to conserved neutralizing epitopes on the complex. We also studied the specificities of antibody responses elicited by immunization with the full-length and soluble E1E2, isolating new MAbs specific to the highly conserved E2 antigenic region from aa 412 to 423 and other immunodominant regions.
MATERIALS AND METHODS
Cloning and expression of E1E2 envelope glycoprotein complex.
The nucleotide DNA sequence of genotype 1a isolate H77 E1E2 (GenBank accession number AF009606) was codon optimized (GenBank accession number KJ701242) (GenScript) and used as a template for generation of the expression plasmids shown in Fig. 1A. To generate soluble E1E2 (sE1E2) constructs, the nucleotide sequences of the ectodomains of E1 and E2 were amplified and joined together by splicing overlapping extension PCR and then cloned into the pCMVtpa expression vector (pCMVtpa_sE1E2) (39). In the pCMVtpa_sE1E2v4 plasmid construct, a tobacco etch virus (TEV) cleavage site and a Strep-tag II were introduced between the E1 and E2 ectodomains. The removal of the transmembrane domains (TMDs) enables the expressed protein to be secreted from the cell. Soluble constructs were generated by cotransfection of 293T cells with pAdvantage (Promega) and the corresponding expression plasmids encoding the engineered E1E2 genes at a 1:1 ratio by polyethylenimine (Polysciences Inc.). Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (Invitrogen) and kifunensine (Callaghan Innovation Research) at a final concentration of 10 μM was used as the growth medium for expression of the soluble constructs. Supernatants were harvested at 72 h posttransfection and analyzed by ELISA and Western blotting for the presence of sE1E2.
FIG 1.
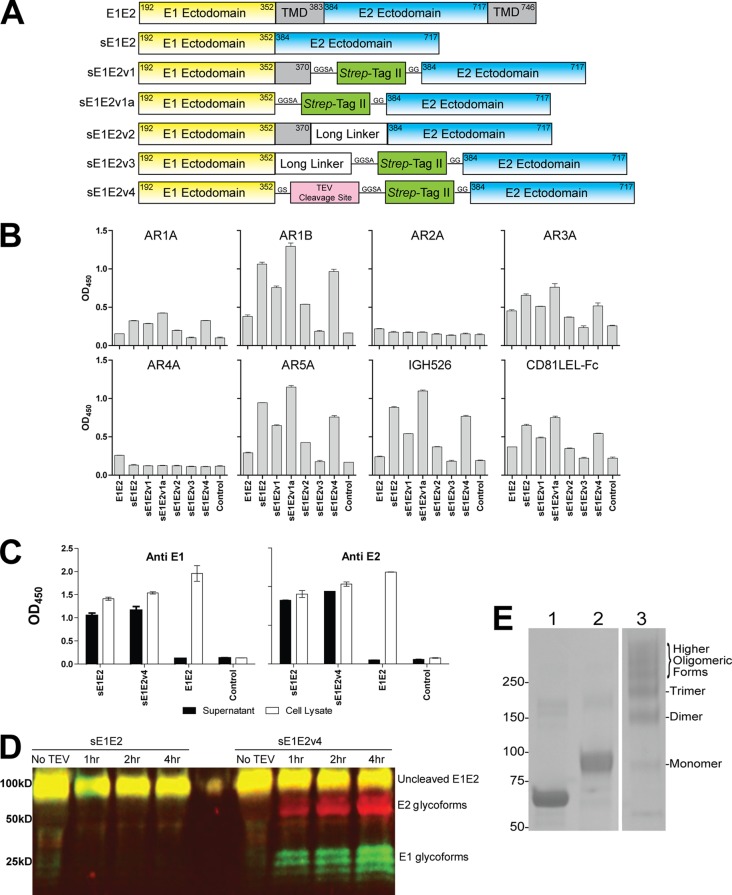
Characterization of sE1E2 constructs. (A) Schematic illustration of 6 sE1E2 constructs. The numbering corresponds to the amino acid positions in the polyprotein of the genotype 1a H77 strain. TMD, transmembrane domain; Strep-Tag II sequence, WSHPQFEK; long linker sequence, GGSSRSSSSGGGGSGGGG; TEV cleavage site sequence, GENLYFQ. (B) Antigenicity of sE1E2. The sE1E2 constructs were expressed in 293T cells by transient transfection in the presence of kifunensine and captured from the cell supernatants onto microwells by MAb AR4A. Captured sE1E2 was probed by MAbs AR1A, AR1B, AR2A, AR3A, AR4A, AR5A, IGH526, and CD81 LEL-Fc. As expected, poor binding was observed for the cell-associated wild-type E1E2 and for detection with MAb AR4A, which was the capture antibody. However, a binding signal was not detected for MAb AR2A, suggesting the absence of this isolate-specific E2 epitope on the engineered sE1E2 complexes. OD450, optical density at 450 nm. (C) sE1E2, sE1E2v4, and E1E2 in cell supernatant and cell lysate. The expressed glycoproteins were captured onto ELISA plates precoated with MAb AR4A and detected with anti-E1 MAb IGH526 and anti-E2 MAb AR1B, as described for panel B. (D) Immunoblot analysis of sE1E2 and sE1E2v4. Soluble E1E2 was treated with TEV protease at 30°C for the specified times, and digested samples were resolved in a reducing SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane for immunoblotting. Mouse anti-E1 MAb A4 and human anti-E2 MAb HCV1 primary antibodies, followed by IRDye800CW anti-mouse (green signal) and IRDye700DX anti-human (red signal) secondary antibodies, were used. Codetection of E1 and E2 generated a yellow signal. (E) SDS-PAGE analysis of purified sE1E2v4. Five micrograms of sE1E2v4 proteins was resolved by reducing (lane 2) and nonreducing (lane 3) SDS-PAGE, and bovine serum albumin (BSA; reduced) (lane 1) was used as a control. The relative abundance of the sE1E2v4 oligomeric forms was determined by densitometry using ImageJ software, and it was found that the sE1E2v4 preparation contained 5%, 19%, 19%, and 37% monomers, dimers, trimers, and higher oligomers, respectively. The numbers on the left are molecular masses (in kilodaltons).
Purification of sE1E2.
sE1E2v4 was expressed by transient transfection of 293F cells (Life Technologies) and was purified from the cell supernatant using an antibody-affinity column based on MAb AR3A (11). The glycoproteins were analyzed by reducing and nonreducing SDS-PAGE, and the gel was stained with Coomassie-based SimplyBlue SafeStain (Life Technologies). The protein species were quantified on the basis of the results for the bovine serum albumin control by densitometry using the ImageJ program (v1.47), obtained from the NIH website (http://rsbweb.nih.gov/ij/download.html).
Experimental animals.
Female specific-pathogen-free inbred BALB/c mice were purchased from The Scripps Research Institute (TSRI) breeding colony at 5 to 6 weeks of age. Animals were housed in ventilated microisolator cages under environmentally controlled conditions at the TSRI animal facility in compliance with AAALAC guidelines and an approved IACUC protocol.
Immunization protocol.
DNA vaccines for E1E2 and sE1E2 were delivered by in vivo electroporation using a TriGrid delivery system (for rodents) developed by Ichor Medical Systems, Inc. (40). A previous in-house study demonstrated that the efficacy of this method in eliciting antibody responses to HIV-1 gp120 is comparable to that of eliciting antibody responses to proteins in adjuvant (39). Blood samples were taken prior to immunization (prebleed samples). Each animal was injected with a 40 μg/dose of DNA plasmid in the two tibialis anterior muscles. Animals were immunized at 2-week intervals, and blood was drawn at 7 days postinjection. A final protein boost with a purified truncated E2 glycoprotein (isolate H77 residues 412 to 645) at 50 μg/animal was administered intraperitoneally without adjuvant, and terminal bleed samples were collected 7 days later while the animals were under anesthesia.
Hybridoma fusion protocol.
The spleens and lymph nodes of immunized mice were removed aseptically, and lymphocytes were harvested after lysing red blood cells by osmotic shock. Hybridomas were produced following standard techniques using SP2/0-AG14 myelomas (ATCC CRL-1581) as fusion partners and polyethylene glycol (molecular mass, 1,500 kDa) as the fusogen. Hybridoma cultures were maintained in RPMI 1640 medium (Life Technologies) supplemented with 1 mM pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM GlutaMAX, 100 μM sodium hypoxanthine, 16 μM thymidine, and 10% fetal calf serum (Invitrogen). Culture supernatant from wells with cell growth was screened by ELISA using an H77 E1E2-transfected cell lysate as the solid-phase antigen. Aliquots of stable H77 E1E2 glycoprotein-specific hybridomas were frozen in a mixture of 10% (vol/vol) dimethyl sulfoxide and 90% low-IgG fetal calf serum for storage in liquid nitrogen.
Antibody isotyping.
