ABSTRACT
Geminivirus AL2/C2 proteins play key roles in establishing infection and causing disease in their plant hosts. They are involved in viral gene expression, counter host defenses by suppressing transcriptional gene silencing, and interfere with the host signaling involved in pathogen resistance. We report here that begomovirus and curtovirus AL2/C2 proteins interact strongly with host geminivirus Rep-interacting kinases (GRIKs), which are upstream activating kinases of the protein kinase SnRK1, a global regulator of energy and nutrient levels in plants. We used an in vitro kinase system to show that GRIK-activated SnRK1 phosphorylates recombinant AL2/C2 proteins from several begomoviruses and to map the SnRK1 phosphorylation site to serine-109 in the AL2 proteins of two New World begomoviruses: Cabbage Leaf Curl Virus (CaLCuV) and Tomato mottle virus. A CaLCuV AL2 S109D phosphomimic mutation did not alter viral DNA levels in protoplast replication assays. In contrast, the phosphomimic mutant was delayed for symptom development and viral DNA accumulation during infection of Arabidopsis thaliana, demonstrating that SnRK1 contributes to host defenses against CaLCuV. Our observation that serine-109 is not conserved in all AL2/C2 proteins that are SnRK1 substrates in vitro suggested that phosphorylation of viral proteins by plant kinases contributes to the evolution of geminivirus-host interactions.
IMPORTANCE Geminiviruses are single-stranded DNA viruses that cause serious diseases in many crops. Dicot-infecting geminiviruses carry genes that encode multifunctional AL2/C2 proteins that are essential for infection. However, it is not clear how AL2/C2 proteins are regulated. Here, we show that the host protein kinase SnRK1, a central regulator of energy balance and nutrient metabolism in plants, phosphorylates serine-109 in AL2 proteins of three subgroups of New World begomoviruses, resulting in a delay in viral DNA accumulation and symptom appearance. Our results support SnRK1's antiviral role and reveal a novel mechanism underlying this function. Phylogenetic analysis suggested that AL2 S109 evolved as begomoviruses migrated from the Old World to the New World and may have provided a selective advantage as begomoviruses adapted to a different environment and different plant hosts. This study provides new insights into the interaction of viral pathogens with their plant hosts at the level of viral protein modification by the host.
INTRODUCTION
Establishment of a host infection by a pathogen is determined by the outcomes of interactions between the virulence factors produced by the pathogen and the defense response of the host. A delicate balance of these interactions is necessary for pathogens to sustain their propagation and for their hosts to survive infection (1). The Geminiviridae is an ancient family of single-stranded DNA viruses that infect diverse plant species around the world and cause serious diseases in many important crops (for reviews, see references 2 to 5). Geminiviruses typically carry genes that encode 5 to 7 proteins that interact with a wide array of host proteins to reprogram plant cell cycle and transcriptional controls, interfere with cell signaling and protein turnover, and suppress defense pathways. Geminiviruses also display high mutation and recombination rates that allow them to adapt rapidly to new environments and new hosts (6, 7). This rapid adaptation in combination with a long evolutionary history with their plant hosts has provided many opportunities for geminiviruses to modulate interactions with their hosts and enhance their success as pathogens (8).
Members of the genus Begomovirus, the largest geminivirus genus, infect dicot plants and are transmitted by whiteflies (9). They are classified as Old or New World lineages and can have monopartite genomes or bipartite genomes, designated DNA-A and DNA-B. Many monopartite begomoviruses are associated with satellites (10). Curtoviruses also infect dicots but are transmitted by leafhoppers and have single genome components. Both genera carry genes that encode 4 overlapping open reading frames (ORFs) on the complementary strand of their genomes. The open reading frames specify the Rep and AL3/C3 proteins, which are involved in viral replication, and AL2/C2 and AL4/C4, which modulate host defenses. (The designation AL indicates proteins of bipartite begomoviruses, while C indicates proteins of monopartite begomoviruses and curtoviruses.)
Begomovirus AL2/C2 proteins are transcriptional activators that are required for expression of coat protein (CP) and nuclear shuttle protein (NSP) genes, which are transcribed late in the infection cycle (11). As a consequence, these AL2/C2 proteins are also called TrAPs, for transcription activation proteins. AL2/C2 proteins do not bind to double-stranded DNA and, instead, are thought to be recruited to viral promoters through interactions with host transcription factors (12). An Arabidopsis thaliana TIFY family transcription factor, PEAPOD2, has been shown to interact with the AL2 proteins and the CP promoters of Tomato golden mosaic virus (TGMV) and Cabbage Leaf Curl Virus (CaLCuV) (13). Another candidate AL2/C2 partner is the JDK transcription factor (14). Because CP and NSP are required for the trafficking of viral DNAs or virions through the nuclear membrane, begomovirus AL2/C2 proteins play an important role in viral movement and the spread of infection. In contrast, there is no evidence linking curtovirus C2 proteins to viral gene transcription.
Begomovirus and curtovirus AL2/C2 proteins interface with host defense responses via several mechanisms. They suppress transcriptional gene silencing (TGS) to reduce host methylation of viral DNA, which interferes with viral replication and transcription (15). AL2/C2 binds to and inhibits plant adenosine kinase (ADK), which is necessary for the synthesis of S-adenosylmethionine (SAM), which provides the methyl group for DNA methylation (16). A curtovirus C2 protein has also been shown to interact with and stabilize S-adenosylmethionine decarboxylase 1 (SAMDC1), an enzyme that reduces the SAM pool in plant cells (17). The importance of these AL2/C2 interactions, which reduce the availability of methyl groups for DNA methylation, was established using a curtovirus C2 mutation that caused increased viral DNA methylation and attenuated symptoms. C2 may also upregulate the expression of the host defense genes ACD6 and GSTF14 by reducing DNA methylation in their promoter regions (18).
AL2/C2 proteins also interact with host signaling pathways involved in nutrient and energy regulation, growth and development, and biotic stress responses. Such interactions may facilitate the establishment and spread of infection or mediate host defense processes. C2 interacts with CSN5, a component of the COP9 signalosome, and inhibits its activity of removing the RUB moiety from cullin (19). Cullin is a member of the ubiquitin E3 ligase complexes, and attachment of RUB is necessary for ligase activity. Perception of several plant hormones is mediated by specific ubiquitin E3 ligases and the consequent degradation of targeted ubiquitinated proteins by the proteosome. There is evidence that AL2/C2 proteins modulate plant jasmonate pathways to suppress host defenses (19) and potentiate cytokinin signaling through interactions with ADK to promote viral DNA synthesis (20). Consistent with this, exogenous application of jasmonate results in increased resistance to geminivirus infection, while exogenous cytokinins lead to increased susceptibility (19).
The plant protein kinase SnRK1 and its homologs in animals and yeasts, AMPK and SNF1, respectively, sense low energy and nutrient levels in the cell and alter the expression of many genes that control energy production and macromolecule synthesis (for reviews, see references 21 to 23). SnRK1 has also been implicated in regulating plant development (24, 25). The geminivirus Rep-interacting kinases (GRIKs), which were first identified as partners of the geminivirus Rep protein and are stabilized in geminivirus-infected cells, phosphorylate SnRK1 to activate its kinase activity (26–29). Plants overexpressing SnRK1 are more resistant to geminivirus infection, while plants carrying SnRK1 antisense constructs are more susceptible than wild-type plants (30, 31). It has been reported that AL2/C2 bind to and directly inactivate SnRK1 (30). It is also possible that AL2/C2 downregulates SnRK1 kinase activity indirectly by inactivating ADK (16), since ADK catalyzes the conversion of ATP to AMP and AMP sustains SnRK1 activity by inhibiting protein phosphatases that dephosphorylate activated SnRK1 (32). SnRK1 also interacts with and phosphorylates the βC1 protein encoded by β satellites associated with many Old World begomoviruses (31). A βC1 phosphomimic delays the establishment of viral infection, providing further evidence of the contribution of the GRIK-SnRK1 kinase cascade to the host defense response during geminivirus infection. AMPK, the SnRK1 counterpart in animals, also impacts viral infection, and AMPK activity is inhibited in mammalian cells infected by human immunodeficiency virus, hepatitis C virus, and human cytomegalovirus through the complex interplay of many cellular pathways (for reviews, see references 33 and 34).
Protein kinases phosphorylate target proteins at serine, threonine, or tyrosine residues to alter their enzymatic activity, cellular localization, interaction with other proteins, and other biochemical properties. Protein phosphorylation plays critical roles in cell signaling for developmental and environmental responses and is thought to be important in modulating plant pathogen-host interactions. Host phosphorylation of plant viral proteins can play positive or negative roles in the infection process (35–37). Geminivirus proteins, including the begomovirus movement proteins MP and NSP, a curtovirus C4 protein, and the satellite βC1 protein, are also substrates of host protein kinases, but less is known about the roles of these phosphorylation events in infection (31, 38–40).
We recently reported the development of an in vitro GRIK-SnRK1 kinase system (28). We describe here the application of this system to better understand the interactions between geminivirus AL2/C2 proteins and the GRIK-SnRK1 kinase cascade and the role of AL2/C2 phosphorylation during infection.
MATERIALS AND METHODS
Plant growth and virus infection.
Arabidopsis thaliana ecotype Col-0 plants were grown at 20°C under an 8-h light/16-h dark cycle. For CaLCuV infection, 6- to 7-weeks-old plants with ca. 30 leaves were bombarded at the center of the rosette with gold particles coated with viral replicon DNA and loaded on Swinnex filters (Millipore) using a handheld microsprayer (Venganza) at a pressure of 200,000 Pa. Each plant was coinoculated with two pUC19-based plasmids (150 ng each) containing partial tandem copies of CaLCuV DNA-A or DNA-B, respectively. Symptom development was scored over time for the leaves in the rosette center using a 5-point scale (0, no symptoms; 1, appearance of mild leaf curling; 2, appearance of chlorosis in 1 to 2 leaves; 3, chlorosis spread to 3 or 4 curly leaves; 4, severe symptoms throughout new growth in the rosette).
Plasmid construction.
The plasmid designations and the oligonucleotide primers used to generate the plasmids are listed in Tables S1 and S2 in the supplemental material, respectively. To construct plasmids producing recombinant glutathione S-transferase (GST)-AL2/C2 fusion proteins in bacteria, the AL2/C2 coding regions were amplified by PCR with Phusion DNA polymerase (New England BioLabs) from plasmids containing viral DNA. The source plasmids were pCPCbLCVA.003 for the CaLCuV DNA-A replicon (41), FS577pGEMX for Tomato mottle virus (ToMoV; a single-copy DNA-A clone, provided by J. Polston of Florida State University), pMON1565 for TGMV (DNA-A replicon [42]), and pTYLC2 for Tomato yellow leaf curl virus (TYLCV; Dominican Republic isolate viral DNA replicon [43]). The Beet curly top virus (BCTV; strain Logan) C2 sequence was amplified from a pBI121-based plasmid containing a BCTV DNA replicon (provided by D. M. Bisaro of Ohio State University). All the PCR products, except for those of TGMV, were digested with BamHI and XhoI and ligated to the vector pGEX-5X-3 that had been cut with the same restriction enzymes. For TGMV AL2, the PCR fragments were digested with XhoI, treated with T4 DNA kinase, and ligated to pGEX-5X-3 that had been digested with BamHI, treated with the Klenow DNA polymerase, and digested with XhoI. Amino acid substitution mutants of AL2/C2 proteins were generated using Pfu Ultra DNA polymerase (Agilent Technologies) and a pair of complementary primers containing the altered sequence to synthesize mutant DNA from plasmids containing the wild-type gene. The DNA products generated by thermal cycles were treated with the DpnI restriction enzyme and used to transform Escherichia coli DH5α cells. Transformants were screened for correct mutations by DNA sequencing. Plasmids of the CaLCuV replicons with the AL2 S109G or S109D mutation were also made with the Pfu Ultra DNA polymerase from pCPCbLCVA.003, which contains CaLCuV DNA-A and an extra copy of the common region in the vector pBluescript SK+ II.
