Abstract
Regulation of the lectin galectin 9 (Gal-9) was investigated for the first time during human cytomegalovirus (HCMV) infection. Gal-9 transcription was significantly upregulated in transplant recipients with reactivated HCMV in vivo. In vitro, Gal-9 was potently upregulated by HCMV independently of viral gene expression, with interferon beta (IFN-β) identified as the mediator of this effect. This study defines an immunoregulatory protein potently increased by HCMV infection and a novel mechanism to control Gal-9 through IFN-β induction.
TEXT
Primary human cytomegalovirus (HCMV) infection is followed by lifelong latency (1). Reactivation from latency is associated with severe morbidity and mortality in the immunocompromised, especially in the allogeneic hematopoietic stem cell transplant (HSCT) setting, where donor or recipient HCMV seropositivity is associated with adverse outcomes. In addition, HCMV is the leading infectious cause of birth defects in the developed world (2).
Galectins are a family of lectins that preferentially bind β-galactosides. Galectin 9 (Gal-9) can modulate diverse biological activities, including cell adhesion, proliferation, apoptosis, and cell cycle progression (3). Despite such varied functions, regulation of Gal-9 is very poorly understood. Functionally, Gal-9 is best characterized as an immunoregulatory molecule controlling T-cell activity via interaction with its receptor, Tim-3 (4), although Tim-3-independent functions have also been described (5). Gal-9 can play an important role in virus life cycles. It modulates human immunodeficiency virus type 1 (HIV-1) entry (6, 7), while Gal-9-knockout mice exhibit more potent antiviral T-cell responses than wild-type mice (8, 9), and infection with Epstein-Barr virus (EBV) modulates Gal-9 expression to evade immune clearance, inducing apoptosis of EBV-specific CD4+ T cells (10, 11). We investigated the expression of Gal-9 in the context of HCMV infection both in vivo and in vitro, identifying a novel, virally induced mechanism to promote Gal-9 expression.
Galectin 9 is upregulated in hematopoietic stem cell transplant recipients with reactivated HCMV infection.
We hypothesized that Gal-9 is upregulated in patients with active HCMV replication. Blood samples were drawn from HSCT recipients before peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation from patients with or without HCMV reactivation at a range of time points posttransplant (detailed in Tables 1 and 2). Patients were monitored for HCMV reactivation by quantitative PCR (qPCR; Roche Cobas Amplicor CMV Monitor test), with a sustained rise in serum HCMV genome copies above assay detection limits over at least two time points used to define HCMV reactivation.
TABLE 1.
Details for patients with HCMV reactivation
| Patient and serostatusa | Time points (days) posttransplant prior to reactivation Early | Time points (days) posttransplant after reactivation Late | Day posttransplant reactivation first detected |
|---|---|---|---|
| Patient 1 (D+/R+) | 25, 38 | 52, 59 | 52 |
| Patient 2 (D−/R+) | 12, 17 | 45, 52 | 38 |
| Patient 3 (D+/R−) | 10, 17 | 31, 47 | 24 |
| Patient 4 (D+/R+) | 12, 18 | 32, 39 | 32 |
| Patent 5 (D+/R+) | 3, 10 | 45, 52 | 45 |
D+/− and R+/− refer to the HCMV serostatus of the donor (D) and recipient (R).
TABLE 2.
Details for patients without HCMV reactivation
| Patient and serostatusa | Matched early time points (days) posttransplant | Matched late time points (days) posttransplant |
|---|---|---|
| Patient 6 (D−/R−) | 11, 18 | 53, 60 |
| Patient 7 (D+/R+) | 10, 17 | 45, 52 |
| Patient 8 (D+/R+) | 12, 17 | 45, 52 |
| Patient 9 (D+/R+) | 10, 17 | 45, 52 |
| Patient 10 (D+/R+) | 4, 11 | 40, 47 |
| Patient 11 (D+/R+) | 3, 10 | 38, 47 |
| Patient 12 (D+/R+) | 12, 19 | 33, 40 |
D+/− and R+/− refer to the HCMV serostatus of the donor (D) and recipient (R).
RNA was then isolated from PBMCs using an RNAqueous kit (Ambion). RNA was converted to cDNA using random primers and SuperScript III reverse transcriptase (Life Technologies). cDNA levels were assayed by qPCR (StepOnePlus real-time PCR system; Life Technologies) using 2× Brilliant II SYBR green qPCR master mix (Agilent Technologies) at 95°C for 10 min and then for 45 cycles at 95°C for 15 s and 60°C for 45 s. Relative transcript levels of Gal-9 were normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the following primers: Gal-9 forward, CTTTCATCACCACCATTCTG; Gal-9 reverse, ATGTGGAACCTCTGAGCACTG; GAPDH forward, TCACCAGGGCTGCTTTTAAC; and GAPDH reverse, GACAAGCTTCCCGTTCTCAG.