Antibodies were isotyped using a commercial isotyping kit (Thermo Scientific). Briefly, ELISA strip wells were precoated with anti-mouse heavy-chain capture antibody (anti-IgG1, IgG2a, IgG2b, IgG3, IgA, and IgM) or anti-mouse light-chain capture antibody (kappa or lambda). Diluted mouse MAb culture supernatant premixed with goat anti-mouse IgG-IgA-IgM-horseradish peroxidase (HRP) conjugate was added to the wells, and the plates were incubated for 1 h and washed before the addition of the chromogenic substrate. The results were recorded by measuring the absorbance of the plates at 450 nm.
Serum and antibody characterization.
To study whether the antibodies recognized continuous or discontinuous epitopes, E1E2 antigens were captured onto ELISA wells precoated with 5 μg/ml Galanthus nivalis lectin. Native proteins or proteins that had been reduced with 0.1% SDS–50 mM dithiothreitol at 100°C for 5 min were captured onto ELISA wells by the lectin. Binding of antibodies to the folded and unfolded proteins was detected by peroxidase-conjugated goat anti-mouse IgG secondary antibody (The Jackson Laboratory) at 10 μg/ml. Mouse MAb A4, specific for the E1 linear epitope from aa 198 to 207 (41), was used as a positive control for unfolded proteins.
The specificity of the antibodies to E1E2 or E2 was determined by incubating the MAbs with E1E2 or E2 antigens without the transmembrane region (soluble E2) precaptured by lectin onto microwells. Soluble E2 antigens were produced in the cell supernatant from 293T cells transfected with an expression plasmid encoding the soluble E2 cDNA (E2 from aa 384 to 717 or E2 with a deletion of the transmembrane region). After 1 h of incubation, the plates were washed, and bound antibodies were detected with HRP-conjugated goat anti-mouse F(ab′)2 fragment antibody (1:2,000; The Jackson Laboratory) and tetramethylbenzidine (TMB) substrate (Pierce).
To study sera and MAb cross-reactivity to other HCV genotypes, ELISA was performed as described above with native and reduced E1E2 cell lysates from isolates H77 (genotype 1a), J6E3 (genotype 2a), UKN3A1.28, UKN4.11.1, UKN5.15.7, and UKN6.5.34.
To map the serum antibody responses to different antigenic regions on E1E2 (12, 16), a saturating concentration of blocking MAbs (20 μg/ml) was added to the E1E2 antigens (isolate H77) that had been captured by precoated G. nivalis lectin (5 μg/ml; Vector Laboratories) for 30 min before the addition of the detecting MAbs. E1E2 antigens were prepared from cell lysates from 293T cells transfected with the H77 E1E2 expression plasmid. Nonfat milk (4%; Bio-Rad) in phosphate-buffered saline (PBS) and 0.05% Tween 20 (PBST-M) was used as a blocker in assays using lectin-captured antigens. The ELISA plates were washed after 1 h incubation, and binding of the detecting antibodies was detected by HRP-conjugated goat anti-mouse IgG Fc antibody (1:2,000; The Jackson Laboratory) in 1% milk–PBST and TMB substrate (Pierce). The level of inhibition by the blocking antibody was calculated as the percent reduction of the optical density signals produced by the detecting antibody in the presence of the blocking antibody.
To study the ability of the MAbs to inhibit E1E2 binding to CD81, the CD81 long external loop (LEL) was immobilized onto ELISA plates at 5 μg/ml to capture 293T cell lysates containing H77 E1E2 in the presence of MAbs. Plates were blocked with PBST-M, and biotinylated MAbs A4 and AR2A, which do not block E2 binding to CD81, were used as detecting MAbs. MAb AR3A, recognizing an epitope overlapping with CD81 binding on E2, was used as a control.
The apparent affinities (50% effective concentrations [EC50s]) of the antibodies were calculated by fitting a dose-response curve to the levels of binding of serially diluted antibodies to lectin-captured E1E2 antigens using GraphPad Prism software.
HCV neutralization assays.
HCV pseudotype particles (HCVpp) were generated by cotransfection of 293T cells with pNL4-3.lucR-E- (42, 43) and the corresponding expression plasmids encoding the E1E2 genes at a 4:1 ratio by polyethylenimine. The E1E2 expression plasmids for isolates H77, UKN1b12.6, and J6E3 have been described previously (44–46). Virus infectivity was detected with the firefly luciferase assay system (Promega), and percent neutralization was calculated as the residual virus infectivity at the antibody concentrations indicated below divided by the infectivity without antibody after background subtraction. The background infectivity of the pseudotype virus was defined by infecting cells with virus made with pNL4-3.lucR-E- only. Pseudotype virus particles displaying the vesicular stomatitis virus envelope glycoprotein G (VSVpp) were a control for nonspecific neutralizing activity. The virus was incubated with the diluted sera/antibodies for 1 h at 37°C before it was added to Huh-7 cell monolayers and was removed after 6 h of incubation. Expression of the luciferase reporter gene in the infected cells was measured at 3 days postinfection.
For cell culture HCV (HCVcc) neutralization, HCV isolate JFH-1 was propagated using Huh-7.5.1 cells as described previously (47). The MAbs were titrated 4-fold from 20 μg/ml and incubated with HCVcc for 1 h at 37°C. The virus-antibody mixture was added to 7 × 103 Huh-7.5.1 cells/well for 6 h and replaced with fresh medium. After 3 days of incubation, cell monolayers were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with PBS–10% fetal calf serum. Virus neutralization was determined by measurement of the reduction of the number of infectious foci. Infectious foci were visualized by indirect immunofluorescent staining using MAb AR3A (2 μg/ml) and Alexa 555-conjugated anti-human IgG (1 μg/ml) (Molecular Probes) with a fluorescence microscope.
Mapping of antibody epitopes by peptide ELISA.
Epitope mapping was done using a series of overlapping peptides covering the E1E2 region of the HCV isolate H77 sequence. The peptides consisted of 18-mers with an overlap of 11 amino acids covering the E1E2 HCV glycoprotein region (HCV type 1a H77 Peptides-Complete Set, catalog no. 7620, lot no. 4, provided by the NIH AIDS Research and Reference Reagent Program). Briefly, plates were coated with 5 μg/ml of peptide, blocked with 4% PBST-M, and incubated with sera/MAbs, and binding was detected in an ELISA format as described above.
RESULTS
Immunogen design.
The E1 and E2 glycoproteins are known to form a complex (E1E2) via noncovalent interactions of their transmembrane domains (48). The E1E2 complex in previous immunization studies has been produced as membrane-associated glycoproteins and was solubilized and purified in the presence of nonionic detergents (25, 28). Recently, we demonstrated that the ectodomains of E1 and E2 are involved in complex formation, with the N-terminal region of E1 interacting with the membrane-proximal external region of E2 (12). Specific mutations in either E1 or E2 can abolish the binding of MAbs recognizing nonoverlapping epitopes present on the quaternary structure of the complex. Given that both flaviviruses and alphaviruses use class II fusion proteins for viral entry, it seems that HCV also likely uses the class II fusion protein for entry. In flaviviruses, the envelope protein E is responsible for both receptor binding and fusion (49). In alphaviruses, two envelope glycoproteins are used instead (50). Since HCV has two envelope glycoproteins forming heterodimers, we propose that the ectodomains of HCV E1 and E2 interact in a manner reminiscent of that of the E2–E1 complexes of the alphaviruses (50, 51). In structural studies of the alphavirus envelope glycoprotein complex, the complex can be expressed as a soluble complex by replacing the transmembrane domain between E1 and E2 with a short linker. Following a similar strategy, we designed a number of soluble E1E2 (sE1E2) constructs (Fig. 1A). The expression of the soluble complexes was analyzed by a capture ELISA with the MAb AR4A and by detection with MAbs specific for the E1 and E2 epitopes and the receptor CD81. MAb AR4A neutralizes HCV by binding to an epitope which is found exclusively on the E1E2 complex and not E1 or E2 per se (12). The constructs sE1E2, sE1E2v1a, and sE1E2v4 expressed soluble E1E2 complexes with more favorable binding properties than the sE1E2 constructs sE1E2v1, sE1E2v2, and sE1E2v3 with a partially truncated E1 transmembrane region or an 18-residue linker inserted between the E1 and E2 ectodomains (Fig. 1B). The secreted forms of E1E2 (sE1E2 and sE1E2v4) were more readily detected in the cell supernatants than full-length E1E2, which required detergent extraction to release the glycoproteins (Fig. 1C). Surprisingly, a folded sE1E2 complex can be obtained by simply deleting the transmembrane domains of E1 and E2 without a flexible linker (construct sE1E2). In the construct sE1E2v4, a tobacco etch virus (TEV) protease cleavage site and a Strep-tag II were inserted between the E1 and E2 ectodomains. Figure 1D shows that sE1E2v4 but not sE1E2 can be cleaved into E1 and E2 by TEV protease. In SDS-PAGE analysis, purified sE1E2v4 showed multiple oligomeric forms under nonreducing conditions. Densitometric analysis showed that sE1E2v4 existed mostly as dimers (19%), trimers (19%), and higher oligomers (37%), with ∼5% being present as monomers (Fig. 1E). Although we think that the quaternary epitopes (AR4A and AR5A) are present on each sE1E2 molecule, the current data do not eliminate the possibility that the epitopes are formed by two or more interacting sE1E2 proteins. Further protein engineering is under way to improve protein homogeneity for studying the presentation of the neutralizing epitopes on the soluble complex and for structural studies.