To make the plasmids for bimolecular fluorescence complementation (BiFC) assays, the coding regions for CaLCuV AL2, Arabidopsis GRIK1 (28), and the kinase domain (KD) of Arabidopsis SnRK1.1 (M1 to Y341) (28) were amplified by PCR and cloned into the Gateway donor vector pDONR221 (Invitrogen). The CaLCuV AL2 ORF was transferred into the binary Gateway cloning-compatible destination vector pSITE-nEYFP-C1 (44) as a C-terminal fusion to the enhanced yellow fluorescent protein (EYFP) N terminus (nEYFP). The ORFs for GRIK1 and the SnRK1.1 kinase domain were transferred into the destination vector pSITE-cEYPF-C1 (44) with the kinases as C-terminal fusions to the EYPF C terminus (cEYFP). Expression of both types of BiFC cassettes was under the control of the Cauliflower mosaic virus (CaMV) E35S promoter and the CaMV 35S terminator.
Recombinant protein production and purification.
The production and purification of recombinant SnRK1 and GRIK proteins in E. coli have been reported previously (28). Plasmids propagated in E. coli strain DH5α were introduced into E. coli strain BL21(DE3) for protein production. Late-log-phase cultures grown at 37°C in 250 ml of LB were induced by 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 16°C for 18 h. Cells were pelleted by centrifugation at 4,000 × g for 15 min and washed in 20 ml of 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM NaH2PO4, pH 7.5 (phosphate-buffered saline [PBS]). Bacteria were disrupted by 12 10-s sonication pulses in 10 ml of PBS containing 0.1% Triton X-100 (PBST). Lysates were cleared by centrifugation at 32,500 × g for 30 min and applied to a 0.4-ml bed volume of glutathione-Sepharose (GE) for batch-wise purification. After three 10-ml washes with PBST, the GST-tagged proteins were eluted in three 400-μl fractions of 10 mM glutathione in 50 mM Tris-HCl, pH 7.5. Purified proteins were dialyzed against 25 mM Tris-HCl, pH 7.5, 140 mM NaCl, and 2.7 mM KCl (Tris-buffered saline [TBS]) and stored in 0.5× TBS, 0.5 mM dithiothreitol (DTT), and 50% (vol/vol) glycerol at −20°C.
Protein quantification, SDS-PAGE, and immunoblotting.
The protein concentration was measured by the Bradford method with protein assay reagent (Bio-Rad). Bovine serum albumin was used as the standard protein. Standard procedures were followed for SDS-PAGE with a stacking gel for protein electrophoresis. Coomassie brilliant blue-stained protein bands were quantified, if necessary, by scanning with a 700-nm laser in a LI-COR Odyssey infrared imager. For immunoblotting, proteins were transferred to a nitrocellulose membrane and probed with goat anti-GST antibodies (1:10,000 dilution; GE) or mouse monoclonal anti-His6 antibodies (1:20,000 dilution; Clontech). Fluorescently labeled secondary antibodies (1:20,000 dilution) of either Alexa Fluor 680 (Life Technologies) or IRdye 680 or 800 (LI-COR) were used to detect the primary antibodies in the Odyssey Imager.
Protein-protein interaction assay.
To examine recombinant protein binding in vitro, 50 pmol of full-length GST-tagged AL2/C2 or GST and an equal amount of the His6-tagged GRIK or SnRK1 kinase domain were mixed in a total volume of 500 μl TBS containing 0.1% Tween 20 (TBST). After incubation at 22°C for 30 min, the proteins were transferred to a 20-μl bed volume of glutathione-Sepharose and incubated for 30 min at 22°C with rotation. The beads were washed 3 times with 1 ml TBST, and the bound proteins were eluted in 100 μl of SDS-PAGE sample buffer at 100°C for 5 min. The eluted proteins were analyzed by SDS-PAGE and immunoblotting and quantified in the Odyssey imager.
To detect protein interactions in vivo by BiFC, the binary plasmids containing expression cassettes of the test proteins fused to the C termini of the split EYFP-coding regions were transferred into Agrobacterium tumefaciens strain LBA4404. Overnight cultures grown in LB medium at 30°C were washed once in 10 mM MES (morpholineethanesulfonic acid)-KOH, pH 5.8, and 10 mM MgCl2. The bacteria were resuspended in the same buffer supplemented with 0.15 mM acetosyringone, incubated at 22°C for 3 h, and diluted to an optical density at 600 nm of 0.6. Equal volumes of strains with expression cassettes for an nEYFP or a cEYFP fusion protein were mixed and used to infiltrate leaves of 4-week-old Nicotiana benthamiana plants grown at 25°C under a 16-h light/8-h dark cycle. Two days after infiltration, the infiltrated leaf area was observed under a Zeiss LSM 710 confocal microscope with excitation at 514 nm for EYFP and an emission window of 519 to 564 nm. Bright-field images were also recorded.
Protein and peptide phosphorylation assays.
Protein phosphorylation reactions were performed in 50 μl of 50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 mM EDTA, 0.1 mM ATP, and 1 μCi [γ-32P]ATP, and the reaction mixtures contained 25 pmol of each kinase and substrate protein and 50 pmol of GST-tagged AL2/C2 protein, as required. After incubation at 30°C for 30 min, the reactions were stopped by adding an equal volume of 2× SDS-PAGE buffer, and the reaction mixtures were boiled for 5 min before being applied to SDS-polyacrylamide gels. The separated proteins were transferred to a nitrocellulose membrane, and the labeled proteins were revealed by autoradiography. To assess SnRK1 T-loop phosphorylation by GRIK, unlabeled ATP was used and the phosphorylated SnRK1 was detected by immunoblotting with antibodies against the human AMPK phospho-T172 peptide (1:10,000 dilution; Cell Signaling Technology). Protein loading concentrations were verified by immunoblotting with antibodies against the protein tags. For some assays, the His6-SnRK1.1 kinase domain was preactivated by incubation with GST-GRIK1 and unlabeled ATP at 30°C for 30 min, followed by incubation with a 20-μl bed volume of glutathione-Sepharose at 22°C for 30 min and centrifugation to remove the bead-bound GST-GRIK1. GST-tagged AL2/C2 proteins and [γ-32P]ATP were then added to the supernatant containing activated His6-SnRK1.1 and analyzed in phosphorylation assays. When the spinach sucrose phosphate synthase (SPS) peptide acetyl-KGRMRRISSVEMMK (GenScript), at 0.2 mM, was used as the substrate, the reactions were stopped by transferring 40-μl aliquots to P81 filter paper discs, which were then rinsed three times in 75 mM H3PO4 and dried. Bound radioactivity was measured in a liquid scintillation counter.
Viral DNA replication in Nicotiana tabacum protoplasts.
Protoplasts were prepared from N. tabacum NT1 cells and electroporated with 50 μg of plasmids containing replicons of wild-type CaLCuV DNA-A or its AL2 mutants using a published protocol (45). The transfected cells were incubated at 25°C for 48 h prior to purification of total DNA and RNase A digestion. Fifty micrograms of total DNA was digested with DpnI and EcoRI for DNA gel blot analysis using a 32P-labeled CaLCuV DNA-A fragment as the probe. Hybridized DNA was visualized by autoradiography.
Viral DNA accumulation in Arabidopsis plants.
Arabidopsis plants infected with wild-type CaLCuV or its AL2 mutants were grown for 6, 7, or 8 days, and the youngest leaves (<1 cm), the rosette center, and the apical meristem tissues were collected, flash-frozen in liquid nitrogen, and stored at −80°C. For genomic DNA extraction, plant tissues were ground in a mortar with a pestle in a buffer containing 100 mM Tris-HCl, pH 8.0, 20 mM EDTA, 1.4 M NaCl, and 2% (wt/vol) cetyltrimethylammonium bromide preheated at 65°C. After centrifugation, the supernatant was extracted once with 1 volume of chloroform, and DNA was precipitated by isopropanol. Dried DNA pellets were dissolved in water containing 10 μg/ml RNase A and incubated at 37°C for 30 min. The DNA was reprecipitated with ethanol and dissolved in water. Five micrograms of DNA was digested with EcoRI and subjected to DNA gel blot analysis as described above.
Phylogenetic analysis.
An AL2/C2 protein sequence from one isolate of each viral species in the begomovirus, curtovirus, and topocuvirus lists assembled previously (46) was selected. The C2 protein of the newly discovered monopartite New World begomovirus Tomato leaf deformation virus (ToLDeV) was included in the list (47). The selected AL2/C2 amino acid sequences were aligned by use of the CLUSTAL program and used to construct a phylogenetic tree by the neighbor-joining method with the software package MEGA (version 4.0).
RESULTS
Geminivirus AL2/C2 proteins interact with the SnRK1-activating kinase GRIK.
A previous study found that the TGMV AL2 protein and BCTV C2 protein interact with the kinase domain of SnRK1 in yeast two-hybrid assays (30). The same study used recombinant kinases produced in insect cells to show that the AL2 and C2 proteins inhibit autophosphorylation of the SnRK1 kinase domain. A potential limitation of the study was the use of eukaryotic systems that express α-, β-, and γ-subunit homologs of SnRK1 and its upstream activating kinase GRIK, which could serve as bridging proteins or impact kinase activity. To address this possibility, we used recombinant proteins produced in E. coli (28), which does not carry genes that encode an SnRK1 homolog, to reexamine the relationship between AL2/C2 proteins and the GRIK-SnRK1 kinase cascade.