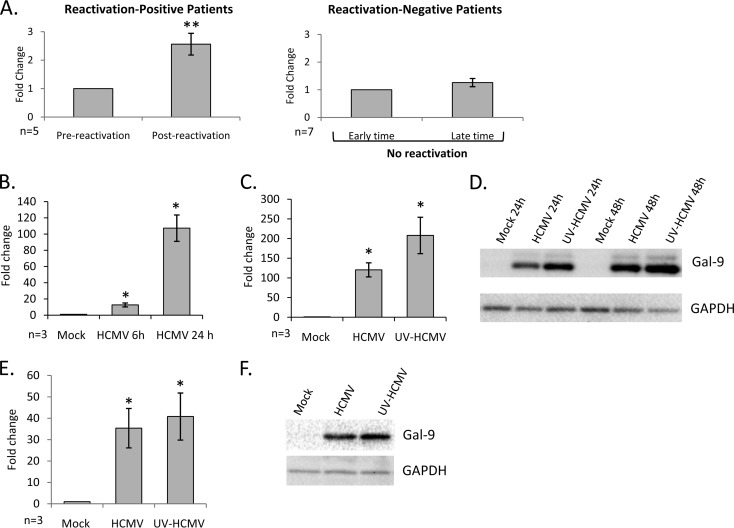
Levels of Gal-9 mRNA (normalized to that of GAPDH transcripts) were determined at two early time points prior to HCMV reactivation and compared to those from two later time points postreactivation (detailed in Table 1), with average serum genome copies/ml of 33,347 ± 17,468 in postreactivation samples. Analysis of samples from five individual HCMV reactivation cases indicated that there was a statistically significant increase in Gal-9 transcription postreactivation compared to that in prereactivation samples (P = 0.003), with no such significant change in seven matched recipients without reactivation (detailed in Table 2) at comparable time points posttransplant (Fig. 1A).
FIG 1.
Modulation of Gal-9 by HCMV infection (A) Relative levels of Gal-9 transcripts were determined by qRT-PCR in PBMCs isolated from peripheral blood drawn from allogeneic HSCT recipients at two early times after transplant (Pre-reactivation) and two time points later, after reactivation (Post-reactivation), for each of five recipients with detectable HCMV reactivation (reactivation-positive patients) and seven patients without detectable HCMV reactivation (reactivation-negative patients). Error bars indicate the standard error of the means. Significant differences in Gal-9 transcript levels at pre- and postreactivation time points were determined using a two-tailed, paired Student's t test (**, P < 0.005). (B) HFs were infected with HCMV at a multiplicity of infection (MOI) of 3 before analysis of Gal-9 transcript levels by qRT-PCR at 6 and 24 h p.i. (C) HFs were infected with HCMV and UV-HCMV at an MOI of 3 before Gal-9 transcript levels were determined by qRT-PCR at 24 h p.i. (D) Representative example of five independent replicates of immunoblotting for Gal-9 (R&D Systems) and GAPDH (Santa Cruz Biotechnology) proteins in mock-, HCMV-, and UV-HCMV-infected HFs at 24 and 48 h p.i. (E and F) Media from mock-, HCMV-, and UV-HCMV-infected HFs was harvested at 24 h p.i. and filtered through a 0.1-μm-pore-size filter before being added to new HF monolayers. Analyses of Gal-9 transcript levels at 24 h posttreatment (E) and protein levels at 48 h posttreatment (F) are depicted. Error bars indicate the standard error of the means. Significant differences in the results of treatments with HCMV and UV-HCMV from the results for mock treatment were determined using a two-tailed, paired Student's t test (*, P < 0.05).
mRNA and protein expression of galectin 9 is upregulated by both viable and UV-inactivated HCMV.