Immunization and antiserum analysis.
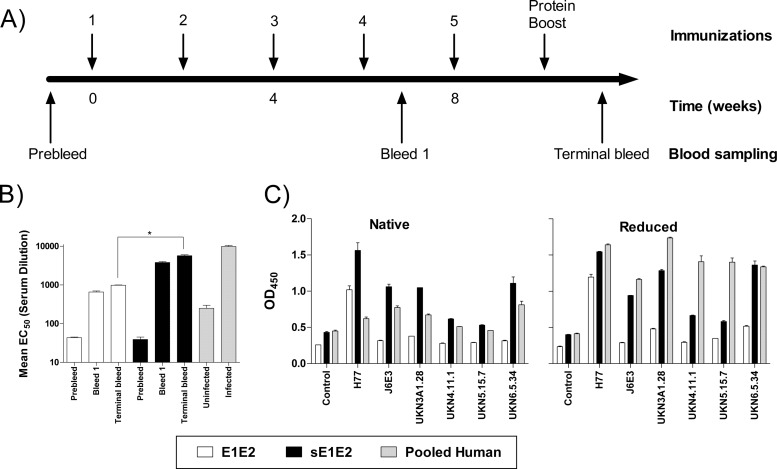
Wild-type E1E2 and sE1E2 were chosen for DNA immunizations. Mice were immunized 4 times biweekly with 40 μg of the DNA constructs by in vivo electroporation (39, 40), followed by a protein boost of 50 μg of a truncated form of soluble E2 (aa 412 to 645). Blood samples were collected 1 week after the fourth and fifth immunizations (Fig. 2A). The serum antibody titers were determined by ELISA, and the titers of antibody to E1E2 were calculated as the half-maximal antibody titer (EC50) (Fig. 2B). The terminal serum titration showed that immunization with sE1E2 and wild-type E1E2 resulted in antibody titers of ∼5,700 and 1,000, respectively. The same assay detected a titer of ∼9,800 in the control pooled human serum (from 5 chronic patients), and the antibody had good virus-neutralizing activity.
FIG 2.
(A) Schematic representation of the study design. Mice were immunized with DNA vaccine at biweekly intervals and boosted with purified E2 protein (arrows), and blood samples obtained at the indicated time points were analyzed for humoral responses. (B) Antibody titers of pooled mouse and human sera to E1E2. Sera were serially diluted, and their reactivity against E1E2 lysate was determined by ELISA. The EC50s of the polyclonal antibody responses were calculated from the titration curve, and values are the means from at least 3 independent experiments. *, P < 0.05). (C) Reactivity of immune sera (diluted 1:300) to native and reduced E1E2. The data shown are the mean values from at least 2 experiments.
Since HCV is genetically diverse, the characterization of epitopes important for protection or antibody escape will provide useful information for the rational design of immunogens for eliciting bNAbs rather than antibodies to immunodominant nonneutralizing epitopes. To address this, we assessed how genotypic variation affected serum reactivity in native and reduced forms of E1E2 glycoproteins (Fig. 2C). As expected, the immune responses were the highest against the homologous H77 E1E2 in both the reduced and the nonreduced forms. E1E2-immunized mice showed restricted reactivity, as they reacted mainly with H77 E1E2. In contrast, antisera from the sE1E2 group had more diverse reactivity across the genotypes. The human sera had moderate binding to native glycoproteins but had noticeably higher reactivity to denatured E1E2. The results suggest that, during chronic HCV infection, unfolded E1 or E2 is presented to the immune system, eliciting antibodies to epitopes that are not found on native E1E2. Overall, the immunization regimen was effective in eliciting anti-E1E2 antibodies in mice.
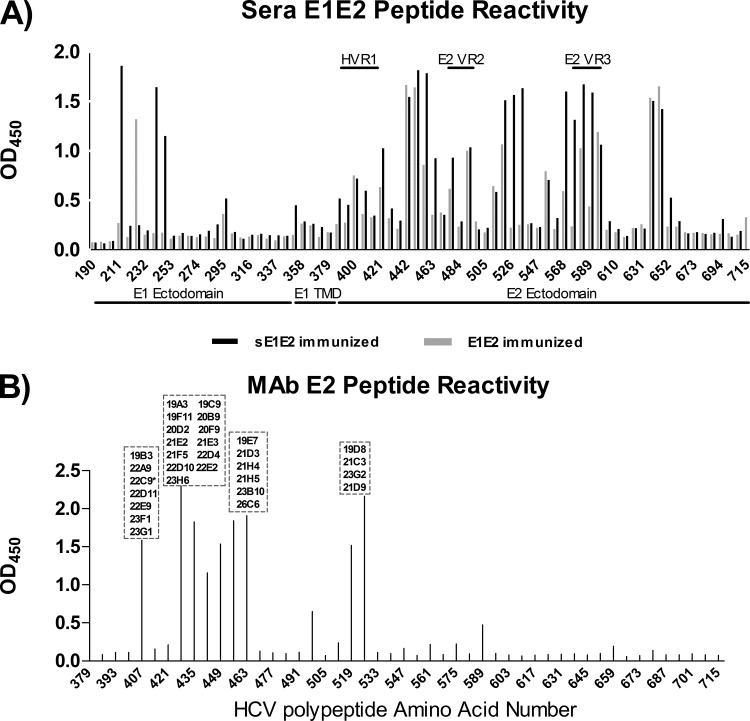
The specificities of the antibody responses to continuous and discontinuous epitopes were mapped by overlapping peptides and competition ELISA. To map antibody responses to continuous epitopes, a library of E1E2 overlapping peptides (18-mers with 11-amino-acid overlaps) was used in an ELISA (Fig. 3A). On the E1 region, wild-type E1E2-immunized mice reacted only with aa 225 to 231, while sE1E2 reacted with aa 211 to 217 and aa 239 to 250. There was little reactivity with the rest of the continuous epitopes in the E1 ectodomain and transmembrane domain in all the immunized groups. The mice showed greater reactivity toward the E2 glycoprotein than the E1 glycoprotein. Wild-type E1E2-immunized mice reacted with aa 442 to 460, aa 582 to 602, and aa 638 to 651, while sE1E2-immunized mice reacted with aa 442 to 469, aa 519 to 539, aa 568 to 59,5 and aa 638 to 651. Both groups had little reactivity toward hypervariable region 1 (HVR1) aa 384 to 411 and E2 variable region 2 (VR2) aa 460 to 485, although sE1E2-immunized mice had reactivity against E2 variable region 3 (VR3) aa 570 to 580.
FIG 3.
Reactivity of sera from E1E2- and sE1E2-immunized animals (A) and MAbs (B) to a library of overlapping E1E2 peptides in ELISA. The MAbs not shown did not bind to the peptides in the ELISA and were assumed to bind discontinuous epitopes. All 7 neutralizing MAbs except 23D9 were mapped to the region from amino acids 412 to 423 of the E1E2 glycoprotein. *, MAb 22C9 was also mapped to this region but was nonneutralizing. Peptides were supplied by the NIH AIDS Research and Reference Reagent Program. Data shown are representative of those from 3 independent experiments performed in duplicate.
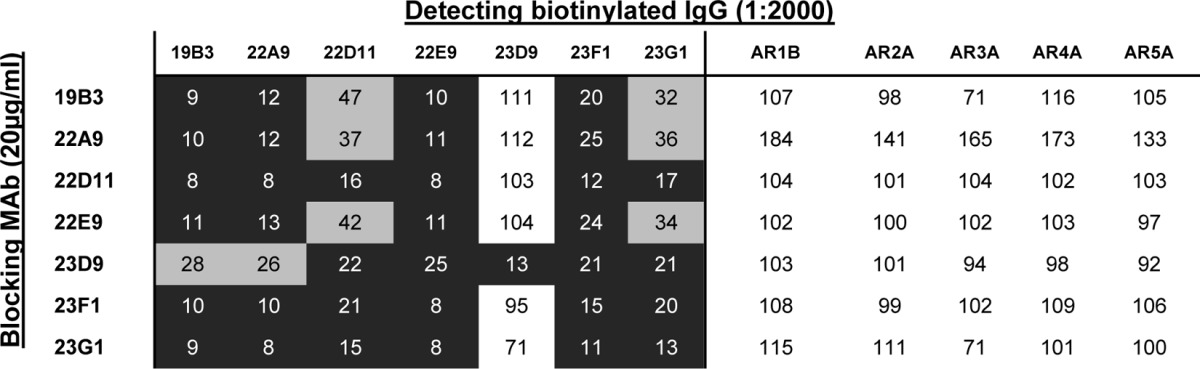
To map antibody specificities to discontinuous epitopes, a previously reported panel of MAbs that recognize 5 distinct antigenic regions (AR1 to AR5) on the HCV E1E2 glycoprotein (12, 16) was used in competition ELISA (Table 1). The neutralizing human sera had the most reactivity toward the AR3A, AR4A, AR2A, and AR5A regions in decreasing order of reactivity. They showed little competition with AR1B antibody. The wild-type E1E2-immunized mice did not compete with the MAb panel, while sE1E2-immunized mice competed with MAbs AR3A and AR4A.