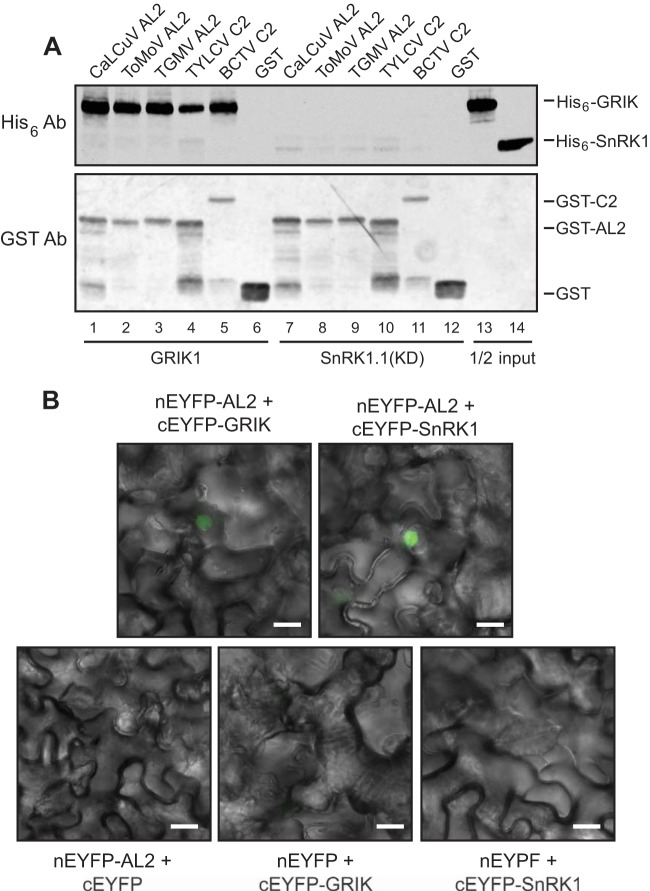
We first asked if His6-tagged versions of the SnRK1.1 kinase domain (M1 to Y341) or full-length GRIK1 cofractionated with GST-tagged AL2/C2 proteins on glutathione-Sepharose. We examined binding to the AL2 proteins from CaLCuV, ToMoV, and TGMV and the C2 proteins of TYLCV and BCTV. The five AL2/C2 proteins are from a diverse group of viruses that included three bipartite New World begomoviruses (CaLCuV, ToMoV, and TGMV), a monopartite Old World begomovirus (TYLCV), and a curtovirus (BCTV). The assay mixtures contained equal molar amounts of the AL2/C2 proteins and the kinases. Immunoblot analysis using an anti-His6 antibody showed that 13 to 34% of the input His6-GRIK1 (Fig. 1A, top, lane 13) cofractionated with the GST-tagged AL2/C2 proteins (lanes 1 to 5). In contrast, less than 1% of the input His6-SnRK1.1 (KD) (lane 14) cofractionated with the GST-AL2/C2 proteins (Fig. 1A, top, lanes 7 to 11), even though immunoblotting with an anti-GST antibody established that equivalent amounts of the AL2/C2 proteins bound to the resin in the presence of GRIK1 or SnRK1 (Fig. 1A, bottom; cf. lanes 1 to 5 and lanes 7 to 11). Neither kinase was detected in binding assays in which the assay mixtures contained GST alone (Fig. 1A, bottom, lanes 6 and 12). These results established that the AL2/C2 proteins bind more strongly to GRIK than to the SnRK1 kinase domain.
FIG 1.
AL2/C2 proteins interact with GRIK and SnRK1. (A) Interaction of recombinant proteins in vitro. GST (lanes 6 and 12) or GST-tagged AL2/C2 proteins from CaLCuV (lanes 1 and 7), ToMoV (lanes 2 and 8), TGMV (lanes 3 and 9), TYLCV (lanes 4 and 10), and BCTV (lanes 5 and 11) were incubated with equal molar amounts of His6-tagged GRIK1 (lanes 1 to 6) or SnRK1.1 (KD) (lanes 7 to 12). Each sample was then incubated with glutathione-Sepharose, and bound proteins were resolved by SDS-PAGE, followed by immunoblotting. An anti-His6 antibody was used to assess if the kinases bound to AL2/C2 proteins (top), while the anti-GST antibody was used to monitor the amounts of AL2/C2 bound to the glutathione-Sepharose (bottom). One half of the input amount of GRIK (lane 13) or SnRK1 (lane 14) in the binding reactions was analyzed in parallel. Ab, antibody. (B) Bimolecular fluorescence complementation in plant cells. Split nEYFP or cEYFP was fused to GRIK1, SnRK1.1 (KD), or CaLCuV AL2, as indicated (top). Constructs containing only a split nEYFP or cEYFP were used as negative controls (bottom). The indicated combinations of the nEYFP and cEYFP constructs were transfected into N. benthamiana leaf epidermal cells. Fluorescence from reconstituted EYFP, shown as the green artificial color, was observed by confocal microscopy. Merged EYFP fluorescent images and bright-field images of cellular structure are shown. Bars, 20 μm.
We also investigated whether CaLCuV AL2 interacts with GRIK1 and/or SnRK1 in plant cells using BiFC assays. The CaLCuV AL2-coding region was fused to the C terminus of the N-terminal part of EYFP (nEYFP-AL2), while the GRIK1-coding region or the SnRK1.1 kinase domain was fused to the C terminus of the C-terminal part of EYFP (cEYFP-GRIK1 and cEYFP-SnRK1.1, respectively). Fluorescence complementation of the split EYFPs was observed in N. benthamiana leaf epidermal cells transiently transformed by Agrobacterium infiltration with the nEYFP-AL2/cEYFP-GRIK1 or nEYFP-AL2/cEYFP-SnRK1.1 combination (Fig. 1B). Fluorescence was observed only in the nucleus in both cases (Fig. 1B). No fluorescence complementation was seen in control transfections containing nEYFP-AL2/cEYFP, nEYFP/cEYFP-GRIK1, or nEYFP/cEYFP-SnRK1.1 (Fig. 1B). These results show that CaLCuV AL2 can interact specifically with both GRIK and SnRK1 in plant nuclei. The strong fluorescent signal detected for nEYFP-AL2/cEYFP-SnRK1.1 in vivo is in contrast to the weak binding activity observed for purified GST-AL2 and His6-SnRK1 in vitro (cf. Fig. 1A, lane 7, and 1B, top right). This difference suggested that AL2-SnRK1 interactions in vivo are mediated by or require an additional plant factor(s) not present in the in vitro assay.
Phosphorylation of a conserved threonine residue in the SnRK1 activation loop is required for its full kinase activity (28). Given the strong interactions between AL2/C2 proteins and GRIK, we asked if the viral proteins alter the efficiency of GRIK-mediated phosphorylation of the SnRK1 kinase domain. We saw no differences in the phosphorylation levels of His6-tagged SnRK1.1 (K48A) or SnRK1.2 (K49A) kinase mutants by GRIK1 (Fig. 2A, top) or GRIK2 (Fig. 2, bottom) either alone (lanes 1 and 6) or in reactions in which the reaction mixtures were supplemented by a 2-fold molar excess of GST-tagged proteins corresponding to CaLCuV AL2 (lanes 3 and 8), TGMV AL2 (lanes 4 and 9), or BCTV C2 (lanes 5 and 10). The presence of the GST tag also had no detectable effect on SnRK1 phosphorylation (Fig. 2A, lanes 2 and 7).
FIG 2.
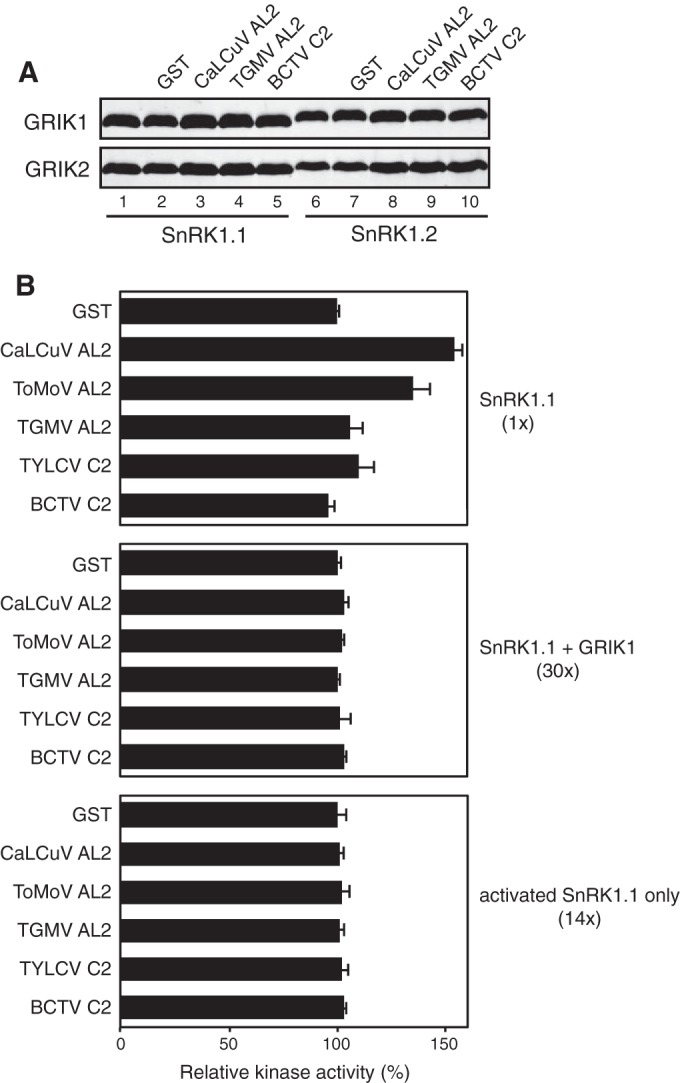
AL2/C2 proteins do not inhibit GRIK or SnRK1 kinase activity. (A) GRIK phosphorylation of the SnRK1 T loop in the presence of AL2/C2 proteins. His6-tagged kinase domain mutants of SnRK1.1 (K48A) (lanes 1 to 5) and SnRK1.2 (K49A) (lanes 6 to 10) were phosphorylated by GST-GRIK1 (top) or GST-GRIK2 (bottom) either alone or in reaction mixtures supplemented with GST (lanes 2 and 7) or GST-tagged AL2/C2 from CaLCuV (lanes 3 and 8), TGMV (lanes 4 and 9), or BCTV (lanes 5 and 10). After SDS-PAGE, SnRK1 phosphorylation was detected by immunoblotting with antibodies against the human AMPK phospho-T172 peptide. (B) Phosphorylation of the SPS peptide by the unactivated SnRK1.1 kinase domain alone (top), GRIK1-activated SnRK1.1 with the presence of GRIK1 (middle, SnRK1.1 plus GRIK1), or GRIK1-activated SnRK1.1 with GRIK removed (bottom). [γ-32P]ATP was used as the phosphate donor. The kinase reactions were performed in the presence of GST or the GST-tagged AL2/C2 protein from CaLCuV, ToMoV, TGMV, TYLCV, or BCTV, as indicated on the left. The molar ratios of the effector proteins to SnRK1.1 were 2:1 for all proteins except GST-tagged BCTV C2, for which the molar ratio was 1:1. Kinase activities are expressed as the counts per minute (cpm) of the radioactivity incorporated into the peptide relative to that in the reactions with GST as the effector protein. The average radioactivity and SD from three experiments are shown. The relative activities of the different reaction conditions with GST are indicated on the right.