The regulation of Gal-9 by HCMV in vitro was then investigated. Gal-9 transcription in HFF-1 cells (HFs) either mock infected or infected with the bacterial artificial chromosome (BAC)-derived clinical HCMV strain Merlin (UL128− RL13−) (12) at 6 and 24 h postinfection (p.i.) was assayed by reverse transcription-quantitative PCR (qRT-PCR) and normalized to the levels of GAPDH housekeeping transcripts. There was a potent upregulation of Gal-9 mRNA following infection that was ∼100-fold more than that for mock-infected cells at 24 h p.i. (Fig. 1B). A similar increase was detected during infection with HCMV strains AD169, Towne, TB40/E, and FIX (data not shown). To further examine upregulation of Gal-9 by HCMV, we mock infected or infected HFs with viable HCMV or UV-inactivated virus (UV-HCMV; UV inactivated as described previously [13]) before qRT-PCR for Gal-9 mRNA. Similar to those infected with viable virus, HFs infected with UV-HCMV potently increased Gal-9 transcription, indicating that de novo viral gene expression was not required (Fig. 1C). In parallel, mock-, HCMV-, or UV-HCMV-infected HFs were immunoblotted for Gal-9 protein (Fig. 1D). Both HCMV and UV-HCMV strongly upregulated Gal-9 protein expression in HFs compared to that in HFs that were mock infected.
Upregulation of galectin 9 by HCMV is via induction of interferon-beta (IFN- β).
Secretion of a number of molecules is induced by both HCMV and UV-HCMV infections (14); therefore, we hypothesized that a soluble factor induced by virion binding/entry may mediate Gal-9 upregulation. Supernatants from mock-, HCMV-, and UV-HCMV-infected HFs were incubated with uninfected HFs. There was a potent upregulation of Gal-9 mRNA and protein in HFs treated with supernatants from HCMV- or UV-HCMV-treated cells compared with that in HFs treated with supernatants from mock-infected cells (Fig. 1E and F). These data demonstrate that a soluble factor is responsible for Gal-9 upregulation by HCMV.
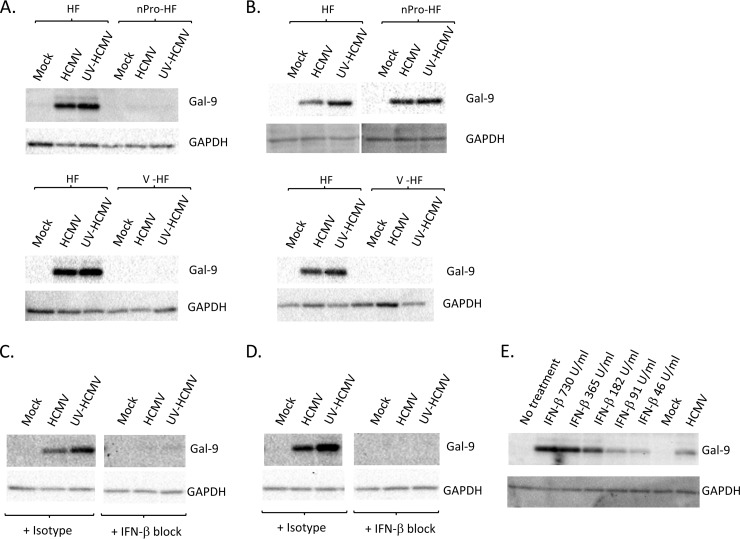
The regulation of Gal-9 is poorly understood, although a number of proinflammatory stimuli can promote its expression (15). The best characterized of these is IFN-γ, which can induce Gal-9 expression in several cell types (16–18). To test if the IFN-responsive pathway was involved in the upregulation of Gal-9 by HCMV, we generated HFs that stably express the V protein of parainfluenza virus type 5 (V-HF) or the nPro protein of bovine viral diarrhea virus (nPro-HF) by lentiviral expression. Lentivirus was generated from transfected 293T cells as described previously (19) before supernatants were added to HFs and selected with 1 μg/ml puromycin. nPro-expressing cells cannot produce type I IFN due to interferon regulatory factor 3 (IRF3) targeting (19) but are able to respond to exogenous IFNs, while cells expressing the V protein are unresponsive to type I/II IFNs, as the V protein targets signal transducers and activators of transcription 1 (STAT1) for degradation (20). HCMV or UV-HCMV infection did not upregulate Gal-9 in either V-HF or nPro-HF cells, implicating the IFN pathway as essential in Gal-9 upregulation (Fig. 2A). Supernatants from parental HFs infected with either HCMV or UV-HCMV did not upregulate Gal-9 when incubated with IFN-unresponsive V-HF but potently increased Gal-9 in IFN-responsive but production-incapable nPro-HF (Fig. 2B), consistent with a requirement for both IFN production and IFN responsiveness to upregulate Gal-9.
FIG 2.