TABLE 1.
Reactivity of sera to AR1 to AR5 on E1E2a
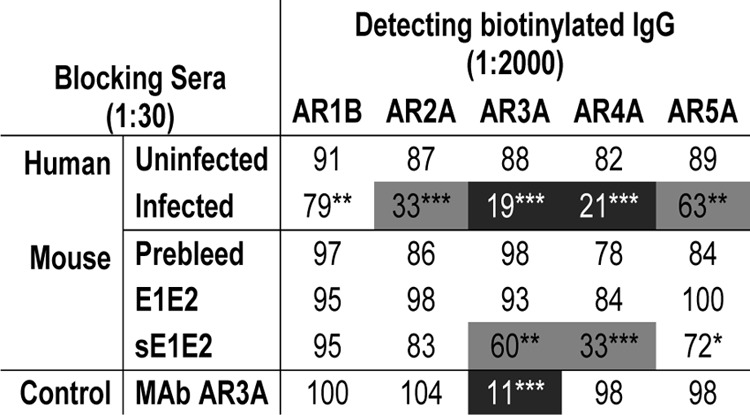
The data shown are representative of those from at least 3 experiments. Numbers indicate the percentage of the residual binding signals of biotinylated human MAbs in the presence of blocking sera, and the extent of competition (residual binding) is highlighted by shading: black background, 0 to 25%; gray background, 26 to 70%; and white background, >70%. Statistical analyses were performed using a two-tailed Student's t test, P values of <0.1 were considered significant. *, P values of between 0.05 and 0.1; **, P values of between 0.01 and 0.05; ***, P values of <0.01). Antibodies to antigenic regions 1 to 5 have been described previously (11, 52).
To assess neutralizing activity, preimmune and terminal bleed sera (diluted 1:20) were tested against a panel of HCV pseudotype virus particles (HCVpp). Weak to moderate neutralizing activity (up to 46% neutralization) was detected in the immune sera of E1E2- and sE1E2-immunized mice (preimmune sera had no noticeable neutralization relative to that for the negative control) (Table 2). In comparison, at a 1:100 dilution pooled human sera from infected individuals neutralized HCVpp >70%.
TABLE 2.
Serum neutralization of HCVppa
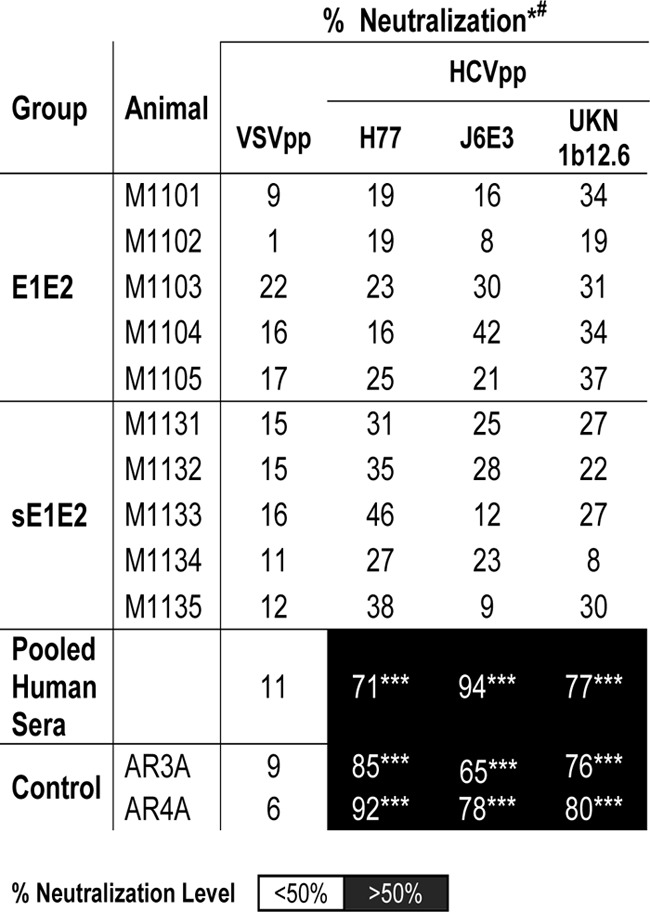
*, sera were assayed at a 1:20 dilution for mice and a 1:100 dilution for human. The data shown are the mean percent neutralization values from 3 independent experiments with duplicate measurements. White background, neutralization level of <50%; black background, neutralization level of >50%. Statistical analyses were done using a two-tailed Student's t test. P values of <0.1 were considered significant. ***, P values of <0.01. #, there was no statistically significant difference between the neutralization levels before and after the E2 protein boost (data not shown).
Overall, the results indicate that sE1E2 elicits a stronger antibody response than wild-type E1E2 in DNA vaccination, yet the serum neutralizing activities are similar for both antigens. The results are consistent with those of previous studies where animals immunized with E2 engineered to be expressed on the cell surface or secreted had antibody responses better than those of rodents and macaques to wild-type E1E2 produced by DNA immunizations (30, 52, 53).
MAb analysis.
The antibody responses were studied further at the monoclonal level. Splenocytes and lymphocytes in lymph tissues from the animals with the highest titers were used in a classical polyethylene glycol fusion with mouse SP2/0-AG14 myeloma cells to generate MAbs. A total of 37 hybridoma clones were obtained and produced MAbs with κ light chains; 33 of these secreted IgG1 antibodies, 3 secreted IgG2a antibodies, and 1 secreted IgG2b antibodies. All MAbs bound E2: 27 of them reacted only with the immunizing antigen (H77, genotype 1a), whereas 10 cross-reacted with heterologous viral genotypes (Table 3). Eleven MAbs recognized discontinuous epitopes, and 26 MAbs recognized continuous epitopes. In pepscan analysis, 7 MAbs bound to peptides corresponding to E2 aa 407 to 424, 19 bound to the immunodominant region from aa 428 to 469, and 4 bound to aa 512 to 539 (Fig. 3B). MAb 21B3 bound to multiple peptides from aa 428 to 445, aa 456 to 480, and aa 589 to 606, while MAb 23D9 bound to peptides aa 428 to 452 and aa 589 to 606. Five MAbs (19F11, 20F9, 21E2, 21E3, and 21F5) that bound poorly to denatured E2 were reactive in the pepscan assay, indicating that their epitopes are partly discontinuous. In terms of conformational epitope specificities, the MAb panel did not compete strongly with our in-house human MAbs specific for AR1 to AR5 (data not shown). The MAbs in hybridoma supernatants were screened for neutralization, and 20% of them neutralized >50% of the infectivity of HCVpp displaying the homologous H77 E1E2 (Table 3). Several MAbs were also found to neutralize heterologous HCV strains UKN1b12.6 (genotype 1b) and J6E3 (genotype 2a) but not the negative control, VSVpp. All the neutralizing MAbs except MAb 23D9 reacted with multiple genotypes in ELISA and bound both reduced and nonreduced forms of E1E2.
TABLE 3.