We then asked if the AL2/C2 proteins produced in E. coli directly impact SnRK1 kinase activity in phosphorylation assays with a commonly used SnRK1 substrate, the spinach SPS peptide KGRMRRISSVEMMK (48). The peptide was incubated with either the His6-tagged SnRK1.1 kinase domain that was not activated but might have residual kinase activity (Fig. 2B, top), the SnRK1.1 kinase domain in the presence of GST-GRIK1 for activation (Fig. 2B, middle), or the activated SnRK1.1 kinase domain with GRIK1 removed (Fig. 2B, bottom). SnRK1 activity in the presence of GST was 30- and 14-fold greater for reaction mixtures containing GRIK1 plus SnRK1.1 or preactivated SnRK1.1, respectively, than reaction mixtures containing unactivated SnRK1.1. In all these assays, there were no significant decreases in the amount of SPS phosphorylation in reaction mixtures containing a 2-fold molar excess of GST-tagged CaLCuV, ToMoV, or TGMV AL2, GST-tagged TYLCV C2, or an equal amount of GST-BCTV C2 compared to that in reaction mixtures containing GST only (Fig. 2). Thus, we concluded that although both GRIK and SnRK1 interact with geminivirus AL2/C2 proteins, the interactions do not inhibit GRIK or SnRK1 kinase activities in in vitro assays in which the assay mixtures contained well-defined eukaryotic components.
An increase in phospholabeling was detected in reaction mixtures containing unactivated SnRK1.1 supplemented with CaLCuV or ToMoV AL2 but not in equivalent reaction mixtures containing GRIK1 plus SnRK1.1 or preactivated SnRK1.1. This increase may reflect a low level of phosphorylation of the GST-AL2 proteins that can be detected only with unactivated SnRK1.1 because of the much lower level of SPS peptide phosphorylation. We did not detect the phosphorylation of GST-tagged CaLCuV AL2 by unactivated SnRK1.1 in assays using SDS-PAGE and autoradiography for analysis (not shown).
SnRK1 phosphorylates geminivirus AL2 proteins.
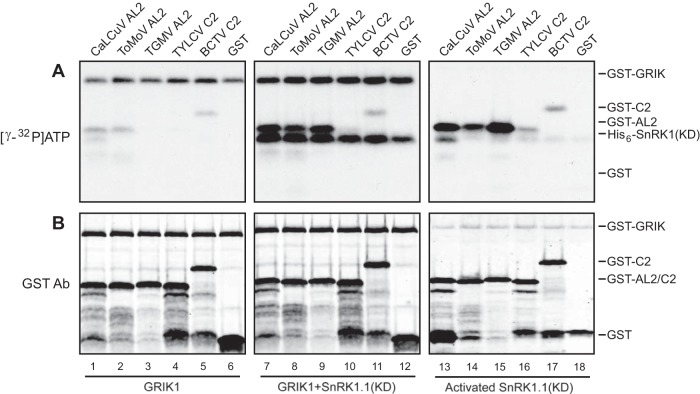
The binding of AL2/C2 proteins to GRIK and SnRK1 raised the possibility that the viral proteins are substrates of either or both kinases. To address this idea, GST-tagged AL2/C2 proteins were incubated in kinase reaction mixtures containing either GST-GRIK1 alone, the His6-SnRK1.1 kinase domain in the presence of GST-GRIK1 (GRIK1 plus SnRK1), or the GRIK1-activated SnRK1.1 with GRIK1 removed. CaLCuV AL2 (Fig. 3A, lane 1), ToMoV AL2 (lane 2), TYLCV C2 (lane 4), and BCTV C2 (lane 5) were weakly phosphorylated in kinase reaction mixtures containing GRIK1, while no labeling was detected for TGMV AL2 (lane 3) or GST (lane 6). Weak phosphorylation was also observed in reaction mixtures containing GRIK1 plus SnRK1.1 and activated SnRK1.1 for TYLCV C2 (Fig. 3A, lane 10), BCTV C2 (lane 11), and GST (lane 12), indicating that they are poor substrates of both kinases in vitro. In contrast, the AL2 proteins from CaLCuV (Fig. 3A, lane 7), ToMoV (lane 8), and TGMV (lane 9) were strongly phosphorylated in reaction mixtures containing GRIK1 plus SnRK1.1 or activated SnRK1.1. Immunoblotting with an anti-GST antibody established that the assay mixtures with GRIK1 alone, GRIK1 plus SnRK1.1, and activated SnRK1.1 kinase contained equivalent amounts of the AL2/C2 proteins (Fig. 3B; cf. lanes 1 to 6 and 7 to 12), ruling out the possibility that the higher levels of phosphorylation of the AL2 proteins in the reaction mixtures with GRIK1 plus SnRK1.1 were due to the increased substrate concentration. Instead, the results suggested that the AL2 proteins of the bipartite geminiviruses but not the C2 proteins of monopartite geminiviruses are good targets for SnRK1 phosphorylation. This observation is consistent with data showing that the AL2 and C2 proteins are only partial functional homologs (49, 50).
FIG 3.
AL2 proteins are phosphorylated efficiently by SnRK1. GST (lanes 6 and 12) or GST-tagged AL2/C2 proteins from CaLCuV (lanes 1 and 7), ToMoV (lanes 2 and 8), TGMV (lanes 3 and 9), TYLCV (lanes 4 and 10), and BCTV (lanes 5 and 11) were incubated with equal molar amounts of GST-GRIK1, with GST-GRIK1 and His6-SnRK1.1 (KD), or with GRIK1-activated SnRK1 with GRIK1 removed in phosphorylation assays in which the assay mixtures contained [γ-32P]ATP. The reactions were resolved by SDS-PAGE followed by autoradiography to detect 32P-labeled proteins (A). Total GST-tagged AL2/C2 proteins in the kinase reactions were monitored by immunoblotting with anti-GST antibodies (B).
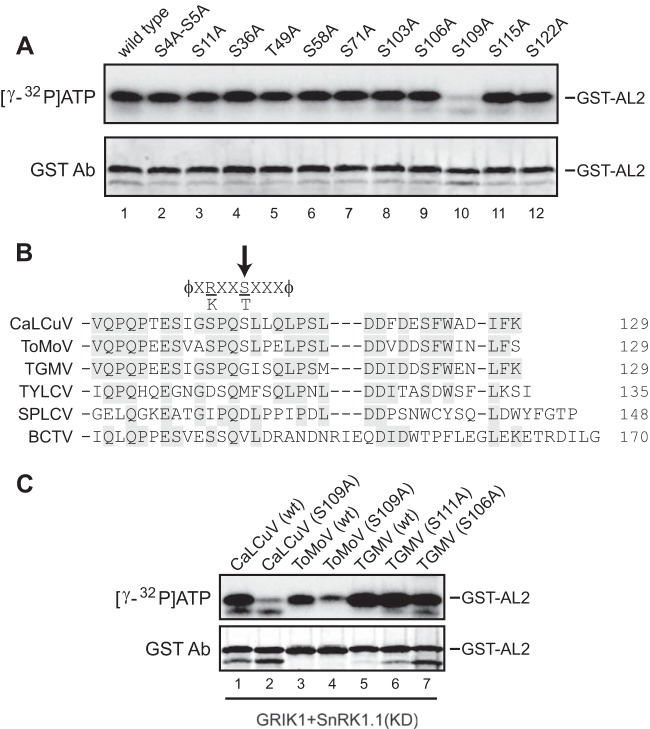
To identify the CaLCuV AL2 residue(s) that is phosphorylated by SnRK1, we selected 12 serine and threonine residues with the highest probability of being SnRK1 targets on the basis of the amino acid occurrence score at flanking positions for further investigation (51). The residues included serines at positions 4, 5, 11, 36, 58, 71, 103, 106, 109, 115, and 122 and a threonine at position 49. Recombinant proteins with alanine substitutions at these positions were analyzed in SnRK1 phosphorylation assays. To avoid interference by background bands from the radiolabeled kinases in the autoradiograph, His6-SnRK1.1 was preactivated by GST-GRIK1 in the presence of cold ATP, and GRIK1 was subsequently removed by binding to glutathione-Sepharose. The S109A mutation caused a substantial reduction in CaLCuV AL2 phosphorylation (Fig. 4A, lane 10). None of the other substitutions, including the S106A (Fig. 4A, lane 9) and S115A (lane 11) mutations in the immediate vicinity of S109, altered the intensity of AL2 labeling. Thus, we concluded that S109 is the major SnRK1 phosphorylation site in CaLCuV AL2. The results also indicated that SnRK1 phosphorylation of CaLCuV AL2 does not require the presence of GRIK. Phosphorylation of wild-type CaLCuV AL2 and the S109A mutant showed no differences in kinase assays in which the reaction mixtures contained only GRIK1 (data not shown), establishing that S109 is specifically phosphorylated by SnRK1.
FIG 4.
SnRK1 phosphorylates S109 in the CaLCuV and ToMoV AL2 proteins. (A) Identification of the SnRK1 phosphorylation site in CaLCuV AL2. GST-tagged wild-type CaLCuV AL2 or Ala substitution mutants with mutations at the selected serine and threonine residues indicated at the top were incubated in reaction mixtures containing [γ-32P]ATP and SnRK1.1 (KD) that was preactivated by GRIK1 and unlabeled ATP. The reactions were resolved by SDS-PAGE, followed by autoradiography to detect 32P-labeled GST-AL2 (top). The total amount of AL2 protein in each reaction mixture was monitored by immunoblotting with anti-GST antibodies (bottom). (B) Alignment of the C-terminal amino acid sequences from AL2/C2 proteins representative of New World begomoviruses (CaLCuV, ToMoV, and TGMV), an Old World begomovirus (TYLCV), a sweet potato begomovirus (sweet potato leaf curl virus [SPLCV]), and a curtovirus (BCTV), corresponding to some of the major branches in the phylogenetic tree shown in Fig. 5. The arrow marks S109, which is conserved in most of the New World begomovirus AL2 proteins, represented by CaLCuV and ToMoV. The consensus SnRK1 phosphorylation sequence is shown on top of the alignment (ϕ, hydrophobic amino acids; X, unspecified amino acids). (C) SnRK1 phosphorylates S109 in ToMoV AL2 but not S106 or S111 in TGMV AL2. GST-tagged wild-type (wt) AL2 or Ala substitution mutants of CaLCuV, ToMoV, and TGMV AL2 were incubated with preactivated SnRK1.1 (KD) and analyzed as described in the legend to panel A. The autoradiograph (top) shows the 32P-labeled AL2 protein in each reaction, and the immunoblot (bottom) shows the total GST-tagged AL2 protein in each reaction.
Alignment of AL2/C2 amino acid sequences from CaLCuV, ToMoV, TGMV, TYLCV, and BCTV uncovered a serine residue at the position equivalent to CaLCuV AL2 S109 only in ToMoV AL2. In contrast, the equivalent position is a glycine residue (position 109) in TGMV AL2, a methionine residue (position 114) in TYLCV C2, and a valine residue (position 142) in BCTV C2 (Fig. 4B). The absence of an equivalent serine residue in TYLCV and BCTV C2 proteins is consistent with their poor phosphorylation (Fig. 3A, lanes 10 and 11) but not with the efficient phosphorylation of TGMV AL2 (lane 9) in GRIK1-SnRK1.1 kinase assays. TGMV AL2 contains a total of 19 serine and threonine residues, with S106 and S111 being in close proximity to G109. Based on the sequence alignments, we generated mutants with alanine substitutions at S109 in GST-tagged ToMoV AL2 and at S106 and S111 in GST-tagged TGMV AL2 and compared their phosphorylation to that of the corresponding wild-type proteins in GRIK1-SnRK1.1 kinase assays. Analogous to the results for CaLCuV AL2 (Fig. 4C, lanes 1 and 2), phosphorylation of the ToMoV AL2 S109A mutant was greatly reduced compared to the level of phosphorylation of the wild-type protein (cf. lanes 3 and 4), confirming that ToMoV AL2 S109 is also a target of SnRK1. In contrast, neither of the TGMV AL2 mutations resulted in a reduced phosphorylation intensity relative to that of the wild type (Fig. 4C; cf. lanes 5 to 7), indicating that the region around G109 in TGMV AL2 is not targeted by SnRK1 (Fig. 4C) and that the SnRK1 phosphorylation site is located elsewhere in the protein.