Regulation of galectin 9 expression by HCMV (A) Parental HF, V-HF, or nPro-HF were infected at an MOI of 3 with HCMV or UV-HCMV before cell lysates were harvested at 48 h p.i. and immunoblotted for Gal-9 (R&D Systems) and GAPDH (Santa Cruz Biotechnology). (B) Media from mock-, HCMV-, or UV-HCMV-infected parental HFs were harvested at 24 h p.i. and filtered through a 0.1-μm-pore-size filter before being added to fresh HF, V-HF, or nPro-HF monolayers and incubated for 48 h before cell lysates were immunoblotted for Gal-9 and GAPDH. Blots are representative of at least 3 independent biological replicates. (C) HFs were mock, HCMV, or UV-HCMV infected in the presence of an IFN-β-blocking Ab or an isotype control. Cell lysates were harvested at 48 h p.i. before immunoblotting for Gal-9 and GAPDH. (D) HFs were treated with media from mock-, HCMV-, or UV-HCMV-infected cultures (harvested at 24 h p.i. and filtered through a 0.1-μm-pore-size filter) in the presence of an IFN-β-blocking Ab or an isotype control. Cell lysates were generated at 48 h postinfection/treatment before immunoblotting for Gal-9 and GAPDH. (E) HFs were treated with recombinant IFN-β at the concentrations indicated before immunoblotting for Gal-9 and GAPDH at 48 h posttreatment. HFs were also mock and HCMV infected in parallel as indicated. Blots are representative of at least 3 independent biological replicates.
Given that IFN-γ is a lymphocyte-specific cytokine and HFs are potent producers of IFN-β, whether type I IFNs upregulated Gal-9 during HCMV infection was investigated. A microarray study indicated that IFN-β could increase Gal-9 transcriptionally (21); however, another report suggested that IFN-β did not modulate Gal-9 (16). HFs were mock infected or infected with HCMV or UV-HCMV or treated with supernatants from mock-infected or infected cells in the presence of 200 neutralization units/ml of an anti-human polyclonal IFN-β-blocking antibody (Ab) (Millipore) or isotype control. In both HCMV- and UV-HCMV-infected and supernatant-treated cells, blocking IFN-β ablated Gal-9 upregulation (Fig. 2C and D), identifying IFN-β as a molecule essential for HCMV upregulation of Gal-9. Recombinant IFN-β (PBL Interferon Source) alone also potently increased Gal-9 (Fig. 2E), which was the first demonstration of type I IFN controlling Gal-9 protein expression. While we cannot exclude the possibility that other secreted factors produced during HCMV infection may contribute to Gal-9 upregulation, the production of IFN-β appears to be essential, and given the ability of IFN-β alone to upregulate Gal-9 it is likely that IFN-β is the major driving factor in HCMV-mediated Gal-9 upregulation.
This identification of IFN-β as a mediator of Gal-9 expression during HCMV infection may help to inform other situations where higher Gal-9 levels have been reported. Indeed, individuals infected with hepatitis C virus (HCV), HIV-1, dengue virus, and influenza virus have higher serum levels of Gal-9 (6, 22–24), yet the mechanism responsible for such increases has not been identified. Given the critical role of type I IFNs in controlling viral infections, it is tempting to speculate that IFN-β may be an essential driver of Gal-9 upregulation. Thus, Gal-9 may be a biomarker of viral infection and perhaps also has potential as a prognostic marker. Indeed, Gal-9 levels are extremely high in patients suffering from either dengue hemorrhagic fever or dengue fever, decline rapidly following recovery, and are correlated with dengue virus load during the critical phase of infection (23).
Gal-9 has been primarily studied as an immunoregulatory molecule controlling a number of virus-specific immune responses (8–10), yet immunostimulatory functions have also been described (25, 26). Interestingly, a recent report also indicates that hepatic NK cells from Gal-9-knockout mice produce significantly more IFN-γ in response to murine cytomegalovirus (MCMV) infection than hepatic NK cells from wild-type mice, indicating the immunomodulatory potential of this molecule in the context of CMV infection (27). Future studies will examine the functional effects of HCMV-mediated Gal-9 upregulation on the antiviral immune response. Gal-9 can be both secreted and expressed at the cell surface, so functional studies will include examination of the importance of posttranslational regulation of Gal-9 on controlling the HCMV-specific immune response.
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance of members of the Bone Marrow Transplant Unit at Westmead Hospital for assistance in collection of blood samples from hematopoietic stem cell transplant recipients and wish to thank Rick Randall from the University of St. Andrews, Scotland, for providing V-protein and nPro expression constructs.