MAbs to E1E2 isolated from immunized animalsa
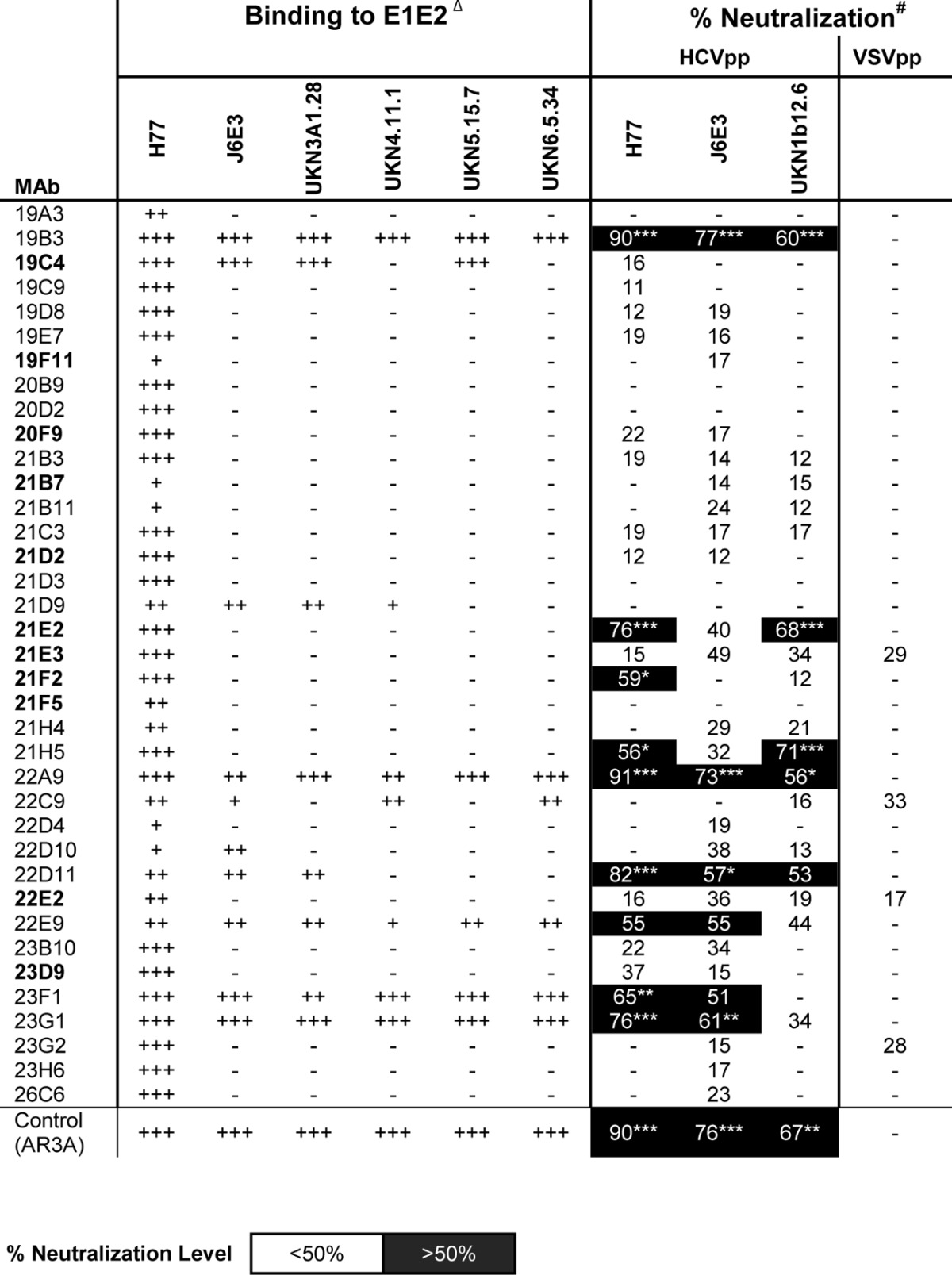
Δ, MAbs in bold recognize discontinuous epitopes on E2. +++, strong binding (optical density at 450 nm, >1.5) to E1E2; ++, moderate binding (optical density at 450 nm, <1.5 and >0.6) to E1E2; +, weak binding (optical density at 450 nm, <0.6) to E1E2; −, no appreciable binding/neutralization detected. Data are representative of those from at least 2 experiments in duplicate. #, neutralization assay against a pseudotype virus panel consisting of vesicular stomatitis virus envelope glycoprotein G (control) and E1E2 of HCV isolates H77 (genotype 1a), UKN1B12.16 (genotype 1b), and J6E3 (genotype 2a) that produce signals consistently at least 10-fold higher than the background signal induced by the control pseudotype virus generated without HCV Env cDNA. Antibodies that did not neutralize more than 50% at a 1:5 supernatant dilution were considered negative. The data shown are the mean percent neutralization values from 3 independent experiments with duplicate measurements. Neutralization levels of less than 50% were considered statistically insignificant in this study. Statistical analyses were done using a two-tailed Student's t test, and P values of <0.1 were considered significant. *, P values of between 0.05 and 0.1; **, P values of between 0.01 and 0.05; ***, P values of <0.01).
Seven MAbs (19B3, 22A9, 22D11, 22E9, 23D9, 23F1, and 23G1) with neutralization potential in the preliminary assessment were chosen for further characterization. The binding properties of the purified antibodies are summarized in Tables 4 and 5. All 7 MAbs were E2 specific, and all except 23D9 cross-reacted with other genotypes. Interestingly, 6 of the NAbs recognized the conserved E2 antigenic site 412 to 423 (Fig. 3B). In a competition ELISA, the MAbs indeed competed for the same E2 binding site for bNAbs HCV1 and AP33 (10, 18, 54–56). The MAbs did not compete with MAbs AR1 to AR5 but competed against each other, indicating that they bind to the same antigenic region on E2 (Table 4). Interestingly, MAb 23D9 competed strongly only when used as a blocking antibody, suggesting that it could bind E2 outside aa 412 to 423 when it was blocked by other MAbs. The exact discontinuous epitope has yet to be determined. At high concentrations (∼10 μg/ml), the MAbs interfered with E1E2 binding to CD81.
TABLE 4.
Competition ELISA of murine MAbs with human MAbs recognizing five antigenic regionsa
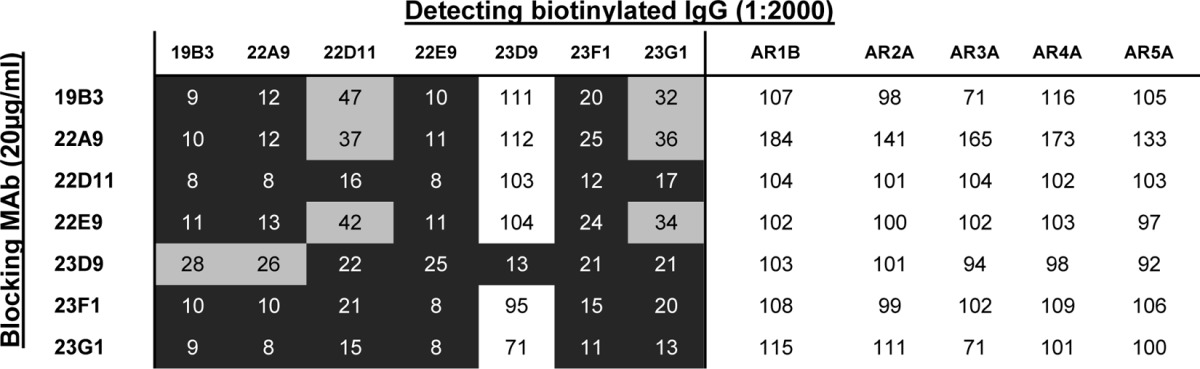
Numbers indicate the percent residual binding signals of biotinylated MAbs in the presence of blocking mouse MAbs, and the extent of competition (residual binding) is highlighted by shading: black background, 0 to 25%; gray background, 26 to 70%; and white background, >70%.
TABLE 5.
HCV-neutralizing MAbsa
| MAb | Isotype | Specificity | Genotype reactivity | Epitope | Apparent affinity (nM) |
|---|---|---|---|---|---|
| 19B3 | IgG2a | E2 | Broad | Continuous | 0.2 |
| 22A9 | IgG1 | E2 | Broad | Continuous | 0.4 |
| 22D11 | IgG1 | E2 | Broad | Continuous | 1.3 |
| 22E9 | IgG1 | E2 | Broad | Continuous | 0.7 |
| 23D9 | IgG1 | E2 | 1a only | Discontinuous | 6.6 |
| 23F1 | IgG1 | E2 | Broad | Continuous | 1.2 |
| 23G1 | IgG1 | E2 | Broad | Continuous | 3.2 |
No competition with AR1-5 MAbs was detected (Table 4), and E1E2 binding to CD81 was weakly blocked.
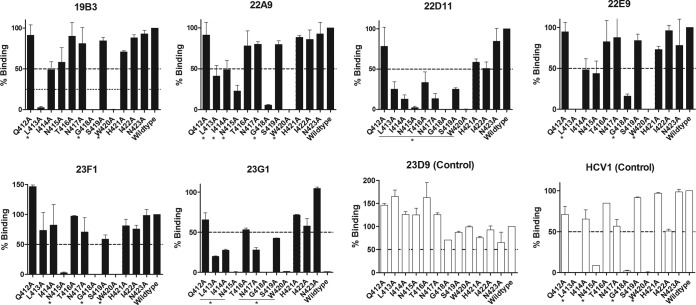
The E2 antigenic site 412 to 423 is highly conserved and a known target for the cross neutralization of HCV genotypes by NAbs (10, 18). Alanine scanning mutagenesis was performed in this site to map the epitopes of the new MAbs (Fig. 4 and Table 6). The results showed that the MAbs 19B3 and 22E9 required L413, G418, and W420 for binding, whereas the MAbs 22A9 and 23F1 mainly required N415, G418, and W420 for binding. The MAbs 22D11 and 23G1 were sensitive to most mutations between L413 and W420. The results showed distinct patterns on the residues critical for binding, though all MAbs required G418 and W420. The data show that these MAbs recognize distinct yet overlapping neutralizing epitopes in this antigenic site originally defined by the bNAbs HCV1 and AP33 (10, 18, 21, 54–56).