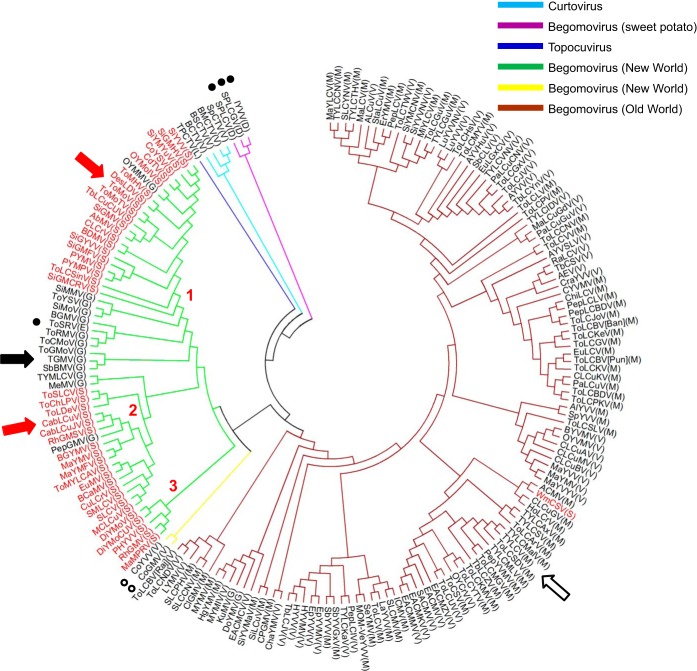
The AL2 S109 site is immediately flanked by conserved amino acid residues in all geminivirus AL2/C2 proteins, with an invariant upstream glutamine and a downstream hydrophobic residue (Fig. 4B). To gain further insight into the conservation of the S109 site, we constructed a phylogenetic tree using AL2/C2 amino acid sequences of single isolates representing each species of begomovirus, topocuvirus, and curtovirus annotated previously (46) (Fig. 5; see also Table S3 in the supplemental material). The tree revealed that AL2 S109 occurs in 3 subgroups of New World begomoviruses that contain a total of 42 species, including ToMoV and CaLCuV (Fig. 5). Only 14 species of New World begomoviruses, including TGMV, lack S109. These species, which tend to cluster in the AL2/C2 tree, have glycine residues at position 109, with the exception of Tomato severe rugose virus (ToSRV). In contrast, there are only three examples of Old World begomoviruses with S109 (Watermelon chlorotic stunt virus [WmCSV]) or G109 (Dolichos yellow mosaic virus [DoYMV], Kudzu mosaic virus [KuMV]) in their AL2/C2 proteins. The other 123 Old World begomoviruses have a methionine or valine residue at the equivalent position in their AL2/C2 proteins. Interestingly, the C2 proteins of the 3 sweet potato begomoviruses in the tree have an aspartic acid residue at the equivalent position (Fig. 4B) and the New World begomovirus ToSRV has a glutamic acid residue; both of these residues are mimics for a phosphoserine.
FIG 5.
Conservation of AL2 S109 in subgroups of the New World begomoviruses. A phylogenetic tree was constructed by the neighbor-joining method based on the AL2/C2 protein sequence of one selected isolate from each geminivirus species listed in Table S3 in the supplemental material. Virus groups are indicated by different branch colors. The amino acid residue of each AL2/C2 protein at the equivalent CaLCuV AL2 S109 site is indicated in parentheses. The species with AL2 S109 are designated in red, and the three groups in which they cluster are numbered. Open circles, a valine residue at the equivalent position; closed circles, an aspartic acid or glutamic acid residue at the equivalent position; arrows, the viral species whose AL2/C2 proteins were characterized in this study.
Phosphorylation of CaLCuV AL2 at Ser-109 could delay symptom development during infection.
Due to the low number of virus-positive cells in CaLCuV-infected leaves (52), it is not feasible to directly assess the phosphorylation of AL2 S109 in vivo and its effect on viral infection and the host response. Therefore, we employed an indirect approach using CaLCuV AL2 S109 phosphomimic and phosphorylation-null mutants. We used a glycine substitution for S109 as the phosphorylation-null mutation for two reasons. First, an S109G AL2 mutation does not alter the amino acid sequence of the overlapping AL3 open reading frame. Second, glycine occurs in the AL2 proteins of the New World begomoviruses that do not have a serine, aspartic acid, or glutamic acid at position 109 (Fig. 4B and 5). We used an aspartic acid substitution for S109 as the phosphomimic mutation because it occurs at position 109 in the C2 proteins of all sweet potato begomoviruses (Fig. 5).
The nucleotide sequence changes for the S109D mutation inevitably result in an A64T mutation in the AL3 protein, which is required for efficient viral replication (43). Thus, we first assessed the impact of the mutations on AL3 function in transient replication assays. N. tabacum protoplasts were electroporated with a wild-type CaLCuV DNA-A replicon or with replicons carrying the AL2 S109G or S109D mutation, and viral DNA accumulation was analyzed at 48 h posttransfection on DNA gel blots using a probe that did not contain the mutated sequence. High levels of double-stranded DNA-A were detected for the wild-type (Fig. 6A, lanes 4 to 6), S109G (lanes 7 to 9), and S109D (lanes 10 to 12) replicons. Typically, a defective AL3 mutation results in 50- to 100-fold less viral DNA accumulation in transient assays (43). Based on these results, we concluded that the A64T mutation does not impair the AL3 replication function and that neither the AL2 phosphorylation-null S109G nor the phosphomimic S109D mutation had an impact on viral DNA replication.
FIG 6.
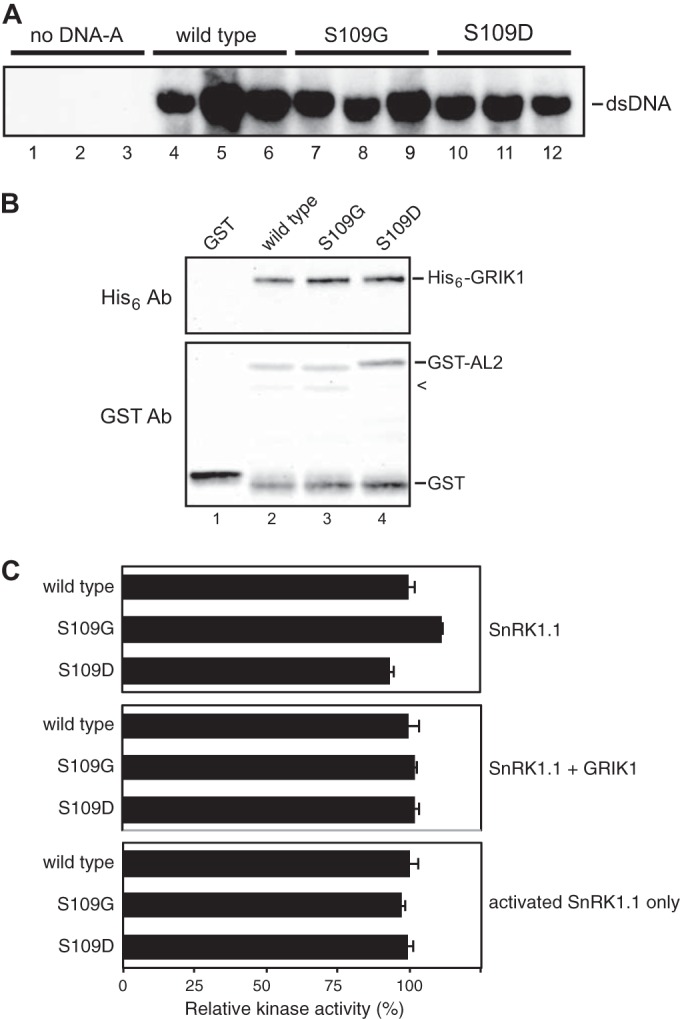
Mutations at CaLCuV AL2 S109 are neutral to viral DNA replication, AL2 interaction with GRIK, and SnRK1 kinase activity. (A) The CaLCuV AL2 S109G and S109D mutations have no impact on viral DNA replication. Tobacco protoplasts were electroporated with wild-type CaLCuV DNA-A replicons (lanes 4 to 6) or the AL2 S109G (lanes 7 to 9) or S109D (lanes 10 to 12) mutant in triplicate. Total DNA was extracted at 48 h posttransfection and analyzed by DNA gel blotting using a 32P-labeled CaLCuV DNA-A probe. dsDNA, double-stranded DNA. (B) The S109G and S109D mutations have no impact on the AL2 interaction with GRIK. GST (lane 1) or GST-tagged wild-type CaLCuV AL2 (lanes 2) or its S109G and S109D mutants (lanes 3 and 4, respectively) were subjected to the His6-GRIK1 binding assay, as described in the legend to Fig. 1. Bound His6-GRIK1 was monitored by the use of anti-His6 antibodies (top), and the GST-AL2 protein was monitored by the use of anti-GST antibodies (bottom). (C) The AL2 S109 mutants, like wild-type AL2, do not inhibit SnRK1 kinase activity. A twofold molar excess of GST-AL2 or its S109G and S109D mutants relative to SnRK1.1 (KD) was included in SPS peptide phosphorylation assays. The kinases used were SnRK1.1 (KD) alone (top), SnRK1.1 (KD) in the presence of GRIK1 (middle; SnRK1.1 + GRIK1), or GRIK1-activated SnRK1.1 with GRIK removed (bottom; activated SnRK1.1 only). [γ-32P]ATP was used as the phosphate donor. Kinase activities are expressed as the amount of radioactivity incorporated into the peptide relative to that in the reactions with wild-type AL2 as the effector protein. The average relative radioactivity and standard deviations from three experiments are shown.
We also determined if the mutations at AL2 S109 have any impact on its interaction with GRIK or the kinase activity of SnRK1. GST-tagged, wild-type CaLCuV AL2 or its S109G or S109D mutant protein was incubated with His6-GRIK1, and glutathione-Sepharose was used to fractionate AL2-bound His6-GRIK1, which was then visualized by the use of anti-His6 antibodies on immunoblots. Figure 6B shows that neither mutation altered the AL2 interaction with GRIK1 (cf. lanes 3 and 4 with lane 2). Excess GST-tagged AL2 or its S109G or S109D mutant protein was also included in peptide phosphorylation assays in which the assay mixtures contained SnRK1.1 alone (Fig. 6C, top), SnRK1.1 plus GRIK1 (Fig. 6C, middle), or activated SnRK1.1 with GRIK1 removed (Fig. 6C, bottom). Phosphorylation of the SnRK1 substrate SPS peptide was not altered by wild-type or mutant AL2 proteins (Fig. 6C).