This work was funded by Australian National Health and Medical Research Council Project Grant funding awarded to B.S. and A.A.
Footnotes
Published ahead of print 9 July 2014
REFERENCES
- 1.Slobedman B, Cao JZ, Avdic S, Webster B, McAllery S, Cheung AK, Tan JC, Abendroth A. 2010. Human cytomegalovirus latent infection and associated viral gene expression. Future Microbiol. 5:883–900. 10.2217/fmb.10.58 [DOI] [PubMed] [Google Scholar]
- 2.Griffiths PD. 2012. Burden of disease associated with human cytomegalovirus and prospects for elimination by universal immunisation. Lancet Infect. Dis. 12:790–798. 10.1016/S1473-3099(12)70197-4 [DOI] [PubMed] [Google Scholar]
- 3.Wiersma VR, de Bruyn M, Helfrich W, Bremer E. 2013. Therapeutic potential of galectin-9 in human disease. Med. Res. Rev. 33(Suppl 1):E102–E126. 10.1002/med.20249 [DOI] [PubMed] [Google Scholar]
- 4.Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. 2005. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol. 6:1245–1252. 10.1038/ni1271 [DOI] [PubMed] [Google Scholar]
- 5.Su EW, Bi S, Kane LP. 2011. Galectin-9 regulates T helper cell function independently of Tim-3. Glycobiology 21:1258–1265. 10.1093/glycob/cwq214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elahi S, Niki T, Hirashima M, Horton H. 2012. Galectin-9 binding to Tim-3 renders activated human CD4+ T cells less susceptible to HIV-1 infection. Blood 119:4192–4204. 10.1182/blood-2011-11-389585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi S, Hong PW, Lee B, Baum LG. 2011. Galectin-9 binding to cell surface protein disulfide isomerase regulates the redox environment to enhance T-cell migration and HIV entry. Proc. Natl. Acad. Sci. U. S. A. 108:10650–10655. 10.1073/pnas.1017954108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma S, Sundararajan A, Suryawanshi A, Kumar N, Veiga-Parga T, Kuchroo VK, Thomas PG, Sangster MY, Rouse BT. 2011. T cell immunoglobulin and mucin protein-3 (Tim-3)/galectin-9 interaction regulates influenza A virus-specific humoral and CD8 T-cell responses. Proc. Natl. Acad. Sci. U. S. A. 108:19001–19006. 10.1073/pnas.1107087108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sehrawat S, Reddy PB, Rajasagi N, Suryawanshi A, Hirashima M, Rouse BT. 2010. Galectin-9/TIM-3 interaction regulates virus-specific primary and memory CD8 T cell response. PLoS Pathog. 6:e1000882. 10.1371/journal.ppat.1000882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klibi J, Niki T, Riedel A, Pioche-Durieu C, Souquere S, Rubinstein E, Le Moulec S, Guigay J, Hirashima M, Guemira F, Adhikary D, Mautner J, Busson P. 2009. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood 113:1957–1966. 10.1182/blood-2008-02-142596 [DOI] [PubMed] [Google Scholar]
- 11.Keryer-Bibens C, Pioche-Durieu C, Villemant C, Souquere S, Nishi N, Hirashima M, Middeldorp J, Busson P. 2006. Exosomes released by EBV-infected nasopharyngeal carcinoma cells convey the viral latent membrane protein 1 and the immunomodulatory protein galectin 9. BMC Cancer 6:283. 10.1186/1471-2407-6-283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanton RJ, Baluchova K, Dargan DJ, Cunningham C, Sheehy O, Seirafian S, McSharry BP, Neale ML, Davies JA, Tomasec P, Davison AJ, Wilkinson GW. 2010. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J. Clin. Invest. 120:3191–3208. 10.1172/JCI42955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung AK, Abendroth A, Cunningham AL, Slobedman B. 2006. Viral gene expression during the establishment of human cytomegalovirus latent infection in myeloid progenitor cells. Blood 108:3691–3699. 10.1182/blood-2005-12-026682 [DOI] [PubMed] [Google Scholar]