FIG 4.
E2 residues required for MAb binding to the conserved E2 antigenic site 412 to 423. Expressed proteins with the indicated amino acid mutated to alanine were analyzed in an E1E2 capture ELISA. Bound antibodies were detected by peroxidase-conjugated goat anti-mouse IgG secondary antibody. The binding of antibody to each mutant is expressed as a percentage of the binding to H77 wild-type protein. Residual binding of <50% was defined as positive, and the critical residues are denoted with asterisks. Note the clear dependence of the contacting residues L413, N415, G418, and W420 for the binding of MAb HCV1, as revealed in the X-ray structure (55). Data shown are representative of those from 3 independent experiments performed in duplicate.
TABLE 6.
Summary of HCV-neutralizing MAbsa
Serial dilutions of MAbs from 20 to 0.02 and 50 to 1 μg/ml were tested for neutralization of HCVcc and HCVpp, respectively. The amount of antibody required for 50% and 90% (in parentheses) neutralization was calculated from titration data. −, the MAb did not neutralize >50% at 50 μg/ml. The 50% inhibitory concentrations of MAbs are highlighted by shading: black background, <1 μg/ml; gray background, 1 to 10 μg/ml; light gray background, >10 μg/ml. None of the MAbs neutralized the control (VSVpp). Residues G418 and W420 were critical for binding by all the MAbs. Data are representative of those from at least 3 independent experiments done in duplicate. N.D., not determined.
DISCUSSION
An effective vaccine against HCV will be one of the most effective and economical tools to combat this important human pathogen. Previous studies have demonstrated that eliciting NAbs to HCV by immunization can prevent virus infection. However, the protection was restricted to homologous virus and ineffective against heterologous virus challenge (28). Despite the extreme genetic diversity of the virus, bNAbs to HCV have recently been shown to protect the humanized mouse and the chimpanzee models against heterologous virus in passive transfer experiments (12, 16, 24), suggesting that it may be possible to develop a vaccine with broad coverage of HCV genotypes by targeting highly conserved immune epitopes.
Elicitation of bNAbs against highly variable viruses remains a great challenge in vaccine research. Early immunization studies in chimpanzees using E1E2 glycoproteins expressed by HeLa cells infected with recombinant vaccinia virus expression vector (28, 57) and with E1E2 expressed in Chinese hamster ovary cell lines (27, 58) induced antibodies that mediated various levels of neutralization in different animal models (30, 59). In human vaccine trials, the vaccine candidate E1E2 glycoproteins have been found to be only mildly immunogenic, inducing weak to modest NAb responses (25–27, 60). Clearly, a better understanding of the quality and specificity of antibody responses following immunization is warranted to inform further improvement of the vaccine antigens. With the recent development of humanized mouse models potentially useful for HCV vaccine research (34–38), we aimed at elucidating the specificity of murine antibody responses to E1E2 immunization in this study.
Our laboratory has developed a large panel of E1E2-specific MAbs and alanine scanning mutants (12, 16) that enables us to study polyclonal and monoclonal antibody specificities in detail. DNA immunization provides a convenient platform that can be used to study immune responses to HCV glycoproteins expressed endogenously. Here we assessed the ability of the cell-associated form and a soluble form of E1E2 to elicit NAbs by DNA immunization. Plasmids expressing full-length E1E2 and soluble versions of E1E2 were constructed and used to immunize mice. Previously, it has been shown that cell surface-expressed E1 and E2 glycoproteins are better immunogens than the wild-type antigens (52, 61, 62). It was also true in this study that sE1E2 had a greater breath of reactivity than wild-type E1E2, probably due to the better chance of it being captured by both local and distal antigen presentation cells, thus increasing its immunogenicity (30, 52, 53, 62). The antibody titers observed here were quite high in comparison to those seen in an earlier study which induced anti-HCV seroconversion at a lower rate (53). This is likely a result of the use of in vivo electroporation technology. Serum responses to E1 glycoprotein were limited, in agreement with the poor immunogenicity of E1 observed in previous studies (63). After boosting with E2 glycoprotein, the overall titers against E1E2 did not increase significantly, suggesting that the low E1 responses were not caused by boosting with E2. On the basis of the results, it appears that E1E2 immunization can elicit NAbs but not at levels sufficient to mediate effective serum neutralization of HCV.
E2 HVR1 (aa 384 to 410) has been suggested to be an immunodominant region involved in viral escape of antibodies in infected chimpanzees and humans (27, 64–66). We did not observe a high reactivity toward this region in the mouse sera, similar to the results of previous mouse studies (52, 53). It seems that HVR1 may not be as immunogenic as was previously thought and that strong antibody responses might not be required to select for mutations in this region. Perhaps its main role is to shield other conserved E2 epitopes underneath (67, 68).
The sera competed significantly with MAbs AR3A (16) and AR4A (12) in binding to E1E2, suggesting that antibodies to the discontinuous antigenic regions were elicited. However, only weak neutralization of HCV was observed in the E1E2- and sE1E2-immunized groups, suggesting that those antibodies were either of low affinity or bound to sites near but outside the neutralizing epitopes.
Despite moderate serum reactivity toward the E2 antigenic site from aa 412 to 423, 7 of the 37 MAbs bound to this site and 6 of them were neutralizing. This is a surprising result, as these antibodies were expected to be relatively rare on the basis of the serum analysis (Fig. 3A). The results suggest a discrepancy between the level of serum antibodies (which are produced mainly by plasma cells in bone marrow) and the frequency of peripheral memory B cells encoding the specified antibodies used in hybridoma fusion. The results are in agreement with the report by Purtha et al. that the memory B cell compartment can possess antigen specificities different from those of the long-lived plasma cells in the host and the two cell compartments may serve two different and nonredundant roles in B cell immunity (69). Comparing the E1E2- and sE1E2-immunized mice, sE1E2 elicited antibodies with a broader epitope coverage than E1E2. The immune sera reacted dominantly with the E1 region 211 to 250 and the E2 regions 421 to 469, 512 to 539, 568 to 609, and 638 to 651. It was previously reported that antibodies to the so-called epitope II at aa 434 to 446 could inhibit virus neutralization by antibodies specific to aa 412 to 426 (70). However, recent studies reported a contradictory conclusion that antibodies to this E2 region can effectively neutralize HCV (20, 71, 72). In this study, MAbs to this region did not neutralize HCV (e.g., 20B9, 20F9, 21F5, 22E2, and 23H6; Table 3).
Previously, mapping experiments suggested that the CD81-binding site on E2 is formed by at least three E2 regions (aa 412 to 424, 436 to 447, and 523 to 540) (16, 21, 73). Interestingly, MAbs to the last two E2 regions are nonneutralizing, suggesting that neither region was presented in the correct conformation during immunization, thus eliciting only nonneutralizing antibodies. Nevertheless, bNAbs specific to discontinuous epitopes in the E2 CD81-binding site have frequently been isolated by us and others (13, 15, 16), suggesting that the correct conformation of this site is immunogenic during an infection. Therefore, an E2 or E1E2 engineered to stabilize the three E2 regions forming the CD81-binding site may promote the production of NAbs after vaccination.
Lastly, murine and human antibodies to the HCV E2 antigenic site 412 to 423 have been isolated previously (10, 18, 74). The human MAb HCV1 has received recent attention, as it neutralizes a broad range of HCV genotypes (10) and was used to prevent and treat HCV infection in chimpanzees (24). Its structure has been solved, demonstrating that the E2 residues L413, N415, G418, and W420 are critical for antibody binding (10, 55). Another bNAb MAb, AP33, recognizing the same antigenic site requires similar binding residues and I414 (55, 56, 74), whereas N415, W420, and H421 are required by the mildly neutralizing MAb 3/11 (11, 74). In alanine scanning mutagenesis analysis, our MAb panel had diverse binding patterns to E2 412 to 424, although G418 and W420 were required for all MAbs, indicating that multiple distinct neutralizing epitopes are present in this highly conserved E2 antigenic site. Of note, W420 is 100% conserved in all HCV isolates, and it is a critical residue for virus binding to CD81 (21).
The results presented above provide useful information on the antigenicity of E1E2. The sE1E2 construct designed in this study, which does not require detergent extraction of cells during production, can be a useful template for rational engineering for improved presentation of conserved neutralizing epitopes.
ACKNOWLEDGMENTS
We thank Ichor Medical Systems, Inc., for providing the in vivo electroporation TriGrid system, Takaji Wakita for clone JFH-1, Frank Chisari for Huh-7.5.1 cells, Georg Lauer for human HCV-neutralizing sera, and Dennis Burton for advice and support of this study.
Support for this work was provided by NIH grants R01AI079031 and R01AI106005.
Footnotes
Published ahead of print 25 June 2014
This is TSRI manuscript number 24067.