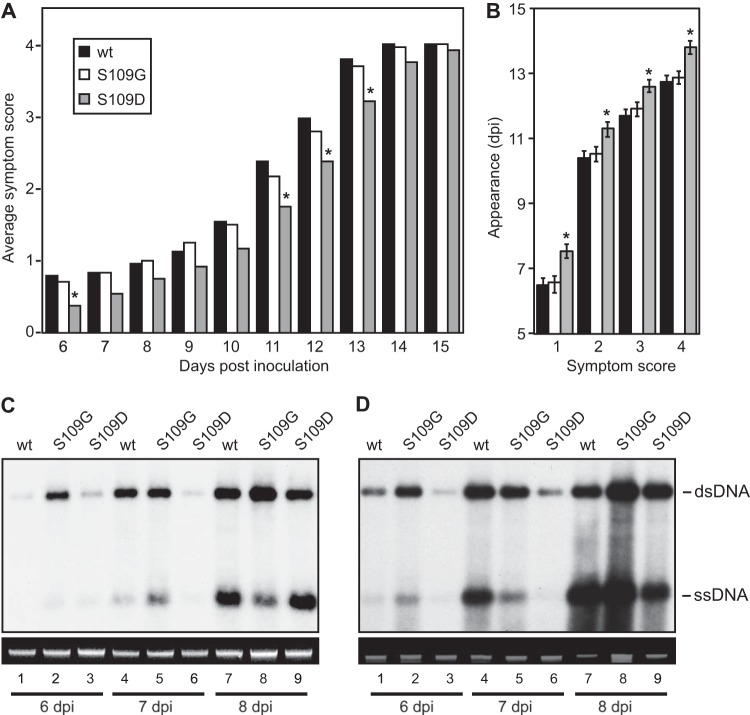
We next investigated whether the CaLCuV AL2 phosphorylation-null S109G or phosphomimic S109D mutation alters CaLCuV infection in planta. Arabidopsis rosette centers were bombarded with a wild-type or mutant CaLCuV DNA-A replicon in combination with a wild-type DNA-B replicon. Plants inoculated with either of the mutants developed symptoms characterized by curly leaves and chlorosis, similar to those observed for plants infected with wild-type CaLCuV. Using a symptom scale of 0 to 4, we scored symptom development on newly emerged leaves around the apical meristem between 6 and 15 days postinoculation (dpi). Plants started to display curly leaves (symptom score, 1) approximately 1 week after inoculation, followed by the appearance of leaf chlorosis (symptom score, 2). These symptoms later spread out to 3 or 4 young leaves (symptom score, 3), indicating firm establishment of the infection. By approximately 2 weeks postinoculation, all the plants displayed leaf curling and severe chlorosis in a wide area around the center of the rosette (symptom score, 4) (Fig. 7A).
FIG 7.
The S109D phosphomimic mutation in CaLCuV AL2 delays symptom development and viral DNA accumulation in Arabidopsis. (A and B) The CaLCuV AL2 S109D phosphomimic mutation delays symptom appearance in Arabidopsis plants. Arabidopsis plants were infected with wild-type CaLCuV or the AL2 S109G or S109D mutant by cobombardment with the corresponding DNA-A replicons and a wild-type DNA-B replicon. Symptom scores were recorded as described in Materials and Methods. The average symptom scores from a representative experiment containing 24 plants infected by each virus genotype (A) and the average time required to reach a symptom score (B) (the SEMs are shown as error bars) are shown. *, P < 0.05 from a Wilcoxon rank sum test of comparisons of plants infected with the AL2 mutants versus wild-type virus. (C and D) Accumulation of viral DNA from the CaLCuV mutant with the AL2 S109D phosphomimic mutation is delayed in Arabidopsis plants. Total DNA was extracted from shoot apical tissues and the youngest leaves (<1 cm) of four plants at 6, 7, and 8 days postinoculation and analyzed on DNA gel blots (top) as described in the legend to Fig. 6. Data from two independent experiments that had similar results are shown. (Bottom) Ethidium bromide-stained undigested total genomic DNAs after agarose gel electrophoresis to control for input DNA quantity. wt, wild type; dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
The average symptom scores of plants infected by the AL2 S109D mutant were always lower than those of plants infected by wild-type CaLCuV, from the earliest symptoms at 6 to 8 dpi until the development of severe symptoms at 14 to 15 dpi (Fig. 7A). The difference in the average symptom scores was statistically significant at 6, 11, 12, and 13 dpi for the phosphomimic mutant versus wild-type CaLCuV (Fig. 7A). In contrast, there was no difference in the average symptom scores over time for the S109G AL2 mutant and the wild-type virus (Fig. 7A). The average time of symptom appearance was also later for the AL2 S109D mutant than wild-type CaLCuV. This was the case at every stage of symptom development, with the phosphomimic mutant showing a 1-day delay that was statistically significant relative to the time of symptom appearance for the wild-type virus for each score (Fig. 7B). In contrast, the average time for symptom appearance did not differ between plants infected with the AL2 S109G mutant and those infected with the wild-type virus across the symptom scores. Together, these data suggest that viral infection is established more slowly in Arabidopsis plants inoculated with the AL2 S109D phosphomimic mutant than in plants infected with either wild-type CaLCuV or the AL2 S109G mutant, in which the times to establishment of infection were similar to each other.
We also compared viral DNA accumulation in plants infected with wild-type CaLCuV or the AL2 mutants. The shoot apical meristem and young leaves (length, <1 cm) were collected from Arabidopsis plants bombarded with wild-type and mutant DNA-A replicons in combination with a wild-type DNA-B replicon at 6, 7, and 8 dpi, and viral DNA accumulation was monitored on DNA gel blots using a 32P-labeled DNA-A probe. Figures 7C and D show the data from two independent experiments. Low levels of double-stranded DNA-A were detected at 6 dpi in the leaves of plants infected with the wild-type virus (Fig. 7C and D, lanes 1) and the AL2 S109D phosphomimic mutant (lanes 3), but plants infected with the AL2 S109G phosphorylation-null mutant showed much higher levels of DNA-A accumulation (lanes 2). At 7 dpi, the levels of viral DNA detected in plants infected with the wild-type virus (lanes 4) resembled those detected in S109G-infected plants (lanes 5). In contrast, the level of viral DNA was still very low in plants infected with the S109D mutant (lanes 6). By 8 dpi, plants infected with the wild-type virus (lanes 7), the S109G mutant (lanes 8), or the S109D (lanes 9) mutant contained significant amounts of DNA-A at similar levels. The initial appearance and accumulation of single-stranded DNA-A in the leaves of infected plants, with a slight delay, had a pattern similar to that of double-stranded DNA-A (Fig. 7C and D). The delay in viral DNA accumulation for the S109D mutant relative to the time of viral DNA accumulation for wild-type CaLCuV resembled the delay in symptom appearance, but earlier infection by the S109G mutant was detected only for DNA accumulation.
Together, the infection data support a model in which SnRK1 phosphorylation of AL2 S109 slows the establishment of infection but does not prevent it from fully developing. The model predicts that the establishment of infection is inversely related to the level of AL2 phosphorylation, as we observed for the CaLCuV AL2 mutants; e.g., S109G showed no phosphorylation and the earliest infection, S109D was perceived to be fully phosphorylated and showed the latest infection, and wild-type S109 showed partial phosphorylation and an intermediate time of infection.
DISCUSSION
Because of their limited coding capacities, geminiviruses must rely on plant pathways and proteins to establish infection (3). The extent of these interactions is illustrated by host transcriptome studies that have identified thousands of plant genes that are differentially expressed in response to geminivirus infection (52, 53). In contrast, less is known about posttranscriptional regulation in the infection process. Phosphorylation is a common posttranslational modification that can alter protein functions. Like all eukaryotes, plants carry genes that encode a large array of protein kinases, only a few of which have been linked to geminivirus infection. The NIK receptor-like kinases are involved in the host defense response (54), a PERK-like receptor kinase enhances infection (38), and shaggy-like kinases have been implicated in plant hormone signaling during infection (40). In this study, we examined the role of SnRK1, a major regulator of plant responses to nutrient starvation and energy limitation (25), during geminivirus infection. Previous studies showed that overexpression of SnRK1 delays infection of both Old and New World begomoviruses and curtoviruses (30, 31). SnRK1 may delay many Old World begomoviruses by phosphorylating and interfering with the βC1 protein encoded by β satellite DNAs, but this mechanism does not explain the delay seen for New World begomoviruses and curtoviruses, which are not associated with β satellites. In the studies described here, we showed that SnRK1 phosphorylates CaLCuV and ToMoV AL2 proteins at S109 (Fig. 4A and C) and that this phosphorylation event delays infection (Fig. 7). However, only about 3/4 of New World begomoviruses and a single Old World begomovirus contain AL2 S109 (Fig. 5), indicating that SnRK1 interactions vary across the Geminiviridae and may be evolving as members of the family move and adapt to new environments.
It was reported that AL2/C2 proteins interact with the SnRK1 kinase domain and inhibit its kinase activity in vitro (30). We also observed an interaction between AL2/C2 and the SnRK1 kinase domain (Fig. 1) but were unable to detect any inhibition of SnRK1 autophosphorylation or SPS peptide phosphorylation activity by the AL2/C2 proteins from CaLCuV, ToMoV, TGMV, TYLCV, or BCTV (Fig. 2). CaLCuV AL2 S109 mutants also showed no inhibition of SnRK1, indicating that the phosphorylation status of AL2 does not influence inhibition (Fig. 6C). A key difference between our studies and those reported earlier is the source of recombinant proteins. All of the proteins included in our kinase assays were produced in E. coli, which does not carry genes that encode a SnRK1 homolog or a kinase that can activate SnRK1. In contrast, AL2/C2 inhibition of SnRK1 activity was detected when the SnRK1 kinase domain was produced in insect cells (30) or N. benthamiana leaves (S. Li and D. M. Bisaro, personal communication). By virtue of their eukaryotic nature, both expression systems contain proteins that could activate and/or bind to the recombinant SnRK1 kinase domain. Our results showing that AL2/C2 proteins do not impair SnRK1 activity in the presence of GRIK indicated that the viral proteins do not interfere with SnRK1 activation (Fig. 2B). Instead, it is more likely that an endogenous insect or plant protein that copurifies with the recombinant SnRK1 kinase domain plays a role in AL2/C2 inhibition of SnRK1 activity. Our observation that CaLCuV interacts strongly with SnRK1 in vivo but not in vitro (Fig. 1) suggests that an unidentified plant factor stabilizes or bridges their interaction in vivo and may facilitate AL2/C2 inhibition of SnRK1. Moreover, SnRK1 is part of a trimeric complex in vivo (23), and a β or γ regulatory subunit of the SnRK1 catalytic subunit may be involved in AL2/C2 inhibition. We also cannot rule out the possibility that AL2/C2 inhibition depends on a posttranslational modification of the SnRK1 kinase domain that is added in eukaryotic cells and is distinct from the GRIK-catalyzed phosphorylation of its activation loop.