- 14.Taylor RT, Bresnahan WA. 2006. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J. Virol. 80:920–928. 10.1128/JVI.80.2.920-928.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heusschen R, Griffioen AW, Thijssen VL. 2013. Galectin-9 in tumor biology: a jack of multiple trades. Biochim. Biophys. Acta 1836:177–185. 10.1016/j.bbcan.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 16.Asakura H, Kashio Y, Nakamura K, Seki M, Dai S, Shirato Y, Abedin MJ, Yoshida N, Nishi N, Imaizumi T, Saita N, Toyama Y, Takashima H, Nakamura T, Ohkawa M, Hirashima M. 2002. Selective eosinophil adhesion to fibroblast via IFN-gamma-induced galectin-9. J. Immunol. 169:5912–5918. 10.4049/jimmunol.169.10.5912 [DOI] [PubMed] [Google Scholar]
- 17.Imaizumi T, Kumagai M, Sasaki N, Kurotaki H, Mori F, Seki M, Nishi N, Fujimoto K, Tanji K, Shibata T, Tamo W, Matsumiya T, Yoshida H, Cui XF, Takanashi S, Hanada K, Okumura K, Yagihashi S, Wakabayashi K, Nakamura T, Hirashima M, Satoh K. 2002. Interferon-gamma stimulates the expression of galectin-9 in cultured human endothelial cells. J. Leukoc. Biol. 72:486–491 http://www.jleukbio.org/content/72/3/486.long [PubMed] [Google Scholar]
- 18.Alam S, Li H, Margariti A, Martin D, Zampetaki A, Habi O, Cockerill G, Hu Y, Xu Q, Zeng L. 2011. Galectin-9 protein expression in endothelial cells is positively regulated by histone deacetylase 3. J. Biol. Chem. 286:44211–44217. 10.1074/jbc.M111.242289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilton L, Moganeradj K, Zhang G, Chen YH, Randall RE, McCauley JW, Goodbourn S. 2006. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J. Virol. 80:11723–11732. 10.1128/JVI.01145-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrejeva J, Young DF, Goodbourn S, Randall RE. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159–2167. 10.1128/jvi.76.5.2159-2167.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaman DW, Chawla-Sarkar M, Jacobs B, Vyas K, Sun Y, Ozdemir A, Yi T, Williams BR, Borden EC. 2003. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J. Interferon Cytokine Res. 23:745–756. 10.1089/107999003772084860 [DOI] [PubMed] [Google Scholar]
- 22.Mengshol JA, Golden-Mason L, Arikawa T, Smith M, Niki T, McWilliams R, Randall JA, McMahan R, Zimmerman MA, Rangachari M, Dobrinskikh E, Busson P, Polyak SJ, Hirashima M, Rosen HR. 2010. A crucial role for Kupffer cell-derived galectin-9 in regulation of T cell immunity in hepatitis C infection. PLoS One 5:e9504. 10.1371/journal.pone.0009504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chagan-Yasutan H, Ndhlovu LC, Lacuesta TL, Kubo T, Leano PS, Niki T, Oguma S, Morita K, Chew GM, Barbour JD, Telan EF, Hirashima M, Hattori T, Dimaano EM. 2013. Galectin-9 plasma levels reflect adverse hematological and immunological features in acute dengue virus infection. J. Clin. Virol. 58:635–640. 10.1016/j.jcv.2013.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh S, Ikeda M, Shimizu H, Mouri K, Obase Y, Kobashi Y, Fukushima K, Hirashima M, Oka M. 2014. Increased levels of plasma galectin-9 in patients with influenza virus infection. Tohoku J. Exp. Med. 232:263–267. 10.1620/tjem.232.263 [DOI] [PubMed] [Google Scholar]
- 25.Gooden MJ, Wiersma VR, Samplonius DF, Gerssen J, van Ginkel RJ, Nijman HW, Hirashima M, Niki T, Eggleton P, Helfrich W, Bremer E. 2013. Galectin-9 activates and expands human T-helper 1 cells. PLoS One 8:e65616. 10.1371/journal.pone.0065616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gleason MK, Lenvik TR, McCullar V, Felices M, O'Brien MS, Cooley SA, Verneris MR, Cichocki F, Holman CJ, Panoskaltsis-Mortari A, Niki T, Hirashima M, Blazar BR, Miller JS. 2012. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood 119:3064–3072. 10.1182/blood-2011-06-360321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golden-Mason L, McMahan RH, Strong M, Reisdorph R, Mahaffey S, Palmer BE, Cheng L, Kulesza C, Hirashima M, Niki T, Rosen HR. 2013. Galectin-9 functionally impairs natural killer cells in humans and mice. J. Virol. 87:4835–4845. 10.1128/JVI.01085-12 [DOI] [PMC free article] [PubMed] [Google Scholar]