REFERENCES
- 1.Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 17:107–115. 10.1111/j.1469-0691.2010.03432.x [DOI] [PubMed] [Google Scholar]
- 2.Shepard CW, Finelli L, Alter MJ. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558–567. 10.1016/S1473-3099(05)70216-4 [DOI] [PubMed] [Google Scholar]
- 3.Wasley A, Miller JT, Finelli L. 2007. Surveillance for acute viral hepatitis—United States, 2005. MMWR Surveill. Summ. 56(SS-03):1–24 [PubMed] [Google Scholar]
- 4.Leuchner L, Lindstrom H, Burstein GR, Mulhern KE, Rocchio EM, Johnson G, Schaffzin J, Smith P. 2008. Use of enhanced surveillance for hepatitis C virus infection to detect a cluster among young injection-drug users—New York, November 2004-April 2007. (Reprinted from MMWR 57:517–521, 2008). JAMA 300:34–36. 10.1001/jama.300.1.34 [DOI] [PubMed] [Google Scholar]
- 5.Kwo PY. 2012. Boceprevir: a novel nonstructural 3 (NS3) protease inhibitor for the treatment of chronic hepatitis C infection. Ther. Adv. Gastroenterol. 5:179–188. 10.1177/1756283X11436317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwo PY, Lawitz EJ, McCone J, Schiff ER, Vierling JM, Pound D, Davis MN, Galati JS, Gordon SC, Ravendhran N, Rossaro L, Anderson FH, Jacobson IM, Rubin R, Koury K, Pedicone LD, Brass CA, Chaudhri E, Albrecht JK, SPRINT-1 Investigators 2010. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet 376:705–716. 10.1016/S0140-6736(10)60934-8 [DOI] [PubMed] [Google Scholar]
- 7.McHutchison JG, Manns MP, Muir AJ, Terrault NA, Jacobson IM, Afdhal NH, Heathcote EJ, Zeuzem S, Reesink HW, Garg J, Bsharat M, George S, Kauffman RS, Adda N, Di Bisceglie AM. 2010. Telaprevir for previously treated chronic HCV infection. N. Engl. J. Med. 362:1292–1303. 10.1056/NEJMoa0908014 [DOI] [PubMed] [Google Scholar]
- 8.Gane E. 2012. Future perspectives: towards interferon-free regimens for HCV. Antivir. Ther. 17:1201–1210. 10.3851/IMP2431 [DOI] [PubMed] [Google Scholar]
- 9.Kuiken C, Simmonds P. 2009. Nomenclature and numbering of the hepatitis C virus. Methods Mol. Biol. 510:33–53. 10.1007/978-1-59745-394-3_4 [DOI] [PubMed] [Google Scholar]
- 10.Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD, Jr, Szabo G, Finberg RW, Ambrosino DM, Babcock GJ. 2009. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J. Virol. 83:12473–12482. 10.1128/JVI.01138-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating JA. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. 2012. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 109:6205–6210. 10.1073/pnas.1114927109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson DX, Voisset C, Tarr AW, Aung M, Ball JK, Dubuisson J, Persson MA. 2007. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 104:16269–16274. 10.1073/pnas.0705522104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keck ZY, Li TK, Xia JM, Gal-Tanamy M, Olson O, Li SH, Patel AH, Ball JK, Lemon SM, Foung SKH. 2008. Definition of a conserved immunodominant domain on hepatitis C virus E2 glycoprotein by neutralizing human monoclonal antibodies. J. Virol. 82:6061–6066. 10.1128/JVI.02475-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, Li AY, Patel AH, Lemon SM, Bukh J, Rey FA, Foung SK. 2012. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 8:e1002653. 10.1371/journal.ppat.1002653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14:25–27. 10.1038/nm1698 [DOI] [PubMed] [Google Scholar]
- 17.Meunier JC, Russell RS, Goossens V, Priem S, Walter H, Depla E, Union A, Faulk KN, Bukh J, Emerson SU, Purcell RH. 2008. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the E1 glycoprotein of hepatitis C virus. J. Virol. 82:966–973. 10.1128/JVI.01872-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owsianka A, Tarr AW, Juttla VS, Lavillette D, Bartosch B, Cosset FL, Ball JK, Patel AH. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79:11095–11104. 10.1128/JVI.79.17.11095-11104.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schofield DJ, Bartosch B, Shimizu YK, Allander T, Alter HJ, Emerson SU, Cosset FL, Purcell RH. 2005. Human monoclonal antibodies that react with the E2 glycoprotein of hepatitis C virus and possess neutralizing activity. Hepatology 42:1055–1062. 10.1002/hep.20906 [DOI] [PubMed] [Google Scholar]
- 20.Keck Z, Wang WY, Wang Y, Lau P, Carlsen THR, Prentoe J, Xia JM, Patel AH, Bukh J, Foung SKH. 2013. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J. Virol. 87:37–51. 10.1128/JVI.01941-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owsianka AM, Timms JM, Tarr AW, Brown RJ, Hickling TP, Szwejk A, Bienkowska-Szewczyk K, Thomson BJ, Patel AH, Ball JK. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695–8704. 10.1128/JVI.00271-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabo MC, Luca VC, Prentoe J, Hopcraft SE, Blight KJ, Yi M, Lemon SM, Ball JK, Bukh J, Evans MJ, Fremont DH, Diamond MS. 2011. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J. Virol. 85:7005–7019. 10.1128/JVI.00586-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keck ZY, Sung VMH, Perkins S, Rowe J, Paul S, Liang TJ, Lai MMC, Foung SKH. 2004. Human monoclonal antibody to hepatitis C virus E1 glycoprotein that blocks virus attachment and viral infectivity. J. Virol. 78:7257–7263. 10.1128/JVI.78.13.7257-7263.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morin TJ, Broering TJ, Leav BA, Blair BM, Rowley KJ, Boucher EN, Wang Y, Cheslock PS, Knauber M, Olsen DB, Ludmerer SW, Szabo G, Finberg RW, Purcell RH, Lanford RE, Ambrosino DM, Molrine DC, Babcock GJ. 2012. Human monoclonal antibody HCV1 effectively prevents and treats HCV infection in chimpanzees. PLoS Pathog. 8:e1002895. 10.1371/journal.ppat.1002895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frey SE, Houghton M, Coates S, Abrignani S, Chien D, Rosa D, Pileri P, Ray R, Di Bisceglie AM, Rinella P, Hill H, Wolff MC, Schultze V, Han JH, Scharschmidt B, Belshe RB. 2010. Safety and immunogenicity of HCV E1E2 vaccine adjuvanted with MF59 administered to healthy adults. Vaccine 28:6367–6373. 10.1016/j.vaccine.2010.06.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law JL, Chen C, Wong J, Hockman D, Santer DM, Frey SE, Belshe RB, Wakita T, Bukh J, Jones CT, Rice CM, Abrignani S, Tyrrell DL, Houghton M. 2013. A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS One 8:e59776. 10.1371/journal.pone.0059776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray R, Meyer K, Banerjee A, Basu A, Coates S, Abrignani S, Houghton M, Frey SE, Belshe RB. 2010. Characterization of antibodies induced by vaccination with hepatitis C virus envelope glycoproteins. J. Infect. Dis. 202:862–866. 10.1086/655902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choo QL, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C. 1994. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 91:1294–1298. 10.1073/pnas.91.4.1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ray R, Khanna A, Lagging LM, Meyer K, Choo QL, Ralston R, Houghton M, Becherer PR. 1994. Peptide immunogen mimicry of putative E1 glycoprotein-specific epitopes in hepatitis C virus. J. Virol. 68:4420–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamataki Z, Coates S, Evans MJ, Wininger M, Crawford K, Dong C, Fong YL, Chien D, Abrignani S, Balfe P, Rice CM, McKeating JA, Houghton M. 2007. Hepatitis C virus envelope glycoprotein immunization of rodents elicits cross-reactive neutralizing antibodies. Vaccine 25:7773–7784. 10.1016/j.vaccine.2007.08.053 [DOI] [PubMed] [Google Scholar]
- 31.Elmowalid GA, Qiao M, Jeong SH, Borg BB, Baumert TF, Sapp RK, Hu ZY, Murthy K, Liang TJ. 2007. Immunization with hepatitis C virus-like particles results in control of hepatitis C virus infection in chimpanzees. Proc. Natl. Acad. Sci. U. S. A. 104:8427–8432. 10.1073/pnas.0702162104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrone P, Fluckiger AC, Mangeot PE, Gauthier E, Dupeyrot-Lacas P, Mancip J, Cangialosi A, Du Chene I, LeGrand R, Mangeot I, Lavillette D, Bellier B, Cosset FL, Tangy F, Klatzmann D, Dalba C. 2011. A prime-boost strategy using virus-like particles pseudotyped for HCV proteins triggers broadly neutralizing antibodies in macaques. Sci. Transl. Med. 3:94ra71. 10.1126/scitranslmed.3002330 [DOI] [PubMed] [Google Scholar]