Although we could find no evidence of AL2/C2 inhibition of SnRK1, our data clearly showed that some AL2/C2 proteins are SnRK1 substrates in vitro (Fig. 3). SnRK1 efficiently phosphorylated the AL2 proteins from three New World begomoviruses, CaLCuV, ToMoV, and TGMV, in vitro. TGMV AL2 is also phosphorylated in insect cells, which express AMPK, the homolog of SnRK1 (12). We identified S109 as the primary SnRK1 phosphorylation site in the CaLCuV and ToMoV AL2 proteins. TGMV AL2 does not have a serine residue at this position, and the two nearby serine residues are not phosphorylated by SnRK1 (Fig. 4B and C). Thus, SnRK1 phosphorylation of AL2 proteins differs among New World begomoviruses, opening up the possibility that AL2 activity is modulated differently by SnRK1 phosphorylation depending on the viral species.
It was not feasible to directly examine the SnRK1 phosphorylation of AL2 S109 during infection because of the low number of infected cells and the concomitant low levels of viral proteins that are characteristic of geminivirus infection. To overcome these limitations, we created AL2 S109 phosphorylation-null (S109G) and phosphomimic (S109D) mutations and examined their effects on CaLCuV infection in Arabidopsis. The two mutations had opposite effects, with the phosphorylation-null mutant accumulating viral DNA earlier and the phosphomimic showing delayed viral DNA accumulation and symptoms relative to the times of appearance of DNA accumulation and symptoms of the wild-type virus (Fig. 7C and D). The effects of the S109 mutations on infection are modest, involving changes in the timing of viral DNA accumulation and symptoms of 1 to 2 days. The moderate phenotypes are consistent with only a fraction of wild-type AL2 protein being phosphorylated in infected plants.
AL2 is a multifunctional protein that interacts with a number of host proteins to overcome transcriptional gene silencing, mediate transcriptional activation of viral gene expression, and alter host ubiquitination and hormone signaling pathways (for a review, see reference 3). Our data showing that the S109 mutants and wild-type CaLCuV accumulate to similar high levels in protoplast replication assays (Fig. 6A) suggested that S109 phosphorylation does not impact the ability of AL2 to block DNA methylation, which inhibits viral replication and transcription (55, 56). This idea is supported by results indicating that the S109 mutations do not compromise AL2-ADK interactions (S. Li and D. M. Bisaro, personal communication), which play a key role in suppressing host DNA methylation pathways (16). Interestingly, the S109 site is located near the AL2 transcriptional activation domain (12) and might impact its ability to activate expression of the viral CP or NSP gene, both of which are required for efficient infection. Again, we were unable to detect any effect of the S109 mutations on AL2 activation of a CP promoter reporter in transient-transcription assays (unpublished results). However, the mechanism of AL2-mediated transcriptional activation is complex, involving both activation and derepression via different upstream regulatory regions and different transcription factor partners (13, 57, 58), and our tobacco protoplast system may not fully reconstitute the situation in planta. Furthermore, it may be difficult to detect an effect of S109 phosphorylation on a specific AL2 function if subtle changes to multiple functions cause the delay. Lastly, S109 phosphorylation may alter an AL2 function that was not tested here or a yet-to-be-identified function.
GRIK is stabilized during geminivirus infection specifically in virus-infected cells and may contribute to host interactions by activating SnRK1 (26, 27). Moreover, GRIK binds strongly to the AL2/C2 proteins, even though they are only weakly phosphorylated by GRIK in vitro, if at all (Fig. 1 and 3). Given that the catalytic subunits of SnRK1 and GRIK bind to each other (28), one possibility is that GRIK acts as a scaffold protein to recruit AL2 to SnRK1 for phosphorylation. However, preactivated SnRK1 was able to efficiently phosphorylate the AL2 proteins in the absence of GRIK (Fig. 3), suggesting that this is not likely the case. In addition, TYLCV and BCTV C2 proteins are poor SnRK1 substrates, indicating that binding to GRIK is not sufficient for phosphorylation by SnRK1 (Fig. 1 and 3).
Phylogenetic analysis based on the amino acid sequences of the AL2/C2 proteins from representative isolates of begomovirus, curtovirus, and topocuvirus species indicated that the occurrence of a serine residue at the AL2/C2 position equivalent to the CaLCuV AL2 S109 clusters almost exclusively in three subgroups of the New World begomoviruses, which together account for 73% of the New World species in the tree (Fig. 5). Interestingly, the context of S109 in CaLCuV and ToMoV AL2 conforms to the SnRK1 substrate consensus sequence (a hydrophobic amino acid)X(R/K)XX(S/T)XXX(a hydrophobic amino acid) (51), with the exception that a serine residue appears in place of the basic residue at the −3 position relative to the phosphoserine (Fig. 4B). The other New World begomovirus AL2 proteins with a serine residue at position 109 have similar sequence contexts, and we predict that they are also phosphorylated by SnRK1 at the same position. It is important to note that protein kinases in the SnRK2, SnRK3, and CDPK families have recognition sequences similar to the SnRK1 recognition sequence (51), and it is possible that AL2 S109 is also phosphorylated by one or more members of these kinase families in vivo. However, the BiFC assay indicated that SnRK1 can interact with CaLCuV AL2 in leaves (Fig. 1B) where geminivirus-positive cells occur (27) and SnRK1 is expressed during infection (28). Hence, it is likely that SnRK1 interacts with CaLCuV AL2 and phosphorylates S109 in vivo in infected plant cells.
Valine, methionine, glycine, and serine are the most common residues at position 109 or the equivalent site in AL2/C2 (Fig. 5). The occurrence of these amino acids in AL2/C2 is not randomly distributed geographically (Fig. 8). The majority of New World begomoviruses contain an S109 residue, with the remainder having a glycine residue at position 109. S109 and G109 are specified by AGC and GGC codons, respectively (Fig. 8), most likely reflecting a constraint to maintain the overlapping AL3 open reading frame. In contrast, nearly all Old World begomoviruses contain a valine or a methionine residue at the equivalent position, and these are specified by GTG and ATG codons, respectively. Two begomoviruses, Corchorus yellow vein virus (CoYVV) and Corchorus golden mosaic virus (CoGMV), which cluster with New World begomoviruses but were isolated in Southeast Asia (59, 60), carry genes that encode a C2 protein that also has a valine specified by the GTG codon at the equivalent position (Fig. 5). Given that begomoviruses emerged first in the Old World and then migrated to the New World between 20 million and 30 million years ago (8), CoYVV and CoGMV might be close relatives of the ancestral New World begomoviruses. On the basis of this, the most likely evolution at position 109 involving the fewest number of nucleotide changes is from GTG (valine) to GGC (glycine) to AGC (serine) (Fig. 8).
FIG 8.
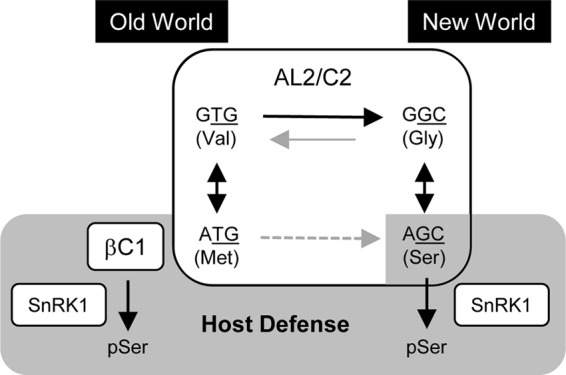
Model for the emergence and selection of S109 in New World begomovirus AL2 proteins. Underlined codon letters, first and second positions of the codons that specify amino acid residues of the AL3/C3 protein; dashed arrow line, a less probable route; pSer, a phosphorylated serine residue.
SnRK1 has been implicated in the host defense response against geminiviruses. Modulating SnRK1 expression and altering the SnRK1 phosphorylation status of βC1 have been associated with strong delays and attenuated symptoms, consistent with SnRK1's involvement in defense (30, 31). In contrast, the effect of the SnRK1 phosphorylation of AL2 on infection is subtler, indicating that it is not a major component of the defense response. Instead, the evolution of AL2 position 109 toward serine in New World begomoviruses suggests that SnRK1 phosphorylation of AL2 provided a selective advantage for these viruses as they adapted to a new environment and to plant hosts that did not exist in the Old World. One possibility is that the modest delay in infection associated with AL2 S109 phosphorylation contributed to increased virus-host compatibility, ensuring both plant survival and virus propagation in the New World, supporting a hypothesis that the reduced virulence of a viral pathogen is beneficial for sustaining its association with its host (1). SnRK1 phosphorylation of TGMV AL2, which lacks S109, may reflect a different evolutionary path to the same endpoint, a possibility that can be addressed by future studies to identify the SnRK1 target site in TGMV AL2 and examine the impact of phosphorylation at this site on infection.
Our results support a role for posttranslational regulation, including the phosphorylation of viral proteins by host kinases, in sustaining host-pathogen interactions over time. They also underscore the variability of geminivirus-host interactions between pathosystems and the possibility that SnRK1 may play different roles during infection, depending on the geminivirus-plant combination (Fig. 8).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Science Foundation (MCB-1052218) to L.H.-B (principal investigator) and M.B.G. (co-principal investigator).
We thank David M. Bisaro and Sizhun Li (Ohio State University, Columbus, OH) for providing the BCTV (Logan) replicon clone and sharing unpublished data. We also thank Devarshi Selote, Terri Long, and Eva Johannes (North Carolina State University, Raleigh, NC) for their help with the BiFC assays and Dominique Robertson and José Trinidad Ascencio-Ibáñez (North Carolina State University, Raleigh, NC) for critical comments on the manuscript.
Footnotes
Published ahead of print 2 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00761-14.