- 33.Reyes-del Valle J, de la Fuente C, Turner MA, Springfeld C, Apte-Sengupta S, Frenzke ME, Forest A, Whidby J, Marcotrigiano J, Rice CM, Cattaneo R. 2012. Broadly neutralizing immune responses against hepatitis C virus induced by vectored measles viruses and a recombinant envelope protein booster. J. Virol. 86:11558–11566. 10.1128/JVI.01776-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, Kay MA, Finegold M, Grompe M. 2007. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat. Biotechnol. 25:903–910. 10.1038/nbt1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, Law M, Rice CM, Ploss A. 2011. A genetically humanized mouse model for hepatitis C virus infection. Nature 474:208–211. 10.1038/nature10168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7:927–933. 10.1038/90968 [DOI] [PubMed] [Google Scholar]
- 37.Meuleman P, Libbrecht L, De Vos R, de Hemptinne B, Gevaert K, Vandekerckhove J, Roskams T, Leroux-Roels G. 2005. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology 41:847–856. 10.1002/hep.20657 [DOI] [PubMed] [Google Scholar]
- 38.Washburn ML, Bility MT, Zhang L, Kovalev GI, Buntzman A, Frelinger JA, Barry W, Ploss A, Rice CM, Su L. 2011. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology 140:1334–1344. 10.1053/j.gastro.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law M, Cardoso RM, Wilson IA, Burton DR. 2007. Antigenic and immunogenic study of membrane-proximal external region-grafted gp120 antigens by a DNA prime-protein boost immunization strategy. J. Virol. 81:4272–4285. 10.1128/JVI.02536-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luxembourg A, Hannaman D, Ellefsen B, Nakamura G, Bernard R. 2006. Enhancement of immune responses to an HBV DNA vaccine by electroporation. Vaccine 24:4490–4493. 10.1016/j.vaccine.2005.08.014 [DOI] [PubMed] [Google Scholar]
- 41.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. 10.1006/viro.1995.1016 [DOI] [PubMed] [Google Scholar]
- 43.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartosch B, Dubuisson J, Cosset FL. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J. Exp. Med. 197:633–642. 10.1084/jem.20021756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. U. S. A. 100:7271–7276. 10.1073/pnas.0832180100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavillette D, Tarr AW, Voisset C, Donot P, Bartosch B, Bain C, Patel AH, Dubuisson J, Ball JK, Cosset FL. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265–274. 10.1002/hep.20542 [DOI] [PubMed] [Google Scholar]
- 47.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299. 10.1073/pnas.0503596102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Op De Beeck A, Montserret R, Duvet S, Cocquerel L, Cacan R, Barberot B, Le Maire M, Penin F, Dubuisson J. 2000. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J. Biol. Chem. 275:31428–31437. 10.1074/jbc.M003003200 [DOI] [PubMed] [Google Scholar]
- 49.Modis Y, Ogata S, Clements D, Harrison SC. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79:1223–1231. 10.1128/JVI.79.2.1223-1231.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468:705–708. 10.1038/nature09546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712. 10.1038/nature09555 [DOI] [PubMed] [Google Scholar]
- 52.Forns X, Emerson SU, Tobin GJ, Mushahwar IK, Purcell RH, Bukh J. 1999. DNA immunization of mice and macaques with plasmids encoding hepatitis C virus envelope E2 protein expressed intracellularly and on the cell surface. Vaccine 17:1992–2002. 10.1016/S0264-410X(98)00448-4 [DOI] [PubMed] [Google Scholar]
- 53.Tedeschi V, Akatsuka T, Shih JWK, Battegay M, Feinstone SM. 1997. A specific antibody response to HCV E2 elicited in mice by intramuscular inoculation of plasmid DNA containing coding sequences for E2. Hepatology 25:459–462. 10.1002/hep.510250234 [DOI] [PubMed] [Google Scholar]
- 54.Kong L, Giang E, Nieusma T, Robbins JB, Deller MC, Stanfield RL, Wilson IA, Law M. 2012. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J. Virol. 86:13085–13088. 10.1128/JVI.01939-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong L, Giang E, Robbins JB, Stanfield RL, Burton DR, Wilson IA, Law M. 2012. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc. Natl. Acad. Sci. U. S. A. 109:9499–9504. 10.1073/pnas.1202924109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potter JA, Owsianka AM, Jeffery N, Matthews DJ, Keck ZY, Lau P, Foung SK, Taylor GL, Patel AH. 2012. Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J. Virol. 86:12923–12932. 10.1128/JVI.02052-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo QL. 1993. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J. Virol. 67:6753–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brazzoli M, Helenius A, Foung SKH, Houghton M, Abrignani S, Merola M. 2005. Folding and dimerization of hepatitis C virus E1 and E2 glycoproteins in stably transfected CHO cells. Virology 332:438–453. 10.1016/j.virol.2004.11.034 [DOI] [PubMed] [Google Scholar]
- 59.Meunier JC, Gottwein JM, Houghton M, Russell RS, Emerson SU, Bukh J, Purcell RH. 2011. Vaccine-induced cross-genotype reactive neutralizing antibodies against hepatitis C virus. J. Infect. Dis. 204:1186–1190. 10.1093/infdis/jir511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meyer K, Banerjee A, Frey SE, Belshe RB, Ray R. 2011. A weak neutralizing antibody response to hepatitis C virus envelope glycoprotein enhances virus infection. PLoS One 6:e23699. 10.1371/journal.pone.0023699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cocquerel L, Meunier JC, Op de Beeck A, Bonte D, Wychowski C, Dubuisson J. 2001. Coexpression of hepatitis C virus envelope proteins E1 and E2 in cis improves the stability of membrane insertion of E2. J. Gen. Virol. 82:1629–1635 [DOI] [PubMed] [Google Scholar]
- 62.Forns X, Emerson SU, Tobin GJ, Mushahwar IK, Purcell RH, Bukh J. 1999. DNA immunization of mice and macaques with plasmids encoding hepatitis C virus envelope E2 protein expressed intracellularly and on the cell surface. Vaccine 17:1992–2002 [DOI] [PubMed] [Google Scholar]
- 63.Fournillier A, Wychowski C, Boucreux D, Baumert TF, Meunier JC, Jacobs D, Muguet S, Depla E, Inchauspe G. 2001. Induction of hepatitis C virus E1 envelope protein-specific immune response can be enhanced by mutation of N-glycosylation sites. J. Virol. 75:12088–12097. 10.1128/JVI.75.24.12088-12097.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A, Purcell RH, Alter HJ. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339–344. 10.1126/science.288.5464.339 [DOI] [PubMed] [Google Scholar]
- 65.Lu L, Tatsunori N, Li CH, Waheed S, Gao FX, Robertson BH. 2008. HCV selection and HVR1 evolution in a chimpanzee chronically infected with HCV-1 over 12 years. Hepatol. Res. 38:704–716. 10.1111/j.1872-034X.2008.00320.x [DOI] [PubMed] [Google Scholar]
- 66.Shimizu YK, Hijikata M, Iwamoto A, Alter HJ, Purcell RH, Yoshikura H. 1994. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 68:1494–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, Zeisel MB, Baumert TF, Keck ZY, Foung SKH, Pecheur EI, Pietschmann T. 2010. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J. Virol. 84:5751–5763. 10.1128/JVI.02200-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prentoe J, Jensen TB, Meuleman P, Serre SBN, Scheel TKH, Leroux-Roels G, Gottwein JM, Bukh J. 2011. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J. Virol. 85:2224–2234. 10.1128/JVI.01594-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Purtha WE, Tedder TF, Johnson S, Bhattacharya D, Diamond MS. 2011. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J. Exp. Med. 208:2599–2606. 10.1084/jem.20110740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang P, Wu CG, Mihalik K, Virata-Theimer ML, Yu MYW, Alter HJ, Feinstone SM. 2007. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc. Natl. Acad. Sci. U. S. A. 104:8449–8454. 10.1073/pnas.0703039104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krey T, Meola A, Keck ZY, Damier-Piolle L, Foung SK, Rey FA. 2013. Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog. 9:e1003364. 10.1371/journal.ppat.1003364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tarr AW, Urbanowicz RA, Jayaraj D, Brown RJ, McKeating JA, Irving WL, Ball JK. 2012. Naturally occurring antibodies that recognize linear epitopes in the amino terminus of the hepatitis C virus E2 protein confer noninterfering, additive neutralization. J. Virol. 86:2739–2749. 10.1128/JVI.06492-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drummer HE, Boo I, Maerz AL, Poumbourios P. 2006. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J. Virol. 80:7844–7853. 10.1128/JVI.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tarr AW, Owsianka AM, Timms JM, McClure CP, Brown RJ, Hickling TP, Pietschmann T, Bartenschlager R, Patel AH, Ball JK. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592–601. 10.1002/hep.21088 [DOI] [PubMed] [Google Scholar]