REFERENCES
- 1.Sacristan S, Garcia-Arenal F. 2008. The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9:369–384. 10.1111/j.1364-3703.2007.00460.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fondong VN. 2013. Geminivirus protein structure and function. Mol. Plant Pathol. 14:635–649. 10.1111/mpp.12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11:777–788. 10.1038/nrmicro3117 [DOI] [PubMed] [Google Scholar]
- 4.Navas-Castillo J, Fiallo-Olive E, Sanchez-Campos S. 2011. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49:219–248. 10.1146/annurev-phyto-072910-095235 [DOI] [PubMed] [Google Scholar]
- 5.Mansoor S, Briddon RW, Zafar Y, Stanley J. 2003. Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 8:128–134. 10.1016/S1360-1385(03)00007-4 [DOI] [PubMed] [Google Scholar]
- 6.Martin DP, Lefeuvre P, Varsani A, Hoareau M, Semegni JY, Dijoux B, Vincent C, Reynaud B, Lett JM. 2011. Complex recombination patterns arising during geminivirus coinfections preserve and demarcate biologically important intra-genome interaction networks. PLoS Pathog. 7:e1002203. 10.1371/journal.ppat.1002203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffy S, Holmes EC. 2008. Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus tomato yellow leaf curl virus. J. Virol. 82:957–965. 10.1128/JVI.01929-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefeuvre P, Harkins GW, Lett JM, Briddon RW, Chase MW, Moury B, Martin DP. 2011. Evolutionary time-scale of the begomoviruses: evidence from integrated sequences in the Nicotiana genome. PLoS One 6:e19193. 10.1371/journal.pone.0019193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. 2005. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43:361–394. 10.1146/annurev.phyto.43.040204.135939 [DOI] [PubMed] [Google Scholar]
- 10.Nawaz-ul-Rehman MS, Fauquet CM. 2009. Evolution of geminiviruses and their satellites. FEBS Lett. 583:1825–1832. 10.1016/j.febslet.2009.05.045 [DOI] [PubMed] [Google Scholar]
- 11.Sunter G, Bisaro DM. 1992. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321–1331. 10.1105/tpc.4.10.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartitz MD, Sunter G, Bisaro DM. 1999. The tomato golden mosaic virus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263:1–14. 10.1006/viro.1999.9925 [DOI] [PubMed] [Google Scholar]
- 13.Lacatus G, Sunter G. 2009. The Arabidopsis PEAPOD2 transcription factor interacts with geminivirus AL2 protein and the coat protein promoter. Virology 392:196–202. 10.1016/j.virol.2009.07.004 [DOI] [PubMed] [Google Scholar]
- 14.Lozano-Duran R, Rosas-Diaz T, Luna AP, Bejarano ER. 2011. Identification of host genes involved in geminivirus infection using a reverse genetics approach. PLoS One 6:e22383. 10.1371/journal.pone.0022383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM. 2009. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83:5005–5013. 10.1128/JVI.01771-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM. 2005. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 79:7410–7418. 10.1128/JVI.79.12.7410-7418.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Chen H, Huang X, Xia R, Zhao Q, Lai J, Teng K, Li Y, Liang L, Du Q, Zhou X, Guo H, Xie Q. 2011. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23:273–288. 10.1105/tpc.110.081695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang LP, Fang YY, An CP, Dong L, Zhang ZH, Chen H, Xie Q, Guo HS. 2013. C2-mediated decrease in DNA methylation, accumulation of siRNAs, and increase in expression for genes involved in defense pathways in plants infected with beet severe curly top virus. Plant J. 73:910–917. 10.1111/tpj.12081 [DOI] [PubMed] [Google Scholar]
- 19.Lozano-Duran R, Rosas-Diaz T, Gusmaroli G, Luna AP, Taconnat L, Deng XW, Bejarano ER. 2011. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 23:1014–1032. 10.1105/tpc.110.080267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baliji S, Lacatus G, Sunter G. 2010. The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virology 402:238–247. 10.1016/j.virol.2010.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardie DG, Ross FA, Hawley SA. 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13:251–262. 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghillebert R, Swinnen E, Wen J, Vandesteene L, Ramon M, Norga K, Rolland F, Winderickx J. 2011. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS J. 278:3978–3990. 10.1111/j.1742-4658.2011.08315.x [DOI] [PubMed] [Google Scholar]
- 23.Polge C, Thomas M. 2007. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12:20–28. 10.1016/j.tplants.2006.11.005 [DOI] [PubMed] [Google Scholar]
- 24.Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM. 2007. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 19:2484–2499. 10.1105/tpc.105.037887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448:938–942. 10.1038/nature06069 [DOI] [PubMed] [Google Scholar]
- 26.Kong LJ, Hanley-Bowdoin L. 2002. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14:1817–1832. 10.1105/tpc.003681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen W, Hanley-Bowdoin L. 2006. Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol. 142:1642–1655. 10.1104/pp.106.088476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen W, Reyes MI, Hanley-Bowdoin L. 2009. Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol. 150:996–1005. 10.1104/pp.108.132787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hey S, Mayerhofer H, Halford NG, Dickinson JR. 2007. DNA sequences from Arabidopsis, which encode protein kinases and function as upstream regulators of Snf1 in yeast. J. Biol. Chem. 282:10472–10479. 10.1074/jbc.M611244200 [DOI] [PubMed] [Google Scholar]
- 30.Hao L, Wang H, Sunter G, Bisaro DM. 2003. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15:1034–1048. 10.1105/tpc.009530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Q, Liu Z, Song F, Xie Q, Hanley-Bowdoin L, Zhou X. 2011. Tomato SlSnRK1 protein interacts with and phosphorylates betaC1, a pathogenesis protein encoded by a geminivirus beta-satellite. Plant Physiol. 157:1394–1406. 10.1104/pp.111.184648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugden C, Crawford RM, Halford NG, Hardie DG. 1999. Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J. 19:433–439. 10.1046/j.1365-313X.1999.00532.x [DOI] [PubMed] [Google Scholar]
- 33.Hardie DG. 2011. Adenosine monophosphate-activated protein kinase: a central regulator of metabolism with roles in diabetes, cancer, and viral infection. Cold Spring Harbor Symp. Quant. Biol. 76:155–164. 10.1101/sqb.2011.76.010819 [DOI] [PubMed] [Google Scholar]
- 34.Mankouri J, Harris M. 2011. Viruses and the fuel sensor: the emerging link between AMPK and virus replication. Rev. Med. Virol. 21:205–212. 10.1002/rmv.687 [DOI] [PubMed] [Google Scholar]
- 35.Stork J, Panaviene Z, Nagy PD. 2005. Inhibition of in vitro RNA binding and replicase activity by phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus. Virology 343:79–92. 10.1016/j.virol.2005.08.005 [DOI] [PubMed] [Google Scholar]
- 36.Waigmann E, Chen MH, Bachmaier R, Ghoshroy S, Citovsky V. 2000. Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 19:4875–4884. 10.1093/emboj/19.18.4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Champagne J, Laliberte-Gagne ME, Leclerc D. 2007. Phosphorylation of the termini of Cauliflower mosaic virus precapsid protein is important for productive infection. Mol. Plant Microbe Interact. 20:648–658. 10.1094/MPMI-20-6-0648 [DOI] [PubMed] [Google Scholar]
- 38.Florentino LH, Santos AA, Fontenelle MR, Pinheiro GL, Zerbini FM, Baracat-Pereira MC, Fontes EP. 2006. A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. J. Virol. 80:6648–6656. 10.1128/JVI.00173-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleinow T, Nischang M, Beck A, Kratzer U, Tanwir F, Preiss W, Kepp G, Jeske H. 2009. Three C-terminal phosphorylation sites in the Abutilon mosaic virus movement protein affect symptom development and viral DNA accumulation. Virology 390:89–101. 10.1016/j.virol.2009.04.018 [DOI] [PubMed] [Google Scholar]
- 40.Piroux N, Saunders K, Page A, Stanley J. 2007. Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKeta, a component of the brassinosteroid signalling pathway. Virology 362:428–440. 10.1016/j.virol.2006.12.034 [DOI] [PubMed] [Google Scholar]
- 41.Turnage MA, Muangsan N, Peele CG, Robertson D. 2002. Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30:107–114. 10.1046/j.1365-313X.2002.01261.x [DOI] [PubMed] [Google Scholar]
- 42.Elmer JS, Sunter G, Gardiner WE, Brand L, Browning CK, Bisaro DM, Rogers SG. 1988. Agrobacterium-mediated inoculation of plants with tomato golden mosaic virus DNAs. Plant Mol. Biol. 10:225–234. 10.1007/BF00027399 [DOI] [PubMed] [Google Scholar]
- 43.Settlage SB, See RG, Hanley-Bowdoin L. 2005. Geminivirus C3 protein: replication enhancement and protein interactions. J. Virol. 79:9885–9895. 10.1128/JVI.79.15.9885-9895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59:150–162. 10.1111/j.1365-313X.2009.03850.x [DOI] [PubMed] [Google Scholar]
- 45.Fontes EP, Eagle PA, Sipe PS, Luckow VA, Hanley-Bowdoin L. 1994. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 269:8459–8465 [PubMed] [Google Scholar]
- 46.Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X. 2008. Geminivirus strain demarcation and nomenclature. Arch. Virol. 153:783–821. 10.1007/s00705-008-0037-6 [DOI] [PubMed] [Google Scholar]
- 47.Melgarejo TA, Kon T, Rojas MR, Paz-Carrasco L, Zerbini FM, Gilbertson RL. 2013. Characterization of a New World monopartite begomovirus causing leaf curl disease of tomato in Ecuador and Peru reveals a new direction in geminivirus evolution. J. Virol. 87:5397–5413. 10.1128/JVI.00234-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang JZ, Huber SC. 2001. Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase. Influence of proline and basic amino acid residues at selected positions. Plant Cell Physiol. 42:1079–1087. 10.1093/pcp/pce137 [DOI] [PubMed] [Google Scholar]
- 49.Sunter G, Sunter JL, Bisaro DM. 2001. Plants expressing tomato golden mosaic virus AL2 or beet curly top virus L2 transgenes show enhanced susceptibility to infection by DNA and RNA viruses. Virology 285:59–70. 10.1006/viro.2001.0950 [DOI] [PubMed] [Google Scholar]
- 50.Hormuzdi SG, Bisaro DM. 1995. Genetic analysis of beet curly top virus: examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 206:1044–1054. 10.1006/viro.1995.1027 [DOI] [PubMed] [Google Scholar]
- 51.Vlad F, Turk BE, Peynot P, Leung J, Merlot S. 2008. A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 55:104–117. 10.1111/j.1365-313X.2008.03488.x [DOI] [PubMed] [Google Scholar]
- 52.Ascencio-Ibanez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L. 2008. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148:436–454. 10.1104/pp.108.121038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pierce EJ, Rey ME. 2013. Assessing global transcriptome changes in response to South African cassava mosaic virus [ZA-99] infection in susceptible Arabidopsis thaliana. PLoS One 8:e67534. 10.1371/journal.pone.0067534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J. 2004. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 18:2545–2556. 10.1101/gad.1245904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brough CL, Gardiner WE, Inamdar NM, Zhang XY, Ehrlich M, Bisaro DM. 1992. DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 18:703–712. 10.1007/BF00020012 [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez-Negrete EA, Carrillo-Tripp J, Rivera-Bustamante RF. 2009. RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J. Virol. 83:1332–1340. 10.1128/JVI.01474-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sunter G, Bisaro DM. 1997. Regulation of a geminivirus coat protein promoter by AL2 protein (TrAP): evidence for activation and derepression mechanisms. Virology 232:269–280. 10.1006/viro.1997.8549 [DOI] [PubMed] [Google Scholar]
- 58.Sunter G, Bisaro DM. 2003. Identification of a minimal sequence required for activation of the tomato golden mosaic virus coat protein promoter in protoplasts. Virology 305:452–462. 10.1006/viro.2002.1757 [DOI] [PubMed] [Google Scholar]
- 59.Briddon RW, Patil BL, Bagewadi B, Nawaz-ul-Rehman MS, Fauquet CM. 2010. Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol. Biol. 10:97. 10.1186/1471-2148-10-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ha C, Coombs S, Revill P, Harding R, Vu M, Dale J. 2006. Corchorus yellow vein virus, a New World geminivirus from the Old World. J. Gen. Virol. 87:997–1003. 10.1099/vir.0.81631-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.